Abstract
Intestinal homeostasis and integrity are important factors for maintaining host health. This study established intestinal epithelial cell lines and organoids from the same swine jejunal crypts to develop seamless swine intestinal in vitro evaluation systems. The study evaluated the proliferative capacity and tight junction formation of the epithelial cell line and characterized the cell differentiation potential of the intestinal organoids. The evaluation systems were subsequently exposed to the Toll-like receptor 3 (TLR3) agonist poly(I:C) to simulate viral infections and assess the antiviral responses. The results demonstrated no differences in the response to type I interferons. There were, however, significant differences in the expression of interferon-stimulated genes. This study collectively introduced a flexible evaluation system using cell lines and organoids and revealed notable differences in the expression of interferon-stimulated genes, highlighting the complexity of the immune responses in these in vitro systems and the importance of intestinal heterogeneity in assessing viral responses.
Keywords: wine small intestine, swine intestinal epithelial cell, swine jejunal organoid, antiviral response, poly(I:C) response, TLR-3 agonist
INTRODUCTION
The world population is projected to reach 9.3 billion by 2050 [1], posing significant challenges, particularly in ensuring a stable and secure food supply. Swine farming is a major contributor to global food production, accounting for an estimated 33.4% of meat production in 2023 [2]. The importance of the swine farming industry is expected to increase; however, it faces significant challenges not only related to the health concerns of pigs arising from various infections but also related to the issue of antibiotic overuse [3]. Therefore, there is a need for a novel and comprehensive rearing approach that does not depend solely on conventional hygiene management and medication. Maintaining homeostasis in the intestinal tract is crucial for pig health and productivity. The intestinal tract is involved in host growth and feed efficiency through nutrient absorption and is also a target for bacterial and viral infections [4]. The integrity of the intestinal tract and the related barrier and absorptive functions are directly linked to host health. The intestinal epithelium has been widely studied for its significant role in the induction and regulation of the local innate immunity [5,6,7]. Therefore, investigation and evaluation of the effects of microbiota, pathogens, nutrients, and functional components on the swine intestinal tract are crucial to developing tools that help to improve pig health and productivity.
Unlike rodents, which are commonly used in life science research, in vivo evaluations of large and medium-sized animals, such as pigs, involve various costs that limit access to the gut tissue. Ethical considerations encourage avoidance of animal experiments whenever possible. In this regard, cell culture systems have been used for the study of swine intestinal physiology. The swine intestinal cell line model IPEC-J2 has allowed scientific research contributing to a better understanding of the responses to viral and bacterial infections and allowing the evaluation of functional ingredients [8,9,10]. In addition, our group has previously established porcine intestinal epitheliocyte (PIE) cells to study the immune responses of swine intestinal epithelial cells via Toll-like receptors (TLRs) and reported on their innate immune responses [11]. Considering that the intestinal tract is composed of various cell types, including absorptive epithelial cells, goblet cells, Paneth cells, enteroendocrine cells, BEST4 cells, and intestinal stem cells [12, 13], it may be difficult to accurately reproduce the response and function of the intestinal tract when relying solely on cell lines composed of a single cell type. Intestinal organoids, which can self-organize in three dimensions and reproduce the heterogeneity and behavior of the intestinal mucosa, are attractive tools to study in vitro the complex interactions between the different cell types and foreign agents, such as commensal or pathogenic microorganisms [14, 15]. Several swine intestinal organoids have been constructed, which has allowed functional evaluations and viral infection studies [16,17,18], bridging the gap between in vitro and in vivo studies in pigs. However, the heterogeneity of cell lines and organoids established to date in terms of pig age, origin of the tissue, and genetic background makes comparisons between the distinct evaluation systems difficult. Therefore, establishing both intestinal epithelial cell lines and intestinal organoids from identical tissues/an identical genetic background can provide a seamless in vitro evaluation system.
This study aimed to establish a single type of intestinal epithelial cell and intestinal organoid from the same host’s tissue and construct a continuous intestinal evaluation platform with an identical genetic background. Furthermore, the study characterized the functionality of the established evaluation systems in terms of their response to the TLR3 agonist poly(I:C) to investigate their potential to be used in the research of the intestinal antiviral responses in the swine host.
MATERIALS AND METHODS
Animals and ethical statement
The animal experiment was approved and performed as per the regulations and guidelines of the Animal Ethics Committee at the National Institute of Animal Health, National Agriculture and Food Research Organization (approval number 19-083). A two-week-old male Duroc piglet, which was purchased from a specific pathogen-free farm located in Miyazaki prefecture (Japan), was sedated and anesthetized via intravenous injection of propofol (5 mg/kg) and euthanized by exsanguination.
Isolation of swine jejunal crypt
All the procedures followed the methodology described by Khalil et al. [16], with minor modifications. Briefly, the upper jejunum was cut into 5–10 cm segments and stored in ice-cold 10% phosphate buffered saline (PBS; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 1% antibiotic-antimycotic (100×, Thermo Fisher Scientific). The intestinal segment was cut open along its long axis, and the mucosal surface was vigorously washed multiple times by vortexing with ice-cold PBS (40 mL) for 10 min. The tissues were shredded in PBS and washed by vortexing with ice-cold PBS (40 mL) for 10 min until the supernatant became clear. Then, the mucosa and villi were removed from the segment using a glass slide and cut into small pieces (about 5 × 5 mm in size) in ice-cold PBS. Minced tissues were again suspended in 40 mL of ice-cold PBS and washed thoroughly by vigorous vortexing until the supernatant became clear. Next, 10 mM ethylenediaminetetraacetic acid (EDTA)/1 mM dithiothreitol (DTT)/PBS (40 mL) was added to a 50 mL tube containing the minced tissue pieces and were gently shaken at 4°C at 50 rpm for 30 min. The supernatant containing the villi and debris was discarded by decanting. The settled minced tissue was suspended in ice-cold PBS (40 mL), and intestinal crypts were detached by vigorous and intermittent vortexing (3 × 10 sec). The supernatant containing detached crypts were collected and labeled as fraction 1 (Fx1). This step was repeated eight times, and eight fractions (Fx1–8) were prepared. The number of crypt-like structures in individual fractions was counted using a microscope. The five fractions rich in crypt-like structures (Fx4 to 8) were pooled and centrifuged at 4°C at 200 × g for 5 min. The pellets were then resuspended in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Thermo Fisher Scientific) and used to establish swine intestinal epithelial cell lines and a culture of swine jejunal organoids.
Establishment of swine intestinal epithelial cell lines
The isolated intestinal crypts were resuspended in the growth medium (GM) consisting of DMEM/F12 containing 1× Insulin-Transferrin-Selenium (ITS, Merck/Millipore Sigma, Burlington, MA, USA), 5 ng/mL recombinant human epidermal growth factor (EGF) (Merck/Millipore Sigma), 10% heat inactivated fetal bovine serum (FBS), and 1% Penicillin-Streptomycin (Thermo Fisher Scientific) and seeded into a collagen-type I-C-coated 6-well plate (Cell-tight C-1, Sumitomo Bakelite, Tokyo, Japan). Cell attachment and colony formation derived from the crypt-like structures were confirmed by microscopy 24 hr later. Multiple passages were performed to remove adherent crypt-like structures and obtain a primary culture free of tissue debris. Lenti-SV40 (tsA58, 2.33 × 106 IU/mL, Applied Biological Materials, Richmond, BC, Canada) was used for cell immortalization. The cells were seeded at a density of 1 × 104 cells/well in a collagen-type I-C-coated 12-well plate and incubated overnight. The medium was aspirated, and the cells were washed with PBS. Then, 1 mL of virus solution containing 8 µg/mL polybrene was added to a well of the plate (multiplicity of infection (MOI)=100) and incubated at 37°C for 6–8 hr. A GM (0.5 mL) was then added to the wells and incubated overnight. The medium containing the virus solution was replaced with 1.5 mL of GM, and the cells were incubated for 2–3 days. The medium was then replaced with a GM containing a final concentration of 2 µg/mL puromycin, and the immortalized cells were selected. To obtain a population of cells derived from the progeny of a single cell, 200 cells were seeded into a 100 mm dish. Individual colonies were encircled with cloning cylinders in the dish, and the cells within the cylinders were harvested. Immortalized clonal swine intestinal epithelial cells (SIECs) were ultimately generated.
The established SIEC line was maintained in the GM described above using a type I collagen-coated flask and plate at 37°C in a humidified incubator supplied with 5% CO2. The SIEC line was passaged once every three days.
Establishment of swine jejunal organoids
The isolated swine jejunal crypts were subjected to the establishment of swine jejunal organoids (SJOs) in IntestiCult™ Organoid Growth Medium Human (StemCell Technologies, Vancouver, BC, Canada) as described by the manufacturer’s instruction, with slight modification. Briefly, the isolated jejunal crypts were centrifuged and resuspended in ice-cold Matrigel (Corning, Corning, NY, USA) at the concentration of 500 crypts/25 µL/well, and the suspension was placed on a prewarmed 48-well plate. After solidification of Matrigel domes in a 37°C incubator, SJOs were cultured in 250 µL/well of organoid growth medium (OGM + Y), which was a 1:1 mixture of the IntestiCult™ OGM basal medium and the Organoid Supplement containing 0.1% of Y27632 (Wako, Osaka, Japan) and 1% of penicillin-streptomycin (Thermo Fisher Scientific). Starting three days after seeding, the medium was replaced with 250 µL of OGM medium (without Y27632) every other day. SJOs were passaged every 5 to 7 days.
For passaging, SJOs in a Matrigel dome were collected by pipetting with 500 µL/well of TrypLE Express (Thermo Fisher Scientific) following removal of the culture medium. After incubation at 37°C in a water bath for 3 min, 10 mL of Advanced DMEM (Thermo Fisher Scientific) supplemented with 4-(2-Hydroxyethyl)-1-piperazinyl ethanesulfonic acid (HEPES) (1 M, Thermo Fisher Scientific), 1% Gluta-Max (×100, Thermo Fisher Scientific), and 1% penicillin-streptomycin (×100, Thermo Fisher Scientific; ADF+++) was added, and digested SJOs were filtrated through a cell strainer (70 μm). The suspension was centrifuged (300 × g, 3 min, 4°C), and the cell clumps were resuspended in ice-cold Matrigel at the concentration of 500 clumps/25 uL/well. The culture was continued as described above. SJOs at the third passage were stocked for further experiments, and those at passage levels 7–15 were employed in this study.
For monolayer cultures, the SJOs were digested for another 6 min in a 37°C water bath following collection and digestion with TrypLE Express as mentioned above. SJOs were dissociated into single cells by pipetting after digestion, followed by filtration through a 40 µm cell strainer. After centrifugation (400 × g, 4°C, 5 min) and removal of the supernatant, the cells were suspended in OGM + Y supplemented with HEPES (1 M). Cells were seeded at 2.5 × 105 cells/mL in 24-well plates coated with 2.5% Matrigel. The medium was replaced with a differentiation medium (DM) consisting of 50% IntestiCult Basal, 50% ADF+++, 1% penicillin-streptomycin (100×), and 1× HEPES (100×) on the third day of incubation.
Immunofluorescence staining, hematoxylin and eosin staining, and EdU staining
The cells were cultured in chamber slides (Thermo Fisher Scientific) or well plates, and the supernatant was aspirated. To fix the cells, 100% methanol precooled to −20°C was added to each well, and the cells were incubated for 15 min at −20°C. They were then air-dried for approximately 1 hr after removing the methanol. The cells were then blocked using the serum of the species used in the secondary antibodies for 1 hr. The primary antibody was added, and the cells were incubated for 2 hr, washed three times with PBS-T (PBS containing 0.05% Tween-20), and incubated with a secondary antibody containing DAPI solution at room temperature (RT) for 45 min. The cells were then washed with PBS-T and mounted. The stained cells were observed using a BZ-X800 all-in-one microscope (KEYENCE, Osaka, Japan).
Hematoxylin and eosin (HE) staining was employed to evaluate the morphology of SJOs. Briefly, SJOs cultured for six days in a Matrigel dome were harvested using TrypLE Express. Subsequently, the SJOs were embedded in OCT compound (Sakura Finetek, Tokyo, Japan) at −20°C and sectioned at a thickness of 6 µm. HE staining was then conducted.
EdU (5-ethynyl 2´-deoxyuridine) staining of the SJOs was performed using a Click-iT Plus EdU Alexa Fluor™ 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific) as per the manufacturer’s instructions. Briefly, the SJOs were cultured in a Matrigel dome for 4, 6, and 14 days, and the medium was changed every three days. Next, 10 mM EdU stock solution was diluted in OGM medium to 20 µM, and 250 µL of the dilution was added to each well. After six hours of incubation, the medium was removed, 250 µL of 4% PFA was added, and the SJOs were fixed at RT for 15 min. After washing three times with 3% bovine serum albumin (BSA) in PBS, the cells were permeabilized with 0.1% Triton-X 100 in PBS at RT for 20 min. Then, 250 µL of Click-iT Plus reaction cocktail was added to the well and incubated for 30 min. After washing with PBS, Hoechst 33342 diluted 2,000× in PBS was added to each well and incubated at RT for 10 min. The stained cells were observed using a BZ-X800 all-in-one microscope (KEYENCE).
Proliferative activity of the swine intestinal epithelial cells line
Cell proliferation activity was analyzed every 7 days after seeding with the trypan blue dye exclusion method in triplicate using a hemocytometer, as previously described [19]. The cells were seeded in three wells of a 6-well plate (Sumilon) at 0.5 or 1.0 × 104 cells/cm2. The doubling time (DT) was calculated using the formula DT = (t − t0) log2/(log N − log N0), where (t − t0) was the cell growth period (days) and N and N0 were the cell numbers at t and t0, respectively.
Cell monolayer integrity
The integrity of the cell monolayer was analyzed by measuring the transepithelial electrical resistance (TEER) as previously described [19]. Cells were seeded onto Transwell inserts (0.4 μm pore size; Falcon cell culture insert PET membrane, Corning, Corning, NY, USA) at 1.0 × 105 cells/0.3 cm2/well. The TEER was measured at multiple time points up to day 15 using a Millicell ERS (Merck/Millipore Sigma, MA, USA). The TEER values were then calculated based on the formula Ω • cm2 = (TEER − blank TEER) × well area (cm2). On day 15, the cells were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer at 37°C for 10 min. The cell-attached PET membranes were excised and embedded in paraffin. The paraffin-embedded membranes were sectioned at a thickness of 4.5 µm and subjected to periodic acid-Schiff (PAS) staining.
Western blot
The cells were cultured for 4, 8, or 13 days. They were washed with PBS and lysed with lysis buffer containing 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.5% (w/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, and 2 mM EDTA. The supernatant was collected after 2 min of centrifugation at 10,000 × g at 4°C. Protein concentrations were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The lysate was mixed with an equal volume of 2 × lithium dodecyl sulfate (LDS) sample loading buffer (Thermo Fisher Scientific) and boiled for 5 min at 95°C. The prepared samples (15 µg/ lane) were electrophoresed on NuPAGE Novex 12% Bis-Tris gels with NuPAGE 3-Morpholinopropanesulfonic acid (MOPS)-sodium dodecyl sulfate (SDS) running buffer following the manufacturer’s instructions (Thermo Fisher Scientific). Proteins were transferred onto Immobilon-P membranes (Merck/Millipore Sigma). The blotted membranes were incubated with a blocking reagent (Blocking One, Nacalai Tesque, Kyoto, Japan) for 30 min and then incubated with the appropriate first antibody (Villin, Santa Cruz, Dallas, TX, SOX9, Merck/Millipore Sigma, claudin 4 (CLD4), Thermo Fisher Scientific, β-actin, Merck/Millipore Sigma) overnight at 4°C. After washing the membrane using PBS-T three times, the corresponding secondary antibody conjugated with HRP (Jackson ImmunoResearch, West Grove, PA, USA) was added and incubated for 2 hr at RT. The signals were developed using a chemiluminescent substrate (SuperSignal, Thermo Fisher Scientific) after washing with PBS-T. For semi-quantification, blots were imaged using a FluorChem system (Alpha Innotech, San Leandro, CA, USA) and analyzed using an image reader software (AlphaEaseFC, Alpha Innotech) as per the manufacturer’s instructions.
Scanning electron microscopy
SIEC morphology was observed using scanning electron microscopy (SEM) [19]. The cells were seeded on polycarbonate membrane Transwell inserts (0.4 μm pore size; Corning) at 9 × 105 cells/1.12 cm2/well. After 3, 6, and 10 days of incubation, the cells were washed with 0.1 M phosphate buffer (pH 7.4) and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 1 hr. After washing three times with 0.1 M phosphate buffer, the cells were dehydrated through serially diluted ethanol, which was subsequently replaced with t-butyl alcohol. The cells were then lyophilized, coated with platinum-palladium, and observed using SEM (S4200, Hitachi, Tokyo, Japan).
Real-time quantitative polymerase chain reaction
Total RNA extraction and cDNA synthesis were performed using TRI Reagent (Molecular Research Center, Cincinnati, OH, USA) and a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time; Takara Bio, Kusatsu, Shiga, Japan) as per the manufacturer’s instruction. A CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA) and TB Green Premix Ex Taq II (Takara Bio) were used to perform real-time quantitative polymerase chain reaction (RT-qPCR). The primers used in this study are listed in Table 1. The amplification conditions were 95°C for 30 sec, 40 cycles at 95°C for 5 sec, and then 60°C for 30 sec. Beta-actin was used as an internal control to normalize mRNA expression levels for differences in total cDNA levels between samples.
Table 1. Primers used in this study.
| Target | Forward (5′–3′) | Reverse (5′–3′) | References |
|---|---|---|---|
| Villin | CTCCCCTAGACAGGCTCATC | CTGCCATAGGTGCTGGAAGAA | This study |
| Lgr5 | CCTTGGCCCTGAACAAAATA | ATTTCTTTCCCAGGGAGTGG | [16] |
| Lyz | GCAAGACACCCAAAGCAGTT | ATGCCACCCATGCTTTAACG | [16] |
| Muc2 | GCTGGCCGACAACAAGAAGA | TGGTGGGAGGATGGTTGGAA | [16] |
| ChgA | TGAAGTGCATCGTCGAGGTC | GAGGATCCGTTCATCTCCTCG | [16] |
| IFN-β | AGTTGCCTGGGACTCCTCAA | CCTCAGGGACCTCAAAGTTCAT | [46] |
| Mx1 | GAGGTGGACCCCGAAGGA | CACCAGATCCGGCTTCGT | [46] |
| OAS1 | GAGCTGCAGCGAGACTTCCT | TGCTTGACAAGGCGGATGA | [46] |
Primers for Villin were designed based on the sequence XM_001925167.
Poly(I:C) stimulation
Poly(I:C) stimulation was performed as described previously [20]. Briefly, the SIEC line (3.0 × 104 cells/mL) was seeded onto 24-well plates coated with type I collagen (SUMILON) and incubated for five days. The SJO monolayer culture was incubated for five days after medium replacement. Poly(I:C) (final concentration 5 µg/mL; Sigma-Aldrich, St. Louis, MO, USA) was then added to each well and incubated for 6 or 12 hr. After incubation, total RNA extraction and RT-qPCR were performed as described above.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v8 (GraphPad Software, San Diego, CA, USA). All data are presented as the mean with the standard deviation (SD). The dots in each graph represent biological replicates.
RESULTS
Establishment and characterization of swine intestinal epithelial cells
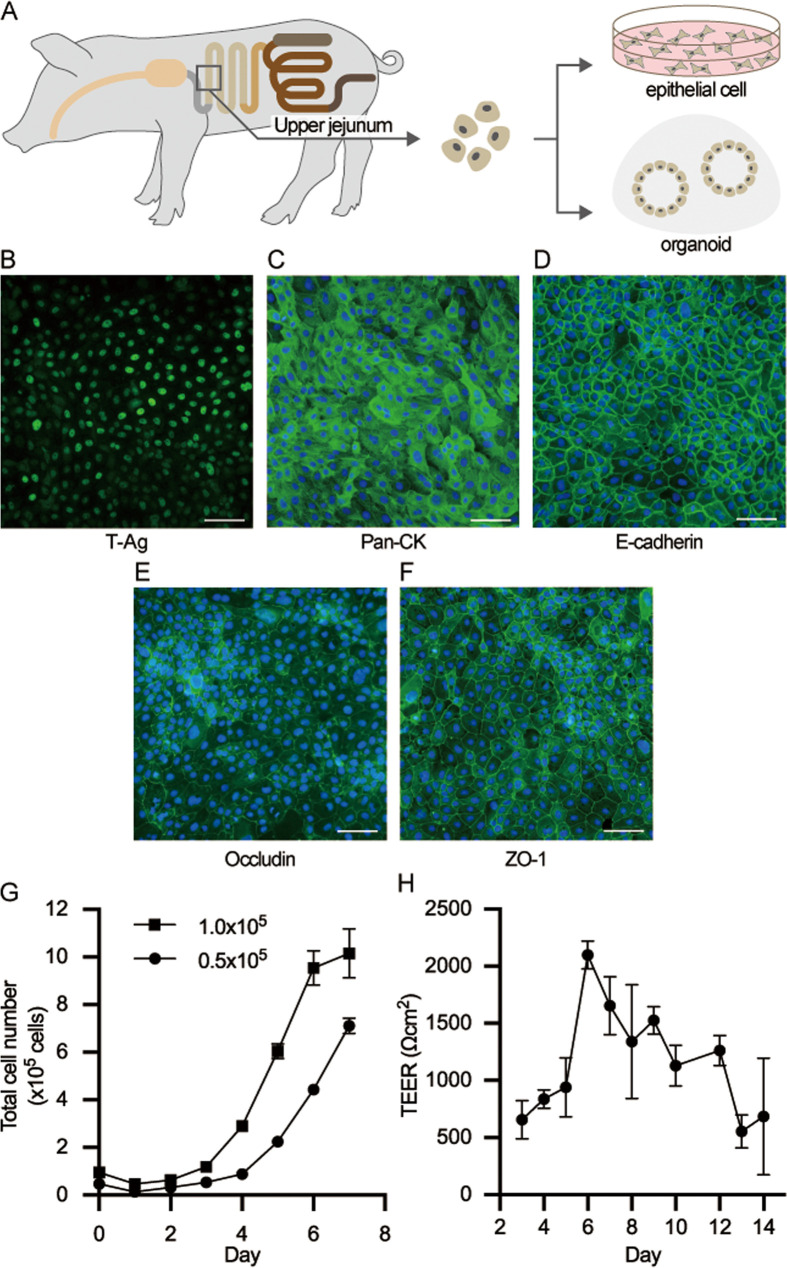
To establish a seamless in vitro evaluation system, cells were isolated from the swine small intestine, and an epithelial cell line and intestinal organoids were established (Fig. 1A). The isolated cells were immortalized using Lenti-simian virus 40 (SV40) and SV40 large T-antigen expression in all nuclei, and this was confirmed by immunostaining (Fig. 1B). Strong expression of cytokeratin, a well-known cytoskeletal protein in epithelial cells, was observed in SIECs (Fig. 1C) in addition to the expression of E-cadherin, which is involved in the adhesion of epithelial cells (Fig. 1D). In terms of the expression of tight junction-associated proteins, expression of occludin and ZO-1 was observed in SIECs (Fig. 1E, 1F). When the proliferation rate of SIECs was analyzed, an exponential increase was detected until day 6 of culture when SIECs were seeded at 1.0 × 105 cells/cm2, and then the proliferation rate plateaued (Fig. 1G). The doubling time was approximately 25.7 hr. Proliferation was slower when SIECs were seeded at 0.5 × 105 cells/cm2 and was still observed on day 7 of culture (Fig. 1G). The SIECs were cultured for 14 days to evaluate the functional integrity of epithelial cells, and the time course of the TEER was examined (Fig. 1H). TEER was approximately 700 Ωcm2 initially but increased to over 2,000 Ωcm2 by day 6 of culture. Subsequently, it decreased as the culture progressed. However, the cells still maintained approximately 700 Ωcm2 until day 14.
Fig. 1.
Establishment of the swine intestinal epithelial cells (SIECs). A Scheme of the epithelial cells and organoid establishment. Cells isolated from the jejunal crypt were immortalized and cultured to establish intestinal epithelial cells or cultured using OGM medium in a Matrigel dome to establish organoids. B–F Immunofluorescence staining images of established SIECs showing the immortalized cell marker SV40 large T tsA58 antigen (B), the epithelial marker pan cytokeratin (C), E-cadherin (D), and the tight junction markers occludin (E) and ZO-1 (F). G Cell proliferation rate of SIECs. H Monolayer integrity of SIEC. The scale bar is 100 µm. Data represent means with standard deviation.
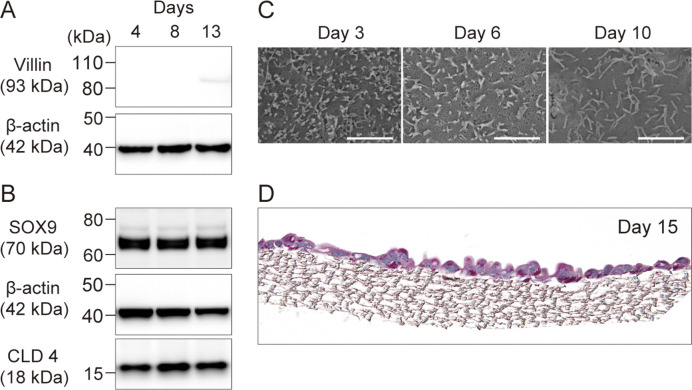
The proteins associated with the epithelial cell phenotype were validated using western blot analysis. Following cell culture for 4, 8, and 13 days, the expression of villin, SOX9, and CLD4 proteins was assessed using their respective antibodies (Fig. 2A, 2B). Beta-actin served as the housekeeping gene and was consistently detected across all samples. Villin exhibited no signals on days 4 and 8; however, a weak signal was noted on day 13 (Fig. 2A). In contrast, SOX9 and CLD4 were consistently detected throughout the culture period (Fig. 2B).
Fig. 2.
Characterization of the established swine intestinal epithelial cells (SIECs). A, B Expression of proteins associated with epithelial cells villin, SOX9, and CLD4 and the housekeeping gene (β-actin) at each culture time (day 4, 8, and 13). C Microvilli development was observed by scanning electron microscopy at each incubation time point (days 4, 6, and 10). The scale bar is 2 µm. D Periodic acid-Schiff (PAS) staining image of SIECs on day 15 of culture.
The morphological and surface characteristics of SIECs were examined using SEM after culture for 3, 6, and 10 days. Fine microvilli were initially observed on day 3; however, over time, the microvilli developed, with longer extensions noted by day 10 (Fig. 2C). Additionally, an accumulation of mucosal polysaccharides was observed after culturing the cells for 15 days and performing PAS staining on the sectioned samples (Fig. 2D).
Establishment of swine jejunal organoids
Cells isolated from the swine intestinal tract were cultured in a Matrigel dome to establish organoids. The cultured organoids were monitored for cell development over time. Initially, the organoids exhibited cell clustering on day 0 (Fig. 3A). Three-dimensional growth occurred as the culture progressed, and the formation of crypt-like structures was observed by the sixth day (Fig. 3A). HE staining confirmed the three-dimensional growth of organoids and revealed the presence of detached cells (Fig. 3B). Fluorescence immunostaining highlighted the expression of ZO-1 between cells inside the organoids and the expression of Lgr5, the intestinal epithelial stem cell marker, in the organoids (Fig. 3C, 3D).
Fig. 3.
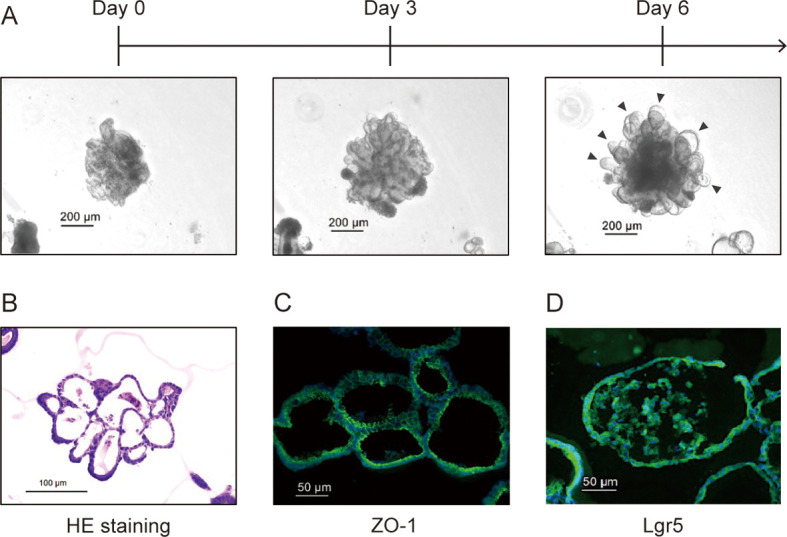
Establishment of the swine jejunal organoids (SJOs) and observation of morphological characteristics. A Observations of SJOs cultured over time in a Matrigel dome. The scale bar is 200 µm. The black arrows indicate the crypt-like structures. B HE-stained images of sliced organoids cultured for six days. The scale bar is 100 µm. Immunofluorescence staining images against ZO-1 (C) and Lgr5 (D).
Analysis of cell differentiation of swine intestinal epithelial cells and swine jejunal organoids
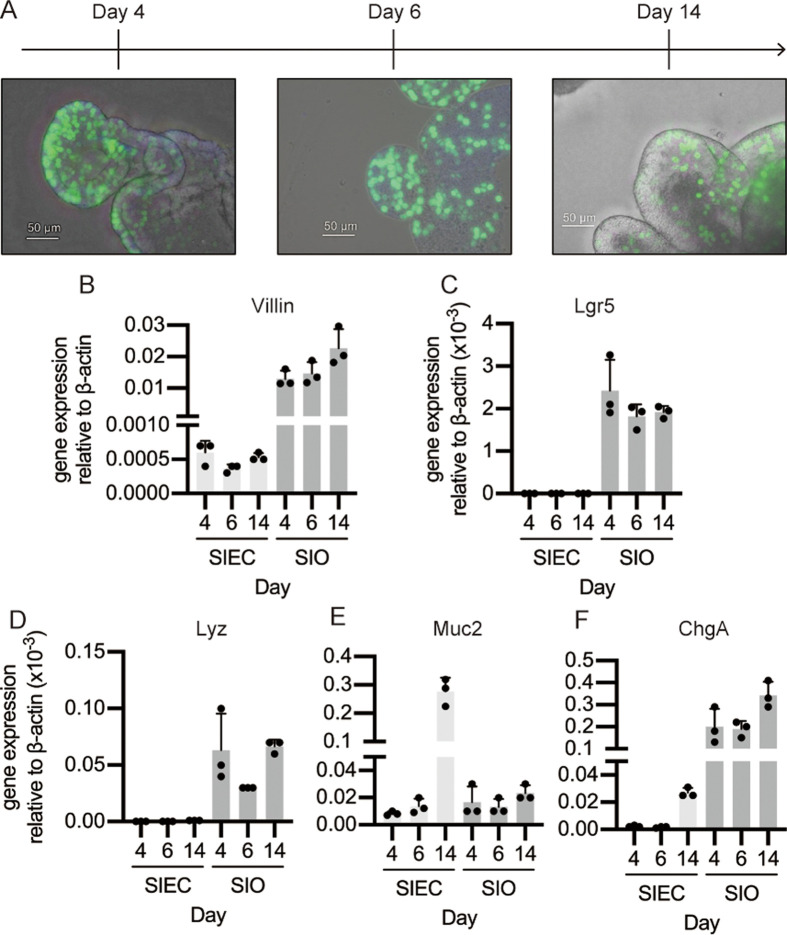
The levels of cell differentiation markers were measured in SIECs and SJOs. The localization of proliferating cells was visualized using EdU staining. The proliferating cells were distributed throughout the organoids in the early stage of culture (day 4). In contrast, the proportion of proliferating cells decreased as the culture progressed, and EdU-positive cells were often observed at the edges of the crypt-like structures (Fig. 4A).
Fig. 4.
Cell differentiation analysis of the swine intestinal epithelial cells (SIECs) and swine jejunal organoids (SJOs). A EdU assay for SJOs incubated for 4, 6, and 14 days and observed using a BZ-X800 all-in-one microscope. The scale bar is 50 µm. B–F Quantitative polymerase chain reaction (RT-qPCR) for analysis of the expression of gene cell differentiation-related markers. Changes in expression of the epithelial cell marker villin (B), intestinal stem cell marker Lgr5 (C), Paneth cell marker Lyz (D), goblet cell marker Muc2 (E), and enteroendocrine cell marker ChgA (F) were measured over time. Gene expression is presented relative to the expression level of β-actin. Data represent means with standard deviation, and the dots in each graph represent biological replicates.
The expression levels of cell differentiation markers were assessed using RT-qPCR. Total RNA was collected from SIECs or SJOs incubated for 4, 6, and 14 days, and the mRNA expression of villin, Lgr5, Lyz, Muc2, and ChgA was quantified. While no change was observed in the gene expression level of villin in SIECs over time, the villin expression level increased with the culture time in SJOs. Furthermore, compared to SIECs, SJO cells exhibited higher overall expression levels of villin (Fig. 4B). Neither Lgr5 nor Lyz gene expression was detected in SIECs. On the other hand, high levels of Lgr5 and Lyz gene expression were detected in SJOs, and there were no expression differences throughout the incubation periods (Fig. 4B, 4D). A high level of Muc2 gene expression was observed in SIECs maintained for 14 days. In contrast, there were no differences between SIECs cultured for 4 and 6 days, and there was also no change in SJOs throughout the culture periods (Fig. 4E). ChgA gene expression increased in SJOs upon culture. Also, the expression in SIECs was detected in day 6; however, the expression level was quite lower than that in SJOs (Fig. 4F).
Antiviral response in swine intestinal epithelial cells and swine jejunal organoids
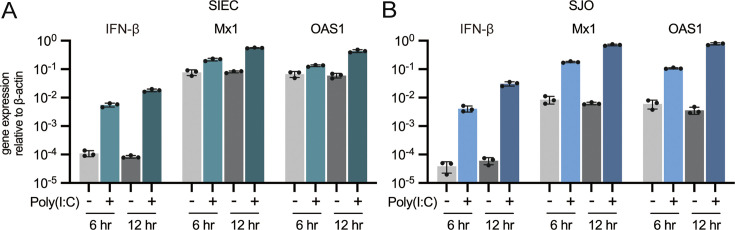
The SIECs and monolayered SJOs were stimulated with poly(I:C), a synthetic analog of double-stranded RNA, to assess the TLR3-mediated antiviral response. The antiviral gene expression was evaluated after 6 and 12 hr. Interferon (IFN)-β expression increased 50-fold and 216-fold with 6 and 12 hr of poly(I:C) stimulation, respectively, in SIECs (Fig. 5A). For SJOs, IFN-β expression increased 57-fold and 270-fold with 6 and 12 hours of stimulation, respectively (Fig. 5B). Additionally, the expression of Mx1 and OAS1 in SIECs increased approximately two- to three-fold after 6 hr of poly(I:C) stimulation and approximately seven-fold after 12 hr of stimulation (Fig. 5A). In contrast, the expression of Mx1 and OAS1 in the SJOs increased approximately 20-fold after 6 hr of stimulation. Of note, after 12 hr of poly(I:C) stimulation, Mx1 increased 80-fold, while OAS1 increased 155-fold (Fig. 5B).
Fig. 5.
Antiviral-related response in the swine intestinal epithelial cells (SIECs) and swine jejunal organoids (SJOs). SIECs (A) and SJOs (B) were stimulated with poly(I:C) for 6 or 12 hr, and the antiviral-related gene response (mRNA expression of IFN-β, Mx1, and OAS1) was investigated using quantitative polymerase chain reaction (RT-qPCR). Data represent means with standard deviation, and the dots in each graph represent biological replicates.
DISCUSSION
The intestinal tract serves as the frontline in distinguishing self- from non-self-antigens, and its homeostasis is tightly linked to host health. Frequent diarrhea during weaning and infectious diseases in the intestine lead to growth retardation and economic losses in swine production. During the short rearing period of pigs, functional feed additives and probiotics can be used to maintain and improve intestinal integrity without the need for antibiotics or drugs [21]. This approach has been helpful for promoting economically, environmentally, and ethically sustainable pig-farming practices. For this reason, the mechanisms through which functional feed additives and probiotics exert their beneficial effects are being actively investigated. In this regard, efficient in vitro evaluation systems capable of elucidating the impact of these functional materials on intestinal homeostasis as well as in the response to pathogenic bacteria and viruses are required.
Intestinal organoids derived from intestinal stem cells are attractive tools for evaluating the complexity of the intestinal tract due to their ability to undergo cell differentiation and exhibit diverse cell type configurations [14]. However, there are several challenges associated with the use of organoids. First, they are expensive to maintain because they require the addition of various factors, including growth factors, signal inhibitory factors, and hormones [22]. Secondly, it has also been noted that the reproducibility of organoid phenotypes remains a concern because of their cell differentiation [23, 24]. In contrast, an epithelial cell line based on a single type of cell is affordable and has a stable response. Therefore, an attractive strategy for flexible and efficient evaluation and screening of functional materials aimed at modulating the intestinal response to pathogens is the combination of intestinal organoids and cell line systems. In this study, a seamless in vitro intestinal evaluation platform utilizing an epithelial cell line and intestinal organoids derived from identical intestinal tissues was introduced that was aimed at reducing genetic background influences.
The crypts from the upper jejunum were isolated, and an immortalized SIEC line expressing the epithelial cell marker cytokeratin and tight junction-related proteins was established. The SIEC line exhibited a significant increase in MUC2 expression after 14 days of incubation, consistent with the observations from PAS staining conducted after 15 days of incubation. In contrast, a previous study reported that IPEC-J2 had no MUC2 detectable by RT-qPCR, thereby emphasizing the differences between the SIEC and IPEC-J2 lines [25]. This finding indicates the ability of SIECs to differentiate into mucin-producing cells within a week period, which represents an advantage for assessing interactions between microbiota and the intestinal tract using SIECs. The mucosal layer serves as the primary interface between the intestinal tract and the members of the microbiota or probiotics before direct contact with the intestinal epithelium. For example, Lacticaseibacillus rhamnosus GG, the most studied probiotic lactic acid bacterium, is known to bind to mucin via glyceraldehyde-3-phosphate dehydrogenase (GAPDH) on its cell wall surface, and it has been reported that the binding strength varies depending on the amount of GAPDH [26]. Furthermore, the mucin within the intestinal mucosa not only functions as a scaffold for microbial adhesion but also serves as a nutrient source for bacterial proliferation [27, 28]. The HT-29 cell line, derived from human colon cancer, is also recognized for its mucus-secreting properties and has previously been utilized to assess intestinal interactions with probiotics [29]. However, it has been noted that the HT-29 cells exhibit abnormally high glucose metabolism as cancer cells and that their origin from the colon tissue imposes limitations on their suitability as an evaluation system for the small intestine [30].
The SIECs formed a monolayer after 6 days of incubation, and barrier integrity was maintained throughout this period (approximately 1,000 Ωcm2 of TEER). E-cadherin and tight junction-related proteins, such as occludin and ZO-1, seem to be expressed in SIECs without gaps. Considering that the intestinal TEER has been reported to be 100–500 Ωcm2 in ex vivo tests using an Ussing chamber, the barrier integrity of SIECs is stronger than that observed in vivo [31]. The TEER in Caco-2, the most used intestinal epithelial cell line, is also around 700–2,000 Ωcm2 [31, 32], indicating that SIECs are suitable for the evaluation of membrane permeability and tight junction structure. Similar TEER values were reported for the IPEC-J2 after 4–9 days of incubation, suggesting that the SIEC line is a comparable evaluation system [33]. Additionally, the PIE cells previously established by our group for assessing innate immunity require 4–5 weeks to form a TEER of 500 Ωcm2 or higher [11, 34]. Therefore, in evaluating intestinal integrity, SIECs would likely be superior to PIE cells.
SOX9 expression in SIECs remained high throughout the incubation period. SOX9 plays an important role in the maintenance of pluripotency and differentiation in the intestine, and it has been reported that intestinal stem cells in the crypts express low levels of SOX9, though high levels are found in intestinal progenitor cells [35]. The high SOX9 levels, therefore, suggest that SIECs have characteristics of progenitor cells rather than stem cells or enterocytes. In support of this, undetectable expression of Lgr5, low villin expression, and sparse microvilli development were observed in SIECs. However, more detailed characterization is needed to discuss the cell types and characteristics of SIECs. In addition, SOX9 expression in SIECs could be associated with the high Muc2 expression, since SOX9 knockout mice are known to show reduced mucin production [36] and SOX9 is known to regulate muc2 expression [37].
To ensure genetic and histological consistency, SJOs were derived from the same host and tissues used to establish the SIECs. Cells isolated from the small intestinal crypts were cultured in Matrigel to form spheroids. As observed in previous studies on intestinal organoids, these spheroids develop crypt-like structures in culture [38]. Tight junctions were visible on the luminal side, and Lgr5-positive cells were detected. In the EdU assay for detecting proliferating cells, most cells were positive at the initiation of culture. However, the positive cells concentrated within the crypt-like structures as the culture progressed. This observation suggested the induction of cell differentiation. Gene expression analysis revealed an increase in markers associated with differentiated cells such as villin and chgA over time, validating the accuracy of the organoid model. The organoids were composed of diverse cells, as expected, including absorptive epithelial cells (vill), Paneth cells (Lyz), and enteroendocrine cells (ChgA). However, the unexpected absence of Muc2 expression in the SJOs suggests the absence of goblet cell in the organoids. The explanation of this phenomenon needs further investigation. In addition, it needs to be tested whether modifications of the culture conditions can promote the presence of goblet cells in the SJOs.
Finally, the antiviral responses in the two evaluation systems, SIECs and SJOs, were evaluated by using poly(I:C). This molecule is a synthetic dsRNA and is known to be a potent TLR3 activator that induces IFN expression [39, 40]. IFN-β expression induced by poly(I:C) stimulation was comparable in SIECs and SJOs, indicating no significant differences in TLR3 signaling. Interestingly, the expression levels of the interferon-stimulated genes (ISGs) Mx1 and OAS1 differed between the two assay systems. Poly(I:C) stimulation of SIECs for 12 hr increased the expression of Mx1 and OAS1 by approximately 7-fold. In contrast, similar poly(I:C) stimulation of SJOs increased Mx1 and OAS1 expression by 100- and 200-fold, respectively. Given that type I interferon, represented by IFN-β, binds to a heterodimer of IFN-α receptor 1 and 2 subunits and initiates the expression of ISGs, it was suggested that there were differences in IFN signaling, such as in the signaling cascade through the Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling between the two evaluation systems [41, 42]. In fact, previous studies have shown that poly(I:C) stimulation shifted gene expression in intestinal organoids from differentiation to defense and inflammation, such as chemokine expression [43, 44]. Therefore, poly(I:C) stimulation induces changes in the inflammatory reactivity of SJOs, and the different IFN signaling responses of the diverse cell types in SJOs may have caused stronger responses in ISGs. The use of vanguard techniques such as single-cell RNA-seq is necessary for a more accurate understanding of the response to poly(I:C) in each cell type in both SIECs and SJOs. In addition, evaluation of the epithelial cell response to challenges with real viruses like rotaviruses is necessary to further characterize the SIECs and SJOs in vitro systems.
Establishment of the two evaluation systems from the same individuals and tissues, thus minimizing genetic background, was also an advantage. Indeed, single nucleotide polymorphisms in pattern recognition receptors have been shown to critically influence innate immune responses [45], and the importance of the two evaluation systems established in this study for genetic identity should be further emphasized in future studies.
This study successfully established an intestinal epithelial cell line and intestinal organoids derived from same swine to propose an evaluation system that connects in vitro and in vivo results. Each system was characterized, and its responsiveness to the activation of TLR3 signaling was compared. SIECs were found to have mucin-like secretions, suggesting that they are useful in evaluating probiotic microorganisms that exert their beneficial effects through the attachment to mucins, and SJOs showed diverse cell differentiation, including intestinal epithelial cell types that cannot be used to generate cell lines. In addition, both systems responded to poly(I:C) stimulation, generating typical innate antiviral responses. The results suggest that combining the two systems would allow for a more complete evaluation of functional materials aimed at beneficially modulating antiviral immunity in the swine intestinal tract. While our study provides valuable insights, the in vitro nature of our models may not fully capture the complexity of in vivo conditions. Future studies should aim to validate these findings in farm-level settings to ensure their applicability. In conclusion, the developed systems could function as a platform for evaluating probiotics, functional additives, and viruses and for generating cellular and molecular information that can contribute to improve the healthy breeding of pigs.
FUNDING
This study was supported by a Grant-in-Aid for Scientific Research (A; 23H00354 H.K.), Grant-in-Aid for Scientific Research (B; 23H02358, K.N.), Grant-in-Aid for Research Activity Start-up (23K19327 F.N.), and Grant-in-Aid for Challenging Research (Exploratory, 23K18072, H.K.). This research was also supported by a Research Program on Development of Innovative Technology Grant (JPJ007097) from the Project of the Bio-Oriented Technology Research Advancement Institution (BRAIN) and by the Japan Racing Association (to H.K.), as well as by AMED Grant Number JP21zf0127001. Kohtaro Fukuyama was supported by a Japan Science and Technology Agency university fellowship towards the creation of science and technology innovation (Grant Number JPMJFS2102).
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Lee R. 2011. The outlook for population growth. Science 333: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FAO2023. Food Outlook—biannual report on global food markets. Food Outlook. [Google Scholar]
- 3.Lekagul A, Tangcharoensathien V, Yeung S. 2019. Patterns of antibiotic use in global pig production: a systematic review. Vet Anim Sci 7: 100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O’Ryan M, Kang G, et al. 2017. Rotavirus infection. Nat Rev Dis Primers 3: 17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda K, Takeda K. 2009. Regulatory mechanisms of immune responses to intestinal bacteria. Mucosal Immunol 2: 187–196. [DOI] [PubMed] [Google Scholar]
- 6.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14: 141–153. [DOI] [PubMed] [Google Scholar]
- 7.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 8.Han Q, Liu R, Wang H, Zhang R, Liu H, Li J, Bao J. 2023. Gut microbiota-derived 5-hydroxyindoleacetic acid alleviates diarrhea in piglets via the aryl hydrocarbon receptor pathway. J Agric Food Chem 71: 15132–15144. [DOI] [PubMed] [Google Scholar]
- 9.Kissova Z, Mudronova D, Link R, Tkacikova L. 2023. Immunomodulatory effect of probiotic exopolysaccharides in a porcine in vitro co-culture model mimicking the intestinal environment on ETEC infection. Vet Res Commun 48: 705–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Hu N, Jiang Q, Zhu L, Zhang M, Jiang J, Xiong M, Yang M, Yang J, Shen L, et al. 2021. Protective effects of sodium butyrate on rotavirus inducing endoplasmic reticulum stress-mediated apoptosis via PERK-eIF2α signaling pathway in IPEC-J2 cells. J Anim Sci Biotechnol 12: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moue M, Tohno M, Shimazu T, Kido T, Aso H, Saito T, Kitazawa H. 2008. Toll-like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim Biophys Acta 1780: 134–144. [DOI] [PubMed] [Google Scholar]
- 12.Wiarda JE, Becker SR, Sivasankaran SK, Loving CL. 2023. Regional epithelial cell diversity in the small intestine of pigs. J Anim Sci 101: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W, Zhong Y, Wei Y, Deng Z, Mao J, Liu J, Valencak TG, Liu J, Xu H, Wang H. 2022. Ileum tissue single-cell mRNA sequencing elucidates the cellular architecture of pathophysiological changes associated with weaning in piglets. BMC Biol 20: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taelman J, Diaz M, Guiu J. 2022. Human intestinal organoids: promise and challenge. Front Cell Dev Biol 10: 854740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flood P, Hanrahan N, Nally K, Melgar S. 2024. Human intestinal organoids: modeling gastrointestinal physiology and immunopathology—current applications and limitations. Eur J Immunol 54: e2250248. [DOI] [PubMed] [Google Scholar]
- 16.Khalil HA, Lei NY, Brinkley G, Scott A, Wang J, Kar UK, Jabaji ZB, Lewis M, Martin MG, Dunn JC, et al. 2016. A novel culture system for adult porcine intestinal crypts. Cell Tissue Res 365: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Lv L, Cai H, Li Y, Gao F, Yu L, Jiang Y, Tong W, Li L, Li G, et al. 2022. Long-term expansion of porcine intestinal organoids serves as an in vitro model for swine enteric coronavirus infection. Front Microbiol 13: 865336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SA, Lee HJ, Gu NY, Park YR, Kim EJ, Kang SJ, Hyun BH, Yang DK. 2023. Evaluation of porcine intestinal organoids as an in vitro model for mammalian orthoreovirus 3 infection. J Vet Sci 24: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazawa K, Hondo T, Kanaya T, Tanaka S, Takakura I, Itani W, Rose MT, Kitazawa H, Yamaguchi T, Aso H. 2010. Characterization of newly established bovine intestinal epithelial cell line. Histochem Cell Biol 133: 125–134. [DOI] [PubMed] [Google Scholar]
- 20.Zelaya H, Arellano-Arriagada L, Fukuyama K, Matsumoto K, Marranzino G, Namai F, Salva S, Alvarez S, Agüero G, Kitazawa H, et al. 2023. Lacticaseibacillus rhamnosus CRL1505 peptidoglycan modulates the inflammation-coagulation response triggered by poly(I:C) in the respiratory tract. Int J Mol Sci 24: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao SF, Nyachoti M. 2017. Using probiotics to improve swine gut health and nutrient utilization. Anim Nutr 3: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Co JY, Margalef-Català M, Monack DM, Amieva MR. 2021. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat Protoc 16: 5171–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen KB, Little MH. 2023. Organoids are not organs: sources of variation and misinformation in organoid biology. Stem Cell Reports 18: 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou WY, Blutt SE, Crawford SE, Ettayebi K, Zeng XL, Saxena K, Ramani S, Karandikar UC, Zachos NC, Estes MK. 2019. Human intestinal enteroids: new models to study gastrointestinal virus infections. Methods Mol Biol 1576: 229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schierack P, Nordhoff M, Pollmann M, Weyrauch KD, Amasheh S, Lodemann U, Jores J, Tachu B, Kleta S, Blikslager A, et al. 2006. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem Cell Biol 125: 293–305. [DOI] [PubMed] [Google Scholar]
- 26.Ishida M, Namai F, Shigemori S, Kajikawa S, Tsukagoshi M, Sato T, Ogita T, Shimosato T. 2020. Ribosome-engineered Lacticaseibacillus rhamnosus strain GG exhibits cell surface glyceraldehyde-3-phosphate dehydrogenase accumulation and enhanced adhesion to human colonic mucin. Appl Environ Microbiol 86: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glover JS, Ticer TD, Engevik MA. 2022. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci Rep 12: 8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama K, Sugiyama M, Mukai T. 2016. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudík B, Kiňová Sepová H, Bilka F, Pašková Ľ, Bilková A. 2020. Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie van Leeuwenhoek 113: 1191–1200. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Maqueda D, Miralles B, Recio I. 2015. HT29 Cell Line. In The Impact of Food Bioactives on Health: in vitro and ex vivo models, Verhoeckx K, Cotter P, Lopez-Exposito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds), Cham (CH), pp. 113–124. [PubMed] [Google Scholar]
- 31.Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. 2015. TEER measurement techniques for in vitro barrier model systems. J Lab Autom 20: 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergauwen H. 2015. The IPEC-J2 Cell Line. In The Impact of Food Bioactives on Health: in vitro and ex vivo models, Verhoeckx K, Cotter P, Lopez-Exposito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds), Cham (CH), pp. 125–134. [PubMed] [Google Scholar]
- 33.Geens MM, Niewold TA. 2011. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnology 63: 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato N, Yuzawa M, Aminul MI, Tomokiyo M, Albarracin L, Garcia-Castillo V, Ideka-Ohtsubo W, Iwabuchi N, Xiao JZ, Garcia-Cancino A, et al. 2021. Evaluation of porcine intestinal epitheliocytes as an in vitro immunoassay system for the selection of probiotic bifidobacteria to alleviate inflammatory bowel disease. Probiotics Antimicrob Proteins 13: 824–836. [DOI] [PubMed] [Google Scholar]
- 35.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. 2009. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, et al. 2007. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 178: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. 2004. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 166: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merenda A, Fenderico N, Maurice MM. 2020. Wnt signaling in 3D: recent advances in the applications of intestinal organoids. Trends Cell Biol 30: 60–73. [DOI] [PubMed] [Google Scholar]
- 39.Field AK, Tytell AA, Lampson GP, Hilleman MR. 1967. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci USA 58: 1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim CS, Jang YH, Lee GY, Han GM, Jeong HJ, Kim JW, Lee JO. 2022. TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand. Nat Commun 13: 6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piret J, Boivin G. 2022. Viral interference between respiratory viruses. Emerg Infect Dis 28: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jose SS, De Zuani M, Tidu F, Hortová Kohoutková M, Pazzagli L, Forte G, Spaccapelo R, Zelante T, Frič J. 2020. Comparison of two human organoid models of lung and intestinal inflammation reveals Toll-like receptor signalling activation and monocyte recruitment. Clin Transl Immunology 9: e1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies JM, Santaolalla R, von Furstenberg RJ, Henning SJ, Abreu MT. 2015. The viral mimetic polyinosinic: polycytidylic acid alters the growth characteristics of small intestinal and colonic crypt cultures. PLoS One 10: e0138531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jozaki K, Shinkai H, Tanaka-Matsuda M, Morozumi T, Matsumoto T, Toki D, Okumura N, Eguchi-Ogawa T, Kojima-Shibata C, Kadowaki H, et al. 2009. Influence of polymorphisms in porcine NOD2 on ligand recognition. Mol Immunol 47: 247–252. [DOI] [PubMed] [Google Scholar]
- 46.Fukuyama K, Zhuang T, Toyoshi E, Raya Tonetti F, Saha S, Zhou B, Ikeda-Ohtsubo W, Nishiyama K, Aso H, Villena J, et al. 2023. Establishment of a porcine bronchial epithelial cell line and its application to study innate immunity in the respiratory epithelium. Front Immunol 14: 1117102. [DOI] [PMC free article] [PubMed] [Google Scholar]