Abstract
P143 is a DNA helicase that tightly binds both double-stranded and single-stranded DNA. DNA-protein complexes rapidly dissociated in the presence of ATP and Mg2+. This finding suggests that ATP hydrolysis causes a conformational change in P143 which decreases affinity for DNA. This supports the model of an inchworm mechanism of DNA unwinding.
The P143 protein of Autographa californica nuclear polyhedrosis virus is an essential protein that was predicted to be a DNA helicase based on the presence of an ATP binding motif that is conserved among DNA helicases (3, 9). Subsequent site-directed mutagenesis studies on seven putative helicase motifs revealed that mutations in motifs I, Ia, II, and III blocked DNA replication, while mutations in the other motifs had no effect (7). This result supports the classification of P143 as a member of superfamily III, which is a relatively uncharacterized group of enzymes that lack recognizable sequences corresponding to motifs IV to VI (2).
P143 is required for transient replication of a plasmid containing a baculovirus origin of DNA replication in addition to IE1, DNApol, LEF-1, LEF-2, and LEF-3 (5, 10). P143 copurifies with LEF-3, the baculovirus single-stranded DNA (ssDNA) binding protein (SSB) (1, 4). This interaction between P143 and LEF-3 is not surprising because many helicases specifically bind their cognate SSBs, and this interaction has been shown to stimulate DNA unwinding (11). In the case of P143, however, the LEF-3 interaction also has an unexpected function because LEF-3 is required for nuclear localization of P143 (15).
Recently, we showed that P143 has both ATPase and DNA unwinding activities (12). As expected for a helicase, P143 binds to double-stranded DNA (dsDNA) and ssDNA in a sequence-nonspecific manner (6, 12). In electrophoretic mobility shift assays (EMSAs), ladders of shifted bands are observed in an enzyme-dependent manner, suggesting that multiple monomers or oligomers bind to both probes in a noncooperative manner.
ATP inhibits the DNA binding activity of P143.
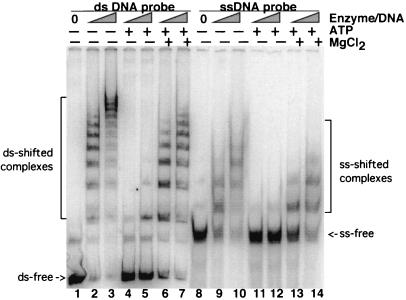
Our aim in this study was to further analyze the DNA binding activity of P143 in order to gain insight into the mechanism of DNA binding and DNA unwinding activities. As a first step, we compared the DNA binding activity of P143 in a standard EMSA buffer and in a buffer optimized for ATPase activity. We found that DNA binding was considerably lower in the ATPase buffer (data not shown). Analysis of each of the different components revealed that the formation of stable protein-DNA complexes was decreased approximately fivefold by the addition of ATP and MgCl2 (Fig. 1, lanes 6, 7, 13, and 14). This experiment was performed with two different concentrations of enzyme in order to cover a wider range of activity. Furthermore, addition of ATP alone had an even more dramatic effect on DNA binding (Fig. 1, lanes 4, 5, 11, and 12). This was true for both dsDNA (Fig. 1, lanes 1 to 7) and ssDNA (Fig. 1, lanes 8 to 14).
FIG. 1.
Effects of ATP and MgCl2 on the binding of P143 to 252-bp hr5 ss- and dsDNA. The conditions for purification of P143, radiolabeling of hr5 probe, and EMSAs have been described previously (12). In this experiment, the molar ratios of enzyme to probe were 10:1 and 50:1. The presence or absence (+ or −) of 2 mM ATP and 6 mM MgCl2 is indicated at the top. Lanes 1 to 7 contain dsDNA probe; lanes 8 to 14 contain ssDNA probe. ssDNA probe was generated from the double-stranded probe by dilution in water followed by boiling and rapid cooling on ice. Free and protein-bound DNAs were separated on a 3.5% acrylamide–Tris-borate-EDTA gel, dried, and exposed to PhosphorImager plates. ds, double-stranded; ss, single-stranded.
Inhibition is specific for ATP.
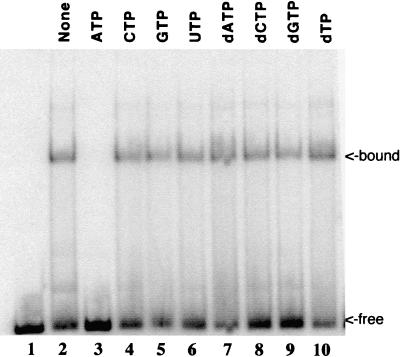
A trivial explanation for the inhibition of DNA binding is that ATP altered the pH or ionic strength of the buffer, even though the nucleotide solution was neutralized to pH 7.0 before use. We reasoned that if inhibition were due simply to the presence of a negatively charged nucleotide that competed with DNA, then other nucleoside triphosphates (NTPs) should have the same effect. To test this, we added eight different NTPs individually to identical reaction mixtures, each at the same concentration. For these experiments, a shorter DNA probe, corresponding to an 88-bp fragment of hr5, was used because it produced only one or two shifted bands. This allowed for better quantitation than did the larger probe, which produced multiple bands containing unknown amounts of protein in each shifted band. The EMSA results showed that DNA binding was inhibited only in the reactions containing ATP (Fig. 2). The other seven nucleotides did not significantly alter the binding of P143 to DNA. Previously, we showed that ATP was the only NTP that could support the DNA unwinding activity of P143 (12). Therefore, this suggests that inhibition is due to binding of the enzyme to ATP and not to nonspecific ionic effects.
FIG. 2.
Effect of NTPs on P143 DNA binding in the absence of divalent cation. Lane 1 contains probe alone; lanes 2 to 10 contain purified P143 in fivefold-molar excess of enzyme over an 88-nucleotide radiolabeled DNA probe. Lanes 3 to 10 contain a 2 mM concentration of the indicated NTP. Free and protein-bound DNAs were fractionated on a 5% acrylamide–Tris-borate-EDTA gel, dried, and exposed to a PhosphorImager plate. An 88-nucleotide probe, also containing hr5 sequences, was cut from the same plasmid, first with EcoRI and then, after kinase labeling, with MluI.
These results suggest that P143 can bind ATP even in the absence of added Mg2+. This was surprising because ATP binding proteins are believed to interact with the metal-bound form of ATP. We therefore conducted filter binding assays in the presence and absence of MgCl2. We found that the enzyme bound ATP in the presence and absence of added magnesium (data not shown); however, we cannot rule out the possibility that the purified enzyme and the ATP solution contained bound metal. The use of ATP analogs like adenylyl-imidodiphosphate and adenosine 5′-0-(3-thiotriphosphate) also gave inconclusive results in DNA binding assays. Subsequent analyses suggested that P143 did not bind these analogs. We found that addition of analogs did not compete with radiolabeled ATP in ATPase assays (data not shown).
Only divalent cations that support ATP hydrolysis restore DNA binding.
We then wanted to determine whether the ability of MgCl2 to reverse the ATP inhibition of DNA binding was related to the catalytic activity of the enzyme. If MgCl2 were only acting as a counterion, then other divalent cation salts should have the same effect. Alternatively, if MgCl2 restored DNA binding because it supported ATPase activity, then only cations that also function as cofactors should relieve inhibition.
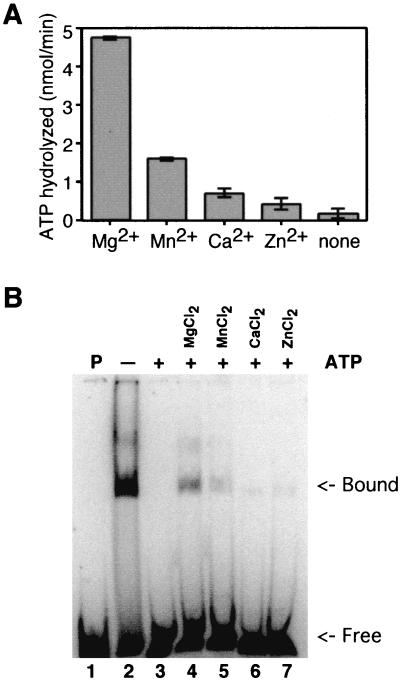
To test this, equal concentrations of four different divalent cation salts were added to individual ATPase reactions (Fig. 3A). Reaction mixtures containing MgCl2 hydrolyzed the most ATP, which was not surprising as most ATPases function best in the presence of magnesium rather than that of another divalent cation (12). The next best divalent cofactor was manganese. Reaction mixtures containing calcium and zinc produced negligible ATP hydrolysis, and there was no hydrolysis in the absence of divalent cation.
FIG. 3.
Effects of divalent cations. (A) ATPase. Each reaction mixture contained a 6 mM concentration of the indicated divalent cation salt. ATPase reaction mixtures contained 2 mM ATP, 0.1 μCi of [γ-32P]ATP, a 6 mM concentration of the indicated divalent cation chloride salt, 20 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 15 mM NaCl, 1 mM dithiothreitol, 140 fmol of P143, and 0.25 μg of activated calf thymus DNA in a final volume of 20 μl. Samples were incubated at 30°C for 30 min and were then terminated by the addition of 25 mM EDTA. Polyethyleneimine thin-layer plates were spotted with 1 μl of each reaction, and the hydrolyzed, radiolabeled phosphate was separated from the input triphosphate by development in 1 M formic acid and 0.5 M LiCl. Thin-layer chromatography plates were dried and exposed to PhosphorImager screens. Results were quantitated using a Storm PhosphorImager. Background levels of hydrolysis in the absence of enzyme were subtracted. An assay was performed in triplicate, and standard error is indicated. (B) DNA binding. Lane 1 contains probe without enzyme. Lanes 2 to 7 contain P143 in a fivefold-molar excess over the 88-nucleotide radiolabeled probe. Lanes 3 to 7 contain 2 mM ATP (+), while lane 2 does not (−). Lanes 4 to 7 contain a 6 mM concentration of the indicated divalent cation salt. Reaction mixtures were fractionated on a 5% acrylamide–Tris-borate-EDTA gel, dried, and exposed to a PhosphorImager plate.
We then added the same divalent cations used in the ATPase assay to EMSA reaction mixtures containing ATP and P143 (Fig. 3B). ATP inhibited the DNA binding activity of P143, and the addition of MgCl2 restored 18% of normal binding, as previously seen. MnCl2 restored binding to a lesser extent, only 7% of the level obtained in the absence of ATP. CaCl2 and ZnCl2 (Fig. 3B, lanes 5 and 6) did not significantly relieve ATP inhibition. Therefore, we concluded that only divalent cations that support ATPase activity supported DNA binding in the presence of ATP, indicating that this effect was related to the catalytic activity of the enzyme and not due to nonspecific buffer effects. Qualitatively similar results were obtained with the larger 252-bp probe and with a single-stranded 88-bp probe (data not shown).
ATP and MgCl2 increase the rate of dissociation.
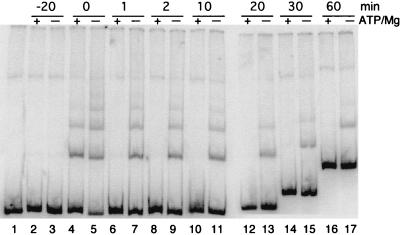
We hypothesized that the DNA binding affinity of P143 was higher in the absence of its substrate and cofactor because it was not engaged in ATP hydrolysis. We tested this idea by assembling two binding reactions, one in the presence of ATP and MgCl2 and one in the absence. The enzyme was incubated on ice for 20 min to allow binding to the radiolabeled double-stranded 252-bp probe. Then, a 100-fold-molar excess of the same unlabeled DNA fragment was added (aliquots were removed at the indicated times) and was loaded onto a gel with the current running continuously. This allowed us to compare the rate of dissociation of P143 from the DNA under both sets of conditions (Fig. 4).
FIG. 4.
The effect of ATP and MgCl2 on the dissociation rate of P143 DNA binding. Lanes 2 to 21 contain a fivefold-molar excess of P143 over a radiolabeled 252-bp dsDNA probe in the absence or presence (+ or −) of 2 mM ATP and 6 mM MgCl2 as indicated at the top. After 20 min of incubation, a 100-fold molar excess of unlabeled probe was added to each reaction mixture, and 20 μl was removed at the times indicated at the top of the gel and was loaded on a 3.5% nondenaturing polyacrylamide gel with the current running continuously (lanes 6 to 21). Lane 1 contains the probe in the absence of enzyme; lanes 2 and 3 contain probe and competitor added together; lanes 4 and 5 had no competitor added. Free probe shifts up with time because these samples were loaded later and, therefore, electrophoresed for a shorter period of time.
Before the addition of competitor DNA, four shifted bands were clearly visible in the absence of cofactors, while only two were detected in the presence of cofactors, as previously shown. By 1 min after the addition of competitor DNA, P143 had dissociated from the probe in the reaction mixture containing ATP and MgCl2. In the absence of cofactors, however, the rate of dissociation was much slower, and at least two shifted bands were present throughout the 60-min time course. Because the stoichiometry of enzyme to DNA in each band is unknown, we were unable to calculate an exact rate of dissociation. But, from this experiment it seems that the rate is in seconds in the presence of cofactors and in minutes in the absence of cofactors.
A model for DNA binding and ATPase activities of P143.
The DNA binding data presented here suggests a model in which P143 undergoes conformational changes upon binding and hydrolysis of ATP. These conformational changes alter its affinity for DNA and presumably involve translocation and/or DNA unwinding. This suggests that helicase does not unwind DNA by the rolling mechanism because a fundamental requirement of that model is that a subunit of the helicase is in contact with the DNA at all times (13, 14). The inchworm model (16) is more consistent with our findings because it proposes that helicases undergo conformational changes that advance the enzyme forward into the double-stranded region of DNA before ATP hydrolysis. In order to slide forward on the DNA, the enzyme must release the nucleotides in the DNA binding pocket. This is consistent with the finding that P143 binds DNA more efficiently in the absence of ATP and MgCl2; the enzyme is static in the absence of cofactors and therefore binds DNA with high affinity. In the presence of ATP and MgCl2, P143 goes through cycles of hydrolysis and translocation. EMSA captures only those molecules that are bound with high affinity, while those in the process of translocation temporarily dissociate from DNA and so do not produce shifted bands.
The observation that the DNA binding activity of a helicase is inhibited by ATP and MgCl2 is unusual but has also been reported for hepatitis C virus helicase NS3 (8). Those authors showed that 2 mM ATP inhibited DNA binding of the wild-type helicase but not of a mutant unable to hydrolyze ATP. They did not speculate on the possible causes of this ATP-induced decrease in binding. The fact that it was not seen in an ATPase-deficient enzyme suggests that it is not an artifact but a part of the helicase mechanism. It seems likely that the binding of ATP alters the conformation of the helicase, modifying the DNA binding affinity.
Further experiments are needed to corroborate the model of P143 helicase activity proposed here. One approach would be to mutagenize the ATP binding domain of P143. Our model would be supported by the finding that ATP no longer inhibited P143 DNA binding in the mutated enzyme, confirming that ATP binding was the cause of the inhibition. Unfortunately we have not been able to express P143 in any system other than the baculovirus system, and since P143 is essential for viral replication, we cannot express proteins with mutations that inhibit function.
Acknowledgments
We thank Wen Dong for technical assistance.
This research was supported by grant MCB-9874532 from the National Science Foundation.
REFERENCES
- 1.Evans J T, Rohrmann G F. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J Virol. 1997;71:3574–3579. doi: 10.1128/jvi.71.5.3574-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 3.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 4.Hang X, Dong W, Guarino L A. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J Virol. 1995;69:3924–3928. doi: 10.1128/jvi.69.6.3924-3928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kool M, Voeten J T, Goldbach R W, Vlak J M. Functional mapping of regions of the Autographa californica nuclear polyhedrosis viral genome required for DNA replication. Virology. 1994;198:680–689. doi: 10.1006/viro.1994.1080. [DOI] [PubMed] [Google Scholar]
- 6.Laufs S, Lu A, Arrell K, Carstens E B. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology. 1997;228:98–106. doi: 10.1006/viro.1996.8361. [DOI] [PubMed] [Google Scholar]
- 7.Liu G, Carstens E B. Site-directed mutagenesis of the AcMNPV p143 gene: effects on baculovirus DNA replication. Virology. 1999;253:125–136. doi: 10.1006/viro.1998.9485. [DOI] [PubMed] [Google Scholar]
- 8.Lohman T M, Bjornson K P. Mechanisms of helicase catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 9.Lu A, Carstens E B. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1991;181:336–347. doi: 10.1016/0042-6822(91)90500-b. [DOI] [PubMed] [Google Scholar]
- 10.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 12.McDougal V V, Guarino L A. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J Virol. 2000;74:5273–5279. doi: 10.1128/jvi.74.11.5273-5279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stasiak A, Tsaneva I R, West Benson S C, Yu X, Engelman E H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong I, Lohman T M. Allosteric effects of nucleotide cofactors on Escherichia coli rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Carstens E B. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology. 1998;247:32–40. doi: 10.1006/viro.1998.9235. [DOI] [PubMed] [Google Scholar]
- 16.Yarronton G T, Gefter M L. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc Natl Acad Sci USA. 1979;76:1658–1662. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]