Abstract
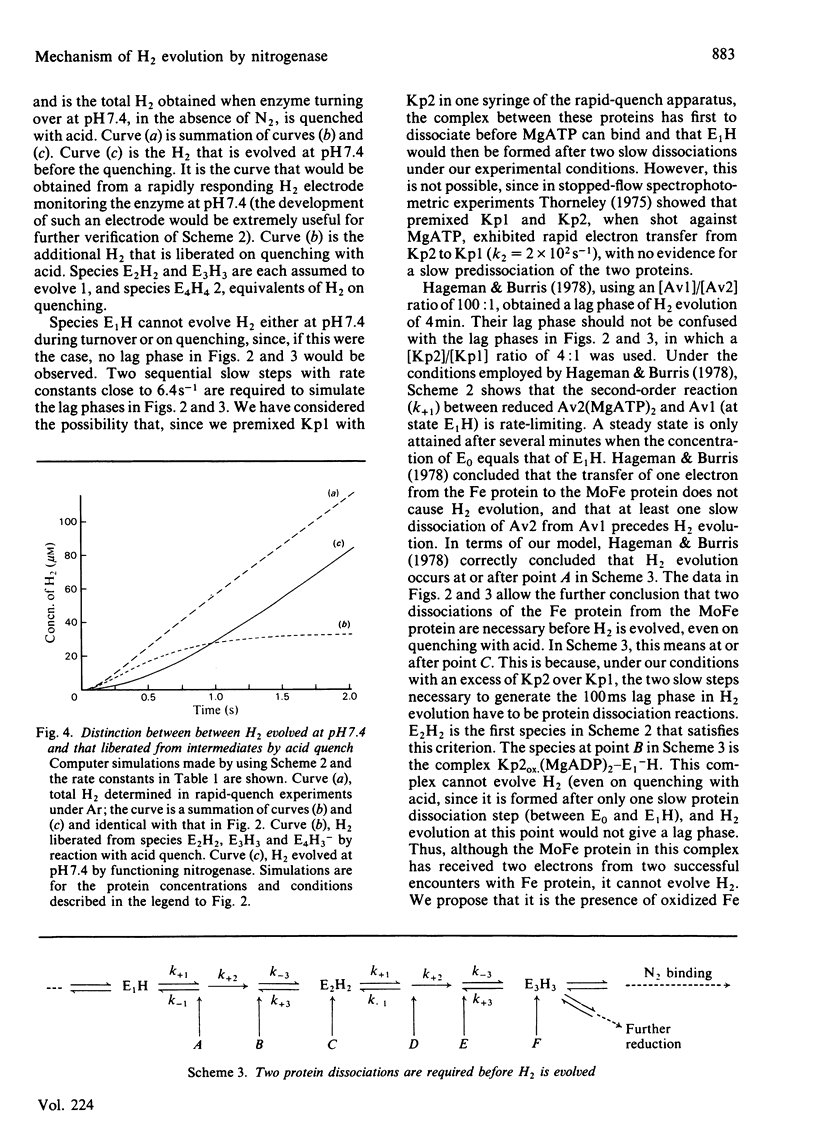
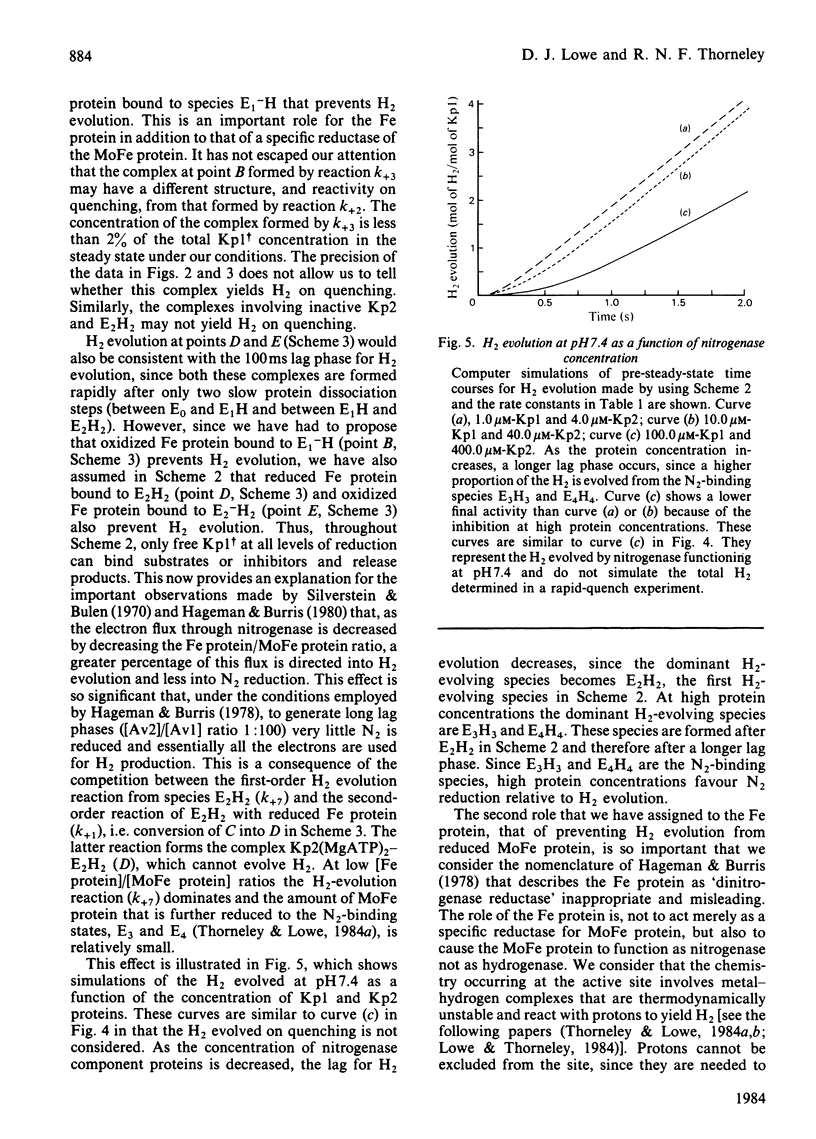
A comprehensive model for the mechanism of nitrogenase action is used to simulate pre-steady-state kinetic data for H2 evolution in the presence and in the absence of N2, obtained by using a rapid-quench technique with nitrogenase from Klebsiella pneumoniae. These simulations use independently determined rate constants that define the model in terms of the following partial reactions: component protein association and dissociation, electron transfer from Fe protein to MoFe protein coupled to the hydrolysis of MgATP, reduction of oxidized Fe protein by Na2S2O4, reversible N2 binding by H2 displacement and H2 evolution. Two rate-limiting dissociations of oxidized Fe protein from reduced MoFe protein precede H2 evolution, which occurs from the free MoFe protein. Thus Fe protein suppresses H2 evolution by binding to the MoFe protein. This is a necessary condition for efficient N2 binding to reduced MoFe protein.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burgess B. K., Wherland S., Newton W. E., Stiefel E. I. Nitrogenase reactivity: insight into the nitrogen-fixing process through hydrogen-inhibition and HD-forming reactions. Biochemistry. 1981 Sep 1;20(18):5140–5146. doi: 10.1021/bi00521a007. [DOI] [PubMed] [Google Scholar]
- Dilworth M. J., Thorneley R. N. Nitrogenase of Klebsiella pneumoniae. Hydrazine is a product of azide reduction. Biochem J. 1981 Mar 1;193(3):971–983. doi: 10.1042/bj1930971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady R. R., Lowe D. J., Thorneley R. N. Nitrogenase of Klebsiella pneumoniae: a pre-steady state burst of ATP hydrolysis is coupled to electron transfer between the component proteins. FEBS Lett. 1978 Nov 15;95(2):211–213. doi: 10.1016/0014-5793(78)80995-8. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Smith B. E., Cook K. A., Postgate J. R. Nitrogenase of Klebsiella pneumoniae. Purification and properties of the component proteins. Biochem J. 1972 Jul;128(3):655–675. doi: 10.1042/bj1280655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth J. H., Burris R. H. Inhibition of nitrogenase-catalyzed NH3 formation by H2. Biochemistry. 1983 Oct 25;22(22):5111–5122. doi: 10.1021/bi00291a010. [DOI] [PubMed] [Google Scholar]
- Gutteridge S., Tanner S. J., Bray R. C. The molybdenum centre of native xanthine oxidase. Evidence for proton transfer from substrates to the centre and for existence of an anion-binding site. Biochem J. 1978 Dec 1;175(3):869–878. doi: 10.1042/bj1750869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman R. V., Burris R. H. Electron allocation to alternative substrates of Azotobacter nitrogenase is controlled by the electron flux through dinitrogenase. Biochim Biophys Acta. 1980 Jun 10;591(1):63–75. doi: 10.1016/0005-2728(80)90220-0. [DOI] [PubMed] [Google Scholar]
- Hageman R. V., Burris R. H. Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2699–2702. doi: 10.1073/pnas.75.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman R. V., Orme-Johnson W. H., Burris R. H. Role of magnesium adenosine 5'-triphosphate in the hydrogen evolution reaction catalyzed by nitrogenase from Azotobacter vinelandii. Biochemistry. 1980 May 27;19(11):2333–2342. doi: 10.1021/bi00552a009. [DOI] [PubMed] [Google Scholar]
- Lowe D. J., Thorneley R. N. The mechanism of Klebsiella pneumoniae nitrogenase action. The determination of rate constants required for the simulation of the kinetics of N2 reduction and H2 evolution. Biochem J. 1984 Dec 15;224(3):895–901. doi: 10.1042/bj2240895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. W., Robson R. L., Yates M. G., Eady R. R. Catalysis of exchange of terminal phosphate groups of ATP and ADP by purified nitrogenase proteins. Can J Biochem. 1980 Jul;58(7):542–548. doi: 10.1139/o80-074. [DOI] [PubMed] [Google Scholar]
- Rivera-Ortiz J. M., Burris R. H. Interactions among substrates and inhibitors of nitrogenase. J Bacteriol. 1975 Aug;123(2):537–545. doi: 10.1128/jb.123.2.537-545.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein R., Bulen W. A. Kinetic studies of the nitrogense-catalyzed hydrogen volution and nitrogen reduction reactions. Biochemistry. 1970 Sep 15;9(19):3809–3815. doi: 10.1021/bi00821a021. [DOI] [PubMed] [Google Scholar]
- Thorneley R. N., Lowe D. J. Nitrogenase of Klebsiella pneumoniae. Kinetics of the dissociation of oxidized iron protein from molybdenum-iron protein: identification of the rate-limiting step for substrate reduction. Biochem J. 1983 Nov 1;215(2):393–403. doi: 10.1042/bj2150393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneley R. N., Lowe D. J. The mechanism of Klebsiella pneumoniae nitrogenase action. Pre-steady-state kinetics of an enzyme-bound intermediate in N2 reduction and of NH3 formation. Biochem J. 1984 Dec 15;224(3):887–894. doi: 10.1042/bj2240887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneley R. N., Lowe D. J. The mechanism of Klebsiella pneumoniae nitrogenase action. Simulation of the dependences of H2-evolution rate on component-protein concentration and ratio and sodium dithionite concentration. Biochem J. 1984 Dec 15;224(3):903–909. doi: 10.1042/bj2240903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneley R. N. Nitrogenase of Klebsiella pneumoniae. A stopped-flow study of magnesium-adenosine triphosphate-induce electron transfer between the compeonent proteins. Biochem J. 1975 Feb;145(2):391–396. doi: 10.1042/bj1450391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt G. D., Bulen W. A., Burns A., Hadfield K. L. Stoichiometry, ATP/2e values, and energy requirements for reactions catalyzed by nitrogenase from Azotobacter vinelandii. Biochemistry. 1975 Sep 23;14(19):4266–4272. doi: 10.1021/bi00690a019. [DOI] [PubMed] [Google Scholar]


