Abstract
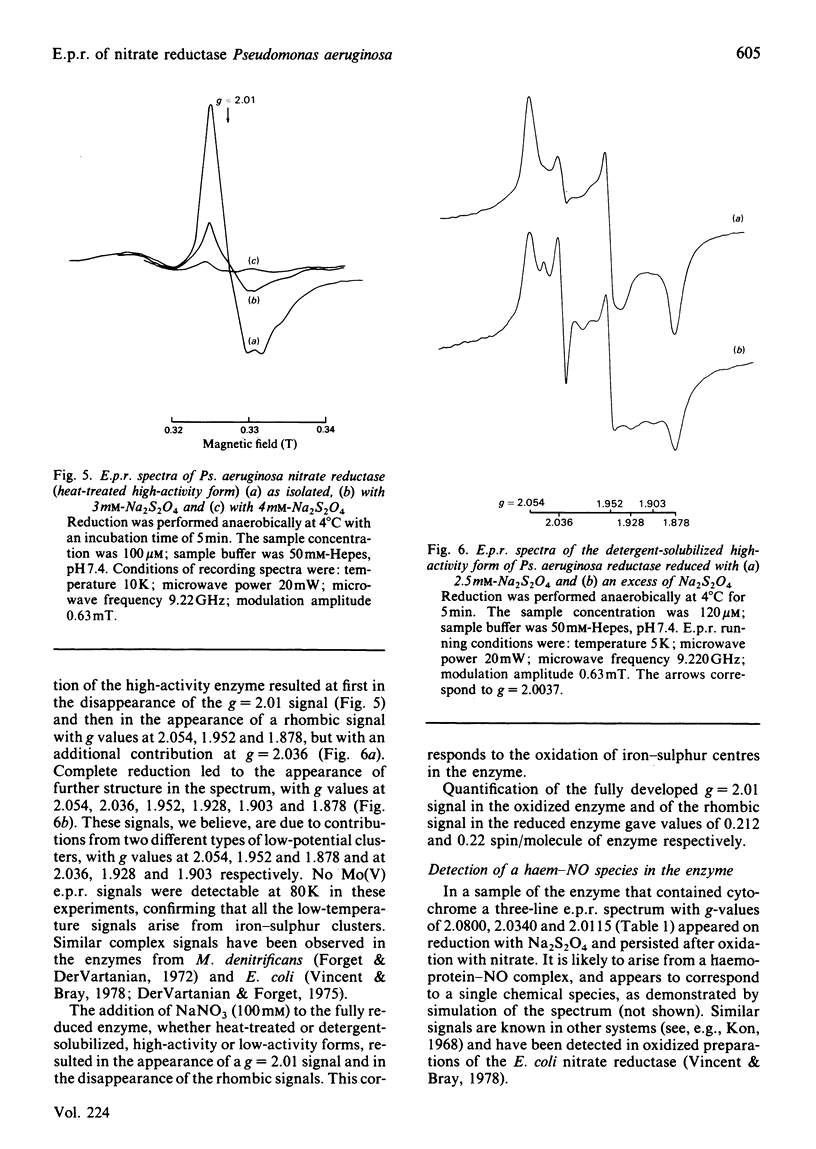
Preparations of nitrate reductase in the resting state from Pseudomonas aeruginosa exhibit an Mo(V) e.p.r. signal. Progressive reduction of the enzyme results at first in the intensification and then in the disappearance of the signal. Three different species of Mo(V) were detected by e.p.r. These are the high-pH species (g1 = 1.9871; g2 = 1.9795; g3 = 1.9632) and nitrate and nitrite complexes of a low-pH species (respectively g1 = 2.0004; g2 = 1.9858; g3 = 1.9670; and g1 = 1.9975; g2 = 1.9848; g3 = 1.9652). These signals are closely analogous to those for the enzyme from Escherichia coli described by Vincent & Bray [(1978) Biochem. J. 171, 639-647]. Signals typical of iron-sulphur clusters were also detected. In the oxidized enzyme these are believed to arise from a [3Fe-4S] cluster (g = 2.01) and in the reduced enzyme from an unusual low-potential [4Fe-4S]+ cluster (g1 = 2.054; g2 = 1.952; g3 = 1.878). The iron-sulphur centres were also studied in a 'high-catalytic-activity' form of the enzyme. Reduction with Na2S2O4 resulted in the formation of a complex signal with g values at 2.054, 1.952, 1.928, 1.903 and 1.878. The signal could be deconvoluted by reductive titration of the enzyme into two species (g1 = 2.054; g2 = 1.952; g3 = 1.878; and g1 = 2.036; g2 = 1.928; g3 = 1.903). The degradation of a [4Fe-4S] into a [3Fe-4S] cluster in the enzyme is suggested by these studies, the process being dependent on the method used to purify the enzyme. The addition of nitrate to the reduced enzyme results in the oxidation of Mo(IV) to Mo(V) and of all the iron-sulphur centres.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonio M. R., Averill B. A., Moura I., Moura J. J., Orme-Johnson W. H., Teo B. K., Xavier A. V. Core dimensions in the 3Fe cluster of Desulfovibrio gigas ferredoxin II by extended X-ray absorption fine structure spectroscopy. J Biol Chem. 1982 Jun 25;257(12):6646–6649. [PubMed] [Google Scholar]
- Beinert H., Emptage M. H., Dreyer J. L., Scott R. A., Hahn J. E., Hodgson K. O., Thomson A. J. Iron-sulfur stoichiometry and structure of iron-sulfur clusters in three-iron proteins: evidence for [3Fe-4S] clusters. Proc Natl Acad Sci U S A. 1983 Jan;80(2):393–396. doi: 10.1073/pnas.80.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H., Thomson A. J. Three-iron clusters in iron-sulfur proteins. Arch Biochem Biophys. 1983 Apr 15;222(2):333–361. doi: 10.1016/0003-9861(83)90531-3. [DOI] [PubMed] [Google Scholar]
- Blum H., Poole R. K. The molybdenum and iron-sulphur centres of Escherichia coli nitrate reductase are non-randomly oriented in the membrane. Biochem Biophys Res Commun. 1982 Aug;107(3):903–909. doi: 10.1016/0006-291x(82)90608-8. [DOI] [PubMed] [Google Scholar]
- Bosma H. J., Wever R., van 't Riet J. Electron paramagnetic resonance studies on membrane-bound respiratory nitrate reductase of Klebsiella aerogenes. FEBS Lett. 1978 Jun 1;90(1):107–111. doi: 10.1016/0014-5793(78)80308-1. [DOI] [PubMed] [Google Scholar]
- Bray R. C., Barber M. J., Lowe D. J. Electron-paramagnetic-resonance spectroscopy of complexes of xanthine oxidase with xanthine and uric acid. Biochem J. 1978 Jun 1;171(3):653–658. doi: 10.1042/bj1710653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray R. C. The reactions and the structures of molybdenum centers in enzymes. Adv Enzymol Relat Areas Mol Biol. 1980;51:107–165. doi: 10.1002/9780470122969.ch3. [DOI] [PubMed] [Google Scholar]
- Cammack R., Rao K. K., Hall D. O., Moura J. J., Xavier A. V., Bruschi M., Le Gall J., Deville A., Gayda J. P. Spectroscopic studies of the oxidation-reduction properties of three forms of ferredoxin from Desulphovibrio gigas. Biochim Biophys Acta. 1977 Feb 22;490(2):311–321. doi: 10.1016/0005-2795(77)90006-x. [DOI] [PubMed] [Google Scholar]
- Emptage M. H., Dreyers J. L., Kennedy M. C., Beinert H. Optical and EPR characterization of different species of active and inactive aconitase. J Biol Chem. 1983 Sep 25;258(18):11106–11111. [PubMed] [Google Scholar]
- FEWSON C. A., NICHOLAS D. J. Nitrate reductase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1961 May 13;49:335–349. doi: 10.1016/0006-3002(61)90133-0. [DOI] [PubMed] [Google Scholar]
- Forget P., Dervartanian D. V. The bacterial nitrate reductases: EPR studies on nitrate reductase A from Micrococcus denitrificans. Biochim Biophys Acta. 1972 Feb 28;256(2):600–606. doi: 10.1016/0005-2728(72)90089-8. [DOI] [PubMed] [Google Scholar]
- Fritz J., Anderson R., Fee J., Palmer G., Sands R. H., Tsibris J. C., Gunsalus I. C., Orme-Johnson W. H., Beinert H. The iron electron-nuclear double resonance (ENDOR) of two-iron ferredoxins from spinach, parsley, pig adrenal cortex and Pseudomonas putida. Biochim Biophys Acta. 1971 Nov 2;253(1):110–133. doi: 10.1016/0005-2728(71)90239-8. [DOI] [PubMed] [Google Scholar]
- Gutteridge S., Bray R. C., Notton B. A., Fido R. J., Hewitt E. J. Studies by electron-paramagnetic-resonance spectroscopy of the molybdenum centre of spinach (Spinacia oleracea) nitrate reductase. Biochem J. 1983 Jul 1;213(1):137–142. doi: 10.1042/bj2130137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R., Yoshida T., Tarr G. E., Williams C. H., Jr, Ludwig M. L., Fee J. A., Kent T. A., Huynh B. H., Münck E. Studies of the ferredoxin from Thermus thermophilus. J Biol Chem. 1983 Nov 10;258(21):13008–13013. [PubMed] [Google Scholar]
- Huynh B. H., Moura J. J., Moura I., Kent T. A., LeGall J., Xavier A. V., Münck E. Evidence for a three-iron center in a ferredoxin from Desulfovibrio gigas. Mössbauer and EPR studies. J Biol Chem. 1980 Apr 25;255(8):3242–3244. [PubMed] [Google Scholar]
- Kon H. Paramagnetic resonance study of Nitric Oxide hemoglobin. J Biol Chem. 1968 Aug 25;243(16):4350–4357. [PubMed] [Google Scholar]
- Moura J. J., Moura I., Kent T. A., Lipscomb J. D., Huynh B. H., LeGall J., Xavier A. V., Münck E. Interconversions of [3Fe-3S] and [4Fe-4S] clusters. Mössbauer and electron paramagnetic resonance studies of Desulfovibrio gigas ferredoxin II. J Biol Chem. 1982 Jun 10;257(11):6259–6267. [PubMed] [Google Scholar]
- Mullinger R. N., Cammack R., Rao K. K., Hall D. O., Dickson D. P., Johnson C. E., Rush J. D., Simopoulos A. Physicochemical characterization of the four-iron-four-sulphide ferredoxin from Bacillus stearothermophilus. Biochem J. 1975 Oct;151(1):75–83. doi: 10.1042/bj1510075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson L. P., Barber M. J., Howard W. D., Johnson J. L., Rajagopalan K. V. Electron paramagnetic resonance studies on the molybdenum center of assimilatory NADH:nitrate reductase from Chlorella vulgaris. J Biol Chem. 1984 Jan 25;259(2):849–853. [PubMed] [Google Scholar]
- Stout C. D., Ghosh D., Pattabhi V., Robbins A. H. Iron-sulfur clusters in Azotobacter ferredoxin at 2.5 A resolution. J Biol Chem. 1980 Mar 10;255(5):1797–1800. [PubMed] [Google Scholar]
- Vincent S. P., Bray R. C. Electron-paramagnetic-resonance studies on nitrate reductase from Escherichia coli K12. Biochem J. 1978 Jun 1;171(3):639–647. doi: 10.1042/bj1710639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. P. Oxidation--reduction potentials of molybdenum and iron--sulphur centres in nitrate reductase from Escherichia coli. Biochem J. 1979 Feb 1;177(2):757–759. doi: 10.1042/bj1770757. [DOI] [PMC free article] [PubMed] [Google Scholar]


