Abstract
Naturally occurring hepatitis C virus (HCV) infection has long been thought to induce a weak immunity which is insufficient to protect an individual from subsequent infections and has cast doubt on the ability to develop effective vaccines. A series of intrahepatic genetic inoculations (IHGI) with type 1a HCV RNA were performed in a chimpanzee to determine whether a form of genetic immunization might stimulate protective immunity. We demonstrate that the chimpanzee not only developed protective immunity to the homologous type 1a RNA after rechallenge by IHGI but was also protected from chronic HCV infection after sequential rechallenge with 100 50% chimpanzee infectious doses of a heterologous type 1a (H77) and 1b (HC-J4) whole-virus inoculum. These results offer encouragement to pursue the development of HCV vaccines.
Approximately 15 to 20% of hepatitis C virus (HCV)-positive individuals, an estimated 170 million people worldwide (48), progress to chronic infections, of which approximately 20 to 30% manifest clinical liver disease (2, 21). Long-term survival of human immunodeficiency virus (HIV)-positive individuals are also increasing the number of HIV/HCV-coinfected patients due to improved antiviral therapies for HIV (33). Antiviral treatments for HCV are currently limited to alpha interferon and ribavirin, which have a maximum combined reported efficacy of ∼40% of treated individuals (12, 29, 34). The lack of vaccines or highly effective antiviral treatments is a major medical concern.
HCV (10) is a member of the genus Hepacivirus, within the family Flaviviridae, and is composed of six major genotypes (40). Type 1 HCV, which comprises the major subtypes 1a and 1b, represents the most common genotype around the world (6). The viral genome is a positive-stranded RNA (10) and has been shown to circulate as a quasispecies (28), which is typical of many RNA viruses. A long open reading frame (ORF) is cotranslationally processed into the structural and nonstructural (NS) proteins by cellular signal peptidase or viral proteases (21). NS3 encodes protease and helicase enzymatic activities, both of which have been proven to be essential to viral replication in vivo (26). The 5′- and 3′-terminal untranslated regions (UTR) (25, 42), which are required for replication and translation functions, flank the long ORF (9). The structural proteins include the nucleocapsid or core (C), envelope 1 (E1), and envelope 2 (E2) glycoproteins. A hypervariable domain of 30 amino acids located at the NH terminus of E2 (HVR1) (23, 47) has been shown to encode virus-neutralizing antibodies (17, 38) and has been implicated in generating neutralization escape mutants (reviewed in reference 46).
The prospect of developing HCV vaccines has been vigorously debated. The lack of strong natural immunity in humans and chimpanzees (14, 35), the only reproducible animal model for HCV infection (1, 4, 41), has been cited as evidence against successful vaccines (16). Despite this concern, studies in chimpanzees have demonstrated that animals can develop immunity to an isolate-specific challenge (8). The variability of the viral genome, especially in the envelope proteins, raises questions as to the host's ability to mount an adequate cross-protective immune response. We investigated the ability of a chimpanzee to develop immunity to both homologous and heterologous type 1 HCV rechallenges using intrahepatic genetic inoculation (IHGI) of HCV RNA. These results indicate for the first time that protection from both homologous and heterologous HCV challenge can be achieved.
MATERIALS AND METHODS
Molecular clones of HCV.
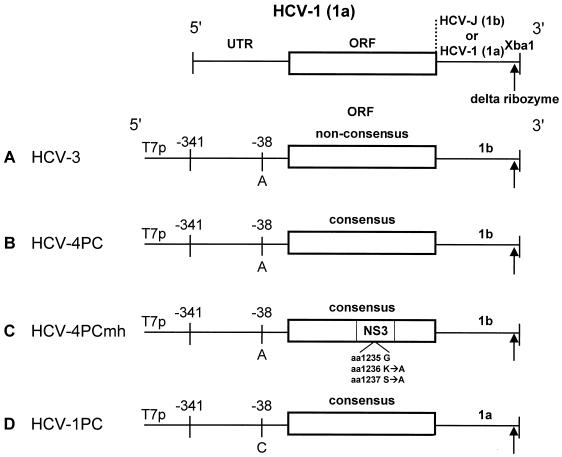
RNA extracted from plasma obtained from three different chimpanzees infected with HCV-1 was converted into cDNA and subjected to reverse transcription (RT)-PCR essentially as described by Yanagi et al. (50). The predicted amino acid sequence from the consensus nucleotide sequence of approximately 6.5 genomes corresponded exactly with the published HCV-1 sequence (9) with one exception (valine to glycine substitution at amino acid 2021). pHCV-4PC (Fig. 1B) was generated by recloning pTMHCV-3 (13) (Fig. 1A), a chimeric cDNA clone containing a nonconsensus HCV-1 (1a) 5′UTR through NS5B and a 1b 3′UTR, into a pUC19 plasmid backbone and using site-directed mutagenesis to create a consensus coding region. Amino acids 1236-Lys and 1237-Ser were converted to Ala in order to mutate the GKS motif (45) of the NS3 helicase domain in HCV-4PCmh (Fig. 1C). HCV-4PC was also modified to contain the authentic HCV-1 3′UTR and consensus 5′UTR using overlapping synthetic oligonucleotides, generating a full-length HCV-1 (1a) genome (Fig. 1D). Two versions of each construct were made so that the extreme 3′ terminus would either be exactly as expected in the genome due to cleavage by the hepatitis D virus ribozyme (49) (Fig. 1) or have a 4-nucleotide overhang after digestion by XbaI. All clones contained the T7 polymerase promoter immediately upstream from the 5′UTR to facilitate synthesis of HCV RNA with the exact 5′ terminus of the genome by in vitro transcription of XbaI-linearized plasmid DNA.
FIG. 1.
Molecular clones of HCV. A chimeric molecular clone (HCV-3) containing the bacteriophage T7 promoter upstream from the 5′ UTR and coding region of HCV-1, type 1a, and the 3′UTR of HCV-J, type 1b, was recloned into pUC19 and modified to create clones: (B) HCV-4PC, (C) HCV-4P Cmh (mutant helicase), and (D) HCV-1PC. All clones except the parent clone HCV-3 had the consensus amino acid sequence for HCV-1 according to Choo et al. and either one of two different extreme 3′ ends (XbaI with or without the hepatitis delta virus ribozyme [↑] [49]). The aa 1236 and 1237 had K-to-A and S-to-A substitutions, respectively, introduced into the helicase domain of NS3 in HCV-4PCmh (C). HCV-1PC (D) represents the full-length HCV-1 genome.
Intrahepatic inoculation of HCV RNA.
Equal amounts of RNA transcribed from either the delta ribozyme/XbaI or XbaI-only templates described in Fig. 1 were each injected directly into three sites in the liver of a chimpanzee (4X0202) by ultrasound-guided procedures described by Yanagi et al. (50). Intrahepatic inoculation of a total of ∼1 mg of RNA was performed according to the schedule indicated by arrows in Fig. 2.
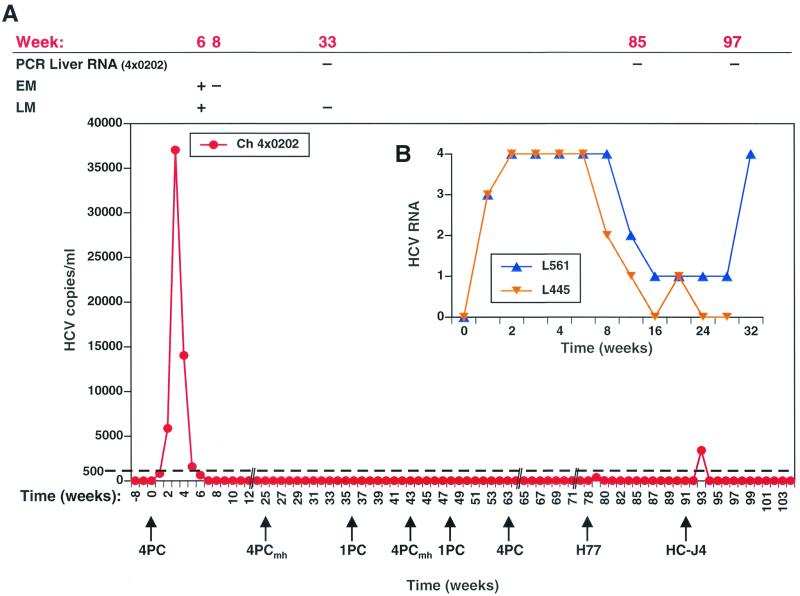
FIG. 2.
HCV RNA in challenged chimpanzees and histology in 4X0202. HCV RNA copy number was measured by Taqman RT-PCR from week −8 to +104 in chimpanzee (Ch) 4X0202, who was given intrahepatic genetic inoculations of HCV-4PC, HCV-4PCmh, and HCV-1PC as indicated (arrows in A). The i.v. challenge of heterologous viral inocula (100 CID50) was performed on weeks 78 (H77, 1a) and 92 (HC-J4, 1b). RNA extracted from liver was also tested by PCR on weeks 33, 85, and 97. Light (LM) and electron microscopic (EM) analysis of liver biopsy tissue was scored positive or negative for histological abnormalities typical of viral hepatitis, as indicated. Semiquantitative RT-PCR was performed on two naive control animals, L445 and L561, who were inoculated IV with 100 CID50 of H77. Scores of 4+, 3+, 2+, and 1+ represent approximately ≥1,000, 500, 250, and 50 copies of HCV RNA, respectively, in the semiquantitative RT-PCR assay (8).
RT-PCR assays.
RNA was extracted from plasma using the Qiagen viral extraction kit and from liver using the RNeasy kit (Qiagen). Taqman quantitative RT-PCR was performed exactly as described by Pileri et al. (32) using primers from the 5′UTR. The maximum sensitivity of the assay was seven copies of HCV, based on the fluorescent signal after RT-PCR on RNA extracted from serial dilutions of an HCV-containing plasma quantitated by Quantiplex 2.0 bDNA (Bayer) assay. A sensitivity of ≤30 copies/ml was achieved by concentrating RNA from 1 ml of plasma on Qiagen columns. Only samples that were negative in the standard 100-μl assay were subjected to 1-ml extraction followed by RT-PCR. Semiquantitative RT-PCR was performed according to Choo et al. (8). Approximately ≥1,000, 500, 250, and 50 molecules of HCV correspond to scores of 4+, 3+, 2+, and 1+, respectively.
Chimpanzees.
Chimpanzees (Pan troglodytes) housed at the Southwest Foundation for Biomedical Research (SFBR; San Antonio, Tex.). Chimpanzee 4X0202 had fluctuating alanine aminotransferase (ALT) values prior to challenge with HCV RNA, which is not uncommon in chimpanzees, compromising the interpretation of these values. ALT values did not correlate with HCV RNA levels in this animal. Chimpanzee L497 resolved a previous HCV infection prior to being inoculated with acute-phase plasma from 4X0202. All studies were approved by the Institutional Animal Care and Use Committees of Chiron Corporation and of SFBR and were performed under the NIH Guidelines for Care and Use of Laboratory Animals.
Proteins and rVV.
The HCV-1a proteins SOD-C22-3 (amino acids [aa] 12 to 120), SOD-C25 (aa 2 to 120 and 1192 to 1935), CHO E2 (aa 384 to 715), CHO E1E2 (aa 192 to 746); SOD-C33c (aa 1192 to 1457), SOD-C200 (aa 1192 to 1931); SOD-C100 (aa 1569 to 1931), and SOD-NS5a (aa 2054 to 2995) were produced in yeast or CHO cells and were approximately 90% pure. The recombinant vaccinia viruses (rVV) expressing core/E1 (aa 1 to 384), E1/E2 (aa 134 to 966), E2-NS3 (aa 364 to 1648), NS3/4 (aa 1590 to 2050), NS5a (aa 2005 to 2396), and NS5b (aa 2396 to 3011) of HCV-1a as well as wild-type VV (VVwt) have been described previously (11).
Cell lines and cellular assays.
The B-lymphoblastoid cell line (B-LCL) line was derived using supernatants from the Epstein-Barr virus producer cell line B95-8. Peripheral blood mononuclear cells (PBMC) purified by centrifugation over a Ficoll-Hypaque gradient were directly entered at 2 × 105 per well into a standard lymphoproliferation assay as described (20) or cultured at 5 × 106 cells per well in the presence of 106 psoralen-treated, rVV-infected autologous B-LCLs in 2 ml of RPMI 1640 culture medium containing 10% heat-inactivated fetal bovine serum and 1% antibiotics and supplemented with 5% interleukin-2-containing supernatant (T-STIM without phytohemagglutinin [PHA]; Collaborative Biomedical Products, Bedford, Mass.) and 50 U of recombinant interleukin-2 (IL-2) per ml (Chiron). Cultures were fed every 3 to 4 days. After 8 to 12 days in culture, CD8+ T cells were purified using anti-CD8 antibodies bound to magnetic beads (Dynal, Oslo, Norway) according to the manufacturer's specification. Purified CD8+ cells (>94% pure as determined by flow cytometry) were cultured for another 2 to 3 days prior to being assayed for cytotoxic activity against a panel of VV-infected autologous B-LCLs by standard 51Cr release assay as described (11). Briefly, B-LCLs were infected at a multiplicity of infection of 10:1 with rVV expressing HCV antigens or VVwt for 1 h, washed, and cultured overnight prior to labeling with 51Cr. CD8+ cells were plated in duplicate at three different effector-to-target cell (E:T) ratios (40:1, 13:1, and 4:1) and incubated with target cells for 4 h in the presence of 2 × 105 unlabeled target cells per well.
Intrahepatic lymphocytes were prepared as described (11) and restimulated in vitro in the presence of 2 × 106 irradiated human feeder cells, IL-2 (50 U/ml), 5% IL-2-containing supernatant, and PHA (1 μg/ml). After the cultures reached a cell number sufficient to allow testing, CD8+ and CD8− (CD4+) T cells were isolated using anti-CD8 antibodies bound to magnetic beads. CD8+ T cells were tested for cytolytic activity as described above. CD8− (CD4+) cells were plated at 5 × 104 cells per well in the presence of 5 × 104 irradiated autologous B-LCLs and HCV proteins (5 μg/ml) as well as a CHO control (0.5 μg/ml) and a yeast control (0.5 μg/ml). After 72 h, plates were pulsed with 1 μCi of [3H]-thymidine per well and harvested 6 to 8 h later. A stimulation index (SI), calculated as [(mean experimental cpm)/(mean cpm in the presence of either the CHO or SOD control)], equal to or greater than 3.0 was considered positive.
HCV antibody assays.
Serum levels of antibodies against structural and nonstructural HCV proteins were quantified by enzyme-linked immunosorbent assay (ELISA) as described (7). HVR 1 ELISA plates were prepared by binding 1 μg of biotinylated peptide (HCV-1, ETHVTGGSAGHTVSGFVSLLAPGAKQN; H77, ETHVTGGNAGRTTAGLVGLLTPGAKQN or ETHVTGGSAGRTTAGLVGLLTPGAKQN) to a steptavidin-coated high-binding Costar 96-well plate, washing with distilled water followed by coat buffer (0.0525 M Na2HPO4, 0.175 M KH2PO4, 0.075 M NaCl), and stored dry at −20°C. Serial dilutions of chimpanzee plasma containing 1 μg of CHO cell lysate per ml were incubated with peptide-coated plates for 1 h at 37°C, washed five times with wash buffer (1 × phosphate-buffered saline, 1% bovine serum albumin, 0.02% NaN3), and reacted with goat antiimmunoglobulin G (heavy and light chain)-peroxidase, F(ab′)2 for an additional 1 h at 37°C. After washing five times with wash buffer, the plate was developed with o-phenylenediamine dihydrochloride at room temperature for 30 min. The reaction was stopped with 4 N sulfuric acid and read in a spectrophotometer at 492 and 620 nm. The cutoff was determined as described (7).
IFN assays.
Levels of alpha interferon (IFN-α), IFN-β, and IFN-γ present at various time points in the chimpanzee plasma were determined by specific ELISAs (BioSource, Camarillo, Calif.) according to the manufacturer's recommendations. Due to sample dilution, the sensitivity of these assays was 40 pg/ml, 5 IU/ml, and 40 pg/ml, respectively.
RESULTS
Intrahepatic inoculation of molecular clones of HCV induce protection against a homologous RNA rechallenge in a chimpanzee.
Three molecular clones were assessed for infectivity and the ability to induce immunity to HCV infection in chimpanzee 4X0202 after intrahepatic inoculation of RNA (24, 50) transcribed from the templates shown in Fig. 1B to D. Following challenge with HCV-4PC, a chimeric clone composed of type 1a 5′UTR through the stop codon of the long ORF and a type 1b 3′UTR, an approximately 50-fold increase in HCV RNA was observed over the first 3 weeks postchallenge. The animal remained PCR positive until week 6 (Fig. 2A), at which time hallmarks of HCV infection, including mild inflamation and ultrastructural changes in hepatocytes (5, 22, 37), were identified by light microscopy and electron microscopy. Pooled plasma from weeks 3 and 4 postinoculation transmitted HCV to a second animal (L497) following intravenous (i.v.) challenge, further proving that replicating HCV-4PC RNA was packaged into infectious virus particles (data not shown).
To prove that the RNA detected 1 to 6 weeks postchallenge was not due to the initial inoculum, the equivalent amount of RNA from a putative defective genome, which had mutations in an essential motif in the helicase domain of NS3 (HCV-4PCmh; Fig. 1C), was introduced by IHGI into 4X0202. Since RNA was not detected up to 8 weeks postchallenge, only replication of the HCV-4PC genome could account for the PCR-positive signal recorded after challenge. To our surprise, the subsequent IHGI of RNA from either the putative infectious HCV-1PC or the proven infectious HCV-4PC clone also failed to replicate to detectable levels in 4X0202. (Note: the HCV-1PC clone was verified by DNA sequencing, and expression of HCV proteins was confirmed in both a vaccinia virus and a defective adenovirus infection-transfection expression system [data not shown].) The data clearly demonstrate that although the animal had been infected with HCV-4PC 63 weeks earlier, he developed protective immunity to a homologous RNA rechallenge.
IHGI of infectious and noninfectious HCV RNA protects a chimpanzee against a heterologous i.v. viral rechallenge.
To test whether 4X0202 could be protected from developing a productive infection after inoculation with heterologous viruses, the same animal was sequentially challenged with 100 50% chimpanzee infective doses (CID50) of H77 (18) (gift of Robert Purcell, National Institutes of Health [NIH]) and the same dose of HC-J4 (31) (gift of Jens Bukh and Robert Purcell, NIH) (Fig. 2) by i.v. inoculation. HCV RNA was not detected in 100 μl of plasma up to 8 weeks postinoculation with H77 (type 1a). In contrast, five control animals who were inoculated i.v. with only 64 CID50 of H77 were PCR positive for a minimum of 25 weeks postchallenge (15, 17), and two control animals (L445 and L561) who received 100 CID50 of H77 i.v. were PCR positive for 20 and 48 weeks, respectively (Fig. 2B). To improve the sensitivity of the RT-PCR assay from 500 to ≤30 copies/ml, 1 ml of plasma was concentrated on a column (Qiagen viral extraction kit) and reassayed. Under these conditions, 4X0202 had low but detectable HCV RNA in plasma on weeks 1 and 2 post-H77 challenge and became PCR negative during the following 11 weeks. Challenge with 100 CID50 of HC-J4 resulted in a detectable viremia 1 week postchallenge (3 × 103 copies/ml), which disappeared by the second week postchallenge. The plasma remained PCR negative for the following 3 months (Fig. 2A) In contrast, a naive animal that received a 10-fold higher dose of HC-J4 developed a chronic HCV infection (J. Bukh, personal communication). RNA extracted from liver biopsies taken either on week 7 post-H77 challenge or week 5 post-HC-J4 challenge were PCR negative. The data indicated that chimpanzee 4X0202 was protected from a productive infection after challenge by both type 1a and 1b inocula.
Immune responses in chimpanzee 4X0202.
Initial IHGI with HCV-4PC elicited strong CD4+ T-cell responses in the periphery against the nonstructural HCV genes NS3/4 and NS5 on weeks 6 and 8 postchallenge. These responses declined rapidly but were boosted by the second IHGI of HCV-4PC at week 63 (Fig. 3A). A weak CD4+ T-cell response against NS3/4 was observed at week 78 in the PBMC after challenge with H77. In the liver, HCV-specific CD4+ responses were only detected for NS3/4 6 weeks following IHGI with HCV-4PC (Fig. 3B) and HCV-4PCmh (week 26; not shown). These data indicated that the molecular clones were correctly translated. No CD4+ responses were detected in either the PBMC or the intrahepatic lymphocytes against core or E1-E2 (Fig. 3A and B).
FIG. 3.
Immune responses primed by IHGI. (A and B) CD4+ and CD8+ cell-mediated T-cell responses in the periphery (A) and in the liver (B) were assayed as described in Materials and Methods. CD8+ responses tested against a panel of VV-infected autologous B-LCLs were scored positive when percent specific lysis at the two highest E:T ratios were greater than or equal to the percent lysis of VVwt-infected targets plus 10 (see text for specifics). HCV-specific CD8+ CTL clones were isolated from liver at week 6 (★). CD4+ responses were tested against the following recombinant HCV 1a proteins: C22-3 (core; blue), E1-E2 (orange), C200 (NS3/4; purple), and NS5 (green). An SI of 3.0 or greater was considered positive. (C) Antibody responses against C22-3 (core; blue line), C33c (NS3; red line), C25 (core plus NS3/4; yellow line), C100 (NS3/4; purple line), and NS5 (green line). Signal-to-cutoff ratios were, respectively, 0.109, 0.098, 0.174, 0.142, and 0.094 optical density (OD) units. (D) Antibody (Ab) responses against E2 (turquoise line), E1-E2 (orange line), and H77 HVR1 variant N/S (aa 384 to 410; black line/broken black line). Signal-to-cutoff ratios were 0.097, 0.126, and 0.139/0.121 optical density units, respectively.
Using bulk cytotoxic T-lymphocyte (CTL) assays, HCV-specific CD8+ CTLs against E2-NS3 were detected in the PBMC but not in the liver 6 weeks postchallenge (Fig. 3A and B). However, since the bulk CTL assay is relatively insensitive, we cloned intrahepatic lymphocytes from the week 6 liver biopsy by limiting dilution and found that 40% (53 of 133) of the CD8+ T-cell receptor alpha-beta-positive T-cell clones established were HCV specific. (Nine clones were specific for core, nine for E1-E2, 24 for E2/NS3/NS4, eight for NS3/NS4, two for NS5A, and one for NS5B [data not shown].) Our data are consistent with a previous report showing a multispecific CD8+ CTL response in the liver of chimpanzees with an acute HCV infection (11). NS5A- and NS5B-specific CD8+ CTL were identified in the PBMC and liver, respectively, on the date of challenge with the homologous virus, HCV-4PC RNA, at week 63 (Fig. 3A and B). Interestingly, the magnitude of the CD4 response decreased rather than increased after each subsequent challenge, and the breadth of the CD8 responses in the PBMC and liver became more restricted after rechallenge with HCV-4PC RNA. The cause of these observed changes in the cellular response after multiple challenges with genomic RNA is not understood and deserves further investigation. Unfortunately, since only bulk CTL assays were performed on the date of the H77 challenge and 2 weeks postchallenge, we cannot be certain whether HCV-specific CTLs were present in the PBMC or liver.
The humoral response to nonstructural proteins NS3 and NS4 appeared 7 weeks after the first challenge with HCV-4PC, as expected (Fig. 3C), while anti-NS5 antibodies were initially detected at week 38 after IHGI of HCV-1PC RNA. Seroconversion to the structural proteins (C, E2, and E1-E2) was also delayed until week 38 and was also detected after subsequent challenges with HCV-4PC RNA and H77 (Fig. 3D). A poor humoral response to structural region proteins of HCV is typical for chimpanzees (3, 8, 44). The appearance of antibodies within 1 to 2 weeks after each rechallenge are indicative of anamnestic responses and were observed for all of the antigens tested (Fig. 3C and D).
Humoral response to HVR1.
Although 4X0202 was immune competent, as evidenced by the humoral response to both structural and nonstructural proteins (Fig. 3C and D), no anti-HCV-1 HVR1 antibodies were observed after repeated challenges with HCV-1PC, -4PC, or -4PCmh, all of which encode the HCV-1 HVR1. In contrast, antibodies against the predominant H77 HVR1 variant (384-ETHVTGGNAGRTTAGLVGLLTPGAKQN-410 [17]) were detected 1 week post-i.v. challenge with H77 plasma and lasted 8 weeks (solid black line in Fig. 3D). Anti-H77 HVR1 antibodies against the second most common variant (S substituted for N), which occurs approximately 10-fold less frequently in H77 (17, 30), were also detected from weeks 1 to 5 post-H77 challenge (broken black line in Fig. 3D).
Induction of IFN-α correlates with protection.
We were interested to know whether the rapidly aborted infection in 4X0202 after challenge with 100 CID50 of H77 could also be associated with production of IFNs. No detectable levels of IFN-β or -γ were detected in the plasma of animal 4X0202 by specific ELISA at any of the time points tested (data not shown). In contrast, levels of IFN-α could easily be detected in the plasma 2 to 3 weeks after each challenge except for the mutated genome, HCV-4PCmh (Fig. 4). These data suggest that HCV-1PC (week 48) and HCV-4PC (week 63) replicated following IHGI, although there were no detectable HCV RNA in the plasma in an assay which could detect ≤30 copies of HCV RNA per ml. The rapid induction of IFN-α correlated with clearance of virus in this animal after both homologous (HCV-4PC) challenge at week 63 and heterologous (H77) challenge at week 78 (Fig. 2), but not after the initial IHGI of HCV-4PC at week 0 (Fig. 2 and 4).
FIG. 4.
IFN-α levels in the plasma following IHGI and viral challenge. Due to sample dilution, the sensitivity of the assay was 40 pg/ml.
DISCUSSION
Concerns about the ability to induce an effective immune response to HCV infection have largely been based on studies showing that naturally infected humans and experimentally infected chimpanzees can be reinfected by subsequent exposure to closely related HCV and that the genetic fluidity of the genome, particularly in the envelope proteins, could be problematic for cross-protection from heterologous viruses. Although neither the humoral immune responses to the HCV structural proteins nor the cellular immune responses were characterized in any of these earlier studies, viremia could be detected in chimpanzees for several weeks after challenge with either homologous or heterologous viruses (14, 35). Similarly, cases of humans with multiple episodes of acute viremia after repeated exposure through transfusion (27) or i.v. drug use (36) have been documented. These data were interpreted to mean that individuals developed weak immunity at best to HCV. Although it is difficult to directly compare our results with historical data due to the differences in protocols and assays, the data in Fig. 2 clearly demonstrate that a chimpanzee who had resolved a previous HCV infection developed immunity when rechallenged with the identical genomic RNA (HCV-4PC). This animal had essentially been boosted four times by IHGI of HCV genomes (HCV-4PC, HCV-4PCmh, and HCV-1PC), all of which encode identical HCV-1 proteins (Fig. 1B to D). The presence or absence of IFN-α (Fig. 4), which is known to be induced by the replication of RNA viruses with double-stranded RNA replicative intermediates, including HCV (19, 39), suggested that a subdetectable level of replication most likely did occur after IHGI of all the genomes except the NS3 helicase mutant (HCV-4PCmh), in which the GKS motif (45) was crippled. In addition, the anamnestic humoral and cellular immune response indicated that the HCV proteins were translated after each challenge independent of their ability to replicate. The IHGI of genomic RNA may have some of the characteristics of attenuated viral vaccines. Since the fitness of the HCV-4PC genome is not known, the relatively low level and brief duration of viremia observed in the acute infection of 4X202 might have been due to the chimeric nature of the genome. We speculate that injection of HCV RNA directly into the liver may transfect cells (e.g., Kupffer cells) which may not be infected by virus, and thereby HCV antigens may be presented to the immune system by a pathway not typically used by the host when infected by virus. Whether the route of infection was influential in establishing immunity will require further experiments in an animal model.
Importantly, chimpanzee 4X202 developed protective immunity to both heterologous type 1a (H77) and type 1b (HC-J4) viral challenge after IHGI of HCV RNA encoding HCV-1 (1a) proteins. Protection from the H77 inoculum was observed approximately 1.5 years after resolution of acute viremia (Fig. 2A). Employing ultrasensitive RT-PCR techniques capable of detecting ≤30 copies/ml, an extremely weak viremia was observed for 2 weeks after challenge with H77, compared to six control animals who had viremia for more than 25 weeks and one animal who was PCR positive for 4 months postchallenge. Similarly, the animal challenged with the 1b inoculum was PCR positive for only 1 week postchallenge. The absence of HCV RNA in the liver of both animals 1 to 2 months postchallenge further confirmed the lack of productive infection in this animal.
The humoral and cellular immune responses measured in this study do not definitively point to a single mechanism by which 4X0202 developed protective immunity. New assays such as virus-neutralizing antibody assays and new animal models will be needed to gain a more comprehensive understanding of how protection from chronic HCV infection is established and maintained. Based on the data obtained in this study, the immunity seen in 4X0202 was most likely due to the cellular immune response, which included a multispecific CD4+ response in the PBMC and the presence of HCV-specific CD8+ CTLs in the liver and PBMC at the time of challenge, with a possible contribution of anti-E1-E2 antibodies. The most notable immune response after rechallenge with whole virus (H77) was the unusually rapid seroconversion to anti-HVR1 antibodies 1 week postchallenge. These data suggest an active T-helper response and are striking, since no anti-HCV-1 HVR1 antibodies were ever detected after IHGI. The ability of anti-H77 HVR1 S variant-specific antibodies to neutralize H77 virus and prevent infection in one chimpanzee has been demonstrated previously by Farci et al. (17). In addition, a correlation between anti-HVR1 antibodies 40 to 60 days postchallenge and resolution of acute HCV infection has been reported previously in chimpanzees (43); however, the cellular immune response was not characterized in this study. We cannot exclude the possibility that an H77-specific CD8+ CTL response which was undetectable in the relatively insensitive bulk CTL assay may also have contributed to the control of viremia. The rapid clearance of HC-J4 will require future studies, as the immune responses were not determined.
Interestingly, the rapid induction of IFN-α after i.v. H77 challenge also correlated with viral clearance. It is unlikely that IFN-α alone could account for viral clearance, since the level of IFN-α was only twofold greater after challenge with H77 relative to the amount measured after the initial IHGI of HCV-4PC, in which the animal developed an acute infection. Based on data from the single animal was used in this study, it appears that the humoral and cellular arms of the immune system in concert with IFN-α may have generated protective immunity against a heterologous challenge in this animal. These data also provide evidence, for the first time, to support the concept that a vaccine in conjunction with antiviral therapies such as IFN-α and/or other immune modulators has considerable potential.
ACKNOWLEDGMENTS
We are grateful to Jens Bukh and Robert Purcell for their generous gift of the HCV H77 and HC-J4 inocula and unpublished data used in this study. We also thank Nelle Cronen for graphics and preparation of the manuscript.
This work was supported by Chiron Corporation and, in part, by Pharmacia Corporation.
REFERENCES
- 1.Alter H J, Purcell R H, Holland P V, Popper H. Transmissible agent in non-A, non-B hepatitis. Lancet. 1978;1:459–463. doi: 10.1016/s0140-6736(78)90131-9. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J, Seeff L B. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 3.Bassett S E, Thomas D L, Brasky K M, Lanford R E. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol. 1999;73:1118–1126. doi: 10.1128/jvi.73.2.1118-1126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley D W, Cook E H, Maynard J E, McCaustland K A, Ebert J W, Dolana G H, Petzel R A, Kantor R J, Heilbrunn A, Fields H A, Murphy B L. Experimental infection of chimpanzees with antihemophilic (factor VIII) materials: recovery of virus-like particles associated with non-A, non-B hepatitis. J Med Virol. 1979;3:253–269. doi: 10.1002/jmv.1890030403. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D W, Maynard J E, Cook E H, Ebert J W, Gravelle C R, Tsiquaye K N, Kessler H, Zuckerman A J, Miller M F, Ling C, Overby L R. Non-A/non-B hepatitis in experimentally infected chimpanzees: cross-challenge and electron microscopic studies. J Med Virol. 1980;6:185–201. doi: 10.1002/jmv.1890060302. [DOI] [PubMed] [Google Scholar]
- 6.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 7.Chien D Y, Choo Q L, Tabrizi A, Kuo C, McFarland J, Berger K, Lee C, Shuster J R, Nguyen T, Moyer D L, et al. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc Natl Acad Sci USA. 1992;89:10011–10015. doi: 10.1073/pnas.89.21.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo Q L, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr P J, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo Q L, Weiner A J, Overby L R, Kuo G, Houghton M, Bradley D W. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull. 1990;46:423–441. doi: 10.1093/oxfordjournals.bmb.a072408. [DOI] [PubMed] [Google Scholar]
- 11.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 12.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 13.Eckart M R, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo Q L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 14.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 15.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farci P, Purcell R H. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- 17.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinstone S M, Alter H J, Dienes H P, Shimizu Y, Popper H, Blackmore D, Sly D, London W T, Purcell R H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 19.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2′,5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong K, Greer C E, Ketter N, Van Nest G, Paliard X. Isolation and characterization of human papillomavirus type 6-specific T cells infiltrating genital warts. J Virol. 1997;71:6427–6432. doi: 10.1128/jvi.71.9.6427-6432.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghton M. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. Hepatitis C viruses; pp. 1035–1058. [Google Scholar]
- 22.Kamimura T, Ponzetto A, Bonino F, Feinstone S M, Gerin J L, Purcell R H. Cytoplasmic tubular structures in liver of HBsAg carrier chimpanzees infected with delta agent and comparison with cytoplasmic structures in non-A, non-B hepatitis. Hepatology. 1983;3:631–637. doi: 10.1002/hep.1840030502. [DOI] [PubMed] [Google Scholar]
- 23.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun. 1992;189:119–127. doi: 10.1016/0006-291x(92)91533-v. [DOI] [PubMed] [Google Scholar]
- 24.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 25.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolykhalov A A, Mihalik K, Feinstone S M, Rice C M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M E, Mazzoleni A P, Argiolu F, De Virgilis S, Balestrieri A, Purcell R H, Cao A, Farci P. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet. 1994;343:388–390. doi: 10.1016/s0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- 28.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 30.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 32.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 33.Poles M A, Dieterich D T. Hepatitis C virus/human immunodeficiency virus coinfection: clinical management issues. Clin Infect Dis. 2000;31:154–161. doi: 10.1086/313892. [DOI] [PubMed] [Google Scholar]
- 34.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT) Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 35.Prince A M, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 36.Proust B, Dubois F, Bacq Y, Le Pogam S, Rogez S, Levillain R, Goudeau A. Two successive hepatitis C virus infections in an intravenous drug user. J Clin Microbiol. 2000;38:3125–3127. doi: 10.1128/jcm.38.8.3125-3127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu Y K, Feinstone S M, Purcell R H, Alter H J, London W T. Non-A, non-B hepatitis: ultrastructural evidence for two agents in experimentally infected chimpanzees. Science. 1979;205:197–200. doi: 10.1126/science.451589. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu Y K, Purcell R H. Cytoplasmic antigen in hepatocytes of chimpanzees infected with non-A, non-B hepatitis virus or hepatitis delta virus: relationship to interferon. Hepatology. 1989;10:764–768. doi: 10.1002/hep.1840100503. [DOI] [PubMed] [Google Scholar]
- 40.Simmonds P. Viral heterogeneity of the hepatitis C virus. J Hepatol. 1999;31(Suppl. 1):54–60. doi: 10.1016/s0168-8278(99)80375-4. [DOI] [PubMed] [Google Scholar]
- 41.Tabor E, Gerety R J, Drucker J A, Seeff L B, Hoofnagle J H, Jackson D R, April M, Barker L F, Pineda-Tamondong G. Transmission of non-A, non-B hepatitis from man to chimpanzee. Lancet. 1978;1:463–466. doi: 10.1016/s0140-6736(78)90132-0. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Doorn L J, Capriles I, Maertens G, DeLeys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Doorn L J, van Hoek K, de Martinoff G, Bosman F, Stuyver L, Kos T, Frantzen I, Sillekens P, Maertens G, Quint W. Serological and molecular analysis of hepatitis C virus envelope regions 1 and 2 during acute and chronic infections in chimpanzees. J Med Virol. 1997;52:441–450. doi: 10.1002/(sici)1096-9071(199708)52:4<441::aid-jmv17>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 45.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiner A J. Humoral response to hepatitis C virus. San Diego, Calif: Academic Press; 2000. pp. 125–145. [Google Scholar]
- 47.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. Global surveillance and control of hepatitis C: report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepatitis. 1999;6:35–47. [PubMed] [Google Scholar]
- 49.Wu H N, Lin Y J, Lin F P, Makino S, Chang M F, Lai M M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]