Abstract
Human foamy virus (HFV), a retrovirus of simian origin which occasionally infects humans, is the basis of retroviral vectors in development for gene therapy. Clinical considerations of how to treat patients developing an uncontrolled infection by either HFV or HFV-based vectors need to be raised. We determined the susceptibility of the HFV to dideoxynucleosides and found that only zidovudine was equally efficient against the replication of human immunodeficiency virus type 1 (HIV-1) and HFV. By contrast, zalcitabine (ddC), lamivudine (3TC), stavudine (d4T), and didanosine (ddI) were 3-, 3-, 30-, and 46-fold less efficient against HFV than against HIV-1, respectively. Some amino acid residues known to be involved in HIV-1 resistance to ddC, 3TC, d4T, and ddI were found at homologous positions of HFV reverse transcriptase (RT). These critical amino acids are located at the same positions in the three-dimensional structure of HIV-1 and HFV RT, suggesting that both enzymes share common patterns of inhibition.
Spumaviruses, in addition to oncornaviruses and lentiviruses, constitute the third genus of the Retroviridae family and are known to be widely prevalent in primates (15). The first spumavirus isolate found in humans was obtained in 1971 from a patient with a nasopharingeal carcinoma (1). This isolate was named human foamy virus (HFV). The sequence data of foamy virus isolates from chimpanzees support the hypothesis that HFV is not a human prototype but rather a variant strain of a simian foamy virus (21). A substantial seroprevalence (1.8%) of infection with simian foamy virus among humans occasionally exposed to nonhuman primates was also described. These infections have not as yet resulted in either disease or sexual transmission and might represent benign endpoint infections (7). Due to its apparent lack of pathogenicity and its capacity to induce proviral genome integration in nondividing cells, HFV is used to generate retroviral vectors in development for gene therapy (8, 19, 27). Clinical considerations of how to treat patients developing an uncontrolled infection by either HFV or HFV-based vectors need to be raised. The nucleotide sequence of HFV was determined and showed that the pol gene was divided into three domains: reverse transcriptase (RT), RNase H, and integrase (9, 14, 17). Biological properties of the corresponding HFV pol gene products were determined (9). The RT of HFV might be an attractive target for efficient antiviral chemotherapy, due to the known inhibitory effect of nucleoside analogs on human immunodeficiency virus type 1 (HIV-1) RT and their well-demonstrated activity against HIV-1 infections in vivo. In this study, we analyzed the relationship between the activity of RT dideoxynucleoside inhibitors against HFV, the RT amino acid sequence of HFV RT, and its three-dimensional (3D) structure.
Susceptibility of HFV to dideoxynucleosides.
Virus stocks were obtained by infecting U373MG cells (American Type Culture Collection, Manassas, Va.) with the HFV prototype strain (kindly provided by G. Peries, Saint Louis Hospital, Paris, France) for 10 days until an 80% cytopathic effect (CPE) was observed (1). Five HIV-1 RT inhibitors were tested: azidothymidine (zidovudine [ZDV]; Sigma), dideoxyinosine (didanosine [ddI]; Sigma), dideoxycytidine (zalcitabine [ddC]; Sigma), lamivudine (3TC; Glaxo Wellcome) and stavudine (d4T; Sigma). The susceptibility of HFV to these drugs was determined by classical procedure: 2 × 104 cells per well were seeded in 96-well plates to obtain a subconfluent cell monolayer after culture for 24 h. Cells were infected in quadruplicate with 50 μl of fivefold serial dilutions of virus stock per well. Antiviral drugs were added at the same time. The antiviral drug concentrations were those usually used for HIV-1 (4) and scaled up when 50% inhibitory concentrations (IC50s) (i.e., the concentration reducing viral infectious titers by 50%) were out of range. After 10 days, plates were checked for the presence of a CPE in each well. The infectious titers in the absence and presence of drugs were calculated from the number of wells exhibiting a CPE at each concentration by the Reed and Münch method (4). All four experiments were done with two stocks of HFV. The susceptibility profile of HFV to an antiretroviral drug was expressed as the IC50. The IC50s for HFV were compared to published IC50s for wild-type-sensitive HIV-1 (4) (Table 1). ZDV appeared to be the most effective drug against HFV, and its IC50 was identical to that published for HIV-1. By contrast, IC50s of ddC, 3TC, d4T, and ddI were 3-, 3-, 30-, and 46-fold higher, respectively, than that for HIV-1, reflecting a lower sensitivity of HFV to these drugs.
TABLE 1.
Comparison of sensitivity of HFV and HIV-1 to antiviral drugs
| Antiviral drug | IC50 (μM) for
|
IC50 ratio (HFV/HIV-1) | |
|---|---|---|---|
| HFVa | HIV-1 | ||
| ZDV | 0.01 | 0.01 | 1 |
| ddI | 39 | 0.85 | 46 |
| ddC | 0.5 | 0.15 | 3 |
| 3TC | 0.3 | 0.09 | 3 |
| d4T | 1.2 | 0.04 | 30 |
Median values of four IC50 determinations are shown.
HFV sequence analysis and comparison to HIV-1 RT amino acids.
We sequenced the RT active-site region of our HFV strain. The region was amplified by PCR on proviral DNA with primers RT1 (5′-CTGGTGATTATCCTCCTCGCCC-3′) and RT2 (5′-AAAAGTGTCTGTTAGGCCACGACC-3′) (40 cycles, each cycle at 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min). A fragment of 652 bp was obtained and sequenced with an automated DNA sequencer (ABI; Perkin-Elmer). Sequence analysis with Geneworks software revealed no difference between our strain and the previously published sequence (14). To locate the homologous amino acid residues of HFV and HIV-1 RT, we performed an amino acid alignment of the deduced HFV and HIV-1 sequences with the Multalign algorithm (3), refined manually (data not shown), which confirmed those sequences previously published (14). The similarity between these proteins is low (about 20% identical amino acid residues between the two proteins). However, this limited homology extends over the entire length of both proteins, and several conserved features were revealed by the alignment. In particular, the presence of strictly conserved residues in all RNA-dependent polymerases characterized so far (18) was observed. This permitted us to investigate the relationship between the spectrum of susceptibility to dideoxynucleosides and the amino acid residues present at crucial homologous positions (Table 2). Some of the classical amino acid mutations which have been involved in the HIV-1 resistance to dideoxynucleosides are naturally present in HFV RT amino acid sequences, at the HIV-1 positions 74 and 184 for ddI and ddC (5, 6, 24), position 184 for 3TC (20, 26) and position 75 for d4T (11, 13; D. Shepp, A. Ashraf, C. Millian, and R. Pergolizzi, Abstr. 2nd Natl. Conf. Human Retrovir., abstr. 139, 1995). At other positions, HFV amino acids were different from those present in sensitive HIV-1 strains but also distinct from those observed in resistant HIV-1 strains. In the case of ZDV, which exhibited the same activity level on HFV and HIV-1, we did not find any of the amino acid residues involved in HIV-1 resistance to the drug at positions 41, 67, 70, 215, and 219 (12), corresponding to positions 91, 118, 121, 252, and 256 of the HFV RT. In addition, neither multidrug resistance mutations (Q151 M profile) (22, 23) nor a position 69 insertion (J. J. de Jong, S. Jurriaans, J. Goudsmit, E. Baan, R. Huismans, S. A. Danner, M. E. Hillebrand, J. H. ten Veen, and F. de Wolf, Antivir. Ther., abstr. 18, 1998) of resistant HIV-1 strains was observed in the case of HFV. These results are in accordance to the susceptibility pattern of HFV to dideoxynucleosides depicted in Table 1.
TABLE 2.
Amino acid residues of HIV-1 RT involved in resistance to dideoxynucleosides and correspondence to amino acid residues of HFV RT
| Antiviral drug | HIV-1 residue(s)
|
HFV RT residue
|
|||
|---|---|---|---|---|---|
| Position | Sensitive strain | Resistant strain | Homologous position | Tested straina | |
| AZT | 41 | M | L | 91 | I |
| 67 | D | N | 118 | D | |
| 70 | K | R | 121 | ||
| 215 | T | Y/F | 252 | V | |
| 219 | K | Q/E | 256 | K | |
| ddI | 65 | K | R | 116 | K |
| 74 | L | V | 124 | V | |
| 75 | V | T | 125 | L | |
| 184 | M | V/I | 222 | V | |
| ddC | 65 | K | R | 116 | K |
| 69 | T | D | 120 | R | |
| 74 | L | V | 124 | V | |
| 75 | V | T | 125 | L | |
| 184 | M | V/I | 222 | V | |
| 3TC | 184 | M | V/I | 222 | V |
| d4T | 75 | V | T/M/S/A/L | 125 | L |
Underlined amino acids are identical in resistant HIV-1 strains and HFV.
HFV RT 3D structural modeling.
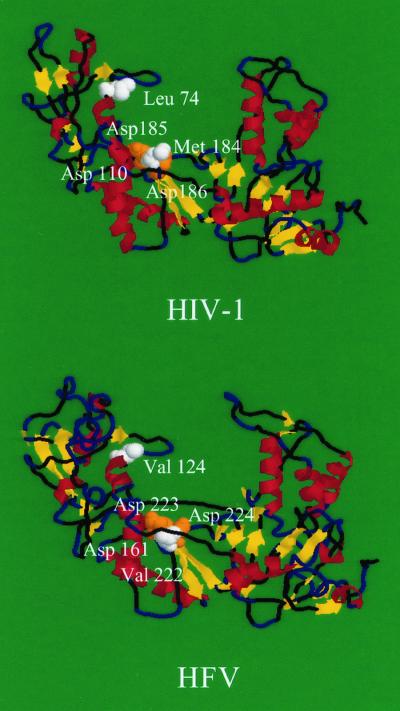
We determined a putative 3D structural modeling of HFV RT to check if the crucial positions defined by sequence alignment were located at the same 3D positions as in HIV-1 RT. The molecular modeling of HFV was calculated by homology with the 3D structure of HIV-1 (BH10 isolate) RT (3HVT.pdb) with the Swiss Model Automated Protein Modelling service at the Glaxo Wellcome Experimental Research Center (Geneva, Switzerland), which makes use of the ProMod software (16). Secondary structure and distribution of hydrophobic clusters were analyzed as previously described (2), and the results suggested that the structural organization of both proteins was similar (data not shown). Therefore, on the basis of the alignment, we calculated the 3D structure of the HFV RT by homology with the 3D structure of HIV-1 enzyme, which has been determined by X-ray diffraction (10). This model markedly suggested that the structural topology was conserved in both proteins (Fig. 1), in particular, the organization in different subdomains (fingers, palm, thumb, and connection regions) (25). Moreover, the amino acids corresponding to the catalytic triad of aspartic acids (Asp 110, Asp 185, and Asp 186 for HIV-1 and Asp 161, Asp 223, and Asp 224 for HFV), as well as those related to dideoxynucleoside resistance (Asp 74, Asp 75, and Asp 184 for HIV-1 and Asp 124, Asp 125, and Asp 222 for HFV), are located at similar positions in the 3D structure of both proteins. This 3D representation permitted us to verify that the amino acids in HFV RT corresponding to those involved in HIV-1 dideoxynucleoside resistance were located at the same 3D positions. The fact that critical amino acid residues involved in resistance are located in the same subdomains in both proteins confirms the previous results identifying the exact localization of HFV RT gene (9).
FIG. 1.
Predicted 3D structure of HFV RT based on the known structure of HIV-1 RT. α-Helices are indicated in red, and β-strands are indicated in yellow. Essential aspartates (Asp 110, Asp 185, and Asp 186 for HIV-1 and Asp 161, Asp 223, and Asp 224 for HFV), which may bind the triphosphate moiety of incoming nucleotide analogs and deoxynucleoside triphosphate substrates via Mg2+, are indicated as orange spheres. White spheres indicate two positions involved in resistance of HIV-1 to dideoxynucleoside (amino acids 74 and 184) and the homologous position in HFV RT.
Our work studied the efficacy and the relationship between dideoxynucleoside analog susceptibility, amino acid sequence, and structure of HFV RT. We evaluated the HFV susceptibility profile of five dideoxynucleoside analogs commonly used in HIV-1 treatment. The IC50 values were shown to be higher than those previously described for HIV-1, except that of ZDV. These results are in accordance with those recently published (28). To correlate function inhibition and structure, we determined the RT amino acid sequence of the HFV strain tested, and we performed alignment with HIV-1 RT. The genomic determinants of the natural resistance pattern of HFV to the dideoxynucleosides tested were found to be homologous to those previously described for HIV-1 resistance, despite a low similarity between HFV and HIV-1 RT. A 3D model of HFV RT confirmed these results, which might have important implications in both the studies of retrovirus phylogeny and the management of HFV infections in vivo.
REFERENCES
- 1.Achong B G, Mansell P, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. [PubMed] [Google Scholar]
- 2.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin C, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol Microbiol. 1999;31:319–330. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 3.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Descamps D, Collin G, Loussert-Ajaka I, Sarragosti S, Simon F, Brun-Vezinet F. HIV-1 group O sensitivity to antiretroviral drugs. AIDS. 1995;9:977–978. [PubMed] [Google Scholar]
- 5.Fitzgibbon J E, Howell R M, Haberzetti C A, Sperber S J, Gocke D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu Z, Gao Q, Li X, Parniak M A, Wainberg M A. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri P, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T M. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 8.Hill C L, Bieniasz P D, McClure M O. Properties of human foamy virus relevant to its development as a vector for gene therapy. J Gen Virol. 1999;80:2003–2009. doi: 10.1099/0022-1317-80-8-2003. [DOI] [PubMed] [Google Scholar]
- 9.Kögel D, Aboud M, Flügel R M. Molecular biological characterization of the foamy virus reverse transcriptase and ribonuclease H domains. Virology. 1995;213:97–108. doi: 10.1006/viro.1995.1550. [DOI] [PubMed] [Google Scholar]
- 10.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 11.Lacey S F, Larder B A. A novel mutation (V75T) in the HIV-1 reverse transcriptase confers resistance to 2′,3′-dehydro-2′,3′-dideoxythymidine (d4T) in cell culture. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine. Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 13.Lin P F, Gonzalez C J, Griffith B, Friedland G, Calvez V, Ferchal F, Schinazi R F, Shepp D H, Ashraf A B, Wainberg M A, Soriano V, Mellors J W, Colonno R J. Stavudine resistance: an update on susceptibility following prolonged therapy. Antivir Ther. 1999;4:21–28. [PubMed] [Google Scholar]
- 14.Maurer B, Bannert H, Darai G, Flügel R M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mergia A, Luciw P A. Replication and regulation of primate foamy viruses. Virology. 1991;184:474–482. doi: 10.1016/0042-6822(91)90417-a. [DOI] [PubMed] [Google Scholar]
- 16.Peitsch M C, Jongeneel V. A 3-D model for the CD40 ligand predicts that it is a compact trimer similar to the tumor necrosis factors. Int Immunol. 1993;5:233–238. doi: 10.1093/intimm/5.2.233. [DOI] [PubMed] [Google Scholar]
- 17.Pfrepper K I, Rackwitz H R, Schnolzer M, Heid H, Lochelt M, Flugel R M. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependant polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schinazi R F, Lloyd R J, Jr, Nguyen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer K O, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. Markers of foamy virus (FV) infection in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected FV prevalence in man. AIDS Res Hum Retrovir. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 22.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirasaka T, Yarchoan R, O'Brien M C, Husson R N, Anderson B D, Kojima E, Shimada T, Broder S, Mitsuya H. Changes in drug sensitivity of human immunodeficiency virus type 1 during therapy with azidothymidine, dideoxycytidine, and dideoxyinosine: an in vitro comparative study. Proc Natl Acad Sci USA. 1993;90:562–566. doi: 10.1073/pnas.90.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St. Clair M H, Martin J L, Tudor W G. Resistance to ddI and sensitivity to AZT induced by mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 25.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 26.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trobridge G D, Russell D W. Helper-free foamy virus vectors. Hum Gene Ther. 1998;9:2517–2525. doi: 10.1089/hum.1998.9.17-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S F, Sullivan M D, Linial M L. Evidence that the human foamy virus genome is DNA. J Virol. 1999;73:1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]