Abstract
Here, we present a method for enrichment of double-stranded cfDNA with an average length of ∼40 bp from cfDNA for high-throughput DNA sequencing. This class of cfDNA is enriched at gene promoters and binding sites of transcription factors or structural DNA-binding proteins, so that a genome-wide DNA footprint is directly captured from liquid biopsies. In short double-stranded cfDNA from healthy individuals, we find significant enrichment of 203 transcription factor motifs. Additionally, short double-stranded cfDNA signals at specific genomic regions correlate negatively with DNA methylation, positively with H3K4me3 histone modifications and gene transcription. The diagnostic potential of short double-stranded cell-free DNA (cfDNA) in blood plasma has not yet been recognized. When comparing short double-stranded cfDNA from patient samples of pancreatic ductal adenocarcinoma with colorectal carcinoma or septic with postoperative controls, we identify 136 and 241 differentially enriched loci, respectively. Using these differentially enriched loci, the disease types can be clearly distinguished by principal component analysis, demonstrating the diagnostic potential of short double-stranded cfDNA signals as a new class of biomarkers for liquid biopsies.
Liquid biopsies are based on various types of analytes, including circulating extracellular nucleic acids like cell-free DNA (cfDNA), extracellular vesicles, or circulating tumor cells, for example (Bronkhorst et al. 2019; Kustanovich et al. 2019). Physiologically, cfDNA is to a large extent released from the hematopoietic system by apoptosis, necrosis, or active secretion from almost all cell types and tissues into the bloodstream (Grabuschnig et al. 2020). In addition to release from normal physiological turnover, cancer cells or microbial pathogens are also known to release their DNA into bloodstream circulation (Heitzer et al. 2020). Released genomic DNA is then degraded by DNA-digesting enzymes (nucleases), producing fragments mainly 147 to 167 bp in size, corresponding to a single nucleosome (Han et al. 2020; Lo et al. 2021). By high-throughput sequencing of cfDNA fragments, nucleosome positioning can be inferred at base pair resolution (Fan et al. 2008; Snyder et al. 2016). The exact positions of nucleosomes and chromatin structure play a key role in regulating gene expression by providing access to DNA for the transcription machinery. Open chromatin structures depleted of nucleosomes make DNA more accessible for key regulators of transcription, including transcription factors, enhancers, or repressors. However, the routine and efficient measurement of genome-wide protection through regulatory DNA-binding proteins (DBPs) is not yet established. Recently, a minor fraction of double-stranded cfDNA that is significantly shorter than normal cfDNA was found, ranging from 35 to 80 nucleotides (nt) using ultradeep sequencing of total cfDNA (Burnham et al. 2016; Snyder et al. 2016). It has been proposed that this short cfDNA might be protected by DNA-binding factors and therefore could represent direct transcription factor binding.
Here, we established an enrichment approach for short double-stranded cfDNA fragments (20–60 bp) from blood plasma (further referred to as “short cfDNA sequencing”). Corresponding short double-stranded cfDNA fragments (further referred to as “short cfDNA”) accumulate at open chromatin as well as gene regulatory elements. Differential enrichment of short cfDNA at genomic loci facilitated discrimination of colorectal and pancreatic cancer patient samples as well as between septic patients and clinical controls, showing potential for diagnostic applications.
Results
Enrichment of cfDNA footprints at regulatory genomic regions
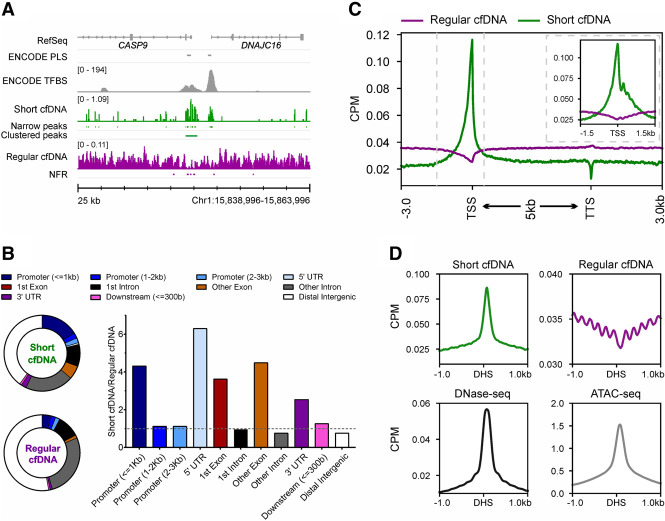
Enrichment of short double-stranded cfDNA comprises extraction of total cfDNA from blood plasma by magnetic beads followed by double-stranded DNA-specific library preparation (Supplemental Fig. S1). To select short double-stranded cfDNA fragments of up to 60 bp, two size-selection steps were performed using a preparative gel electrophoresis device. For this, fragments in the range of 150 bp to 200 bp are enriched, which corresponds to double-stranded cfDNA fragments of up to 60 bp ligated to a sequencing adapter of 140 bp (Supplemental Fig. S2). After high-throughput sequencing of size-selected libraries, reads were quality-checked to ensure a size range between 20 bp and 60 bp (Supplemental Fig. S3). The protocol revealed sequencing reads with a mean read length of 37.9 bp across the patient conditions and comparable read length distributions per condition despite varying sample storage durations (SD = 6.6 bp, n = 2 × 107 reads uniformly sampled from the total reads of 20 individuals: four healthy individuals, four patients with pancreatic ductal adenocarcinoma [PDAC], four patients with colorectal carcinoma [CRC], four patients with sepsis, and four nonseptic postoperative clinical control patients [Post-OP]) (Supplemental Fig. S4; Supplemental Table S1). Short cfDNA reads revealed an elevated average GC content of 57.8% (SD = 1.9%) (Supplemental Fig. S5) in contrast to 40.9% for average human genomic DNA (Piovesan et al. 2019). We mapped short cfDNA to the human genome and compared it with the mapping of regular cfDNA of four additional healthy individuals. Coverage profiles of short cfDNA and regular cfDNA differed considerably as short cfDNA tends to accumulate either in single narrow peaks or in clusters of narrow peaks (clustered peaks), which are frequently located at transcriptional start sites (TSSs) or ChIP-seq-validated transcription factor binding sites (TFBSs) (Fig. 1A). Nucleosome-free regions (NFRs) were analyzed in comparison to clustered peaks, because the absence of regular cfDNA can be an indicator of the presence of other DBPs. Assignment of clustered peaks from short cfDNA and NFRs to annotated functional elements of the genome shows that an approximately four- and a sixfold higher proportion of clustered peaks are found in promoters (>1 kb upstream of the TSS) and 5′ UTRs of genes, respectively. Moreover, the proportion of clustered peaks assigned to exons is about four times higher than the proportion of NFRs (Fig. 1B). A more detailed examination of the genomic coverage for protein-coding genes reveals an opposite profile between short cfDNA and regular cfDNA. Although short cfDNA is enriched at TSSs, regular cfDNA is depleted at these sites. In addition, short cfDNA possesses a reciprocal pattern oscillation compared with regular cfDNA, with short cfDNA exhibiting an inverted pattern 1 kb downstream from the TSS in regular cfDNA (Fig. 1C, inset). In addition to enrichment at the TSS, short cfDNA also exhibits a prominent signal at DNase I hypersensitive sites (DHSs) from a reference annotation of the B cell line GM12878, whereas the regular cfDNA signal oscillates at neighboring genomic locations and is depleted at the DHS (Fig. 1D). Taken together, short cfDNA accumulates at open chromatin or TSS of genes, whereas regular cfDNA, namely, nucleosomes, was clearly depleted in these regions.
Figure 1.
Short cfDNA is enriched in regulatory regions of genes and open chromatin. (A) Coverage profile of short cfDNA (green; S03) and regular cfDNA (violet; average of S05–S08). The short cfDNA profile shows narrow and clustered peaks, whereas regular cfDNA shows nucleosome-free regions (NFRs; depletion of reads). RefSeq genes, ENCODE promoter-like structures (PLSs), and ENCODE transcription factor binding sites (TFBSs) based on ChIP-seq experiments are included as references. (B) Pie charts display the proportions of annotated genomic features for clustered peaks from short cfDNA and NFRs from regular cfDNA. The bar plot shows the ratio between clustered peaks and NFRs for each genomic feature. (C) Average coverage profiles for short cfDNA (S03) and regular cfDNA (S06) along all annotated protein-coding genes. The gene bodies of all genes are scaled to 5 kb. Dashed vertical lines indicate the interval shown in the subplot. The subplot shows average profiles at the transcription start site without rescaling of the gene body. (D) Average coverage profiles for short cfDNA, regular cfDNA, and publicly available ATAC-seq data at DNase I hypersensitive sites (DHSs) derived from publicly available DNase-seq data.
Short cfDNA sequencing detects binding of transcription factors
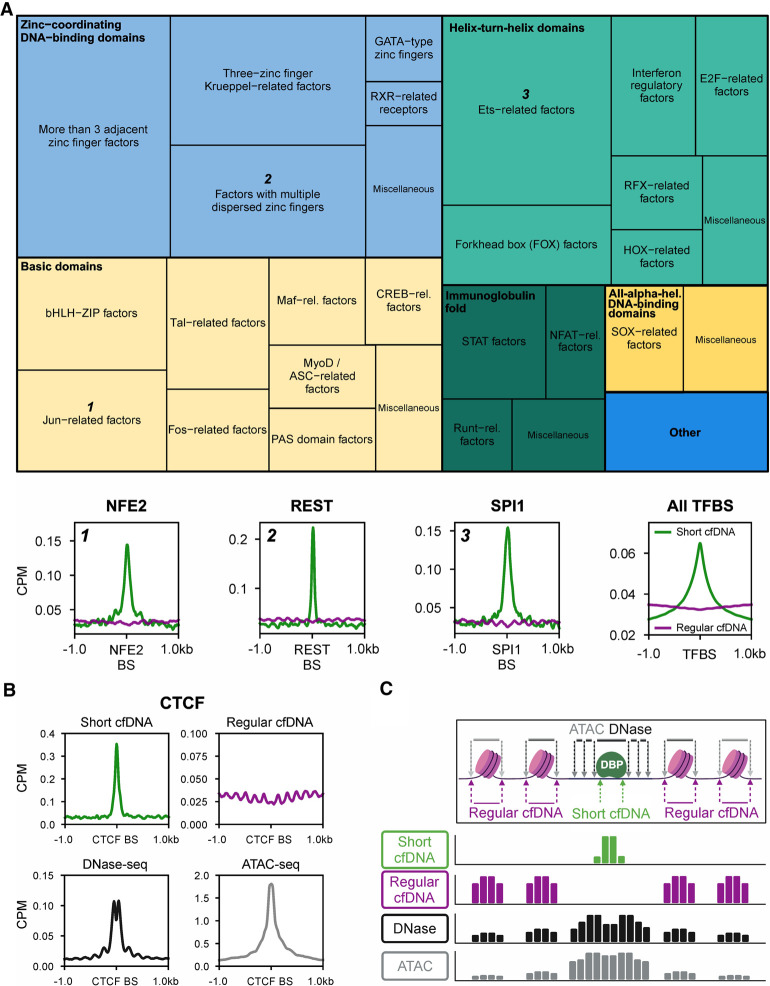
Because short cfDNA could be protected from nuclease digestion by binding to regulatory DBPs, peak regions were examined for encompassing transcription factor binding motifs. Accordingly, a consensus peak set was defined from peaks of short cfDNA sequencing data of four healthy individuals. Transcription factor motif enrichment analysis at the genomic locations of these consensus peaks revealed a significant enrichment of 203 transcription factor binding motifs out of 401 listed in the reference motif database HOCOMOCO (Kulakovskiy et al. 2018). The 203 transcription factor motifs belong to 46 transcription factor families from nine transcription factor superclasses. ChIP-seq-validated TFBSs from ENCODE3 reveal clear enrichment signals for nuclear factor, erythroid 2 (NFE2), RE1 silencing transcription factor (REST), and Spi-1 proto-oncogene (SPI1) in short cfDNA, for example. On the other hand, regular cfDNA is depleted at these binding sites. Overall, the average profile of all ChIP-seq-based TFBSs shows a clear enrichment of short cfDNA in contrast to regular cfDNA independent of the transcription factor (Fig. 2A). ChIP-seq-validated binding sites of CCCTC-binding factor (CTCF) show an even more pronounced signal for short cfDNA, whereas regular cfDNA exhibits a high-frequency oscillation occupancy adjacent to these binding sites (Fig. 2B). In good agreement, DNase-seq data show a combination of a footprint peak with adjacent oscillation patterns, whereas ATAC-seq detects a peak representative for open chromatin at the CTCF binding sites (Fig. 2B). Our data suggest that short cfDNA most specifically reveals protection of DNA through DBPs at regulatory sites with high resolution as exemplified by CTCF, NFE2, REST, or SPI1 (Fig. 2C). Previously, it has been shown that a significant fraction of short cfDNA exists as short single-stranded DNA (ssDNA). To analyze how short ssDNA signals compare to short cfDNA signals, we examined transcription start sites and TFBSs with data from Snyder et al. (2016) for short cfDNA sequencing (single-stranded library preparation). We found a superior signal-to-noise ratio for DBP footprints, allowing the detection of signals, for example, tumor protein p53 binding protein 1 (TP53BP1), that were not detectable in the short ssDNA of Snyder et al. (Snyder et al. 2016; Supplemental Fig. S6). These data suggest that short cfDNA represents a biological entity that captures DBP footprints.
Figure 2.
Short cfDNA sequencing directly captures the protection of DNA across the genome by various transcription factors. (A) Treemap showing transcription factors whose DNA motif was significantly enriched in short cfDNA consensus peaks from four healthy individuals. More than 200 transcription factor motifs of all nine transcription factor superclasses in the database were identified as enriched in short cfDNA peaks. The size of squares in the treemap encodes the relative number of transcription factor motifs per class (thin outlined boxes) and superclass (thick outlined boxes). As examples, average coverage profiles of short cfDNA (S03) and regular cfDNA (S06) are shown for three transcription factors (NFE2, REST, and SPI1) from three different transcription factor superclasses. Average profiles are based on the 1000 most robust binding sites annotated in the Gene Transcription Regulation Database (GTRD). In addition, the average profile of all ChIP-seq TFBSs annotated in ENCODE3 is shown for short cfDNA in comparison to regular cfDNA. (B) Average coverage profile of short cfDNA, regular cfDNA, DNase-seq, and ATAC-seq at CTCF binding sites (CTCF BSs). (C) Inferred molecular model for the formation of short cfDNA at the binding site of a DNA-binding protein (DBP) surrounded by two nucleosomes on either side. Arrows indicate the endpoints of DNA fragments for the respective sequencing technique. The resulting theoretical coverage tracks are depicted for each sequencing technique.
Correlation to epigenetic activation and gene transcription
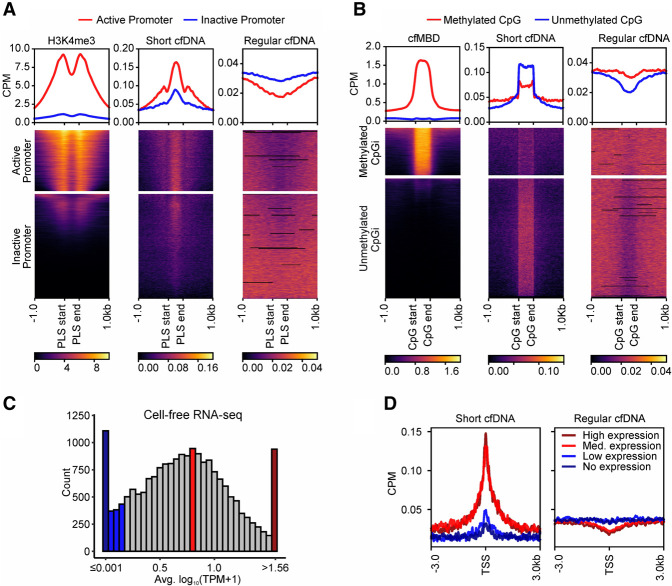
Given that short cfDNA fragments are likely derived from the protection of regulatory DBPs and are overrepresented at open chromatin regions, we investigated a potential relationship between short cfDNA signal strength (i.e., local read enrichment) and promoter activation state. Annotated promoter-like structures were classified as active or inactive promoters based on cell-free histone 3 lysine 4 triple-methylation (H3K4me3) signals from publicly available ChIP-seq data of a healthy individual (Sadeh et al. 2021). Active promoters with a strong H3K4me3 signal showed a higher coverage of short cfDNA than did promoters with weak or no H3K4me3 signal, whereas regular cfDNA shows exactly the opposite behavior (Fig. 3A). Moreover, the strongest signals for H3K4me3, representing the nucleosome positions, occur at local minima of short cfDNA. DNA methylation is known to be an essential regulator of gene activity and is associated with transcription factor binding and thus, potentially, DNA footprint signals. Consequently, we classified CpG islands (CpGis) into methylated and unmethylated CpGis based on the signal strength of cell-free methyl-CpG-binding domain sequencing (cfMBD-seq) data from a healthy individual. Strongly methylated CpGis show a weaker accumulation of short cfDNA compared with unmethylated CpGis, whereas regular cfDNA again behaves the opposite (Fig. 3B), demonstrating the relationship between DNA methylation and short cfDNA. To further analyze a connection between localized short cfDNA signals and downstream gene transcription, we used publicly available cell-free RNA-seq data from five healthy individuals (Zhu et al. 2021). Based on the average transcript abundance level of protein-coding genes, four subsets of genes were defined: “no expression,” “low expression,” “medium expression,” and “high expression” genes (Fig. 3C). Genes with medium or high expression show considerable enrichment of short cfDNA reads at their respective TSSs, whereas genes with low or no expression show no substantial enrichment. Regular cfDNA again behaves contrary to short cfDNA and shows a much less pronounced difference between active expression (high and medium) and low, or no, expression (Fig. 3D). Consistent with the definition of these gene subgroups, H3K4me3 histone modifications increase and DNA methylation levels decrease as expression levels increase at the respective TSS of the genes (Supplemental Fig. S7B,C). For a more detailed analysis of the joint effect of DNA footprint signals and DNA methylation in regulatory elements of genes on its transcription, we used matched sequencing data from four septic patients. Different combinations of short cfDNA and DNA methylation signals also seem to affect the gene expression levels (Supplemental Fig. S8A). Genes with clearly decreased DNA methylation in their proximal CpGis and significantly elevated short cfDNA abundance at their core promoter seem to be associated with higher transcription levels (clusters 2 and 4), whereas genes with increased DNA methylation and reduced short cfDNA abundance seem to be associated with lower transcription levels (clusters 5, 8, and 9). Between these clear cases, the associations of regulatory signals with the resulting transcription levels of the genes are more complex and becoming less clear (Supplemental Fig. S8B). In summary, the signal strength of short cfDNA is higher in active promoters than in inactive promoters, higher in unmethylated CpGis than in methylated CpGis, and higher in TSSs of actively transcribed genes than in TSSs of untranscribed genes. Thus, short cfDNA is enriched at loci with markers for epigenetic activation and transcriptionally active genomic regions.
Figure 3.
Short cfDNA is enriched at loci with markers of epigenetic activation and transcriptionally active regions. (A) Annotated PLSs from ENCODE are clustered based on publicly available cell-free H3K4me3 ChIP-seq signal strength into two clusters, representing active (red) or inactive (blue) promoters. Average coverage profiles for short cfDNA and regular cfDNA at active or inactive promoters demonstrate the influence of the promoter activation status. (B) Annotated CpG islands are clustered based on cell-free methyl-CpG-binding domain (cfMBD) sequencing signal strength into two clusters. Average coverage profiles for short cfDNA and regular cfDNA at methylated or unmethylated CpG islands reveal the influence of methylation levels at CpG islands. (C) Histogram showing average expression levels of protein-coding genes in publicly available cell-free RNA sequencing data. For each category, 5% of all analyzed genes were selected (938 genes each for no expression [dark blue], low expression [blue], medium expression [red], and high expression [dark red]). (D) Average coverage profiles for short cfDNA and regular cfDNA at the transcription start sites of the defined gene groups of C reveal the influence of transcriptional activity. All data in A and D were generated from samples of healthy individuals. S03 was used as the short cfDNA data set, and S06 was used as the regular cfDNA data set.
Disease-specific short cfDNA signatures in liquid biopsies
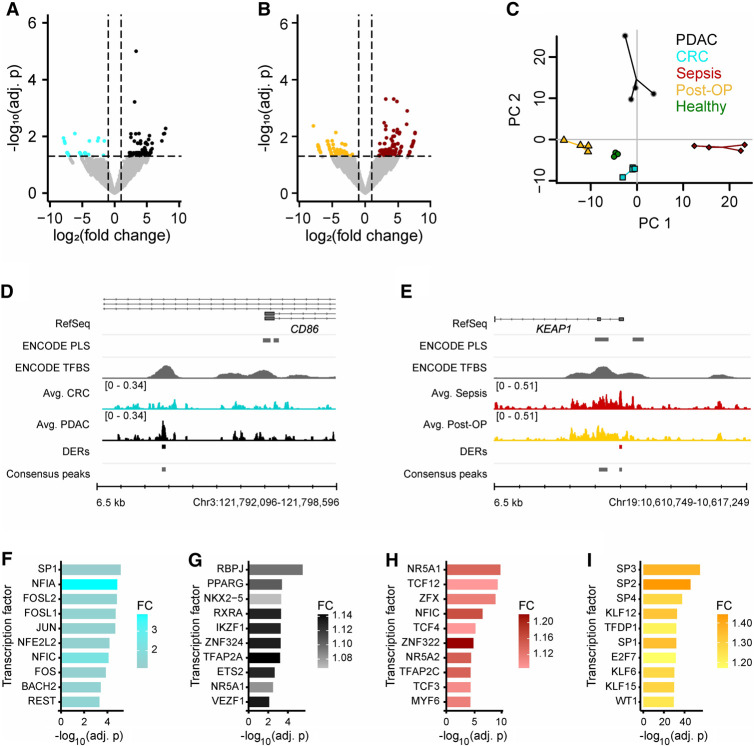
To identify disease-specific signatures, short cfDNA data were generated for four biological replicates of four different clinical indications: two types of gastrointestinal carcinomas (PDAC and CRC), as well as sepsis and Post-OP controls (Supplemental Table S1). Post-OP patients were selected as controls because this patient group is prone to develop a sepsis after surgery, and therefore, a tight control and separation are of clinical relevance (Chen et al. 2019b; Lukaszewski et al. 2022). Comparison of consensus peak sets for PDAC versus CRC revealed 136 differentially enriched loci (Fig. 4A; Supplemental Fig. S10) and 241 loci with a differential enrichment comparing sepsis with Post-OP (Fig. 4B; Supplemental Fig. S10). Principal component analysis (PCA) based on all differentially enriched regions (DERs) demonstrated a clear separation of all four clinical indications as well as the healthy samples by the first two principal components (Fig. 4C). Two exemplary DERs demonstrate a distinct differential DNA footprint near a TSS in the context of a larger locus with TFBSs (Fig. 4D,E). A differentially enriched TFBS was detected near the alternative transcription start site in the promoter of the CD86 gene in PDAC patients compared with CRC patients (Fig. 4D), whereas another protected TFBS was detected near the promoter of the kelch-like ECH associated protein 1 (KEAP1) in sepsis patients in contrast to Post-OP patients (Fig. 4E). In addition to differential analysis of signal strength at defined consensus peaks, differential enrichment of transcription factor binding motifs was also analyzed in all consensus peaks. For this purpose, the relative abundance of transcription factor motifs in the consensus peaks of one condition was compared with the abundance in the consensus peaks of the respective reference condition. Enrichment of 14 different transcription factor motifs was detected for PDAC in comparison to CRC, 19 for CRC in comparison to PDAC, 14 for sepsis in comparison to Post-OP, and 126 for Post-OP in comparison to sepsis (E-value ≤ 10) (Fig. 4F–I). The binding motif with the strongest enrichment in PDAC patients compared with CRC patients belongs to the recombination signal binding protein for immunoglobulin kappa J region (RBPJ) (Fig. 4F), for example. In this context, one of the physiological roles of the transcription factor RBPJ is the regulation of early pancreatic cell development. Overall, short cfDNA sequencing enables the detection of condition-specific enrichment signatures in liquid biopsies, which can be used to discriminate different diseases for diagnostic purposes. In addition, short cfDNA sequencing might also help identify transcription factors that may have physiological relevance to the condition.
Figure 4.
Comparison of short cfDNA data reveals condition-specific protection of TFBSs. (A) Differential enrichment analysis for pancreatic ductal adenocarcinoma (PDAC) samples with colorectal cancer (CRC) samples identifies differentially enriched TFBSs (Adj. P-value ≤ 0.05 and |log2(FC)| ≥ 1). (B) Differential enrichment analysis for sepsis samples with postoperative (Post-OP) samples identifies differentially enriched TFBSs (Adj. P-value ≤ 0.05 and |log2(FC)| ≥ 1). (C) Principal component analysis based on identified differential TFBSs separates all conditions and healthy individuals. Four biological replicates were used per condition. In addition to the samples, a centroid for each group is depicted as a larger data point. Variance explained: PC1 = 29.6%, PC2 = 20.9%. (D) Example for a differentially enriched TFBS in PDAC with novel TF binding near the alternative transcription start site of CD86. (E) Example for a differentially enriched TFBS in sepsis with novel TF binding in the promoter of KEAP1. (F–I) Top 10 differentially enriched TF motifs. (F) CRC consensus peaks in comparison to PDAC consensus peaks. CTCF and CTCFL were identified as well but were not included in the figure. (G) PDAC consensus peaks in comparison to CRC consensus peaks. (H) Sepsis consensus peaks in comparison to Post-OP consensus peaks. (I) Post-OP consensus peaks in comparison to sepsis consensus peaks. CTCF and CTCFL were identified as well but were not included in the figure.
Discussion
In this study, we present a novel approach for comprehensive DNA footprinting in liquid biopsies by analysis of short double-stranded plasma cfDNA. Our short cfDNA sequencing approach comprises preparative gel electrophoresis to specifically enrich cfDNA fragments with a mean length of ∼40 bp in combination with high-throughput sequencing. We observed an enrichment of short cfDNA at the TSS of genes, in which regular cfDNA is depleted. A closer inspection of the average signals at the TSS also showed an oscillation pattern reciprocal to that of regular cfDNA at the 5′ UTR. These findings indicate a regular shift of nucleosomes from the promoter to the 5′ UTR of genes and protection by regulatory DBPs between displaced nucleosomes as revealed by short cfDNA enrichment. In line with the enrichment of short cfDNA at TSS, we also found that short cfDNA fragments showed a higher GC content than the human genome on average (short cfDNA = 57.8%, human genome = 40.9%). Gene regulatory elements in humans possess an increased GC content. Consequently, short cfDNA fragments and TFBSs that are enriched in such regions of the genome have elevated GC contents. Short cfDNA also accumulates at DHSs. At these open chromatin locations, regular cfDNA exhibits a combined signal originating from high-frequency and low-frequency nucleosomal oscillation signals, as described by Ulz et al. (2019). Comparable to this finding, we observed two different types of short cfDNA signals created by DBPs with a narrow binding signal, such as CTCF, and multiple adjacent protein binding events that result in a broader signal. The observed narrow signal of regulatory proteins, like CTCF, SPI1, or REST, can be explained by their ability to initiate nucleosome displacement in closed chromatin (pioneer factors) (Fu et al. 2008; Heinz et al. 2010; Barozzi et al. 2014; Vanzan et al. 2021). Pioneer factors can bind directly to closed chromatin without the need or presence of auxiliary proteins. In contrast, the binding sites of regulatory proteins such as MYC proto-oncogene, bHLH transcription factor (MYC), or TP53BP1 exhibit a much broader signal. These regulatory proteins bind in open chromatin, accompanied by the presence of additional regulatory proteins. Consequently, we also found a striking correlation between DNA footprint signals and markers representative for transcriptional regulation. In this context, we determined that promoters with H3K4me3 histone modifications, a characteristic for active gene transcription, revealed stronger short cfDNA signals than those without. Furthermore, we observed an opposing correlation between methylation levels of CpGis and corresponding enrichment of short cfDNA. Because unmethylated CpGis are considered markers of epigenetic activation, depletion of regular cfDNA and the presence of DNA footprinting signals indicate a protection from regulatory activating factors at such loci. Regarding gene expression, short cfDNA signals at the TSS are increased at actively transcribed genes compared with genes with low expression. However, short cfDNA does not strictly capture continuous dynamics of gene expression levels, rather more it exhibits a binary switch between high and low expression. Comparable findings were obtained through the analysis of the joint effect of DNA footprint signals and DNA methylation in regulatory elements of genes on their transcription. Here, the change between low and high gene expression levels can be detected to a certain extent, but not so precisely to exactly quantify gene expression levels. These aberrations are likely owing to the multitude of other epigenetic regulatory mechanisms that fine-tune gene expression levels. Although this holds also true for other regulatory signals of gene expression, such as DNA methylation, this might reflect a potential limitation of our approach to quantitatively predict an individual's physiology exactly. Further targeted studies are needed to assess the extent of this limitation. Nevertheless, short cfDNA is clearly correlated with DNA-methylation signatures, H3K4me3 histone modifications, and transcriptional activity of downstream gene loci. We found consistent enrichment of short cfDNA where regular cfDNA is depleted. Hence, we propose that the molecular origin of short cfDNA most likely is double-stranded cfDNA, which has been protected from enzymatic digestion by regulatory proteins, including transcription factors. In agreement with this model, we found that short cfDNA consensus peaks comprise a plethora of transcription factor motifs and that known TFBSs revealed clear enrichment for short cfDNA. Taken together, our data provide evidence that short cfDNA represents a characteristic subset of cfDNA that is distinct from nucleosomal cfDNA and therefore most likely does not originate from the degradation of regular cfDNA.
In contrast to short cfDNA, which is characterized by short double-stranded DNA (short dsDNA), publications by Snyder et al. (2016), Hisano et al. (2021), and Hudecova et al. (2022) reported on the presence of short ssDNA. Contrary to short dsDNA, short ssDNA fragments may represent a significant fraction of total cfDNA, comprising as much as 20% of a sequencing library. However, short ssDNA demonstrated that the relative signal strength at transcription start sites or TFBSs of proteins, such as MYC, is very low compared with short dsDNA. Therefore, we assume that cfDNA originally protected by regulatory DBPs is short dsDNA, whereas short ssDNA might be of other biological origin with higher signal-to-noise ratios. Hisano et al. (2021) and Hudecova et al. (2022) suggested that much of the short ssDNA could originate from noncanonical DNA structures, such as G4 quadruplexes, which may largely superimpose signals from regulatory protein DNA-binding events.
Transcription factor motifs from short cfDNA revealed sets of differentially enriched transcription factors between different conditions including samples from septic, CRC, and pancreatic cancer (PDAC) patients, suggesting a diagnostic potential for liquid biopsy applications. Sepsis is characterized by a complex interplay of proinflammatory and anti-inflammatory processes orchestrated by regulators of the immune system. Seven out of the top 10 differentially enriched transcription factor motifs identified in septic samples are linked to regulatory pathways of the immune system. For example, transcription factor 4 (TCF4; also known as E2-2 or ITF2) regulates genes for the differentiation of dendritic cells into interferon-producing plasmacytoid dendritic cells (Reizis 2010). TCF4 also regulates the immune response as a downstream target of toll-like receptor 2 (TLR2) signaling to induce the expression of immunoregulatory genes such as interleukin 10 (IL10) (Manoharan et al. 2014). The identified transcription factor zinc finger protein X-linked (ZFX) is known to be involved in the maintenance of peripheral T cells as well as expansion and maintenance during B cell development and peripheral homeostasis (Arenzana et al. 2009; Smith-Raska et al. 2018). For CRC patients, Sp1 transcription factor (SP1) was identified as the top hit, and several FOS/JUN family transcription factors were identified as differentially enriched. SP1 is a ubiquitous transcription factor and mediator of critical physiological pathways, including cell cycle, proliferation, and metastasis. SP1 plays a key role in regulating genes involved in CRC growth and metastasis (Bajpai and Nagaraju 2017). Two members of the FOS/JUN family—FOS-like 1, AP-1 transcription factor subunit (FOSL1), and FOS-like 2, AP-1 transcription factor subunit (FOSL2)—are known to promote tumorigenesis and metastasis in colon cancer (Li et al. 2018; Liu et al. 2021). For PDAC patients, RBPJ was identified as the corresponding top hit. RBPJ is known to form a heterocomplex with pancreas associated transcription factor 1a (PTF1A), and their interaction is required in the early stage of pancreatic growth, morphogenesis, and lineage fate decision (Masui et al. 2007). In addition, peroxisome proliferator-activated receptor gamma (PPARG) and retinoid X receptor alpha (RXRA) were identified, which are known to form a heterocomplex (Lehrke and Lazar 2005). PPARG is a key regulator of adipocyte differentiation, regulates insulin and adipokine production and secretion, and is associated with PDAC. In addition, RXRA promotes proliferation and inhibits apoptosis of pancreatic cancer cells (Chen et al. 2019a). Moreover, we found disease-specific differentially enriched loci that may enable clear separation of patients with PCA. For example, in PDAC patients, we found differential protection from DBPs near the alternative TSS of the CD86 gene compared with CRC patients. In PDAC, the tumor microenvironment is highly immunosuppressive and characterized by a dense stroma and an abundance of immunosuppressive cells such as myeloid-derived suppressor cells and regulatory T cells, as well as increased levels of monocytes in peripheral blood (Gautam et al. 2023; Hansen et al. 2023). CD86 is an important costimulatory molecule involved in T cell activation. CD86, together with other immune markers such as integrin subunit alpha X (ITGAX; also known as CD11c) and CD274 molecule (CD274; also known as PD-L1), can serve as a prognostic indicator in PDAC, in which increased expression of CD86 on peripheral blood monocytes of PDAC patients significantly correlates with disease severity (Hansen et al. 2023). In septic patients, we found an additional signal of short cfDNA near the TSS of the KEAP1 gene compared with postoperative controls. KEAP1 regulates oxidative stress and inflammation in sepsis via the NFE2-like bZIP transcription factor 2 (NFE2L2; also known as NRF2) signaling pathway. Inhibition of KEAP1 allows NRF2 to activate antioxidant responses, thereby reducing the damage caused by sepsis. Enhanced NRF2 activity is associated with better outcomes, whereas impaired signaling worsens sepsis in mouse studies. The use of protein–protein interaction (PPI) inhibitors targeting the NRF2-KEAP1 signaling pathway are considered and evaluated in various studies for the treatment of sepsis by increasing NRF2 activity (Gunne et al. 2020; Wang et al. 2023).
Taken together, analysis of short double-stranded cfDNA might provide the most accurate picture of a genome-wide transcription factor footprint in liquid biopsies. With the ability to identify disease-specific TFBS enrichment for patient classification, we see considerable potential in the application of short cfDNA sequencing for liquid biopsy applications.
Many studies explore the properties and information to be inferred from various cfDNA fragments and fragmentation patterns. Often the clinical relevance of the obtained information is not yet well evaluated. Although we started to explore the clinical relevance and potential of short cfDNA sequencing with clinical samples, the results presented in this study are based on a limited number of samples. Further studies with more clinical samples are still required to validate the potential of this approach. In addition to validity and utility, cost-effectiveness is an essential factor for clinical diagnostic methods. Although our workflow already represents a resource-efficient alternative to ultradeep high-throughput sequencing of total cfDNA, a targeted and even-more-economical approach might be advantageous for implementation. With the new short cfDNA sequencing approach, which directly maps the protection across the genome via regulatory proteins, we want to add a new tool to liquid biopsy diagnostics that could improve the detection of cancer or enable clinically relevant differential diagnosis of cancer types.
Methods
Blood samples
This study included blood samples from nine healthy individuals, four individuals with PDAC, four individuals with CRC, four individuals with sepsis, and four individuals that underwent major abdominal surgery (Supplemental Table S1). Blood from the healthy individuals was acquired commercially from Biomex. Septic patients participated in a previously published, prospective observational clinical study that was conducted in the surgical intensive care unit of Heidelberg University Hospital, Germany between November 2013 and January 2015 (S13–S16 and S25–S37; German Clinical Trials Register: DRKS00005463) (Decker et al. 2017). Treatment of these sepsis patients included early-goal directed therapy, elimination of the septic focus, and broad-spectrum antibiotic therapy. Identified pathogens of all sepsis patients and blood cell counts are included in Supplemental Table S2. Patients without cancer (nonseptic controls, i.e., Post-OP) that underwent major abdominal surgery and all individuals with cancer were recruited at the University Hospital of Erlangen with the approval of the local ethics committee and the clinical trial number 180_19 B (S09–S12 and S17–S24). All experiments were performed in accordance with the study protocol approved by the ethics committee.
cfDNA isolation
Plasma was prepared by centrifugation of whole blood for 10 min at 1600g and 4°C. Afterward, blood plasma was centrifuged again for 10 min at 16,000g and 4°C. Afterward, 1.1 mL of the supernatant was transferred into a fresh 1.5 mL DNA LoBind tube and stored at −80°C. If necessary, the sample volume was filled up with freshly prepared 1 × phosphate-buffered saline solution. cfDNA was isolated with the QIAsymphony SP DNA preparation system and the QIAsymphony DSP circulating DNA kit (Qiagen) according to the manufacturer's advice. Eluted cfDNA was quantified with the Qubit dsDNA HS assay kit (Thermo Fisher Scientific), and the cfDNA quality was assessed by the fragment analyzer high-sensitivity DNA kit (Agilent).
High-throughput sequencing
Sequencing libraries for regular cfDNA were prepared with the NEBNext Ultra II DNA library prep kit (New England Biolabs) according to the manufacturer's protocol; 0.5 ng isolated cfDNA was used as input. The NEBNext adaptor was diluted 1:25 for all reactions. PCR was performed with 10 PCR cycles and 4 µL primers. Sequencing was performed on a NextSeq 2000 (Illumina) with 100 bp single-end reagent kits.
Sequencing libraries for short double-stranded cfDNA (short cfDNA) were prepared from 3 ng to 15 ng of cfDNA, depending on the clinical condition, using the NEXTFLEX cell free DNA-seq kit (V2; PerkinElmer) according to the manufacturer's advice with one exception: The final library after PCR amplification was eluted in 20 µL nuclease-free water. Library generation was performed with a Biomek FXP workstation (Beckman Coulter). Library quality was assessed by the fragment analyzer high-sensitivity DNA kit (Agilent), and the concentration was measured by the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Size selection of the cfDNA libraries was performed using a BluePippin instrument (Sage Science). To select the short cfDNA portion in the range of 150–200 bp, samples were applied to a 3% agarose BluePippin cassette according to the manufacturer's protocol. Briefly described, samples were filled up with water to 30 µL and were mixed with 10 µL of supplied internal marker (100 bp to 250 bp). Twenty-three microliters of the eluted, size-selected sample was reamplified with 25 µL of NEXTFLEX PCR master mix 2.0 and 2 µL of 1:10 diluted NEXTFLEX primer mix 2.0 as described in the NEXTFLEX cell free DNA-seq kit (V2; PerkinElmer), step C PCR amplification. Afterward, samples were purified with 1.2 × the volume of AMPureXP beads (Beckman Coulter) according to the manufacturer's advice. Size-selection performance was evaluated by the fragment analyzer high-sensitivity DNA kit (Agilent). If the sample still contained fragments outside the target range of 150 bp to 200 bp, size selection was repeated as previously described, with the exception that the input sample was adjusted to 30 µL with water and the reamplification step was performed with five to eight cycles. After size selection, sequencing was performed on a NextSeq 2000 (Illumina) with 100 bp single-end reagent kits.
Methylation enrichment of cfDNA was performed using the EpiMark methylated DNA enrichment kit (New England Biolabs) according to the manufacturer's instructions. Two reactions with 5–15 ng of cfDNA input each were performed in parallel per sample. Methylated cfDNA was eluted sequentially from both reactions using the same 50 µL of nuclease-free water to increase yield. Sequencing libraries were prepared with the NEBNext Ultra II DNA library prep kit (New England Biolabs) according to the manufacturer's protocol. Fifty microliters of methylated cfDNA from the EpiMark procedure was used as input. The NEBNext adaptor was diluted 1:25 for all reactions. PCR was performed with 17 PCR cycles and 3 µL primers. Sequencing was performed on a NextSeq 2000 (Illumina) with 100 bp single-end reagent kits.
For whole-blood RNA-seq, blood of patients was collected in PAXgene blood RNA tubes (BD Biosciences), incubated at room temperature for 2 h to achieve complete lysis of blood cells, and frozen at −80°C until further processing. Before nucleic acid isolation, tubes were thawed at room temperature for 2 h. Nucleic acid isolation was performed using the QIAcube (Qiagen) and the PAXgene blood miRNA kit according to the manufacturer's protocol to extract gene-encoding mRNAs. Nucleic acids were eluted in 2 × 40 µL buffer BR5. The quantity and quality of the isolated RNA were determined with a Qubit fluorometer (Thermo Fisher Scientific) and a fragment analyzer (Agilent), respectively. Library preparation and sequencing were performed using 250 ng RNA with the ScriptSeq kit v2 (Illumina). Sequencing of the libraries was performed with HiSeq 2500 (Illumina), with 100 bp single-end reagent kits.
Sequencing data processing
After initial quality control of short cfDNA sequencing of raw sequencing reads with FastQC (v0.11.8), the following steps were performed sequentially to remove sequencing artifacts: (1) removal of sequencing adapters, terminal poly(G) sequences (min 10 sequential G's), and quality trimming (BBTools bbduk.sh v38.67); (2) removal of terminal single A nucleotide added during library preparation (BBTools bbduk.sh v38.67); (3) size selection of sequenced fragments allowing read lengths >20 bp and <60 bp (BBTools bbduk.sh v38.67); (4) removal of sequencing reads with low complexity, that is, dust scores smaller than seven (prinseq-lite v0.20.4) (Andrews 2010; Schmieder and Edwards 2011; Bushnell et al. 2017); (5) processed reads mapped to the human reference genome assembly GRCh37 using NextGenMap (v0.5.5) with default settings (Sedlazeck et al. 2013); and (6) mapped reads deduplicated with SAMtools rmdup (v1.9), reads in blacklisted regions removed with BEDTools intersect (v2.30.0), and reads with a MAPQ value lower than one removed with SAMtools view (v1.9) (Li et al. 2009; Quinlan and Hall 2010; Amemiya et al. 2019). These reads were converted to bigWig format for visualization and other downstream analyses with deepTools (bin size = 10, normalization = counts per million [CPM], bamCoverage v3.5.1) (Ramírez et al. 2016). This workflow is graphically summarized in Supplemental Figure S3. Realigning the reads to GRCh38 or newer assemblies would not significantly affect the results and conclusions, as only Chromosomes 1–22, X, and Y were analyzed, which are highly conserved across different genome versions. Using the ENCODE blacklist with GRCh37 helps to avoid potential sequencing artifacts in poorly mappable regions of these chromosomes, such as centromeres and telomeres, by excluding such problematic regions.
After initial quality control of regular cfDNA raw sequencing reads with FastQC (v0.11.8), the following steps were performed (Andrews 2010): (1) removal of sequencing adapters, removal of terminal poly(G) sequences (min 10 sequential G's), removal of reads <50 bp, and quality trimming (BBTools bbduk.sh v38.67) (Bushnell et al. 2017); (2) processed reads mapped to the human reference genome assembly GRCh37 using NextGenMap (v0.5.5) with default settings (Sedlazeck et al. 2013); and (3) mapped reads deduplicated with SAMtools rmdup (v1.9), and reads in blacklisted regions removed with BEDTools intersect (v2.30.0) (Li et al. 2009; Quinlan and Hall 2010). Mapped reads were converted to bigWig format for visualization and other downstream analyses with deepTools (bin size = 10, normalization = CPM, bamCoverage v3.5.1) (Ramírez et al. 2016).
cfMBD-seq data were processed as regular cfDNA sequencing data.
Public sequencing data processing
FASTQ files of five publicly available cell-free RNA sequencing data sets of different healthy individuals from Zhu et al. (2021) were obtained from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra). Accession numbers and unique identifiers are listed in Supplemental Table S3. All samples were sequenced in paired-end mode with a read length of 150 bp and about 10 million reads per sample on average. After initial quality control of raw sequencing reads with FastQC (v0.11.8), the following steps were performed to obtain read counts for individual genes (Andrews 2010): (1) removal of sequencing adapters, quality trimming, and removal of reads <100 bp (BBTools bbduk.sh v38.67) (Bushnell et al. 2017); (2) mapping of processed reads to the human reference genome assembly GRCh37 using NextGenMap (v0.5.5) with default settings, keeping only reads with a MAPQ value greater than two (Sedlazeck et al. 2013); (3) gene quantification in raw read counts using featureCounts (v2.0.1) with the UCSC Genes annotation (available at https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/genes/hg19.knownGene.gtf.gz) (Liao et al. 2014), and only protein-coding genes on the autosomal chromosomes included in the final readcount matrix to reduce confounding by the gender of sample donors; and (4) if required, raw read counts converted to transcripts per million (TPM) using R (R Core Team 2024).
FASTQ files of cfDNA sequencing data sets of a healthy individual from Snyder et al. (2016) were obtained from the SRA. Accession numbers and unique identifiers are listed in Supplemental Table S3. After initial quality control of raw sequencing reads with FastQC (v0.11.8), the following steps were performed: (1) removal of sequencing adapters and quality trimming (cutadapt v4.0) (Martin 2011); (2) mapping of trimmed reads to the human reference genome assembly GRCh37 using NextGenMap (v0.5.5) with minimum map quality of 10 (Sedlazeck et al. 2013); (3) in silico size selection of the mapped reads with 35–80 nt for short cfDNA and 120–180 nt for regular cfDNA; (4) removal of low complexity reads and deduplication (prinseq-lite v0.20.4) (Schmieder and Edwards 2011); (5) removal of reads in blacklisted regions with BEDTools intersect (v2.30.0) (Quinlan and Hall 2010); and (6) for downstream analysis and visualization, bigWig files generated with deepTools (bin size = 10, normalization = CPM, bamCoverage v3.5.1) (Ramírez et al. 2016).
ATAC-seq and DNase-seq data from ENCODE (see Supplemental Table S3) were retrieved as processed BAM files and only converted to other data types, like bigWig for downstream analysis. Cell-free H3K4me3 ChIP-seq data from Sadeh et al. (2021) (see Supplemental Table S3) were retrieved as processed BED files and only converted to other data types, like BAM and bigWig for downstream analysis.
Peak calling and NFR calling
For short cfDNA sequencing data, narrow peaks and clustered peaks were called with MACS2 callpeak (narrow: ‐‐nomodel ‐‐extsize 32 ‐‐call-summits ‐‐min-length 30 -q 0.05, clustered: ‐‐broad ‐‐nomodel ‐‐extsize 32 ‐‐max-gap 100 ‐‐min-length 500 ‐‐broad-cutoff 0.05) (Zhang et al. 2008). Consensus peaks of a condition were identified using R and defined as genomic sites where a narrow peak was identified in at least three of four samples (R Core Team 2024). In addition, consensus peaks <31 nt apart were combined. For regular cfDNA, NFRs were called on the merged data set of four biological replicates to increase genome-wide coverage and, thus, reliability. NFRs were identified with the R packages NucDyn, utilizing nucleR, with default settings (Buitrago et al. 2019). Peaks and NFRs were annotated to genomic features in R with the ChIPseeker package and default settings (Yu et al. 2015).
Average coverage profiles and heatmaps
Average coverage profiles and heatmaps were created from bigWig files with deepTools (computeMatrix, plotHeatmap, and plotProfile; v3.5.1) (Ramírez et al. 2016). Different genomic reference locations were used for average coverage profiles. For Figure 2 and Supplemental Figure S2, validated binding sites of transcription factors were retrieved from the Gene Transcription Regulation Database (GTRD) (Kolmykov et al. 2021). Here, TFBSs were ranked according to their robustness across different cell lines and tissues, namely, the number of tissues and cell lines in which the respective binding site was found. Only the top 1000 binding sites were used for average coverage profiles. Promoter-like structures from ENCODE were converted from GRCh38 to GRCh37 with the liftOver tool from UCSC and used as reference regions in Figure 3A (available at https://www.encodeproject.org/files/ENCFF379UDA/) (Hinrichs et al. 2006). CpGis were used as reference regions in Figure 3B (available at UCSC TableBrowser, track name = cpgIslandExt) (Karolchik et al. 2004). The heatmaps in Figure 3, A and B, were clustered in two groups using the implemented k-means clustering based on the line-wise average. Missing data in heatmaps were plotted in black.
Composite effect of DNA methylation and DNA footprint signals on gene transcription
From four sepsis patients, matched short cfDNA, cfMBD, and whole-blood RNA sequencing data were acquired (S26–S37). For a combined analysis, different genomic annotations were combined. The same gene annotation as used with the cfRNA sequencing data from Zhu et al. (2021) was utilized. A core promoter annotation for GRCh37 was retrieved from the eukaryotic promoter database (EPD; version 6; available at https://epd.expasy.org/ftp/epdnew/H_sapiens/006/Hs_EPDnew_006_hg19.bb) (Dreos et al. 2015; Meylan et al. 2020). For each gene, only the best-matching core promoters were kept (“_1” flag). As for CpGis, the aforementioned CpGis from UCSC were used again. CpGis were assigned to genes based on their proximity to the EPD promoter of the gene in the genome, with a maximum absolute distance of 5 kb. With these assignments, from the roughly 20,000 protein-coding genes in the annotation, about 12,000 remained with an associated core promoter and CpGis. Counts of sequencing reads of each sequencing data type and each patient were retrieved for the curated gene bodies, core promoters, and CpGis with featureCounts (v2.0.1) and converted to TPM (Liao et al. 2014). From the patient replicates a mean TPM value was calculated per region and sequencing type. Additionally, a pseudocount of 0.01 was added. For better comparability of the signals, the cfMBD-seq data in CpGis and short cfDNA sequencing data in core promoters were additionally adjusted for GC content. The GC content of the respective regions was extracted from GRCh37 using BEDTools nuc (v2.30.0) (Quinlan and Hall 2010). The adjusted signal was calculated as follows: A locally weighted regression function was fitted with the mean readcount value and GC content for each annotation type (R lowess function). Each mean readcount value (observed) is divided by its fitted value (expected) to obtain an observed over expected (OoE) ratio. For better interpretability and visualization, the values are also log2-scaled. Genes with comparable composite signals in the core promoter and in the CpGis were identified by k-means clustering (k = 10) and visualized in a heatmap using R and ComplexHeatmap (Fig. 3E; Gu et al. 2016). The average gene expression values were added as an annotation but had no influence on clustering. For better visualization and comparison of the distributions of gene expression values, the RNA-seq data were also visualized in a ridge plot in R with ggplot2, according to the clusters from the heatmap, and sorted by descending median value (Fig. 3F). Clusters with fewer than 200 genes were excluded from this visualization as the sample size was considered too small for a representative distribution.
Transcription factor motif enrichment analysis
Enriched transcription factor motifs were identified with the AME tool from MEME suite (‐‐scoring avg ‐‐method fisher; v5.4.1) (McLeay and Bailey 2010). Input DNA sequences were retrieved from consensus peak sets and analyzed for the enrichment of motifs listed in the HOCOMOCOv11_core_HUMAN_mono database (Kulakovskiy et al. 2018). DNA sequences of consensus peaks from short cfDNA sequencing data of healthy individuals were compared with control sequences, generated by shuffling the letters in the input while preserving the frequencies of k-mers (‐‐shuffle). The proportions of identified transcription factor motif classes and their respective superclasses were summarized in a treemap plot using R and the treemap package (v.2.4.3; https://cran.r-project.org/web/packages/treemap/index.html). Short cfDNA sequencing data from other than healthy states were compared with each other. The DNA sequences underlying each consensus peak set were used as control sequences, for example, consensus peak DNA sequences from CRC as input compared with consensus peak DNA sequences from PDAC as control.
Differential enrichment analysis
Identification of DERs was performed with the R package DEBrowser (v1.2.0) using the implemented edgeR method with raw read counts in the combined consensus peak sets of two compared conditions, TMM normalization, a glmLRT, and dispersion = 0 (Robinson et al. 2010; Kucukural et al. 2019). The use of this analysis is based on the observation that the read counts in the consensus peak sets are best modeled with a negative binomial distribution in comparison to a Poisson of geometric distribution (Supplemental Fig. S9). Genes were considered as differentially expressed between two conditions (four biological replicates per condition) with an adjusted P-value smaller than 0.05 and a log2(fold change) ≤ −1 or ≥1. Volcano plots for the differential enrichment analysis were generated with the R package EnhancedVolcano (v1.8.0). The identified DERs of both comparisons were used for a PCA, and the first three principal components were visualized in R with pca3d (v0.10.2). Samples of each condition were linked to the centroid of the respective condition with a line.
Data access
The raw high-throughput sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA1033613. Processed high-throughput sequencing data of samples S01–S33 are available as genome coverage tracks in a UCSC Genome Browser session (https://genome.ucsc.edu/s/jnmllr/Short_cfDNA_seq_manuscript) or as files at Figshare (https://doi.org/10.6084/m9.figshare.25211525.v2). Custom data analysis code created for this work is available on GitHub (https://github.com/janmueller17/short_ds_cfDNA) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Drs. Jens and Susanne Freitag for their valuable support. This work was supported by internal funding from Fraunhofer-Gesellschaft.
Author contributions: J.M. and K.S. conceptualized and designed the study. C.H., M.S., L.B., and K.G. processed blood samples, isolated cfDNA, and established the size-selection method for short double-stranded cfDNA. J.M. performed bioinformatics analysis. C.A. analyzed the effect of library preparation methods on short and regular cfDNA in public data. J.M., K.S., Y.V., and A.v.H. analyzed and interpreted the sequencing data. G.F.W., S.O.D., and T.B. recruited patients and collected samples. J.M., K.S., and M.S. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279326.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
K.S. is a cofounder of Noscendo, a diagnostic company dedicated for detection of pathogens based on high-throughput sequencing. K.S., C.H., M.S., and J.M. are inventors of a patent application in bioinformatics algorithms for analyzing cfDNA fragmentomics.
References
- Amemiya H, Kundaje A, Boyle A. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9: 9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FASTQC. A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- Arenzana T, Smith-Raska M, Reizis B. 2009. Transcription factor Zfx controls BCR-induced proliferation and survival of B lymphocytes. Blood 113: 5857–5867. 10.1182/blood-2008-11-188888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Nagaraju G. 2017. Specificity protein 1: its role in colorectal cancer progression and metastasis. Crit Rev Oncol Hematol 113: 1–7. 10.1016/j.critrevonc.2017.02.024 [DOI] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronkhorst A, Ungerer V, Holdenrieder S. 2019. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif 17: 100087. 10.1016/j.bdq.2019.100087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago D, Codó L, Illa R, de Jorge P, Battistini F, Flores O, Bayarri G, Royo R, Del Pino M, Heath S, et al. 2019. Nucleosome dynamics: a new tool for the dynamic analysis of nucleosome positioning. Nucleic Acids Res 47: 9511–9523. 10.1093/nar/gkz759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham P, Kim M, Agbor-Enoh S, Luikart H, Valantine H, Khush K, De Vlaminck I. 2016. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep 6: 27859. 10.1038/srep27859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B, Rood J, Singer E. 2017. BBMerge: accurate paired shotgun read merging via overlap. PLoS One 12: e0185056. 10.1371/journal.pone.0185056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hu M, Wang X-C, Du S-H, Cheng Z-X, Xu J, Qu Y-K. 2019a. Effects of RXRα on proliferation and apoptosis of pancreatic cancer cells through TGF-β/Smad signaling pathway. Eur Rev Med Pharmacol Sci 23: 4723–4729. 10.26355/eurrev_201906_18053 [DOI] [PubMed] [Google Scholar]
- Chen P-Y, Luo C-W, Chen M-H, Yang M-L, Kuan Y-H. 2019b. Epidemiological characteristics of postoperative sepsis. Open Med (War) 14: 928–938. 10.1515/med-2019-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker S, Sigl A, Grumaz C, Stevens P, Vainshtein Y, Zimmermann S, Weigand M, Hofer S, Sohn K, Brenner T. 2017. Immune-response patterns and next generation sequencing diagnostics for the detection of mycoses in patients with septic shock—results of a combined clinical and experimental investigation. Int J Mol Sci 18: 1796. 10.3390/ijms18081796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R, Ambrosini G, Périer R, Bucher P. 2015. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res 43: D92–D96. 10.1093/nar/gku1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Blumenfeld Y, Chitkara U, Hudgins L, Quake S. 2008. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA 105: 16266–16271. 10.1073/pnas.0808319105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson C, Weng Z. 2008. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138. 10.1371/journal.pgen.1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S, Batra S, Jain M. 2023. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol Cancer 22: 118. 10.1186/s12943-023-01813-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabuschnig S, Bronkhorst A, Holdenrieder S, Rosales Rodriguez I, Schliep K, Schwendenwein D, Ungerer V, Sensen C. 2020. Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int J Mol Sci 21: 8062. 10.3390/ijms21218062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gunne S, Heinicke U, Parnham M, Laux V, Zacharowski K, von Knethen A. 2020. Nrf2-A molecular target for sepsis patients in critical care. Biomolecules 10: 1688. 10.3390/biom10121688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Ni M, Chan R, Chan V, Lui K, Chiu R, Lo Y. 2020. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet 106: 202–214. 10.1016/j.ajhg.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F, David P, Akram M, Knoedler S, Mittelstädt A, Merkel S, Podolska M, Swierzy I, Roßdeutsch L, Klösch B, et al. 2023. Circulating monocytes serve as novel prognostic biomarker in pancreatic ductal adenocarcinoma patients. Cancers (Basel) 15: 363. 10.3390/cancers15020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, Cheng J, Murre C, Singh H, Glass C. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E, Auinger L, Speicher M. 2020. Cell-Free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med 26: 519–528. 10.1016/j.molmed.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Hinrichs A, Karolchik D, Baertsch R, Barber G, Bejerano G, Clawson H, Diekhans M, Furey T, Harte R, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano O, Ito T, Miura F. 2021. Short single-stranded DNAs with putative non-canonical structures comprise a new class of plasma cell-free DNA. BMC Biol 19: 225. 10.1186/s12915-021-01160-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudecova I, Smith C, Hänsel-Hertsch R, Chilamakuri C, Morris J, Vijayaraghavan A, Heider K, Chandrananda D, Cooper W, Gale D, et al. 2022. Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res 32: 215–227. 10.1101/gr.275691.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs A, Furey T, Roskin K, Sugnet C, Haussler D, Kent W. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmykov S, Yevshin I, Kulyashov M, Sharipov R, Kondrakhin Y, Makeev V, Kulakovskiy I, Kel A, Kolpakov F. 2021. GTRD: an integrated view of transcription regulation. Nucleic Acids Res 49: D104–D111. 10.1093/nar/gkaa1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukural A, Yukselen O, Ozata D, Moore M, Garber M. 2019. DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics 20: 6. 10.1186/s12864-018-5362-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy I, Vorontsov I, Yevshin I, Sharipov R, Fedorova A, Rumynskiy E, Medvedeva Y, Magana-Mora A, Bajic V, Papatsenko D, et al. 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustanovich A, Schwartz R, Peretz T, Grinshpun A. 2019. Life and death of circulating cell-free DNA. Cancer Biol Ther 20: 1057–1067. 10.1080/15384047.2019.1598759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrke M, Lazar M. 2005. The many faces of PPARgamma. Cell 123: 993–999. 10.1016/j.cell.2005.11.026 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fang X-D, Wang X-Y, Fei B-Y. 2018. Fos-like antigen 2 (FOSL2) promotes metastasis in colon cancer. Exp Cell Res 373: 57–61. 10.1016/j.yexcr.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth G, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu Y, Yue M, Li Z. 2021. FOSL1 promotes tumorigenesis in colorectal carcinoma by mediating the FBXL2/Wnt/β-catenin axis via Smurf1. Pharmacol Res 165: 105405. 10.1016/j.phrs.2020.105405 [DOI] [PubMed] [Google Scholar]
- Lo Y, Han D, Jiang P, Chiu R. 2021. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 372: eaaw3616. 10.1126/science.aaw3616 [DOI] [PubMed] [Google Scholar]
- Lukaszewski R, Jones H, Gersuk V, Russell P, Simpson A, Brealey D, Walker J, Thomas M, Whitehouse T, Ostermann M, et al. 2022. Presymptomatic diagnosis of postoperative infection and sepsis using gene expression signatures. Intensive Care Med 48: 1133–1143. 10.1007/s00134-022-06769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan I, Hong Y, Suryawanshi A, Angus-Hill M, Sun Z, Mellor A, Munn D, Manicassamy S. 2014. TLR2-dependent activation of β-catenin pathway in dendritic cells induces regulatory responses and attenuates autoimmune inflammation. J Immunol 193: 4203–4213. 10.4049/jimmunol.1400614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Masui T, Long Q, Beres T, Magnuson M, MacDonald R. 2007. Early pancreatic development requires the vertebrate suppressor of hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev 21: 2629–2643. 10.1101/gad.1575207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay R, Bailey T. 2010. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165. 10.1186/1471-2105-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan P, Dreos R, Ambrosini G, Groux R, Bucher P. 2020. EPD in 2020: enhanced data visualization and extension to ncRNA promoters. Nucleic Acids Res 48: D65–D69. 10.1093/nar/gkz1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan A, Pelleri M, Antonaros F, Strippoli P, Caracausi M, Vitale L. 2019. On the length, weight and GC content of the human genome. BMC Res Notes 12: 106. 10.1186/s13104-019-4137-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A, Hall I. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan D, Grüning B, Bhardwaj V, Kilpert F, Richter A, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2024. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reizis B. 2010. Regulation of plasmacytoid dendritic cell development. Curr Opin Immunol 22: 206–211. 10.1016/j.coi.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, McCarthy D, Smyth G. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh R, Sharkia I, Fialkoff G, Rahat A, Gutin J, Chappleboim A, Nitzan M, Fox-Fisher I, Neiman D, Meler G, et al. 2021. ChIP-seq of plasma cell-free nucleosomes identifies gene expression programs of the cells of origin. Nat Biotechnol 39: 586–598. 10.1038/s41587-020-00775-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck F, Rescheneder P, von Haeseler A. 2013. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29: 2790–2791. 10.1093/bioinformatics/btt468 [DOI] [PubMed] [Google Scholar]
- Smith-Raska M, Arenzana T, D'Cruz L, Khodadadi-Jamayran A, Tsirigos A, Goldrath A, Reizis B. 2018. The transcription factor Zfx regulates peripheral T cell self-renewal and proliferation. Front Immunol 9: 1482. 10.3389/fimmu.2018.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Kircher M, Hill A, Daza R, Shendure J. 2016. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164: 57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulz P, Perakis S, Zhou Q, Moser T, Belic J, Lazzeri I, Wölfler A, Zebisch A, Gerger A, Pristauz G, et al. 2019. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat Commun 10: 4666. 10.1038/s41467-019-12714-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzan L, Soldati H, Ythier V, Anand S, Braun S, Francis N, Murr R. 2021. High throughput screening identifies SOX2 as a super pioneer factor that inhibits DNA methylation maintenance at its binding sites. Nat Commun 12: 3337. 10.1038/s41467-021-23630-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang B, Li H, Zheng J, Zhang C, Yang Z, Tan X, Luo P, Ma L, Wang Y, et al. 2023. A small-molecule inhibitor of Keap1–Nrf2 interaction attenuates sepsis by selectively augmenting the antibacterial defence of macrophages at infection sites. EBioMedicine 90: 104480. 10.1016/j.ebiom.2023.104480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, He Q-Y. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nusbaum C, Myers R, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang S, Xi X, Zhang M, Liu X, Tang W, Cai P, Xing S, Bao P, Jin Y, et al. 2021. Integrative analysis of long extracellular RNAs reveals a detection panel of noncoding RNAs for liver cancer. Theranostics 11: 181–193. 10.7150/thno.48206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.