Abstract
Serious infections caused by multidrug-resistant (MDR) organisms (Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii) present a critical need for innovative drug development. Herein, we describe the preclinical evaluation of YU253911, 2, a novel γ-lactam siderophore antibiotic with potent antimicrobial activity against MDR Gram-negative pathogens. Penicillin-binding protein (PBP) 3 was shown to be a target of 2 using a binding assay with purified P. aeruginosa PBP3. The specific binding interactions with P. aeruginosa were further characterized with a high-resolution (2.0 Å) X-ray structure of the compound’s acylation product in P. aeruginosa PBP3. Compound 2 was shown to have concentration > 1 μg/ml at the 6 hour time point when administered intravenously or subcutaneously in mice. Employing a meropenem resistant strain of P. aeruginosa, 2 was shown to have dose-dependent efficacy at 50 and 100 mg/kg q6h dosing in a mouse thigh infection model. Lastly, we showed that a novel γ-lactam and β-lactamase inhibitor (BLI) combination can effectively lower minimum inhibitory concentrations (MICs) against carbapenem resistant Acinetobacter spp. that demonstrated decreased susceptibility to 2 alone.
Keywords: γ-lactam, siderophore, multidrug-resistant Gram-negative pathogens, Penicillin-binding protein
1. Introduction
Multidrug-resistant (MDR) bacterial infections pose an increasing threat to public health, causing significant morbidity, mortality, and economic hardship. In the United States alone, more than three million infections each year are caused by antibiotic-resistant bacteria resulting in approximately 36,000 deaths and more than $20 billion in healthcare costs [1]. The future global impact is even more staggering with an estimated cumulative economic cost of $100 trillion from now until 2050, with an associated 10 million deaths, resulting from MDR infections [2]. Invasive infections with Gram-negative bacterial pathogens in particular have become increasingly problematic and are associated with 28 day mortality rates of 30–70% (https://www.cdc.gov/drugresistance/biggest-threats.html). Resistant strains of Acinetobacter baumannii, Pseudomonas aeruginosa, and carbapenem-resistant Enterobacterales are identified as “urgent” and “serious” threats by the CDC and WHO, illustrating the critical need for new therapeutics [3–7].
The “hard-to-treat” nature of these infections is often caused by extensive antibiotic-resistant phenotypes that allow pathogens to overcome standard antibiotic regimens [8]. Despite the increasing public health threat, few truly novel agents are in development to treat such infections, as most large pharmaceutical companies have withdrawn from antibacterial research [3, 4, 9, 10]. Government agencies are aware of the ever-growing issue of antibacterial resistance and are responding with several initiatives intended to incentivize renewed efforts in antibiotic development [11]. They include public-private partnerships highlighting the urgent need for novel treatments.
Resistance in multidrug-resistant Gram-negative organisms is multifactorial. These causes may include reduced permeability, modification of target proteins, and overexpression of efficient and diverse efflux pumps [12, 13]. For β-lactam antibiotics, a particular concern is the increasing diversity of plasmid-mediated carbapenemases (β-lactamases (blas): NDM-1, KPC, and OXA-type class D carbapenemases) making β-lactam antibiotics ineffective, despite their common coadministration with β-lactamase inhibitors [1].
We recently reported the initial attributes of a new series of antibacterial agents effective against Gram-negative pathogens based on a revitalized non-β-lactam pyrazolidinone scaffold [14 – 21] exemplified by 1 (Figure 1) [22]. The strategy involves inhibiting penicillin-binding proteins (PBPs), which are validated targets for antibacterial discovery, while avoiding susceptibility to β-lactamase inactivation. This is accomplished by using a γ-lactam ring with tunable reactivity as the molecule’s core [18, 20]. Additionally, 1 includes a siderophore moiety which exploits intrinsic bacterial iron transport processes (“Trojan horse approach”) to overcome the decreased permeability of Gram-negative bacteria [23 – 25]. Agent 1, which contains a prototype γ-lactam-siderophore, inactivates PBP3 and possesses excellent in vitro potency against MDR clinical isolates of P. aeruginosa, K. pneumoniae, and E. coli. Herein, we report the discovery and properties of YU253911, 2, a chloroaminothiazole analog of 1 (Figure 1), which possesses enhanced potency versus Acinetobacter spp. and improved pharmacokinetics versus 1 as illustrated by in vivo efficacy in a rodent thigh infection model employing an MDR strain of P. aeruginosa.
Figure 1.
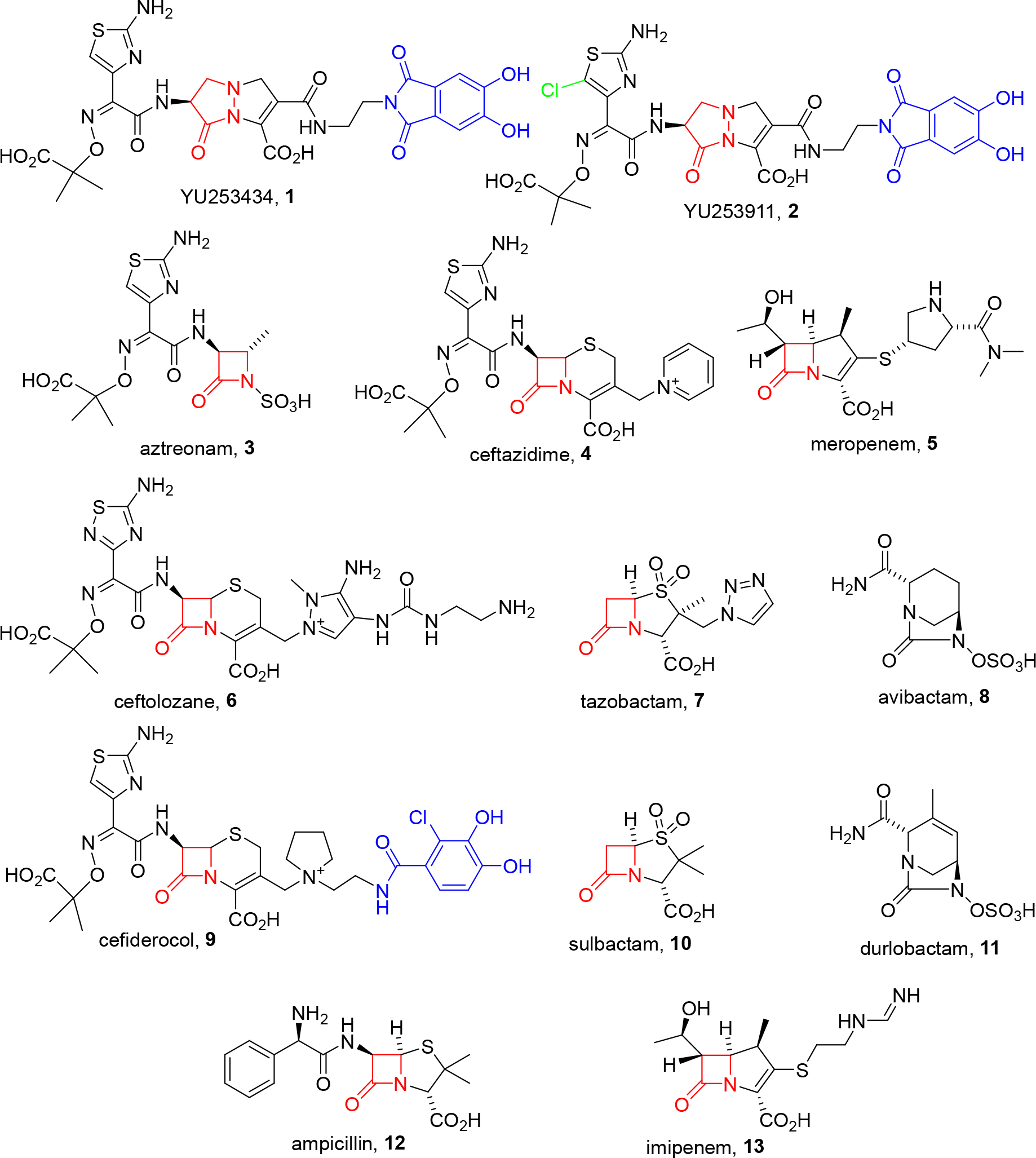
Structures of γ-lactam siderophores 1 and 2 versus comparator β-lactam antibiotics and β-lactamase inhibitors. The β- or γ-lactam core is highlighted in red; iron-binding siderophore moieties in blue.
2. Results and Discussion
2.1. Chemistry
We previously reported the synthesis and characterization of 1 as a prototype of a new class of siderophore-conjugated γ-lactam antibiotic with enhanced activity against MDR Gram-negative bacteria [22]. In an effort to explore this finding, additional analogs with modified side chains were screened, leading to the identification of 2 which contains a chloroaminothiazole group. This group was found to favorably modulate several biological properties (vide infra). The synthesis of 2 parallels the synthesis of 1 and employs a common advanced intermediate dihydroxyphthalimide [26] -appended bicyclic pyrazolidinone, 14 (details of the synthesis of 14 are in the Supporting Information, pages 4 –17). Coupling of 14 using the appropriately protected chloroaminothiazole 15 followed by deprotection and reverse phase chromatography provides compound 2, Scheme 1. The chiral purity of 2 was not assessed after the synthesis was completed though Boc-L-serine was employed as the starting material.
Scheme 1.
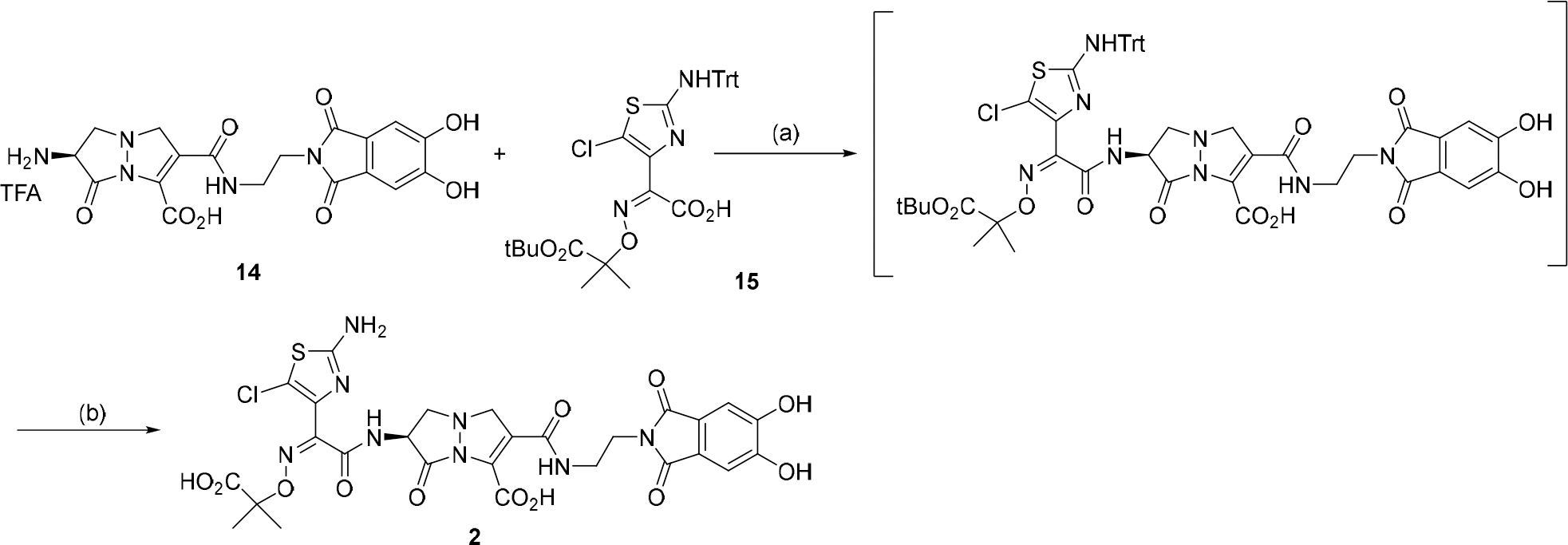
Synthesis of YU253911, 2: (a) i. 15, Oxalyl chloride, catalytic DMF; ii) 14, MSTFA, Hunig’s base, 3.5 h, 100% crude. (b) 2:1 DCM/TFA, triethylsilane, 0 °C to RT, 1.5 h, toluene chase, reverse phase MPLC C18, 0 to 60% acetonitrile 0.1% formic acid/water with 0.1% formic acid, 30%.
2.2. Microbiology
γ-Lactam 2 inhibits the growth of MDR Gram-negative bacilli.
A comparison of minimum inhibitory concentrations (MICs) for 2 and 1 vs. previously described clinical carbapenem-resistant isolates of P. aeruginosa27 and K. pneumoniae28 are provided in Figure 2 [22]. MIC data for both 2 and 1 is provided for comparison; the detailed values for each individual strain are in Supplemental Table 1, page 18. In all but one case (YUKP-39), microbiologic potency of 2 was maintained, although the activity trended to be slightly less active for 2 compared to 1, particularly for K. pneumoniae. Nevertheless, MIC testing of 2 afforded an MIC50 of 0.5 and 1 mg/mL against 23 samples each of MDR P. aeruginosa and K. pneumoniae, respectively. Notably, MIC data of 2 were significantly lower than meropenem in all cases but 1 isolate (YUKP-39). We purposely selected a subset of our P. aeruginosa and K. pneumoniae panels that had the most potent compound 1 MIC values. Using such a narrow comparison means it is possible that 2 may not necessarily have poorer overall activity against larger panels. Similar to what was previously reported for 1 [22], MIC values were dependent on maintaining low iron concentrations in the culture media (data not shown). All results were generated using Chelex® resin-treated media to mimic the low iron concentrations found in vivo, and in accord with CLSI recommendations for siderophore-containing antibiotics.
Figure 2.

Compound 2 and 1 MIC values against representative γ-lactam susceptible/carbapenem-resistant K. pneumoniae and P. aeruginosa strains (23 each). Except for one isolate, YUKP-39, compound 2 maintains microbiologic potency against MDR Gram-negative rods previously described for this γ-lactam-siderophore class (see also Supplemental Table 1, page 18).
The γ-Lactam 2 possesses enhanced antimicrobial activity against MDR A. baumannii.
MICs in low iron media against a 198-member panel of previously described Acinetobacter spp. clinical isolates, (79% A. baumannii, 21% a mixture of A. nosocomialis, and A. pittii) [30] are presented in Figure 3 and Table 1. Activity against a 98-member carbapenem-resistant A. baumannii subset of the panel is also shown in Figure 3 and Table 1. Importantly, the MIC values of 2 compare favorably to all β-lactam classes, including aztreonam 3 (monobactam), ceftazidime 4 (cephalosporin) and meropenem 5 (carbapenem) (Table 1). Applying the breakpoints for cefiderocol 9 (susceptibility <=4 μg/mL and resistance >=16 μg/mL), a similar approved drug utilizing a siderophore transport mechanism [29], to compound 2 results in a susceptibility to 2 of 86% (171 out of 198) of the full Acinetobacter panel and 83% (81 out of 98) of the subset of carbapenem-resistant A. baumannii (CRAB). Detailed values for each individual strain are in Supplemental Table 2, page 19.
Figure 3.
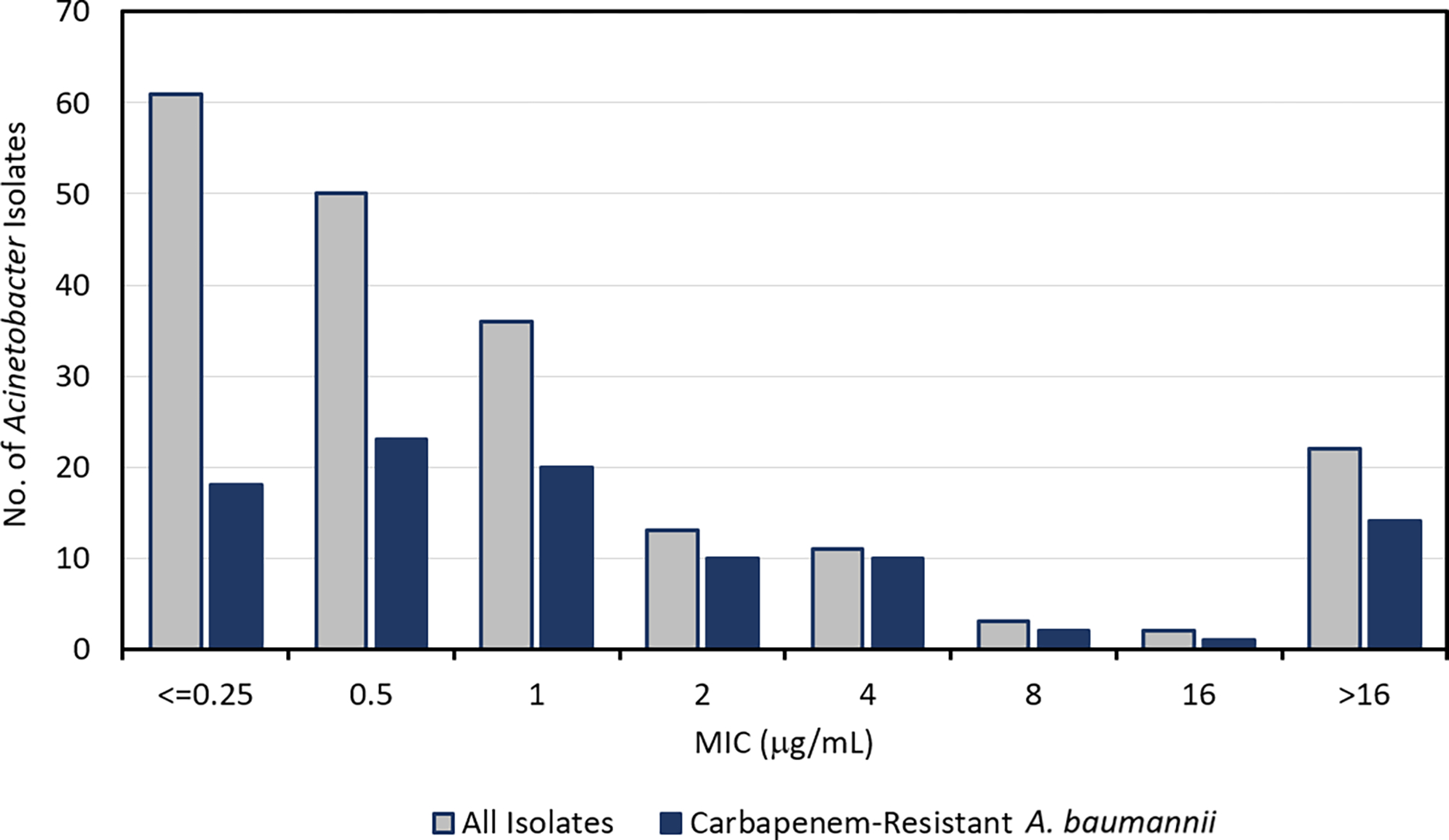
Distribution of MICs in low iron media of 2 (μg/mL) against a 198-member Acinetobacter spp. panel and subset of 98 carbapenem-resistant A. baumannii (CRAB) isolates. See also Supplemental Table 2, page 19.
Table 1.
MIC50 and MIC90 (μg/mL) of 2 and comparator β-lactam antibiotics against a 198-member panel of clinical isolates of Acinetobacter spp., as well as the 98-member subset of carbapenem-resistant A. baumannii (CRAB).30 Chemical structures for all agents are provided in Figure 1.a See also Supplemental Table 2, page 19.
| 198 Acinetobacter spp. | 98 CRAB | |||
|---|---|---|---|---|
| Compound | MIC50 | MIC90 | MIC50 | MIC90 |
|
| ||||
| 2 | 0.5 | >16 | 1 | >16 |
| Aztreonam (3) | >32 | >32 | >32 | >32 |
| Ceftazidime (4) | 64 | >64 | >64 | >64 |
| Meropenem (5) | 8 | >64 | 64 | >64 |
| Ceftazidime (4)/avibactam (8) | 16 | >64 | 64 | >64 |
| Ceftolozane (6)/tazobactam (7) | 8 | >64 | 32 | >64 |
MICs were determined using iron-depleted cation-adjusted Mueller-Hinton broth that was supplemented with iron (as ferric chloride) as indicated. The initial iron-depleted media was prepared by the standard treatment with cation-exchange resin, which has been reported to reduce iron concentrations to 0.02 μg/mL.29
Combination of 2 with sulbactam further enhances growth inhibition of Acinetobacter spp.
We investigated whether partner agents could improve the already potent activity of 2 against resistant Acinetobacter spp. growth. β-lactam antibiotics are often used clinically in combination with β-lactamase inhibitors (BLIs) to protect against enzymatic hydrolysis. Although the γ-lactam core of 2 possesses intrinsic stability to β-lactamase hydrolysis (vide infra), β-lactamase inhibitors (BLIs) were nevertheless evaluated for their ability to augment the breadth of 2 susceptibility against Acinetobacter spp.
Sulbactam 10, a BLI commonly marketed in combination with ampicillin (12, Unasyn®), was selected for study as a potential partner of 2 for two reasons. Firstly, sulbactam has been shown for decades to be safe [32, 32] and well-tolerated in patients [33], with favorable pharmacokinetics [34, 35]; secondly, sulbactam possesses intrinsic antimicrobial activity against Acinetobacter spp. due to postulated inhibition of PBP1 and PBP3 [36]. Furthermore, sulbactam is currently under investigation in combination with durlobactam, 11, for the treatment of Acinetobacter infections. The γ-lactam 2 is believed to target PBP3 (vide infra) and inhibition of two PBP enzymes would be expected to produce enhanced antimicrobial effects.
The 24 Acinetobacter isolates from Figure 3 with higher MICs to compound 2 (MIC >= 16 μg/mL) were tested versus sulbactam alone or in combination with 2. MIC values show 79% (19/24) of the tested isolates were found to have their growth inhibited to some extent by sulbactam (MICs <=16 μg/mL) (Table 2). Furthermore, when 2 and sulbactam were combined in a 1:1 ratio in a typical 2-fold MIC dilution scheme, 50% (12/24) of the isolates previously resistant to 2 showed improved MICs <=4 μg/mL. Published human pharmacologic data for ampicillin-sulbactam and sulbactam-durlobactam demonstrate that serum concentrations of sulbactam of 20 μg/mL are readily attainable from intravenous dosing. Therefore, additional studies were undertaken to evaluate the in vitro effectiveness of 2 under conditions of a constant level of sulbactam coadministration. The MIC values for 2 were determined using sulbactam set at 20 μg/mL (Table 2). Under these conditions, the growth of only 3 of the isolates resistant to 2 were not significantly inhibited, collectively implying that 98% of the full 198 member Acinetobacter spp. panel could be susceptible to 2 when used in combination with a 20 μg/mL coadministration of sulbactam.
Table 2.
MIC (μg/mL) values in low iron media of 2 in combination with sulbactam, 10, when combined 1:1 (by mass), or when 2 was tested with a constant concentration of 10 at 20 μg/mL. Individual MIC values for 2 and 10 are provided for comparison as well as the identity of each Acinetobacter isolate. See text for details.
| Isolate No | Strain Identitya | 2 | 10 | 2 + 10 (1:1) | 2 + 10 (20 μg/mL) |
|---|---|---|---|---|---|
|
| |||||
| PR-314 | CRAB | >16 | 16 | 4 | <=0.25 |
| PR-319 | CRAB | >16 | >32 | 4 | 0.5 |
| PR-325 | CRAB | >16 | 32 | >16 | >16 |
| PR-326 | CRAB | >16 | 2 | 2 | <=0.25 |
| PR-340 | CSAB | >16 | 4 | 1 | <=0.25 |
| PR-342 | CRAB | 16 | >32 | >16 | >16 |
| PR-345 | CRAB | >16 | 16 | 16 | <=0.25 |
| PR-351 | CRAB | >16 | 16 | 16 | <=0.25 |
| PR-362 | AN | >16 | 1 | <=0.25 | <=0.25 |
| PR-365 | AN | >16 | 1 | <=0.25 | <=0.25 |
| PR-380 | CRAB | >16 | 16 | 16 | <=0.25 |
| PR-384 | CRAB | >16 | 16 | 16 | >16 |
| PR-387 | CRAB | >16 | 16 | 16 | <=0.25 |
| PR-399 | CRAB | >16 | 16 | 16 | <=0.25 |
| PR-401 | CRAB | >16 | 16 | 8 | <=0.25 |
| PR-412 | AN | >16 | 2 | 1 | <=0.25 |
| PR-423 | CRAB | >16 | 16 | 8 | <=0.25 |
| PR-434 | CSAB | >16 | 8 | 2 | <=0.25 |
| PR-452 | CSAB | >16 | 1 | <=0.25 | <=0.25 |
| PR-459 | CRAB | 16 | 32 | 8 | 4 |
| PR-464 | CSAB | >16 | 1 | <=0.25 | <=0.25 |
| PR-478 | CSAB | 16 | 4 | 1 | <=0.25 |
| PR-482 | CRAB | >16 | 32 | >16 | <=0.25 |
| PR-491 | AN | >16 | 2 | 0.5 | <=0.25 |
Strain identity: CRAB = carbapenem-resistant A. baumannii; CSAB = carbapenem-susceptible A. baumannii; AN = A. nosocomialis (carbapenem-susceptible).
2.3. PBP Inhibition
Based on the mechanism established previously for γ-lactam 1, 2 was also suspected of inhibiting PBP3, an essential bacterial transpeptidase. This was verified through labeling studies with Bocillin™, a fluorescent penicillin analog, using purified P. aeruginosa PBP3 according to an established protocol [37]. Increasing concentrations of 2 inhibited fluorescent labeling of the protein by Bocillin™, with an IC50 of 5 ± 1 μM (Figure 4). This value is comparable to 1 (2.5 ± 0.5 μM) and a previously published value for doripenem (IC50 = 2.3 ± 0.5 μM, for Acinetobacter spp. PBP3) [37].
Figure 4.
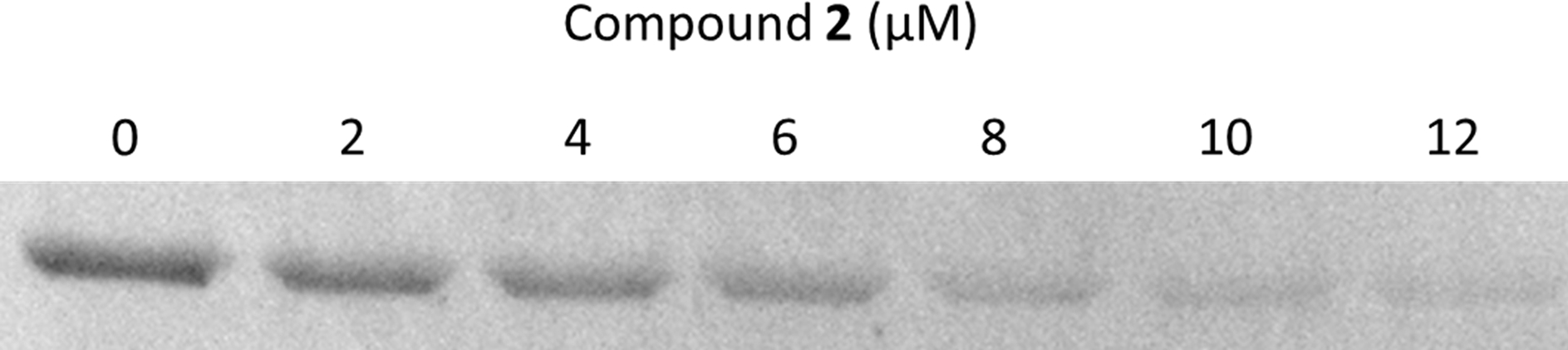
Determination of the IC50 of 2 for P. aeruginosa PBP3 using a competitive assay. Bocillin™, a fluorescent substrate of PBP3 was reacted with enzyme that had been pre-incubated with increasing concentrations of 2. An IC50 was calculated as the concentration of 2 required to reduce the fluorescence intensity of the Bocillin™-labeled protein by 50%.
2.4. Crystal Structure and Molecular Modeling
The Compound 2 Crystal Structure complexed with P. aeruginosa PBP3a.
The crystal structure of P. aeruginosa PBP3 complexed to 2 was determined to 2.0 Å resolution (Table 3). The difference electron density in the active site shows that 2 has formed a covalent bond with the catalytic S294 residue (Figure 5). Density for most of the compound 2 moieties are well resolved. This includes the chlorine substituent on the aminothiazole ring, both amide groups, the carboxyl group and the dihydropyrazole ring of the core. Note that the occupancy for the chlorine atom refined to 0.58 whereas the rest of the aminothiazole ring had an occupancy of 1.0. This decreased occupancy for the chlorine atom is likely due to radiation damage during the synchrotron radiation X-ray experiment. Halogen-aromatic ring bonds are known to be sensitive to X-ray radiation, and their breakage has been used to monitor radiation damage in protein crystals [38]. Density for the 2-carboxypropan-dimethyl moiety extending from the oxime and the 5,6-dihydroxyphthalimide siderophore group of 2 are not well resolved in the electron density map indicating their inherent flexibility (Figure 5).
Table 3.
X-ray diffraction data collection and crystallographic refinement statistics for the P. aeruginosa PBP3 complex with 2.
| Wavelength (Å) | 0.97946 |
| Resolution range (Å) | 50.00 – 2.00 (2.03 – 2.00) |
| Space group | P212121 |
| Unit cell (Å, °) | 67.77 82.66 88.80 90 90 90 |
| Total reflections | 427,156 |
| Unique reflections | 34,157 (1,673) |
| Multiplicity | 12.5 (12.6) |
| Completeness (%) | 99.8 (99.8) |
| Mean I/sigma (I) | 30.5 (4.4) |
| CC1/2 | 0.995 (0.934) |
| R-merge (%) | 15.8 (115.9) |
| Resolution refinement (Å) | 33.91 – 2.00 (2.05 – 2.00) |
| Reflections used in refinement | 32,335 (2,169) |
| Reflections used for R-free | 1,770 (141) |
| R-work | 0.174 (0.202) |
| R-free | 0.227 (0.237) |
| Number of non-hydrogen atoms | 3,863 |
| Macromolecules | 3,621 |
| Ligand | 49 |
| Solvent | 193 |
| Protein residues | 473 |
| RMS(bonds, Å) | 0.010 |
| RMS(angles, °) | 1.65 |
| Ramachandran favored (%) | 97.64 |
| Ramachandran allowed (%) | 2.15 |
| Ramachandran outliers (%) | 0.22 |
Statistics for the highest-resolution shell are shown in parentheses.
Figure 5.

Omit Fo-Fc electron density showing the presence of a covalently bound 2. The compound 2 is shown with cyan-colored carbon atoms. The density is contoured at the 3 σ level.
The ligand 2 forms a number of hydrogen bonds in the active site of P. aeruginosa PBP3 (Figure 6). The chloroaminothiazole ring hydrogen bonds with E291, and main chain oxygen and nitrogen atoms of R489. The chloroaminothiazole also makes hydrophobic interactions with G293, Y409, and A488. In addition, the chlorine substituent makes a special close-to-linear “C-Cl … O” interaction with the carbonyl oxygen of Y407. Such favorable interactions with a halogen atom have previously been observed in protein:ligand interactions [39, 40]. The amide side chain, which connects to the aminothiazole ring, hydrogen bonds with N351 and the backbone oxygen of T487. The carboxyl moiety of the core hydrogen bonds with T487, S485, and conformation 2 of residue S349. The secondary nitrogen atom of the dihydropyrazole ring hydrogen bonds with conformation 1 of S349 (Figure 6). The amide moiety that serves to attach the 5,6-dihydroxyphthalimide siderophore side chain of 2 interacts with a nearby water molecule (W#2). The 5,6-dihydroxyphthalimide does not form many interactions in the active site.
Figure 6.
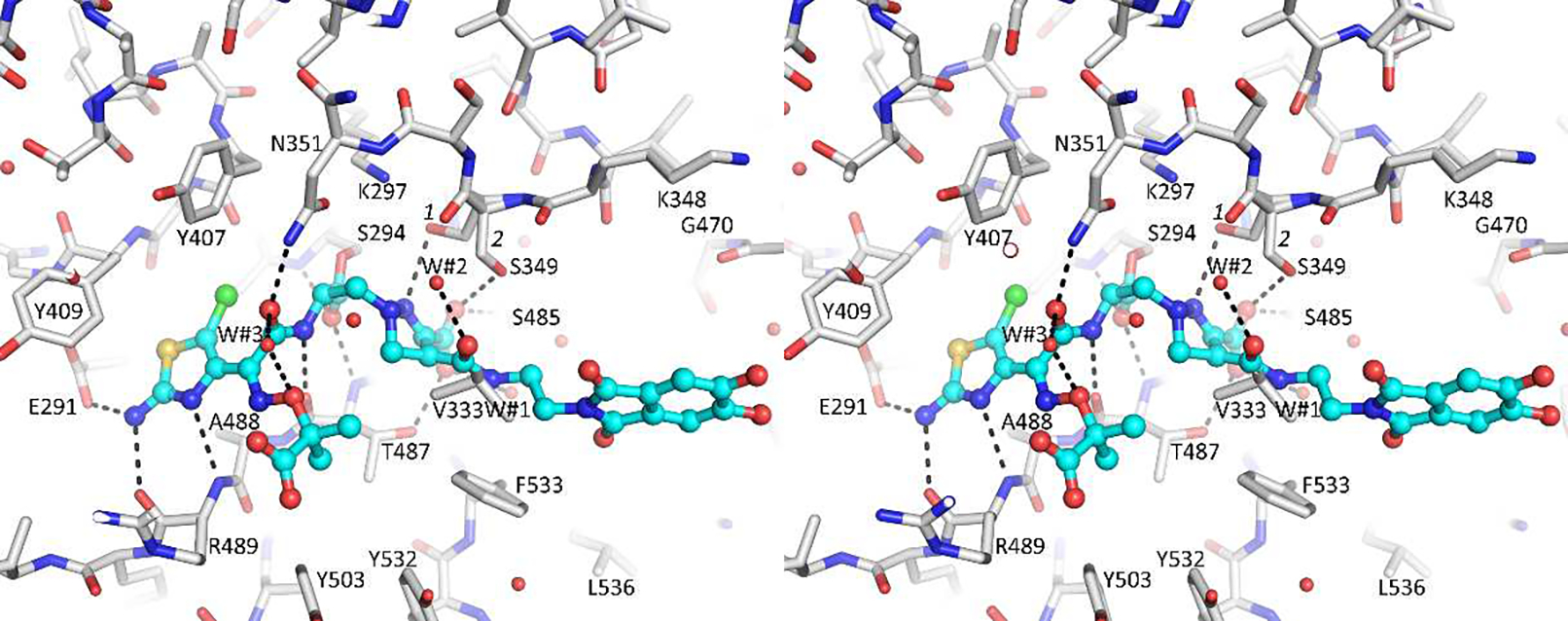
Stereo diagram depicting the interactions of 2 in the active site of P. aeruginosa PBP3. Hydrogen bonds are depicted as dashed lines.
The compound 2 is chemically identical to the previously reported 1, except the 5-position of the aminothiazole in 2 has a chloro- substituent. The binding mode of 1 is very similar to that of 2 when bound to P. aeruginosa PBP3 (Figure 7; PDBid 6VOT) [22]. A difference is an inversion of the tertiary nitrogen in the dihydropyrazole ring of 2 compared to 1. This is likely a consequence of that the 2 PBP3 complex is determined to a higher resolution (2.0 vs. 2.5 Å resolution) allowing for improved refinement of this ring region. A second difference is that residue E291 in the 2 PBP3 complex has moved closer to the aminothiazole ring compared to the 1 PBP3 complex (Figure 7).
Figure 7.
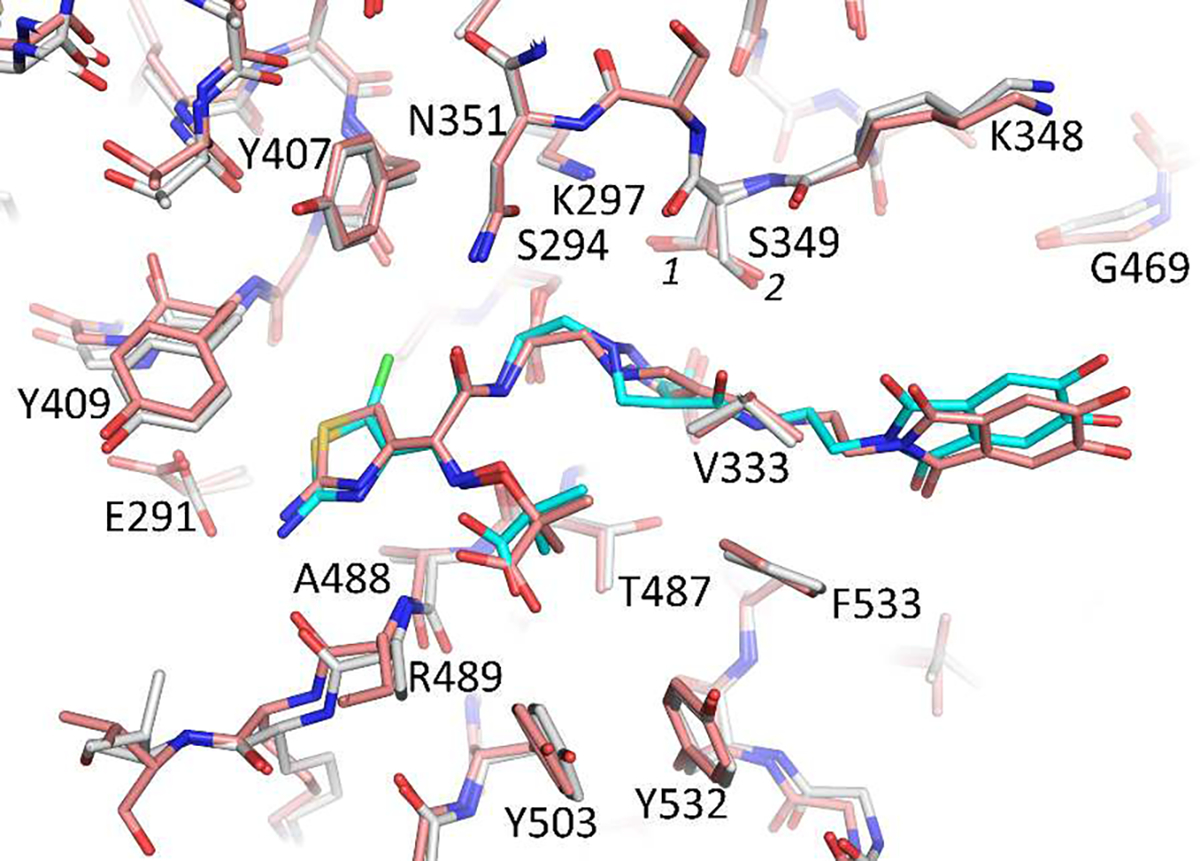
Superpositioning of the 1 and 2 bound structures of P. aeruginosa PBP3. The carbon atoms of the 1:PBP3 complex is shown in salmon color; while 2 is colored cyan with its protein shown in white.
2.5. Pharmacokinetics and In vivo efficacy; drug-like attributes
Pyrazolidinone 2 has many characteristics that are favorable drug-like attributes (details of the data are found in the Supporting Information, pages 33–38) for potential administration either by IV or inhaled routes as 2 has high solubility >100 μM in pH 7.5 phosphate buffer as measured by nephelometry. Though highly soluble, 2 has poor Caco-2 permeability and would not be expected to have oral bioavailability thus IV and sub cutaneous PK was obtained (vide infra). Compound 2 is highly protein-bound as shown by measurements in both mouse and human plasma, 94% and 88%, respectively. Stability of 2 to both human and CD-1 mouse microsomes is notable with a half-life >60 minutes in both preparations. Furthermore, 2 does not inhibit 7 of the 8 CYP enzymes it was tested against at a 30 μM concentration and showed approximately 50% inhibition of CYP2C8 at 100 μM. Compound 2 was also 280 evaluated for cytotoxicity was not found when tested against human primary hepatocytes at a maximal 281 dose of 100 μM.
In a general safety screen, testing 2 against 44 enzymes and receptors the only activity of note was >50% inhibition at 30 μM against cyclooxygenase (COX-1) and phosphodiesterase PDE3A (Supporting Information, page 37). Testing against 6 ion channels, including hERG at a 30 μM maximal dose revealed 2 did not inhibit these channels (Supporting Information, page 38). The pharmacokinetics (PK) of 2 was evaluated both subcutaneously and by intravenous administration at a dose of 50 mg/kg in CD-1 mice (Table 4). Both routes of dosing result in similar PK data. Notably, the plasma levels of 2, at the 6h time point were >1.0 mg/mL by both dosing routes (Figure 8 and Table 4). This value is clearly superior to 1 where at the 6-hour timepoint its plasma concentration was about 0.35 mg/mL [22]. The cause for improved plasma level with 2 is not known, though it might be due to clearance differences caused by a slight increase in protein binding or a small increase in LogP due to the additional chlorine atom. Nevertheless, the >1.0 mg/mL plasma level encouraged us to comparatively test both 2 and 1 in a mouse efficacy model as this plasma level is above the MIC50 values of 2 for both P. aeruginosa and A. baumannii.
Table 4.
Individual plasma concentrations of 2 after sc (50 mg/kg) dosing in mice, each value represents an individual mouse, showing mean blood levels >1μg/mL at the 8-hour time point.
| Time (h) | Sample Conc (ng/ml) | Mean (ng/ml) | SD (ng/ml) | ||
|---|---|---|---|---|---|
| 0.167 | 48690 | 48689 | 44921 | 47433 | 2176 |
| 0.5 | 20245 | 14355 | 14094 | 16231 | 3478 |
| 1 | 3138 | 3515 | 3186 | 3280 | 205 |
| 2 | 3273 | 2876 | 3072 | 3074 | 199 |
| 4 | 1534 | 1732 | 739 | 1751 | 526 |
| 6 | 1426 | 1102 | 1425 | 1318 | 187 |
| 8 | 1001 | 1473 | 1126 | 1200 | 245 |
| 24 | 678 | 545 | 579 | 601 | 69 |
Figure 8.
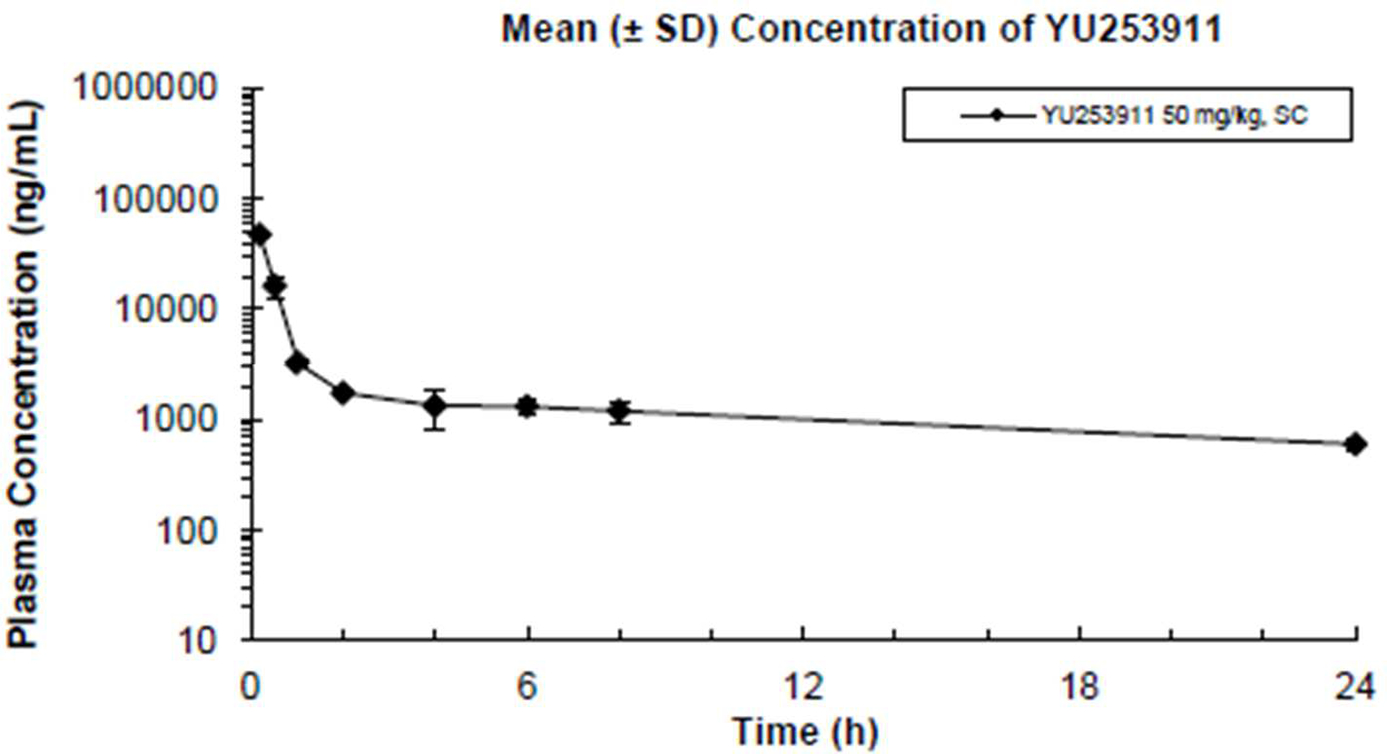
The mean plasma concentration-time profile for YU253911, 2, after sc dosing (50mg/kg) in mice.
We chose AR-BANK#0229 strain of carbapenem-resistant P. aeruginosa as the infectious agent for the neutropenic thigh infection model. This decision was made for several reasons: a) this strain establishes infection in mice; b) both compound 2, and 1 have potent activity against this strain (MIC = 0.5 μg/ml and 0.25μg/mL, respectively); and c) the strain is highly resistant to currently clinically used antibiotics as it is susceptible to only colistin, amikacin, and gentamicin (Supplemental Table 4, page 42).
Neutropenia was induced with cyclophosphamide. Animals were then intramuscularly (IM) inoculated with 1.02 × 106 CFU/mouse (0.1 mL/animal) of the P. aeruginosa AR-BANK#0229 strain. Test antibiotics, 1 and 2, both at 50 and 100 mg/kg, were administered subcutaneously (SC) four times per day (QID) with 6 h intervals (q6h) at 2, 8, 14 and 20 h after infection. The reference control was colistin, which was dosed at 30 mg/kg and administered subcutaneously (SC) twice (BID) with 12 h intervals (q12h) at 2 and 14 h after infection. Animals were sacrificed at 2 or 26 h post-infection, and the thigh tissues were harvested and weighed from each of the test animals. The bacterial counts (CFU/g) of thigh tissue homogenates were compared. Full details of the method and protocol are in the Supplemental Information as well as the individual animal data (Supplemental Information, pages 45–57). Bacterial burden (CFU/g tissue) of test article treated animal groups was compared to the baseline bacterial count at 2 h after infection and the difference in counts (Δ) was reported. The significance of effects was assessed with ANOVA. The burden was also compared to the vehicle control.
As might have been predicted by the pharmacokinetic results, 1 was not found to have efficacy in the animal model against this carbapenem-resistant strain even though the MIC values indicated the strain had good susceptibility of 0.25 μg/mL (Figure 9). Importantly, 2, with superior pharmacokinetics does show a dose-dependent reduction in CFU values against this difficult to treat P. aeruginosa strain. It is notable that the MIC value for 2 of 0.5 μg/mL is below the expected blood levels of 2 at the 6 hour dosing interval and, in spite of the high protein binding of 94%, encouraging activity was seen (Figure 9).
Figure 9.
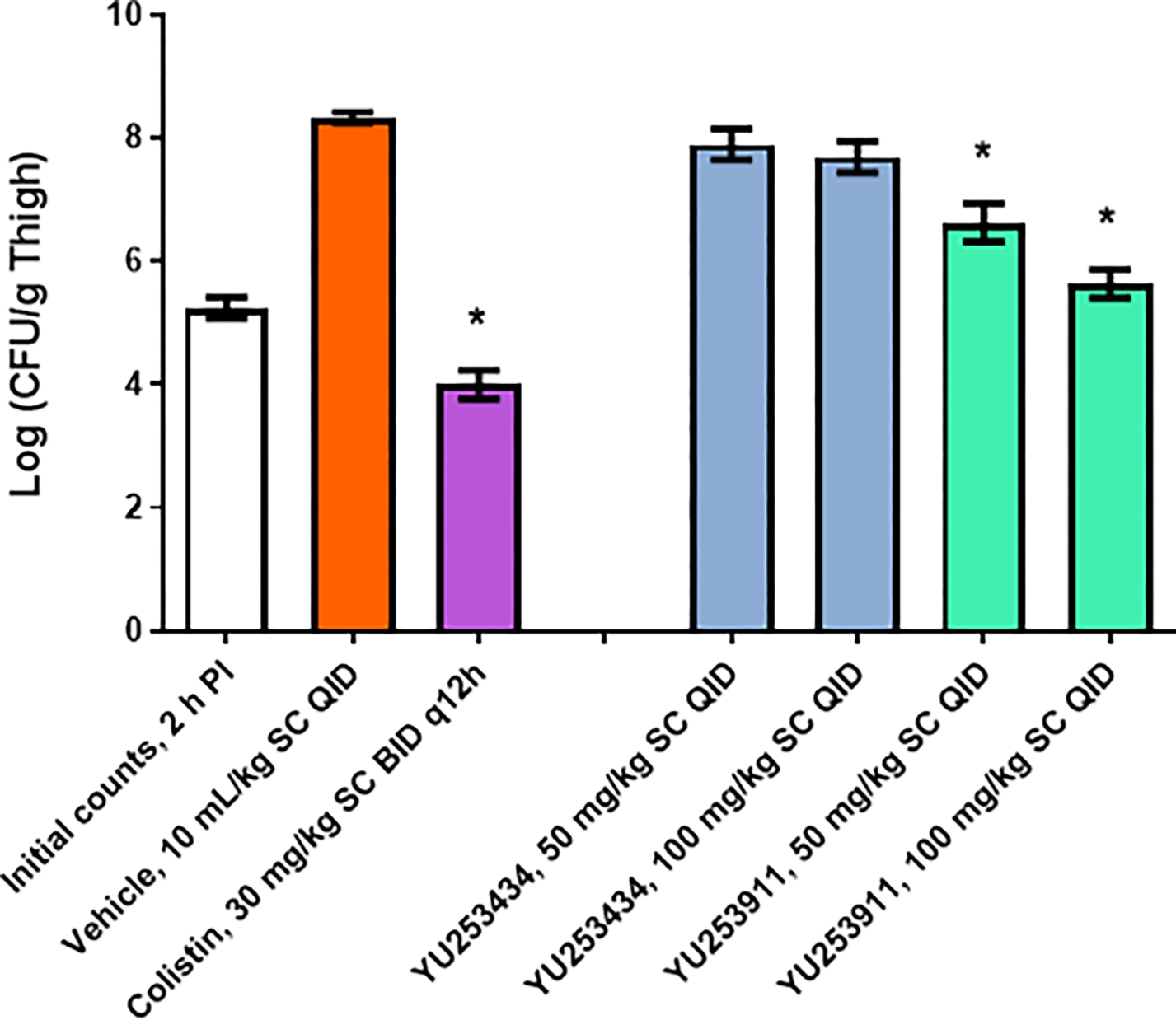
The test compounds YU253434 1, YU253911 2, and colistin efficacy in the P. aeruginosa AR-BANK#0229 thigh infection model.
(*) A significant difference (p < 0.05) between the vehicle control and treatment group was determined by one-way ANOVA followed by Dunnett’s test.
Error bars are SEM values
2.6. Resistance
Although 2 showed broad coverage against a large panel of resistant clinical isolates of Acinetobacter spp. including carbapenem-resistant A. baumannii, a fraction of all tested strains (24 of 198, 12%) and the carbapenem-resistant subgroup (15 of 98, 15%) demonstrated MICs >=16 μg/mL.
Understanding the mechanism or mechanisms of resistance for 2 is important as it allows the optimization of partner agents or alternative dosing schedules to ameliorate potential issues. This knowledge will help predict whether further strains will be susceptible and influence future clinical use, as well as design and synthesis of subsequent analogs.
There are a number of mechanisms that have been identified for resistance to antibiotics for Acinetobacter spp. as well as other Gram-negative pathogens. They fall into 4 general categories: 1) degradation of the antibiotic (e.g., β-lactamase hydrolysis); 2) target modification (e.g., PBP mutations or changes in the expression of PBP genes); 3) efflux of the antibiotic from the periplasm or cytoplasm (e.g., mutations or decreased changes in expression of pump genes such as ErmAB-TolC, macA, UadeA-H, mdtABC); or 4) decreased permeability of the bacterial outer membrane (e.g., TolC, OmpH, OPRD genes). All of these represent potential mechanisms for resistance to 2. Additionally, due to the incorporation of the siderophore moiety as a key component of the antibiotic molecule, changes in the expression of genes related to the bacterial iron transport system could also have an effect on antibiotic susceptibility as was identified as a potential resistance mechanism for cefiderocol, 9 [Ito, A. et al. IDWeek 2018 poster 696. https://idsa.confex.com/idsa/2018/webprogram/Paper69661.html].
2.7. Genomic analysis for resistance genes
Our initial investigation to determine the mechanism of resistance toward 2 seen in Acinetobacter spp., as well as P. aeruginosa and K. pneumoniae, was undertaken by comparing the genomes of susceptible and resistant organisms. Using whole-genome sequencing (WGS), potential resistance genes including β-lactamases, efflux pumps, porin proteins, PBPs, and iron transport were analyzed for potential gene mutations [41] (see supplemental genomic analysis data for details). There were no identified trends in the presence of resistance genes related to β-lactamases, efflux pumps, outer membrane porins, or iron transport when comparing isolates of all analyzed species. When analyzing the PBP genes of P. aeruginosa, a mutation in the PBP3, F533L, previously identified as a gain of function mutation for β-lactam antibiotics [42, 43] may be partially responsible for resistance to 1 and 2 (Table 5). The side chain of F533 in the active site of P. aeruginosa PBP3 makes contacts with the γ-lactam acylation product as seen in the X-ray structure (Figure 6).
Table 5.
MICs (μg/mL) of γ-lactams 1 and 2 against representative susceptible (MIC <=4 μg/mL) and resistant (MIC >= 16 μg/mL) clinical isolates of P. aeruginosa that have been whole genome sequenced and the identity of the amino acid at position 533: phenylalanine (wild-type) or leucine (mutation shown previously to provide gain of function for β-lactam antibiotics in some resistant strains of P. aeruginosa).
| P. aeruginosa ID no. | 1 | 2 | Position 533 residue |
|---|---|---|---|
|
| |||
| PR-545 | <= 0.25 | 0.5 | Phenylalanine |
| PR-548 | <= 0.25 | 1 | Phenylalanine |
| PR-567 | <= 0.25 | <= 0.25 | Phenylalanine |
| PR-672 | <= 0.25 | <= 0.25 | Phenylalanine |
| PR-604 | <= 0.25 | <= 0.25 | Phenylalanine |
| PR-638 | >16 | 0.5 | Phenylalanine |
| PR-676 | 16 | 2 | Phenylalanine |
| PR-503 | 16 | 16 | Phenylalanine |
| PR-564 | >16 | >16 | Leucine |
| PR-606 | 16 | >16 | Phenylalanine |
| PR-627 | >16 | >16 | Leucine |
| PR-635 | 16 | 16 | Phenylalanine |
| PR-636 | >16 | >16 | Leucine |
| PR-651 | >16 | >16 | Leucine |
| PR-668 | >16 | >16 | Leucine |
| PR-673 | >16 | >16 | Leucine |
| PR-677 | 16 | 16 | Phenylalanine |
| PR-680 | >16 | >16 | Leucine |
| PR-688 | >16 | >16 | Leucine |
| PR-699 | >16 | >16 | Phenylalanine |
2.8. β-Lactamase stability
As was previously reported, the dihydroxyphthalimide siderophore mimetic side chain appended to the γ-lactam core was: a) intended to impart stability against β-lactamase hydrolysis; and b) promote periplasm uptake. Thus, γ-Lactam 2 was evaluated as a potential substrate against representative members of all 4 Ambler classes of β-lactamases. We did not observe measurable reactions with KPC-2 (class A K. pneumoniae carbapenemase), ADC-7 (class C Acinetobacter-derived cephalosporinase) or OXA-23 (class D oxacillinase) when tested as purified isolated enzymes. Class B metallo β-lactamases NDM-1, VIM-2, and IMP-1, however, did demonstrate measurable activity (Table 6). Remarkably, the calculated catalytic efficiencies, kcat/Km of 0.003–0.02 μM−1s−1 were orders of magnitude lower than those previously determined for a comparator antibiotic (imipenem 2.7–6.7 μM−1s−1) [22] and similar to what was previously reported for γ-lactam 1 (0.02–0.07 μM−1s−1) [22]. Furthermore, the observed stability to purified β-lactamase enzymes correlates with the MIC data, where a relationship was not noted between microbiologic activity and the presence or absence of any β-lactamase gene. See Supplemental Table 3, page 27, for the full listing of the Acinetobacter spp. β-lactamase genes in this experiment.
Table 6.
Susceptibility of 2 to β-Lactamase hydrolysisa
| Metallo-β-lactamase | kcat (s−1) | Km (μM) | kcat/Km (μM−1s−1) |
|---|---|---|---|
|
| |||
| NDM-1 | 35 ± 4 | 1808 ± 200 | 0.020 ± 0.002 |
| VIM-2 | 54 ± 6 | 4400 ± 450 | 0.012 ± 0.001 |
| IMP-1 | 5 ± 1 | 1490 ± 300 | 0.003 ± 0.001 |
| KPC-2 | NM | NM | NM |
| ADC-7 | NM | NM | NM |
| OXA-23 | NM | NM | NM |
Steady-state reactions of 2 were monitored using purified enzymes: KPC-2, ADC-7, OXA-23, NDM-1, VIM-2, and IMP-1. Kinetic parameters provided were determined from double reciprocal plots (Supplemental Figure 2, page 44). Measurable reaction was not detected for KPC-2, ADC-7, or OXA-23 using 200 nM enzyme concentration and 100 μM 2. NM = not measureable.
2.9. Efflux inhibitor experiments
To further investigate potential resistance mechanisms, the growth inhibition of 2 in combination with efflux pump inhibitors (EPIs) was examined. The MIC (μg/mL) values of 2 in combination with the EPIs carbonyl cyanide 3-chlorophenylhydrazone (CCCP) or phenylalanine-arginine β-naphthylamide (PaβN) were tested against a subgroup of Acinetobacter spp. isolates resistant to 2 (Supplemental Table 4, page 32). The EPIs were kept at a constant concentration at levels commonly used in published reports against Gram-negative bacteria (2 μg/mL for CCCP and 32 μg/mL for PAβN). No significant inhibition effect on growth was observed directly from the EPIs, and no consistent effect on MIC values were observed when combined with 2. Potential synergistic effects were observed for only one isolate, PR-452, when combined with CCCP and since this was not a general effect, a detailed investigation was not pursued.
3. Conclusions
In conclusion, we demonstrate the potency and effectiveness of a novel γ-lactam siderophore antibiotic. Our intent was to build upon previous investigations performed with compound 1. We show potency against MDR strains of K. pneumoniae, Acinetobacter spp., and P. aeruginosa (MIC50 ≤ 0.5 μg/ml vs. > 8 for meropenem). Consistent with these microbiological findings, molecular analyses reveal stability against problematic β-lactamases. The atomic structure of P. aeruginosa PBP-3 at 2.0 Å resolution revealed an important “C-Cl … O” interaction with the carbonyl oxygen of Y407. PK/PD and animal testing established that reductions in CFU were significant when compared to colistin. Importantly, toxicity was not evident in a series of assays. Against CRAB, we lowered MICs significantly by combining 2 with sulbactam, designing a novel γ-lactam BLI combination. Studies are planned to further increase our understanding regarding optimal structure activity relationships (SARs) and dosing to overcome resistant infection.
4. Experimental
4.1. Syntheses
The reagents and solvents used for synthesis were of reagent grade quality. Dry solvents were purchased and used as such. All compounds were individually purified by chromatography on silica gel or by recrystallization and were of >95% purity for characterization purposes as determined by LCMS using UV absorption at 220 or 280 nM and/or NMR integration. In practice, compounds were not always purified to >95% purity prior to using in the next synthetic step, and often crude material was of sufficient purity and was carried forward. Copies of the spectra of 2 are included in the Supplemental Information, page 8.
Synthesis of (S,Z)-6-(2-(((1-(tert-butoxy)-2-methyl-1-oxopropan-2-yl)oxy)imino)-2-(5-chloro-2-(tritylamino)thiazol-4-yl)acetamido)-2-((2-(5,6-dihydroxy-1,3-dioxoisoindolin-2-yl)ethyl)carbamoyl)-5-oxo-6,7-dihydro-1H,5H-pyrazolo[1,2-a]pyrazole-3-carboxylic acid.
(Z)-2-(((1-(tert-butoxy)-2-methyl-1-oxopropan-2-yl)oxy)imino)-2-(5-chloro-2-(tritylamino)thiazol-4-yl)acetic acid [44] 15 (537 mg, 0.89 mmol) in dry dichloromethane (7 mL) and catalytic DMF was cooled in an ice bath and treated with oxalyl chloride (484 μL of 2.0 M solution, 0.97 mmol) and stirred for 1 hour. In a second flask starting amine 14 as the TFA salt (348 mg, 0.81 mmol) in dichloromethane (7 mL) was cooled in an ice bath and MSTFA (600 μL, 3.2 mmol) and Hunig’s base (773 μL, 4.4 mmol) was added. After stirring for 20 minutes all of the starting amine was in solution. During this time the dichloromethane from the acid chloride forming reaction was evaporated and placed on a high vacuum to give a colorless foam. This foam was dissolved in dry DCM (7 mL) and added to the MSTFA treated amine followed by the addition of Hunig’s base (250 μL, 1.4 mmol). The reaction was allowed to warm to room temperature over 1 hour and stirred overnight to give the crude product (S,Z)-6-(2-(((1-(tert-butoxy)-2-methyl-1-oxopropan-2-yl)oxy)imino)-2-(2-(tritylamino)thiazol-4-yl)acetamido)-2-((2-(5,6-dihydroxy-1,3-dioxoisoindolin-2-yl)ethyl)carbamoyl)-5-oxo-6,7-dihydro-1H,5H-pyrazolo[1,2-a]pyrazole-3-carboxylic acid that was not isolated but directly deprotected in situ.
Synthesis of (S,Z)-6-(2-(2-amino-5-chlorothiazol-4-yl)-2-(((2-carboxypropan-2-yl)oxy)imino)acetamido)-2-((2-(5,6-dihydroxy-1,3-dioxoisoindolin-2-yl)ethyl)carbamoyl)-5-oxo-6,7-dihydro-1H,5H-pyrazolo[1,2-a]pyrazole-3-carboxylic acid (2).
To the crude material from the overnight reaction above (0.82 g, 0.81 mmol) was added triethyl silane (0.64 mL, 4.0 mmol). The solution was cooled in and ice bath and trifluoroacetic acid (6.2 mL, 81 mmol) was added. The ice bath was removed after 30 minutes and the reaction was stirred at room temperature for 1 hour. Toluene (25 mL) was added and the reaction was evaporated to dryness. The crude reaction mixture was dissolved in dimethyl sulfoxide, diluted with water and chromatographed using reverse phase C18 MPLC eluting with 0 to 40% acetonitrile with 0.1% formic acid in water with 0.1% formic acid to give the product (S,Z)-6-(2-(2-amino-5-chlorothiazol-4-yl)-2-(((2-carboxypropan-2-yl)oxy)imino)acetamido)-2-((2-(5,6-dihydroxy-1,3-dioxoisoindolin-2-yl)ethyl)carbamoyl)-5-oxo-6,7-dihydro-1H,5H-pyrazolo[1,2-a]pyrazole-3-carboxylic acid, 2, as a yellow powder after lyophilization (139 mg, 23%). 1H NMR (400 MHz, DMSO-d6) δ 12.61 (s, 1H), 10.34 (s, 4H), 8.78 (d, J = 8.34 Hz, 1H), 8.56 – 7.91 (m, 1H), 7.38 (s, 2H), 7.12 (s, 4H), 5.03 (dt, J = 8.05, 10.89 Hz, 1H), 4.11 (d, J = 12.50 Hz, 1H), 3.81 (dd, J = 6.81, 10.26 Hz, 2H), 3.67 – 3.48 (m, 2H), 2.96 (dd, J = 8.71, 10.98 Hz, 1H), 1.66 – 1.36 (m, 6H). Mass spectrum M+H+ = 721.0; HRMS (ESI/QTOF) Calcd for C27H25Cl1N8O12S1 720.1001, found M+H+ 721.1077.
4.2. Minimum Inhibitory Concentrations, MICs
Bacterial strains used were from previously described collections [22, 45]. MICs were determined using the general recommendations of the Clinical and Laboratory Standards Institute (CLSI). Standard broth microdilution methods were followed but with a slightly lower inoculum (6 × 104 cfu/mL) which afforded no difference in MICs in our testing. MICs were performed using iron-depleted cation-adjusted Mueller-Hinton broth, except as mentioned elsewhere, using a standard protocol used with other siderophore-containing antibiotics.
4.3. Protein expression, purification, crystallization, and structure determination of the compound 2 P. aeruginosa PBP3 complex.
The P. aeruginosa PBP3 protein was expressed, purified, and crystallized as previously described [22]. A crystal of P. aeruginosa PBP3 was soaked with 2 mM compound 2 for 43 hours in mother liquor before freezing the crystal in liquid nitrogen prior to data collection. Data were collected at the SSRL synchrotron beamline 12–2 and processed to 2.0 Å resolution using HKL3000 [46]. The structure was solved using molecular replacement using PHASER [47] with the PBP3 ceftazidime complex protein coordinates as the search model (PBD ID 3PBO). The structure was refined using REFMAC 5 [48], and the program Coot [49] was used for model building. Refinement parameter files for compound 2 were generated using ACEDRG [50]. Molecular figures were generated using Pymol (www.pymol.org). The coordinates and structure factors of the P. aeruginosa PBP3 YU253911 complex have been deposited with the Protein Data Bank (PDB ID 7LC4).
4.4. PBP Binding Kinetics
A method was adapted from the work using purified, soluble P. aeruginosa PBP3 and Bocillin™, a fluorescent β-lactam and substrate [37]. Reactions were conducted in 10 mM phosphate-buffered saline at pH 7.4 using 1.6 μM P. aeruginosa PBP3 incubated with increasing concentrations of 2. To ensure that equilibrium between 2 and PBP3 occurred, the enzyme was preincubated with the compound for 20 min at 37 °C before the addition of 50 μM Bocillin™ and then incubated for an additional 20 min. The reactions were stopped by adding SDS-PAGE loading dye and boiling for 2 min. Samples were analyzed by SDS-PAGE and the gel illuminated at λ = 365 nm and imaged with a Fotodyne gel imaging system. ImageJ analysis software was used to assign fluorescence intensity (FI). The IC50 was calculated as the concentration of 2 required to reduce the FI of the Bocillin™-labeled protein by 50%.
4.5. β-lactamase stability testing
Steady-state kinetics were determined with purified enzymes (KPC-2, ADC-7, OXA-23, NDM-1, VIM-2, and IMP-1) using an Agilent model 8453 diode array spectrophotometer as previously described [22]. Assays were performed at 25 °C (room temperature) using 10 mM PBS, pH 7.4 (KPC-2, ADC-7), 50 mM sodium phosphate buffer supplemented with 20 mM sodium bicarbonate (OXA-23) or 10 mM HEPES, pH 7.5, 0.2 M NaCl, 50μg/ml bovine serum albumin, and 50 μM Zn (NDM-1, VIM-2, and IMP-1).
Compound 2 was used as a substrate at excess molar concentrations to establish pseudo-first-order kinetics. The following extinction coefficient was used: 2, Δɛ336 = −4362 M−1 cm−1. For velocity determinations, a 0.2 cm path length quartz cuvette was employed. Hydrolysis of 600 μM 2 was monitored over time until completion. Based on the starting absorbance (λ = 336 nm, 600 μM 2) and final absorbance (λ = 336 nm, 0 μM 2), concentrations of 2 were determined along the progress curve and velocities during a 10 second period for each concentration calculated. A double reciprocal plot ( vs. ) was employed to obtain the steady-state kinetic parameters , , and according to Equations 1 and 2:
| Eq. 1 |
| Eq. 2 |
where = reaction velocity, = Michaelis-Menten constant, = maximum reaction velocity, and [S] = substrate concentration. Compound 1 was retested with NDM-1, VIM-2, and IMP-1 using this same method, resulting in similar values to those previously reported [22] (Supplemental Figure 1, page 43 and Supplemental Table 5, page 44).
4.6. Genomic analysis
Acinetobacter spp., and K. pneumoniae sequences were downloaded from the sequencing read archive project PRJNA384060, PRJNA384065, and PRJNA339843, respectively (National Center for Biotechnology Information, U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA). Pseudomonas aeruginosa were sequenced by the Illumina Hiseq platform with 2×150bp paired-end sequencing. Reads were assembled and annotated using PATRIC, the Pathosystems Resource Integration Center [41]. Antibiotic resistant genes were identified using RES Finder (Center for Genomic Epidemiology; http://www.genomicepidemiology.org/). Amino acid variations were identified by comparison of strains to the Acinetobacter ATCC 17978 and ATCC 19606, Pseudomonas aeruginosa PA01, or Klebsiella pneumoniae MGH 78578 (GenBank Accessions CP000523, ACQB00000000, CP001183, CP000647).
Supplementary Material
Acknowledgements
MSP: Financial support was provided by the Program in Innovative Therapeutics for Connecticut’s Health grant by the Connecticut Bioscience Innovation Fund, administered by the Connecticut Innovations. RAB: Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under Award Numbers R01AI100560, R01AI063517, and R01AI072219. This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Numbers 1I01BX002872 to KMP-W and 1I01BX001974 to RAB, from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10 (RAB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
BNK: Research reported in this publication was supported by a grant from the National Institutes of Health, R01AI090155.
RAB wishes to gratefully thank the Antimicrobial Resistance Leadership Group (ARLG) for the use of the isolates. The research described in this publication is supported, at least in part, by a grant from the National Institutes of Health through Duke University and the findings, opinions, and recommendations expressed herein are those of the authors and not necessarily those of Duke University or the National Institutes of Health. This study is also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under Award Number UM1AI104681.
FVDA: We also thank Stanford Synchrotron Radiation Lightsource (SSRL) for help with data collection. Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Abbreviations used
- MDR
multidrug-resistant
- PBP
penicillin-binding protein
- MIC
minimal inhibitory concentration
- blas
β-lactamases
- SC
subcutaneously
- IM
intramuscularly
- CFU
colony-forming unit
- WGS
whole-genome sequencing
- CRAB
carbapenem-resistant Acinetobacter baumannii
- CSAB
carbapenem-susceptible Acinetobacter baumannii
- BLIs
β-lactamase inhibitors
- LCMS
liquid chromatography mass spectrometry
- NMR
nuclear magnetic resonance
- UV
ultraviolet
- DMSO
dimethylsulfoxide
- MSTFA
N-methyl-N-(trimethylsilyl)-trifluoroacetamide
- AN
Acinetobacter nosocomialis
- FI
fluorescence intensity
- SAR
structure activity relationship
Footnotes
Accession codes
PDB code for P. aeruginosa PBP3 with bound YU253911, 2, is PDB ID 7LC4. Authors will release the atomic coordinates upon article publication.
References
- [1].a) Centers for Disease Control and Prevention (2019) Antibiotic Resistance Threats in the United States, 2019 AR Threats Report https://www.cdc.gov/drugresistance/biggest-threats.html (accessed March 11, 2020); b) Cassini A; Högberg LD; Plachouras D; Quattrocchi A; Hoxha A; Simonsen GS; Colomb-Cotinat M; Kretzschmar ME; Devleesschauwer B; Cecchini M; Ouakrim DA; Oliveira TC; Struelens MJ; Suetens C; Monnet DL; and the Burden of AMR Collaborative Group; Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis, Lancet Infectious Disease 2019, 19, 56–66. Doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].World Health Organization. Antimicrobial Resistance: Global Report on Surveillance, 2014. https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/ (accessed March 11, 2020)
- [3].Spellberg B; Guidos R; Gilbert D; Bradley J; Boucher HW; Scheld WM; Bartlett JG; Edwards J Jr. Infectious Diseases Society of America, The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America, Clin. Infect. Dis. 2008, 46, 155–164. Doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- [4].Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M;Spellberg B; Bartlett J Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America, Clin. Infect. Dis. 2009, 48, 1–12. Doi . [DOI] [PubMed] [Google Scholar]
- [5].World Health Organization. Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics, 2017. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed March 11, 2020)
- [6].Peleg AY; Hooper DC Hospital-acquired infections due to Gram-negative bacteria, N. Engl. J. Med. 2010, 362, 1804–1813. Doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sydnor ER; Perl TM Hospital epidemiology and infection control in acute-care settings, Clin. Microbiol. Rev. 2011, 24, 141–173. Doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hassani M The crisis of Gram-negative bacterial resistance: is there any hope for ESKAPE?, Clinical Research in Infectious Disease. 2014, 1, 1005–1009. [Google Scholar]
- [9].Silver LL Challenges of antibacterial discovery, Clin. Microbiol. Rev. 2011, 24, 71–109. Doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Payne DJ; Gwynn MN;Holmes DJ; Pompliano DL Drugs for bad bugs: confronting the challenges of antibacterial discovery, Nat. Rev. Drug Discov. 2007, 6, 29–40. Doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- [11].So AD; Gupta N; Brahmachari SK; Chopra I; Munos B; Nathan C; Outterson K; Paccaud JP; Payne DJ; Peeling RW; Spigelman M; Weigelt J Towards new business models for R&D for novel antibiotics, Drug Resist. Updat. 2011, 14, 88–94. Doi: 10.1016/j.drup.2011.01.006. [DOI] [PubMed] [Google Scholar]
- [12].Livermore DM Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002, 34, 634–640. Doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- [13].Tomas M; Doumith M; Warner M; Turton JF; Beceiro A; Bou G; Livermore DM; Woodford N Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients, Antimicrob. Agents Chemother. 2010, 54, 2219–2224. Doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ternansky RJ; Draheim SE [4.3.0] Pyrazolidionones as potential antibacterial agents, Tet. Let. 1988, 29, 6569–6572. [Google Scholar]
- [15].Allen NE; Hobbs JN Jr.; Preston DA; Turner JR; Wu CY Antibacterial properties of the bicyclic pyrazolidinones, J. Antibiot. (Tokyo). 1990, 43, 92–99. Doi: 10.7164/antibiotics.43.92. [DOI] [PubMed] [Google Scholar]
- [16].Ternansky RJ; Draheim SE Structure-activity relationship within a series of pyrazolidinone antibacterial agents. 1. Effect of nuclear modification on in vitro activity, J. Med. Chem. 1993, 36, 3219–3223. Doi: 10.1021/jm00074a001. [DOI] [PubMed] [Google Scholar]
- [17].Ternansky RJ; Draheim SE; Pike AJ, Counter FT; Eudaly JA; Kasher JS Structure-activity relationship within a series of pyrazolidinone antibacterial agents. 2 Effect of side-chain modification on in vitro activity and pharmacokinetic parameters, J. Med. Chem. 1993, 36, 3224–3229. Doi: 10.1021/jm00074a002. [DOI] [PubMed] [Google Scholar]
- [18].Boyd DB Application of the hypersurface iterative projection method to bicyclic pyrazolidinone antibacterial agents, J. Med. Chem. 1993, 36, 1443–1449. Doi: 10.1021/jm00062a017. [DOI] [PubMed] [Google Scholar]
- [19].Jungheim LN; Sigmund SK; Fisher JW Bicyclic Pyrazolidinones, a New Class of Antibacterial Agents Based on the β-Lactam Model., Tetrahedron Letters, 1987, 28, 285–288. [Google Scholar]
- [20].Jungheim LN; Sigmund SK; Jones ND; Swartzendruber J Bicyclic Pyrazolidinones, Steric and Electronic Effects on Antibacterial Activity Tetrahedron Letters, 1987, 28, 289–292. [Google Scholar]
- [21].Jungheim LN,.; L.N.Ternansky RJ; Holmes RE. Bicyclic Pyrazolidinone Antibacterial Agents Drugs of the Future, 1990, 15, 149; Doi: 10.1358/dof.1990.015.02.114554. [DOI] [Google Scholar]
- [22].Goldberg JA; Nguyen H; Kumar V; Spencer EJ; Hoyer D; Marshall EK; Cmolik A; O’Shea M; Marshall SH; Hujer AM; Hujer KM; Rudin SD; Domitrovic TN; Bethel CR; Papp-Wallace KM; Logan LK; Perez F; Jacobs MR; van Duin D; Kreiswirth BM; Bonomo RA; Plummer MS; van den Akker F A γ-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli Journal of Medicinal Chemistry 2020, 63 (11), 5990–6002. Doi: 10.1021/acs.jmedchem.0c00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cornelis P; Matthijs S; Van Oeffelen L Iron uptake regulation in Pseudomonas aeruginosa Biometals 2009, 22, 15–22. Doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- [24].a) Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in iron metabolism: from mechanism to therapy potential trends in molecular medicine 2016, 22,12. 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schalk IJ; Mislin GLA; Bacterial iron uptake pathways: gates for the import of bactericide compounds, J. Med. Chem. 2017, 60, 4573–4576. Doi: 10.1021/acs.jmedchem.7b00554. [DOI] [PubMed] [Google Scholar]
- [25].Luscher A; Moynie l.; Auguste PS; Bumann D; Mazza; Pletzer d.; Naismith JH; Kohler T TonB-Dependant receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates, Antimicrob. Agents Chemother. 2018, 63, 6, e00097–e00118. Doi: 10.1128/AAC.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baudart MG; Hennequin LF Synthesis and biological activity of C-3’ ortho dihydroxyphthalimido cephalosporins, The Journal of Antibiotics 1993, 46, 9, 1458–1470. Doi: 10.7164/antibiotics.46.1458. [DOI] [PubMed] [Google Scholar]
- [27].Evans SR; Tran TTT; Hujer AM; Hill CB; Hujer KM; Mediavilla JR; Manca C; Domitrovic TN; Perez F; Farmer M; Pitzer KM; Wilson BM; Kreiswirth BN; Patel R; Jacobs MR; Chem L; Fowler VG; Chambers HF; Bonomo RA Rapid molecular diagnostics to inform empiric use of Ceftazidime/Avibactam and Ceftolozane/Tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin. Infect. Dis. 2019, 68, 1823–1830. Doi: 10.1093/cid/ciy801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Henig O; Cober E; Richter SS; Perez F; Salata RA; Kalayjian RC; Watkins RR; Marshall S; Rudin SD; Domitrovic TN; Hujer AM; Hujer KM; Doi Y; Evans S; Fowler VG Jr.; Bonomo RA; van Duin D; Kaye KS A prospective observational study of the epidemiology, management, and outcomes of skin and soft tissue infections due to carbapenem-resistant Enterobacteriaceae. Open Forum Infect Dis 2017, 4, ofx157. Doi: 10.1093/ofid/ofx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ito A; Nishikawa T; Matsumoto S; Yoshizawa H; Sato T; Nakamura R; Tsuji M; Yamano Y Siderophore cephalosporin Cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa, Antimicrob. Agents Chemother. 2016, 60, 7396–7401. Doi: 10.1128/AAC.01405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Evans SR; Hujer AM; Jiang H; Hill CB; Hujer KM; Mediavilla JR; Manca C; T. Tran, TT; Domitrovic N,.; Higgins PG; Seifert H; Kreiswirth BN; Patel R; Jacobs MR; Chen L; Sampath R; Hall T; Marzan C; Fowler VG; Chambers HF; Bonomo RA Antibacterial Resistance Leadership Group. Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics to identify susceptibility and resistance to carbapenems against Acinetobacter spp. in PRIMERS III. J. Clin. Microbiol. 2017, 55, 134–144. Doi: 10.1128/JCM.01524-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nechifor M; Buic D; Diaconu E; Poiata A; Teslariu E; Filip C; Negru A; Antonescu C Pharmacological studies of sulbactam and its association with semisynthetic beta-lactam antibiotics. Rev. Med. Chir. Soc. Med. Nat. Iasi. 1992, 96, 51–55. [PubMed] [Google Scholar]
- [32].Berezhinskaia VV; Dolgova GV; Egorenko GG; Svinogeeva TP; Shtegel’man LA; Smolkina TV; Nikitin AV Study of general toxic and organotropic properties of ampicillin combined with sulbactam. Antibiot. Khimioter. 1992, 37, 25–28. Doi: 10.1517/17425250903145251. [DOI] [PubMed] [Google Scholar]
- [33].Betrosian AP; Douzinas EE Ampicillin-sulbactam: an update on the use of parenteral and oral forms in bacterial infections. Expert Opin. Drug Metab. Toxicol. 2009, 5, 1099–1112. [DOI] [PubMed] [Google Scholar]
- [34].Benson JM; Nahata MC Sulbactam/ampicillin, a new beta-lactamase inhibitor/beta-lactam antibiotic combination. Drug Intell. Clin. Pharm. 1988, 22, 534–541. Doi: 10.1177/106002808802200702. [DOI] [PubMed] [Google Scholar]
- [35].Hampel B; Lode H; Bruckner G; Koeppe P Comparative pharmokinetics of sulbactam/ampicillin and clavulanic acid/amoxycillin in human volunteers. Drugs. 1988, 35, 29–33. Doi: 10.2165/00003495-198800357-00007. [DOI] [PubMed] [Google Scholar]
- [36].Penwell WF; Shapiro AB; Giacobbe RA; Gu RF; Gao N; Thresher J; McLaughlin RE; Huband MD; DeJonge BLM; Ehmann DE; Miller AA Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 1680–1689. Doi: 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Papp-Wallace KM; Senkfor B; Gatta J; Chai W; Taracila MA; Shanmugasundaram V; Han S; Zaniewski RP; Lacey BM; Tomaras AP; Skalweit MJ; Harris ME; Rice LB; Buynak JD; Bonomo RA Early insights into the interactions of different β-lactam antibiotics and β-lactamase inhibitors against soluble forms of Acinetobacter baumannii PBP1a and Acinetobacter sp. PBP3. Antimicrob. Agents Chemother. 2012, 56, 5687–5692. Doi: 1128/AAC.01027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38).Polsinelli I; Savko M; Rouanet-Mehouas C; Ciccone L; Nencetti S; Orlandini E; Stura EA; Shepard W Comparison of helical scan and standard rotation methods in single-crystal X-ray data collection strategies J. Synchrotron Radiat. 2017. Jan 1, 24(Pt 1),42–52. Doi: 10.1107/S1600577516018488. [DOI] [PubMed] [Google Scholar]
- [39].Bissantz C; Kuhn B; Stahl M A medicinal chemist’s guide to molecular interactions J Med Chem. 2010, 53(14), 5061–84. Doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shinda NK; de Brevern AG; Schmidtke P Halogens in Protein-Ligand Binding Mechanism: A Structural Perspective J Med Chem. 2019, 62(21), 9341–9356. Doi: 10.1021/acs.jmedchem.8b01453. [DOI] [PubMed] [Google Scholar]
- [41].Davis JJ; Wattam AR; Aziz RK; 4,5,Brettin T; Butler R; Butler RM; Chlenski P; Conrad N; Dickerman A; Dietrich EM; Gabbard JL; Gerdes S; Guard A; Kenyon RW; Machi D; Mao C; Murphy-Olson D; Nguyen M; Nordberg EK; Olsen GJ; Olson RD; Overbeek JC; Overbeek R; Parrello B; Pusch GD; Shukla M; Thomas C; VanOeffelen M; Vonstein V; Warren AS; Xia F; Xie D; Yoo H; Stevens R The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, 606–612. Doi: 10.1039/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Han S; Zaniewski RP; Marr ES; Lacey BM; Tomaras AP; Evdokimov A; Miller, R J; Shanmugasundaram V Structural basis for effectiveness of siderophoreconjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. PNAS 2010, 107, 22002–22007. Doi: 10.1073/pnas/1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].López-Causapé C; Cabot G; del Barrio-Tofiño E; Oliver A The versatile mutational resistome of Pseudomonas aeruginosa. Frontiers in Microbiology 2018, 9, 1–9. Doi: 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Preparation of siderophore conjugated pyrazolidinones, and analogs thereof as antibacterial agents; Plummer M; Hoyer D; Spencer E; WO2020018929. [Google Scholar]
- [45].Evans SR; Hujer AM; Jiang H; Hill CB; Hujer KM; Mediavilla JR; Manca C; Tran TT; Domitrovic TN; Higgins PG; Seifert H; Kreiswirth BN; Patel R; Jacobs MR; Chen L; Sampath R; Hall T; Marzan C; Fowler VG Jr; Chambers HF; Bonomo RA Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics to identify susceptibility and resistance to carbapenems against Acinetobacter spp. in PRIMERS III. J Clin Microbiol 2016, 55, 134–144. Doi: 10.1128/JCM.01524-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wladek M; Cymborowski M; Zbyszek O; Chruszcz M HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes Acta Crystallogr D Biol Crystallogr. 2006. Aug;62(Pt 8):859–66. Doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- [47].McCoy AJ; Grosse-Kunstleve RW; Adams PD; Winn MD; Storoni LC; Read RJ Phaser crystallographic software, J Appl Crystallogr. 2007, 40, 658–674. Doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Murshudov GN; Skubak P; Lebedev AA; Pannu NS; Steiner RA; Nicholls RA; Winn MD; Long F; Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures, Acta crystallographica Section D, Biological crystallography. 2011, 67, 355–367. Doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Emsley P; Cowtan KC COOT: model-building tools for molecular graphics, Acta crystallographica Section D, Biological crystallography. 2004, 60, 2126–2132. Doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [50].Long F; Nicholls RA; Emsley P; Graaeulis S; Merkys A; Vaitkus A; Murshudov GN AceDRG: a stereochemical description generator for ligands. Acta Crystallogr D Struct Biol. 2017. Feb 1;73(Pt 2):112–122. Doi: 10.1107/S2059798317000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


