Abstract
Traumatic brain injury (TBI) is an acquired insult to the brain caused by an external mechanical force, potentially resulting in temporary or permanent impairment. Microglia, the resident immune cells of the central nervous system, are activated in response to TBI, participating in tissue repair process. However, the underlying epigenetic mechanisms in microglia during TBI remain poorly understood. ARID1A (AT-Rich Interaction Domain 1 A), a pivotal subunit of the multi-protein SWI/SNF chromatin remodeling complex, has received little attention in microglia, especially in the context of brain injury. In this study, we generated a Arid1a cKO mouse line to investigate the potential roles of ARID1A in microglia in response to TBI. We found that glial scar formation was exacerbated due to increased microglial migration and a heightened inflammatory response in Arid1a cKO mice following TBI. Mechanistically, loss of ARID1A led to an up-regulation of the chemokine CCL5 in microglia upon the injury, while the CCL5-neutralizing antibody reduced migration and inflammatory response of LPS-stimulated Arid1a cKO microglia. Importantly, administration of auraptene (AUR), an inhibitor of CCL5, repressed the microglial migration and inflammatory response, as well as the glial scar formation after TBI. These findings suggest that ARID1A is critical for microglial response to injury and that AUR has a therapeutic potential for the treatment of TBI.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01852-y.
Keywords: Microglia, Arid1a, Migration, Inflammation, Ccl5, TBI
Background
Traumatic brain injury (TBI), one of the leading causes of death and disability globally, has emerged as a predominant public health issue due to its rising incidence, diverse etiological factors, and profound lifelong consequences for families and society [1–3]. The initial injury typically manifests as structural brain abnormalities and intracranial hemorrhages. This is followed by a secondary injury characterized by intricate alterations in the brain microenvironment, resulting in cellular apoptosis, cerebral edema, metabolic disturbances, oxidative stress, and sustained inflammation [4–7]. Unfortunately, there are limited efficacious pharmaceutical interventions or therapeutic modalities available to mitigate the secondary cerebral damage post-TBI [8].
Microglia, as inherent immune effector cells of the central nervous system (CNS), are instrumental in modulating brain ontogeny and preserving neural environmental homeostasis under physiological conditions [9–13]. Following TBI, activation of microglia occurs early after injury and even persist for years in association with tissue damage [14, 15]. In the acute post-injury phase, microglia are triggered to rapidly proliferate and migrate towards sites of damage [16]. This early activation of microglia is thought to support wound healing, providing benefits such as the clearance of cellular debris [17]. However, we still know so little about the mechanisms underlying the transition between an activated state and a homeostatic state of microglia following TBI.
Emerging research underscores the pivotal influence of epigenetic reconfiguration on microglial function [18, 19]. ARID1A (AT-rich interacting domain-containing protein 1 A, also known as BAF250a), categorized as a subunit of mammalian SWI/SNF (mSWI/SNF), binds DNA in a non-sequence-specific manner by its ARID domain to guide the location of the mSWI/SNF complex [20–22]. More and more researches have shown that ARID1A is the most commonly mutated member of the SWI/SNF complex, being aberrant in ∼6% of cancers overall [20–22]. In addition to cancers, ARID1A mutations cause neurodevelopmental disorders like Coffin-Siris syndrome (CSS), a rare congenital malformation syndrome [23, 24]. Our previous research has demonstrated that the deletion of Arid1a in microglia leads to alterations in microglial homeostasis, subsequently causing disruptions in neural progenitor cell differentiation and anxiety-like behaviors [25]. However, the precise function of microglial Arid1a under TBI condition remains largely unknown.
In this study, we investigated the role of Arid1a in microglia using a well-established mouse model of TBI. We found that Arid1a was up-regulated in microglia at the injury site. The absence of Arid1a in microglia exacerbated brain glial scar formation, as evidenced by increased aggregation of microglia and astrocytes to the lesion sites following TBI. We then identified Ccl5, a key cytokine related to cell migration and inflammatory reactivity, as a downstream target of Arid1a in microglia. Finally, we discovered that auraptene (AUR) could inhibit abnormal microglial migration and inflammatory response caused by microglial Arid1a deletion in the injured mice.
Materials and methods
Animals
All animal experiments were performed following guidelines of ethical regulations and approved by the Animal Committee of the Institute of Zoology, Chinese Academy of Sciences. The Arid1afl/fl mice was a kind gift from Dr. Zhong Wang at University of Michigan and Dr. Chun-sheng Han at the Institute of Zoology, Chinese Academy of Sciences. The Arid1afl/fl mice (wildtype, WT) were intercrossed with Cx3cr1-Cre (JAX Stock No. 005628) line to generate animals with microglia-specific deletion of Arid1a, referred to as conditional knockouts (cKO) mice. All mice were housed in specific-pathogen-free facility with a 12 h light/dark cycle and provided with ad libitum access to food and water.
Traumatic brain injury
2-3-month-old mice were subjected to moderate and unilateral TBI as previously described [26, 27]. Briefly, the mice were anesthetized with 200 mg/kg Avertin (T48402, Sigma-Aldrich) and placed in a stereotaxic frame prior to TBI. After opening a cranial window (1 × 4 mm) on the right side of the skull, a surgical blade No. 15 was inserted into the hippocampus at 3.5 mm deep below the dura and stayed in the brain for 1 min before cautiously pulled out. After the wound was cleaned and sutured, the animals were monitored for recovery at 36–37 °C and returned to their home cages.
For identifying dividing cells in vivo, mice were intraperitoneally injected with Bromodeoxyuridine (BrdU, B5002, Sigma-Aldrich) at a dosage of 100 mg/kg. The AUR treatment procedure was performed as described previously [28] with slight modifications. AUR (A868464, Macklin) was dissolved in DMSO/Polyethylene Glycol 300 (1:9) solution and administered subcutaneously at a dosage of 25 mg/kg/day for three consecutive days prior to TBI, continuing for seven consecutive days post-TBI. The control group received injections of the vehicle solvent.
Immunofluorescence staining
Immunofluorescence staining was performed as previously described [25]. Brains from perfused mice were fixed overnight in 4% paraformaldehyde (PFA, P804537, Macklin) and subsequently dehydrated in 30% sucrose (S6212, Macklin). The tissue was then cryo-embedded and sectioned into 40 μm thick coronal slices. For immunofluorescence staining, sagittal brain slices or coverslips were permeabilized with 0.5% Triton X-100 (T8787, Sigma-Aldrich) for 15 min and blocked with 0.3% Triton X-100 and 3% bovine serum albumin (BSA, BE6254, Easybio) in phosphate-buffered saline (PBS) for 1 h at room temperature. Subsequently, the slices were then incubated overnight at 4 °C with primary antibodies against specific markers. Following primary antibody incubation, slices were treated with Alexa Fluor-conjugated secondary antibodies and counterstained with DAPI (D9542, Sigma-Aldrich) to visualize nuclei for 1 h at room temperature. Between each solution step, brain sections were washed with PBS for 10 min, three times. A full list of antibodies is provided in Table S2 (Supplementary Information).
Image analysis
Images were acquired using a Zeiss LSM880 confocal laser-scanning microscope equipped with a digital camera. All images were captured using identical exposure times. Cells were counted only if nuclei were entirely within the frame, on the bottom boundary, or along the right-hand border. Image analysis and quantification were conducted using ImageJ software (V1.54).
Quantitative real-time PCR
Cells or tissues were harvested in TRIZOL Reagent (15596018CN, Invitrogen) and total RNA was isolated following the manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA with reverse transcriptase kit (AT311-03, Transgen). Real-time qPCR was performed on Hieff QPCR SYBR Green Master Mix (11201ES08, Yeasen) as the detection dye. The amplification protocol was 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s using the LightCycler Real-Time PCR System (Roche). riplicate assays were averaged, and relative mRNA levels were normalized to Gapdh expression. A full list of primer sequences was provided in Table S1 (Supplementary Information).
Primary microglia culture
As previously described, primary microglia were isolated and purified from postnatal day 0–3 mice [18, 25]. The obtained tissue was enzymatically dissociated, and the resulting cell suspension was plated onto tissue culture flasks containing Dulbecco’s Modified Eagle Medium (DMEM, C11995500BT, Gibco) supplemented with 10% fetal bovine serum (FBS, 10099141, Gibco) and 1% Penicillin-Streptomycin (SV30010, Hyclone). After a 2-week culture period, microglia were harvested by shaking the flasks at 130–200 revolutions per minute for 2 h.
Microglia were then treated with 100 mM AUR and/or 100 ng/mL lipopolysaccharide (LPS) or 1 μM adenosine triphosphate (ATP). AUR was dissolved in DMSO to prepare the stock solution as previously described [28]. LPS (L8274, Sigma-Aldrich) and ATP (B3304, APExBIO) were dissolved in saline. The plates were incubated at 37 °C and 5% CO2 for 48 h.
For the anti-CCL5 neutralizing antibody procedures, microglia were exposed to 100 ng/mL LPS and anti‐CCL5 or normal rabbit IgG antibodies. The plates were incubated at 37 °C and 5% CO2 for 48 h. A full list of antibodies was provided in Table S2 (Supplementary Information).
Transwell assay
The microglia transwell assay was performed using 8 μm transwell inserts (3422, Corning) in 24-well culture plates. Approximately 50,000 cells in 200 μL of culture medium supplemented with 1% FBS and/or 100 ng/mL LPS were placed in the upper compartment, while the lower compartment was filled with 5% FBS, with or without anti-CCL5 antibody at a concentration of 2 μg/mL and/or IgG. For AUR treatment, the lower compartment was filled with 5% FBS with or without 100 mM AUR. After 48 h, non-migrated microglia on the upper side of the filters were removed and washed with PBS. Migrated microglia on the reverse side were fixed in 4% PFA for 10 min, stained with crystal violet, and counted in five non-overlapping fields under a bright-field microscopy (Ti2-U, Nikon) at 20X magnification.
Flow cytometry
Mice were anesthetized and underwent transcardial perfusion with ice-cold PBS. Tissue digestion was performed using the Papain Dissociation System kit (LK003150, Worthington) according to the manufacturer’s instructions. For cultured primary microglia, cells were digested with trypsin, centrifuged. The cell suspension was collected and thoroughly washed with EBSS buffer containing 2% FBS. Subsequently, cells were stained with anti-CD11b APC and anti-CD45 PE for microglia isolation, and with anti-mouse CD16/32 APC/cy7 and anti-mouse CD206 FITC for analysis. FlowJo software was used to quantify proportion of positively stained cells within specific cell populations. A full list of antibodies was provided in Table S2 (Supplementary Information).
ELISA
The levels of CCL5 in the growth medium of primary microglia of mice were determined using an ELISA kit (AE51709MO, Abebio) according to the manufacturer’s instructions. The absorbance of the samples at a wavelength of 450 nm was measured with a BioTek microplate reader.
Western blotting analysis
Microglial lysates were prepared using RIPA buffer (P0013B, Beyotime) and then mixed with 5X loading buffer containing 2-mercaptoethanol before boiling. The denatured samples were resolved by SDS-PAGE and transferred onto a Nitrocellulose membrane (Bio-Rad). The membrane was blocked with 5% milk solution and incubated overnight at 4 °C with primary antibodies diluted in 5% milk. After washing, the membrane was incubated with HRP-conjugated secondary antibodies in 5% milk for at least 1 h at room temperature. Chemiluminescence signals were quantitatively detected by Tanon-5200. A full list of antibodies was provided in Table S2 (Supplementary Information).
RNA-seq
Microglia were isolated via flow cytometry, followed by the generation of sequencing libraries utilizing the NEBNext® Ultra™ Directional RNA Library Prep Kit® for Illumina as per the manufacturer’s protocols. Subsequently, the cDNA library underwent analysis using the Illumina HiSeq 2500 platform. Index-coded samples were clustered on the HiSeq PE Cluster Kit v4-cBot-HS (Illumina), and the libraries were subsequently sequenced on an Illumina platform. Finally, 150 bp paired-end reads were generated with help of Annoroad Gene Technology (Beijing).
Statistical analysis
Data were represented as mean ± SEM, unless otherwise indicated. Experiments were conducted in at least three biological replicates (n ≥ 3) for each group. For statistical analyses, unpaired Student’s t-test or one-way analysis of variance significant difference test were executed using GraphPad Prism software (V9.5.1).
Results
ARID1A expression is increased in microglia after brain injury
To examine the expression patterns of Arid1a following brain injury, we firstly analyzed GSE129927 dataset in the Gene Expression Omnibus (GEO) database and found that Arid1a, Arid2 and Arid3a transcripts are increased following brain injury (Fig. S1a). However, the baseline expressions of Arid2 and Arid3a are considerably low (Fig. S1b), indicating that only Arid1a may play a crucial role after brain injury.
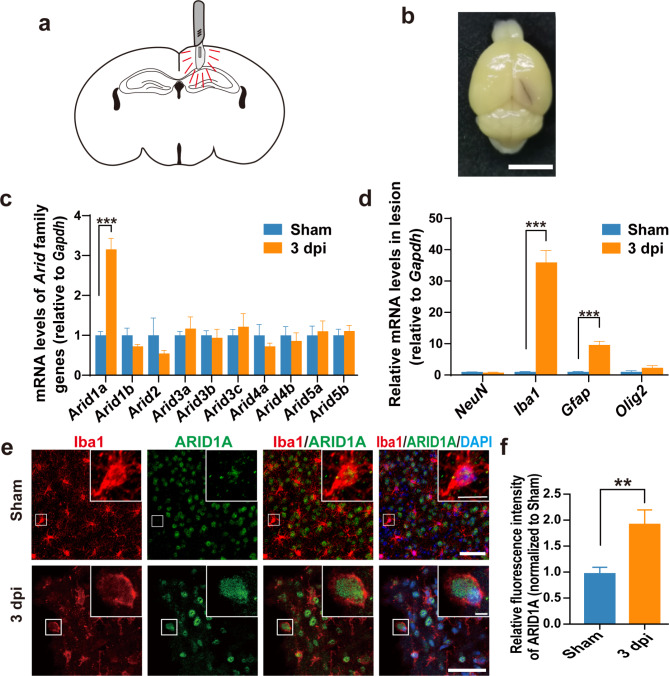
To test whether Arid1a is involved to brain injury, we used the already established TBI model in our lab [26] in which the cortex and hippocampus of mice were pierced with a No. 15 blade, resulting in an acute brain trauma (Fig. 1a-b). Quantitative PCR analysis indicated that Arid1a mRNA levels was significantly elevated in injured brain tissue at 3 days post-injury (dpi) (Fig. 1c), suggesting that Arid1a might be involved in the process of brain injury. To better understand the cell origin of Arid1a up-regulation post-TBI, we conducted RT-PCR analysis of mRNA levels of marker genes for cells within the lesion area and found that the mRNA expression of Aif1 (Iba1, a marker for microglia/macrophages) was the most significantly up-regulated, while NeuN (neuronal cell marker) and Olig2 (oligodendrocyte marker) mRNA levels were not altered in the injurer tissues at 3 dpi, compared to the sham group (Fig. 1d). Therefore, we postulated that the increased Arid1a expression observed after TBI was likely from Iba1+ microglia/macrophages. Macrophage infiltration is widely observed following TBI [29, 30]. To distinguish between microglia and infiltrating macrophages at lesion sites, we conducted IF staining for Tmem119 (a specific marker for microglia) and Iba1 at 3 dpi. We found that over 90% of Iba1+ cells around the injury area were Tmem119+/ Iba1+ cells, suggesting that the majority of Iba1+ cells accumulating in the lesion area after TBI are indeed microglia, rather than infiltrating macrophages (Fig. S2a-b).
Fig. 1.
ARID1A expression is increased in microglia following brain injury in mice. a Schematic diagram of the mouse TBI model. b Photograph of a PBS-perfused mouse brain post-TBI. Scale bars: 5 mm. c qPCR analysis of Arid family genes in the brain tissues at 3 dpi. n = 4. d Relative mRNA levels of marker genes for cells within the lesion area at 3 dpi. n = 4. e-f Representative images (e) and quantification of ARID1A immunostaining (f) in microglia at 3 dpi. n = 4. Scale bars: 50 μm and 10 μm. Data are presented as mean ± SEM; ***P < 0.001, **P < 0.01 by Student’s t-test
To validate the cellular composition at lesion sites, we further performed co-immunofluorescence (IF) staining on brain sections at 3 dpi utilizing anti-ARID1A and anti-Iba1, anti-GFAP (a marker for astrocyte) or anti-NeuN antibodies. Notably, no significant accumulation of astrocytes or neurons within the lesion area was observed, and also no significant alterations in ARID1A expression were observed in astrocytes or in neurons (Fig. S3a-d). However, the quantification results clearly showed ARID1A expression was dramatically increased in Iba1+ cells surrounding the injury core (Fig. 1e-f), suggesting the up-regulation of ARID1A following TBI was primarily attributed to microglia.
Selective deletion of Arid1a in microglia exacerbates severity of glial scar formation
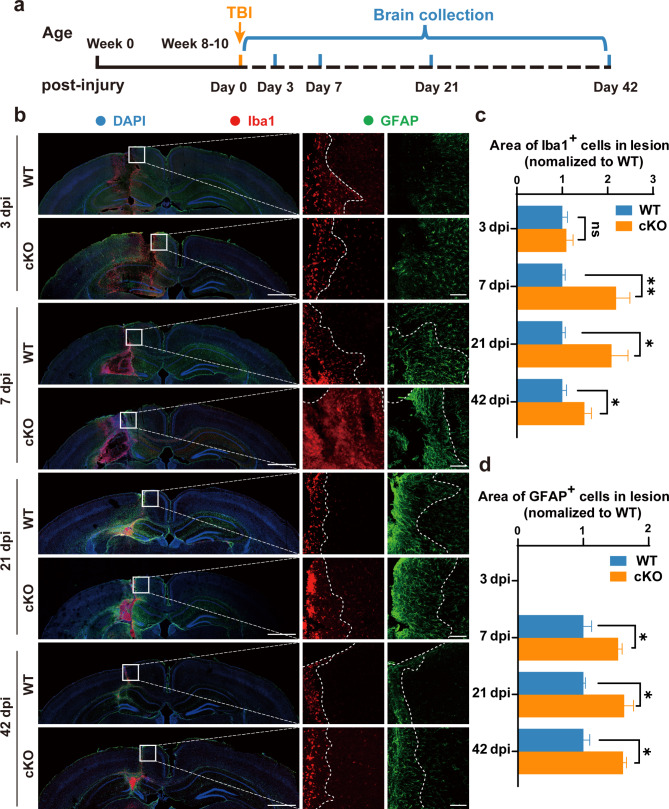
The cells in the CNS orchestrates a complex array of responses after TBI [31, 32]. A pathological feature of TBI is the formation of the glial scar, resulting from the rapid recruitment of microglia, in tandem with astrocytes, to the perilesional region where cellular necrosis is most prominent [33]. To explore whether ARID1A plays a functional role in glial scar formation, we used an Arid1a microglial cKO mice by crossing Arid1afl/fl mice with transgenic Cx3cr1-Cre mice [25] (Fig. S4a), and performed a comprehensive histological analysis at different time points after TBI. As we expected, ARID1A protein expression was nearly undetectable in the primary microglia isolated from Arid1a cKO pups (Fig. S4c-d). According to the experimental design in Fig. 2a, brain tissues were collected at specified time following the injury 3, 7, 21 and 42 dpi after undergoing either TBI or sham procedure. Our IF staining demonstrated that the numbers of both microglia and astrocytes were not significantly increased at lesion sites in either WT and Arid1a cKO mice at 3 dpi, but both microglia and astrocytes dramatically increased at lesion sites in the Arid1a cKO group at 7, 21 and 42 dpi (Fig. 2b).
Fig. 2.
Selective deletion of Arid1a in microglia exacerbates the formation of glial scar after TBI. a Illustration of timeline for brains collection after TBI. b Representative images of IF staining of Iba1 (red), GFAP (green) and DAPI (blue) in brain slices at 3, 7, 21 and 42 dpi, respectively. Mouse brains were sectioned at 40-μm thickness into coronal sections. n = 5 animals per group. Scale bars: left panel, 1,000 μm; right panel, 100 μm. c, d Quantification of Iba1+ area (c) or GFAP+ area (d) at 3, 7, 21 and 42 dpi. n = 5 animals per group. Data are presented as mean ± SEM; ns, no significance; **P < 0.01, *P < 0.05 by Student’s t-test
Next, we carefully mapped and quantified the areas of aggregation for Iba1+ cells and GFAP+ cells near the injury track. The results indicated a significant increase in Iba1+ cells aggregation near the injury track in the Arid1a cKO group after TBI, reaching peak levels at 7 dpi and persisting until 42 dpi, the longest timepoint we tested (Fig. 2c). Additionally, we found that more astrocytes accumulated around lesion sites in the Arid1a cKO group compared to WT group (Fig. 2d). Meanwhile, we assessed neuronal density at 7 dpi and found that Arid1a cKO mice exhibited more severe neuronal loss compared to WT mice (Fig. S5a-b). This finding suggests that the absence of microglial Arid1a exacerbates brain damage during TBI. Taken together, these data indicate that Arid1a ablation in microglia aggravates glial scar formation and brain damage following TBI, underscoring the essential role of ARID1A in regulating microglial responses to brain injury.
Increased microglial migration in Arid1a cKO mice following TBI
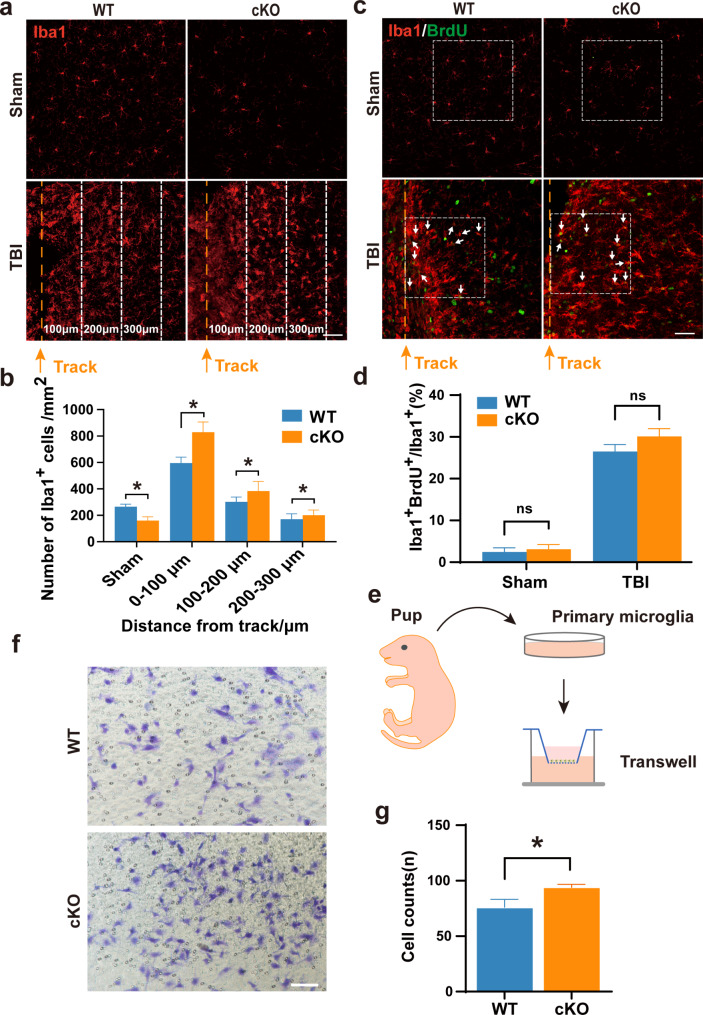
To determine whether the increased number of microglia in the cKO-TBI group is due to enhanced migration and/or proliferation of microglia with Aridla-deficiency under TBI conditions, we firstly quantified numbers of microglia at intervals of 100 μm along the 300 μm distance away from the injury track at 7 dpi. The results revealed a reduced amount of microglia from proximal to distal locations and a notable increase in microglia accumulation at each distance away from injury track in the Aridla cKO group compared to the WT group. (Fig. 3a-b).
Fig. 3.
Selective deletion of Arid1a in microglia results in increased microglial migration. a Representative image of Iba1 (red) staining around the injury sites at 7 dpi. The injury track is indicated by the yellow dotted lines. The distance between adjacent dotted lines is 100 μm. Scale bar: 50 μm. b Quantification of the density of Iba1+ cells at 7 dpi. n = 4 mice per group. c-d Representative images (c) and quantification (d) of Iba1 (red) and BrdU (green) immunostaining around the injury sites at 7 dpi. Scale bar: 50 μm. n = 4 mice per group. e Schematic diagram of the transwell assay for primary cultured microglia. f-g Transwell migration assay measuring primary microglia migration. cKO microglia exhibited increased migratory capability compared with WT group. n = 4. Scale bar: 100 μm. Data are presented as mean ± SEM; ns, no significance; *P < 0.05 by Student’s t-test
In order to compare the proliferative capacity between WT and Aridla cKO microglia, we administered intraperitoneal injections of BrdU to WT and cKO mice at 30 min before TBI. We counted the BrdU+ Iba1+ cells with a specific focus on regions adjacent to the glial scar at 7 dpi. Surprisingly, the quantification results demonstrated that there was no significant difference in BrdU+Iba1+ cells between the Aridla cKO and WT groups after TBI (Fig. 3c-d), suggesting that the increased number of microglia in the cKO-TBI group is not caused by the enhanced proliferation of Aridla cKO microglia.
Next, we isolated primary microglia from neonatal pups and performed a transwell migration assay in vitro (Fig. 3e). The results showed that there were more Aridla cKO microglia migrated to the lower chamber when compared to WT microglia (Fig. 3f-g), supporting that Arid1a cKO microglia exhibit enhanced migratory capacity compared to WT control.
To rule out the influence of macrophages, we performed immunofluorescence co-staining using Tmem119 alongside Iba1. The results showed that the majority of Tmem119+/Iba1+ cells around the injury site post-TBI were also Iba1+ microglia, with no significant differences observed between the WT and cKO groups (Fig. S6a-b). These findings suggested that macrophage infiltration did not significantly impact microglial accumulation following TBI. Altogether, these data indicate that the loss of Arid1a in microglia enhances their migratory tendency, leading to more severe gliosis after TBI.
Enhanced microglia response to injury in Arid1a cKO mice
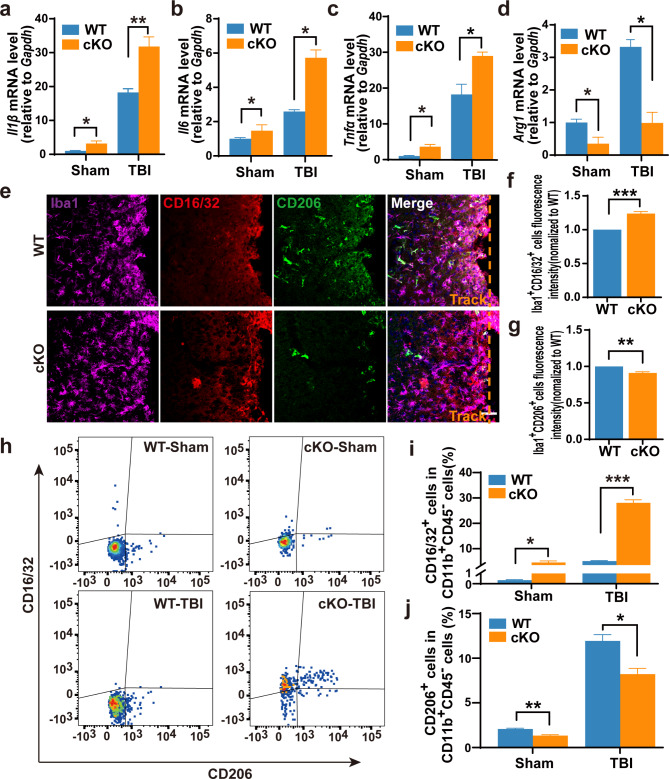
Since microglia plays a pivotal role in neuroinflammation after TBI [32, 34] and our previous research demonstrates that Arid1a-deficient microglia exhibit an enhanced neuroinflammatory response during the developmental stage [25], we then explored whether Aridla loss-of-function could affect the inflammatory responses in microglia following TBI. qRT-PCR analysis showed that the mRNA expression levels of pro-inflammatory cytokines (Il6, TNFα, Il1β) increased, but the anti-inflammatory cytokine Arg1 decreased in the lesion tissues of Aridla cKO mice at 7 dpi (Fig. 4a-d). Consistently, there was a significant increase in the percentage of Iba1+CD16/32+ microglia and a decrease in the percentage of Iba1+CD206+ microglia among Iba1+ cells around the lesion sites in Aridla cKO mice compared to that in WT mice (Fig. 4e-g). These results support that Aridla cKO microglia surrounding the glial scar exhibit a higher-grade pro-inflammatory state after TBI. To further confirm the results, we performed a flow cytometric analysis with the injured brain tissue at 7 dpi. Again, our results validated those microglia (CD11b+CD45−) in the Aridla cKO group at the injury site exhibited a higher proportion of pro-inflammatory state (CD16/32+) and a lower proportion of anti-inflammatory or tissue-repairing state (CD206+) compared to microglia in the WT group (Fig. 4h-j).
Fig. 4.
Increased microglial response to injury in Arid1a cKO mice following TBI. a-d The mRNA expression levels of proinflammatory cytokines, Il1β(a), Il6(b), TNFα(c) and anti-inflammatory cytokine Arg1(d) in brain tissues at 7 dpi. n = 4. e Representative images of Iba1, CD16/32 and CD206 immunostaining of brain sections at 7 dpi. f, g Quantification of microglia at proinflammatory state (f) or anti-inflammatory state (g). n = 3. h-j Representative images (h) and quantifications of flow cytometric analysis of CD16/32+ microglia (i) and CD206+ microglia (j). n = 3 or 4. Data are presented as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test
We next applied ATP and LPS to activate microglia from homeostatic state to activated phenotype in vitro [25], and detected a higher expression of pro-inflammatory cytokines in Aridla cKO microglia with LPS stimulation compared to that with ATP stimulation (Fig. S7a-c). Therefore, we adopted the LPS stimulation protocol for subsequent experiments. Consistent with the in vivo results, the cKO primary microglia subjected to LPS stimulation in vitro did exhibit a higher proportion of pro-inflammatory state (Fig. S7a-f), supporting the enhanced neuroinflammatory response to injury in microglial Arid1a-depleted mice.
ARID1A regulates the injury-induced reactivity of microglia by repressing Ccl5
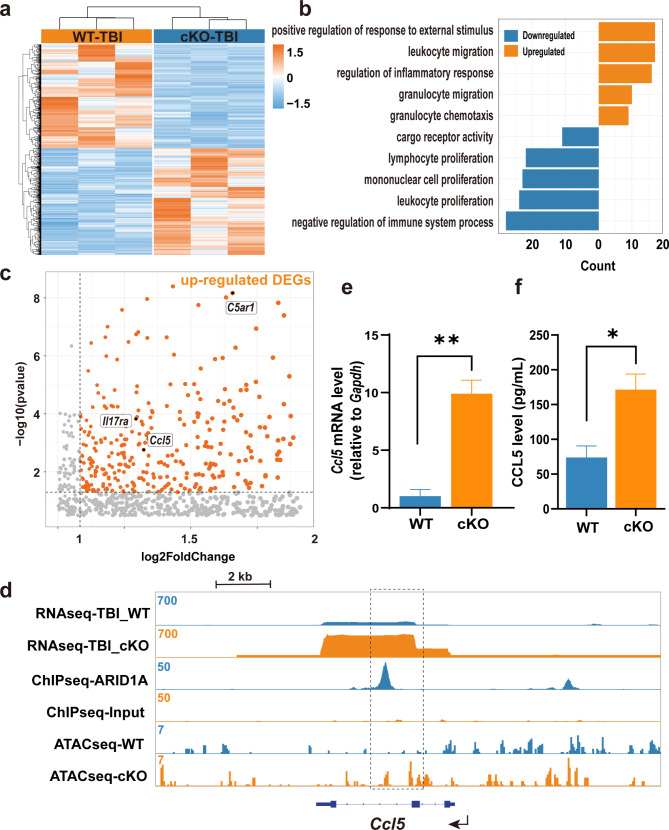
To gain mechanistic insights into how ARID1A regulates microglial migration and reactivity following TBI, we conducted an in-depth transcriptomic comparison between Arid1a cKO and WT microglia. Microglia (CD11b+CD45−) were scrupulously harvested using flow cytometry from the areas adjacent to the TBI lesions in mice from both WT and cKO groups [25]. To assess the similarity between the WT and cKO samples, we conducted Pearson correlation analysis and principal components analysis (PCA) on the transcriptomes. The Pearson correlation coefficients indicated strong positive correlations between the gene expression profiles of the two groups. In PCA, PC1 and PC2 explained 54% and 20% of the variance, respectively. These analyses demonstrated that our samples exhibited significant inter-group differences and intra-group consistency (Fig. S8a-b). The RNA sequencing revealed significant changes in gene expression in Arid1a cKO microglia post-TBI (p-value < 0.01; |log2 fold change| > 1) (Fig. 5a). To understand the roles of these up- and down-regulated differentially expressed genes (DEGs), we performed Gene Ontology (GO) analysis and found that up-regulated DEGs were significantly associated with migration- and inflammatory response-relevant GO terms, such as Ccl5, Il17ra and C5ar1 (Fig. 5b-c).
Fig. 5.
ARID1A directly regulates Ccl5. a Heatmap showing DEGs in Arid1a cKO microglia post-TBI. n = 3. b Top significantly enriched GO terms of up-regulated and down-regulated genes post-TBI. Up-regulated DEGs are significantly associated with GO terms related to migration and inflammatory response. c Volcano plot for DEGs with log2 fold change. Key DEGs of interest, including Ccl5, Il17ra and C5ar1, are highlighted within the black boxes. d Genome-browser view of Ccl5 from the RNA-seq, ChIP-seq, and ATAC-seq datasets. The black dotted rectangle highlights the peak gain. e-f Both qPCR (e) and ELISA (f) assays reveal a significant upregulation of Ccl5 expression in cKO microglia compared to WT group in vitro. n = 4. Data are presented as mean ± SEM; *P < 0.05; **P < 0.01 by Student’s t-test
CCL5 is a well-known chemokine that plays a crucial role in promoting migration of various cells. To investigate whether ARID1A directly regulates Ccl5, we re-analyzed publicly available ChIP-seq and ATAC-seq databases related with ARID1A [25, 35]. We found that ARID1A directly binds the coding regions of the Ccl5 gene, the chromatin accessibility is significantly increased at the Ccl5 loci after Arid1a deletion (Fig. 5d), This supports the idea that Ccl5 may be directly regulated by ARID1A, rather than Il17ra and C5ar1(Fig. S9a-b). To experimentally validate Ccl5 as a downstream target of ARID1A, we isolated microglia from WT and cKO newborn pups, and stimulated them with LPS. The RT-PCR results showed that there was a significant up-regulation of Ccl5 mRNA expression in Arid1a cKO microglia after LPS stimulation (Fig. 5e). Our ELISA assay also detected an elevated CCL5 protein level in cell culture supernatant of Arid1a cKO microglia in response to LPS stimulation (Fig. 5f). These findings support that Ccl5 expression was repressed by ARID1A.
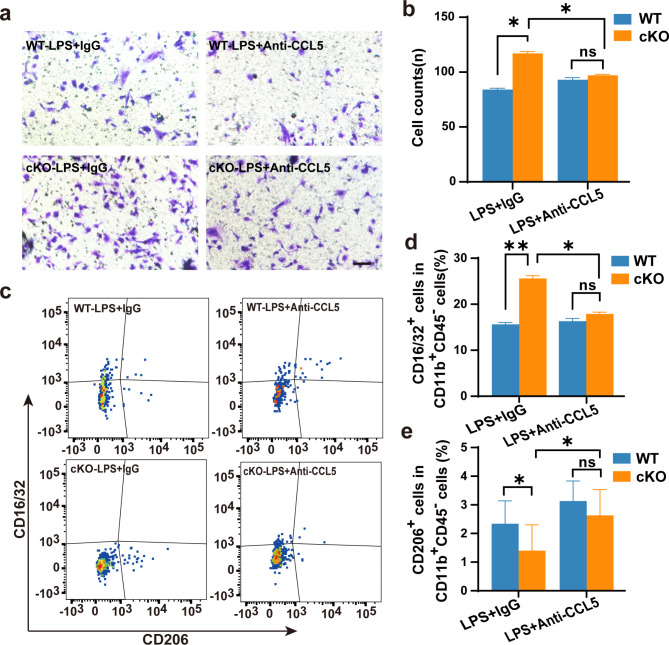
To investigate the potential of Ccl5 in regulating the migration of microglia, we administrated anti-CCL5 neutralizing antibody to treat primary microglia and observed a reduction in the migration capability to the lower chamber (Fig. 6a-b) as well as a phonotypic switching from pro-inflammatory state to tissue-repairing state in cKO microglia (Fig. 6c-d). These results suggest that blocking CCL5 could rescue the migration and pro-inflammation defects in Arid1a cKO microglia.
Fig. 6.
Arid1a regulates microglial migration and state through Ccl5. a-b Representative images (a) and quantification (b) of microglia migration in transwell assay following treatment with anti-CCL5 antibody. n = 4. Scale bar: 100 μm. c-e Representative images (c) and quantification of flow cytometry in vitro. Quantification of flow cytometric analysis of CD16/32+ microglia (d) and CD206+ microglia (e). n = 4. Data are presented as mean ± SEM; ns, no significance; *P < 0.05; **P < 0.01 by Student’s t-test
Auraptene rescues abnormal microglia migration and reactivity in Arid1a cKO mice after TBI
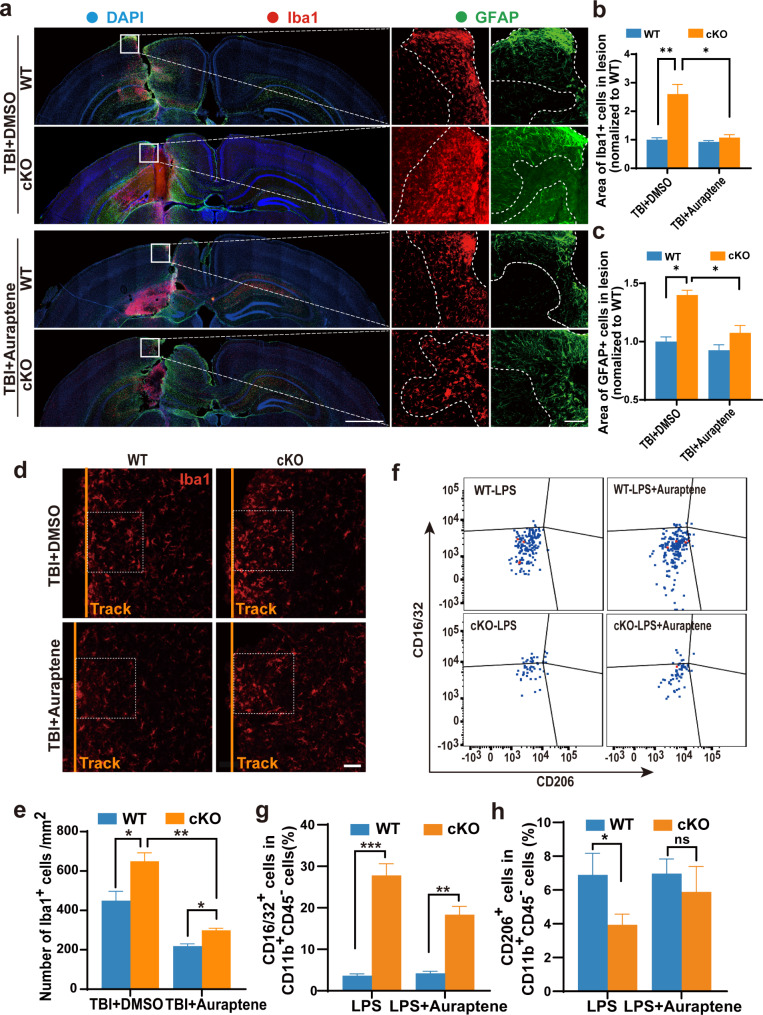
Recent studies have reported that AUR exerts anti-inflammatory effects as evidenced by suppressing inflammatory responses in the ischemic brain and by ameliorating LPS-induced inflammation in the mouse brain [36–38]. Since 0.2 μM AUR can significantly decrease the secretion of CCL5 secreted by LPS-stimulated oral epithelial cells [39], we speculated that AUR might be beneficial to Arid1a cKO mice after TBI. To test this hypothesis, we subcutaneously administered AUR at a dosage of 25 mg/kg/day for 3 consecutive days before TBI and for 7 consecutive days after TBI. Such a dosage of AUR can efficiently act as an anti-inflammatory agent in the mouse brain with ischemic surgery [28]. We then performed IF staining and observed that AUR treatment significantly reduced the accumulation of microglia at lesion sites in the cKO group at 7 dpi (Fig. 7a-b). Similarly, the aggregation of astrocytes in the cKO group was also significantly reduced following AUR treatment (Fig. 7a and c). Subsequent analyses revealed a notable decrease in the density of microglia near the injury site in the cKO group following AUR treatment (Fig. 7d-e). Consistently, transwell migration assay proved that treatment of AUR did reduce the migration of Arid1a cKO microglia (Fig. S10a-b). Additionally, we observed that AUR treatment effectively attenuated the percentage of pro-inflammatory state (CD16/32+ microglia) from 28% down to 19% in Arid1a cKO primary microglia (Fig. 7f-h).
Fig. 7.
Auraptene treatment alleviates the formation of glial scar in Arid1a cKO mice following TBI. a Representative image of IF staining for Iba1(red), GFAP (green) and nuclei (DAPI, blue) in the auraptene-treated brains at 7 dpi. Scale bars: left panel, 1,000 μm; right panel, 100 μm. b, c Quantification of Iba1+ (b) and GFAP+ area (c) at 7 dpi. n = 3. d Representative images of Iba1 (red) immunostaining around the injury sites at 7 dpi. The injury tracks are indicated by the yellow lines. n = 3. Scale bar: 50 μm. e Quantification of the density of Iba1+ cells around the lesion sites at 7 dpi. f-h AUR represses inflammatory response of Arid1a cKO microglia upon LPS-stimulation. Representative images of flow cytometric analysis in vitro (f). Quantification of CD16/32+ microglia (g) and CD206+ microglia (h) in the flow cytometric assay. n = 3 or 4. Data are presented as mean ± SEM; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001
Altogether, this study provides evidence that Arid1a plays a crucial role in regulating microglial migration and reactivity in response to TBI. Microglial ARID1A deficiency leads to exacerbated glial scar formation and elevated inflammation through the upregulation of CCL5. Importantly, treatment with AUR ameliorates the deficits of Arid1a cKO mice after TBI.
Discussion
Traumatic brain injury is the most prevalent of all head injuries. Microglia play critical roles in both the healthy and injured brains. Microglia detect and rapidly respond to any disruption in the status quo of the CNS, including infections or tissue injury, and often act to remove cellular debris. The activation of microglia occurs within minutes of the injury and is critical for recovery. In the present study, we elucidated that the deletion of Arid1a results in enhanced microglial migration, elevated inflammation and enlarged glia scar (Fig. S11) through the up-regulation of Ccl5 after TBI.
TBI triggers a cascade of events in the brain, including the damaged blood-brain barrier, migration of glial cells to the injury site, heightened inflammatory responses, and widespread cell death [6, 40]. Our study reveals that the increased number of microglia at the lesion site in the Aridla cKO mice mainly resulted from the enhanced migration of cKO microglia in the unilateral hippocampal stab wound injury model. Considering that the number of infiltrated macrophages is closely related with the types and time windows of TBI [41, 42], we cannot rule out the possibility that ARID1A also plays an important role in the macrophage infiltration and their inflammatory response. Future studies are needed to explore the function of ARID1A in the infiltrated monocytes/macrophages and to elucidate the mechanism underlying the enhanced migration of Aridla cKO microglia to the injury sites in both diffuse and focal TBI models before translating research evidence into clinical practice.
Our study revealed an enhanced migration capability of microglia in the cKO-TBI group. Specifically, we observed an increased accumulation of microglia near the injury track. However, defining the initial position of microglia near the glial scar in TBI, measuring the migration distance, and distinguishing between migration and proliferation during this process remain challenging. While there are established methods for tracking microglia migration in vivo [43, 44], these approaches often require extended observation periods and the assistance of surrounding cells to accurately trace microglial movement. Such methods are not ideally suited to our TBI model. Instead, we utilized a transwell assay in vitro to demonstrate that the migration ability of microglia is indeed increased in the cKO group.
CCL5 is a chemokine that signals to various cells and plays a crucial role in promoting migration of monocytes [45], lung cancer cells [46], and glioma cells [47]. Our study indicated that the loss of microglial Arid1a increased the expression of the chemokine Ccl5, thereby leading to enhanced migration of microglia towards the lesion of injury after TBI. This finding aligns with a previous study demonstrating that CCL5 attracts monocytes and induces significant monocyte migration in vitro, but its underlying mechanism remains unknown [45]. In cancer cells, CCL5 acts through PI3K/AKT/NF-κB signaling pathway in the upregulation of the adhesive molecule αvβ3 integrin that contributes to the migration of lung cancer cells [46]. In contrast, in response to microglia/macrophages-produced CCL5, glioma cells exhibit an increase in PYK2 phosphorylation and MMP2 activation which is critical for glioma invasion [47]. Since the activation of MMP2 has been reported in various TBI models [48–50], we speculate that microglia at the lesion site release a large amount of CCL5 following TBI, which in turn induce microglia surrounding the lesion area to activate the PI3K/AKT/NF-κB/αvβ3 and/or MMP2 signaling pathways that ultimately lead to the migration of microglia in the distance towards the lesion site. Further investigations are required to confirm the hypothesis here identified.
Our findings suggest that AUR alleviates the inflammatory response by reducing glial scar formation and microglial migration in the Arid1a cKO mice after TBI. Since AUR has an inhibitory effect on CCL5 expression [39], and our study presents multiple lines of evidence highlighting the importance of CCL5, in alleviating the inflammatory response by reducing glial scar formation and microglial migration in the Arid1a cKO mice after TBI, we speculate that AUR is working at least through CCL5-mediated signaling pathways. It is also worth noting that AUR has the ability to suppress the release of cytotoxic agents, such as COX-2 from astrocytes [36], nitric oxide (NO) from macrophages [51], and TNF-α from inflammatory leukocytes [52]. As astrocytes and infiltrated leukocytes are key regulators of neuroinflammation and glial scar formation after TBI, further investigations are required to clarify the molecular interactions between AUR and COX-2/NO/ TNF-α in Arid1a cKO mice following TBI.
In summary, this study demonstrates that the ARID1A-CCL5 axis in the regulation of microglial migration, inflammation and glial scar formation after TBI. More importantly, AUR administration rescues the deficits in the migration and reactivity of Arid1a cKO microglia in response to injury, highlighting its therapeutic potential for TBI.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was financially supported by grants from the National Key Research and Development Program of China Project (2021YFA1101400), the National Science Foundation of China (82271428/ 82301346/82130029/82201540/32170808), and the Open Project Program of the State Key Laboratory of Stem Cell and Reproductive Biology. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Abbreviations
- ATP
Adenosine triphosphate
- AUR
Auraptene
- BrdU
Bromodeoxyuridine
- BSA
Bovine serum albumin
- cKO
Conditional knockout
- CNS
Central nervous system
- CSS
Coffin-Siris syndrome
- DAPI
4’,6-diamidino-2-phenylindole
- DEGs
Differentially expressed genes
- DMEM
Dulbecco’s Modified Eagle Medium
- dpi
Days past injury
- FBS
Fetal calf serum
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- IF
Immunofluorescence
- LPS
Lipopolysaccharide
- mSWI/SNF
Mammalian SWI/SNF
- NO
Nitric oxide
- PBS
Phosphate-buffered saline
- PCA
Principal components analysis
- PFA
Paraformaldehyde
- TBI
Traumatic brain injury
- WT
Wildtype
Author contributions
Jin-Peng Ke, Zhao-Qian Teng and Chang-Mei Liu designed the experiments of the study. Jin-Peng Ke, Bao-Dong He, and Chang-Mei Liu contributed to the execution of experiments and manuscript composition. Bao-Dong He performed animal TBI model. Jin-Peng Ke, Mao-Lei Gong and Zhong-Ze Yan performed sequence analysis. All authors discussed the results, revised the manuscript, and read and approved the final manuscript.
Data availability
All data in this study are included in this published article and its additional information files. The sequencing data have been deposited in Genome Sequence Archive of Beijing Institute of Genomics, Chinese Academy of Sciences (http://gsa.big.ac.cn/) under accession number PRJCA026775.
Declarations
Ethics approval and consent to participate
The experiments were performed according to the Guide for the Care and Use of Laboratory Animals of Institute of Zoology of Chinese Academy of Sciences.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhao-Qian Teng, Email: tengzq@ioz.ac.cn.
Chang-Mei Liu, Email: liuchm@ioz.ac.cn.
References
- 1.Mollayeva T, Mollayeva S, Colantonio A. Traumatic brain injury: sex, gender and intersecting vulnerabilities. Nat Rev Neurol. 2018;14(12):711–22. [DOI] [PubMed] [Google Scholar]
- 2.Quaglio G, et al. Traumatic brain injury: a priority for public health policy. Lancet Neurol. 2017;16(12):951–2. [DOI] [PubMed] [Google Scholar]
- 3.Ponsford J, Spitz G, Hicks AJ. Highlights in traumatic brain injury research in 2021. Lancet Neurol. 2022;21(1):5–6. [DOI] [PubMed] [Google Scholar]
- 4.Li LM, Carson A, Dams-O’Connor. Psychiatric sequelae of traumatic brain injury - future directions in research. Nat Rev Neurol. 2023;19(9):556–71. [DOI] [PubMed] [Google Scholar]
- 5.Jarrahi A et al. Revisiting traumatic brain injury: from molecular mechanisms to therapeutic interventions. Biomedicines. 2020;8(10). [DOI] [PMC free article] [PubMed]
- 6.Karve IP, Taylor JM, Crack PJ. The contribution of astrocytes and microglia to traumatic brain injury. Br J Pharmacol. 2016;173(4):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinn MJ, Povlishock JT. Pathophysiology of traumatic brain injury. Neurosurg Clin N Am. 2016;27(4):397–407. [DOI] [PubMed] [Google Scholar]
- 8.Khellaf A, Khan DZ, Helmy A. Recent advances in traumatic brain injury. J Neurol. 2019;266(11):2878–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cserép C, Pósfai B, Dénes Á. Shaping neuronal fate: functional heterogeneity of direct microglia-neuron interactions. Neuron. 2021;109(2):222–40. [DOI] [PubMed] [Google Scholar]
- 10.Harry GJ. Microglia during development and aging. Pharmacol Ther. 2013;139(3):313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sierra A, et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7(4):483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23(9):1018–27. [DOI] [PubMed] [Google Scholar]
- 13.Prinz M, Jung S, Priller J. Microglia biology: one century of evolving concepts. Cell. 2019;179(2):292–311. [DOI] [PubMed] [Google Scholar]
- 14.Johnson VE, et al. Inflammation and white matter degeneration persist for years after a single traumatic brain injury. Brain. 2013;136(Pt 1):28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witcher KG, et al. Traumatic brain injury causes chronic cortical inflammation and neuronal dysfunction mediated by microglia. J Neurosci. 2021;41(7):1597–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8(6):752–8. [DOI] [PubMed] [Google Scholar]
- 17.Bellver-Landete V, et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat Commun. 2019;10(1):518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YY, et al. Loss of microglial EED impairs synapse density, learning, and memory. Mol Psychiatry. 2022;27(7):2999–3009. [DOI] [PubMed] [Google Scholar]
- 19.Shi Z, et al. Microglia drive transient insult-induced brain injury by chemotactic recruitment of CD8(+) T lymphocytes. Neuron. 2023;111(5):696–e7109. [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro-Silva C, Vermeulen W, Lans H. SWI/SNF: complex complexes in genome stability and cancer. DNA Repair (Amst). 2019;77:87–95. [DOI] [PubMed] [Google Scholar]
- 21.Mashtalir N, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175(5):1272–e128820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullen J, et al. Targeting ARID1A mutations in cancer. Cancer Treat Rev. 2021;100:102287. [DOI] [PubMed] [Google Scholar]
- 23.Kosho T, Okamoto N. Genotype-phenotype correlation of coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am J Med Genet C Semin Med Genet. 2014;166c(3):262–75. [DOI] [PubMed] [Google Scholar]
- 24.Liu PP, et al. Acetate supplementation restores cognitive deficits caused by ARID1A haploinsufficiency in excitatory neurons. EMBO Mol Med. 2022;14(12):e15795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong M, et al. Abnormal microglial polarization induced by Arid1a deletion leads to neuronal differentiation deficits. Cell Prolif. 2022;55(11):e13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He BD, Liu CM, Teng ZQ. A mouse model of neurodegeneration induced by blade penetrating stab wound to the hippocampus. Biology (Basel). 2022;11(9). [DOI] [PMC free article] [PubMed]
- 27.Wang JX et al. Agomir-331 suppresses reactive gliosis and neuroinflammation after traumatic brain injury. Cells. 2023;12(20). [DOI] [PMC free article] [PubMed]
- 28.Okuyama S, et al. Auraptene acts as an anti-inflammatory agent in the mouse brain. Molecules. 2015;20(11):20230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegdekar N, et al. Inhibition of autophagy in microglia and macrophages exacerbates innate immune responses and worsens brain injury outcomes. Autophagy. 2023;19(7):2026–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corps KN, Roth TL, McGavern DB. Inflammation and neuroprotection in traumatic brain injury. JAMA Neurol. 2015;72(3):355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolte AC, Lukens JR. Neuroimmune cleanup crews in brain injury. Trends Immunol. 2021;42(6):480–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mira RG, Lira M, Cerpa W. Traumatic brain injury: mechanisms of glial response. Front Physiol. 2021;12:740939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiweck J, et al. Drebrin controls scar formation and astrocyte reactivity upon traumatic brain injury by regulating membrane trafficking. Nat Commun. 2021;12(1):1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosi S. A polarizing view on posttraumatic brain injury inflammatory response. Brain Circ. 2016;2(3):126–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang XY, et al. ARID1A deficiency weakens BRG1-RAD21 interaction that jeopardizes chromatin compactness and drives liver cancer cell metastasis. Cell Death Dis. 2021;12(11):990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuyama S, et al. Anti-inflammatory and neuroprotective effects of auraptene, a citrus coumarin, following cerebral global ischemia in mice. Eur J Pharmacol. 2013;699(1–3):118–23. [DOI] [PubMed] [Google Scholar]
- 37.Okuyama S, et al. Auraptene in the peels of citrus kawachiensis (Kawachi Bankan) ameliorates lipopolysaccharide-induced inflammation in the mouse brain. Evid Based Complement Alternat Med. 2014;2014:408503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuyama S et al. Auraptene and other prenyloxyphenylpropanoids suppress microglial activation and dopaminergic neuronal cell death in a lipopolysaccharide-induced model of Parkinson’s disease. Int J Mol Sci. 2016;17(10). [DOI] [PMC free article] [PubMed]
- 39.La VD, et al. Anti-inflammatory and wound healing potential of citrus auraptene. J Med Food. 2013;16(10):961–4. [DOI] [PubMed] [Google Scholar]
- 40.Adams KL, Gallo V. The diversity and disparity of the glial scar. Nat Neurosci. 2018;21(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra SK, et al. Early monitoring and quantitative evaluation of macrophage infiltration after experimental traumatic brain injury: a magnetic resonance imaging and flow cytometric analysis. Mol Cell Neurosci. 2017;78:25–34. [DOI] [PubMed] [Google Scholar]
- 42.Alam A, et al. Cellular infiltration in traumatic brain injury. J Neuroinflammation. 2020;17(1):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn SJ, et al. Diverse inflammatory response after cerebral microbleeds includes coordinated microglial migration and proliferation. Stroke. 2018;49(7):1719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casden N, et al. Astrocyte-to-microglia communication via Sema4B-Plexin-B2 modulates injury-induced reactivity of microglia. Proc Natl Acad Sci U S A. 2024;121(22):e2400648121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussen J, et al. The chemokine CCL5 induces selective migration of bovine classical monocytes and drives their differentiation into LPS-hyporesponsive macrophages in vitro. Dev Comp Immunol. 2014;47(2):169–77. [DOI] [PubMed] [Google Scholar]
- 46.Huang CY, et al. CCL5 increases lung cancer migration via PI3K, Akt and NF-kappaB pathways. Biochem Pharmacol. 2009;77(5):794–803. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y-J. CCL5 of glioma-associated microglia/macrophages regulates glioma migration and invasion via calcium-dependent matrix metalloproteinase 2. Neuro Oncol. 2020;22(2):253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdul-Muneer PM, et al. Role of matrix metalloproteinases in the pathogenesis of traumatic brain injury. Mol Neurobiol. 2016;53(9):6106–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdul-Muneer PM, et al. Traumatic brain injury induced matrix metalloproteinase2 cleaves CXCL12α (stromal cell derived factor 1α) and causes neurodegeneration. Brain Behav Immun. 2017;59:190–9. [DOI] [PubMed] [Google Scholar]
- 50.Salehi A, Zhang JH, Obenaus A. Response of the cerebral vasculature following traumatic brain injury. J Cereb Blood Flow Metab. 2017;37(7):2320–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohno S, et al. Methyl galbanate, a novel inhibitor of nitric oxide production in mouse macrophage RAW264.7 cells. J Nat Med. 2011;65(2):353–9. [DOI] [PubMed] [Google Scholar]
- 52.Murakami A, et al. Suppression by citrus auraptene of phorbol ester-and endotoxin-induced inflammatory responses: role of attenuation of leukocyte activation. Carcinogenesis. 2000;21(10):1843–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study are included in this published article and its additional information files. The sequencing data have been deposited in Genome Sequence Archive of Beijing Institute of Genomics, Chinese Academy of Sciences (http://gsa.big.ac.cn/) under accession number PRJCA026775.