Abstract
Background
Proteoglycans are important tumor microenvironment extracellular matrix components. The regulation of key proteoglycans, such as decorin (DCN), by miRNAs has drawn attention since they have surfaced as novel therapeutic targets in cancer. Accordingly, this study aimed at identifying the impact of miR-181a in liver cancer and its regulatory role on the extracellular matrix proteoglycan, DCN, and hence on downstream oncogenes and tumor suppressor genes.
Results
DCN was under-expressed in 22 cirrhotic and HCC liver tissues compared to that in 11 healthy tissues of liver transplantation donors. Conversely, miR-181a was over-expressed in HCC liver tissues compared to that in healthy liver tissues. In silico analysis predicted that DCN 3’UTR harbors two high-score oncomiR-181a binding regions. This was validated by pmiRGLO luciferase reporter assay. Ectopic miR-181a expression into HuH-7 cells repressed the transcript and protein levels of DCN as assessed fluorometrically and by western blotting. DCN siRNAs showed similar results to miR-181a, where they both enhanced the cellular viability, proliferation, and clonogenicity. They also increased Myc and E2F and decreased p53 and Rb signaling as assessed using reporter vectors harboring p53, Rb, Myc, and E2F response elements. Our findings demonstrated that miR-181a directly downregulated the expression of its direct downstream target DCN, which in turn affected downstream targets related to cellular proliferation and apoptosis.
Conclusion
To our knowledge, this is the first study to unveil the direct targeting of DCN by oncomiR-181a. We also highlighted that miR-181a affects targets related to cellular proliferation in HCC which may be partly mediated through inhibition of DCN transcription. Thus, miR-181a could be a promising biomarker for the early detection and monitoring of liver cancer progression. This would pave the way for the future targeting of the oncomiR-181a as a therapeutic approach in liver cancer, where miR-181a-based therapy approach could be potentially combined with chemotherapy and immunotherapy for the management of liver cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12876-024-03413-6.
Keywords: Decorin (DCN), microRNA-181a (miR-181a), Hepatocellular carcinoma (HCC), Liver cancer
Introduction
Hepatocellular carcinoma (HCC) ranks as the third most common cause of cancer-related death worldwide, posing a constant threat in both developed and developing countries [1]. In Egypt, liver cancer has the highest incidence, mortality, and prevalence, according to the Global Cancer Statistics 2020 report [1]. Alarmingly, the burden of HCC in Egypt has been on the rise with a doubling in the incidence rate among patients with chronic liver disease over the period of a decade [2]. Several environmental and biological factors contribute to the etiology or progression of HCC, particularly hepatitis C virus (HCV) infection where, until the recent advent of direct acting antiviral agents against HCV, Egypt has had one of the highest prevalence rates of this viral infection in the world [3, 4]. Moreover, liver cancer is highly resistant and refractory to various therapeutic interventions; a recent prospective study of HCC management concluded the poor survival of the patients with HCV-related liver cancer with the most common therapeutic modalities. HCV-infected HCC patients face several significant treatment challenges. Antiviral resistance in HCV-related HCC, which is often diagnosed at an advanced stage, limits the treatment options. While recent advancements in antiviral therapy have improved the treatment outcomes, the risk of disease recurrence and limited access to care in certain regions remain significant challenges. Although this study was conducted in Egypt, the authors reported findings similar to those reported from high-income countries [5, 6]. Another reason behind this therapeutic challenge is the tumor microenvironment, which can foster recurrent de novo HCC tumors, thus rendering HCC treatment as challenging [7].
The extracellular matrix is an important component of the tumor microenvironment and is primarily composed of proteoglycans, which have a core protein and glycosaminoglycan chains. Proteoglycans maintain tissue homeostasis on the physiological level and show dramatic changes in their expression levels in carcinogenic tissues. Decorin (DCN) is a small multifunctional cellular or pericellular matrix proteoglycan and structural constituent of the hepatic microenvironment. Its name stems from its ability to “decorate” the collagen fibrils, thus regulating matrix assembly and homeostasis by fibrillogenesis [8]. However, there was a paradigm shift in our understanding of the emerging role of DCN in impeding the growth of tumor cells [9, 10], where it was manifested that DCN affects various biological processes, including cell growth, differentiation, proliferation, adhesion, spread and migration, and regulates inflammation and fibrillogenesis [11]. Accordingly, two chief footprints have emerged for DCN on the functional level, namely, preserving the cellular structure and controlling different signal transduction pathways with an eventual culmination of tumor suppressive effects.
Several studies have reported higher levels of DCN in normal liver tissue compared to those in tumorous liver tissues [12, 13]. Interestingly, the sequential evolution of liver cirrhosis into HCC is marked by the progressive reduction of DCN levels, suggesting a critical role for this proteoglycan in cancer progression [14]. Of note, HCC patients with higher expression of DCN have more favorable survival rates [13, 15]. Mechanistically, DCN potently suppresses cell proliferation, survival, migration, and angiogenesis and could thus generate a powerful antitumorigenic signal [16–19]. Moreover, introduction of recombinant human DCN increases apoptosis, inhibits cell proliferation, and induces G0/G1 phase arrest through reactivating the expression of cyclin-dependent kinase inhibitor 1 C (p57Kip2) [20]. In a carcinogen-induced HCC mouse model, DCN knockout mice showed enhanced tumor predominance compared with wild-type mice [21]. Therefore, regulating DCN by noncoding RNAs, specifically miRNAs, might represent a cornerstone in understanding the behavior of DCN in liver cancer.
Several miRNAs are aberrantly expressed in HCC tissues; thus, these small endogenous molecules emerged as potential biomarkers and novel molecules or targets for tumor therapy. The first small-interfering RNA was granted FDA approval as a therapeutic agent in 2018, and several candidate miRNA therapeutics are in clinical development or clinical trials [22]. In this age of rapidly developing RNA technology, the use of miRNA mimics or repressors is continuously gaining traction as antitumor therapeutic molecules. The mechanistic epigenetic role of miRNAs as potential upstream microregulators of DCN in HCC and their subsequent impact on downstream targets and cancer progression pose a significant research gap. Therefore, this study aimed at identifying the mechanistic role of miR-181a as a possible upstream microregulator of DCN in HCC and hence its impact on the regulation of downstream transcription factors and cancer progression.
Patients and methods
Patients
Liver biopsies were obtained from 22 HCC patients and 11 healthy donors during liver transplantation at the Kasr El Ainy and Dar El-Fouad hospitals in Egypt. The clinical data of patients and healthy volunteers is shown in Table 1. The experiments were undertaken with the understanding and written informed consent of each patient. The inclusion criteria of the study patients with HCC included patients with : (1) histologically confirmed diagnosis of HCC; (2) age range of 20–65 years; (3) confirmed HCV infection; and (4) no history of other primary cancers. The exclusion criteria included: (1) the presence of other active malignancies; (2) severe comorbidities that could interfere with study participation or outcomes; and (3) lymph node metastasis. The inclusion criteria for the healthy donors included: (1) age- and sex-matching with the patients with HCC; (2) no history of liver disease or other chronic conditions; and (3) no current or past history of cancer.
Table 1.
Clinical parameters and characteristic features of non-metastatic HCC patients and healthy volunteers
| Parameters | Average ± SDa | |
|---|---|---|
| HCC and Cirrhotic Patients | Age | 49 ± 13.5 |
| Sex (male% / female %) | 95.5% / 4.5% | |
| Ethanol abuse | None | |
| Aspartate aminotransferase (AST) (U/L) | 100.5 ± 65.8 | |
| Alanine aminotransferase (ALT) (U/L) | 85.6 ± 95.6 | |
| Alkaline phosphatase (U/L) | 110.2 ± 60.7 | |
| Serum albumin (g/dL) | 4.6 ± 1.5 | |
| Serum AFP (ng/mL) | 155.7 ± 22.3 | |
| HCV Ab | 100% (22 HCC patients) | |
| HBV Ab | 18.2% (4/22 HCC patients) | |
| Healthy Controls (Liver Donors) | Age | 31 ± 10.5 |
| Sex (male% / female %) | 70% / 30% | |
| Ethanol abuse | None | |
| Diabetic | None | |
| Hypertensive | None | |
| HCV status b | None | |
| HBV status c | None |
a Data is presented as average ± SD
b HCV status was determined using anti-HCV antibody and/or HCV viral RNA quantification
c HBV status was determined using anti-HBc and anti-HBs antibodies and by detection of HBsAg
The study methodologies conformed to the ethical standards set by the declaration of Helsinki. The ethical committees of Cairo University and the German University in Cairo approved the study methodologies. Liver disease assessment for each patient is shown in Table 2.
Table 2.
Liver disease assessment for each patient: Number/size of focal lesions, scoring, and PT-INR of each HCC patient according to the Milan criteria
| Patients | Number of focal lesions | Size of focal lesions | MELDa Score | Child-Pugh Score | PT-INRb |
|---|---|---|---|---|---|
| Patient 1 | 3 focal lesions | 1.5 cm, 1 cm, and 1 cm | 11 | C | 1.4 |
| Patient 2 | Unifocal | 2.5 cm | 15 | C10 | 1.6 |
| Patient 3 | 3 focal lesions | 2 cm, 2.5 cm, and 3 cm | 10 | C11 | 2.4 |
| Patient 4 | 3 focal lesions | 2 cm, 2 cm, and 3.5 cm | 10 | C | 2 |
| Patient 5 | Unifocal | 1.5 × 2 cm | 16 | C | 1.4 |
| Patient 6 | 3 focal lesions | 3 × 4 cm, 1 cm, and 1 cm | 12 | B | 1.5 |
| Patient 7 | Unifocal | 4 cm | 11 | B7 | 1.5 |
| Patient 8 | 3 focal lesions | 4 cm, 1 cm, and 1 cm | 8 | A5 | 1.3 |
| Patient 9 | 3 focal lesions | 1 cm, 1 cm, and 1.5 cm | 17 | C | 1.5 |
| Patient 10 | Unifocal | 2.5 cm | 10 | B8 | 1.4 |
| Patient 11 | 2 focal lesions | 1 cm and 1.7 cm | 15 | B8 | 1.4 |
| Patient 12 | 3 focal lesions | 1 cm each | 17 | C11 | 1.8 |
| Patient 13 | Unifocal | 3 cm | 13 | B7 | 1.6 |
| Patient 14 | 3 focal lesions | 3 cm, 1.5 cm, and 2 cm | 15 | B | 1.6 |
| Patient 15 | 3 focal lesions | 1 cm, 1 cm, and 4 cm | 12 | B7 | 1.14 |
| Patient 16 | 2 focal lesions | 3 cm and 1.5 cm | 19 | C12 | 1.9 |
| Patient 17 | 2 focal lesions | 1.5 cm and 3 cm | 16 | C10 | 1.55 |
| Patient 18 | 3 focal lesions | 2.5 cm, 2.5 cm, 1.5 cm | 9 | B7 | 1.18 |
| Patient 19 | 3 focal lesions | 1.5 cm, 1 cm, and 1 cm | 11 | B8 | 1.19 |
| Patient 20 | Unifocal | 2 cm | 18 | C | 1.7 |
| Patient 21 | Unifocal | 1.5 cm | 17 | C | 1.55 |
| Patient 22 | 3 focal lesions | 3 cm, 2.5 cm, and 1 cm | 12 | B | 1.6 |
a MELD: Model for End-stage Liver Disease score
b PT-INR: international normalized ratio for prothrombin time
Extraction and quantification of microRNA and mRNA from liver tissues and HuH-7 cell line
MirVana microRNA Isolation Kit (Ambion, USA) and BIOZOL RNA Extraction Reagent (Bioer Technology, China) were used, as per the manufacturer’s protocol, for total mRNA extraction from liver biopsies and HuH-7 cells, respectively. Extracted RNA was then reversed transcribed into complementary DNA (cDNA) (Applied Biosystems, USA). cDNA was amplified and quantified using TaqMan quantitative real-time polymerase chain reaction (RTqPCR) (Applied Biosystems). The housekeeping genes, RNU6B and beta-2 microglobulin (B2M) were used to normalize the relative expression of miR-181a and DCN in each sample, respectively. The 2−∆∆Ct method was used to calculate the relative gene expression.
Cell culture and transfection
The HuH-7 cells that were used in all cell culture experiments were kindly provided by Prof. Kai Breuhahn (Institute of Pathology, University Heidelberg, Heidelberg, Germany). The HuH-7 cells were cultured in DMEM supplemented with 10% FBS, 1% mycozap/penicillin/ streptomycin (Lonza, Switzerland). HiPerFect Transfection Reagent (Qiagen, Germany) was used, as per the manufacturer’s protocol, to transfect cells with siRNAs against DCN, miR-181a mimics, or negative control oligonucleotides (Qiagen, Germany).
Bioinformatics
To predict microRNAs that putatively target the 3’untranslated region (3’UTR) of the DCN transcript, the online bioinformatics software microrna.org (http://www.microrna.org) and Target Scan (http://www.targetscan.org/) were used.
MTT cell viability assay
4,5-Dimethyl thiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT) was added to HuH-7 cells 48 h post-transfection. This was followed by incubation for 4 h and adding ethanol/DMSO solution for cell lysis. Optical density was measured at 572 nm, and relative viability was calculated.
BrdU cell proliferation assay
BrdU Cell Proliferation ELISA kit (Roche Applied Biosystems, Germany) was used, as per manufacturer’s protocol, to assess proliferation 48 h post-transfection of HuH-7 cells. Luminescence was measured and expressed as relative light units per second (RLUs/s).
Colony-forming assay
HuH-7 cells were transfected with miR-181a or DCN siRNA. Then, 48 h post-transfection, the cells were detached and imbedded in a layer of 0.36% soft agarose superimposed on another agarose layer of 0.76%. The plates were then incubated for two weeks to colonize. Colonies were stained with 0.05% crystal violet and counted.
Microplate fluorimetry for protein quantification
Transfected HuH-7 cells were fixed using 4% paraformaldehyde. Blocking solution was added followed by overnight incubation with primary mouse monoclonal antibody against DCN (Santa Cruz, USA). After 24 h, Texas Red-conjugated goat anti-mouse secondary antibody (Santa Cruz, USA) solution was added and incubated in the dark for 1 h. Fixed cells were mounted by adding DAPI Fluoroshield Mounting Medium (Abcam, UK) to counterstain the nuclei and prevent rapid photobleaching of TR fluorescence. Fluorescent images were captured using Zeiss Immunofluorescent microscope. Perkin Elmer Wallac 1420 VICTOR2™ fluorometer was used to quantify fluorescence of immunolabeled proteins in each well. Excitation and emission wavelengths of TR fluorochrome (596 and 615 nm, respectively) normalized to the fluorescence of nuclei after DAPI staining using its excitation and emission wavelengths (360 and 460 nm, respectively) were used.
Protein extraction and Western blot
HuH-7 cells were lysed using RIPA buffer (Serva) containing 1X phosphatase inhibitors and 1X protease inhibitors (Thermofisher Scientific). After centrifugation, protein concentrations in the supernatants were measured using Modified Lowry protein assay kit (Thermofisher Scientific). Denaturation at 95 °C for 5 min was performed, then 40 μg of protein lysates were run on a 10% SDS-polyacrylamide gel, and transferred to a nitrocellulose membrane (Serva), using Bio-Rad western blot system (Bio-Rad). The membrane was incubated overnight with primary antibody against DCN (R&D Systems) at 4 °C on a shaker with a concentration of 3 μg in 5% BSA as well as GAPDH (Cell Signaling Technologies) overnight at 4 °C with a dilution of 1:1000 in 5% BSA. After three washes with TBS-T, the membrane was incubated for 1 h with the respective secondary antibodies as follows: secondary anti-mouse antibody (dilution, 1: 2,500) in 5% milk (Santa Cruz Biotechnology) was used for DCN, and secondary anti-rabbit antibody (dilution, 1:10,000) in 5% milk (Cell Signaling Technology) was used for GAPDH. Protein bands were visualized using Pierce ECL Western blotting substate (ThermoFisher Scientific), and relative band intensity was quantified using ImageQuant software.
miR-181a binding confirmation using reporter constructs
Wild-type constructs (WT) were made by designing oligonucleotide inserts containing the miR-181a-binding regions in the 3’UTR of DCN, and ligating these inserts downstream to a luciferase reporter gene in the pmirGLO Dual-Luciferase microRNA Target Expression Vector (Promega, USA). To ensure site-specific binding, mutant constructs (MUT) were also prepared, where the ligated oligonucleotide inserts did not contain the binding region nucleotides. Table 3 shows the oligonucleotide insert sequences that were designed and ligated to form the wild-type and mutant constructs for each of the two miR-181a/DCN 3’UTR binding regions. HuH-7 cells were transfected with 2 μg of either the WT or MUT construct using SuperFect transfection reagent (Qiagen). After 24 h, cells were co-transfected in a 96-well plate with miR-181a mimics (2 μL; ∼22.4 ng) using HiPerFect transfection reagent (0.75 μL; Qiagen) and 25 μL serum-free DMEM. Relative luciferase activity was measured after 48 h using Steady-Glo® Luciferase Reporter Assay Kit (Promega, USA).
Table 3.
Oligonucleotide design for hsa-miR-181a/DCN 3’UTR inserts (wild-type and mutant)
| Insert name | Oligonucleotide sequence* |
|---|---|
| Binding site 1 (wild-type) | Sense: 5’- CCATTACTGGTAAAGCCTCATTTGAATGTGTGAATTT-3’ |
| Antisense: 5’-CTAGAAATTCACACATTCAAATGAGGCTTTACCAGTAATGGAGCT-3’ | |
| Binding site 1 (mutant) | Sense: 5’- CCATTACTGGTAAAGCCTCATTGTGAATTT-3’ |
| Antisense: 5’-CTAGAAATTCACAATGAGGCTTTACCAGTAATGGAGCT-3’ | |
| Binding site 2 (wild-type) | Sense: 5’- CGGTAACTTATGTCATCTATGTTGAATGTAAGCTTAT-3’ |
| Antisense: 5’-CTAGATAAGCTTACATTCAACATAGATGACATAAGTTACCGAGCT-3’ | |
| Binding site 2 (mutant) | Sense: 5’-CGGTAACTTATGTCATCTATGTAAGCTTAT − 3’ |
| Antisense: 5’-CTAGATAAGCTTACATAGATGACATAAGTTACCGAGCT-3’ |
*the miR-181a binding regions are indicated by bold and underlined font
Analysis of downstream oncogenes and tumor suppressor transcription factors
Using SuperFect transfection reagent, HuH-7 cells were transfected with four different vectors containing cis-acting enhancer response elements specific for the cell cycle regulatory proteins, p53, Rb, Myc, and E2F, upstream of a sensitive luciferase reporter gene (pp53-TA-Luc, pRb-TA-Luc, pMyc-TA-Luc, and pE2F-TA-Luc, respectively; Clontech, Germany). This setup allows for the measurement of transcriptional activity driven by these specific regulatory elements. As a control, pLuc vector containing an unspecific binding site (pTAL-Luc; Clontech, Germany) was used. After 24 h, cells were co-transfected with siRNAs against DCN or with miR-181a mimics. Upon binding of the respective protein to its response element, a downstream luciferase gene is expressed and assayed 48 h later using Steady-Glo® Luciferase Assay Kit (Promega, Germany), as per the manufacturer’s instructions. Luminescence was used to calculate the transcriptional activity of each regulatory protein and plotted as percentage transcriptional activity relative to cells transfected with the vector alone. Unspecific luminescence detected by the reagents and empty pTAL-Luc vector (baseline luminescence) was subtracted from all the values before plotting.
Statistical analyses
Gene expression is expressed in relative quantitation (RQ = 2–∆∆Ct). All data are expressed as mean ± standard error of the mean (SEM), unless otherwise stated. Unpaired Student’s t test (parametric and two-tailed) was used for all the experiments to compare between two groups. All experiments were performed in quadruplicates (n = 4) and repeated at least three times, unless otherwise stated. A p-value less than 0.05 was considered as statistically significant, where *** = p < 0.001, ** = p < 0.01, * = p < 0.05 and ns = statistically not significant. All the data were statistically analyzed using GraphPad Prism 5.00 software.
Results
Expression profiling of DCN and miR-181a in liver tissues and HuH-7 cells
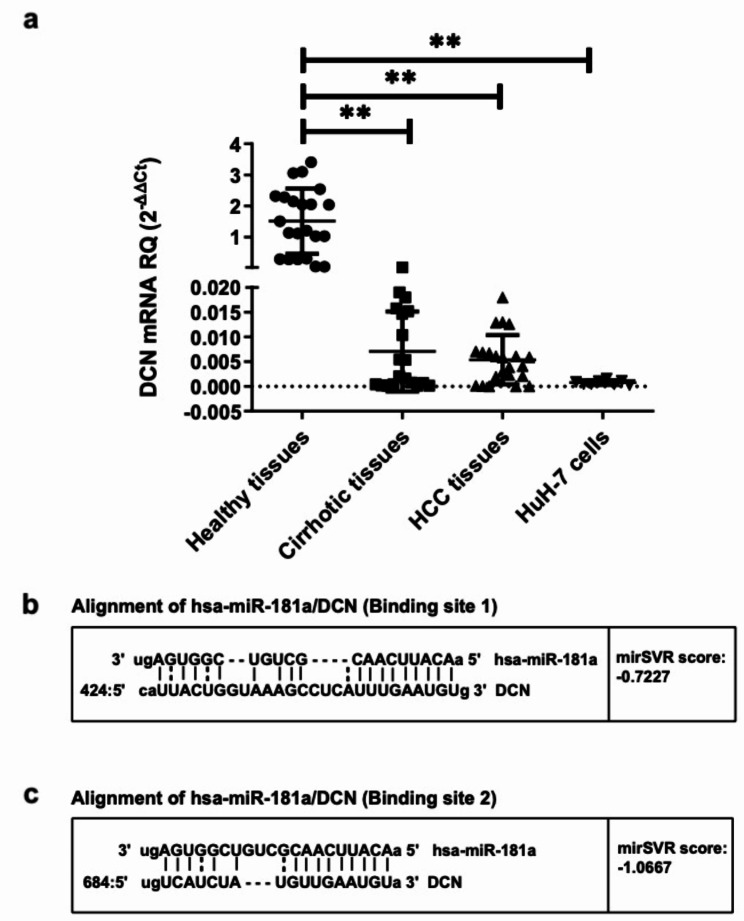
The small leucine-rich pericellular proteoglycan, DCN, an important component of the hepatic microenvironment, acts as a tumor suppressor in a variety of cancers, including HCC [9, 10]. To establish its expression levels in a cohort of Egyptian HCV-infected HCC patients, DCN mRNA was screened in both tumorous and adjacent cirrhotic liver tissues from HCC patients undergoing liver transplantation and compared to normal liver tissues from healthy liver donors. DCN was significantly under-expressed in both cirrhotic (p = 0.0016) and HCC tissues (p = 0.0059) when compared to those taken from healthy livers. Similarly, HCC cell line HuH-7 showed significantly lower expression levels of DCN compared to healthy liver tissues (p = 0.0016, Fig. 1a). Next, it was crucial to screen for the expression levels of miR-181a in the HCC and healthy liver tissues. miR-181a levels were significantly overexpressed in the HCC tissues when compared to those in the healthy liver tissues (p = 0.0244, Supplementary Figure S1).
Fig. 1.
Expression profiling of DCN in liver tissues and predicted targeting of DCN 3’UTR by miR-181a. (a) The expression of DCN was investigated in healthy, cirrhotic, and HCC liver biopsies in addition to HuH-7 cells. Each tissue sample was carried out in duplicates and repeated three times. (b, c) Alignment of the two high-score binding sites on DCN transcript with miR-181a seed sequence, as predicted by microRNA.org. Vertical lines indicate complementarity between the binding region of the mRNA and the seed sequence of the miRNA, while the dots indicate mismatches or GU wobbles. Also shown are the mirSVR scores, which factor in multiple features of the predicted miRNA: mRNA duplex, including (1) duplex features which includes base pairing at the seed region and 3’end of the miRNA; (2) sequence features which include A/U composition near the target sites and secondary structure accessibility; and (3) global features such as length of the UTR, relative position of the target site in the UTR and conservation score. The MirSVR downregulation scores correlate linearly with the extent of downregulation. Asterisks indicates statistically significant differences, where **p < 0.01
Bioinformatics to predict an upstream microregulator of DCN
Since DCN was found to be severely suppressed in cancerous liver tissues and given that miRNAs are predicted to regulate approximately 30% of all protein-coding genes, bioinformatics was performed to identify a miRNA with high potential to be targeting and downregulating this proteoglycan. Using two different computational online algorithms, namely microrna.org (http://www.microrna.org) and Target Scan (http://www.targetscan.org/), hsa-miR-181a was predicted to target the 3’UTR of DCN at two different binding regions with perfect complementarity between the 8-nucleotide seed sequence of miR-181a and its target sites (Fig. 1b, c). Binding scores given by the software represent the hybridization energy exerted for miR-181–DCN mRNA binding, where the lower the energy, the higher the predicted binding [23].
Impact of miR-181a on DCN mRNA and protein expression in HuH-7 cells
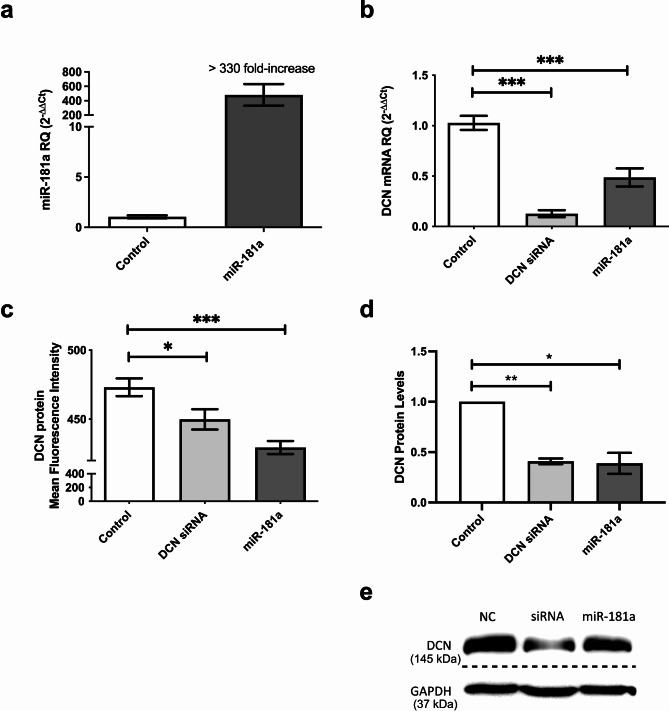
To explore the effect of miR-181a on its predicted downstream target, DCN, synthetic oligonucleotide miR-181a mimics were transfected into HuH-7 cells. Ectopic delivery of mimics led to a more than 330-fold increase in intracellular levels of miR-181a in transfected HuH-7 cells (p = 0.0096), confirming transfection efficiency (Fig. 2a). Overexpression of miR-181a resulted in a significant downregulation of DCN mRNA (p < 0.0001), similar to the decrease caused by knockdown of DCN using specific siRNAs (p < 0.0001, Fig. 2b). Moreover, both DCN siRNAs and miR-181a mimics produced a marked reduction of DCN protein levels, measured both fluorimetrically (p = 0.0240 for DCN siRNAs and p < 0.0001 for miR-181a, Fig. 2c) and by western immunoblotting (p = 0.0023 for DCN siRNAs and p = 0.0278 for miR-181a, Fig. 2d, e). The original uncropped blots of DCN and GAPDH are presented in Additional file 1 (Supplementary Figure S2).
Fig. 2.
Impact of miR-181a on DCN mRNA and protein in HuH-7 cells. (a) HuH-7 cells were transfected with miR-181a mimics and the efficiency of miR-181a delivery was confirmed on the mRNA level using RTqPCR. (b) Relative expression of DCN was determined using RTqPCR in HuH-7 cells transfected with miR-181a mimics or specific siRNAs against DCN as a positive control. (c) Relative fluorescence intensity of DCN protein was quantified using a multilabel counter fluorometer after fluorescent labeling of transfected HuH-7 cells, and readings were plotted as % mean fluorescence intensity (MFI). The experiment was carried out in quadruplets and repeated three times. (d) Relative band intensity was quantified using ImageQuant software after western blotting to determine the DCN protein levels in transfected HuH-7 cells. The experiment was carried out in duplicates and repeated three times. (e) Representative image of the western blot results. The loading controls were run on the same gel. Then, the blot was cropped during the experiment as a different set of secondary antibodies was used for DCN and GAPDH. Thus, the figure represents two grouped blots from different parts of the same blot (the three upper bands represent the blot that was incubated with DCN antibody, while three lower bands represent the blot that was incubated with GAPDH antibody). The dashed line delineates the cropping site. The original uncropped blots of DCN and GAPDH are presented in Additional file 1 (Supplementary Figure S2). Asterisks indicates statistically significant differences, where *p < 0.05, ** p < 0.01, and ***p < 0.001
Binding confirmation of miR-181a to the 3’UTR of DCN
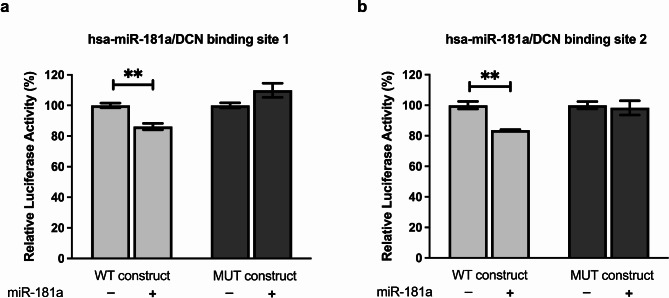
To determine whether the observed effects of miR-181a on DCN are mediated through direct targeting of the DCN 3’UTR, binding confirmation using luciferase reporter constructs was performed for each of the two predicted binding regions. For both WT binding site constructs, luciferase activity was inhibited upon co-transfection with miR-181a mimics compared to cells transfected with the WT constructs alone (p = 0.0060 and p = 0.0029 for binding site 1 and 2, Fig. 3a and b, respectively). Luciferase activity, however, was unaffected in cells co-transfected with miR-181a and MUT constructs compared to cells transfected with MUT construct alone, confirming the specificity of these miR-target gene interactions. These observations validate the independent and site-specific direct binding of miR-181a to two binding regions within the DCN 3’UTR (Fig. 3a, b).
Fig. 3.
miR-181a targeting of DCN. Huh-7 cells were co-transfected with miR-181a mimics and either the wildtype (WT) or mutant (MUT) construct for each of the two predicted binding regions on DCN 3’UTR. Luciferase reporter assay was used to validate the independent and site-specific binding of miR-181a to (a) the first and (b) the second predicted binding regions on DCN 3’UTR. Asterisks indicates statistically significant differences, where ** p < 0.01
Impact of miR-181a on viability, proliferation, and clonogenicity in HuH-7 cells
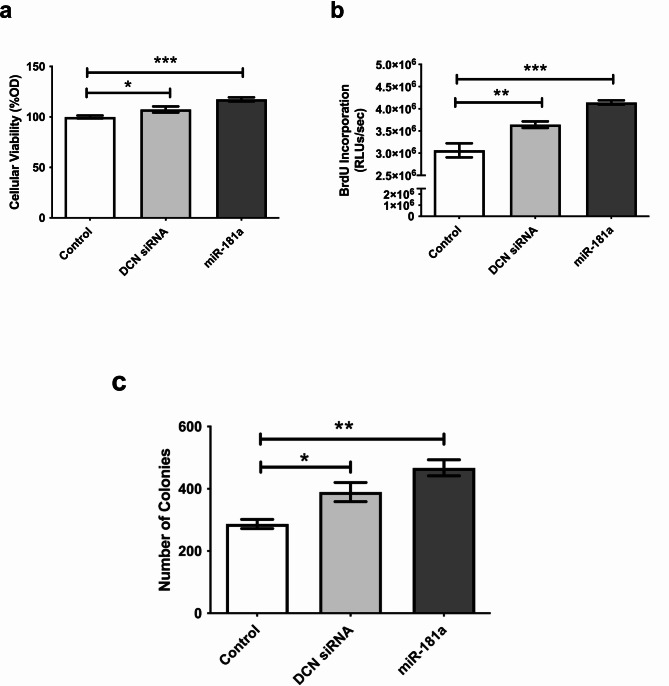
Having validated the direct targeting of DCN by miR-181a, it was important to assess how the regulation of this tumor suppressor proteoglycan would affect various characteristic properties of tumor cells. MTT, BrdU, and colony-formation assays were performed after transfection of miR-181a mimics. Ectopic expression of miR-181a into HuH-7 cells significantly increased cellular viability (p < 0.0001), proliferation (p < 0.0001), and clonogenicity (p = 0.0037, Fig. 4a–c). These findings were similar to those incurred by knockdown of DCN (p = 0.0391, p = 0.0064, and p = 0.0395, for viability, proliferation, and clonogenicity, respectively) signifying the oncogenic role of miR-181a in liver cancer, which is in part mediated through targeting of DCN. Representative images for the colony forming assay are shown in Supplementary Figure S3.
Fig. 4.
Impact of miR-181a on viability, proliferation, and clonogenicity of HuH-7 cells. HuH-7 cells were transfected with miR-181a mimics or specific siRNAs against DCN as a positive control, and (a) MTT assay for cellular viability, (b) BrdU incorporation assay for cellular proliferation, and (c) colony-forming assay for clonogenicity were performed. Representative images for the colony forming assay are shown in Supplementary Figure S3. Asterisks indicates statistically significant differences, where *p < 0.05, ** p < 0.01, and ***p < 0.001
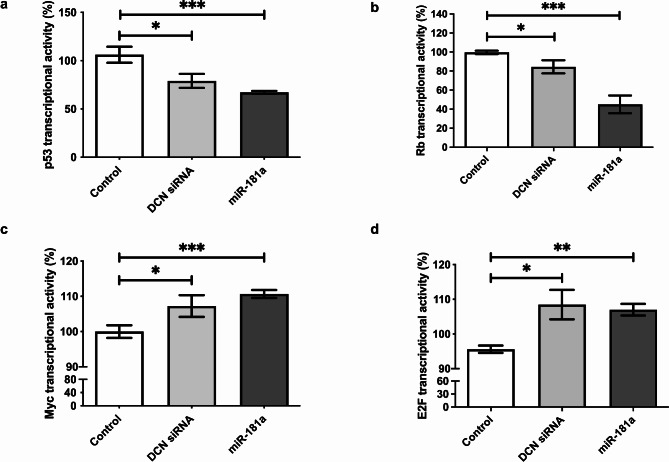
Effect of miR-181a on oncogenes and tumor suppressor transcription factors in HuH-7 cells
Given the role of DCN in regulating the cell cycle, it was interesting to verify the effect of its upstream microregulator, miR-181a, on the induction of key signaling pathways controlling apoptosis and proliferation. Various reporter vectors containing cis-acting enhancer elements specific to p53, Rb, Myc, or E2F, upstream of a sensitive luciferase reporter gene were used. These vectors allow for the measurement of transcriptional activity driven by these specific regulatory elements. After treatment of cells with miR-181a mimics, binding of these transcription factors to their enhancer elements was monitored via luciferase activity to quantify the activation or repression of each signaling pathway. Transfection of miR-181a markedly repressed signaling through the tumor suppressors p53 (p = 0.0007) and Rb (p < 0.0001, Fig. 5a and b, respectively). Moreover, miR-181a mimicking significantly activated the signaling pathways of proto-oncogenes Myc (p < 0.0001) and E2F (p = 0.0018, Fig. 5c and d, respectively). The effects on all four key transcription factors were paralleled in cells in which DCN was knocked down (p = 0.0401, p = 0.0234, p = 0.0476, and p = 0.0264, for p53, Rb, Myc, and E2F, Fig. 5a–d, respectively).
Fig. 5.
Impact of miR-181a on various oncogenes and tumor suppressor transcription factors. HuH-7 cells were transfected with reporter vectors containing cis-acting enhancer elements specific to cell cycle regulating transcription factors (a) p53, (b) Rb, (c) Myc, or (d) E2F, upstream of a sensitive luciferase reporter gene, and luciferase activity was used to quantify the activation or repression of each signaling pathway after knockdown of DCN or miR-181a mimicking. Asterisks indicates statistically significant differences, where *p < 0.05, ** p < 0.01, and ***p < 0.001
Discussion
DCN represents a potent inhibitor of tumor growth and migration by modulating the cell-signaling pathways and deposition of tumor stroma. Moreover, the lack of DCN allows for the development of tumors [24]. Being an essential macromolecule of the tumor microenvironment, DCN is downregulated in numerous cancers, including endometrial carcinoma, colorectal cancer, breast cancer, and giant cell tumor of bone [25–28]. Furthermore, DCN was reported to be the most downregulated gene during HCC progression, where its transcript expression levels showed a stepwise decline from cirrhosis to adjacent non-tumor and HCC tumor tissues [14]. Consistent with this, we also found that DCN was significantly under-expressed in liver cancer. This is supported by several other studies reporting the downregulation of DCN in HCC using various methodologies and confirmed by studies analyzing publicly available gene expression datasets for cirrhotic, HCC, and non-tumorous liver samples [13–16, 29–31]. While our data did not reveal any further reduction in DCN levels between the adjacent cirrhotic liver tissues and tumor samples, both cirrhotic and HCC tissues had significantly lower DCN expression when compared to healthy liver tissues. It is possible, however, that expanding the sample size would unveil a similar pattern of gradual decline in DCN levels throughout the process of cirrhosis progression to HCC.
In the present study, using bioinformatics algorithms, we identified miR-181a as a promising miRNA which may be regulating DCN. Most studies indicate that miRNAs associate with specific sequences at the 3′ UTR of their target mRNAs to cause either translational repression or target mRNA cleavage, where fully complimentary targets are cleaved while targets with central mismatches to the miRNA are translationally silenced [32]. Similar to the findings reported by Brockhausen et al., Chang et al., and Yang et al. [33–35], our study revealed that miR-181a is overexpressed in HCC liver tissues compared to that in healthy liver tissues. On the other hand, the study by Korhan et al. revealed that mir-181a is downregulated in HCC and suppresses motility, invasion, and branching-morphogenesis. However, that study used human liver tissue microarray samples containing 12 normal and 19 cirrhotic liver tissues, and 48 HCC tissues to detect the relative miR-181a-5p expression levels in the respective liver tissues by in situ hybridization [36]. Conversely, we used primary tissues from Egyptian patient cohorts. Thus, the different experimental models used might have influenced the observed discrepancy in the miR-181a expression pattern, potentially leading to different functional outcomes.
For further confirmation, we retrieved TCGA data regarding the differential expression pattern of miR-181a and DCN in HCC and normal liver tissues. The findings revealed the overexpression of miR-181a in 370 HCC tissues compared to that in 50 normal liver tissues. They also revealed the down expression of DCN in 374 HCC tissues compared to that in 50 normal liver tissues (Supplementary Figure S4 A and B).
Next, we revealed that miR-181a decreases DCN not only at the protein level, but also on the mRNA level, indicating that the miR-181a–DCN interaction most likely results in degradation of the DCN transcript. Indeed, bioinformatics software predicted two perfectly complementary binding regions within the 3’UTR of DCN with high binding potential to miR-181a. The site-specific binding of this miRNA to both regions was independently confirmed with luciferase reporter assays. Thus, the tumor suppressor proteoglycan DCN is shown for the first time to be directly targeted by miR-181a in HCC. Despite the scarce literature about the epigenetic regulation of DCN, our findings conform with hypertrophic scar studies which showed that miR-181b, a miRNA closely related to miR-181a, directly targets DCN thereby decreasing its levels in hypertrophic scar fibroblasts [37, 38]. Interestingly, multiple previous studies have reported miR-181a to be up-regulated in cirrhotic and HCC liver tissues compared to that in normal liver tissues [35, 39–41]; this pattern of miR-181a expression is the inverse of what we have observed for its target gene, DCN, which further endorses the repressive action of miR-181a on the DCN transcript.
Next, we assessed the impact of miR-181a-mediated DCN repression on cancer progression and whether the effects would simulate those caused by silencing of DCN via siRNAs. Functionally, repressing DCN by miR-181a or siRNAs resulted in increased cell viability, proliferation, and colony-forming ability of the human HCC cell line HuH-7. These data are in agreement with previous studies that have characterized the antitumorigenic role of DCN where cellular proliferation was inhibited, while apoptosis was enhanced in a time- and dose-dependent manner in vitro by culturing HuH-7 cells with DCN [42]. Moreover, DCN downregulated integrin β1 protein expression thereby inhibiting HCC cell invasion and migration [12]. These findings were also reflected in vivo where several studies revealed that DCN gene delivery diminished tumor occurrence, while genetic ablation of DCN led to increased tumor formation [10, 29, 43]. The reported function of miR-181a in HCC has been fairly controversial. miR-181a directly inhibited Egr1, reduced Gab2 protein levels, and repressed c-Met activation, thereby hindering HCC cell proliferation and migration, and diminishing branching-morphogenesis and invasion [36, 44, 45]. Thus, miRNA may be a tumor suppressor miRNA. However, the findings reported by multiple previous studies that used in vitro and in vivo HCC models go in line with our findings, attributing tumor-promoting properties to miR-181a; liver-specific knockout of the miR-181 family in primary liver cancers in mice decreased liver tumor size and inhibited tumor progression [46]. Moreover, miR-181a mimics induced an in vitro EMT-like change in human hepatocyte cell lines which could be reversed by a miR-181a inhibitor [39]. Moreover, miR-181a decreased apoptosis and increased cell proliferation of HCC cells in vitro and enhanced tumor growth in vivo, through targeting various genes including Atg5, Fas, RASSF1, PTEN, CDKN1β, and E2F7 [35, 47–50]. Hence, from our study, the tumor suppressor proteoglycan DCN can be added to the repertoire of target genes directly regulated by miR-181a, offering another mechanism by which this miRNA mediates its oncogenic effects in HCC.
DCN is a natural pan-inhibitor of cell surface receptor tyrosine kinases (RTKs), thus affecting downstream signaling pathways which regulate the cell cycle. Remarkably, in DCN-deficient mice, tumorigenesis was reported to be associated with the downregulation of the cyclin-dependent kinase inhibitors p21WAF1 and p27Kip1 and a concurrent upregulation of beta-catenin [51]. Other studies have highlighted the impact of DCN on cell cycle proteins where, in experimental animal models of hepatocarcinogenesis with DCN deficiency, multiple RTKs were constitutively phosphorylated and activated with a simultaneous increase in phosphorylation and inactivation of the tumor suppressor protein Rb, as well as higher protein levels of the proto-oncogene c-Myc [10, 43]. On the other hand, when DCN was overexpressed in mouse models of hepatocarcinogenesis, the activity of several RTKs was suppressed while the tumor suppressor p53 function was enhanced [29, 52]. Mechanistically, in this study we provide evidence for how the oncogenic miR-181a-mediated repression of DCN may affect the regulation of key cell cycle proteins and hence cancer progression. Similar to the effect of siRNAs, repression of DCN through the ectopic delivery of miR-181a into HuH-7 cells harboring reporter vectors with response elements for various cell cycle regulating proteins resulted in distinctly repressed signaling of the tumor suppressors p53 and Rb, and markedly activated signaling of the oncogenes Myc and E2F. Taken together, these findings suggest that DCN deficiency caused by miR-181a can be permissive for tumorigenesis.
While our expression profiling of DCN and MiR-181a was performed on human tissue samples which have precious clinical value in research, as a limitation to our study all other work was performed in vitro; therefore, it would be recommended to investigate the effect of the conditional knockout or knock-in of miR-181a in animal models to confirm the effects of this miRNA on its multiple downstream direct target genes including DCN, as well as oncogenes and tumor suppressor genes downstream of DCN.
Conclusions
The findings of this study revealed that oncomiR-181a directly targets the tumor suppressor proteoglycan DCN, leading to enhancing the oncogenic properties of HCC cells. Hence, it would be of clinical significance to explore its diagnostic and therapeutic potential in liver cancer. miR-181a could be a promising biomarker for the early detection and monitoring of liver cancer progression. In addition, measuring the miR-181a levels in liver biopsies could be incorporated into existing diagnostic and surveillance strategies to assess HCC progression. Finally, potential therapeutic strategies based on targeting miR-181a could include developing miRNA inhibitors or mimics to modulate miR-181a expression and its downstream effects on tumorigenesis. This miR-181a-based therapy approach could be potentially combined with chemotherapy and immunotherapy for the management of liver cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
None.
Abbreviations
- cDNA
Complementary DNA
- DCN
Decorin
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- RLUs
Relative light units
Author contributions
RAA contributed to the study concept and design, execution of experiments, acquisition of data, statistical analysis, interpretation of data, and writing of the manuscript. RBEAE optimized, performed, and analyzed the western blot experiments. RAY assisted in the execution of functional analysis experiments and acquisition of their data. MMA assisted in the execution of functional analysis experiments. HZ assisted in optimization of the western blot experiments. The surgeons, KAH and GE, provided tissue samples and clinical data. KB contributed to the study concept and provided critical revision of the manuscript for important intellectual content. NE and IOF contributed to the western blot experiments and writing of the manuscript. AIA contributed to the study concept, design, supervision, and provided critical revision of the manuscript for important intellectual content. All authors have contributed to and agreed on the content of the final version of the manuscript.
Funding
Part of the work of this study was based upon work supported by the Egyptian Science, Technology & Innovation Funding Authority (STDF) under Basic & Applied Research Grants number BARG-37096 and YRG-33464 and the Deutsche Forschungsgemeinschaft (DFG) grant number 412294938. The funder had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available to protect the patients’ privacy, but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study methodologies conformed to the ethical standards set forth by the declaration of Helsinki. The ethical committees of Cairo University and the German University in Cairo approved the study methodologies. The experiments were undertaken with the understanding and written informed consent of each patient.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/CAAC.21660 [DOI] [PubMed] [Google Scholar]
- 2.El-Zayadi AR, Badran HM, Barakat EMF, Attia MED, Shawky S, Mohamed MK, Selim O, Saed A. Hepatocellular carcinoma in Egypt: a single center study over a decade. World J Gastroenterol. 2005;11:5193–8. 10.3748/WJG.V11.I33.5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweed D, Sweed E, Moaz I, Mosbeh A, Fayed Y, Elhamed SMA, Sweed E, Macshut M, Abdelsattar S, Kilany S, et al. The clinicopathological and prognostic factors of hepatocellular carcinoma: a 10-year tertiary center experience in Egypt. World J Surg Oncol. 2022;20. 10.1186/S12957-022-02764-2 [DOI] [PMC free article] [PubMed]
- 4.Omran D, Alboraie M, Zayed RA, Wifi MN, Naguib M, Eltabbakh M, Abdellah M, Sherief AF, Maklad S, Eldemellawy HH, et al. Towards hepatitis C virus elimination: Egyptian experience, achievements and limitations. World J Gastroenterol. 2018;24:4330–40. 10.3748/WJG.V24.I38.4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waziry R, Gomaa A, Waked I, Dore GJ. Determinants of survival following hepatocellular carcinoma in Egyptian patients with untreated chronic HCV infection in the pre-DAA era. Arab J Gastroenterol. 2018;19:26–32. 10.1016/J.AJG.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Shiina S, Tateishi R, Arano T, Uchino K, Enooku K, Nakagawa H, Asaoka Y, Sato T, Masuzaki R, Kondo Y, et al. Radiofrequency ablation for hepatocellular carcinoma: 10-year outcome and prognostic factors. Am J Gastroenterol. 2012;107:569–77. 10.1038/AJG.2011.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka Y, Tateishi R, Koike K. Proteoglycans are attractive biomarkers and therapeutic targets in hepatocellular carcinoma. Int J Mol Sci. 2018;19. 10.3390/IJMS19103070 [DOI] [PMC free article] [PubMed]
- 8.Schaefer L, Iozzo RV. Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem. 2008;283:21305–9. 10.1074/JBC.R800020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang Y, Du Z, Wang Q, Wu M, Wang X, Wang L, Cao L, Hamid AS, Zhang G. Recombinant human decorin suppresses liver HepG2 carcinoma cells by p21 upregulation. Onco Targets Ther. 2012;5:143–52. 10.2147/OTT.S32918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horváth Z, Kovalszky I, Fullár A, Kiss K, Schaff Z, Iozzo RV, Baghy K. Decorin deficiency promotes hepatic carcinogenesis. Matrix Biol. 2014;35:194–205. 10.1016/J.MATBIO.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Ge Y, Cheng Q, Zhang Q, Fang L, Zheng J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget. 2018;9:5480–91. 10.18632/oncotarget.23869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Wang P, Li L, Yu J, Yu C, Xu L, Li L, Dai F, Feng L, Zou H, et al. Cancer-associated fibroblasts promote vascular invasion of hepatocellular carcinoma via downregulating decorin-integrin β1 signaling. Front cell Dev Biol. 2021;9. 10.3389/FCELL.2021.678670 [DOI] [PMC free article] [PubMed]
- 13.Lin T, Lin Z, Mai P, Zhang E, Peng L. Identification of prognostic biomarkers associated with the occurrence of portal vein tumor thrombus in hepatocellular carcinoma. Aging. 2021;13:11786–807. 10.18632/AGING.202876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang YK, Li F, Zhang Y, Liu ZK, Wang ZL, Bian H, Chen ZN. Systems analysis of key genes and pathways in the progression of hepatocellular carcinoma. Med (Baltim). 2018;97. 10.1097/MD.0000000000010892 [DOI] [PMC free article] [PubMed]
- 15.Li Y, Yuan SL, Yin JY, Yang K, Zhou XG, Xie W, Wang Q. Differences of core genes in liver fibrosis and hepatocellular carcinoma: evidence from integrated bioinformatics and immunohistochemical analysis. World J Gastrointest Oncol. 2022;14:1265–80. 10.4251/WJGO.V14.I7.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Ezura Y, Chervoneva I, Robinson PS, Beason DP, Carine ET, Soslowsky LJ, Iozzo RV, Birk DE. Decorin regulates assembly of collagen fibrils and acquisition of biomechanical properties during tendon development. J Cell Biochem. 2006;98:1436–49. 10.1002/JCB.20776 [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Chen S, Goldoni S, Calder BW, Simpson HC, Owens RT, McQuillan DJ, Young MF, Iozzo RV, Birk DE. Genetic evidence for the coordinated regulation of collagen fibrillogenesis in the cornea by decorin and biglycan. J Biol Chem. 2009;284:8888–97. 10.1074/JBC.M806590200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keene DR, San Antonio JD, Mayne R, McQuillan DJ, Sarris G, Santoro SA, Iozzo RV. Decorin binds near the C terminus of type I collagen. J Biol Chem. 2000;275:21801–4. 10.1074/JBC.C000278200 [DOI] [PubMed] [Google Scholar]
- 19.Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol. 1997;136:729–43. 10.1083/JCB.136.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamid AS, Li J, Wang Y, Wu X, Ahmed Ali HA, Du Z, Bo L, Zhang Y, Zhang G. Recombinant human decorin upregulates p57KIP2 expression in HepG2 hepatoma cell lines. Mol Med Rep. 2013;8:511–6. 10.3892/MMR.2013.1510 [DOI] [PubMed] [Google Scholar]
- 21.Horvath Z, Kovalszky I, Fullar A, Kiss K, Schaff Z, Iozzo RV, Baghy K. Decorin deficiency promotes hepatic carcinogenesis. Matrix Biol. 2014;35:194–205. 10.1016/j.matbio.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristen AV, Ajroud-Driss S, Conceição I, Gorevic P, Kyriakides T, Obici L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis Manag. 2019;9:5–23. 10.2217/NMT-2018-0033 [DOI] [PubMed] [Google Scholar]
- 23.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11. 10.1186/GB-2010-11-8-R90 [DOI] [PMC free article] [PubMed]
- 24.Iozzo RV, Chakrani F, Perrotti D, McQuillan DJ, Skorski T, Calabretta B, Eichstetter I. Cooperative action of germline mutations in decorin and p53 accelerates lymphoma tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieveld M, Bodson E, De Boeck G, Nouman B, Cleton-Jansen AM, Korsching E, Benassi MS, Picci P, Sys G, Poffyn B, et al. Gene expression profiling of giant cell tumor of bone reveals downregulation of extracellular matrix components decorin and lumican associated with lung metastasis. Virchows Arch. 2014;465:703–13. 10.1007/S00428-014-1666-7 [DOI] [PubMed] [Google Scholar]
- 26.Colas E, Perez C, Cabrera S, Pedrola N, Monge M, Castellvi J, Eyzaguirre F, Gregorio J, Ruiz A, Llaurado M, et al. Molecular markers of endometrial carcinoma detected in uterine aspirates. Int J cancer. 2011;129:2435–44. 10.1002/IJC.25901 [DOI] [PubMed] [Google Scholar]
- 27.Eshchenko TY, Rykova VI, Chernakov AE, Sidorov SV, Grigorieva EV. Expression of different proteoglycans in human breast tumors. Biochem (Mosc). 2007;72:1016–20. 10.1134/S0006297907090143 [DOI] [PubMed] [Google Scholar]
- 28.Bi X, Tong C, Dockendorff A, Bancroft L, Gallagher L, Guzman G, Iozzo RV, Augenlicht LH, Yang W. Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis. 2008;29:1435–40. 10.1093/CARCIN/BGN141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reszegi A, Horváth Z, Fehér H, Wichmann B, Tátrai P, Kovalszky I, Baghy K. Protective role of decorin in primary hepatocellular carcinoma. Front Oncol. 2020;10. 10.3389/FONC.2020.00645 [DOI] [PMC free article] [PubMed]
- 30.Chung EJ, Sung YK, Farooq M, Kim Y, Im S, Tak WY, Hwang YJ, Kim Y, Il, Han HS, Kim J-C, et al. Gene expression profile analysis in human hepatocellular carcinoma by cDNA microarray. Mol Cells. 2002;14:382–7. [PubMed] [Google Scholar]
- 31.Miyasaka Y, Enomoto N, Nagayama K, Izumi N, Marumo F, Watanabe M, Sato C. Analysis of differentially expressed genes in human hepatocellular carcinoma using suppression subtractive hybridization. Br J Cancer. 2001;85:228–34. 10.1054/BJOC.2001.1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. 10.1038/NRG2936 [DOI] [PubMed] [Google Scholar]
- 33.Brockhausen J, Tay SS, Grzelak CA, Bertolino P, Bowen DG, d’Avigdor WM, Teoh N, Pok S, Shackel N, Gamble JR, Vadas M, McCaughan GW. miR-181a mediates TGF-β-induced hepatocyte EMT and is dysregulated in cirrhosis and hepatocellular cancer. Liver Int. 2015;35:240–53. 10.1111/liv.12517. PMID: 24576072. [DOI] [PubMed] [Google Scholar]
- 34.Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. 10.1186/s12885-017-3216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, He Y, Zhai N, Ding S, Li J, Peng Z. MicroRNA-181a inhibits autophagy by targeting Atg5 in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2018;23:388–96. 10.2741/4596 [DOI] [PubMed] [Google Scholar]
- 36.Korhan P, Erdal E, Atabey N. MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-Met. Biochem Biophys Res Commun. 2014;450:1304–12. 10.1016/j.bbrc.2014.06.142 [DOI] [PubMed] [Google Scholar]
- 37.Liu B, Guo Z, Gao W. miR-181b-5p promotes proliferation and inhibits apoptosis of hypertrophic scar fibroblasts through regulating the MEK/ERK/p21 pathway. Exp Ther Med. 2019;17. 10.3892/ETM.2019.7159 [DOI] [PMC free article] [PubMed]
- 38.Kwan P, Ding J, Tredget EE. MicroRNA 181b regulates decorin production by dermal fibroblasts and may be a potential therapy for hypertrophic scar. PLoS ONE. 2015;10. 10.1371/JOURNAL.PONE.0123054 [DOI] [PMC free article] [PubMed]
- 39.Brockhausen J, Tay SS, Grzelak CA, Bertolino P, Bowen DG, d’Avigdor WM, Teoh N, Pok S, Shackel N, Gamble JR, et al. miR-181a mediates TGF-β-induced hepatocyte EMT and is dysregulated in cirrhosis and hepatocellular cancer. Liver Int. 2015;35:240–53. 10.1111/LIV.12517 [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Yao X, Zhang D, Sheng J, Wen X, Wang Q, Chen G, Li Z, Du Z, Zhang X. Analysis of transcription factor-related regulatory networks based on bioinformatics analysis and validation in hepatocellular carcinoma. Biomed Res Int. 2018;2018. 10.1155/2018/1431396 [DOI] [PMC free article] [PubMed]
- 41.Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17. 10.1186/S12885-017-3216-6 [DOI] [PMC free article] [PubMed]
- 42.Shangguan J, Dou K, Li X, Hu X, Zhang F, Yong Z, Ti Z. Effects and mechanism of decorin on the proliferation of HuH7 hepatoma carcinoma cells in vitro. Chin J Cell Mol Immunol. 2009;25:780–2. [PubMed] [Google Scholar]
- 43.Baghy K, Horváth Z, Regos E, Kiss K, Schaff Z, Iozzo RV, Kovalszky I. Decorin interferes with platelet-derived growth factor receptor signaling in experimental hepatocarcinogenesis. FEBS J. 2013;280:2150–64. 10.1111/FEBS.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi Jgang, Zheng Jfeng, Li Q, Bao S, yun, Yu X fang, Xu P, Liao Cxian. MicroRNA-181a-5p suppresses cell proliferation by targeting Egr1 and inhibiting Egr1/TGF-β/Smad pathway in hepatocellular carcinoma. Int J Biochem Cell Biol. 2019;106:107–16. 10.1016/J.BIOCEL.2018.11.011 [DOI] [PubMed]
- 45.Huang L, Liu R, Zhou P, Tian Y, Lu Z. miR-9 and miR-181a target Gab2 to inhibit the proliferation and migration of hepatocellular carcinoma HepG2 cells. Genes (Basel). 2022;13. 10.3390/GENES13112152 [DOI] [PMC free article] [PubMed]
- 46.Chen J, Zhao Y, Zhang F, Li J, Boland JA, Cheng NC, Liu K, Tiffen JC, Bertolino P, Bowen DG, et al. Liver-specific deletion of miR-181ab1 reduces liver tumour progression via upregulation of CBX7. Cell Mol Life Sci. 2022;79. 10.1007/S00018-022-04452-6 [DOI] [PMC free article] [PubMed]
- 47.Zou C, Li Y, Cao Y, Zhang J, Jiang J, Sheng Y, Wang S, Huang A, Tang H. Up-regulated MicroRNA-181a induces carcinogenesis in hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer. 2014;14. 10.1186/1471-2407-14-97 [DOI] [PMC free article] [PubMed]
- 48.Zou C, Chen J, Chen K, Wang S, Cao Y, Zhang J, Sheng Y, Huang A, Tang H. Functional analysis of miR-181a and Fas involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Exp Cell Res. 2015;331:352–61. 10.1016/J.YEXCR.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 49.Azumi J, Tsubota T, Sakabe T, Shiota G. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci. 2016;107:1256–62. 10.1111/CAS.13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan JYL, Habib NA, Chuah YW, Yau YH, Geifman-Shochat S, Chen WN. Identification of cellular targets of MicroRNA-181a in HepG2 cells: a new approach for functional analysis of MicroRNAs. PLoS ONE. 2015;10. 10.1371/JOURNAL.PONE.0123167 [DOI] [PMC free article] [PubMed]
- 51.De Luca A, Santra M, Baldi A, Giordano AIozzoRV. Decorin-induced growth suppression isassociated with upregulation of p21, an inhibitor ofcyclin-dependent kinases. J Biol Chem. 1996;271:18961–5. [DOI] [PubMed] [Google Scholar]
- 52.Reszegi A, Horváth Z, Karászi K, Regős E, Postniková V, Tátrai P, Kiss A, Schaff Z, Kovalszky I, Baghy K. The protective role of decorin in hepatic metastasis of colorectal carcinoma. Biomolecules. 2020;10:1–17. 10.3390/BIOM10081199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available to protect the patients’ privacy, but are available from the corresponding author on reasonable request.