Abstract
Lumpy skin disease virus (LSDV), a member of the capripoxvirus genus of the Poxviridae, is the etiologic agent of an important disease of cattle in Africa. Here we report the genomic sequence of LSDV. The 151-kbp LSDV genome consists of a central coding region bounded by identical 2.4 kbp-inverted terminal repeats and contains 156 putative genes. Comparison of LSDV with chordopoxviruses of other genera reveals 146 conserved genes which encode proteins involved in transcription and mRNA biogenesis, nucleotide metabolism, DNA replication, protein processing, virion structure and assembly, and viral virulence and host range. In the central genomic region, LSDV genes share a high degree of colinearity and amino acid identity (average of 65%) with genes of other known mammalian poxviruses, particularly suipoxvirus, yatapoxvirus, and leporipoxviruses. In the terminal regions, colinearity is disrupted and poxvirus homologues are either absent or share a lower percentage of amino acid identity (average of 43%). Most of these differences involve genes and gene families with likely functions involving viral virulence and host range. Although LSDV resembles leporipoxviruses in gene content and organization, it also contains homologues of interleukin-10 (IL-10), IL-1 binding proteins, G protein-coupled CC chemokine receptor, and epidermal growth factor-like protein which are found in other poxvirus genera. These data show that although LSDV is closely related to other members of the Chordopoxvirinae, it contains a unique complement of genes responsible for viral host range and virulence.
Capripoxviruses (CaPVs) represent one of eight genera within the chordopoxvirus (ChPV) subfamily of the Poxviridae. The capripoxvirus genus is currently comprised of lumpy skin disease virus (LSDV), sheeppox virus (ShPV), and goatpox virus (GPV). These viruses are responsible for some of the most economically significant diseases of domestic ruminants in Africa and Asia (18). CaPV infections are generally host specific and they have specific geographic distributions (10, 14, 15). CaPVs are, however, serologically indistinguishable from each other, able to induce heterologous cross-protection, and able in some instances to experimentally cross-infect (8, 10, 15, 16). Restriction fragment analysis and limited DNA sequence data support a close relationship between CaPVs (5, 25, 26, 33). The molecular basis of CaPV host range restriction and virulence remains to be elucidated.
LSD is a subacute to acute cattle disease in Africa. It is characterized by extensive cutaneous lesions and signs typical of generalized poxvirus diseases (14, 15). Transmission of LSD between cattle is inefficient, and arthropod-vectored transmission may be significant in epizootic outbreaks and in the spread of LSD into nonenzootic regions (4, 10–12, 15, 36, 54). Attenuated LSDV strains and ShPV have been successfully used as LSD vaccines in enzootic and outbreak areas; however, vaccine failure and restrictions on the use of live virus vaccines create the need for a safe and effective, live attenuated vaccine (4, 13, 15, 53).
Current molecular data on the LSDV genome consists of restriction endonuclease analysis, cross-hybridization studies, and limited transcriptional and DNA sequence analysis (5, 19, 20, 26, 27, 33). Given the economic significance of LSD, its potential for spread into nonenzootic regions, and the interest in developing more effective LSDV-based vaccines and expression vectors, we have sequenced and analyzed the genome of a pathogenic LSDV. These data provide the first view of a CaPV genome, and they define the gene complement that underlies LSDV virulence and host range.
MATERIALS AND METHODS
LSDV DNA isolation, cloning, sequencing, and sequence analysis.
LSDV genomic DNA was extracted from primary lamb testicle (LT) cells infected with LSDV Neethling type strain 2490 (9). The virus was originally isolated in Kenya in 1958, passed 16 times in LT cells, and subsequently reisolated in 1987 from lesions of an experimentally infected cow (U.S. Department of Agriculture Animal Plant Health Inspection Service, Greenport, N.Y.). Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509 I endonuclease (New England Biolabs, Beverly, Mass.), and DNA fragments of 1.0 to 6.0 kbp were cloned and used in dideoxy sequencing reactions as previously described (2). Reaction products were run on a Applied Biosystems PRISM 3700 automated DNA sequencer (PE Biosystems, Foster City, Calif.). Sequence data were assembled, and gaps were closed as described previously (1) with confirmatory assemblies performed using CAP3 (30). The final DNA consensus sequence represented on average eightfold redundancy at each base position.
Genome DNA composition, structure, repeats and restriction enzyme patterns were analyzed as previously described (1) using the GCG v.10 software package (17). Open reading frames (ORFs) longer than 30 codons were evaluated for coding potential as previously described (2). All potentially coding ORFs and ORFs greater than 60 codons were subjected to homology searches as previously described (1, 2). Based on these criteria, 156 ORFs were annotated as potential genes. Gene families and promoter regions were analyzed and annotated as previously described (1, 2) and with additional use of Geanfammer (44) and GCG MEME programs. Vaccinia virus (VV) A52R-like protein family was clustered from a nonredundant peptide database of all known poxvirus sequences using the CLUS program (34) and BLASTP2 scores greater than 110. Phylogenetic comparisons were done with the PHYLO_WIN software package (23).
Nucleotide sequence accession number.
The LSDV genome sequence has been deposited in GenBank under accession no. AF325528.
RESULTS AND DISCUSSION
Organization of the LSDV genome.
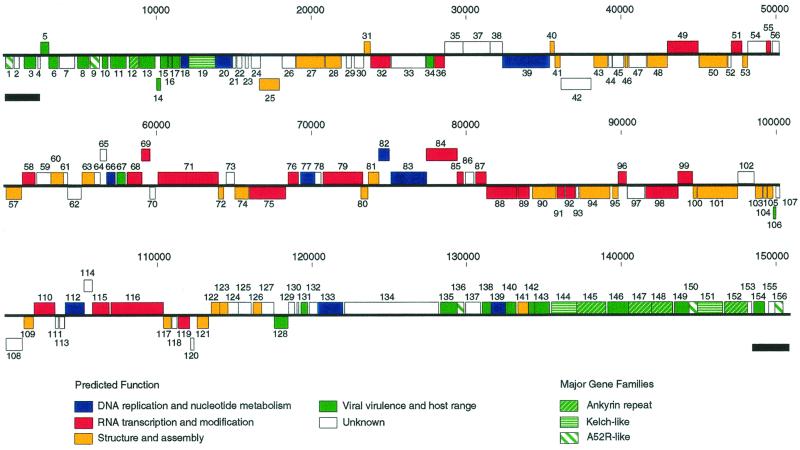
LSDV genome sequences were assembled into a contiguous sequence of 150,773 bp which is in accordance with previous size estimates of 145 to 152 kbp (19, 26, 27). Because the hairpin loops were not sequenced, the leftmost nucleotide was arbitrarily designated base 1. The nucleotide composition is 73% A+T and is uniformly distributed. As seen for other poxviruses, the LSDV genome contains a central coding region bounded by two identical inverted terminal repeat (ITR) regions which contain at least 2,418 bp at both termini (Fig. 1). The most terminal 241 nucleotides of the assembled sequence contain 7.5 copies of a 24-bp imperfect tandem repeat and four copies of a 15-bp imperfect tandem repeat and are similar to those described in ShPV (25). Comparison with published restriction fragment analysis of the genome indicates there may be additional terminal sequences of less than 200 bp present (27, 33).
FIG. 1.
Linear map of the LSDV genome. ORFs are numbered from left to right based on the position of the methionine initiation codon. ORFs transcribed to the right are located above the horizontal line; ORFs transcribed to the left are below. Genes with similar functions and members of gene families are colored according to the figure key. ITRs are represented as black bars below the ORF map.
LSDV contains 156 ORFs which have been annotated here as putative genes. These genes represent a 95% coding density and encode proteins of 53 to 2,025 amino acids (Fig. 1, Table 1). Similar to other poxviruses, many of the 41 putative early genes are members of gene families and/or putative host range genes, while the 46 genes containing the VV late promoter sequence (TAAATG) at the ATG codon (41) include many of the conserved virion-associated poxviral genes (Table 1).
TABLE 1.
LSDV ORFs
| ORF | Position (length in codons) | CaPV accession no.a | Best match
|
Predicted structure and/or functiond | Promoter typee | MYX
|
VV
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Speciesb | Accession no. | BlastP2 score | % Identity | Length (aa)c | ORFf | % Identity | ORFg | % Identity | |||||
| LSDV001 | 713–237 (159) | P18387 | RFV | P25949 | 308 | 46 | 143 | A52R-like family protein, SP | M003.1 | 42 | B15R | 34 | |
| LSDV002 | 1179–787 (131) | P18388 | MYX | AJ012282 | 242 | 43 | 131 | M003.2 | 43 | ||||
| LSDV003 | 2151–1432 (240) | P18386 | MYX | AF002684 | 378 | 37 | 231 | ER-localized apoptosis regulator, SP, TM | E | M004 | 37 | B9R | 37 |
| LSDV004 | 2394–2224 (57) | SPV | P32230 | 125 | 47 | 57 | M004.1 | 41 | |||||
| LSDV005 | 2446–2955 (170) | Ovis aries | U11421 | 357 | 43 | 174 | IL-10, SP, TM | ||||||
| LSDV006 | 3664–2972 (231) | Mus musculus | P27931 | 179 | 33 | 144 | IL-1 receptor, SP | B16R | 26 | ||||
| LSDV007 | 4753–3689 (355) | Yaba-like DV | AJ293568 | 552 | 35 | 355 | IFN-γ | C10L | 34 | ||||
| LSDV008 | 5664–4840 (275) | SPV | P32226 | 435 | 37 | 257 | family receptor, SP | L | M007 | 26 | B8R | 31 | |
| LSDV009 | 6389–5700 (230) | Yaba-like DV | AJ293568 | 342 | 36 | 233 | α-amanitin-sensitive protein. A52R-like protein | E | MI39R | 27 | N2L | 25 | |
| LSDV010 | 6929–6444 (162) | SPV | P32225 | 310 | 36 | 150 | LAP/PHD-finger protein, TM | E | M153R | 38 | |||
| LSDV011 | 8118–6976 (381) | S78201 | Homo sapiens | AY016370 | 683 | 43 | 311 | G protein-coupled CC chemokine receptor, TM | E | ||||
| LSDV012 | 8860–8228 (211) | S78201 | Yaba-like DV | AJ293568 | 436 | 40 | 210 | Ankyrin repeat protein | E | M149R | 25 | B4R | 28 |
| LSDV013 | 9924–8902 (341) | S78201 | Equus caballus | AB033415 | 200 | 27 | 340 | IL-1 receptor, SP | E | B16R | 24 | ||
| LSDV014 | 10253–9987 (89) | SPV | P32224 | 248 | 49 | 85 | IF2α-like PKR inhibitor | M156R | 34 | K3L | 42 | ||
| LSDV015 | 10725–10243 (161) | SPV | P32223 | 308 | 44 | 129 | IL-18 binding protein | ||||||
| LSDV016 | 11031–10765 (89) | Yaba-like DV | AJ293568 | 129 | 50 | 48 | EGF-like growth factor | M010L | 32 | C11R | 50 | ||
| LSDV017 | 11552–11025 (176) | Yaba-like DV | AJ293568 | 173 | 34 | 173 | Integral membrane protein, apoptosis regulator, TM | E | M011L | 29 | |||
| LSDV018 | 12034–11597 (146) | SPV | P32208 | 499 | 68 | 141 | dUTPase | E | M012L | 63 | F2L | 63 | |
| LSDV019 | 13790–12084 (569) | SPV | P32206 | 1022 | 40 | 535 | Kelch-like protein, TM | M014L | 32 | F3L | 25 | ||
| LSDV020 | 14820–13858 (321) | Yaba-like DV | AJ293568 | 1361 | 79 | 320 | Ribonucleotide reductase, small subunit, TM | E | M015L | 76 | F4L | 75 | |
| LSDV021 | 15121–14864 (86) | SPV | P32220 | 179 | 38 | 86 | TM | E,L | M016L | 34 | |||
| LSDV022 | 15500–15165 (112) | E | |||||||||||
| LSDV023 | 15949–15734 (72) | MYX | AF170726 | 169 | 51 | 72 | E | M018L | 51 | F8L | 42 | ||
| LSDV024 | 16676–16029 (216) | SPV | P32207 | 754 | 64 | 216 | TM | L | M019L | 50 | F9L | 46 | |
| LSDV025 | 17997–16657 (447) | SPV | P32216 | 1996 | 80 | 435 | Ser/Thr protein kinase; virus assembly | L | M020L | 77 | F10L | 72 | |
| LSDV026 | 18941–18036 (302) | VV | P21052 | 241 | 28 | 293 | F11L | 28 | |||||
| LSDV027 | 20866–18953 (638) | Yaba-like DV | AJ293568 | 1298 | 45 | 626 | EEV maturation, TM | M021L | 39 | F12L | 38 | ||
| LSDV028 | 21985–20876 (370) | AF199595 | Yaba-like DV | AJ293568 | 1515 | 77 | 370 | Palmitylated EEV envelope protein | L | M022L | 73 | F13L | 57 |
| LSDV029 | 22624–22190 (145) | Yaba-like DV | AJ293568 | 518 | 68 | 148 | E | M024L | 54 | F15L | 56 | ||
| LSDV030 | 23360–22704 (219) | Yaba-like DV | AJ293568 | 391 | 39 | 215 | E | M025L | 35 | F16L | 34 | ||
| LSDV031 | 23434–23745 (104) | Yaba-like DV | AJ293568 | 410 | 74 | 105 | DNA-binding virion core phosphoprotein | L | M026L | 75 | F17L | 61 | |
| LSDV032 | 25176–23755 (474) | RFV | AF170722 | 1813 | 75 | 474 | Poly(A) polymerase PAPL | E | M027L | 74 | E1L | 66 | |
| LSDV033 | 27380–25176 (735) | MYX | AF170726 | 1839 | 46 | 728 | TM | M028L | 46 | E2L | 39 | ||
| LSDV034 | 27925–27395 (177) | Yaba-like DV | AJ293568 | 464 | 48 | 183 | PKR inhibitor, host range | E | M029L | 40 | E3L | 36 | |
| LSDV035 | 28590–29795 (402) | RFV | AF170722 | 667 | 39 | 379 | M031R | 39 | E5R | 27 | |||
| LSDV036 | 28591–27989 (201) | Yaba-like DV | AJ293568 | 738 | 69 | 181 | RNA polymerase subunit RPO30 | E | M030L | 68 | E4L | 62 | |
| LSDV037 | 29807–31504 (566) | MYX | AF170726 | 2190 | 70 | 566 | M032R | 70 | E6R | 62 | |||
| LSDV038 | 31514–32311 (266) | Yaba-like DV | AJ293568 | 1144 | 79 | 263 | TM | M033R | 79 | E8R | 68 | ||
| LSDV039 | 35343–32314 (1010) | MYX | AF170726 | 4061 | 74 | 1010 | DNA polymerase | M034L | 74 | E9L | 66 | ||
| LSDV040 | 35377–35661 (95) | RFV | AF170722 | 418 | 77 | 95 | Potential redox protein, virus assembly | M035R | 73 | E10R | 69 | ||
| LSDV041 | 36053–35664 (130) | Yaba-like DV | AJ293568 | 375 | 54 | 130 | Virion core protein | L | E11L | 46 | |||
| LSDV042 | 38094–36043 (684) | Yaba-like DV | AJ293568 | 1448 | 43 | 678 | M036L | 41 | O1L | 35 | |||
| LSDV043 | 39144–38203 (314) | MYX | AF170726 | 1221 | 75 | 309 | DNA-binding virion core protein, virus assembly | L | M038L | 75 | I1L | 66 | |
| LSDV044 | 39369–39154 (72) | MYX | AF170726 | 191 | 50 | 72 | TM | L | M039L | 50 | 12L | 44 | |
| LSDV045 | 40200–39373 (276) | Yaba-like DV | AJ293568 | 848 | 62 | 266 | DNA-binding phosphoprotein | E | M040L | 59 | I3L | 49 | |
| LSDV046 | 40482–40249 (78) | Yaba-like DV | AJ293568 | 263 | 68 | 76 | IMV membrane protein, SP, TM | L | M041L | 50 | I5L | 35 | |
| LSDV047 | 41684–40503 (394) | RFV | AF170722 | 1140 | 53 | 394 | TM | M042L | 53 | I6L | 51 | ||
| LSDV048 | 42978–41680 (433) | Yaba-like DV | AJ293568 | 1779 | 75 | 433 | Virion core protein | L | M043L | 75 | I7L | 66 | |
| LSDV049 | 42984–45011 (676) | Yaba-like DV | AJ293568 | 2359 | 65 | 676 | NPH-II, RNA helicase | M044R | 60 | I8R | 59 | ||
| LSDV050 | 46801–45014 (596) | RFV | AF170722 | 1976 | 62 | 595 | Metalloprotease, virion morphogenesis | L | M045L | 61 | G1L | 55 | |
| LSDV051 | 47124–47789 (222) | MYX | AF170726 | 737 | 62 | 225 | Putative transcriptional elongation factor | M047R | 62 | G2R | 50 | ||
| LSDV052 | 47130–46801 (110) | RFV | AF170722 | 332 | 59 | 105 | TM | L | M046L | 58 | G3L | 48 | |
| LSDV053 | 48136–47759 (126) | MYX | AF170726 | 527 | 77 | 123 | Glutaredoxin 2, virion morphogenesis, SP | L | M048L | 77 | G4L | 49 | |
| LSDV054 | 48139–49449 (437) | Yaba-like DV | AJ293568 | 1144 | 52 | 438 | M049R | 49 | G5R | 47 | |||
| LSDV055 | 49453–49641 (63) | Yaba-like DV | AJ293568 | 286 | 85 | 63 | RNA polymerase subunit RPO7 | E,L | M050R | 85 | G5.5R | 79 | |
| LSDV056 | 49644–50165 (174) | RFV | AF170722 | 541 | 58 | 172 | M051R | 58 | G6R | 44 | |||
| LSDV057 | 51303–50185 (373) | Yaba-like DV | AJ293568 | 1128 | 60 | 374 | Virion core protein, TM | L | M052L | 60 | G7L | 55 | |
| LSDV058 | 51333–52112 (260) | Yaba MTV | AB015885 | 1214 | 86 | 260 | Late transcription factor VLTF-1, TM | I | M053R | 85 | G8R | 87 | |
| LSDV059 | 52142–53149 (336) | Yaba-like DV | AJ293568 | 1125 | 61 | 336 | Myristylated protein | M054R | 52 | G9R | 46 | ||
| LSDV060 | 53153–53887 (245) | Yaba-like DV | AJ293568 | 1069 | 81 | 244 | Myristylated IMV envelope protein, TM | L | M055R | 76 | L1R | 66 | |
| LSDV061 | 53928–54203 (92) | Yaba MTV | AB015885 | 187 | 48 | 75 | E | M056R | 33 | L2R | 35 | ||
| LSDV062 | 55172–54219 (318) | Yaba-like DV | AJ293568 | 1170 | 67 | 318 | L | M057L | 61 | L3L | 55 | ||
| LSDV063 | 55197–55955 (253) | RFV | AF170722 | 1075 | 82 | 253 | DNA-binding virion core protein VP8 | L | M058R | 81 | L4R | 61 | |
| LSDV064 | 55974–56366 (131) | Yaba-like DV | AJ293568 | 393 | 59 | 127 | TM | L | M059R | 55 | L5R | 51 | |
| LSDV065 | 56326–56766 (147) | P19746 | Yaba-like DV | AJ293568 | 505 | 67 | 146 | L | M060R | 61 | J1R | 55 | |
| LSDV066 | 56797–57327 (177) | P16600 | SPV | P23335 | 640 | 68 | 175 | Thymidine kinase | M061R | 70 | J2R | 66 | |
| LSDV067 | 57402–57995 (198) | A06139 | SPV | P23333 | 467 | 48 | 186 | Host range protein | E | M062R | 41 | C7L | 33 |
| LSDV068 | 58056–59054 (333) | MYX | P18628 | 1451 | 81 | 333 | Poly(A) polymerase PAPs | M065R | 81 | J3R | 72 | ||
| LSDV069 | 58972–59526 (185) | Yaba-like DV | AJ293568 | 756 | 76 | 185 | RNA polymerase subunit RPO22 | M066R | 73 | J4R | 68 | ||
| LSDV070 | 59936–59538 (133) | MYX | AF170726 | 536 | 67 | 132 | M067L | 67 | J5L | 59 | |||
| LSDV071 | 60022–63876 (1285) | RFV | AF170722 | 5838 | 86 | 1286 | RNA polymerase subunit RPO147 | E | M068R | 85 | J6R | 80 | |
| LSDV072 | 64399–63887 (171) | AF124517 | MYX | L31960 | 715 | 76 | 171 | Protein-tyrosine phosphatase, virus assembly | L | M069L | 76 | H1L | 63 |
| LSDV073 | 64415–64984 (190) | AF124517 | RFV | AF170722 | 764 | 70 | 190 | TM | M070R | 69 | H2R | 65 | |
| LSDV074 | 65952–64987 (322) | AF124516 | MYX | AF170726 | 986 | 54 | 319 | IMV envelope protein p35, TM | M071L | 54 | H3L | 38 | |
| LSDV075 | 68378–65985 (798) | Yaba-like DV | AJ293568 | 3289 | 78 | 798 | RNA polymerase-associated protein RAP94 | L | M072L | 77 | H4L | 70 | |
| LSDV076 | 68522–69190 (223) | Yaba MTV | AB015885 | 419 | 46 | 223 | Late transcription factor VLTF-4 | E | M073R | 44 | H5R | 37 | |
| LSDV077 | 69235–70185 (317) | Yaba-like DV | AJ293568 | 1171 | 68 | 317 | DNA topoisomerase | M074R | 70 | H6R | 65 | ||
| LSDV078 | 70208–70648 (147) | MYX | AF170726 | 482 | 60 | 146 | L | M075R | 60 | H7R | 43 | ||
| LSDV079 | 70682–73207 (842) | Yaba-like DV | AJ293568 | 3258 | 72 | 842 | mRNA capping enzyme, large subunit | E | M076R | 68 | D1R | 64 | |
| LSDV080 | 73639–73175 (155) | Yaba-like DV | AJ293568 | 269 | 38 | 151 | Virion protein | M077L | 21 | D2L | 41 | ||
| LSDV081 | 73641–74375 (245) | Yaba MTV | AB015885 | 477 | 39 | 245 | Virion protein | M078R | 33 | D3R | 34 | ||
| LSDV082 | 74375–75028 (218) | RFV | P32941 | 926 | 74 | 218 | Uracil DNA glycosylase | M079R | 74 | D4R | 66 | ||
| LSDV083 | 75074–77431 (786) | Yaba-like DV | AJ293568 | 3394 | 78 | 786 | NTPase; DNA replication | M080R | 76 | D5R | 67 | ||
| LSDV084 | 77431–79335 (635) | Yaba MTV | AB015885 | 2944 | 88 | 635 | Early transcription factor VETFa+ TM | L | M081R | 88 | D6R | 81 | |
| LSDV085 | 79363–79851 (163) | Yaba-like DV | AJ293568 | 725 | 80 | 160 | RNA polymerase subunit RPO18 | M082R | 80 | D7R | 68 | ||
| LSDV086 | 79895–80533 (213) | Yaba-like DV | AJ293568 | 742 | 68 | 211 | mut T motif | E | M084R | 61 | D9R | 57 | |
| LSDV087 | 80536–81294 (253) | Yaba-like DV | AJ293568 | 776 | 64 | 237 | mut T motif; gene expression regulator | M085R | 57 | D10R | 5 | ||
| LSDV088 | 83210–81306 (635) | Yaba-like DV | AJ293568 | 2559 | 75 | 632 | NPH-I; transcription termination factor | M086L | 74 | D11L | 71 | ||
| LSDV089 | 84100–83240 (287) | SPV | Q08512 | 1259 | 80 | 287 | mRNA capping enzyme, small subunit; VITF | E,L | M087L | 77 | D12L | 74 | |
| LSDV090 | 85789–84143 (549) | Yaba-like DV | AJ293568 | 2400 | 80 | 549 | Rifampin resistance protein, IMV assembly | M088L | 76 | D13L | 69 | ||
| LSDV091 | 86268–85819 (150) | Yaba-like DV | AJ293568 | 563 | 67 | 150 | Late transcription factor VLTF-2 | I,L | M089L | 67 | A1L | 63 | |
| LSDV092 | 86996–86301 (232) | RFV | AF170722 | 1049 | 88 | 225 | Late transcription factor VLTF-3 | I | M090L | 88 | A2L | 84 | |
| LSDV093 | 87220–86996 (75) | MYX | AF170726 | 297 | 72 | 74 | L | M091L | 72 | 8.9kd | 52 | ||
| LSDV094 | 89214–87232 (661) | MYX | AF170726 | 2597 | 73 | 661 | Virion core protein P4b, TM | L | M092L | 73 | A3L | 65 | |
| LSDV095 | 89824–89342 (161) | Yaba-like DV | AJ293568 | 245 | 39 | 158 | Virion core protein, virion morphogenesis | M093L | 35 | A4L | 29 | ||
| LSDV096 | 89865–90374 (170) | Yaba-like DV | AJ293568 | 537 | 62 | 170 | RNA polymerase subunit RPO19 | M094R | 62 | A5R | 62 | ||
| LSDV097 | 91501–90377 (375) | MYX | AF170726 | 1434 | 75 | 371 | L | M095L | 75 | A6L | 56 | ||
| LSDV098 | 93666–91525 (714) | MYX | AF170726 | 3131 | 82 | 712 | Early transcription factor VETFL | L | M096L | 82 | A7L | 71 | |
| LSDV099 | 93723–94592 (290) | U95121 | MYX | AF170726 | 1090 | 72 | 288 | Intermediate transcription factor VITF-3 | E | M097R | 72 | A8R | 62 |
| LSDV100 | 94855–94622 (78) | RFV | AF170722 | 323 | 75 | 78 | IMV membrane protein, SP, TM | L | M098L | 73 | A9L | 68 | |
| LSDV101 | 97570–94859 (904) | Yaba-like DV | AJ293568 | 3193 | 67 | 904 | Virion core protein P4a | L | M099L | 65 | A10L | 50 | |
| LSDV102 | 97585–98535 (317) | MYX | AF170726 | 1222 | 73 | 317 | TM | L | M100R | 73 | A11R | 53 | |
| LSDV103 | 99107–98538 (190) | Yaba-like DV | AJ293568 | 446 | 56 | 189 | Virion core protein | M101L | 56 | A12L | 51 | ||
| LSDV104 | 99375–99175 (67) | RFV | AF170722 | 223 | 65 | 67 | IMV membrane protein, TM | M102L | 60 | A13L | 39 | ||
| LSDV105 | 99744–99460 (95) | Yaba-like DV | AJ293568 | 391 | 78 | 95 | IMV membrane protein, SP, TM | L | M103L | 66 | A14L | 55 | |
| LSDV106 | 99922–99764 (53) | Yaba-like DV | AJ293568 | 223 | 79 | 53 | Virulence factor | L | M104L | 79 | A14.5 | 58 | |
| LSDV107 | 100199–99915 (95) | Yaba MTV | AB015885 | 297 | 57 | 95 | E,L | M105L | 49 | A15L | 47 | ||
| LSDV108 | 101316–100186 (377) | Yaba-like DV | AJ293568 | 1367 | 62 | 381 | Myristylated protein, TM | L | M106L | 61 | A16L | 51 | |
| LSDV109 | 101922–101335 (196) | Yaba-like DV | AJ293568 | 606 | 62 | 196 | Phosphorylated IMV membrane protein, TM | L | M107L | 55 | A17L | 42 | |
| LSDV110 | 101937–103376 (480) | RFV | AF170722 | 1516 | 59 | 479 | DNA helicase; transcriptional elongation | M108R | 59 | A18R | 55 | ||
| LSDV111 | 103584–103363 (74) | RFV | AF170722 | 315 | 81 | 74 | L | M109L | 81 | A19L | 62 | ||
| LSDV112 | 103931–105220 (430) | MYX | AF170726 | 1140 | 50 | 426 | DNA polymerase processivity factor | E | M111R | 50 | A20R | 46 | |
| LSDV113 | 103932–103588 (115) | MYX | AF170726 | 374 | 58 | 115 | SP, TM | M110L | 58 | A21L | 55 | ||
| LSDV114 | 105192–105695 (168) | Yaba-like DV | AJ293568 | 578 | 69 | 156 | M112R | 55 | A22R | 62 | |||
| LSDV115 | 105723–106877 (385) | MYX | AF170726 | 1291 | 64 | 384 | Intermediate transcription factor VITF-3 | E | M113R | 64 | A23R | 60 | |
| LSDV116 | 106911–110378 (1156) | Yaba-like DV | AJ293568 | 5476 | 87 | 1156 | RNA polymerase subunit RPO132 | E | M114R | 88 | A24R | 83 | |
| LSDV117 | 110841–110398 (148) | P16717 | Yaba MTV | AB018404 | 323 | 46 | 145 | Fusion protein, virus assembly | L | M115L | 48 | A27L | 33 |
| LSDV118 | 111264–110845 (140) | P16718 | RFV | AF170722 | 520 | 66 | 139 | TM | L | M116L | 65 | A28L | 52 |
| LSDV119 | 112173–111268 (302) | MYX | AF170726 | 1059 | 63 | 300 | RNA polymerase subunit RPO35 | E | M117L | 63 | A29L | 60 | |
| LSDV120 | 112366–112145 (74) | Yaba MTV | AB018404 | 228 | 61 | 75 | L | M118L | 60 | A30L | 49 | ||
| LSDV121 | 113309–112548 (254) | MYX | AF170726 | 1168 | 86 | 252 | DNA packaging, virus assembly | M120L | 86 | A32L | 60 | ||
| LSDV122 | 113441–114028 (196) | Yaba-like DV | AJ293568 | 364 | 42 | 193 | EEV glycoprotein, TM | M121R | 40 | A33R | 30 | ||
| LSDV123 | 114061–114573 (171) | MYX | AF170726 | 569 | 56 | 171 | EEV protein, SP, TM | L | M122R | 56 | A34R | 42 | |
| LSDV124 | 114604–115176 (191) | RFV | AF170722 | 418 | 46 | 181 | M123R | 44 | A35R | 33 | |||
| LSDV125 | 115216–116079 (288) | RFV | AF170722 | 657 | 41 | 288 | E | M124R | 41 | ||||
| LSDV126 | 116141–116683 (181) | Yaba-like DV | AJ293568 | 164 | 33 | 155 | EEV glycoprotein, TM | M125R | 31 | A36R | 25 | ||
| LSDV127 | 116697–117515 (273) | RFV | AF170722 | 620 | 43 | 273 | M126R | 43 | A37R | 26 | |||
| LSDV128 | 118424–117525 (300) | MYX | AF170726 | 489 | 38 | 271 | CD47-like protein, TM | M128L | 38 | A38L | 26 | ||
| LSDV129 | 118522–118890 (123) | MYX | AF170726 | 126 | 35 | 116 | M130R | 35 | |||||
| LSDV130 | 118962–119204 (81) | Yaba-like DV | AJ293568 | 156 | 45 | 80 | |||||||
| LSDV131 | 119263–119745 (161) | MYX | AF170726 | 528 | 60 | 158 | Superoxide dismutase-like protein | L | M131R | 60 | A45R | 35 | |
| LSDV132 | 119783–120310 (176) | SP | |||||||||||
| LSDV133 | 120343–122019 (559) | MYX | AF170726 | 1728 | 61 | 553 | DNA ligase | M133R | 61 | A50R | 53 | ||
| LSDV134 | 122176–128250 (2025) | MYX | AF170726 | 5102 | 53 | 1987 | VAR B22R homologue, TM | M134R | 53 | ||||
| LSDV135 | 128323–129402 (360) | Yaba-like DV | AJ293568 | 475 | 37 | 316 | IFN-α/β binding protein, SP | M135R | 28 | B19R | 32 | ||
| LSDV136 | 129453–129911 (153) | MYX | AF170726 | 292 | 40 | 145 | A52R-like family protein | E | M136R | 40 | K7R | 24 | |
| LSDV137 | 129980–130984 (335) | Yaba-like DV | AJ293568 | 721 | 43 | 338 | E,L | M137R | 39 | A51R | 30 | ||
| LSDV138 | 131017–131574 (186) | Yaba-like DV | AJ293568 | 285 | 51 | 106 | Ig domain, OX-2-like protein, SP, TM | M141R | 48 | ||||
| LSDV139 | 131616–132530 (305) | MYX | AF170726 | 1054 | 66 | 300 | Ser/Thr protein kinase, DNA replication | M142R | 66 | B1R | 50 | ||
| LSDV140 | 132565–133284 (240) | RFV | L26342 | 556 | 43 | 230 | N1R/p28-like host range RING finger protein | M143R | 43 | ||||
| LSDV141 | 133336–134010 (225) | Yaba-like DV | AJ293568 | 362 | 38 | 204 | EEV host range protein, SP, TM | M144R | 35 | B5R | 27 | ||
| LSDV142 | 134015–134416 (134) | MYX | AF170726 | 222 | 43 | 109 | Secreted virulence factor | M146R | 43 | N1L | 24 | ||
| LSDV143 | 134456–135361 (302) | RFV | JQ1743 | 776 | 52 | 262 | Tyrosine protein kinase-like protein | M147R | 51 | ||||
| LSDV144 | 135533–137173 (547) | VV | P24768 | 456 | 28 | 516 | Kelch-like protein | M140R | 26 | A55R | 28 | ||
| LSDV145 | 137222–139123 (634) | RFV | AF170722 | 1113 | 36 | 662 | Ankyrin repeat protein | M148R | 34 | B4R | 25 | ||
| LSDV146 | 139255–140493 (413) | VV | U94848 | 1030 | 52 | 388 | Phospholipase D-like protein | M022L | 29 | K4L | 52 | ||
| LSDV147 | 140557–142050 (498) | RFV | AF170722 | 973 | 40 | 498 | Ankyrin repeat protein | M149R | 39 | B4R | 25 | ||
| LSDV148 | 142101–143441 (447) | Yaba-like DV | AJ293568 | 670 | 35 | 454 | Ankyrin repeat protein | E | M148R | 29 | B4R | 26 | |
| LSDV149 | 143465–144475 (337) | Yaba-like DV | AJ293568 | 771 | 44 | 334 | Serpin, SP, TM | E,L | M151R | 38 | C12L | 38 | |
| LSDV150 | 144517–144999 (161) | Yaba-like DV | AJ293568 | 125 | 27 | 155 | A52R-like family protein | E | M139R | 27 | A52R | 24 | |
| LSDV151 | 145045–146694 (550) | Yaba-like DV | AJ293568 | 519 | 30 | 515 | Kelch-like protein | E | M140R | 26 | A55R | 28 | |
| LSDV152 | 146764–148230 (489) | RFV | AF170722 | 467 | 27 | 481 | Ankyrin repeat protein | E | M005 | 24 | B4R | 25 | |
| LSDV153 | 148278–148550 (91) | SPV | P32230 | 217 | 46 | 91 | SP | M004.1 | 43 | ||||
| LSDV154 | 148623–149342 (240) | P18386 | MYX | AF002684 | 378 | 37 | 231 | ER-localized apoptosis regulator, SP, TM | E | M004 | 37 | B9R | 37 |
| LSDV155 | 149595–149987 (131) | P18388 | MYX | AJ012282 | 242 | 43 | 131 | M003.2 | 43 | ||||
| LSDV156 | 150061–150537 (159) | P18387 | RFV | P25949 | 308 | 46 | 143 | A52R-like family protein, SP | M003.1 | 42 | B15R | 34 | |
Accession numbers are from the GenBank or SwissProtein database.
Yaba MTV, Yaba monkey tumor virus.
aa, amino acids.
Function was deduced either from the degree of similarity to known genes or from the presence of Prosite signatures. TM, a Z score of > 1.96 was used for the prediction of transmembrane (TM) domains with the MEMSAT computer program (32); SP, N-terminal signal peptide (Z score of >3.5 within 40 amino acids of the N terminus using the SIGCLEAVE computer program (ftp://ftp.ebi.ac.uk/pub/software/unix/EMBOSS/) [51]). ER, endoplasmic reticulum.
Putative promoters (E, early; I, intermediate; L, late) were identified as previously described (2).
Best-matching ORF from the MYX genome (accession no. AF170726).
Nucleic acid biogenesis, virion structure, and virion assembly.
LSDV contains the majority of conserved poxviral genes involved in basic replicative mechanisms, including at least 26 genes encoding RNA polymerase subunits, mRNA transcription initiation, elongation, and termination factors, and enzymes which direct posttranscriptional processing of viral mRNA (41) (Table 1). Also present in LSDV are seven homologues of ChPV genes necessary for, or potentially involved in, DNA replication, including LSDV039, LSDV077, LSDV082, LSDV083, LSDV112, LSDV133, and LSDV139 (41). LSDV proteins potentially involved in nucleotide metabolism include homologues of thymidine kinase, dUTP pyrophosphatase, and the small subunit of ribonucleotide reductase (Table 1). LSDV contains the same complement of nucleotide metabolism genes found in the leporipoxviruses and, like the leporipoxviruses, it lacks a large subunit of ribonucleotide reductase (52). This shared complement likely reflects phylogenetic relatedness but may also be significant in cell and/or tissue tropism.
LSDV encodes at least 30 homologues of poxviral proteins known to be structural or involved in virion morphogenesis and assembly (Table 1). These include proteins present in the virion core; proteins present in the intracellular mature virus (IMV) and associated membranes; potential enzymes involved in protein modification, DNA packaging, and redox activity; and at least four VV proteins found in or associated with the release of extracellular enveloped virions (EEV) (Table 1). Additionally, LSDV095, LSDV126, and LSDV141, although significantly different from VV A4L core protein, A36R EEV protein, and B5R EEV protein, respectively, were annotated here as putative structural protein homologues based on similar genomic position and other conserved features. LSDV, like molluscum contagiosum virus (MCV) and fowlpox virus (FPV), lacks an obvious homologue of the VV IMV membrane protein D8L, a cell surface binding protein which is also present in the leporipoxviruses.
Host-related functions.
LSDV contains a number of potential host range genes with likely functions in modulation or evasion of host immune responses, in modulation or inhibition of host cell apoptosis, and in aspects of cell and/or tissue tropism. Many potential LSDV host range genes are similar in sequence and in terminal genomic location to genes present in other poxviruses. However, LSDV encodes a unique complement genes which dictate its specific host range properties.
Six LSDV proteins are potentially secreted and are likely involved in the disruption or modulation of host immune responses, as indicated by the presence of potential signal peptide sequences and/or similarity to other secreted immunomodulators. These include homologues of cellular and viral interleukin-10 (IL-10), gamma interferon (IFN-γ) receptor (R), IL-1R, IFN-α/β binding protein, and IL-18 binding protein (Table 1). Similar to other IL-10 homologues present in orf virus and some herpesviruses, LSDV005 strongly resembles cellular IL-10 in the carboxyl terminus and likely has similar immunoregulatory and immunosuppressive activities (22, 40). Notably, phylogenetic analysis indicates that LSDV005 is divergent from both cellular IL-10 (43% amino acid identity) and orf virus IL-10 (48% amino acid identity), which is very similar to ovine IL-10 (81% amino acid identity). This suggests an independent and more recent acquisition of host IL-10 into orf virus than into LSDV. LSDV is the first poxvirus known to encode two proteins, in addition to poxvirus IFN-α/β binding proteins, with similarity to IL-1 R (LSDV013 and LSDV006). LSDV013 contains the three immunoglobulin (Ig) domains common to IL-1R and likely functions as an IL-1 binding protein. LSDV006 lacks a third Ig domain in the carboxyl terminus and may perform a similar or perhaps alternative immunomodulatory function.
LSDV contains four potentially membrane localized, immunomodulatory proteins. Homologues of a G protein-coupled CC chemokine receptor (GPCR), CD47, and poxvirus OX-2-like proteins potentially bind extracellular factors and/or influence intracellular signal transduction mechanisms to affect immune mechanisms or host range (7, 35, 37, 45) (Table 1). LSDV010 and homologues in swinepox virus (SPV), Yaba-like disease virus (Yaba-like DV), and leporipoxviruses are similar to several immunomodulatory proteins found in gammaherpesviruses. All contain the cysteine-rich amino-terminal motif (CWICX10–11CXCX4–7HX2CX3WX8–16CX2C) previously noted as similar to the C4HC3 LAP/PHD finger motif and two positionally conserved transmembrane domains located in central to carboxyl-terminal regions (data not shown) (43). The gammaherpesvirus proteins affect virus-induced inhibition of class I major histocompatibility antigen (MHC-I)-mediated antigen presentation through decreased cell surface expression of MHC-I and can downregulate the expression of natural killer (NK) cell activation ligands to effectively inhibit NK cell-mediated cytotoxicity (31, 50). LSDV010, like the gammaherpesvirus proteins, may function in viral immune evasion.
Several LSDV proteins are likely to have intracellular roles in immune modulation or immune evasion. These include homologues of VV PKR inhibitors (LSDV014 and LSDV034) which confer resistance to the antiviral effects of IFN (Table 1). Poxviral serine proteinase inhibitors (serpins) are known to perform anti-inflammatory roles, and the single serpin encoded in LSDV (LSDV149) is similar to Yaba-like DV 149R, myxoma virus (MYX) M151R, and the single serpin in SPV (37; C. L. Afonso et al., unpublished data). Notably, LSDV001, LSDV009, LSDV136, LSDV150, and LSDV156 genes are similar to a group of poxviral genes which includes VV A52R and others previously described as a gene family (Family 5 [48]) (data not shown). Although the function of most genes in this group is not known, VV A52R functions as an antagonist for host cell IL-1 and Toll-like receptor-mediated intracellular signaling, including IL-1R, Toll-like receptor 4, and IL-18R-mediated induction of NF-κB activation (6). The potential for IL-1/Toll-like receptor inhibition by a family of poxvirus proteins is significant considering the role of IL-1/Toll-like receptor signaling in the induction of innate immune responses and inflammation (21).
LSDV encodes six homologues of other poxviral proteins known to affect virus virulence, virus growth in specific cell types, and/or cellular apoptotic responses (Table 1). These include homologues of epidermal growth factor (EGF), VV C7L host range, N1L virulence, and A14.5L virulence proteins, MYX M004 and M011L anti-apoptosis proteins, and the rabbit fibroma virus (RFV) N1R/ectromelia virus p28 host range factor. LSDV also encodes five proteins containing ankyrin repeat motifs, two of which (LSDV145 and LSDV147) appear to be orthologues of proteins encoded in leporipoxviruses and SPV based on genomic position, amino acid similarity, and phylogenetic analysis (7, 52; Afonso et al., unpublished) (Table 1). Poxviral ankyrin repeat genes have been associated with host range functions in MYX, cowpox virus, and VV and may inhibit virally induced apoptosis (28, 42, 49). It has been suggested that specific complements of ankyrin genes dictate poxvirus host range, and the same is likely for LSDV (3, 47).
Three LSDV genes are homologues of poxvirus genes resembling cellular enzymes (Table 1). These include LSDV146, which resembles the VV K4L phospholipase D-like protein thus far found only in VV and LSDV. Notably, LSDV proteins similar to Cu-Zn superoxide dismutase (LSDV131) and tyrosine protein kinase (LSDV143) resemble other poxvirus homologues (in leporipoxviruses and orthopoxviruses and in leporipoxviruses and FPV, respectively) in that they lack residues that would predict enzymatic activity.
In terminal genomic regions, LSDV encodes several homologues of poxvirus proteins with unknown function, including VV C10L and 8.9 kD proteins, which interact with VV host range and morphogenesis proteins, respectively, a yatapoxvirus protein (LSDV130), and a homologue of the variola virus B22R putative membrane protein (Table 1) (39). LSDV encodes three proteins (LSDV019, LSDV144, and LSDV151) that contain four to five imperfect carboxyl-terminal repeats similar to those found in the Drosophila kelch protein and other poxvirus kelch-like proteins (Table 1). Notably, LSDV potentially encodes two proteins (LSDV022 and LSDV132) that lack homology to other known proteins.
Comparison LSDV to other ChPV.
LSDV is very similar to other ChPVs in overall genome structure and composition, including the presence of a central conserved core of genes, adjacent variable region containing many genes with host related functions, and ITRs (2, 3, 7, 29, 38, 46, 52). Most of the LSDV genome is highly colinear with those of other ChPV (Table 1) (24). Sixty-five percent of the LSDV genome (LSDV024 to LSDV123) consists of a central core of genes conserved across divergent ChPV genera (2, 29, 46). LSDV gene colinearity is most conserved, however, with Yaba-like DV and leporipoxviruses (83% of the LSDV genome, from LSDV016 to LSDV143) (Table 1). Overall amino acid identity is higher between LSDV and MYX proteins (56% average) and between LSDV and Yaba-like DV proteins (57% average) than between LSDV and VV proteins (49% average). Thus, the genomes of LSDV, Yaba-like DV, and leporipoxviruses appear to be relatively well conserved in gene content, gene arrangement, and amino acid identity (Table 1).
The terminal genomic regions of LSDV encode many of the proteins with probable functions involving host range, virulence, and immune modulation. At the amino acid level, many of these LSDV proteins are less similar to their homologues than are proteins encoded in the conserved central core region, and several are most similar to cellular proteins (Table 1). Although terminal regions are similar to leporipoxviruses, yatapoxviruses, and SPV in gene content, several LSDV genes have homologues in other ChPV genera (Table 1). For instance, LSDV homologues of IL-1 binding protein, IL-10, GPCR, and VV C10L are absent in the closely related leporipoxviruses, and LSDV homologues of IL-10, IFN-γR, MYX M004, DNA ligase, superoxide dismutase-like protein, tyrosine protein kinase-like protein, and phospholipase D are absent in Yaba-like DV. LSDV lacks many genes for virulence and/or host range proteins found in other poxviruses. These include the 35-kDa secreted chemokine binding protein (leporipoxviruses and orthopoxviruses), tumor necrosis factor receptor homologues (leporipoxviruses and orthopoxviruses), MDA-7 cytokine-like protein (Yaba-like DV), MHC-I-like proteins (Yaba-like DV, SPV, and MCV), semaphorin-like protein (orthopoxviruses, FPV), glutathione peroxidase (MCV and FPV), hydroxysteroid dehydrogenase (Yaba-like DV, orthopoxviruses, MCV, and FPV), CPD photolyase (leporipoxviruses and FPV), lysophospholipase (Yaba-like DV and orthopoxviruses), and sialyltransferase (leporipoxviruses). LSDV contains only one serpin-like protein and one GPCR-like protein, while other poxviruses contain multiple distinct serpin proteins (Yaba-like DV, leporipoxviruses, orthopoxviruses, and FPV) and GPCR proteins (Yaba-like DV and FPV). LSDV also lacks homologues of poxviral A type inclusion proteins (orthopoxviruses, MCV, and FPV) (24).
Finally, LSDV genes were nearly identical (97 to 100% amino acid identity) to 16 genes previously sequenced from either LSDV or ShPV (Table 1). The terminal regions of LSDV strain 2490 were highly similar to regions sequenced from two ShPV isolates (25). Interestingly, greater conservation was seen between LSDV strain 2490 and a nonpathogenic Kenya ShPV (KS) isolate than was observed between KS and a pathogenic India ShPV isolate whose homologue of LSDV002 is disrupted (25). Comparative analysis of the LSDV genome sequence with those of ShPV and GPV will help define the genetic basis of CaPV host range.
Conclusions.
LSDV gene content and organization indicates a close structural and functional relationship to other ChPV, particularly to yatapoxviruses and leporipoxviruses. The highest conservation occurs with genes involved in basic replicative mechanisms, including mRNA biogenesis, DNA replication, and virion structure and assembly. Terminal genomic sequences contain a unique complement of at least 34 genes which are in gene families or likely function in virulence, host range, and/or immune evasion. An improved understanding of how these genes affect LSDV/host interactions will permit the engineering of novel vaccine viruses and expression vectors with enhanced efficacy and greater versatility. Additionally, the LSDV genomic sequence provides a basis from which comparisons with other CaPVs may be made, thus contributing to our understanding of the genetic basis of CaPV virulence and host range.
ACKNOWLEDGMENTS
We thank J. Lubroth for providing the 2490 strain of LSDV and A. Zsak and A. Ciupryk for excellent technical assistance.
REFERENCES
- 1.Afonso C A, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 4.Barnard B J, Munz E, Dumbell K, Prozesky L. Lumpy skin disease. In: Coetzer J A W, Thomson G R, Tustin R C, editors. Infectious diseases of livestock. Vol. 1. Cape Town, South Africa: Oxford University Press; 1994. pp. 604–612. [Google Scholar]
- 5.Black D N, Hammond J M, Kitching R P. Genomic relationship between capripoxviruses. Virus Res. 1986;5:277–292. doi: 10.1016/0168-1702(86)90024-9. [DOI] [PubMed] [Google Scholar]
- 6.Bowie A, Kiss-Toth E, Symons J A, Smith G L, Dower S K, O'Neill L A. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc Natl Acad Sci USA. 2000;97:10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao J X, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 8.Capstick P B. Lumpy skin disease—experimental infection. Bull Epizoot Dis Afr. 1959;7:51–62. [Google Scholar]
- 9.Capstick P B, Coackley W. Protection of cattle against lumpy skin disease. Res Vet Sci. 1961;2:362–375. [Google Scholar]
- 10.Carn V M. Control of capripoxvirus infections. Vaccine. 1993;11:1275–1279. doi: 10.1016/0264-410x(93)90094-e. [DOI] [PubMed] [Google Scholar]
- 11.Carn V M, Kitching R P. The clinical response of cattle experimentally infected with lumpy skin disease (Neethling) virus. Arch Virol. 1995;140:503–513. doi: 10.1007/BF01718427. [DOI] [PubMed] [Google Scholar]
- 12.Carn V M, Kitching R P. An investigation of possible routes of transmission of lumpy skin disease virus (Neethling) Epidemiol Infect. 1995;114:219–226. doi: 10.1017/s0950268800052067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carn V M, Timms C P, Chand P, Black D N, Kitching R P. Protection of goats against capripox using a subunit vaccine. Vet Rec. 1994;135:434–436. doi: 10.1136/vr.135.18.434. [DOI] [PubMed] [Google Scholar]
- 14.Coetzer J A W, Thomson G R, Tustin R C. Poxviridae. In: Coetzer J A W, Thomson G R, Tustin R C, editors. Infectious diseases of livestock. Vol. 1. Cape Town, South Africa: Oxford University Press; 1994. pp. 601–603. [Google Scholar]
- 15.Davies F G. Lumpy skin disease, an African capripox virus disease of cattle. Br Vet J. 1991;147:489–503. doi: 10.1016/0007-1935(91)90019-J. [DOI] [PubMed] [Google Scholar]
- 16.Davies F G. Observations on the epidemiology of lumpy skin disease in Kenya. J Hyg. 1982;88:95–102. doi: 10.1017/s002217240006993x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenner F. Poxviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2673–2702. [Google Scholar]
- 19.Fick W C, Viljoen G J. Early and late transcriptional phases in the replication of lumpy-skin-disease virus. Onderstepoort J Vet Res. 1994;61:255–261. [PubMed] [Google Scholar]
- 20.Fick W C, Viljoen G J. Identification and characterisation of an early/late bi-directional promoter of the capripoxvirus, lumpy skin disease virus. Arch Virol. 1999;144:1229–1239. doi: 10.1007/s007050050582. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald K A, O'Neill L A. The role of the interleukin-1/Toll-like receptor superfamily in inflammation and host defence. Microbes Infect. 2000;2:933–943. doi: 10.1016/s1286-4579(00)00396-8. [DOI] [PubMed] [Google Scholar]
- 22.Fleming S B, Haig D M, Nettleton P, Reid H W, McCaughan C A, Wise L M, Mercer A. Sequence and functional analysis of a homolog of interleukin-10 encoded by the parapoxvirus orf virus. Virus Genes. 2000;21:85–95. doi: 10.1023/b:viru.0000018443.19040.99. [DOI] [PubMed] [Google Scholar]
- 23.Galtier N, Gouy M, Gautier C. SEA VIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. CABIOS. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 24.Gershon P D, Ansell D M, Black D N. A comparison of the genome organization of capripoxvirus with that of the orthopoxviruses. J Virol. 1989;63:4703–4708. doi: 10.1128/jvi.63.11.4703-4708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gershon P D, Black D N. A capripoxvirus pseudogene whose only intact homologs are in other poxvirus genomes. Virology. 1989;172:350–354. doi: 10.1016/0042-6822(89)90138-4. [DOI] [PubMed] [Google Scholar]
- 26.Gershon P D, Black D N. A comparison of the genomes of capripoxvirus isolates of sheep, goats, and cattle. Virology. 1988;164:341–349. doi: 10.1016/0042-6822(88)90547-8. [DOI] [PubMed] [Google Scholar]
- 27.Gershon P D, Black D N. Physical characterization of the genome of a cattle isolate of capripoxvirus. Virology. 1987;160:473–476. doi: 10.1016/0042-6822(87)90019-5. [DOI] [PubMed] [Google Scholar]
- 28.Gillard S, Spehner D, Drillien R, Kirn A. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc Natl Acad Sci USA. 1986;83:5573–5577. doi: 10.1073/pnas.83.15.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishido S, Choi J K, Lee B S, Wang C, DeMaria M, Johnson R P, Cohen G B, Jung J U. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity. 2000;13:365–374. doi: 10.1016/s1074-7613(00)00036-4. [DOI] [PubMed] [Google Scholar]
- 32.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 33.Kitching R P, Bhat P P, Black D N. The characterization of African strains of capripoxvirus. Epidemiol Infect. 1989;102:335–343. doi: 10.1017/s0950268800030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin E V, Tatusov R L, Rudd K E. Protein sequence comparison at genome scale. Methods Enzymol. 1996;266:295–322. doi: 10.1016/s0076-6879(96)66020-0. [DOI] [PubMed] [Google Scholar]
- 35.Lalani A S, Masters J, Zeng W, Barrett J, Pannu R, Everett H, Arendt C W, McFadden G. Use of chemokine receptors by poxviruses. Science. 1999;286:1968–1971. doi: 10.1126/science.286.5446.1968. [DOI] [PubMed] [Google Scholar]
- 36.MacOwen K D S. Observations on the epizootiology of lumpy skin disease during the first year of its occurence in Kenya. Bull Epizoot Dis Afr. 1959;7:7–20. [Google Scholar]
- 37.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 38.Massung R F, Liu L-I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 39.McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore K W, O'Garra A, de Waal Malefyt R, Vieira P, Mosmann T R. Interleukin-10. Annu Rev Immunol. 1993;11:165–190. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 41.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 42.Mossman K, Lee S F, Barry M, Boshkov L, McFadden G. Disruption of M-T5, a novel myxoma virus gene member of poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J Virol. 1996;70:4394–4410. doi: 10.1128/jvi.70.7.4394-4410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J, Teichmann S A. DIVCLUS: an automatic method in the GEANFAMMER package that finds homologous domains in single- and multi-domain proteins. Bioinformatics. 1998;14:144–150. doi: 10.1093/bioinformatics/14.2.144. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson C M, Parkinson J E, Hollinshead M, Smith G L. Overexpression of the vaccinia virus A38L integral membrane protein promotes Ca2+ influx into infected cells. J Virol. 1996;70:905–914. doi: 10.1128/jvi.70.2.905-914.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of Molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 47.Shchelkunov S N, Safronov P F, Totmenin A V, Petrov N A, Ryazankina O I, Gutorov V V, Kotwal G J. The genome sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- 48.Smith G L, Chan Y S, Howard S T. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 49.Spehner D, Gillard S, Drillien R, Kirn A. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J Virol. 1988;62:1297–1304. doi: 10.1128/jvi.62.4.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson P G, Efstathiou S, Doherty P C, Lehner P J. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci USA. 2000;97:8455–8460. doi: 10.1073/pnas.150240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willer D O, McFadden G, Evans D H. The complete genome sequence of shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 53.Yeruham I, Perl S, Nyska A, Abraham A, Davidson M, Haymovitch M, Zamir O, Grinstein H. Adverse reactions in cattle to a capripox vaccine. Vet Rec. 1994;135:330–332. doi: 10.1136/vr.135.14.330. [DOI] [PubMed] [Google Scholar]
- 54.Yeruham I, Nir O, Braverman Y, Davidson M, Grinstein H, Haymovitch M, Zamir O. Spread of lumpy skin disease in Israeli dairy herds. Vet Rec. 1995;137:91–93. doi: 10.1136/vr.137.4.91. [DOI] [PubMed] [Google Scholar]