Abstract
The product of adenovirus (Ad) type 5 gene IX (pIX) is known to actively participate in the stability of the viral icosahedron, acting as a capsid cement. We have previously demonstrated that pIX is also a transcriptional activator of several viral and cellular TATA-containing promoters, likely contributing to the transactivation of the Ad expression program. By extensive mutagenesis, we have now delineated the functional domains involved in each of the pIX properties: residues 22 to 26 of the highly conserved N-terminal domain are crucial for incorporation of the protein into the virion; specific residues of the C-terminal leucine repeat are responsible for pIX interactions with itself and possibly other proteins, a property that is critical for pIX transcriptional activity. We also show that pIX takes part in the virus-induced nuclear reorganization of late infected cells: the protein induces, most likely through self-assembly, the formation of specific nuclear structures which appear as dispersed nuclear globules by immunofluorescence staining and as clear amorphous spherical inclusions by electron microscopy. The integrity of the leucine repeat appears to be essential for the formation and nuclear retention of these inclusions. Together, our results demonstrate the multifunctional nature of pIX and provide new insights into Ad biology.
Replication-deficient adenoviruses (Ad) (20) efficiently transfer and express candidate therapeutic genes into a variety of dividing and postmitotic cell types (6, 27, 51, 57). For these reasons such viruses constitute effective vectors for direct in vivo gene therapy (5, 16, 19, 24). However, several drawbacks, such as toxicity, host inflammatory response (14), or transient in vivo transgene expression (18, 41), impair the full success of Ad vectors in human gene therapy protocols. Multiple factors are involved, among which some viral proteins whose functions are often not fully understood.
We focused our attention on the study of the product of gene IX (pIX) from Ad serotypes 2 and 5 (Ad2 and Ad5) (3, 8). Protein pIX is a small polypeptide of 140 residues (14.3 kDa) that is incorporated into the mature viral capsid. It is associated with hexon proteins to form group-of-nine hexons (GON) that make up the central region of each facet of the icosahedron (8, 10). Precise determination of the stoichiometry of this assembly has revealed that there are 12 molecules of pIX, organized as four trimers per GON, and therefore 240 molecules per virion (56, 59, 60). The protein acts as a capsid cement and thereby enhances the thermal stability of the virions (17, 23). It is essential for packaging 100% and more of the full-length Ad DNA (25). By themselves, these properties of pIX appear to be important enough to be taken into consideration during the design of Ad vectors.
Additional observations strongly suggest that pIX is more than a capsid protein and may serve additional functions during the infectious cycle (44): (i) gene IX is the only structural protein coding gene which is uncoupled from the Ad major late promoter (MLP); (ii) its expression pattern follows a different time course and begins at intermediate times postinfection (p.i.), much earlier than that of all the other structural proteins; (iii) finally, pIX accumulates in the infected cell nuclei with a speckled distribution. In agreement with this nuclear localization, we have previously shown that pIX is a transcriptional activator of several viral and cellular TATA-containing promoters, among which are the Ad E1a, E4, and MLP promoters (44). We therefore hypothesized that pIX could be involved in the transactivation of Ad genome expression.
To precisely delineate the functional domains of pIX responsible for the structural and transcriptional properties, we performed an extensive mutational analysis of the pIX coding sequence. We show that the highly conserved N-terminal part of the protein is essential for the structural properties of the capsid, whereas the C-terminal leucine repeat (putative coiled-coil domain) is critical for the transactivating function. Accumulation of pIX results in the formation of specific nuclear structures (the clear amorphous [c.a.] inclusions), the function of which is presently unknown. Our results suggest that formation of these structures involves self-assembly of pIX through its coiled-coil domain.
MATERIALS AND METHODS
Cells and viruses.
Monolayer human A549 (54) cells were grown in Dulbecco medium supplemented with 10% fetal calf serum (FCS). 293 cells (28) were grown in Dulbecco modified Eagle medium with 2% FCS. A549 cells (at 80% confluence) were infected with wild-type (wt) Ad2 or Ad5 at a multiplicity of infection (MOI) of 50 PFU per cell. Mutant viral genomes were constructed as infectious plasmids by homologous recombination in Escherichia coli, as described elsewhere (12). All vectors contain, in addition to alterations of gene IX (see below), a deletion in E1 (between nucleotides 459 and 3331) and in E3 (between nucleotides 28592 and 30470) (Ad E1° E3°) (41). Nucleotide numbering throughout this study conforms to that of Chroboczek et al. (15). Mutant viruses were amplified in 293 cells. Viral growth, purification, titration, and storage were previously described (27, 38, 41).
Recombinant eukaryotic expression vectors.
The sequence encoding wt pIX was derived from the Ad5 genome by PCR amplification as previously described (44) and inserted into three types of expression vectors: (i) the pAT4 vector (gift from M. Vigneron), in a site located 3′ to the sequences encoding the F domain of the human estrogen receptor (hER) (4), generating a wt pIX fusion protein tagged at its N terminus (F/IX); (ii) the pXJ41 vector (61), which yields an untagged protein; (iii) the pXX vector, a pSG5-derived vector (29), in which the sequence encoding wt pIX was inserted in a site located 5′ to the sequences encoding the F domain of the hER, generating a pIX fusion protein tagged at its C terminus (IX/F). In these vectors, the expression of the wt or mutated pIX sequences was directed by the cytomegalovirus enhancer and herpes simplex virus type 1 (HSV-1) thymidine kinase gene promoter (pAT4 and pXJ41), or the simian virus 40 promoter (pSG5-derivative pXX).
The Ad E1a promoter sequence (positions +100 to +560, with numbering according to the study of Chroboczek et al.) was subcloned in front of the chloramphenicol acetyltransferase (CAT) reporter gene of the promoterless pBLCAT6 vector, as previously described (44).
Point mutations and deletions in the pIX coding sequence (as indicated in Fig. 2A) were generated by following the protocol of the QuickChange Site-Directed Mutagenesis system (Stratagene). All plasmids were verified by sequencing.
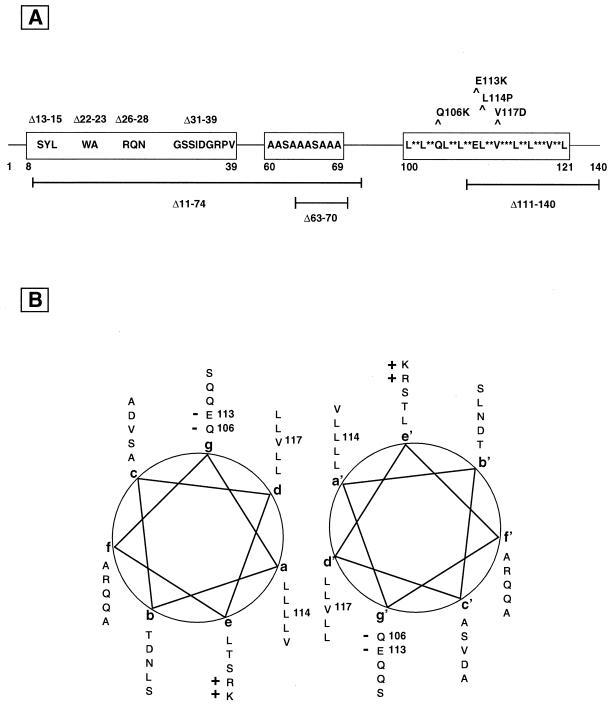
FIG. 2.
Schematic representation of specific subdomains of pIX. (A) The conserved pIX sequence domains, including the central human-specific polyalanine stretch, are represented as boxes, with relevant peptidic elements and coordinates from the Ad2 or Ad5 wt sequence. Point mutations or small deletions are indicated above, while larger deletions mentioned in the text are depicted below. (B) The predicted helical-wheel representation of the C-terminal leucine repeat of Ad2 or Ad5 pIX, from residues 100 (first L at position a) to 134 (S at position g) is shown next to a symmetrically positioned wheel of the same region. The potential hydrophobic interactions (a-d′ and d-a′) between the two helices are suggested by the alignments of the residues at positions a and d with positions d′ and a′, respectively, in a putative coiled-coil structure. The charged residues (“+” or “−”), which presumably stabilize these interactions, are indicated. Residues (with corresponding coordinates) that are altered by site-directed mutagenesis are indicated.
Transfections, cell extracts, and Western blotting.
A549 cells were transfected by calcium phosphate coprecipitation (13). For CAT assays, the cells were harvested 36 h after transfection, extracts were prepared, and aliquots, normalized by protein concentration, were assayed for CAT activity as described earlier (7). CAT activities were determined from at least three independent experiments and quantitated with a Bioimaging analyser (Fuji Photo Film Co.).
For immunoprecipitations, the cells were harvested 36 h after transfection by three cycles of freeze-thaw in buffer A (50 mM Tris-HCl, pH 7.9; 20% glycerol; 1 mM dithiothreitol; 0.1% NP-40) containing 0.4 M KCl. The expression of recombinant proteins was verified by Western blotting. After an additional clearing step on protein G-Sepharose to adsorb nonspecific binding proteins, cell extracts were incubated for 2 h with 1 μg of the anti-F antibody, after which 30 μl of protein G-Sepharose beads were added and incubation was continued for an additional 2 h. The beads were then washed three times at room temperature with buffer A containing 200 mM KCl and 0.5% NP-40 (mild-salt conditions). The resin was then dissociated by boiling for 5 to 10 min in sodium dodecyl sulfate (SDS) sample buffer. The bound proteins were detected on Western blots with specific antibodies using the ECL System (Amersham), as previously described (7). Anti-pIX rabbit polyclonal antibodies were raised against purified recombinant glutathione S-transferase–IX fusion protein (anti-pIX) (44). Monoclonal antibodies against the F domain of the hER (Mab3A6) have been described (44). Monoclonal antibodies against the Ad5 penton base were provided by Transgene (Strasbourg).
Electron microscopy.
A549 cells near confluence were infected at an MOI of 5 to 10 PFU of Ad5 per cell for 30 min. Monolayers were then rinsed with phosphate-buffered saline (PBS), fresh medium was added, and the cells were reincubated for 16 and 30 h before fixation. A549 cells were transfected with the vector generating the untagged wt pIX and cultured for 36 h. The cells were fixed with 4% formaldehyde (Merck) in 0.1 M Sörensen's phosphate buffer (pH 7.2) at 4 to 8°C for 1 h. During the fixation step, the cells were scraped from their plastic substrate and centrifuged. The resulting pellets were dehydrated in increasing concentrations of methanol and embedded in Lowicryl K4M (Polysciences Europe GmbH). Polymerization was performed at −30°C for 5 days under long-wavelength UV light (Philips fluorescence tubes, TL 6W) and subsequently at room temperature for 1 day. Ultrathin sections were collected on Formvar-carbon-coated copper grids (mesh 200).
For identifying structures containing the pIX viral protein, grids bearing Lowicryl sections were floated for 2 min over drops of Aurion BSA-C (purchased from Biovalley) (0.01% in PBS) in order to prevent background, with prior incubation (30 min at room temperature) in the presence of anti-pIX polyclonal antibody diluted 1/50 in PBS. After a rapid washing in PBS, grids were incubated at room temperature for 30 min in the presence of goat anti-rabbit immunoglobulin G (IgG) conjugated to gold particles (10-nm diameter; British Biocell International, Ltd., Cardiff, United Kingdom) and stained with uranyl acetate prior to observation with a Philips 400 transmission electron microscope at 80 kV and 6,000- to 22,000-fold magnification. To make sure that secondary antibody did not bind nonspecifically to biological material, we verified that no labeling occurred when primary antibody was omitted.
Immunofluorescence.
Immunofluorescence staining experiments were carried out as previously described (44). A549 cells were fixed with formaldehyde (2% [vol/vol] in PBS) and permeabilized with 0.1% Triton X-100 in PBS. The primary antibodies were diluted in PBS containing 0.1% Triton X-100. The anti-pIX rabbit polyclonal antibody was used as described elsewhere (44) and the Mab3A6 anti-F antibody was used at a 1/5,000 dilution in PBS containing 0.1% Triton X-100. After incubation for 1 h, the coverslips were washed several times in PBS–0.1% Triton X-100 and then incubated with goat Texas red-conjugated anti-rabbit IgG and/or donkey fluorescein isothiocyanate (FITC)-labeled anti-mouse IgG (Sigma) at concentrations recommended by the suppliers. Nuclei were counterstained with Hoechst 33258. After the staining, the coverslips were mounted and analyzed using a confocal laser scanning microscope (Leica). Image enhancement software was used to balance signal strength, and eightfold scanning was used to separate signal from noise.
RESULTS
Peptide sequence and functional domains of pIX.
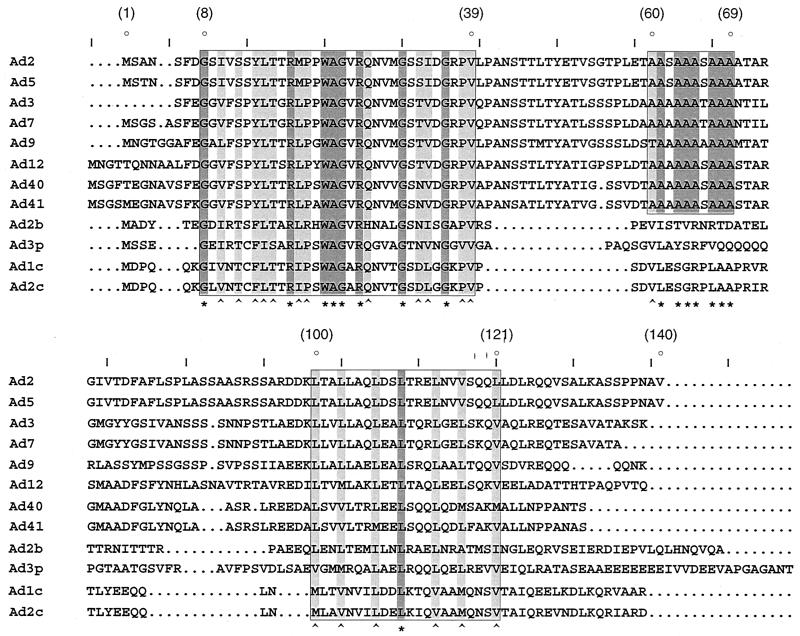
Multiple sequence alignments between pIX proteins from human and animal Ad serotypes (Fig. 1) revealed a high degree of identity (95%) over the entire length of pIX from serotypes belonging to the same subgenus. Although the extent of homology between serotypes from different species was lower, two conserved domains could be identified when human and animal serotypes were compared: upon referring to the coordinates of pIX residues (Aa) from the human Ad2 serotype, these domains can be located at the N-terminal (Aa8-39) and C-terminal (Aa100-121) ends of pIX, respectively. An additional, alanine-rich domain, specific to the human serotypes, could be delineated (Aa60-69).
FIG. 1.
Conserved sequence elements in pIX. Amino acid sequence alignments (CLUSTAL X) (58) of pIX from several human (top 8 sequences) and animal (bovine [b], porcine [p], and canine [c]) Ad serotypes, as indicated on the left, were performed. Accession numbers are as follows: Ad2 (p03282), Ad5 (p03285), Ad3 (J01962), Ad7 (03283); Ad9 (q9yl97), Ad12 (03284), Ad40 (p48312), Ad41 (p32539), Ad2b (q65377), Ad3p (q9w9x3), Ad1c (q65944), and Ad2c (p14268). Dots correspond to gaps inserted by the program to optimize alignments. Conserved sequence elements are boxed. The symbols “∗” and “^” (bottom) denote identical or related amino acid residues, respectively. Numbers in parentheses (top) refer to coordinates of amino acids in Ad2 and Ad5 pIX, relative to the starting methionine.
No particular structural motif could be identified within the N-terminal domain. The alanine-rich stretch, which is unlikely to adopt any particular structure, may serve as a flexible link between the two halves of the pIX molecule. In contrast, the C-terminal domain clearly revealed features of a leucine-repeat (or coiled-coil domain), as suggested by the helical wheel representation (Fig. 2B): 10 nonpolar amino acid residues (leucine and valine), spaced every three and four residues, align at positions a and d on one side of the wheel; these residues presumably provide a hydrophobic interface to interact with similar residues, symmetrically positioned (a′ and d′) on a second monomer (Fig. 2B), thereby potentially adopting a coiled-coil conformation (36, 37, 39, 40). Moreover, in the case of Ad2 and Ad5, ionized residues of opposite charge, located on either side of the helical wheel at positions e and g, may further stabilize protein assembly by symmetrically interacting with corresponding residues at positions e′ and g′ (35).
To determine the functional significance of these domains, the effects of a series of deletions or point mutations altering the conserved sequence elements were examined (Fig. 2A). Within the N-terminal half of pIX, amino acid stretches that are conserved between all (Aa13-15, Aa22-23, Aa26-28, and Aa31-39) or only human Ad serotypes (Aa63-70) were deleted (Δ). The C-terminal leucine repeat was disrupted by changing, either separately or simultaneously, leucine-114 and valine-117 to proline and aspartate, respectively, to generate mutant L114P, V117D, or L114P-V117D (L-V). Interruptions of the apolar series at positions a and d by these residues were indeed expected to disturb the correct alignment (proline) or hydrophobic bonding (aspartate) (36, 37, 39, 40). Two mutated forms of pIX were also constructed in which the net charge at position e was inverted by exchanging Aa106 or Aa113 with a lysine residue (mutants Q106K and E113K, respectively), thereby triggering electrostatic repulsion between protein monomers (26, 35).
The N-terminal part of pIX critically contributes to its incorporation into the capsid.
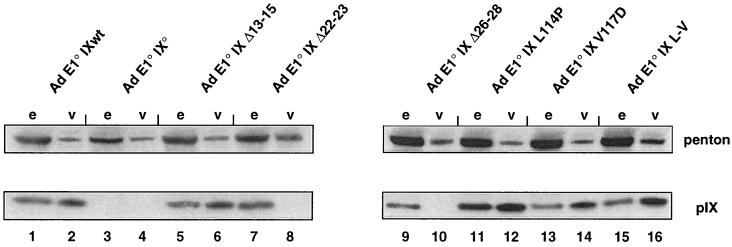
Earlier immunoelectron microscopy studies with purified Ad virions and pIX-specific antisera (2) revealed that only the C-terminal portion of pIX was accessible to the antibodies. The authors of that study concluded that the C-terminal part of the protein was exposed at the surface of the virions, whereas its N-terminal domain was hidden inside the viral capsid. To further define the pIX elements involved in capsid assembly, we introduced a set of mutations into the E1-deleted viral genome by homologous recombination to generate viruses expressing the pIX variants during viral production. The mutations were designed to alter sequence elements that are most strictly conserved within both human and animal serotypes (see above). Because all recombinant viruses are E1 defective, they were produced on the 293 cells which constitutively express the E1a and E1b Ad5 genes (28). Each pIX mutant virus was grown on 293 cells, viral particles (Fig. 3, v) were purified, and equal quantities were submitted to SDS-PAGE, in parallel with equal quantities of the corresponding infected crude extracts (Fig. 3, e). As a control, viral particles and extracts were assayed by Western blotting for the presence of a viral protein (penton base) that was expressed to equal levels by all variants, irrespective of pIX alteration. The presence of pIX was examined by Western blot analysis, using anti-pIX antibodies. As an additional control, we verified that the wt pIX protein was present both in infected-cell extracts and in purified Ad E1° IXwt virions (Fig. 3, lanes 1 and 2), whereas it was absent from both fractions, in the case of a virus (Ad E1° IX°) lacking gene IX (Fig. 3, lanes 3 and 4).
FIG. 3.
Integrity of the conserved N-terminal domain of Ad2 pIX is required for incorporation into the capsid. CsCl-purified Ad5 E1° virions (2 × 1010 particles of each recombinant virus) expressing either wt pIX (Ad E1° IXwt), no pIX (Ad E1° IX°), or specific pIX variants (as indicated on the top) were disrupted by boiling in SDS sample buffer, fractionated by SDS–10% PAGE, and analyzed by immunoblotting using monoclonal anti-penton (top) and polyclonal anti-pIX (bottom) antibodies (even “v” lanes). Extracts were prepared (44) from 293 cells that had been infected by the same viruses (MOI of 20 PFU per cell) and collected at 36 h pi. Aliquots were analyzed by immunoblotting (odd “e” lanes) next to the corresponding virions. Signals corresponding to penton and pIX are indicated.
Mutations altering pIX within its C-terminal part (L114P, V117D, and L-V) did not prevent incorporation of the mutant protein into the capsid (Fig. 3, lanes 12, 14, and 16), indicating that the integrity of the leucine repeat is not required for this function. Moreover, capsid stability (assessed as in references 17 and 22) appeared not to be altered (data not shown). In contrast, deletions within the conserved N-terminal domain of pIX, such as in mutants Δ22-23 and Δ26-28, completely abolished incorporation of pIX into the capsid (Fig. 3, lanes 8 and 10, respectively), despite nearly normal levels of pIX synthesis (Fig. 3, lanes 7 and 9, respectively). Mutation (Δ13-15) did not impair virion insertion of pIX (Fig. 3, lane 6), but the resulting capsid was less stable, as indicated by thermolability measurements (assessed as in references 17 and 22) (data not shown).
Together, these results define the N-terminal region spanning Aa22-28 as crucial for the correct and stable recruitment of pIX into the viral capsid, whereas the C-terminal region is not involved at all.
The integrity of the C-terminal leucine repeat and central alanine stretch of pIX are essential for its transcriptional activity.
We previously showed that pIX exhibits transcriptional properties (44). Recombinant pIX efficiently stimulated, in a dose-dependent manner, the activity of several viral and cellular TATA-containing promoters. To precisely delineate the transactivating domain of pIX, we examined the effect of the complete set of pIX mutations on E1a promoter activation (Fig. 4). To this end, vectors expressing wt or mutated pIX sequences as proteins fused at their N termini (F/IX) or C termini (IX/F) to the F epitope tag were transfected together with a CAT reporter gene driven by the E1a promoter. After we verified that equal levels of pIX were expressed, as revealed by immunoblotting with antibodies against the F epitope (data not shown), the CAT activities were measured. Under these conditions, relative CAT activities will reflect the intrinsic transcriptional activating capacity of each recombinant protein (44).
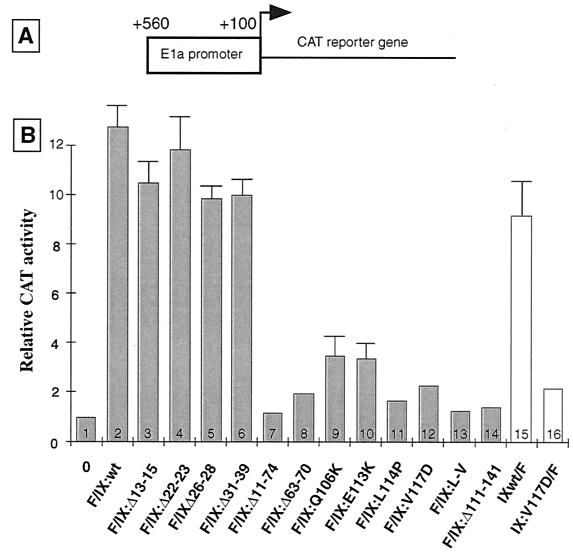
FIG. 4.
The leucine repeat and central hinge region are crucial for Ad2 pIX transcriptional activity. (A) Structure of the chimeric pE1a-CAT reporter plasmid in which the promoter region of the Ad5 E1a transcription unit (44) was fused to the CAT gene. (B) A549 cells were transfected with 1 μg of the pE1a-CAT reporter plasmid, either alone (column 1) or together with plasmids expressing the wt or mutated pIX as F-tagged fusion proteins: F/IX derivatives (0.1 μg; column 2 and columns 3 to 14, respectively) or IX/F derivatives (0.5 μg; columns 15 and 16, respectively). Cells were collected 36 h later, and extracts were prepared. Relative CAT activities (means from three independent experiments) are represented with corresponding standard deviations.
In agreement with our earlier structural analysis (44), truncated versions of pIX lacking half of the leucine repeat (F/IX:Δ111-140) or most of the N-terminal half (F/IX:Δ11-74) of the protein, lost their transactivating property (compare in Fig. 4B, columns 2 and 14 or 7, respectively)
Point mutations within the sequence encoding the leucine repeat in the C-terminal end of pIX severely reduced reporter stimulation: single (L114P and V117D) or double (L-V) alterations had effects similar to complete deletion (Δ111-140) of the C-terminal part of the protein (Fig. 4B, compare column 2 with columns 11 to 14). Point mutations Q106K and E113K also reduced transactivation (Fig. 4B, compare column 2 and columns 9 to 10), stressing the contribution of electrostatic interactions in functional assembly of pIX monomers. The transactivation function of pIX also depends on the integrity of the central domain, since deletion of the corresponding polyalanine stretch (Δ63–70) led to a strong reduction of reporter stimulation (Fig. 4B, compare columns 2 and 8). In contrast, deletions within the N-terminal part of pIX had no detectable effect on its intrinsic stimulatory activity, since very similar levels of transactivation were obtained with F/IX:Δ13-15, F/IX:Δ22-23, F/IX:Δ26-28, and F/IX:Δ31–39 pIX variants and the wt protein (Fig. 4B, compare column 2 and columns 3 to 6). Very similar results were obtained with vectors expressing C-terminally instead of N-terminally tagged pIX derivatives (Fig. 4B, compare columns 15 and 16 with columns 2 and 12, respectively; also data not shown).
Together, our results suggest that the pIX transactivating function is dependent on the integrity of the C-terminal leucine repeat, as well as on the central alanine-rich element. Interestingly, the N-terminal region, critically involved in the capsid integration of pIX, is not involved in this function.
The integrity of the C-terminal leucine repeat and central alanine stretch of pIX are essential for its self-interaction.
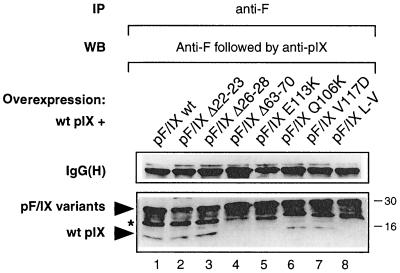
The presence of a leucine repeat type of structure at the C-terminal end of pIX suggests that the protein may dimerize (or multimerize) by interacting through this element (36, 37). To test this possibility, a vector expressing the nontagged wt pIX was cotransfected into A549 cells with vectors expressing F epitope-tagged wt or mutant pIX proteins (F/IX). As revealed by Western blot analysis of cell extracts with monoclonal anti-F or polyclonal anti-pIX antibodies, the transfected vectors were expressed to very similar levels (data not shown).
When these extracts were immunoprecipitated with the monoclonal anti-F antibody under mild-salt conditions and subjected to SDS-polyacrylamide gel electrophoresis (PAGE), a single band of nontagged wt pIX protein, with the expected size, was revealed with the anti-pIX antibody, as indicated in Fig. 5, in addition to the F-tagged protein variants. As expected, the band corresponding to the nontagged pIX was not revealed by the anti-F antibody which was applied first on the blot (see legend to Fig. 5), ruling out the possibility that it may correspond to a proteolytic degradation product of the F-tagged pIX. As an additional control, we verified that, when expressed in the absence of F-tagged pIX, the nontagged pIX was not detected in the anti-F immunoprecipitate (not shown). The results indicate, therefore, that a detectable fraction of the nontagged wt pIX was coprecipitated together with the F epitope-tagged wt pIX (F/IXwt; Fig. 5, lane 1). The relative weakness of the band corresponding to nontagged pIX, compared to that of the F-tagged protein, could be due in part to the fact that self-association of the F-tagged proteins may compete for heteromeric interactions. That this coprecipitation involved the leucine repeat domain was demonstrated by our observation that single point mutations (E113K, Q106K, and V117D) or a double point mutation (L-V) disrupting this structure reduced or abolished the carrying effect (Fig. 5, compare lane 1 with lanes 5 to 8). Similarly, a mutant lacking the C-terminal mid-part of the leucine repeat (Δ111–140) did not coprecipitate wt pIX (data not shown). As expected, mutations within either N-terminal domain (Δ22-23 and Δ26-28 [Fig. 5, compare lane 1 with lanes 2 and 3] or Δ13-15 and Δ31-39 [not shown]) had no effect on the interaction. In contrast, the central alanine-rich element (Δ63–70) of pIX was essential for coprecipitation (Fig. 5, compare lanes 1 and 4) and is therefore also involved in the oligomerization process. Furthermore, our observation that the same mutations affect both transactivation and interaction properties of pIX suggests that the two activities may be linked.
FIG. 5.
Self-interaction of pIX. A549 cells were transfected with vectors expressing the wt pIX, together with vectors expressing N-terminally F-tagged wt or mutated pIX proteins, as indicated. Cell extracts were prepared, and aliquots were immunoprecipitated (IP) with monoclonal antibodies directed against the F epitope. The immunoprecipitates were subjected to Western blot analysis (WB), with probing first with the anti-F monoclonal antibody. After exposure, the same blot was washed and reprobed with polyclonal antibodies against pIX. The position of bands corresponding to the untagged and F-tagged wt or mutated pIX are indicated. The bands marked by the asterisk likely correspond to proteolytic breakdown products of the F-tagged derivatives. The position of the IgG heavy subunit [IgG(H)] is indicated to show that equal amounts of antibody were used in the immunoprecipitation reaction. Numbers on the right correspond to the positions of molecular size markers (in kilodaltons).
Protein pIX accumulates in virus-induced clear amorphous inclusions.
We have previously shown (44), by immunofluorescence staining with pIX-specific antibodies, that pIX was predominantly associated with infected cell nuclei, in accordance with the transcriptional properties of the protein. In addition, we observed that the nuclear staining of pIX was dynamic, showing a speckled distribution at later times of infection (unpublished observation). To study more precisely the intranuclear distribution of pIX protein, Ad5-infected A549 cells were examined by immunoelectron microscopy at different times p.i. from 16 to 30 h p.i., in order to observe the accumulation of pIX as a function of the successive steps of nuclear alteration during infection (46–49).
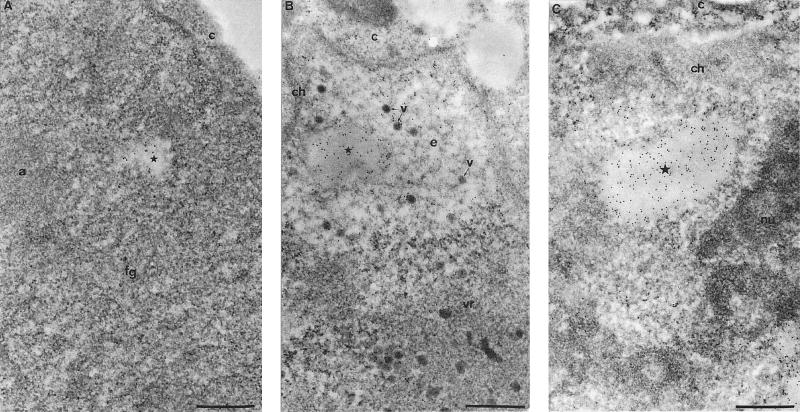
No significant labeling was observed with the anti-pIX antibody before 16 h p.i. (not shown). At this time, the infected cells were mainly at the intermediate stage of nuclear alteration (49). Among several virus-induced structures (including sites of viral DNA replication or transcription and sites of viral genome or single-stranded viral DNA accumulation) (49), pIX was detected in small, irregularly shaped or spherical c.a. inclusions (Fig. 6A).
FIG. 6.
Protein pIX actively induces specific nuclear c.a. inclusions. (A) Ad2-infected A549 cells at the intermediate stage of nuclear transformation (14 to 16 h pi) (49) were processed for immunogold labeling with anti-pIX polyclonal antibody on Lowicryl sections of formaldehyde-fixed cells. Gold particles are scattered over the fibrillogranular network (fg), one component of the viral region, and accumulate over an enclosed small irregularly shaped clear amorphous inclusion (c.a. inclusion; star). The accumulation site of viral single-stranded DNA (a), the other compartment of the viral region, is entirely devoid of pIX protein. c, cytoplasm. Bar, 0.5 μm. (B) Late stage of Ad-mediated nuclear transformation (24 to 30 h p.i.) (49) are characterized by the presence of progeny viruses. The roughly spherical c.a. inclusion (star) is intensely and homogeneously labeled. It is located in the electron-translucent region (e) which separates the perinuclear layer of host chromatin (ch) from the large, centrally located viral region (vr). Some viruses (v), both scattered in the electron-translucent region and clustered within the viral region are labelled. c, cytoplasm. Bar, 0.5 μm. (C) Overexpression of recombinant pIX protein induces the accumulation of the protein within newly formed c.a. inclusions: A549 cells were transfected with the vector expressing the untagged wt pIX. Gold particles accumulate over the entire surface of an ovoid c.a. inclusion present in the nucleoplasm. It clearly appears that the labelled inclusion (star) is similar to those observed in panel B following Ad infection. c, cytoplasm; ch, perinuclear layer of condensed chromatin; nu, nucleolus. Bar, 0.5 μm.
At a later stage (24 to 30 h p.i.), when a central viral compartment and a perinuclear electron-translucent area with protein crystals and isolated viruses were apparent (47, 49), pIX remained concentrated within the c.a. inclusions, which became more frequent and spherical (Fig. 6B). Sometimes two or three of these inclusions were juxtaposed (unpublished data). In addition, pIX was observed over the crystalline arrays of viruses and the isolated viruses particles (Fig. 6B).
Hence, since pIX is efficiently neosynthesized and belongs the late phase of infection, the protein is predominantly associated with c.a. inclusions that are dynamic in their shape and location in the nucleus. They grow in size as the infection progresses, as the pIX protein accumulates into the nucleus. Irrespective of their shape, size, and location within the nucleus, they were always intensely and homogeneously labeled with the anti-pIX antibody.
Overexpression of pIX induces by itself the formation of c.a. inclusions.
Our ultrastructural data suggest that pIX is the main component of the virus-induced c.a. inclusions. To determine whether pIX was directly responsible for their occurrence, we attempted to localize the protein in cells transfected with a wt pIX expression vector, i.e., in the absence of any other viral protein. The morphology of pIX-expressing cells was similar to that of nontransfected cells except for the additional presence in the nucleoplasm of c.a. inclusions (Fig. 6C) identical to those observed in lytically infected cells (Fig. 6B). Depending on the amount of expressed pIX (i.e., as a function of time posttransfection), c.a. inclusions were variable in size and frequency but always showed the same amorphous aspect. Immunogold detection of pIX protein resulted in an intense labeling of each c.a. inclusion and in a slight labeling of the surrounding nucleoplasm and cytoplasm. Therefore, in the absence of other viral proteins, pIX is able to induce the formation of c.a. inclusions similar to those induced by Ad infection.
Formation of c.a. inclusions requires the integrity of the pIX leucine repeat.
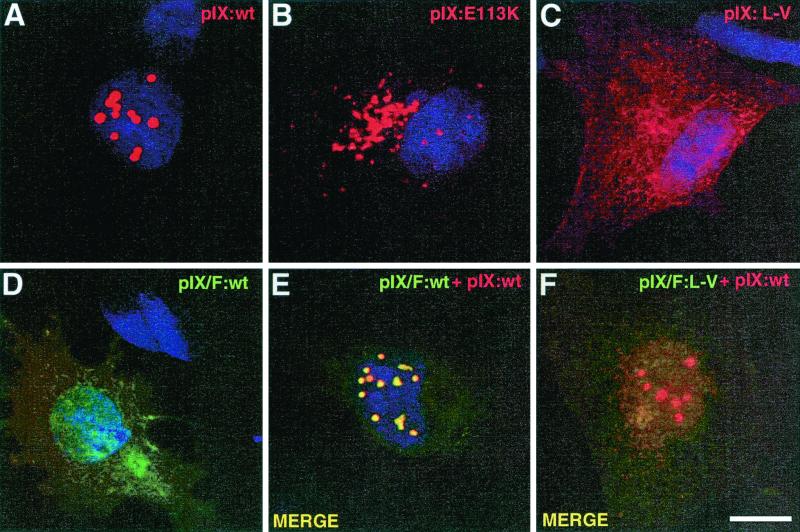
Specific immunofluorescence staining of cells infected with wt Ad5 (unpublished data) or transfected with the wt pIX-expressing vector revealed a speckled distribution of the protein in the nucleus (Fig. 7A), most likely corresponding to the accumulation of pIX within the c.a. inclusions observed by electron microscopy. To identify peptidic domains of pIX which may be responsible for the formation of c.a. inclusions, we examined the effect of our set of mutations on the nuclear distribution of pIX in transfected cells. Mutations affecting either the N-terminal (Δ13-15, Δ22-23, Δ26-28, and Δ31-39) or central (Δ63-70) domains of pIX did not affect the formation and nuclear location of the c.a. inclusions, since the corresponding mutant proteins all yielded the same speckled distribution as had the wt pIX upon transfection (data not shown). In contrast, alteration of the leucine repeat of pIX by modification of the net charge of specific residues (mutations E113K or Q106K) drastically changed the intracellular distribution of the corresponding pIX variants which were confined to the cytoplasm, as revealed by immunofluorescence staining (Fig. 7B and data not shown). Furthermore, although the overall level of mutant expression was similar to that of the wt protein (as revealed by Western blotting [data not shown]), these variants accumulated with a microspeckled pattern.
FIG. 7.
Formation of the nuclear inclusions is dependent on the integrity of the pIX leucine repeat. A549 cells were transfected with plasmids encoding the untagged and/or F-tagged wt or mutated pIX, as indicated. At 36 h posttransfection, cells were processed for immunofluorescence staining with polyclonal anti-pIX antibodies and goat Texas red-conjugated anti-rabbit IgG (A to C and E and F), and/or with mouse anti-F antibody and donkey FITC-labeled anti-mouse IgG (D to F). Nuclei were counterstained with Hoechst 33258. Panels E and F are the merged confocal images obtained with each fluorochrome separately. Bar, 10 μm.
Similarly, both point mutations (L114P, V117D, and L-V) and deletions (Δ111-140) affecting the integrity of the leucine repeat abolished formation of the c.a. inclusions and resulted in a diffuse distribution of the altered pIX throughout the nucleus and cytoplasm (Fig. 7C and data not shown). Fusion of the F epitope tag at the C-terminal end of wt pIX (IX/F) also prevented the nuclear accumulation of the protein and induced the same diffuse pattern, despite the integrity of the leucine repeat (Fig. 7D). This effect, most likely related to the steric hindrance imposed by the tag, suggests that free access to the leucine repeat was essential for nuclear retention of pIX and c.a. inclusion formation. Interestingly, when cells were cotransfected with vectors expressing the untagged (wt pIX) and C-terminally F-tagged (IX/F) proteins, both types of proteins accumulated within the same inclusion bodies, as revealed by the merged immunofluorescence staining (Fig. 7, compare panels D and E). It appears, therefore, that the unfocused IX/F protein distribution was redirected by the coexpressed wt pIX into its corresponding nuclear accumulation sites. In contrast, when the F-tagged partner of the cotransfection carried an alteration within the leucine repeat (IX/F:L-V), this recruitment was nearly abolished, as shown by the persistent diffuse distribution pattern of the F-tagged mutant (Fig. 7F).
Together, our results clearly indicate that the leucine repeat of pIX is chiefly involved in the formation and nuclear retention of the c.a. inclusions. Furthermore, our data strongly suggest that the assembly of the pIX-specific inclusion bodies is an active process driven by pIX itself, most likely through self-assembly.
DISCUSSION
Viruses, as obligatory cell parasites, usually evolved toward the highest possible degree of simplification of their structure and components to reach at minimal expense the most efficient rates of proliferation. Ad comply with this rule, not only in exploiting the coding capacity of their genome by using alternative reading frames but also in producing proteins with multiple biological activities. The product of the Ad gene IX is an example of such multifunctional proteins: pIX (140 residues) is a structural component of the viral capsid, acts as a transcriptional activator, and accumulates in infected cell nuclei as specific structures (c.a. inclusions), the function of which remains to be established. In the present study, we have performed extensive site-directed mutagenesis to define the corresponding functional domains of the protein.
Structural involvement of pIX in viral capsid assembly.
The pIX protein has previously been described as a capsid cement between the viral hexons (8, 10), thereby optimizing DNA packaging capacity and thermal stability of Ad virions (17, 23). Based on immunochemical approaches, it had been suggested that the N-terminal portion of pIX sticks inside the capsid, while its C-terminal part points outward (2). Our present results indicate that residues 22 to 28 are essential for pIX incorporation into the capsid and for virion thermostability. The importance of these residues is further supported by their complete conservation among human and animal Ad serotypes. Additional residues within the N-terminal region of pIX likely contribute to this function, as suggested by the effect of the Δ13-15 deletion, which did not impair pIX integration into the capsid but affected virion stability.
Interestingly, our results rule out any contribution of the putative coiled-coil element of pIX to these capsid properties. This clearly indicates that the corresponding capsid interactions occur through elements other than the coiled-coil domain and suggests that the N-terminal element defined above fulfills this function. Since coiled-coil elements have been shown to be responsible for the trimerization or oligomerization of other proteins (36, 37, 39, 40), like the well-documented yeast GCN4 (30, 31) or human immunodeficiency virus type 1 gp41 (53), it is likely that the coiled-coil element of pIX might also be involved in the trimerization of this protein in the GON complexes, as in fact suggested by our immunoprecipitation experiments. This raises the question of whether the pIX molecules are still organized as trimers in the leucine repeat mutants or whether the N-terminal domain of pIX, in close contact with the hexon molecules, is primarily responsible for the integration of pIX trimers into the virion capsid. Clearly, additional experiments, including coimmunoprecipitation assays and three-dimensional structure analyses, will be required to solve these questions. Our conclusions (i) that only the N-terminal part of pIX is implicated in its integration into the virion capsid, (ii) that its C-terminal end points outward, and (iii) that this extremity could be modified without altering the overall structural properties of the molecule open up the potential to hook ligands of interest onto this C-terminal end in order to modify the cellular tropism of the virus.
Transcriptional activity of pIX.
We have previously reported that recombinant pIX exhibits properties of a transcriptional activator when assayed in transfection experiments or in a reconstituted in vitro transcription system (44). Here we show that the integrity of the leucine repeat element of pIX is mandatory for this transactivating function, while the highly conserved residues within the N-terminal half of the molecule are not associated to this activity. Larger deletions in the N-terminal region (Δ11-74) or just removing the alanine-rich element (Δ63-70) also impaired pIX transcriptional activity, despite preserving the coiled-coil domain. As suggested by predictive structure analyses (data not shown), the polyalanine stretch may serve a hinge function in the pIX molecule. It is therefore possible that mutations targeting this element disrupt the global structure of pIX and thereby affect its essential functions.
Additional evidence (M. Rosa-Calatrava et al., unpublished data) revealed that, soon after cell entry and virion decapsidation, the released capsid pIX accumulates in the cell nucleus, as soon as 45 min p.i. until 6 h p.i. These pIX molecules may then activate the highly responsive E1a promoter, thus behaving as a ready-to-use transactivator, much like the VP16 factor in the case of HSV-1 infection (1). Furthermore, it is likely that the newly synthesized pIX, which starts accumulating at intermediate times p.i., also contributes to the activation of the late phase of Ad infection by stimulating MLP activity. Thus, it will be of interest to decipher the molecular mechanism of pIX-mediated transactivation.
As expected from the absence of basic residues flanking its leucine repeat (to make up a bona fide basic-leucine-zipper) (36, 37) and from the lack of any other DNA-recognizing motif, pIX has no DNA-binding activity (data not shown). In contrast, as revealed by our coimmunoprecipitation experiments, the leucine repeat is involved in the interaction of pIX with itself, allowing its homodimerization or oligomerization via a coiled-coil structure. The fact that the same mutations affect both pIX self-interaction and transactivation properties suggests that the two activities are directly correlated. Alternatively, or in addition, pIX might interact through this leucine repeat element with components of the transcription apparatus: preliminary results suggest indeed that pIX contacts specific RNA polymerase II subunits and general transcription factors (unpublished data), thus mimicking other viral transactivators such as Ad E1a (34), HSV-1 pX (50), or VP16 (32).
Interestingly, one feature that is shared by all pIX-responsive promoters assayed so far, whether from a viral or a cellular origin, is the presence of a canonical TATA box (44). It will be of interest to identify the mechanism of this specific promoter targeting. It is tempting to speculate that such a promoter preference for pIX is of some advantage for Ad propagation since all viral promoters, with the exception of the E2 promoter, contain a TATA box. This hypothesis is further supported by the fact that only episomal genes (i.e., either in plasmidic or viral form) but not chromosome-integrated reporters have been found to be responsive to pIX (unpublished observation).
Nuclear accumulation structures of pIX.
Despite the absence of any detectable homology with consensus nuclear localization signals (9) in its peptide sequence, pIX concentrates within the cell nucleus (44). We show here that the leucine repeat of pIX is essential for this nuclear accumulation, as well as for the formation of the c.a. inclusions. Together, these observations indicate that pIX, by virtue of its low molecular weight, freely diffuses from the cytoplasm to the nucleus, where it is retained, most likely through interactions involving the leucine repeat and specific nuclear components. Preliminary biochemical evidence suggests that pIX actually associates with fractions of the nuclear matrix (unpublished data). It is likely, therefore, that the targeting of the nuclear matrix by pIX constitutes the initial step in the assembly of the c.a. inclusions, providing a nucleation point for pIX oligomerization. Since these nuclear structures are built up in the absence of any other viral protein except pIX, we conclude that they reflect an intrinsic property of pIX.
The leucine repeat plays a central role in both pIX transcriptional activity and the ability to form c.a. inclusions. However, the following lines of evidence clearly indicate that these functions are two independent properties of pIX: (i) during the late phase of infection, c.a. inclusions were always found to be excluded from the viral transcription sites; (ii) neither RNA polymerase II nor primary transcripts could be detected within the c.a. inclusions from Ad-infected cells or cells transfected with wt pIX-expressing vectors (unpublished data); (iii) a mutant lacking part of its alanine-rich element (Δ63-70) lost its capacity to transactivate the E1a promoter (Fig. 4) and to self-associate under our immunoprecipitation conditions (Fig. 5) but retained its ability to accumulate into nuclear c.a. inclusions (data not shown); and (iv) conversely, fusion of the F epitope to the C-terminal end of pIX (IX/F), completely abolished the ability of the protein to form c.a. inclusions in the nucleus (Fig. 7D) but did not affect its transcriptional properties (Fig. 4, lane 15), unless the structural integrity of the leucine repeat was altered (Fig. 4, lane 16). The most simple interpretation to account for the apparent unrelatedness of these pIX functions is that pIX might exert its transcriptional properties only at low concentrations (i.e., at initial times of infection and just after the onset of pIX synthesis), while it starts forming nuclear inclusions at higher concentrations by accumulating on nuclear matrix structures, as observed later in infection or after transfection. Our finding that addition of the F tag at the C-terminal end of pIX differentially affects pIX transcriptional activity and c.a. inclusion formation may therefore merely reflect the preferential impairment of contacts implicated in the assembly of c.a. inclusions compared to those required for promoter transactivation.
The physiological role of these c.a. inclusions remains puzzling. An interesting indication might be provided by the earlier observation (47) that the host cell PML protein was confined, late in Ad infection, inside virus-induced nuclear structures, now identified as the pIX-induced c.a. inclusions (unpublished data). Thus, like the Ad early E4orf3 product which has previously been shown to relocalize PML into viral “fibrous-like” structures (11, 21), pIX might also contribute to the viral process of alteration of the nuclear PML oncogenic domains (33, 45, 52, 55), but during the late phase of infection.
In conclusion, it appears that pIX plays multiple functions during infection. Interestingly, it shares some of these functions with the product of the other Ad intermediate gene, pIVa2: both proteins are transcriptional activators (42, 44), take part in the virus-induced alterations of the host cell by accumulating as specific nuclear inclusions (43), and are present in the mature virion particles (8, 43, 44, 59, 62). No doubt these multifaceted viral entities have yet additional secrets to reveal and thus clearly deserve attention when designing Ad-based vectors for gene therapy protocols.
ACKNOWLEDGMENTS
We thank M. Courtney, M. Methali, and M. Lusky for help and stimulating discussions; A. Bahr for sequence alignment; N. Messaddeq for crucial technical assistance; the IGBMC cell culture staff for providing cells; and the chemistry staff for preparing oligonucleotides and sequencing DNA.
This work was supported by Transgene S.A., the Convention Industrielle pour la Formation par la Recherche CIFRE, and by funds from the French Ministry of Science, the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the University Louis Pasteur, the Alsace Region, the Fondation pour la Recherche Médicale, and the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akalu A, Liebermann H, Bauer U, Granzow H, Seidel W. The subgenus-specific C-terminal region of protein IX is located on the surface of the adenovirus capsid. J Virol. 1999;73:6182–6187. doi: 10.1128/jvi.73.7.6182-6187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alestrom P, Akusjarvi G, Perricaudet M, Mathews M B, Klessig D F, Pettersson U. The gene for polypeptide IX of adenovirus type 2 and its unspliced messenger RNA. Cell. 1980;19:671–681. doi: 10.1016/s0092-8674(80)80044-4. [DOI] [PubMed] [Google Scholar]
- 4.Ali S, Lutz Y, Bellocq J P, Chenard-Neu M P, Rouyer N, Metzger D. Production and characterization of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 5.Bellon G, Michel-Calemard L, Thouvenot D, Jagneaux V, Poitevin F, Malcus C, Accart N, Layani M P, Aymard M, Bernon H, Bienvenu J, Courtney M, Doring G, Gilly B, Gilly R, Lamy D, Levrey H, Morel Y, Paulin C, Perraud F, Rodillon L, Sene C, So S, Touraine-Moulin F, Pavirani A, et al. Aerosol administration of a recombinant adenovirus expressing CFTR to cystic fibrosis patients: a phase I clinical trial. Hum Gene Ther. 1997;8:15–25. doi: 10.1089/hum.1997.8.1-15. [DOI] [PubMed] [Google Scholar]
- 6.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 7.Bocco J L, Reimund B, Chatton B, Kedinger C. Rb may act as a transcriptional co-activator in undifferentiated F9 cells. Oncogene. 1993;8:2977–2986. . (Erratum, 9:999, 1994.) [PubMed] [Google Scholar]
- 8.Boulanger P, Lemay P, Blair G E, Russell W C. Characterization of adenovirus protein IX. J Gen Virol. 1979;44:783–800. doi: 10.1099/0022-1317-44-3-783. [DOI] [PubMed] [Google Scholar]
- 9.Boulikas T. Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem. 1994;55:32–58. doi: 10.1002/jcb.240550106. [DOI] [PubMed] [Google Scholar]
- 10.Burnett R M, Grutter M G, White J L. The structure of the adenovirus capsid. I. An envelope model of hexon at 6 Å resolution. J Mol Biol. 1985;185:105–123. doi: 10.1016/0022-2836(85)90186-x. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho T, Seeler J S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli J. Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christ M, Louis B, Stoeckel F, Dieterle A, Grave L, Dreyer D, Kintz J, Ali Hadji D, Lusky M, Mehtali M. Modulation of the inflammatory properties and hepatotoxicity of recombinant adenovirus vectors by the viral E4 gene products. Hum Gene Ther. 2000;11:415–427. doi: 10.1089/10430340050015888. [DOI] [PubMed] [Google Scholar]
- 15.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 16.Clayman G L, el-Naggar A K, Lippman S M, Henderson Y C, Frederick M, Merritt J A, Zumstein L A, Timmons T M, Liu T J, Ginsberg L, Roth J A, Hong W K, Bruso P, Goepfert H. Adenovirus-mediated p53 gene transfer in patients with advanced recurrent head and neck squamous cell carcinoma. J Clin Oncol. 1998;16:2221–2232. doi: 10.1200/JCO.1998.16.6.2221. [DOI] [PubMed] [Google Scholar]
- 17.Colby W W, Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981;39:977–980. doi: 10.1128/jvi.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connelly S, Smith T A, Dhir G, Gardner J M, Mehaffey M G, Zaret K S, Mclelland A, Kaleko M. In vivo gene delivery and expression of physiological levels of functional human factor VIII in mice. Hum Gene Ther. 1995;6:185–193. doi: 10.1089/hum.1995.6.2-185. [DOI] [PubMed] [Google Scholar]
- 19.Crystal R G, Hirschowitz E, Lieberman M, Daly J, Kazam E, Henschke C, Yankelevitz D, Kemeny N, Silverstein R, Ohwada A, Russi T, Mastrangeli A, Sanders A, Cooke J, Harvey B G. Phase I study of direct administration of a replication deficient adenovirus vector containing the E. coli cytosine deaminase gene to metastatic colon carcinoma of the liver in association with the oral administration of the pro-drug 5-fluorocytosine. Hum Gene Ther. 1997;8:985–1001. doi: 10.1089/hum.1997.8.8-985. [DOI] [PubMed] [Google Scholar]
- 20.Davis A R, Wivel N A, Palladino J L, Tao L, Wilson J M. Construction of adenoviral vectors. Methods Mol Biol. 2000;135:515–523. doi: 10.1385/1-59259-685-1:515. [DOI] [PubMed] [Google Scholar]
- 21.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 22.Fallaux F J, Bout A, van der Velde I, van den Wollenberg D J, Hehir K M, Keegan J, Auger C, Cramer S J, van Ormondt H, van der Eb A J, Valerio D, Hoeben R C. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum Gene Ther. 1998;9:1909–1917. doi: 10.1089/hum.1998.9.13-1909. [DOI] [PubMed] [Google Scholar]
- 23.Furcinitti P S, van Oostrum J, Burnett R M. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989;8:3563–3570. doi: 10.1002/j.1460-2075.1989.tb08528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gahery-Segard H, Molinier-Frenkel V, Le Boulaire C, Saulnier P, Opolon P, Lengagne R, Gautier E, Le Cesne A, Zitvogel L, Venet A, Schatz C, Courtney M, Le Chevalier T, Tursz T, Guillet J G, Farace F. Phase I trial of recombinant adenovirus gene transfer in lung cancer. Longitudinal study of the immune responses to transgene and viral products. J Clin Investig. 1997;100:2218–2226. doi: 10.1172/JCI119759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh-Choudhury G, Haj-Ahmad Y, Graham F L. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987;6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez L, Jr, Woolfson D N, Alber T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 27.Graham F L, Prevec L. Adenovirus-based expression vectors and recombinant vaccines. Biotechnology. 1992;20:363–390. doi: 10.1016/b978-0-7506-9265-6.50022-1. [DOI] [PubMed] [Google Scholar]
- 28.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 29.Green S, Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988;4:309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- 30.Harbury P B, Kim P S, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 31.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi F, Ishima R, Liu D, Tong K I, Kim S, Reinberg D, Bagby S, Ikura M. Human general transcription factor TFIIB: conformational variability and interaction with VP16 activation domain. Biochemistry. 1998;37:7941–51. doi: 10.1021/bi9801098. [DOI] [PubMed] [Google Scholar]
- 33.Hodges M, Tissot C, Howe K, Grimwade D, Freemont P S. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horikoshi N, Maguire K, Kralli A, Maldonado E, Reinberg D, Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 1991;88:5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J C, Newell N E, Tidor B, Sauer R T. Probing the roles of residues at the e and g positions of the GCN4 leucine zipper by combinatorial mutagenesis. Protein Sci. 1993;2:1072–1084. doi: 10.1002/pro.5560020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurst H C. Transcription factors. 1:bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- 37.Hurst H C. Transcription factors. 1:bZIP. Protein Profile. 1994;1:123–168. [PubMed] [Google Scholar]
- 38.Legrand V, Spehner D, Schlesinger Y, Settelen N, Pavirani A, Mehtali M. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J Virol. 1999;73:907–919. doi: 10.1128/jvi.73.2.907-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 40.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 41.Lusky M, Grave L, Dieterle A, Dreyer D, Christ M, Ziller C, Furstenberger P, Kintz J, Hadji D A, Pavirani A, Mehtali M. Regulation of adenovirus-mediated transgene expression by the viral E4 gene products: requirement for E4 ORF3. J Virol. 1999;73:8308–8319. doi: 10.1128/jvi.73.10.8308-8319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutz P, Kedinger C. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J Virol. 1996;70:1396–1405. doi: 10.1128/jvi.70.3.1396-1405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutz P, Puvion-Dutilleul F, Lutz Y, Kedinger C. Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J Virol. 1996;70:3449–3460. doi: 10.1128/jvi.70.6.3449-3460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997;71:5102–5109. doi: 10.1128/jvi.71.7.5102-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 46.Puvion-Dutilleul F, Bachellerie J P, Visa N, Puvion E. Rearrangements of intranuclear structures involved in RNA processing in response to adenovirus infection. J Cell Sci. 1994;107:1457–1468. doi: 10.1242/jcs.107.6.1457. [DOI] [PubMed] [Google Scholar]
- 47.Puvion-Dutilleul F, Chelbi-Alix M K, Koken M, Quignon F, Puvion E, de The H. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res. 1995;218:9–16. doi: 10.1006/excr.1995.1125. [DOI] [PubMed] [Google Scholar]
- 48.Puvion-Dutilleul F, Puvion E. Replicating single-stranded adenovirus type 5 DNA molecules accumulate within well-delimited intranuclear areas of lytically infected HeLa cells. Eur J Cell Biol. 1990;52:379–388. [PubMed] [Google Scholar]
- 49.Puvion-Dutilleul F, Roussev R, Puvion E. Distribution of viral RNA molecules during the adenovirus type 5 infectious cycle in HeLa Cells. J Struct Biol. 1992;108:209–220. doi: 10.1016/1047-8477(92)90021-2. [DOI] [PubMed] [Google Scholar]
- 50.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell W C. Update on adenovirus and its vectors. J Gen Virol. 2000;81:2573–2604. doi: 10.1099/0022-1317-81-11-2573. [DOI] [PubMed] [Google Scholar]
- 52.Seeler J S, Dejean A. The PML nuclear bodies: actors or extras? Curr Opin Genet Dev. 1999;9:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 53.Shu W, Ji H, Lu M. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- 54.Smith B T. Cell line A549: a model system for the study of alveolar type II cell function. Am Rev Respir Dis. 1977;115:285–293. doi: 10.1164/arrd.1977.115.2.285. [DOI] [PubMed] [Google Scholar]
- 55.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 56.Stewart P L, Burnett R M, Cyrklaff M, Fuller S D. Image reconstruction reveals the complex molecular organization of adenovirus. Cell. 1991;67:145–154. doi: 10.1016/0092-8674(91)90578-m. [DOI] [PubMed] [Google Scholar]
- 57.Trapnell B C, Gorziglia M. Gene therapy using adenoviral vectors. Curr Opin Biotechnol. 1994;5:617–625. doi: 10.1016/0958-1669(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided byquality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Oostrum J, Burnett R M. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Oostrum J, Smith P R, Mohraz M, Burnett R M. The structure of the adenovirus capsid. III. Hexon packing determined from electron micrographs of capsid fragments. J Mol Biol. 1987;198:73–89. doi: 10.1016/0022-2836(87)90459-1. [DOI] [PubMed] [Google Scholar]
- 61.Xiao J H, Davidson I, Matthes H, Garnier J M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 62.Zhang W, Imperiale M J. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J Virol. 2000;74:2687–2693. doi: 10.1128/jvi.74.6.2687-2693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]