Abstract
The coxsackie B virus and adenovirus (Ad) receptor (CAR) functions as an attachment receptor for multiple Ad serotypes. Here we show that the Ad serotype 9 (Ad9) fiber knob binds to CAR with much reduced affinity compared to the binding by Ad5 and Ad12 fiber knobs as well as the knob of the long fiber of Ad41 (Ad41L). Substitution of Asp222 in Ad9 fiber knob with a lysine that is conserved in Ad5, Ad12, and Ad41L substantially improved Ad9 fiber knob binding to CAR, while the corresponding substitution in Ad5 (Lys442Asp) significantly reduced Ad5 binding. The presence of an aspartic acid residue in Ad9 therefore accounts, at least in part, for the reduced CAR binding affinity of the Ad9 fiber knob. Site-directed mutagenesis of CAR revealed that CAR residues Leu73 and Lys121 and/or Lys123 are critical contact residues, with Tyr80 and Tyr83 being peripherally involved in the binding interaction with the Ad5, Ad9, Ad12, and Ad41L fiber knobs. The overall affinities and the association and dissociation rate constants for wild-type CAR as well as Tyr80 and Tyr83 CAR mutants differed between the serotypes, indicating that their binding modes, although similar, are not identical.
Binding of virus particles to specific cell surface receptors is an essential step in infection and represents a key determinant of host cell tropism. Adenovirus (Ad) attachment and uptake into cells are separate but cooperative events that result from the interaction of the viral fiber and penton base proteins with specific cell surface receptors. The fiber knob domain mediates the primary event, attachment. Each human Ad serotype has its own fiber protein variant, but Ad40 and Ad41 have two versions, long and short, that can be found together in a single virus particle (5). The penton base protein mediates the second step, virus internalization (15), and is also involved in cell membrane permeabilization (14). The coxsackie B virus and Ad receptor (CAR) (2, 3, 13) binds to the fiber knob of several Ad serotypes but not to subgroup B Ad fibers such as serotypes 3 or 7 or to the short fiber of subgroup F Ad (Ad40s and Ad41s) (10, 11). The recent structure of the Ad12 knob-human CAR complex revealed residues that interact with CAR (4). Comparison of the Ad12 fiber knob sequence with the sequence of Ad5, Ad9, and Ad41L fiber knobs shows that while some contact residues identified in the structure of the Ad12-CAR complex are totally conserved, others differ among the serotypes (Fig. 1). In this study, the kinetics of binding of Ad5, Ad9, Ad12, and Ad41L fibers for CAR was assessed using surface plasmon resonance (SPR) (7, 8) to establish for the first time whether differences in binding kinetics and affinity correlate with functional differences. SPR allows the association and dissociation kinetics to be determined in real time without labeling of either component. Studying association and dissociation events is important, since affinity measurements alone may not reflect differences in the kinetics of the interactions.
FIG. 1.
Alignment of Ad12, Ad5, Ad9, and Ad41L fiber knob sequences. Residues in contact with CAR, based on the structure of the Ad12/CAR complex (4), are shown in bold. The CAR residues mutated in this study are in contact with the following residues in the Ad12 fiber knob: Leu73 in CAR interacts with Pro418; Tyr83 interacts with Leu426; Lys121 as well as Lys123 interact with Asp415 (4); Tyr80 interacts with the region around Leu426 in the AB loop of the Ad12 fiber knob (4). Lys451 and Gln487 in Ad12 are highlighted (*), and the corresponding residues in Ad9 that had been mutated in this study are underlined.
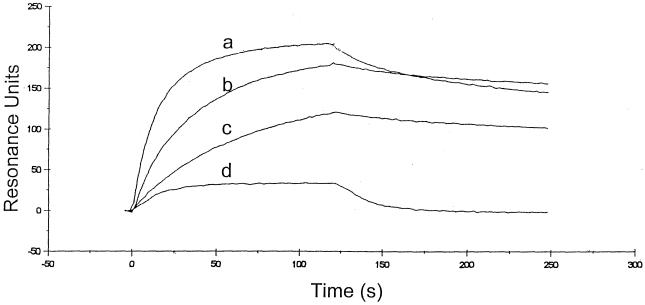
We expressed histidine-tagged wild-type fiber knobs of Ad3, Ad5, Ad9, Ad12, and Ad41L in bacteria and purified each fiber knob by nickel affinity chromatography and size exclusion. Each protein accumulated as a stable trimer as assessed by native gel electrophoresis, size exclusion, and boiled and nonboiled sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The kinetics of binding of wild-type Ad fibers to immobilized CAR was determined by SPR on a BIAcore biosensor, as previously described (7, 8). The association (ka) and dissociation (kd) rate constants for a monophasic model of binding were obtained using the BIAevaluation analysis package (version 2.1). The ability of this model to describe the experimental data was determined by examination of the residual plots, which were calculated by subtracting the experimental data points from the fitted curve. Residuals were small and randomly distributed around zero (range, 0.5 to −0.5). Nonspecific binding measured on a blank surface (<1%) was subtracted from specific binding prior to kinetic analysis. We found that the Ad3 fiber knob did not bind to immobilized CAR at any concentration (20 nM to 5 μM), which is in keeping with previous findings (9, 11). Ad5 and Ad41L fiber knobs bound to CAR with similar overall affinity but with different association and dissociation rate constants (Fig. 2; Table 1). The Ad12 fiber knob showed slightly reduced affinity for CAR, due to a lower association rate constant (ka). Our most striking observation, however, was that the Ad9 fiber knob bound to CAR with markedly lower affinity, due to altered association and dissociation rate constants (Fig. 2; Table 1).
FIG. 2.
SPR analysis of wild-type Ad41L (a), Ad5 (b), Ad12 (c), and Ad9 (d) fiber knob binding to immobilized CAR. The fiber knob of each Ad serotype was injected over the sensor surface at different concentrations (200 nM, 400 nM, 40 μM, and 100 μM). A 2-min association phase was followed by a 2-min dissociation phase with HEPES-buffered saline flowing over the sensor surface at 25, 30, 35, 40, and 50 μl/min. Representative sensograms for each wild-type Ad fiber knob protein used as analyte are shown.
TABLE 1.
Summary of the kinetic data for the interaction of Ad5, Ad9, Ad12, and Ad41L fiber knob proteins with wild-type sCARa
| Serotype | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| Ad5 | 1.27 × 105 ± 4.4 × 103 | 1.01 × 10−3 ± 1.2 × 10−4 | 7.9 × 10−9 ± 1.8 × 10−9 |
| Ad9 | 1.61 × 103 ± 2.1 × 102 | 4.10 × 10−2 ± 1.2 × 10−4 | 6.4 × 10−6 ± 6.0 × 10−5 |
| Ad12 | 7.31 × 104 ± 5.6 × 103 | 1.1 × 10−3 ± 6.0 × 10−4 | 1.5 × 10−8 ± 6.1 × 10−9 |
| Ad41L | 2.60 × 105 ± 3.0 × 104 | 1.9 × 10−3 ± 2.3 × 10−4 | 7.3 × 10−9 ± 2.6 × 10−9 |
SPR was used to obtain the data. Sensorgrams were analyzed using a monophasic model of interaction. Results are expressed as the mean ± standard deviation for six separate determinations.
Recent studies have shown that Ad9, as well as a chimeric virus composed of the Ad5 capsid with its fiber knob replaced by the Ad9 fiber knob, bound to CAR-expressing cells with lower efficiency than did wild-type Ad5 (12). The fact that the Ad9 fiber knob binds to CAR with markedly reduced affinity provides, at least in part, an explanation for these observations. Our results also explain the earlier finding that the Ad9 fiber knob was less efficient than the Ad5, Ad12, and Ad41L fiber knobs in inhibiting Ad5 infection (9). It has also been shown that Ad41 binds to differentiated enterocytes more efficiently than Ad5 does (6) and that Ad41 but not Ad5 binding to A549 cells was dependent on contact time (1), implying that Ad5 and Ad41 attachment to cells is not identical. These differences in infectivity between Ad5 and Ad41 cannot be explained by kinetic differences between Ad5 and Ad41L fiber knobs for CAR, as is the case with Ad9, since the Ad41L fiber knob bound to CAR with similar affinities to Ad5 fiber knob (Table 1). Since only the long fiber of Ad41 binds to CAR (9), we speculate that the proportion of long and short fibers in the Ad41 viral capsid may influence Ad41 infectivity.
Although some of the contact residues identified in the structure of the Ad12-CAR complex are totally conserved among the four serotypes, others differ among them (Fig. 1). In particular, Lys451 in Ad12 is conserved in Ad5 (Lys442) and Ad41L (Lys429) but is an aspartic residue in Ad9 (Asp222) (Fig. 1). Moreover, the region adjacent to Asp222 in Ad9 is highly conserved among Ad5, Ad9, Ad12, and Ad41L fiber knob domains. This, along with the conservation of the adjacent glycine (Gly223 in Ad9), suggests that the local conformation in this region is identical among all four serotypes. A nonconservative substitution such as this may therefore account for the observed difference in the kinetics for Ad9. Another charge difference in Ad9 relative to the other serotypes occurs at another peripheral residue (Gln487 in the Ad12 fiber knob; Lys260 in the Ad9 fiber knob) (Fig. 1), which may also contribute to the observed difference in kinetics. This hypothesis was evaluated by mutating Asp222 and Lys260 in the Ad9 fiber knob and Lys442 in the Ad5 fiber knobs. We generated mutant Ad9 fiber knob domains Asp222Lys (substitution of Asp222 with the corresponding lysine in Ad5), Lys260Pro (substitution of Lys260 with the corresponding proline in Ad5), and Lys260Gln (substitution of Lys262 with the corresponding glutamine in Ad12). We also generated Ad5 fiber knob mutant Lys442Asp (substitution of Lys442 with the corresponding aspartic acid in Ad9). Each mutant fiber knob was expressed as histidine-tagged protein in bacteria as described above. The proteins were purified by gel filtration and size exclusion chromatography. All mutant fiber knobs accumulated as soluble trimers, and their kinetics of binding to soluble CAR was assessed by SPR.
The Asp222Lys Ad9 fiber knob bound to CAR with much increased affinity (KD = 3.1 × 10−7 M versus 6.4 × 10−6 M for the wild-type Ad9 fiber knob) (Table 1). The Ad5 fiber knob with the corresponding substitution (Lys442Asp) bound to CAR with much reduced affinity compared to the wild-type Ad5 fiber knob (KD = 2.8 × 10−6 M versus 7.9 × 10−9 M) (Table 2). These findings demonstrate that the presence of this aspartic acid residue in Ad9, instead of the conserved lysine in the Ad5, Ad12, and Ad41L fiber knobs, accounts, at least in part, for the reduced CAR binding affinity of Ad9. Our findings also suggest that a lysine at this position must function similarly in Ad5 and the mutated Ad9 fiber knob domains. In contrast, Lys260Pro and Lys260Glu bound to CAR with Ad9 wild-type affinity (data not shown). Sequence alignment shows a lack of sequence similarity in the immediate region of four residues around Lys260 (Fig. 1), and this would indicate that the modes of binding of the Ad5, Ad9, Ad12, and Ad41L fiber knobs to CAR are likely to be distinct in this particular region. This may explain why substitution of Lys260 in Ad9 with the corresponding residues in Ad5 and Ad12 failed to alter the Ad9 fiber knob binding kinetics to resemble that of Ad5 and Ad12 fiber knobs.
TABLE 2.
Summary of the kinetic data for the interaction of the Ad5 mutant fiber knob (Lys442Asp) and the Ad9 mutant fiber knob (Asp222Lys) with CAR, and comparison to wild-type Ad5 and Ad9 fiber knob bindinga
| Fiber knob | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| Ad5 Lys442Asp | 2.01 × 104 ± 6.4 × 103 | 5.7 × 10−2 ± 2.8 × 10−2 | 2.8 × 10−6 ± 7.4 × 10−6 |
| Ad5 Wild type | 1.27 × 105 ± 4.4 × 103 | 1.01 × 10−3 ± 1.2 × 10−4 | 7.9 × 10−9 ± 1.8 × 10−9 |
| Ad9 Asp222Lys | 2.6 × 103 ± 5.5 × 102 | 7.9 × 10−4 ± 3.4 × 10−3 | 3.1 × 10−7 ± 1.3 × 10−7 |
| Ad9 Wild type | 1.61 × 103 ± 2.1 × 102 | 4.10 × 10−2 ± 1.2 × 10−1 | 6.4 × 10−6 ± 6.0 × 10−5 |
SPR was used to obtain the data. Sensorgrams were analyzed using a monophasic model of interaction. Results are expressed as the mean ± standard deviation for six separate determinations.
To assess further the binding interaction of Ad5, Ad9, Ad12, and Ad41L with CAR, we constructed a number of CAR mutants, based on the structure of the Ad12-CAR complex (4). We mutated Leu73, Tyr80, Tyr83, Lys121, and Lys123 to create mutant CAR proteins Leu73Lys, Tyr80Ala, Tyr83Lys, and Lys121Ala/Lys123Ala, respectively (residue numbers are based on CAR sequence accession number P78310 and are referred to as residues Leu54, Tyr61, Tyr64, Lys102, and Lys104 by Bewley et al. [4]). According to the structure of the complex, Tyr80 and Tyr83 lie at the periphery of the binding site whereas Leu73, Lys121, and Lys123 constitute part of the major binding interface (4). Each CAR mutant was expressed as a FLAG-tagged recombinant protein in insect cells and was then purified by FLAG affinity chromatography and size exclusion. Leu73Lys, Tyr80Ala, Tyr83Lys, and Lys121Ala/Lys123Ala CAR mutants were coupled to separate CM5 sensor chips using the amine coupling reaction as specified by the manufacturer (BIACORE). Immobilization densities of 600-800 RU were used in all experiments. The Ad5, Ad9, Ad12, and Ad41L fiber knobs failed to bind to immobilized Leu73Lys and the double mutant Lys121Ala/Lys123Ala at any of the concentrations used (20 nM to 100 μM). These findings confirm that CAR residues Leu73 and Lys121 and/or Lys123 are contact residues for the CAR-Ad12 fiber knob interaction (4) and demonstrate for the first time that they are also critical for CAR binding to the Ad5, Ad9, and Ad41L fiber knobs. Although the binding of Ad5, Ad12, and Ad41L fiber knobs to CAR mutants Tyr80Ala and Tyr83Lys was reduced, the overall affinities and the association and dissociation rate constants differed among the serotypes (Tables 3 and 4). Tyr80Ala and Tyr83Lys CAR mutants recognized the Ad12 fiber knob with significantly lower affinity than they recognized the Ad5 and Ad41L fiber knobs, mainly due to an effect on the association rate constant (Tables 3 and 4). This would suggest that these two CAR residues are more critically involved in the Ad12-CAR binding interaction than in the binding interaction of Ad5 and Ad41L with CAR. Moreover, the Ad5 and Ad41L fiber knobs bound to Tyr80Ala and Tyr83Lys with wild-type association kinetics but increased dissociation kinetics (Tables 3 and 4), further indicating that each serotype interacts differently with this CAR region. These data show that the binding interface between CAR and different Ad serotypes, although similar, is not identical, a conclusion also supported by our previous studies. For example, Gln494, Pro519, and Asn520 in the Ad12 fiber knob are all contact residues (4), but the corresponding residues in Ad5 fibre knob (Asn482, Ser507 and His508) are not involved in the binding interaction since their substitution resulted in Ad5 fiber knob proteins that bound to CAR with wild-type affinity (7).
TABLE 3.
Summary of the kinetic data for the interaction of Ad5, Ad9, Ad12, and Ad41L Fiber Knob proteins with mutant CAR Tyr80Alaa
| Serotype | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| Ad5 | 1.20 × 105 ± 5.7 × 104 | 4.44 × 10−3 ± 2.0 × 10−2 | 3.8 × 10−8 ± 2.2 × 10−9 |
| Ad9 | NBb | NB | NB |
| Ad12 | 4.94 × 104 ± 2.3 × 103 | 6.80 × 10−3 ± 3.2 × 10−4 | 1.4 × 10−7 ± 5.4 × 10−8 |
| Ad41L | 3.16 × 105 ± 7.8 × 104 | 4.9 × 10−3 ± 1.9 × 10−4 | 1.6 × 10−8 ± 4.0 × 10−9 |
SPR was used to obtain the data. Sensorgrams were analyzed using a monophasic model of interaction. Results are expressed as the mean ± standard deviation for six separate determinations.
NB, no specific binding.
TABLE 4.
Summary of the kinetic data for the interaction of Ad5, Ad9, Ad12, and Ad41L fiber knob proteins mutant CAR Tyr83Lysa
| Serotype Fiber Knob | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| Ad5 | 1.42 × 105 ± 5.7 × 103 | 4.10 × 10−3 ± 2.1 × 10−4 | 2.9 × 10−8 ± 2.8 × 10−9 |
| Ad9 | NBb | NB | NB |
| Ad12 | 4.2 × 104 ± 5.5 × 103 | 6.80 × 10−3 ± 4.4 × 10−4 | 1.7 × 10−7 ± 2.9 × 10−8 |
| Ad41L | 4.10 × 105 ± 1.6 × 104 | 5.52 × 10−3 ± 1.5 × 10−4 | 1.4 × 10−8 ± 3.8 × 10−9 |
SPR was used to obtain the data. Sensorgrams were analyzed using a monophasic model of interaction. Results are expressed as the mean ± standard deviation for six separate determinations.
NB, no specific binding.
The Asp222Lys Ad9 fiber knob bound to Tyr80Ala and Tyr83Lys with fourfold-lower affinity (KD = 1.1 × 10−6 M for both interactions versus 3.1 × 10−7 M for wild-type CAR). However, the Ad9 fiber knob failed to bind to these mutants at any of the concentrations used. Since the wild-type affinity of the Ad9 fiber knob for CAR is low, a similar decrease in Ad9 binding to Tyr80Ala and Tyr83Ala may have reduced the kinetics of their interaction below the threshold of this assay.
Taken together, our findings demonstrate important structural differences in the interaction of the Ad5, Ad9, Ad12, and Ad41L fiber knob domains with CAR and indicate that the binding interaction between various Ad serotypes and CAR, although similar, is not identical. Our findings also demonstrate that knowledge not only of receptor specificity but also of the binding kinetics and affinity of Ad fibers with their corresponding cell receptor may contribute to our understanding of Ad tropism. The existence of distinct internalization receptors would suggest that a similar understanding of the protein-protein interactions involved in this process will also be required to determine fully the contribution of attachment and internalization steps to the life cycle of different Ad serotypes.
Acknowledgments
This work was funded by grants from the Wellcome Trust and the Charitable Foundation of Guy's and St. Thomas' Hospitals to G. Santis. B. J. Sutton also thanks the Wellcome Trust for its support.
REFERENCES
- 1.Albinsson B, Kidd A H. Adenovirus type 41 lacks an RGD α-integrin binding motif on the penton base and undergoes delayed uptake in A459 cells. Virus Res. 1999;64:125–136. doi: 10.1016/s0168-1702(99)00087-8. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Krithivas A, Celi L, Droguett G, Horwitz M S, Wickham T, Crowell R L, Finberg R W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bewley M C, Springer K, Zhang Y-B, Freimuth P, Flanagan J M. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor CAR. Science. 1999;286:1579–1586. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 5.Chroboczek J, Ruigrok R W, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 6.Croyle M A, Stone M, Amidon G L, Roessler B J. In vitro and in vivo assesment of adenovirus 41 as a vector for gene delivery to the intestine. Gene Ther. 1998;5:645–654. doi: 10.1038/sj.gt.3300645. [DOI] [PubMed] [Google Scholar]
- 7.Kirby I, Davison E, Beavil A J, Soh C P C, Wickham T J, Roelvink P W, Kovesdi I, Sutton B J, Santis G. Identification of contact residues and definition of the CAR binding site of adenovirus type 5 fiber protein. J Virol. 2000;74:2804–2813. doi: 10.1128/jvi.74.6.2804-2813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirby I, Davison E, Beavil A J, Soh C P C, Wickham T J, Roelvink P W, Kovesdi I, Sutton B J, Santis G. Mutations in the DG loop of adenovirus 5 fiber protein abolish high affinity binding to its cellular receptor CAR. J Virol. 1999;73:9508–9514. doi: 10.1128/jvi.73.11.9508-9514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roelvink P, Kovesdi I, Wickham T. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilise the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackie-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santis G, Legrand V, Hong S, Davison E, Kirby I, Chartier C, Pavirani A, Bergelson J, Mehtali M, Boulanger P. Molecular determinants and serotype specificity of Ad5 fiber binding to its high affinity receptor CAR. J Gen Virol. 1999;80:1519–1527. doi: 10.1099/0022-1317-80-6-1519. [DOI] [PubMed] [Google Scholar]
- 12.Shayakhmetov D Y, Lieber A. Dependence of adenovirus infectivity on length of fiber shaft domain. J Virol. 2000;74:10274–10286. doi: 10.1128/jvi.74.22.10274-10286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomko R, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackie viruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin ανβ5 selectively promotes adenovirus cell membrane permeability. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]