Abstract
Background:
NUP98 gene fusions in acute myeloid leukemia (AML) have recently attracted much interest. Despite substantial research illuminating the roles of NUP98 fusions in the course of AML, their impacts on the outcome of patients with AML should be explored in more detail. As a result, this meta-analysis was designed to provide further light on the prognostic implications of NUP98 fusions in AML.
Methods
We completed an extensive search in PubMed, Scopus, and Web of Science to identify papers evaluating the prognostic effects of NUP98 rearrangements in patients with AML until August 22, 2022. In total, 15 publications with 6142 participants fulfilled the requirements for the current meta-analysis. All the qualified studies were examined for information regarding HRs and 95% confidence interval (95%CI) for overall survival (OS) and event-free survival (EFS). In addition, we utilized Comprehensive Meta-analysis software version 2 (CMA2) for calculating pooled HRs and 95% CI.
Section Title
Our Results:
analyses for NUP98-NSD1 indicated that this fusion could significantly impact the outcome of patients with AML (pooled HR: 2.84; 95% CI: 2.49–3.24, P=0.000). Additionally, we observed a strong correlation between NUP98-KDM5A rearrangement and poor prognosis in AML (pooled HR: 2.65; 95% CI: 2.5-2.81; P=0.000). A subgroup analysis also showed that the NUP98-NSD1 and FLT3-ITD together confer a poor prognostic effect (pooled HR: 2.60, 95% CI: 1.61-4.18; P=0.000).
Conclusions:
NUP98 fusions could significantly impact the outcome of patients with AML. The use of these fusions as prognostic indicators in AML seems rational.
Key Words: Acute myeloid leukemia, Prognosis, Nucleoporin98, NUP 98
Acute myeloid leukemia (AML) is one of the several hematological cancers with uncontrollable proliferation in myeloid precursors in the bone marrow (BM) (1, 2). It is well known that various genetic abnormalities have essential roles in the clinical presentation of AML (3). Accordingly, fusions in the nucleoporin 98 (NUP98) gene have received considerable attention. Numerous studies have shown that NUP98 fusions participate in the development and advancement of AML (4). NUP98 is a part of the nuclear pore complex, transporting proteins between the cytoplasm and the nucleus. NUP98 also functions in the regulation of gene transcription by interacting with histone-modifying enzymes CBP/p300 and HDAC1 (5). To date, over 30 genes that can fuse with NUP98 have been recognized. These partner genes encompass two main categories of homeobox (such as HOXA9) and non-homeobox genes (such as NSD1) (6). The NUP98-NSD1 fusion protein, produced by t (5;11) (q35; p15.5) translocation, is one of the fusion proteins that are more frequently seen in pediatric AML. It is noteworthy that adult patients also exhibit these fusions (7, 8). However, the rate of NUP98 translocations in adult patients is much lower compared to the pediatric group. Previous research highlighted NUP98-NSD1 as an indicator of inferior outcome and invasiveness in pediatric AML (7, 8).
However, the rate of NUP98 translocations in adult patients is much lower compared to the pediatric group. Previous research highlighted NUP98-NSD1 as an indicator of inferior outcome and invasiveness in pediatric AML (7, 8). However, adult patients with AML seldom show this fusion (9). Interestingly, the research on animals has demonstrated that NUP98-NSD1 overexpression is potent enough to initiate an AML-like condition in mice independently (10). The NUP98-NSD1 has also been suggested for routine screening, MRD monitoring, and assessing patients' responses to therapy in patients with AML. However, due to its cryptic nature, it cannot be recognized by conventional karyotyping, which necessitates employing other methods, such as RT-PCR and sequencing (11, 12).
The KDM5A is another well-known fusion partner for NUP98, frequently found in pediatric patients with acute megakaryoblastic leukemia (AMKL). The KDM5 subfamily comprises four members (KDM5A-D) involved in recognizing and regulating histone methylation (13-15). Overexpression of NUP98-KDM5A has been suggested in maturation arrest and elevated proliferation rate in hematopoietic stem cells or progenitors (16). Also, KDM5A has been identified as one of the retinoblastoma-binding protein-2 (RBP2) involved in cell cycle regulation. According to earlier research, KDM5A may impact the expansion of various malignancies, including AML (17, 18). Moreover, the N-terminal fusions of the NUP98 with PBX heterodimerization domains of HOXA9 generate the translocation t (7;11) (p15, p15). NUP98-HOXA9 (NHA9) fusion has been suggested as a rare cytogenetic abnormality in AML and MDS (Myelodysplastic syndromes). This translocation can lead to epigenetic disruptions in myeloid progenitor cells. HOXA9 also contributes to leukemogenesis by inducing proliferation and inhibiting the differentiation of hematopoietic stem cells (11, 19-21). As the prognostic value of NUP98 translocations seems helpful in managing AML patients with these translocations, the present meta-analysis focuses on assessing the relationship between common NUP98 fusions with outcomes in patients with AML.
Methods
2.1. Search Strategy: In the current study, we searched thoroughly in PubMed, Scopus, and Web of Science repositories to discover original articles investigating the influence of NUP98 fusions on the prognosis of patients with AML until August 22, 2022. For this, we utilized the following keywords: "Acute Myeloid Leukemia" OR "Acute Myelocytic Leukemia," OR "Acute Myeloblastic Leukemia" OR "AML" AND "Nucleoporin98" OR "NUP98" OR "nuclear pore complex protein 98".
2.2. Inclusion and exclusion criteria: The present meta-analysis used all available original articles in the English language with enough data about the effects of NUP98 translocations on the prognosis of patients with AML, including hazard ratios (HR) and 95% confidence intervals (CI) or any other data by which estimating HR and 95% CI was possible. We eliminated studies with inadequate data for HR with 95% CI, review articles, case reports, letters, conference articles, in-vitro studies, animal research, and articles in any language except English.
2.3. Data Extraction and Quality Assessment: Two investigators (MR and ZKH) removed duplicate studies and elicited all the necessary information from the eligible studies. These data encompass the name of the first author, the publication year, the name of the journal, the country, the number of patients in each study, criteria for the classification of AML, patients' demographic, and clinical data (e.g., age, sex, median WBC count), rate of NUP98 fusions in each study, and HR with their 95% CI for overall survival (OS) and event-free survival (EFS). The Newcastle-Ottawa (NOS) tool presents a form with pre-defined criteria to determine the quality of each study. Two researchers (ZKH and MSH) scored all the selected articles by filling out the standard form. A third reviewer was consulted in case of any discrepancy in the article selection or quality assessment process. Noteworthy, the NOS score for each study is depicted in table 1.
Table 1.
Main characteristics of all selected studies
| No | Authors (Ref) | Year (Country) | Patients | Type | Criteria | Number of Nup98 fusions | Age (year) * +/- | Sex (M/F) * +/- | WBC, *10 9 /L +/- | methods | NOS Score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Hollink I (11) | 2011 (USA) |
1101 | AML | FAB | NUP98-NSD1+ (n: 23) |
16.8 | 40 | %65.2 / 34.8% |
%52.0/ 48% |
146.5 | 26.2 | RT-qPCR | 7 |
| 2 | ShibaN (22) | 2013 (Japan) |
124 | de novo AML | FAB | NUP98-NSD1+ (n: 24) |
8.8 | 6.7 | 14/10 | 57/43 | 74.4 | 49.7 | RT-qPCR sequencing |
7 |
| 3 | Shiba N (23) |
2015 (Japan) |
369 | de novo AML | FAB | NUP98-NSD1+ (n: 11) |
- | - | - | RT-PCR | 5 | |||
| 4 | S Struski (24) |
2016 (France) |
212 | de novo AML | FAB | NUP98-NSD1+ (n: 22) |
11.8 | 9.1 | 5/17 | 82/108 | 194 | 12.2 | RT-qPCR | 7 |
| 5 | Marceau-Renaut A (25) |
2018 (France) |
385 | de novo AML | WHO | NUP98-NSD1+ (n: 9) |
8.6 Pos: 9.9 |
210/175* | *16.6 Pos: 179.8 |
RT-qPCR | 6 | |||
| 6. | Miyamura T (26) | 2018 (Japan) |
443 | AML | FAB | NUP98-NSD1+ (n: 7) |
6.1 | - | 28/15 | - | 43 | 96 | RT-PCR | 6 |
| 7. | Niktoreh N (27) |
2019 (Germany) |
246 | de novo AML | FAB | NUP98-NSD1+/Flt3+ (n:15) |
9.09 | - | 183/170 | - | 24.5 | - | RT-qPCR | 6 |
| 8. | Ostronoff F (12) |
2014 (USA) |
253 | de novo AML | - | Nup98-NSD1+/Flt3+ (n:37) |
11 | - | 26/11 | - | 173 | - | RT-PCR sequencing |
7 |
| NUP98-NSD1+/Flt3- (n=26) |
12 | - | 19/7 | - | 79 | - | ||||||||
| 9. | Shimada Y (28) |
2017 (Japan) |
44 | de novo AML | FAB & WHO | NUP98-NSD1+/Flt3+ (n:8) |
* 10 | * 27/17 | * 71.230 | RT-PCR | 6 | |||
| 10. | Rooij JDE (29) |
2016 (USA) |
153 | AMKL | FAB | NUP98-KDM5A+ (n:14) | *1.6 Pos: 1.9 |
* 70/83 Pos: 6/8 |
*13.73 Pos: 14.03 |
RT-PCR sequencing |
6 | |||
| 11. | Rooij J (30) |
201 (Netherlands) |
93 | non-DS-AMKL | FAB | NUP98-KDM5A+ (n:10) | - | - | - | sequencing | 5 | |||
| 12. | Hara Y (31) |
2019 (Japan) |
132 | de novo AML | FAB | NUP98-KDM5A+ (n:6) | * <3 | *68/ 64 | * 30.5 | RT-PCR sequencing |
6 | |||
| 13 | Iacobucci L (32) |
2019 (Australia) |
147 | AML | FAB &WHO |
NUP98-KDM5A+ (n:7) | - | - | - | RT-PCR | 6 | |||
| 14. | Bertrums EJM (33) |
2020 (Different ethnicities) |
2396 | AML | FAB | NUP98-KDM5A+ (n:34) | 2.8 | - | 17/17 | - | 11.3 | - | sequencing | 8 |
| NUP98-NSD1+ (n: 110) |
10.0 | - | 70/40 | - | 173 | - | ||||||||
| 15. | Hara Y (34) |
2017 (Japan) |
44 | non-DS-AMKL | FAB | NUP98-KDM5A+ (n:4) | * 1 | * 21/23 | * 22.0 | RT-PCR sequencing | 6 | |||
No, Number; M, Male; F, Female; FAB, French-American-British; WHO, Word Health Organization; Ref, references; AML, Acute Megakaryoblastic Leukemia; non-DS-AMKL, acute megakaryoblastic leukemia with non-Down syndrome; qPCR, Quantitative Polymerase Chain Reaction
* Data in all patients are reported, Pos: Data is reported for patients with NUP-98 Rearrangement only.
2.4 Data Synthesis and Analysis: Those NUP98 fusions evaluated in at least two or more studies were used for statistical analysis employing the Comprehensive Meta-analysis software (Version 2). The effects of NUP98 translocations on the OS or EFS of patients with AML were assessed using HRs and 95% CI. In our meta-analysis, HR>1 and P-value<0.05 was interpreted as unfavorable prognosis. In addition, the I2 statistics was used to determine the heterogeneity. We used the random-effects model for significant heterogeneity (I2 > 50%). Moreover, table 2 represents a summary of all statistical data in our study.
Table 2.
Summary of meta-analysis statistical data for prognostic implications of NUP98 fusions
| Parameters | No. studies | No. patients | Heterogeneity | Model | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I 2 | P | T-au 2 | T-au |
HR
( 95%CI ) |
Z | P | ||||
| OS | ||||||||||
| Multivariable (NUP98-NSD1) | 4 | 4010 | 96.65 | 0.018 | 0.69 | 0.83 | Random | 2.84 (2.49-3.24) |
15.632 | 0.000 |
| Univariable (NUP98-NSD1) | 2 | 581 | 0.00 | 0.322 | 0.000 | 0.000 | Fixed | 3.67 (2.80-4.82) |
9.418 | 0.000 |
| Multivariable (NUP98-KDM5A) | 6 | 2835 | 0.00 | 0.553 | 0.000 | 0.000 | Fixed | 2.63 2.50-2.81) |
32.89 | 0.000 |
| NUP98-NSD1+FLT3-ITD1 + | 3 | 543 | 60.71 | 0.078 | 0.467 | 0.684 | Random | 2.6 (1.61-4.18) |
3.95 | 0.000 |
| EFS | ||||||||||
| Multivariable (NUP98-NSD1) | 2 | 1486 | 0.00 | 0.335 | 0.00 | 0.00 | Fixed | 2.79 (1.87-4.16) |
5.03 | 0.000 |
| Multivariable (NUP98-KDM5A) | 4 | 422 | 39.86 | 0.173 | 0.146 | 0.382 | Fixed | 2.35 (1.59-3.47) |
4.28 | 0.000 |
| NUP98-NSD1+FLT3-ITD1 + | 3 | 543 | 29.42 | 0.242 | 0.089 | 0.299 | Fixed | 2.82 (1.86-4.26) |
4.93 | 0.000 |
Results
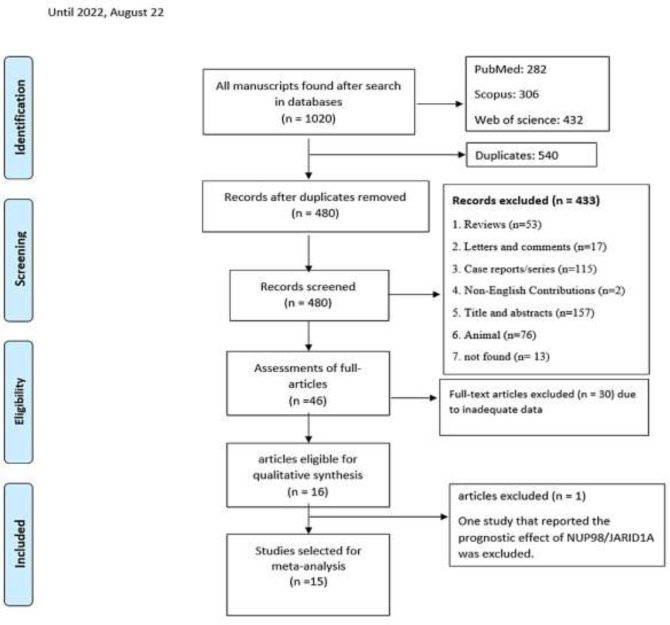
3.1. Study Selection and Characteristics of included Studies: Figure 1 depicts the flow diagram of the study selection for NUP98 fusions. In our preliminary search, we found 1020 articles, of which 282, 306, and 432 studies were acquired from PubMed, Scopus, and Web of Science repositories. Following initial screening and eliminating duplicated articles, 480 articles remained. We eliminated 434 out of 480 articles after screening the abstracts because they had at least one of the exclusion criteria—finally, 46 articles were retrieved in full text. Of those, 15 papers with 6142 cases showed eligibility for the current meta-analysis. One study that reported the prognostic effect of NUP98/JARID1A in patients with AML was not analyzed because the availability of at least two original articles is required for a single fusion to perform a meta-analysis. A comprehensive list of all included studies' characteristics, such as region of study, AML diagnostic criteria, and the number of participants, is shown in table 1.
Figure 1.
Flowchart representing the selection strategy for identifying qualified studies
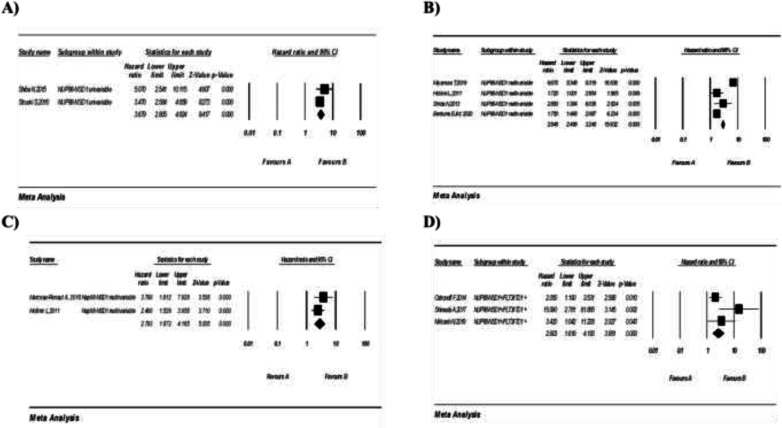
3.2. Prognostic importance of NUP98-NSD1 in patients with AML: Six studies explored the impacts of NUP98-NSD1 translocation on OS of patients with AML, including two with 581 patients reporting univariate HRs and four with 4010 participants reporting multivariate HRs. In addition, two studies with a total of 1486 cases investigated the effects of NUP98-NSD1 on the EFS of patients with AML by reporting multivariable HRs.
A meta-analysis of studies with univariate HRs for OS showed that the NUP98-NSD1 rearrangement confers a poor prognosis in patients with AML (pooled HR: 3.67; 95% CI: 2.80–4.82, P=0.000), with no heterogeneity (I2=00%, P=0.322) (figure 2, A). Similarly, the results of a meta-analysis of studies with multivariate HRs for OS showed a poor outcome (pooled HR: 2.84; 95% CI: 2.49–3.24, P=0.000) with high heterogeneity (I2=96.65%, P=0.018) (figure 2, B). Moreover, NUP98-NSD1 was associated with a shorter EFS (pooled HR: 2.79; 95% CI: 1.87-4.16; P=0.000), with no sign of heterogeneity (I2=00%, P=0.335) (figure 2, C).
Figure 2.
Forest plot of the HRs and 95% CI for OS and EFS in AML patients with NUP98-NSD1 in AML fusion. (A) OS univariable analysis, (B) OS multivariable analysis, (C) EFS multivariable analysis. (D) Forest plot of the HRs and 95% CI for OS in AML patients with NSD1+/FLT3-ITD1 + OS.
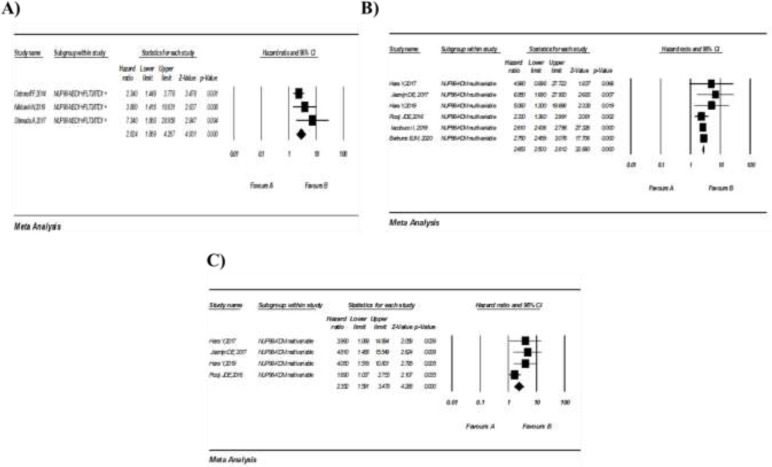
3.3. Prognostic importance of NUP98-NSD1 in the presence of FLT3-ITD: Three studies evaluated the impact of NUDP98-NSD1 and FLT3-ITD co-occurrence on the prognosis of patients with AML. A meta-analysis of these studies showed that the simultaneous presence of both NUP98-NSD1 and FLT3-ITD was correlated with a poor OS in AML (pooled HR: 2.60, 95% CI: 1.61-4.18; P=0.000), with high heterogeneity (I2=60.71%, P=0.078) (figure 2, D). Similarly, the co-occurrence was associated with a shorter EFS (pooled HR: 2.82; 95% CI: 1.86-4.26; P=0.000), with a moderate heterogeneity (I2=29.42%, P=0.242) (figure 3, A).
Figure 3.
(A) Forest plot of the HRs and 95% CI for and EFS in AML patients with NUP98 NSD1+/FLT3-ITD1 + in AML fusion. (A) and (C) Forest plot of the HRs and 95% CI for OS and EFS in AML patients with NUP98-KDM5A in AML fusion, respectively.
3.4. Prognostic importance of NUP98-KDM5A in patients with AML: Six studies consisting of 2835 patients investigated the relationship between the NUP98-KDM5A fusion and OS in AML patients. All these studies demonstrated multivariate HRs.
According to our meta-analysis, NUP98-KDM5A was significantly associated with a lower OS in patients with AML (pooled HR: 2.65; 95% CI: 2.5-2.81; P=0.000), without heterogeneity (I2=00%, P=0.553) (figure 3, B). Correspondingly, a meta-analysis of four studies demonstrated a significant relationship between NUP98-KDM5A and shorter EFS (pooled HR: 2.35; 95% CI: 1.59-3.47; P=0.000), with moderate heterogeneity (I2=39.86%, P=0.173) (figure 3, C).
3.5. Publication bias assessment: Begg's and Egger's tests and funnel plots were employed to assess the possible sources of bias. As shown in table 3, no significant publication bias exists for studies evaluating OS in patients with AML and NUP98 fusions. However, Egger's test revealed some evidence of bias for studies assessing EFS in patients with NUP98-KDM5 fusion. In addition, funnel plots for OS in AML Patients with NUP98-NSD1 fusion and patients with NUP98-KDM5A fusion, and EFS in AML Patients with NUP98-KDM5A fusion were sent in supplementary file.
Table 3.
Summary of statistical data for the Egger’s and the Begg’s test
| Group |
OS
NUP98-KDM+ multivariable |
OS
NUP98-NSD1 + multivariable |
OS
NUP98-NSD1 + FLT3-ITD1 + |
EFS
NUP98-KDM5A+ multivariable |
EFS
NUP98-NSD1+ FLT3-ITD1 + |
|---|---|---|---|---|---|
| Begg’s test (P-value) |
0.259 | 0.734 | 0.296 | 0.734 | 0.296 |
| Egger’s test (P-value) |
0.108 | 0.962 | 0.223 | 0.035 | 0.097 |
Discussion
There have been reports of NUP98 fusion in numerous hematological cancers, including acute myeloid leukemia, infrequent occurrences of myelodysplastic syndromes, and some cases with acute biphenotypic leukemia (11, 35-37). A plethora of previous studies highlighted the distinct roles of NUP98 fusions in initiating and developing acute myeloid leukemia (8, 35). A meaningful link was found between NUP98 fusions and the prognosis of individuals with AML (6). The findings of the current meta-analysis also demonstrated that patients with AML and NUP98-NSD1 or NUP98-KDM5A fusions showed significantly lower OS and EFS than patients without these fusions. Our study results parallel several previous original research articles.
According to a study by Hollink et al. (11), NUP98-NSD1 was significantly correlated with gloomy prognosis in both pediatric and adult cases with cytogenetically normal (CN)-AML. They also found that NUP98-NSD1 was considerably more prevalent in the younger age group and was significantly correlated with higher WBC counts. In parallel with this study, our meta-analyses also showed poor outcomes for AML cases with NUP-NSD1. Based on our review of the studies, most included articles focused on the pediatric group with an average age of 9.1 ± 4.25 years for patients included. Therefore, the prognostic effect of NUP98-NSD1 fusion is more relevant to the pediatric group. However, we couldn't conduct a separate analysis of children versus adults due to a lack of access to detailed information. Additionally, we were unable to perform subgroup meta-analyses to evaluate the effects of other variables, such as the WBC count and FAB subtype, on the prognosis of NUP98-NSD1, and this was mainly due to a need for more comprehensive data in the literature. In addition, the study of Hollink et al. showed the presence of additional genetic lesions, such as WT1, in a substantial number of patients with positive NUP98-NSD1 and suggested that mutant NPM1 is related to a more favorable outcome than wild-type NPM1 in childhood cases of AML harboring both FLT3-ITD and NUP98-NSD1.
On the contrary, we could not conduct subgroup meta-analyses of the independent effects WT1 and NPM1 on the prognosis of NUP98 fusions in AML. A growing body of literature demonstrates that FLT3-ITD mutations frequently occur in individuals with NUP98-NSD1 fusions (38). It has been demonstrated that FLT3-ITD alone is insufficient for leukemia initiation (39), but rather it must be accompanied by other mutations, such as mutant NPM1(40), fusions of SMMHC-CBF, or deletions of TET2 (41). According to a study by Ostronoff F et al. (12), while the coexistence of both NUP98-NSD1 and FLT3-ITD could result in an elevated rate of therapy failure, a higher MRD following induction regimen and shorter survival in AML, NUP98-NSD1 alone does not necessarily induce a poor outcome when FLT3-ITD is absent. Based on this study, a cooperative relationship between these two genetic aberrations appears necessary for chemotherapy resistance in leukemic clones. Noteworthy, they observed a high concurrent occurrence of WT1 in AML patients with both FLT3-ITD and NUP98-NSD1, which may be another potential factor for poor response to therapy.
Similarly, we also performed a meta-analysis on the prognostic effects of NUP98-NSD1 and FLT3-ITD co-occurrence in patients with AML. Based on our findings, their co-occurrence could result in significantly shorter OS and EFS. However, we could not carry out a meta-analysis on AML cases carrying NUP98-NSD1 without FLT3-ITD because we did not find enough studies in the literature. In our search, we found only one study (12) that evaluated the prognostic effects of NUP98-NSD1 in AML patients without FLT3-ITD. Interestingly, the results of this study showed that NUP98-NSD1 has a better prognostic effect in patients without FLT3-ITD than those harboring this mutation (HR,0.34, CI: 0.048-2.4). In this regard, we strongly recommend more studies evaluating the prognostic effects of NUP98-NSD1 in AML patients without FLT3-ITD to shed more light on this issue.
Several studies found a distinctive gene expression pattern in patients with NUP98-NSD1 fusions. Overexpression of HOX genes, PRDM 16, Meis 1, mir-196b, and mir-10a, has been reported in these patients (10, 11, 42, 43). Further studies focusing on gene expression patterns of patients with AML and NUP98-NSD1 can result in promising insights into the leukemogenic mechanisms of this fusion in AML, which can lead to the development of novel therapies (11). Considering the poor outcomes of NUP98-NSD1, some researchers previously highlighted shifting to more efficient therapies for these patients as first- or second-line treatment (27, 33).
Furthermore, occurrences of NUP98-KDM5A have been demonstrated in nearly 10-15% of pediatric cases with non-Down syndrome acute megakaryoblastic leukemia and approximately 20% of pediatric acute erythroleukemia. According to Miller J et al., NUP98-KDM5A could substantially affect the proliferation and differentiation of human hematopoietic stem and progenitor cells in-vitro. Also, this fusion was associated with significant splenomegaly in grafted mice (44). In separate studies by Hara Y et al. (34) and Noort S et al. (18), NUP98-KDM5A was associated with poor OS and EFS in AML. Similarly, our meta-analysis indicated a poor prognostic effect for NUP98-KDM5A in patients with AML. Despite these findings, the small number of patients with NUP98-KDM5A caused some previous original studies to fail to reach statistical significance for the prognostic effects of NUP98-KDM5A. Therefore, studies with larger sample sizes are suggested to broaden our horizon on its exact prognostic effects. Overall, despite our best endeavors to conduct a thorough meta-analysis, the current research should be viewed with caution due to some shortcomings, including:
1. The simultaneous coexistence of multiple other genetic lesions, such as FLT3-ITD, NPM1, and WT1, in individuals carrying NUP98-NSD1 could lead to heterogeneity in our findings.
2. Patients' therapy protocols and their clinical and demographic factors, such as age, gender, and hematological parameters, could also affect their prognosis, which is not considered in the current meta-analysis.
3. There needed to be more data regarding the prognostic effects of other NUP 98 fusions, such as NUP98-Hoxa9.
To sum up, our findings showed that two of the most frequent NUP98 fusion, including NUP98-NSD1 and NUP98-KDM5A, were correlated with worse OS and EFS in patients with AML. Our results highlight these fusions as possible biomarkers for prognosis in AML. Despite these results, further large cohorts in various ethnicities are warranted to clarify the exact prognostic roles of NUP98 fusions in AML.
Acknowledgments
We would like to thank all the authors who participated in the study.
Funding:
This work was supported by Mashhad University of Medical Sciences, Mashhad, Iran under Grant number 4011274.
Ethics Approval:
IR.MUMS.MEDICAL.REC.1402.301.
Conflict of Interests:
All authors declare that there is no possible conflict of interest.
Authors’ contribution:
MSH, Z KH, HA, MR and PS conceived, analyzed, interpreted the results and wrote the first draft of manuscript. AA, E Z, MH S and M GH reviewed the manuscript. MSH, RH and Z KH conceived the presented idea, supervised the findings of this work, paper drafting and review. All authors approved the contents of the manuscript.
Consent for publication:
Not applicable.
Availability of data and materials:
Not applicable.
References
- 1.Zhou HS, Carter BZ, Andreeff M. Bone marrow niche-mediated survival of leukemia stem cells in acute myeloid leukemia: Yin and Yang. Cancer Biol Med. 2016;13:248–59. doi: 10.20892/j.issn.2095-3941.2016.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundu S, Park ES, Chung YJ, et al. Thymic precursor cells generate acute myeloid leukemia in NUP98-PHF23/NUP98-HOXD13 double transgenic mice. Sci Rep. 2019;9:1–17. doi: 10.1038/s41598-019-53610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou HA, Tien HF. Genomic landscape in acute myeloid leukemia and its implications in risk classification and targeted therapies. J Biomed Sci. 2020;27:81. doi: 10.1186/s12929-020-00674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmoellerl J, Barbosa IAM, Eder T, et al. CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood. 2020;136:387–400. doi: 10.1182/blood.2019003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai XT, Gu BW, Yin T, et al. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006;66:4584–90. doi: 10.1158/0008-5472.CAN-05-3101. [DOI] [PubMed] [Google Scholar]
- 6.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–57. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panarello C, Rosanda C, Morerio C. Cryptic translocation t (5; 11)(q35; p15 5) with involvement of the NSD1 and NUP98 genes without 5q deletion in childhood acute myeloid leukemia. Genes, Chromosomes Cancer. 2002;35:277–81. doi: 10.1002/gcc.10119. [DOI] [PubMed] [Google Scholar]
- 8.Mohanty S, Jyotsana N, Sharma A, et al. Targeted inhibition of the NUP98-NSD1 fusion oncogene in acute myeloid leukemia. Cancers (Basel) 2020;12:2766. doi: 10.3390/cancers12102766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaju RJ, Fidler C, Haas OA, et al. A novel gene, NSD1, is fused to NUP98 in the t (5; 11)(q35; p15 5) in de novo childhood acute myeloid leukemia. Blood. 2001;98:1264–7. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- 10.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 11.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–56. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 12.Ostronoff F, Othus M, Gerbing RB, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124:2400–7. doi: 10.1182/blood-2014-04-570929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10:413–24. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beshiri ML, Holmes KB, Richter WF, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci. 2012;109:18499–504. doi: 10.1073/pnas.1216724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenblatt SM, Nimer SD. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia. 2014;28:1396–406. doi: 10.1038/leu.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slape C, Lin YW, Hartung H, et al. NUP98-HOX translocations lead to myelodysplastic syndrome in mice and men. J Natl Cancer Inst Monogr. 2008;2008:64–8. doi: 10.1093/jncimonographs/lgn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shokri G, Doudi S, Fathi-Roudsari M, Kouhkan F, Sanati MH. Targeting histone demethylases KDM5A and KDM5B in AML cancer cells: a comparative view. Leuk Res. 2018;68:105–11. doi: 10.1016/j.leukres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Noort S, Wander P, Alonzo TA, et al. The clinical and biological characteristics of NUP98-KDM5A pediatric acute myeloid leukemia. Haematologica. 2021;106:630–4. doi: 10.3324/haematol.2019.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rio-Machin A, Gómez-López G, Muñoz J, et al. The molecular pathogenesis of the NUP98-HOXA9 fusion protein in acute myeloid leukemia. Leukemia. 2017;31:2000–5. doi: 10.1038/leu.2017.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe A, Yamamoto Y, Iba S, et al. ETV6‐LPXN fusion transcript generated by t (11; 12)(q12 1; p13) in a patient with relapsing acute myeloid leukemia with NUP98‐HOXA9. Genes Chromosomes Cancer. 2016;55:242–50. doi: 10.1002/gcc.22327. [DOI] [PubMed] [Google Scholar]
- 21.Yanagisawa H, Mizuta S, Kawabata H, et al. Faggot cells in acute myeloid leukemia with t (7; 11)(p15; p15) and NUP98-HOXA9 fusion. Ann Hematol. 2021;100:2121–3. doi: 10.1007/s00277-020-04122-2. [DOI] [PubMed] [Google Scholar]
- 22.Shiba N, Ichikawa H, Taki T, et al. NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes, Chromosomes Cancer. 2013;52:683–93. doi: 10.1002/gcc.22064. [DOI] [PubMed] [Google Scholar]
- 23.Shiba N, Ohki K, Kobayashi T, et al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol. 2020;189:581–91. doi: 10.1111/bjh.13869. [DOI] [PubMed] [Google Scholar]
- 24.Struski S, Lagarde S, Bories P, et al. NUP98 is rearranged in 3 8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31:565–72. doi: 10.1038/leu.2016.267. [DOI] [PubMed] [Google Scholar]
- 25.Marceau-Renaut A, Duployez N, Ducourneau B, et al. Molecular profiling defines distinct prognostic subgroups in childhood AML: a report from the French ELAM02 study group. HemaSphere. 2018;2:e31. doi: 10.1097/HS9.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamura T, Moritake H, Nakayama H, et al. Clinical and biological features of paediatric acute myeloid leukaemia (AML) with primary induction failure in the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 study. Br J Haematol. 2019;185:284–8. doi: 10.1111/bjh.15799. [DOI] [PubMed] [Google Scholar]
- 27.Niktoreh N, Walter C, Zimmermann M, et al. Mutated WT1, FLT3-ITD, and NUP98-NSD1 fusion in various combinations define a poor prognostic group in pediatric acute myeloid leukemia. J Oncol. 2019;2019:1609128. doi: 10.1155/2019/1609128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada A, Iijima-Yamashita Y, Tawa A, et al. Risk-stratified therapy for children with FLT3-ITD-positive acute myeloid leukemia: results from the JPLSG AML-05 study. Int J Hematol. 2018;107:586–95. doi: 10.1007/s12185-017-2395-x. [DOI] [PubMed] [Google Scholar]
- 29.de Rooij JDE, Masetti R, Van Den Heuvel-Eibrink MM, et al. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: A retrospective intergroup study. Blood. 2016;127:3424–30. doi: 10.1182/blood-2016-01-695551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Rooij JDE, Branstetter C, Ma J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49:451–6. doi: 10.1038/ng.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara Y, Shiba N, Yamato G, et al. Patients aged less than 3 years with acute myeloid leukaemia characterize a molecularly and clinically distinct subgroup. Br J Haematol. 2020;188:528–39. doi: 10.1111/bjh.16203. [DOI] [PubMed] [Google Scholar]
- 32.Iacobucci I, Wen J, Meggendorfer M, et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet. 2019;51:694–704. doi: 10.1038/s41588-019-0375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertrums EJ, Smith JL, Ries RE, et al. The molecular characteristics and clinical Relevance of NUP98-Other translocations in pediatric acute myeloid leukemia. Blood. 2020;136:36–7. [Google Scholar]
- 34.Hara Y, Shiba N, Ohki K, Tabuchi K, Yamato G, Park Mj, et al. Prognostic impact of specific molecular profiles in pediatric acute megakaryoblastic leukemia in non‐Down syndrome. Genes, Chromosomes and Cancer. 2017;56(5):394–404. doi: 10.1002/gcc.22444. [DOI] [PubMed] [Google Scholar]
- 35.Cardin S, Bilodeau M, Roussy M, Aubert L, Milan T, Jouan L, et al. Human models of NUP98-KDM5A megakaryocytic leukemia in mice contribute to uncovering new biomarkers and therapeutic vulnerabilities. Blood Advances. 2019;3(21):3307–21. doi: 10.1182/bloodadvances.2019030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Starza R, Gorello P, Rosati R, Riezzo A, Veronese A, Ferrazzi E, et al. Cryptic insertion producing two NUP98/NSD1 chimeric transcripts in adult refractory anemia with an excess of blasts. Genes, Chromosomes and Cancer. 2004;41(4):395–9. doi: 10.1002/gcc.20103. [DOI] [PubMed] [Google Scholar]
- 37.Romana S, Radford-Weiss I, Ben Abdelali R, et al. NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogenetique Hematologique. Leukemia. 2006;20:696–706. doi: 10.1038/sj.leu.2404130. [DOI] [PubMed] [Google Scholar]
- 38.Krock B, Oberley MJ. Molecular genetics of pediatric acute myeloid leukemia. C Clin Lab Med. 2021;41:497–515. doi: 10.1016/j.cll.2021.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Garg M, Nagata Y, Kanojia D, et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126:2491–501. doi: 10.1182/blood-2015-05-646240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dovey OM, Cooper JL, Mupo A, et al. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood. 2017;130:1911–22. doi: 10.1182/blood-2017-01-760595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer cell. 2015;27:502–15. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiba N, Ohki K, Kobayashi T, et al. High PRDM 16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT 3‐ITD, KMT 2A‐PTD, and NUP 98‐NSD 1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML‐05 trial. Br J Haematol. 2016;172:581–91. doi: 10.1111/bjh.13869. [DOI] [PubMed] [Google Scholar]
- 43.Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol. 2016;23:354–61. doi: 10.1097/MOH.0000000000000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller J, Hiltenbrand R, Lamprecht T, et al. NUP98-KDM5A fusion induces hematopoietic cell pliferation and alters myelo-erythropoietic differentiation. Blood. 2019;134:3775. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.