Abstract
Asia remains vulnerable to new and emerging infectious diseases. Understanding how to improve next generation sequencing (NGS) use in pathogen surveillance is an urgent priority for regional health security. Here we developed a pathogen genomic surveillance assessment framework to assess capacity in low-resource settings in South and Southeast Asia. Data collected between June 2022 and March 2023 from 42 institutions in 13 countries showed pathogen genomics capacity exists, but use is limited and under-resourced. All countries had NGS capacity and seven countries had strategic plans integrating pathogen genomics into wider surveillance efforts. Several pathogens were prioritized for human surveillance, but NGS application to environmental and human–animal interface surveillance was limited. Barriers to NGS implementation include reliance on external funding, supply chain challenges, trained personnel shortages and limited quality assurance mechanisms. Coordinated efforts are required to support national planning, address capacity gaps, enhance quality assurance and facilitate data sharing for decision making.
Subject terms: Policy and public health in microbiology, Comparative genomics
Surveying next generation sequencing capabilities in 13 Asian countries identifies challenges to be met for improved implementation, pathogen surveillance and integration into public health decision-making.
Main
Asia is particularly vulnerable to emerging infectious disease outbreaks. Factors such as dense populations with high rates of mobility, poor water and sanitation, abundant wildlife with frequent human–animal interaction, climate stress and a rapidly changing environment combine to concentrate outbreak risk1. In recent years, Asia has witnessed outbreaks ranging from Nipah Virus (1998), SARS-CoV (2003), Influenza A H1N1 (2009), MERS-CoV (2015), Zika virus (2016) and SARS-CoV-2 (2019). Furthermore, endemic pathogens, such as dengue virus, Chikungunya virus, Japanese encephalitis virus, tuberculosis and malaria continue to pose major threats to population health and economies. Strengthening early detection through infectious disease surveillance remains a central pillar of regional outbreak preparedness2.
Pathogen genomics using next generation sequencing (NGS) has emerged as a powerful tool to enhance early pathogen detection3,4. NGS played a key role during the COVID-19 pandemic, contributing to the initial identification of SARS-CoV-2 and allowing for detection and monitoring of new variants of concern5. Phylogenetic analyses of SARS-CoV-2 sequences combined with epidemiologic data allowed public health officials and policymakers to understand geographic and temporal spread within and across borders, providing critical evidence to inform public health interventions6,7. Genomic data are also crucial for the development of outbreak response and prevention tools, such as diagnostics, therapeutics (monoclonal antibodies) and vaccines (including tailored seasonal influenza and SARS-CoV-2 vaccines)8–10. Furthermore, its applications to human–animal interface and environmental surveillance are increasingly important complementary strategies for detecting early signals of outbreak risk11,12. During the COVID-19 pandemic, NGS was utilized for pathogen tracking in wastewater and other environmental samples to monitor community infection dynamics, assess the effectiveness of existing control measures and serve as an early warning strategy13,14.
Accelerating the application of genomic sequencing to infectious disease surveillance among lower-resourced countries in Asia is a priority. Despite NGS having been employed for several years as a surveillance tool in high-income settings, global disparities exist in its application among low- and middle-income countries (LMICs)15,16. During the COVID-19 pandemic, high-income countries submitted 10-fold more sequences per COVID-19 case than LMICs17. Where NGS has been established in resource-constrained settings, it has primarily been used to support outbreak investigation with limited integration into routine surveillance systems18–20. In response, the Asia Pathogen Genomics Initiative (Asia PGI) was established in 2021 with the aim of enhancing the use of NGS for pathogen genomic surveillance in Asia. The initiative builds on positive learnings from a parallel effort, the Africa PGI, established in 2018 as a platform to effectively implement and translate pathogen genomics into public health action21.
The pathogen genomic surveillance landscape in Asia has rapidly evolved, especially during the COVID-19 pandemic. Generating a deeper understanding of country capacity, perspectives and priorities is essential for detecting and responding to novel, emerging and endemic pathogens. As part of the Asia PGI efforts, we assessed the status of pathogen genomic surveillance across countries in South and Southeast Asia from mid-2022 to mid-2023. Findings from this assessment aim to inform national surveillance planning, future resource allocation and regional technical assistance requirements to advance pathogen genomic surveillance as a key component of regional pandemic preparedness and response efforts.
Results
Summary indicators of NGS capabilities in Asia
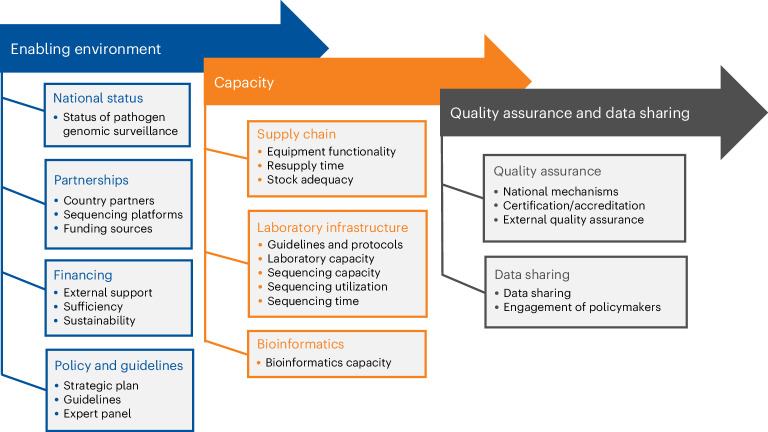
Between June 2022 and June 2023, institutions from 13 Asian countries responded to a survey (Supplementary Table 2), providing country-level data on the status of pathogen genomic surveillance. The survey was developed in line with a system-wide assessment framework for pathogen genomic surveillance capacity, consisting of three thematic focus areas: enabling environment, capacity and quality assurance and data sharing (Fig. 1). Responses were from 42 local institutions comprising governmental agencies, academic and research institutes, and NGOs.
Fig. 1.
A system-wide assessment framework of pathogen genomic surveillance capacity.
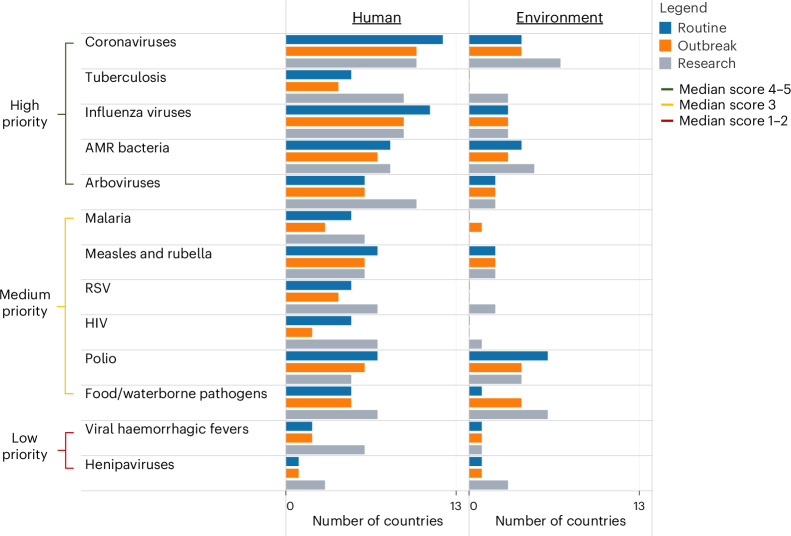
Among known pathogens, coronaviruses, tuberculosis, influenza viruses, antimicrobial resistant (AMR) bacteria and arboviruses were reported as the highest priority for genomic surveillance across countries (Fig. 2). These pathogen groups were also the most highly sequenced in the past 5 years. NGS was used by most countries for routine surveillance of coronaviruses (12/13), influenza viruses (11/13) and AMR bacteria (8/13), and for outbreak investigation of coronaviruses (10/13) and arboviruses (10/13). Pathogens most frequently sequenced by countries for research purposes included coronaviruses (10/13) and influenza viruses (9/13). Pathogen genomics was more widely conducted for human surveillance than for environmental or animal surveillance.
Fig. 2. Pathogens for which genomic surveillance was conducted in the past 5 years across 13 countries in Asia.
For each pathogen, countries reported use of NGS for routine surveillance, outbreak or research purposes, as applied to human or environmental surveillance. Pathogens are listed in order of their median Likert score (1–2 = low priority, 3 = medium priority, 4–5 = high priority). RSV, respiratory syncytial virus; HIV, human immunodeficiency virus.
Summary indicators for each thematic area are profiled in Table 1, organized under the framework categories of Enabling environment, Capacity, and Data quality, sharing and impact. Findings indicate that all countries are conducting pathogen surveillance using NGS. The assessment of unknown pathogens also took place in the context of human surveillance in nearly all countries (11/13), though less frequently on environmental (7/13) or animal samples (7/13).
Table 1.
Summary indicators on the status of pathogen genomic surveillance across 13 countries in Asia
| Framework Category | Thematic area | Indicator name | Definition | Value | Standard deviation (±s.d.), Interquartile range (IQR) |
|---|---|---|---|---|---|
| Enabling environment | National status | Status of pathogen genomic surveillance | Proportion of countries having used NGS for pathogen genomic surveillance in the past 2 years | 13/13 (100%) | N/A |
| NGS for unknown pathogens | Proportion of countries using NGS to detect ‘unknown pathogens’ |
Human = 11/13 (85%) Animal = 7/13 (54%) Environment = 7/13 (54%) |
N/A | ||
| Partnerships | Country partners | Average proportion of NGS capacity for pathogen genomic surveillance in the past 2 years, by sector |
Public = 43% Academic = 39% Private =6% Others = 12% |
±32% ±37% ±15% ±30% |
|
| Sequencing platform | Proportion of countries using each platform for genomic surveillance, by manufacturer |
ONT = 11/13 (85%) Illumina = 11/13 (85%) Thermo Fisher = 4/13 (31%) MGI/BGI = 2/13 (15%) |
N/A | ||
| Financing | Funding sources | Average proportion of funds provided for pathogen genomic surveillance in the past year, by funding source |
External = 57% Public = 32% Academic = 6% Private = 4% |
±43% ±40% ±14% ±9% |
|
| External support | Proportion of countries where over-reliance on external support is low/ not a barrier for NGS | 3/13 (23%) | N/A | ||
| Sufficient funding | Proportion of countries that perceive sufficient funding for pathogen genomic surveillance systems over the coming 5-year cycle | 2/13 (17%) | N/A | ||
| Sustainable funding | Proportion of countries that perceive sustainable funding for genomic surveillance systems for the coming 5-year cycle | 1/13 (8%) | N/A | ||
| Policy and guidelines | Strategic plan | Proportion of countries where a national strategic plan exists that includes pathogen genomic surveillance | 7/13 (54%) | N/A | |
| Guidelines | Proportion of countries where national guidelines exist for pathogen genomic surveillance | 6/13 (46%) | N/A | ||
| Expert panel | Proportion of countries where a national expert panel or technical advisory group exists to advise government interpretation/use of pathogen genomic surveillance data | 9/13 (69%) | N/A | ||
| Capacity | Supply chain | Equipment repair lead time | Proportion of countries that perceive equipment repair lead time as low/no barrier to sequencing capacity | 6/13 (46%) | N/A |
| Resupply time length | Median resupply time between order and receipt of reagents and consumables | 8 weeks | IQR: 6–9 | ||
| Stock adequacy - reagents and consumables | Proportion of countries reporting no stock out of reagents/consumables in the past 6 months | 10/13 (77%) | N/A | ||
| Laboratory infrastructure | Laboratory guidelines and protocols | Proportion of countries where laboratory guidelines and protocols exist for genomic sequencing of one or more pathogens | 9/13 (69%) | N/A | |
| Laboratory capacity | Median number of laboratories in country performing NGS for public health surveillance, per million population | 0.12 per million pop | IQR: 0.04–0.27 | ||
| Sequencing capacity | Median monthly pathogen sequences generated in the past year, per million population | 6.8 per million pop | IQR: 1.7–13.5 | ||
| Sequencing utilization | Average monthly sequencing output relative to maximum monthly sequencing capacity for the past year | 51% | ±35% | ||
| Sequencing time | Median estimated time required for NGS surveillance between specimen collection, sequence generation and reporting | 18 days | IQR: 13–25 | ||
| Bioinformatics | Bioinformatics capacity | Proportion of countries with in-country bioinformatics expertise (defined as the ability to utilize published workflows (containerized or locally installed) or in-house pipelines for >75% of genomic data analysis) | 6/13 (46%) | N/A | |
| Data quality, sharing and impact | Quality assurance | National quality assurance mechanism | Proportion of countries where national quality assurance mechanisms exist for governance of national laboratory quality (not specific to NGS) | 9/13 (69%) | N/A |
| Laboratory certification or accreditation | Proportion of countries where >75% of laboratories conducting NGS have been certified or accredited by any local or internationally recognized body | 1/13 (8%) | N/A | ||
| External quality assurance | Proportion of countries where >75% of laboratories have participated in any proficiency testing or external quality assurance audits for NGS | 2/13 (15%) | N/A | ||
| Data sharing and impact | Data sharing | Proportion of countries reporting >75% of total sequences are shared on public databases | 9/13 (69%) | N/A | |
| Engagement of policymakers | Proportion of countries reporting regularly sharing genomic data to policymakers to inform decision making | 10/13 (77%) | N/A |
Strategic planning and national integration of NGS
Regarding the enabling environment for NGS, 7/13 countries had a national strategic plan that integrated some component of pathogen genomics within infectious disease surveillance programmes. Less than half the countries (6/13) had developed guidelines for pathogen genomics for surveillance use, although many (9/13) had established expert panels to advise policymakers and programme managers on using genomic data for decision making. A range of formal and informal partner coordination mechanisms had been established, with pathogen genomic surveillance capacity residing across the public sector (43%), academic institutions (39%), the private sector (6%) and other partners (12%).
Financial and other resources to support pathogen genomic sequencing for surveillance were primarily from donors and external sources (57%), followed by contributions from the public sector (32%) and academic institutions (6%). Most countries (10/13) indicated a reliance on external funders for pathogen sequencing as a major barrier.
Over 100 laboratories across 13 countries were reported as contributing towards pathogen genomic surveillance. These represented a median of 0.12 labs per million population (IQR: 0.04 – 0.27), which together contribute a median of 6.8 pathogen sequences per million population (IQR: 1.7–13.5) each month. While two-thirds of countries reported having developed laboratory guidelines and protocols for NGS for one or more pathogens, countries reported utilizing only 51% of their maximum monthly sequencing capacity in the past year. In addition, the median turnaround time between sample collection and sequence reporting was estimated to be 18 days.
Barriers to NGS implementation
A diverse range of NGS technologies was deployed across countries. The most common platforms were Illumina and Oxford Nanopore Technologies (ONT), followed by Ion Torrent (by Thermo Fisher Scientific) and MGI. Seven countries reported equipment repair delays as a major barrier to sequencing. Although most countries (10/13) reported no stock out of reagents/consumables in the past 6 months, the resupply time for consumables and reagents was a median of 8 weeks (IQR 6–9). Bioinformatics capacity was largely reliant on proprietary software or solutions provided by NGS manufacturers, with the utilization of published workflows or in-house pipelines for genomic data analysis reported in only 46% of countries (6/13).
In terms of quality assurance, nine countries had national quality assurance mechanisms for governance of laboratory quality, not specific to NGS. However, only one country reported having >75% of laboratories certified or accredited by a local or internationally recognized programme, and only two countries had >75% of laboratories that participated in external quality assurance (EQA) programmes for NGS. Many countries (9/13) reported data sharing using publicly available platforms for at least 75% of pathogen samples sequenced. In terms of the impact of these data on decision making, 10/13 countries reported regularly sharing genomic findings to policymakers to inform public health policy.
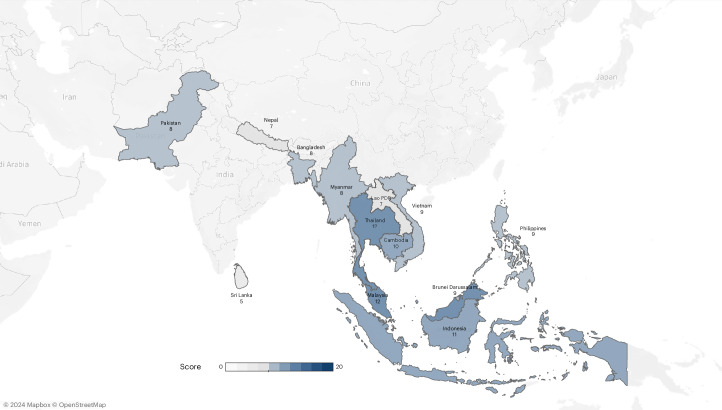
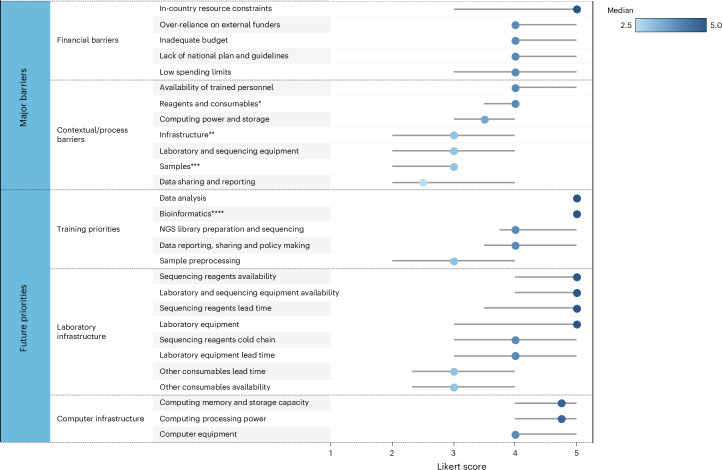
Country summary scores (Fig. 3) reflect the overall status of pathogen genomic surveillance across thematic areas, using binary scoring applied to 20 indicators (Supplementary Table 5). Nine countries that scored below 10 out of 20 were assessed as having limited system-requirements to support NGS implementation for national pathogen genomic surveillance, while four countries scoring 10 or above (Cambodia, Indonesia, Malaysia and Thailand) demonstrated more fully developed pathogen genomic surveillance capacity. Major barriers and forward-looking priorities for genomic surveillance are profiled in Fig. 4. The highest ranked barriers across all categories were financial constraints. The highest ranked process barriers included the availability and lead time of reagents/consumables, the lack of human resources, and inadequate computing power and storage. The most highly ranked future priorities included training needs in data analysis and bioinformatics; improved laboratory equipment service and maintenance, and availability of sequencing reagents; and increased computing memory and storage capacity.
Fig. 3. Country summary score for pathogen genomic surveillance status.
Total scores for each country are aggregated across thematic areas within the system-wide assessment framework (Enabling environment, Capacity and data quality, Sharing and impact) and include binary scoring for 20 indicators. A higher score indicates a more robust system supporting NGS implementation. It reflects the presence of supportive policies, investment, infrastructure and expertise, as well as good data dissemination practices essential for enhancing genomic surveillance.
Fig. 4. Major barriers and future priorities for pathogen genomic surveillance across 13 countries.
Median Likert scores (1–5) and interquartile ranges (IQR) are displayed, with scores for major barriers ranging from 1 (not a barrier) to 5 (always a barrier), and future priorities from 1 (not a priority) to 5 (essential). Median Likert score is displayed by a dot, and IQR by a grey bar, where the lowest and highest of the grey bar marks the 25th and 75th percentile, respectively (*includes availability, lead time and expiry date on arrival; **includes electricity and internet connection; ***includes transportation time and quality; ****includes data processing, quality assurance and storage).
Discussion
We assessed the status of pathogen genomic surveillance across 13 South and Southeast Asian countries with a focus on low-resource settings. Findings show that all countries are conducting in-country pathogen genomic sequencing and sharing data through publicly available platforms. However, wide variation exists between countries, with up to a seven-fold difference in the number of laboratories and sequences generated per million population. While national public health institutions have the mandate for surveillance efforts, a substantial portion of NGS capacity resides in academic/research entities.
Sufficient and sustainable financing were identified as key barriers for pathogen genomic surveillance. While costs have come down substantially in recent years, genomic surveillance remains relatively expensive, and countries rely heavily on external partner support. National investment cases to align domestic and external contributions should be informed by updated national plans that integrate pathogen genomics into wider surveillance efforts. Leveraging global financing mechanisms such as The Global Fund and The World Bank’s Pandemic Fund have the potential to provide additional resources to support the use of genomics for early detection of novel and endemic pathogens, particularly in LMICs22. Supporting countries in the cost-efficient use of NGS through effective surveillance planning, sampling strategy and system design is a key priority. This includes guidance on embedding pathogen genomics as a complementary strategy alongside conventional and molecular diagnostics and defining which pathogens would be most beneficial for routine sequencing through national surveillance programmes.
While LMICs in Asia have deployed a diversity of NGS technologies for pathogen sequencing, major procurement and supply chain challenges exacerbate inefficiencies and drive up costs. Similar challenges have been faced by other LMICs globally, especially during the scale up of genomic surveillance of SARS-CoV-2 (refs. 21,23–25). A recent market assessment suggests that resource-constrained countries are paying up to tenfold more per sequence than high-income countries26. Long lead times averaging 8 weeks for procurement of reagents and consumables, alongside equipment repair delays, were identified as common constraints by most countries included in this assessment. Higher prices in LMICs result from a range of issues including small volume purchases, limited numbers of suppliers and distributors, and delays in customs clearance that affect the ability to use time-sensitive biological reagents efficiently. To respond to procurement challenges, NGS equipment and reagents have recently been listed on global supply catalogues27. These mechanisms of global market shaping efforts enable price reductions through aggregating demand across countries.
Laboratory capacity and quality assurance were identified as major challenges by country respondents. Although national guidelines for pathogen genomics were identified in six countries, few facilities were certified or accredited, and EQA for NGS was largely absent. Working with national bodies to develop accreditation standards for laboratories conducting genomic surveillance and developing low-cost regional EQA hubs to stress-test surveillance systems from sample collection through to sequence generation and reporting are essential.
Improving the quality and timeliness of pathogen genomics data is critical for public health decision-making. Countries remain reliant on bioinformatics tools provided by NGS manufacturers, resulting in limited reproducibility of bioinformatics pipelines. Few countries employed containerized (that is, a consistent virtual environment) or locally installed (that is, on local hardware) public workflows. While most countries in this assessment reported sharing sequencing data on publicly accessible sites (such as NCBI, EBI, GISAID), global analysis indicates that GISAID was the main sharing platform during the SARS-CoV-2 pandemic, and that for many countries, available data represent only a small subset of overall sequences28. In addition, although countries report having technical committees to feedback the results of genomic surveillance to decision makers, the 18-day turnaround time between sample collection and reporting limits the utility of pathogen genomic surveillance to inform timely public health responses. Linking genomic data to clinical and epidemiological information is essential for real-time decision-making. This will require agreements on meta-data standards for human surveillance alongside efforts to bridge monitoring systems between human, animal and environmental health17.
The country summary scores generated in this assessment provide useful insights into overall pathogen genomics capacity. Findings suggest that most countries in South and Southeast Asia remain in the process of introducing system-wide capacities for NGS use for pathogen surveillance and will require support across the full range of thematic areas. Countries with more well-established systems such as Cambodia, Indonesia, Malaysia and Thailand have the potential to play an important leadership role, sharing lessons to support cross-country learning. However, the existence of well-established partnerships, financing and policies were not always associated with strong pathogen genomic surveillance capacity. In these settings, more-targeted capacity development will be required to strengthen systems.
This assessment provides a regional snapshot of NGS capacities and priorities across Asia. Except for Myanmar, country institutions contributing data to this review represent an average of 87% of each country’s SARS-CoV-2 sequences submitted to GISAID in the past year (January to December 2022). As this study focuses on public health, Myanmar GISAID data were excluded since 98% of SARS-CoV-2 sequences submitted to GISAID in 2022 were from the Defense Services Medical Research Center (DSMRC) under the Ministry of Defense. Nonetheless, this assessment also has several limitations. First, responding partner institutions were predominantly from Ministries of Health and National Public Health Laboratories which focus primarily on the human health sector. Expanding future efforts to the animal and environmental health sectors and including national environmental agencies, the UN Food and Agricultural Organization and the World Organization for Animal Health are essential for data use across sectors, adopting a One Health approach. Second, our methodology relied on expert perceptions which may only partially capture realities on the ground. This study does not cover sampling methodology which influences the accuracy and representativeness of genomic surveillance. Finally, the cross-sectional nature of the survey can be enhanced through a standardization of metrics and periodic assessments to examine shifts in NGS capacity, barriers and priorities over time. Given the rapid pace of innovation in this space, documenting country experience around specific use-cases with active cross-country knowledge sharing has the potential to accelerate adoption and scale.
In summary, among Asian countries there exists a wide range of capacity and application of genomic sequencing for infectious disease surveillance. The findings from this assessment inform a set of recommendations (Table 2) for accelerating early pathogen detection through NGS across the region. These efforts aim to advance an emerging global agenda to institutionalize genomics for novel and endemic pathogens, as laid out by the World Health Organization’s Global Surveillance Strategy for Pathogens with Pandemic and Epidemic Potential (2022–2032)29 and through the establishment of the International Pathogen Surveillance Network30. The pathogen-specific utility of genomic surveillance has been articulated elsewhere31. Recommendations highlighted in this assessment respond to key challenges including the need for sustainable financing, strengthening national surveillance planning, addressing procurement and supply chain issues, improving laboratory capacity and quality assurance, and supporting training and advances in bioinformatics and cross-country data sharing. Coordinated efforts that draw from the full range of national partners, leverage the expertise of regional and global partners, and optimize support from manufacturers will be essential to accelerate early pathogen detection through NGS and strengthen regional health security.
Table 2.
Recommendations to accelerate pathogen genomic surveillance in Asia
| Key constraints | Recommendations |
|---|---|
| FINANCING: insufficient and unsustainable domestic financing; over-reliance on donors/external partners. |
∙ Develop national investment cases for pathogen genomic surveillance. ∙ Prioritize genomic surveillance in country applications to global financing mechanisms (The Global Fund, the Pandemic Fund). ∙ Pooled procurement support for genomic surveillance commodities through established global procurement catalogues. |
| POLICY and GUIDELINES: few LMICs in Asia have updated comprehensive national strategic plans that integrate pathogen genomics into wider surveillance efforts. |
∙ Establish multipartner national coordination mechanisms that leverage capacity between national public health institutions, academic bodies and other stakeholders. ∙ Support national planning in the design of cost-efficient systems for pathogen genomic surveillance that optimize public health impact. ∙ Define where pathogen genomics should take place in routine systems vs research. ∙ Enable cross-sectoral collaborations for One Health surveillance that include pathogen genomics. |
| SUPPLY CHAIN: procurement, supply and distribution bottlenecks for NGS equipment, consumables and reagents limit the timeliness of response and impact of pathogen genomic surveillance. |
∙ Enhance regional supply chains to support regional manufacturing, warehousing and distribution of genomics commodities. ∙ Track procurement lead times. ∙ Address customs/tax-exemption challenges through coordinated national engagement. ∙ Establish mechanisms for supply chain problem solving between manufacturers and country partners. |
| LABORATORY INFRASTRUCTURE: pathogen genomics remains a novel and rapidly evolving technology with on-going training needs. Timeliness of sequencing and reporting remains constrained among most LMICs in Asia. |
∙ Coordinate regional efforts to enhance laboratory capacity in genomic sequencing for endemic and novel pathogens. ∙ Facilitate joint capacity development efforts between human and animal laboratories. ∙ Design, test and share system-level innovations that reduce the time between specimen collection, pathogen sequencing and reporting. |
| QUALITY ASSURANCE: laboratories undergo limited national/international accreditation and are not undergoing External Quality Assessments (EQA). |
∙ Define national accreditation standards for pathogen genomics. ∙ Establish low-cost regional EQA. |
| BIOINFORMATICS AND DATA SHARING: bioinformatics capacity remains limited. Data quality standards need to be strengthened to ensure high utility of sequences shared regionally and globally. |
∙ Enhance in-country bioinformatics infrastructure and capacity. ∙ Work with global partners to develop and implement meta-data standards for samples used in pathogen genomic surveillance. |
Methods
A cross-sectional assessment was conducted with partners directly contributing to pathogen genomic sequencing efforts in 13 countries: Bangladesh, Brunei Darussalam, Cambodia, Indonesia, Lao PDR, Malaysia, Myanmar, Nepal, Pakistan, Philippines, Sri Lanka, Thailand and Vietnam. Major local institutions conducting pathogen genomic surveillance were identified through national stakeholder consultation with public health and government institutes. Participating institutions included those from government, academia, public/private laboratories and NGOs (Supplementary Table 2). Participation was voluntary. The study was exempted from full Institutional Review Board approval on the basis of minimal risk to participants (NUS-IRB-2022-373).
System-wide assessment framework and survey tool
We held consultations with a reference group of global and regional experts in pathogen genomic surveillance and conducted an in-depth scoping review of pre-existing tools for assessing genomic surveillance capacity (Supplementary Table 3). These activities contributed to the development of a system-wide framework, which assesses the major influencing elements affecting the national adoption of pathogen genomic surveillance. This assessment framework then guided the development and refinement of a survey tool to capture the status of pathogen genomic surveillance at the national level (Supplementary Table 4). The system-wide framework and survey tool have three thematic focus areas: (1) enabling environment, (2) capacity and (3) data quality, sharing and impact. The final survey tool contained over 90 questions and was shared with respondents in electronic Microsoft Word document format.
Data collection, validation and analysis
Introductory sessions were held with country respondents to review the objectives, format and contents of the survey. Respondents, generally comprising heads of institutions as well as teams of pathogen genomic experts, completed the survey through self-assessment. Responses were compiled at the national level or submitted separately for each reporting institution. In the event of multiple completed surveys per country, responses were merged and validated with all participating country partners. Survey data were transferred from Microsoft Word (v.16.86) to Excel (v.16.86) through double entry, and data were analysed using Tableau software (v.2023.1). Summarized findings for each country were presented and validated with the respective country respondents. Inconsistencies, missing responses or responses that required clarification were discussed during validation calls. During these calls, study participants had the opportunity to provide further qualitative insights on major challenges and bottlenecks.
From survey responses, 25 cross-country summary indicators were selected and calculated to assess the regional status of pathogen genomic surveillance, partnerships, financing, policy and guidelines, supply chain, laboratory infrastructure, bioinformatics, quality assurance, and data sharing and impact. Indicator definitions and values are provided in Table 1. Survey responses were captured as Likert scores (scale of 1–5), binary (Yes/No) or continuous data. Proportions are expressed as values and percentages. Likert data are shown as proportions of countries scoring above an indicated threshold (for example, score of 4–5 on the Likert scale), with higher scores consistently reflecting greater capacity. Continuous data are displayed as cross-country means with standard deviations (s.d.) for normally distributed data, or as cross-country medians with interquartile ranges (IQR) for non-normally distributed data. Variations between countries are expressed on the basis of the s.d. or IQR due to outlier effects.
Country summary score and recommendations
To depict country status across domains, 20 of the 25 summary indicators were converted to binary scores, with 5 descriptive indicators omitted as they were not appropriate for binary scoring (Supplementary Table 5). Scores were summed per country to generate an aggregate metric to illustrate the comparative status of pathogen genomic surveillance across countries.
A synthesis of key barriers and priorities supported by follow-up discussions with country partners was used to inform a set of recommendations with the aim of informing a regional agenda for action.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
All supplementary tables.
Acknowledgements
We thank all of the Asia PGI country teams for their time and dedication in completing the system-wide pathogen genomic surveillance assessment; our regional and global partners for their guidance and support, including the members of the Asia PGI external reference group: X. Xuanhao Chan (Industry), A. Christoffels (SANBI/PHA4GE), R. Lin (NPHL, Singapore), D. Lye (NCID, Singapore), N. Mulder (NGS Academy), D. Naidoo (WHO SEARO), A. Suresh (FIND), S. Tessema (Africa PGI) and T. W. Yeo (Singapore PREPARE). Funding was provided by The Bill and Melinda Gates Foundation (INV-037608), and the Duke-NUS Signature Research Programme funded by the Ministry of Health, Singapore. Regarding this study’s authors from the World Health Organization, this work represents the personal opinion of the authors and not that of the World Health Organization.
Author contributions
M.G., R.d.A. and P.P. contributed to conceptualization, investigation, methodology, formal analysis, validation, writing the original draft, reviewing and editing. S.W. contributed to investigation, data curation, formal analysis, validation and visualization. S.A. contributed to conceptualization, investigation, methodology, validation and project administration. Y.K.K. and T.-M.M. contributed to conceptualization, investigation, validation and writing. J.P. contributed to investigation, validation and writing. L.M. and A.-C.S. contributed to investigation and validation. M.H.F.H.A.M., A.A., L.R.A., G. Azzam, S.C., T.C., G. Arunkumar, D.T.H., A.I., R.J., E.A.K., M.Q.L.T., S.M., G.N.M., J.E.M., S.L.M., N.V.T., I.N., F.Q., F.N.Q., M.T.R., S.S., C.P.S., T.S., L.V.T., T.J.R.D., R.T., H.M.T., H.T., P.X. and Z.Z. contributed to the survey implementation and data gathering. S.M.-S., G.J.D.S., L.-F.W. and J.C.W.L. contributed to conceptualization, and writing, review and editing of the manuscript. All authors approved the final version of the manuscript.
Peer review
Peer review information
Nature Microbiology thanks Marvin Hsiao, Stephanie Lo, Samuel Scarpino and Sara Tomczyk for their contribution to the peer review of this work.
Data availability
Study data will be available upon request and following clearance from country teams. For access to data, please email the corresponding author. Expect a response within 2 weeks.
Code availability
No custom code was developed or used to analyse the data presented in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marya Getchell, Suci Wulandari.
A full list of members and their affiliations appears in the Supplementary Information.
Change history
10/16/2024
A Correction to this paper has been published: 10.1038/s41564-024-01848-x
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-024-01809-4.
References
- 1.Allen, T. et al. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun.8, 1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Regional Office for the Western Pacific (eds). Asia Pacific strategy for emerging diseases and public health emergencies (APSED III): advancing implementation of the International Health Regulations (2005) : working together towards health security (WHO, 2017).
- 3.Armstrong, G. L. et al. Pathogen genomics in public health. N. Engl. J. Med381, 2569–2580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockdale, J. E., Liu, P. & Colijn, C. The potential of genomics for infectious disease forecasting. Nat. Microbiol.7, 1736–1743 (2022). [DOI] [PubMed] [Google Scholar]
- 5.John, G. et al. Next-generation sequencing (NGS) in COVID-19: a tool for SARS-CoV-2 diagnosis, monitoring new strains and phylodynamic modeling in molecular epidemiology. Curr. Issues Mol. Biol.43, 845–867 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robishaw, J. D. et al. Genomic surveillance to combat COVID-19: challenges and opportunities. Lancet Microbe2, e481–e484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genomic Sequencing of SARS-CoV-2: A Guide to Implementation for Maximum Impact on Public Health (WHO, 2021).
- 8.Quer, J. et al. Next-generation sequencing for confronting virus pandemics. Viruses14, 600 (2022). [DOI] [PMC free article] [PubMed]
- 9.Van Poelvoorde, L. A. E. et al. Whole-genome-based phylogenomic analysis of the Belgian 2016–2017 influenza A(H3N2) outbreak season allows improved surveillance. Microb. Genom.7, 000643 (2021). [DOI] [PMC free article] [PubMed]
- 10.Pollett, S. et al. Genomic epidemiology as a public health tool to combat mosquito-borne virus outbreaks. J. Infect. Dis.221, S308–s318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardy, J. L. & Loman, N. J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet.19, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wille, M., Geoghegan, J. L. & Holmes, E. C. How accurately can we assess zoonotic risk? PLoS Biol.19, e3001135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dharmadhikari, T. et al. High throughput sequencing based direct detection of SARS-CoV-2 fragments in wastewater of Pune, West India. Sci. Total Environ.807, 151038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao, L. et al. Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chem. Soc. Rev.50, 3656–3676 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Colson, P. & Raoult, D. Global discrepancies between numbers of available SARS-CoV-2 genomes and human development indexes at country scales. Viruses13, 775 (2021). [DOI] [PMC free article] [PubMed]
- 16.SARS-CoV-2 Genomic Surveillance Global Capacity Mapping (FIND, 2023); https://www.finddx.org/covid-19/covid-19-genomic-surveillance/covid-19-next-generation-sequencing-global-capacity-mapping/
- 17.Brito, A. F. et al. Global disparities in SARS-CoV-2 genomic surveillance. Nat. Commun.13, 7003 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohl, J. A. et al. Discovering disease-causing pathogens in resource-scarce Southeast Asia using a global metagenomic pathogen monitoring system. Proc. Natl Acad. Sci. USA119, e2115285119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoenen, T. et al. Nanopore sequencing as a rapidly deployable ebola outbreak tool. Emerg. Infect. Dis.22, 331–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyakarahuka, L. et al. First laboratory confirmation and sequencing of Zaire ebolavirus in Uganda following two independent introductions of cases from the 10th Ebola Outbreak in the Democratic Republic of the Congo, June 2019. PLoS Negl. Trop. Dis.16, e0010205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inzaule, S. C., Tessema, S. K., Kebede, Y., Ogwell Ouma, A. E. & Nkengasong, J. N. Genomic-informed pathogen surveillance in Africa: opportunities and challenges. Lancet Infect. Dis.21, e281–e289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Pandemic Fund (World Bank, 2022); https://www.worldbank.org/en/programs/financial-intermediary-fund-for-pandemic-prevention-preparedness-and-response-ppr-fif
- 23.Khan, W. et al. Building up a genomic surveillance platform for SARS-CoV-2 in the middle of a pandemic: a true North-South collaboration. BMJ Glob. Health8, e012589 (2023). [DOI] [PMC free article] [PubMed]
- 24.Merhi, G. et al. SARS-CoV-2 genomic epidemiology: data and sequencing infrastructure. Future Microbiol.17, 1001–1007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahadeo, N. S. D. et al. Implementation of genomic surveillance of SARS-CoV-2 in the Caribbean: lessons learned for sustainability in resource-limited settings. PLOS Glob. Public Health3, e0001455 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertholet, A. FIND Sequencing Market Analysis, prepared for ACT-A Sequencing Task Force (FIND, 2021).
- 27.Partnership for Supply Chain Management Product Catalogue 2022 v3 (PFSCM, 2022).
- 28.Chen, Z. et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat. Genet.54, 499–507 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter, L. L. et al. Global genomic surveillance strategy for pathogens with pandemic and epidemic potential 2022–2032. Bull. World Health Organ.100, 239–239A (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO Launches Global Network to Detect and Prevent Infectious Disease Threats (WHO, 2023). https://www.who.int/news/item/20-05-2023-who-launches-global-network-to--detect-and-prevent-infectious-disease-threats
- 31.Pronyk, P. M. et al. Advancing pathogen genomics in resource-limited settings. Cell Genom.3, 100443 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All supplementary tables.
Data Availability Statement
Study data will be available upon request and following clearance from country teams. For access to data, please email the corresponding author. Expect a response within 2 weeks.
No custom code was developed or used to analyse the data presented in this study.