Abstract
The nematode Caenorhabditis elegans is a powerful experimental setting for uncovering fundamental tenets of nervous system organization and function. Its nearly invariant and simple anatomy, coupled with a plethora of methodologies for interrogating single-gene functions at single-cell resolution in vivo, have led to exciting discoveries in glial cell biology and mechanisms of glia–neuron interactions. Findings over the last two decades reinforce the idea that insights from C. elegans can inform our understanding of glial operating principles in other species. Here, we summarize the current state-of-the-art, and describe mechanistic insights that have emerged from a concerted effort to understand C. elegans glia. The remarkable acceleration in the pace of discovery in recent years paints a portrait of striking molecular complexity, exquisite specificity, and functional heterogeneity among glia. Glial cells affect nearly every aspect of nervous system development and function, from generating neurons, to promoting neurite formation, to animal behavior, and to whole-animal traits, including longevity. We discuss emerging questions where C. elegans is poised to fill critical knowledge gaps in our understanding of glia biology.
The nematode Caenorhabditis elegans is a powerful experimental setting in which to uncover biological principles in molecular detail (Brenner 1974; Goldstein 2016). An extensive genetic toolkit coupled with optical transparency, enabling facile in vivo microscopy and optogenetics, has allowed the nervous system of this animal to be probed at unprecedented single-gene and single-synapse resolution. C. elegans uses its structurally invariant neural network to perform complex and flexible behaviors, including sensory preference choice, locomotion, sleep, mating, and decision-making, and to store information of different qualities and on different timescales (White et al. 1986; Bargmann 1993; Sengupta and Samuel 2009; Bargmann and Marder 2013; Emmons 2018; Schafer 2018; Cook et al. 2019; Goodman and Sengupta 2019; Witvliet et al. 2021).
C. elegans glia are molecularly and anatomically diverse (Cao et al. 2017; Singhvi and Shaham 2019; Purice et al. 2023), arising primarily from ectodermal precursors, and associating with sense organs and with the brain neuropil, the nerve ring. One glial class is mesodermally derived and also abuts the nerve ring (White et al. 1986). Gene expression and functional studies of C. elegans glia have revealed extensive similarities to vertebrate glia (Shaham 2005; Heiman and Shaham 2007; Katz et al. 2019; Singhvi and Shaham 2019; Purice et al. 2023).
C. elegans glia, however, differ from vertebrate glia in one important aspect: while vertebrate glia provide neurons with trophic support, C. elegans glia do not (Shaham 2005; Wagner et al. 2006; Barres 2008; Singhvi and Shaham 2019; Chiareli et al. 2021). Trophic support in vertebrates may serve to ensure precision in neural cell number and circuit formation during development (Barres 2008; Clarke and Barres 2013; Farhy-Tselnicker and Allen 2018). In C. elegans, however, development and circuit formation are stereotyped, pre-determined by cell lineage (Sulston and Horvitz 1977; Sulston et al. 1980, 1983; White et al. 1986; Varshney et al. 2011; Jarrell et al. 2012; Doroquez et al. 2014). Further, neuronal cell bodies are not known to be metabolically privileged by a restrictive blood–brain barrier. Thus, glial support of neuron survival may be unnecessary (Shaham 2015; Singhvi et al. 2016; Singhvi and Shaham 2019; Rapti 2021). C. elegans, therefore, provides a unique in vivo arena for perturbing glial cell functions without the concern that associated neuron survival is affected (Singhvi and Shaham 2019; Rapti 2021).
GENERAL PROPERTIES OF C. elegans GLIA
Glial Cell Types
Like C. elegans neurons, whose numbers are invariant between individuals (302/387 in hermaphrodites/males), C. elegans glia numbers and developmental origins are constant, with 50 sex-shared glia, 36 male-specific neuroectoderm-derived glia, and six sex-shared, mesoderm-derived glial cells (Fig. 1; Table 1; Sulston and Horvitz 1977; Sulston et al. 1980, 1983; White et al. 1986). Whole-animal, single-cell transcriptome profiling (RNA-seq) initially suggested that even the handful of glia interrogated in those studies were not molecularly identical (Cao et al. 2017; Packer etal. 2019; Fung etal. 2020; Tayloret al. 2021). This has now been conclusively demonstrated in a complete molecular atlas of C. elegans glia across the male and hermaphrodite nervous systems (Purice et al. 2023). Single-nuclear RNA-seq transcriptome profiling, coupled with custom computational/machine-learning analytics and in vivo validation studies, show that glia are molecularly heterogeneous, with even anatomically similar glia having distinct molecular signatures.
Figure 1.
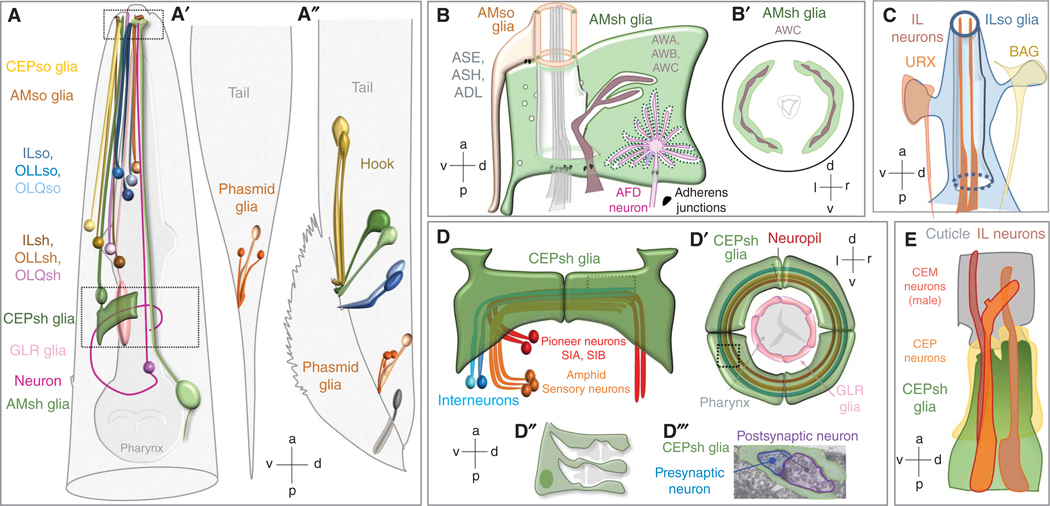
Subtypes and anatomy of Caenorhabditis elegans glia. (A) A schematic representation of each glia type in the head, (A′) hermaphrodite tail, (A′′) and male tail. Anterior and posterior deirid glia (ADEsh/so, PDEsh/so) are not depicted. Glia–neuron associations are magnified in B–E as follows: amphid glia (B), IL socket glia (C ), cephalic sensilla (E), and cephalic posterior membrane sheaths enveloping the brain neuropil (D). (B–B′): (B) Amphid sensilla schematic showing AMso–AMsh sense organ glia forming a channel lumen associating with dendrite tips, neuron-receptive endings (NREs) that traverse the glial channel (ASE, ASH, ADL), and embedded NREs (AFD and AWA/B/C neurons). (B′) Cross section of bilateral AMsh glia–AWC pairs, (C ) ILso glia interact with NREs of different neurons (URX, IL, BAG) at distinct contact sites, with only IL neurons traversing a channel made by the glia. (D) Schematic of CEPsh glial processes (green) ensheathing different axon commissures. (D′) A schematic cross-section view of the brain neuropil shows the relative location of axon commissures with CEPsh and GLR glia. (D′′) Glial processes also infiltrate between neuron processes in the neuropil. (D′′′) Electron micrograph showing the CEPsh glia–ALA neuron–AVE neuron tripartite synapse. (E) Schematic of the cephalic sensilla of a male animal, noting relative localization and glia–neuron contacts of the sex-shared CEP neuron and male-specific CEM neuron. Sensory NREs in B, C, and E are depicted without dendrites and cell bodies for simplicity. Neurons in D are depicted without dendrites for simplicity. (Schematics in A, B, and D are reprinted from Singhvi and Shaham 2019 with permission from the author. Electron microscope [EM] image in D′′′ is adapted with permission from White et al. 1986. Schematic in C is based on data in White et al. 1986, Ward et al. 1975, and Cebul et al. 2020. Panel E is based on data in Wang et al. 2015 and Sulston et al. 1980.)
Table 1.
Caenorhabditis elegans glia, associated neurons, and behaviors affected by respective glia–neuron interactions
| Organ | Glia | Associated neuron(s) | Number of sensilla | Documented functions of glia-neuron interactions | Animal behavior regulated by glia-neuron interactions |
|---|---|---|---|---|---|
| Sensilla glia | |||||
| Amphid | AMso, AMsh | Contact AMsh only: AFD, AWA, AWB, AWC AMsh-AMso Channel neurons: ASE, ASG, ASH, ASI, ASK, ASJ, ADF, ADL |
2 (L/R) | AMsh: NRE shape and plasticity, glial compartment, dendrite outgrowth, ion regulation, engulfment, interaction with epithelia AMso: adult neurogenesis |
Olfaction, gustation, tactile sensation, thermosensation |
| Phasmid | PHsh, PHsol, PHso2 | PHA, PHB, PHC Contact PHso only: PQR | 2 (L/R) | PHso: adult neurogenesis | n.d. |
| Anterior deirid | ADEsh, ADEso | ADE | 2 (L/R) | n.d. | n.d. |
| Posterior deirid | PDEsh, PDEso | PDE | 2 (L/R) | n.d. | n.d. |
| Inner labial | ILsh,ILso | IL1, IL2 Contact ILso only: BAG, URX, FLP | 6 (L/R, D/V/L) | IL/OL nerve outgrowth, neuropil placement | Mechanosensation |
| Outer labial | OL(Q/L)sh, OL(Q/L)so | OLQ or OLL | 6 (L/R, D/V/L) | n.d. | Mechanosensation |
| Cephalic | CEPsh, CEPso | CEP (±CEM in males) | 4 (D/V, L/R) | CEPso: aECM pore formation | n.d. |
| Hook | HOsh, HOso | HOA, HOB | 1 | n.d. | Male mating |
| Ray | R(N)st | R(N)A, R(N)B | 18 (L/R), (N) = 1–9 | n.d. | Male mating |
| Postcloacal | PCsh, PCso | PCA PCsh only: PCB, PCC | 2 (L/R) | n.d. | Male mating |
| Spicules | SPsh syncytium (two cells), SPso syncytium (four cells) | SPD, SPV | 2 (L/R) | NRE development. SPso: extrasynaptic source of dopamine | Male mating |
| Neuropil glia | |||||
| Neuropil | CEPsh | Many neuropil axons | 4 (D/V, L/R) | Neuropil assembly, AIY synapse positioning, neurotransmitter clearance | Locomotion during sleep, repetitive behavior, swimming-induced paralysis |
| Mesodermal-lineage glia | |||||
| Mesodermal | GLR | RME, others (?) | 6 (D/V/L, L/R) | RME axon specification | Locomotion |
Reproduced from Singhvi and Shaham 2019 with permission from the author.
Glia associate with every level of information transfer in C. elegans circuits (Ward et al. 1975; White et al. 1986). Ten bilateral sheath-glia pairs (ADEsh, AMsh, ILsh, ILshD/V, OLLsh, OLQshV/D, PDEsh, PHsh) and 13 bilateral socket glia pairs (ADEso, AMso, CEPsoD/V, ILso, ILsoD/V, OLLso, OLQsoD/V, PDEso, PHso1/2) fasciculate with neuron dendrites, and enwrap their sensory tips, forming environment-accessible compartments (Fig. 1A). Like epithelia, socket glia also secrete cuticles (Chisholm and Hsiao 2012). Four CEPsh glia and six mesoderm-derived GLR glia associate with both dendrites and axons. Anterior CEPsh glia processes fasciculate with dendrites and form compartments around sensory tips of CEP and male-specific CEM neurons. Distal GLR glia processes fasciculate with IL1 neuron dendritic bundles without ensheathing their endings. CEPsh glia posterior processes surround the outer aspect of the nerve ring (brain neuropil) and penetrate it to contact synapses. GLR glia proximal processes expand sheet-like structures that surround the inner aspect of the nerve ring. Here, GLR glia form gap junctions with GABAergic RME neurons and muscles. GLR glia sheets may create a seal between the nerve ring and the pseudocoelom, perhaps resembling a blood–brain barrier.
Thirty six male-specific neuroectodermal glia—seven sheath, 11 socket, and 18 sheath–socket hybrid structural cells (HOsh/HOso, bilateral pairs of PCsh, PCso, R1–9, SPso1–4, SPshD/V)—associate with the cloaca and with sensory rays and hook sensillum copulation structures (Sulston et al. 1980; Emmons 2005; Cook et al. 2019).
Sexual Dimorphism
Several C. elegans glial cells exhibit structural sexual dimorphism. The four CEPsh and CEPso glia form a compartment around dendritic endings of CEP and CEM neurons in males, but only around CEP neurons in hermaphrodites, as CEMs die during development (Sulston et al. 1983; White et al. 1986). CEPso glial cells control the formation of this pore through a male-specific transcriptional switch visualized by secretion of a Hedgehog-related protein, to differentially pattern the apical extracellular matrix (ECM) (Fung et al. 2023). In the tail phasmid sense organ, the phasmid channel is built by PHso1 glia in hermaphrodites, while PHso2 serves this function in males (Hall 1977; Sulston et al. 1980). Finally, AMso glia and PHso1 glia are neurogenic only in males, as described in detail below.
Molecular profiling of glia from sexually mature day 1 adults using shRNA-seq also reveals sex dimorphism (wormglia.org). Specifically, profiles of functionally analogous hermaphrodite PHso1 and male PHso2 socket glia are different (Purice et al. 2023; wormglia.org). Furthermore, some anatomically sex-shared glia like PHsh, OLsh, and OLso glia also exhibit divergent identities. In contrast, there is no discernable molecular sex dimorphism in either CEPsh, CEPso, or AMso glia that persists into the adult stage after dimorphic development is complete.
Other Heterogeneity
Despite physical association and similar contributions to forming sensory organ channels, sheath and socket glia, exhibit both morphological diversity (White et al. 1986) and distinct molecular signatures (Purice et al. 2023). Sheath glia of different sensory organs resemble each other more than lineally or anatomically related socket glia, implying functional convergence during development. Within each class (sheath/socket), glia are also different across sense organs (Bacaj et al. 2008; Katz et al. 2019; Fung et al. 2020; Purice et al. 2023). These differences likely reflect the identities of neurons with which they associate. For example, ILso and AMsh glia promote dendrite elongation of associated neurons through retrograde extension, but using distinct physical structures and molecular mechanisms (Cebul et al. 2020). Molecular heterogeneity between and within glial cell types is also evident (Oikonomou and Shaham 2011; Mizeracka and Heiman 2015; Singhvi and Shaham 2019). For example, ventral and dorsal CEPsh glia develop and specify their gene expression through molecularly distinguishable pathways (see below) (Yoshimura et al. 2008).
Glial Membrane Subdomains
Each C. elegans glial cell associates with a defined number of neurons, whose identities and contact sites are invariant, allowing for studies of glia–neuron interactions at the resolution of single contact sites. Often, the glial membranes apposing a given neuron are enriched for specific proteins. For example, the apical membranes of a single AMsh glia can be divided into at least three molecularly distinct domains contacting different sensory neurons (Ray et al. 2024). Membranes associated with AFD neurons accumulate the KCC-3 K/Cl transporter (Singhvi et al. 2016; Rayet al. 2024), domains associated with amphid channel neurons accumulate DAF-6/Patched-related, VAP-1/secreted protein, and LIT-1/Nemo-like kinase (Perens and Shaham 2005; Oikonomou et al. 2011), and those around the AWC neuron are devoid of these proteins (Fig. 1B). Cilia of non-AFD neurons dictate KCC-3 localization to a microdomain around AFD, revealing cross talk across these molecularly distinct glia–neuron contacts (Rayet al. 2024). Likewise, URX and BAG neuron dendrite endings contact ILso glia at distinct sites, suggesting molecularly distinct membrane contact regions (Fig. 1C; Cebul et al. 2020). Such separation of function is revealed in the extreme for CEPsh glia; CEPsh anterior processes wrap around dendritic endings of CEP sensory neurons (also of CEM in males), while posterior ramifications envelop the ∼180 neurite processes of the nerve-ring neuropil and penetrate the neuropil to contact synaptic sites and engage in tripartite synapses (Fig. 1D,E; White et al. 1986; Doroquez et al. 2014; Katz et al. 2019).
Similarities to Vertebrate Glia
CEPsh glia development suggests homology with vertebrate radial glia and astrocytes. Embryonically, CEPsh glia guide midline-crossing of nerve-ring axons using Netrin, just as radial glia pial branches direct axon guidance in the vertebrate spinal cord (Dominici et al. 2017; Rapti et al. 2017; Varadarajan et al. 2017). Vertebrate radial glia eventually transform into astrocytes (Schmechel and Rakic 1979; Noctor et al. 2008), and CEPsh glia undergo similar remodeling (Rapti et al. 2017). In both settings, glia abut synapses (White et al. 1986) and direct synapto-genesis (Christopherson et al. 2005; Colón-Ramos et al. 2007; Eroglu et al. 2009; Allen et al. 2012; Shao et al. 2013). Furthermore, astrocytes and CEPsh glia cover nonoverlapping neural domains, respecting unknown tiling rules (White et al. 1986; Bushong et al. 2002). Gene expression profiles reveal that CEPsh glia are more similar to mouse astrocytes than to any other murine brain cell (Katz et al. 2019). Finally, astrocytes exhibit Ca2+transients, and gap junctions allow Ca2+ flow between astrocytes (Shigetomi et al. 2016; Khakh 2019; Nagai et al. 2021). CEPsh glia exhibit similar responses (M Katz and S Shaham, unpubl. data). Finally, multiple glia express gap junction proteins like mammalian glia (Cuadras et al. 1985; White et al. 1986; Nedergaard 1994), whose functions are not yet explored (Altun et al. 2015).
Sensory organ socket and sheath glia are related in function and molecules to vertebrate sensory organ glia and to astrocytes. For example, similar to glia-like retinal pigment epithelium cells in the eye and astrocytes, AMsh glia regulate contacting neurons by pruning their endings, regulating their ionic milieu, and deploying thrombospondin-domain proteins (Bacaj et al. 2008; Singhvi et al. 2016; Allen and Eroglu 2017; Raiders et al. 2021a,b; Ray and Singhvi 2021). Like olfactory epithelium sustentacular cells, they also express xenobiotic metabolism gene batteries (Wallace et al. 2021). Lastly, sensory cues that activate neurons ensheathed by AMsh glia promote changes in AMsh glia intracellular Ca2+ concentration, similar to glia in other animals, suggesting that this property may be a conserved glial feature (Rousse and Robitaille 2006; Han et al. 2013; Shigetomi et al. 2016; Duan et al. 2020; Fernandez-Abascal et al. 2022).
GLR glia, like vertebrate microglia, arise mesodermally. Like microglia, GLRs express GABAergic signaling effectors and regulators and may also engulf dying cell debris (Nass et al. 2002; Gendrel et al. 2016; Wilton et al. 2019; Favuzzi et al. 2021).
SPECIFICATION OF C. elegans GLIA
Pan-glia Cell-Fate Specification
Most C. elegans glia are born embryonically and diversify using transcription factors expressed either after or before progenitor cell division. The Zn-finger transcription factor LIN-26 is expressed in neuroectoderm-derived glia using enhancer elements embedded within the lin-26 genomic locus (Landmann et al. 2004). LIN-26 loss promotes glial cell degeneration and/or adoption of nonglial fates (Ferguson et al. 1987; Labouesse et al. 1996). Although there is no obvious sequence similarity, LIN-26 functions similarly to Drosophila glial-cells-missing (Gcm), a Zn-finger protein required for glia specification (Hosoya et al. 1995; Jones et al. 1995; Vincent et al. 1996). In both lin-26 and gcm mutants, cells slated to become glia can become transformed into neurons. More direct molecular conservation is evident with the C. elegans transcription factor PROS-1, a homolog of Prospero/Prox1 that specifies Drosophila and vertebrate glia, respectively (Bunk et al. 2016; Peco et al. 2016). PROS-1 is expressed in a large subset of glia to regulate the expression of many glial genes, and defects in pros-1 animals are rescued by the expression of human Prox1 (Kage-Nakadai et al. 2016; Wallace et al. 2016).
While they are expressed in most, if not all glia, both embryonic LIN-26 and postembryonic MIR-228, a microRNA expressed in most glia, are also present in nonglial cells. Indeed, RNA-seq studies suggest that there are no panglial genes expressed exclusively in glia (Purice et al. 2023).
Specification of Glial Subtypes
AMsh Glia
AMsh glial fate specification requires a cascade of transcription factors. Earlyon, AMsh glia express and require the gliogenic transcription factor LIN-26/Zn finger and the UNC-130/FKHD repressor (Labouesse et al. 1996; Mizeracka et al. 2021). UNC-130 specifies the identities of AMsh and other glia and epidermal cells (but not neurons) that arise from a discrete developmental lineage. The roles of UNC-130 in AMsh glia can also be executed by human FOXD3, a neural crest glial lineage regulator, suggesting possibly conserved programs of peripheral glia fate specification (Kastriti and Adameyko 2017; Dawes and Kelsh 2021; Mizeracka et al. 2021).
To ensure that only two AMsh glia are generated, this fate is suppressed in other lineages. Conserved Atoh/NeuroD family proneural bHLH genes restrict AMsh glial fate expression. Loss of lin-32/Atonal/Atoh results in misexpression of AMsh markers in CEPsh glia, and possibly other cells, leading to supernumerary AMsh glia. LIN-32 acts in parallel to CND-1/NeuroG1 and NGN-1/NeuroD1 (Zhang et al. 2020c) to prevent ectopic expression of AMsh glia fate. LIN-32 also regulates AMso glia specification, although its roles here are unknown (Zhang et al. 2020c). Finally, once AMsh glial fate is specified, PROS-1/Prox1/Prospero acts to maintain expression of the AMsh glia secretome until the adult stage (Kage-Nakadai et al. 2016; Wallace et al. 2016).
AMso/PHso
AMso and PHso glia express ALR-1, the C. elegans ortholog of Aristaless, a Paired homeo-domain transcription factor that drives neural fate and sense organ specification in flies and mammals (Meijlink et al. 1999). ALR-1 regulates glial cell shape and adhesion to overlying epithelia (Tucker et al. 2005). AMso, PHso, and other glial cells express hedgehog-related genes like GRL-12, although their roles in glial development await inquiry (Melkman and Sengupta 2005; Hao et al. 2006).
CEPsh Glia
CEPsh glia express the transcription factor HLH-17 (McMiller and Johnson 2005; Yoshimura et al. 2008), the C. elegans protein most similar to vertebrate Olig2, which is expressed in precursors of oligodendrocytes and motor neurons, and in subsets of astrocytes (Masahira et al. 2006; Tatsumi et al. 2018). VAB-3/Pax6/7 cell-autonomously controls HLH-17 expression in CEPsh glia. With the Nkx/Hmx-related protein MLS-2, VAB-3 activates HLH-17/Olig2 in ventral CEPsh glia; but does so independently of MLS-2 in dorsal CEPsh glia (Yoshimura et al. 2008). This gene-expression pathway is reminiscent of specification events in the mouse spinal cord, where Olig2 expression depends on Nkx6 in ventral regions and Pax6/Pax7 in ventral/dorsal regions (Rowitch 2004; Miller 2005). HLH-17 expression in dorsal CEPsh glia is also affected by the loss of LIN-32/Atoh1 (see above) (Zhang et al. 2020c).
ILso Glia
Like CEPsh glia, the six ILso glia arise from distinct lineages. UNC-130/FoxD3, acts as a repressor, promoting generation of the two dorsal, but not ventral or lateral, ILso glia (Mizeracka et al. 2021). UNC-130, acting through its DNA-binding domain, functions in progenitor and newly born terminal cells to also specify other cells related by lineage to ILsoD glia. These studies reveal that different transcriptional programs operate in different ILso developmental lineages to specify similar cell fates.
Ray Structural Glia
Ray neuroblast progenitors give rise to RnB neurons and to their sister glia—the ray structural cells. This asymmetric division requires asymmetric Wnt and POP-1/LEF-β-catenin signaling to specify anterior–posterior neuron–glia fates. Loss of the Wnt receptor LIN-17/Frizzled transforms posterior glial cell daughters into anterior neurons through the aberrant expression of LIN-32/Atoh in these cells (Sulston et al. 1980; Portman and Emmons 2000; Emmons 2005; Miller and Portman 2011). Thus, LIN-32/Atoh1/Atonal can have context-specific pro- or antigliogenic fate specification roles (see above). Finally, VAB-3/Pax6/7 regulates fate specification here through interactions between a subset of developing Rnst and their contacting neurons and epithelia (Zhang and Emmons 1995), highlighting context-specific gene functions.
GLR Glia
The six GLR glia derive from the MS mesodermal lineage (Sulston et al. 1983; White et al. 1986). GLRs express the C. elegans myoD homolog HLH-1; type IV collagen, like muscles; and mesoderm-lineage-enriched genes like DIG-1 and EMB-9 (Krause et al. 1994; Graham et al. 1997), and contain GABA and the GABA transporter SNF-10 (Gendrel et al. 2016). Recent transcriptome studies suggest that GLR glia merge astrocytic and endothelial characteristics relegated to separate cell types in vertebrates (Stefanakis et al. 2024). Combined fate acquisition is orchestrated by LET-381/FoxF, a fate specification/maintenance transcription factor expressed in glia and endothelia of other animals. Among LET-381/FoxF targets, UNC-30/Pitx2 transcription factor controls GLR glia morphology and represses an alternative mesodermal fate of the HMC cell, a GABA-containing, muscle-related cell. LET-381 and UNC-30 coexpression in naive cells is sufficient for GLR glia gene expression.
MORPHOGENESIS AND CELL BIOLOGY OF C. elegans
GLIA Glial Cell Polarity
C. elegans neuroectodermal glia are lineally related to epithelia and neurons, which are both polarized. Sheath glia make tight junctions with socket glia and with associated neurons (Ward et al. 1975; Perkins et al. 1986). For AMsh glia, whose apical domain faces neuron contact sites, apical–basolateral domain segregation can be visualized by expression of subcellular domain markers, including ERM-1/ERM and PIP2 (Low et al. 2019; Martin et al. 2024; Ray et al. 2024). Like vertebrate ZP proteins, DYF-7/ZP is also an apical ECM component of these glia (Fig. 2A; Low et al. 2019). Similar organization likely characterizes all sense organ–ensheathing processes of C. elegans (Lee et al. 2021b).
Figure 2.
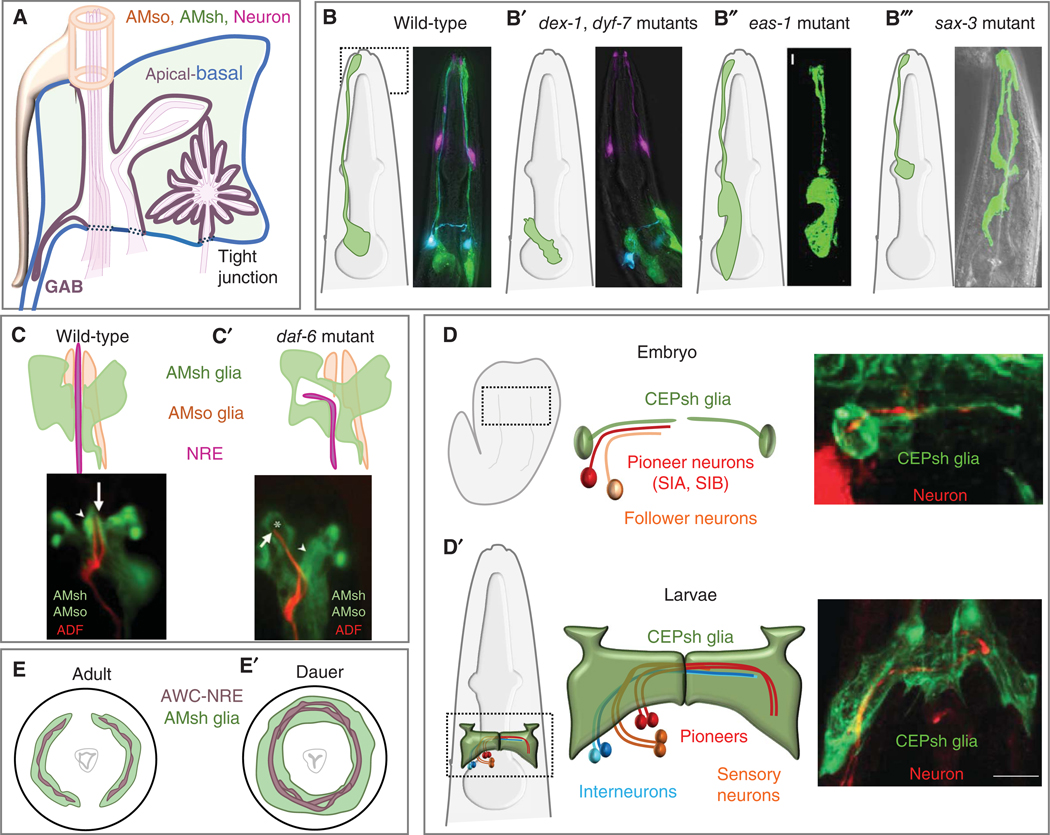
Glial cell development and morphogenesis. (A) Schematic depicting apical–basal polarity of AMsh and AMso glia at the amphid channel lumen. (B–B′′′) Diagram and images of AMsh glia in wild-type (B) and different mutant backgrounds (B′–B′′′). AMsh glia anterior processes are collapsed in dyf-7 and dex-1 mutant (B′), glial cell size is enlarged in eas-1 mutant (B′′) and glial cell body migration is disrupted in sax-3 mutant (B′′′) animals.(C–C′) Diagram and images of AMsh glia in wild-type (C ) and daf-6 (C′) mutant animals showing aberrant sensory compartment lumen in mutants, which impedes ADF-NRE from accessing the outside environment. (D–D′) Diagram and image of CEPsh and pioneer/follower neurons growing into the neuropil for brain assembly in the embryo (D) and larvae (D′). Scale bar: 10 μm. Micrograph shows CEPsh glia process guiding pioneer axon processes. (E, E′) Schematic of AMsh glia and AWC neuron remodeling in non-dauer (E) and post-dauer animals (E′). (AMsh) Amphid sheath glia, (AMso) amphid socket glia, (CEPsh) cephalic sheath glia, (NRE) neuronal receptive ending. (Fluorescence images as follows: B,B′ reprinted from Heiman and Shaham 2009 with permission; B′′ from Zhang et al. 2020b, reprinted under the terms of the Creative Commons CC BY 4.0 License; C,C′′ from Qu et al. 2020, reprinted under the terms of the Creative Commons Attribution License; D,D′ from Rapti et al. 2017, reprinted with permission from the author. Schematics from Singhvi and Shaham 2019, reprinted with permission from the author.)
Glial Cell Size Control
The size of AMsh glia cells is regulated by the conserved cis-Golgi membrane protein EAS-1/GOLT1B, the loss of which causes enlarged AMsh glial cell bodies (Fig. 2B; Zhang et al. 2020b). EAS-1/GOLT1B, through the E3 ubiquitin-ligase gene rnf-145/RNF145, promotes nuclear activation of sbp-1/SREBP, a sterol and fatty-acid synthesis regulator. Long-chain polyunsaturated fatty acids may be relevant products of this pathway, although how they affect cell size is not understood.
Glial Cell Morphogenesis
AMsh Glia Process Extension
Adult amphid sensory dendrites and glial processes extend ∼100 μm toward the nose tip (Ward et al. 1975; White et al. 1986). Elongation of these processes occurs in embryogenesis through a mechanism termed retrograde extension (Sulston et al. 1983; Heiman and Shaham 2009; Lamkin and Heiman 2017). Newly born amphid sensory neurons and AMsh glia extend short projections, anchored at the nose tip with an ECM composed of the zona-pellucida (ZP) domain protein DYF-7, secreted by the neurons, and DEX-1, a zonadhesin domain-containing protein secreted by nonneuronal neighboring cells (Fig. 2B′; Heiman and Shaham 2009; Oikonomou and Shaham 2011). DEX-1 and DYF-7 resemble α- and β-tectorins, proteins comprising the tectorial membrane that anchors hair cell cilia in the inner ear (Legan et al. 1997). Subsequent migration of neuron and glia cell bodies extends dendrites and glial processes, respectively. dyf-7 or dex-1 mutations result in short dendrites and AMsh glia processes. AMso glia, which connect anteriorly to AMsh glia, are generally unaffected; however, abnormal AMso glia posterior processes attached to AMsh glia are occasionally observed, suggesting that AMsh–AMso adhesion is independent of DYF-7 and DEX-1 (Heiman and Shaham 2009).
Genes controlling glial cell body migration, and presumably relevant for retrograde extension, are known. Lesions in the VAB-3/Pax-6 transcription factor block posterior migration of several anterior glia and neuron cell bodies (Yoshimura et al. 2008). A cleaved, secreted form of SAX-3/Robo can interact with SYG-1/Neph, expressed in AMsh glia, to promote glia migration (Qu et al. 2020). Likewise, dietary vitamin B12, acting through PTP-3/LAR PRTP and NID-1/Nidogen on glia, controls glial migration (Zhang et al. 2020a).
AMsh Glia Process Tip Morphogenesis
Eight amphid neurons extend ciliated dendrites passing through a matrix-filled channel open to the environment (Ward et al. 1975; Perkins et al. 1986). AMso and AMsh glial membranes comprise the anterior and posterior channel sections, respectively, and are joined by tight junctions to form a continuous tube (Fig. 1B; Martin et al. 2024). This compartment resembles synaptic compartments surrounded by astrocyte end-feet (Shaham 2010). The anterior region of AMsh glia in this compartment is decorated by a web of apical βH-spectrin (Martin et al. 2024). Glial compartment formation occurs in the embryo before neuronal cilia enter it (Oikonomou and Shaham 2011). Subsequent morphogenesis requires DAF-6/Patched-related and the secreted DAF-6-binding protein DYF-4, both of which restrict amphid channel expansion (Fig. 2C–C′; Perens and Shaham 2005; Oikonomou et al. 2011; Hong et al. 2021). Bloated channels in daf-6 or dyf-4 mutants can be rescued by mutations in the lit-1, snx-1, or igdb-2 genes (Oikonomou and Shaham 2011; Oikonomou et al. 2012; Wang et al. 2017; Hong et al. 2021), suggesting that these genes antagonize DAF-6 and DYF-4 functions and promote channel growth. LIT-1/NEMO-like kinase, SNX-1, a retromer component, and IGBD-2, an Ig/FNIII protein, act in parallel. IGDB-2 functions in AMso glia and its loss can be partially compensated for by the loss of LGC-34, a predicted ligand-gated ion channel (Wang et al. 2017), indicating that extracellular-ion levels and/or glia:glia coordination influence amphid channel dimensions. daf-6, lit-1, and snx-1 also interact genetically with che-14/Dispatched-related, required for apical secretion from AMsh glia (Michaux et al. 2000; Perens and Shaham 2005; Oikonomou and Shaham 2011; Oikonomou et al. 2012). All identified channel size regulators function in AMsh/so glia; nonetheless, sensory-neuron cilia defects affect DAF-6 localization (Perens and Shaham 2005), suggesting cross talk between these cell types (Martin et al. 2024).
CEPsh Glia Morphogenesis
CEPsh glia are born in the embryo (Sulston et al. 1983) where, in addition to extending a process associated with CEP dendrites, they also extend nonbranching processes that mark the location of the presumptive nerve ring and that promote nerve-ring assembly. Later, CEPsh glia processes become highly ramified, extending membrane sheets that envelop the nerve ring, and finer processes that penetrate it and abut synapses (Fig. 2D–D′; White et al. 1986; Rapti et al. 2017; Katz et al. 2019). The CEPsh glial cell morphology does not appear disrupted after ablation of nerve-ring pioneer neurons SIA and SIB, suggesting that these neurons may be dispensable for the development of CEPsh glia membrane sheaths.
Glia–Epithelia Interactions in Glial Shape Maintenance
C. elegans glia are mechanically coupled to overlying epithelia, Briefly, epithelial UNC-23/ BAG2, an Hsp cochaperone, maintains epithelial cell shape against mechanical stress (Martin et al. 2024). Loss of unc-23, through misregulated HSP-1-DNJ-13 chaperone cycling, induces progressive age-dependent deformation in the epithelia cell shape of adult animals. This, through FGFR signaling, disrupts the glial apical cytoskeleton protein SMA-1/β-spectrin and F-actin, resulting in the loss of shape of AMsh glial apical domains that contact neuron endings. This leads to the consequent loss of neuron-ending shapes and function (Martin et al. 2024). This epithelia–AMsh glia coupling only occurs at the L4-adult critical developmental window (Martin et al. 2024). Further, this coupling only affects glia in the anterior head region, like AMsh and CEPsh glia (Martin et al. 2024), but tail PHsh glia or phasmid neurons are unaffected (Martin et al. 2024). Thus, epithelia–glia mechanical coupling is regulated with exquisite spatial and temporal specificity (Martin et al. 2024). UNC-23 also affects tissue viscoelasticity and integrity of the Perlecan ECM, in relation to temperature and mechanical stress, to affect the architecture of CEPsh glia sheaths in an age-progressive manner (Rahmani et al. 2015; Coraggio et al. 2023; Martin et al. 2024).
Similarly, loss of the ECM protein DIG-1, secreted by muscle and epithelia to regulate basement membrane architecture, induces fragmentation of AMsh glia anterior endings and abnormal cell position, shape, and fasciculation of neurons associated with ILso glia, and other neuronal processes (Bénard et al. 2006; Chong et al. 2021). Loss of either UNC-23 or DIG-1 results in progressive defects in adult animals, indicating lifelong roles for epithelia in glia-shape maintenance and neural aging (Chong et al. 2021; Martin et al. 2024).
Morphology of postembryonic CEPsh glia posterior processes is also maintained by CIMA-1/SLC17A5 transporter, which controls EGL-15/FGFR levels in epithelia, and MIG-17/ADAMTS protease, which influences basement membrane composition (Fig. 3A; Shao et al. 2013; Fan et al. 2020).
Figure 3.
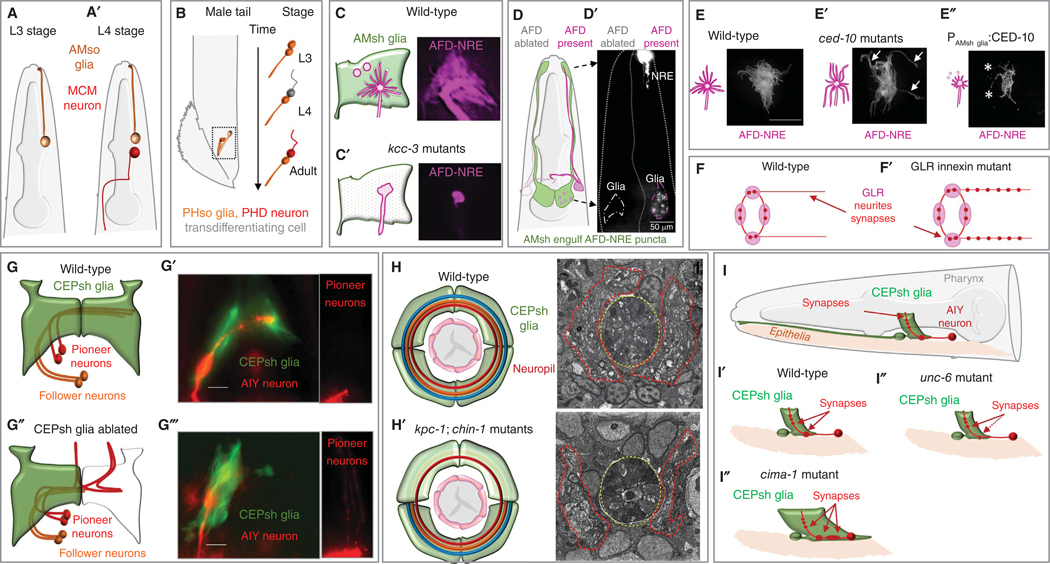
Caenorhabditis elegans glial cells in neuronal generation and morphogenesis. (A–A′) Schematic of AMso glia in males in L3 (A) and L4 (A′) larval stages showing its cell division to generate the MCM neuron. (B) Schematic of the male PHso glia transdifferentiating into a PHD neuron during developmental L3–L4-adult transition stages. (C–C′) Diagram and image of AFD-NRE in wild-type animals with intact AMsh glial ensheathment (C), and in kcc-3 mutant animals (C′). (D–D′) Schematic (D) and image (D′) of bilateral AMsh glia–AFD. AFD-NRE staining (top arrow) is also seen as punctate fragments in the AMsh glia cell body (bottom arrow) on the side with AFD neurons present and lost in the AMsh glial cell body on the side with AFD neuron ablated. (E–E′′) Pruning by AMsh glia regulates AFD-NRE shape. Reduced pruning (ced-10 mutants) causes elongated AFD-NRE, and excess pruning (overexpress CED-10 in AMsh) causes shorter AFD-NRE. (F–F′) RME synapses localized to a specific region of the neurite in wild-type (F) are misrouted along the neurite processes in GLR innexin mutant animals (F′). (G–G′′′) Diagram and image of CEPsh glial posterior membrane sheaths directing pioneer and follower neuronal axons (G, G′), and their misdirection in glia-ablated-animals (G′–G′′′). (H–H′) Diagram and image of glial cell and axon processes in the brain neuropil of wild-type animals (H), which become truncated or misguided in kpc-1; chin-1 double mutant animals with abnormal trafficking in CEPsh glial cells, leading to reduced brain neuropil size. (I–I′′) Schematic of epithelia and AIY neuron synapses within CEPsh glia posterior process zone. Densities of AIY synapses apposing specific CEPsh glia posterior membrane sheath process regions in wild-type animals (I, I′) are decreased in animals defective for glia-secreted UNC-6/Netrin (I′) and ectopically positioned in cima-1 animals with aberrant CEPsh posterior sheath processes. (Panel C is reprinted, with permission, from Singhvi et al. 2016; D, E reprinted from Raiders et al. 2021a under the terms of the Creative Commons Attribution License; G,H reprinted from Rapti et al. 2017 with permission from the author. Schematics adapted from Singhvi and Shaham 2019 with permission from the author.)
Finally, RAM-5/ZP domain protein and DPY-18/prolyl-4-hydroxylase subunit are expressed in the male tail-specific Rnst glia and epithelia, respectively, and may act in a glycosylation-dependent pathway to mediate epithelia–glial interactions and Rnst cell shape (Yu et al. 2000).
Recurrent epithelia–glia juxtaposition across species raises the possibility that analogous signals may broadly maintain glial cell shape and polarity (Salzer 2003; Derouiche et al. 2012).
Glia Remodeling in Dauer Animals
Upon stress, C. elegans larvae enter a developmental state called dauer through steroid hormone signaling. In dauers, the bilateral AMsh glia expand, fuse, and exchange cytoplasm (Fig. 2E–E′; Cassada and Russell 1975; Albert and Riddle 1983; Procko et al. 2011). AMsh glia expansion occurs concomitantly with the expansion of AWC neuron dendritic endings ensheathed by these glia. Behavior studies reveal that remodeling facilitates dauer exit upon exposure to favorable conditions (Lee et al. 2021a). Dauer-induced glial fusion is driven by AMsh glia-expressed REMO-1/G-protein-coupled receptor; transcription factors TTX-1/Otx and ZTF-16/Ikaros; fusogen AFF-1; and the stress-responsive factor VER-1/RTK (Procko et al. 2011, 2012; Lee et al. 2021a). REMO-1 localizes to AMsh glia anterior tips and, with TTX-1 and ZTF-16, is required for the expression of VER-1/RTK (Procko et al. 2011, 2012; Lee et al. 2021a). While glial REMO-1 impacts the remodeling of AWC neurons, loss of AWC neurons does not alter glia remodeling, suggesting that glial cues dictate downstream neuron remodeling events (Procko et al. 2011, 2012; Lee et al. 2021a). Nonetheless, some dauer-neuron remodeling events are independent of the REMO-1 glial pathway (Lee et al. 2021a).
GLIAL REGULATION OF NEURON GENERATION AND DEGENERATION
Glia as Neuronal Progenitors
The embryonic and postembryonic lineages of C. elegans elucidated four decades ago, report that glia are terminally differentiated cells (Sulston and Horvitz 1977; Sulston et al. 1983). Recent studies, however, reveal that some glial cell divisions were missed. Indeed, male AMso glia are a postembryonic source of neurons (Fig. 3A). During male sexual maturation, each AMso divides asymmetrically in a budding-like division to generate an AMso cell and a male-specific MCM neuron that expresses neuronal gene batteries, forms brain synapses, and regulates male-specific behaviors (Sammut et al. 2015). Remarkably, the AMso daughter retains glial markers, cilia-ensheathing projections, and polarized morphology of the precursor cell through the division, reminiscent of vertebrate radial glia divisions. Concomitantly, during sexual maturation, two other (trans)differentiation events occur in the male. The male glial cell PHso2 transforms to make the pore for tail sensory sensilla, formed by socket glia PHso1 in hermaphrodites, while male PHso1 socket glia trans-differentiate to generate sensory PHD neurons, which contribute to male copulation circuits (Fig. 3B; Sulston et al. 1980; Molina-García et al. 2020). This glia-to-neuron fate change generally happens without cell division and is molecularly distinct from Y-to-PDA differentiation, a different trans-differentiation event (Jarriault et al. 2008; Rashid et al. 2022).
Glial Roles in Neurodegeneration
Mutations in the gene swip-10 result in swimming-induced paralysis, mediated by hyperexcitability of dopaminergic CEP neurons (Hardaway et al. 2015; Gibson et al. 2018). SWIP-10, a metallo-β-lactamase-domain protein, is expressed in CEP-ensheathing CEPsh glia, and other glia. Its action may be indirect and mediated through its effects on glutamate signaling. Of note, swip-10 loss promotes age-dependent dopaminergic-neuron degeneration, suggesting that glia play important roles in maintaining cell numbers by preventing neurodegeneration.
GLIAL CONTROL OF NEURON MORPHOLOGY
Dendrite Outgrowth
Studies of retrograde extension in amphid development (see above) suggest important roles for glia in sensory-neuron dendrite extension (Fig. 2B–B′). AMsh glia ablation during embryogenesis results in short dendrites, resembling those of dyf-7 or dex-1 mutants (Singhal and Shaham 2017). Similarly, CEPsh glia ablation results in CEP neuron dendrite extension defects (Yoshimura et al. 2008). The URX and BAG sensory dendrites also form through retrograde extension (Chong et al. 2021). Unlike the amphid, dendritic tips here contact a protrusion of ILso glia to form a dendritic anchor independent of DYF-7/DEX-1. Instead, SAX-7/L1CAM, acting in neurons and ILso glia, and GRDN-1/Girdin/CCDC88C, acting in ILso glia, anchor BAG and URX neurite tips as the neuron cell bodies migrate (Lamkin and Heiman 2017).
Neuron-Receptive-Ending (NRE) Shape
Sensory NREs are specialized subcellular domains housing sensory-transduction machinery and are proposed as analogs of postsynaptic dendritic spines (Shaham 2010). C. elegans sense-organ glia regulate NRE shape, sensory-neuron function, or both; resulting in behavioral deficits (Singhvi and Shaham 2019; Ray and Singhvi 2021; Martin et al. 2024; Ray et al. 2024). Glial regulation of NRE shape has been predominantly investigated for AMsh glia, which associate with sensory NREs of 12 neurons and regulate the shape of many (Bacaj et al. 2008). Molecular studies reveal that asingle AMsh glial cell uses distinct mechanisms to regulate each neuron.
The glial-secretome regulator PROS-1/Prox regulates the shapes of multiple amphid NREs (Wallace et al. 2016). In contrast, the glial K/Cl cotransporter KCC-3 regulates only AFD neuron-receptive-endings shape (Fig. 3C–C′; Singhvi et al. 2016; Yoshida et al. 2016). KCC-3 controls chloride levels in the glia–neuron intercellular milieu. Chloride inhibits the AFD neuron receptor-guanylyl-cyclase GCY-8. GCY-8-dependent control of neuronal cGMP signaling, in turn, regulates NRE shape through the actin-polymerization factor WASP-1/nWSP (Singhvi et al. 2016). kcc-3 mutants exhibit impaired AFD-dependent thermosensory behavior.
AMsh glia also regulate NRE shape by phagocytosis of AFD neuron NRE fragments (Fig. 3D–E′′; Raiders et al. 2021a). Briefly, AMsh glia dynamically tune pruning rates based on neuron activity. This pruning requires molecular cues regulating exposure of phosphatidylserine on neurons similar to apoptotic cells and recognized by glia using apoptotic cell engulfment components (Raiders et al. 2021a). Similar pruning is documented at Drosophila and vertebrate synapses, supporting the notion that synapses and sensory endings are functionally related (Shaham 2010; Wilton et al. 2019; Hilu-Dadia and Kurant 2020; Raiders et al. 2021b).
AMsh glia also uptake NRE-derived extracellular vesicles (Ohkura and Bürglin 2011; Razzauti and Laurent 2021). This may impact NRE shape and function for some neurons, but the physiological significance remains to be determined.
Finally, maintenance of NRE shape also requires appropriate glial cell cytoskeleton maintenance downstream from epithelia–glia signaling and epithelial UNC-23/BAG2 Hsp cochaperone activity (see glia-shape section above) (Martin et al. 2024).
Neurite Specification
The mesodermal-lineage GLR glia regulate the specification of RME motoneuron neurites. Through gap junctions with RME neurons, GLR glia regulate calcium concentration, a CDK-5 pathway, and microtubule polarity to control the placement of synaptic proteins (Fig. 3F–F′; Meng et al. 2016). Glia–neuron gap junctions are also reported in other species including humans, suggesting that these mechanistic insights may be broadly relevant (Cuadras et al. 1985; White et al. 1986; Nedergaard 1994).
Axon Guidance and Brain Assembly
In the embryo, CEPsh glia extend processes along the dorsoventral axis, demarcating the presumptive nerve ring. These processes guide sublateral commissure pioneer neurons (primarily SIA and SIB) into the nerve ring, and a combination of CEPsh glia and pioneer neuron signals directs follower neuron entry (Fig. 3G–H′; Rapti et al. 2017). CEPsh glia ablation, mutations inactivating axon-guidance factors released from these cells or blocking trafficking of these cues in Chimaerin/Furin double mutants disrupts pioneer axon pathfinding and brain assembly (Rapti et al. 2017). CEPsh glia communicate with pioneers via UNC-6/Netrin and with follower axons using MAB-20/Semaphorin and FMI-1/CELSR (Rapti et al. 2017). Since the loss of MLS-2/Nkx/Hmx and VAB-3/Pax6/7 affects CEPsh fate specification and nerve-ring axons guidance defects (Yoshimura et al. 2008), it is possible that these transcription factors regulate the expression of guidance genes in CEPsh glia.
GLR glia, which line the inner surface of the nerve ring, are required for maintaining the nerve-ring position between the anterior and posterior pharyngeal bulbs. Embryonic ablation of GLR-glia precursor cells causes postembryonic anterior displacement and defasciculated of the nerve ring (Shah et al. 2017).
Synapse Formation and Maintenance
Like astrocytes, which mediate synapses formation and maturation (Ullian et al. 2001; Christopherson et al. 2005; Allen et al. 2012; Chung et al. 2013, 2016), CEPsh glia may also control synaptic placement. UNC-6/Netrin expressed in these glia appears to promote the enrichment of its receptor, UNC-40/DCC, in presynaptic regions of AIY neurons, as well as process guidance of its postsynaptic partner RIA (Fig. 3I–I′′; Colón-Ramos et al. 2007). Whether CEPsh glia affect synaptogenesis directly or by fine-tuning AIY/RIA process guidance remains unclear, since their processes in the neuropil contact axons of these neurons but not their synapses directly (White et al. 1986; Witvliet et al. 2021).
CEPsh glia may also affect synapse maintenance (Fig. 3I′′). Aberrant elongation of CEPsh glia upon disruption of epithelial CIMA-1/SLC17A5, EGL-15/FGFR, and MIG-17/ADAMTS, promotes ectopic presynaptic sites in AIY axons (Shao et al. 2013). These sites are adjacent to mispositioned postsynaptic RIA axons, correlate with ectopic axon–glia contacts, and are independent of Netrin (Shao et al. 2013; Fan et al. 2020). Whether these synapses result from new synapse assembly or abnormal density due to altered axon morphology remains unclear. Post-embryonic maintenance of axon shape and AIY synaptic sites also requires maintenance of CEPsh glia sheath integrity by UNC-23/BAG2 (Shao et al. 2013; Coraggio et al. 2023). By contrast, CDC-42 GTPase and its effector PAS-7/IQGAP act downstream from glia, within the neuron, to promote ectopic sites, without regulating glia morphology (Dong et al. 2020).
Ray Structural Glia in Ray Sensilla Morphogenesis
The male Rnst structural glial cells express RAM-5, a transmembrane protein that acts with epidermal MAB-7/SNED during tissue remodeling that leads to ray neuron morphogenesis (Yu et al. 2000).
GLIA IN NEURAL CIRCUIT FUNCTION AND ANIMAL BEHAVIOR
Glia as Sensory Cells
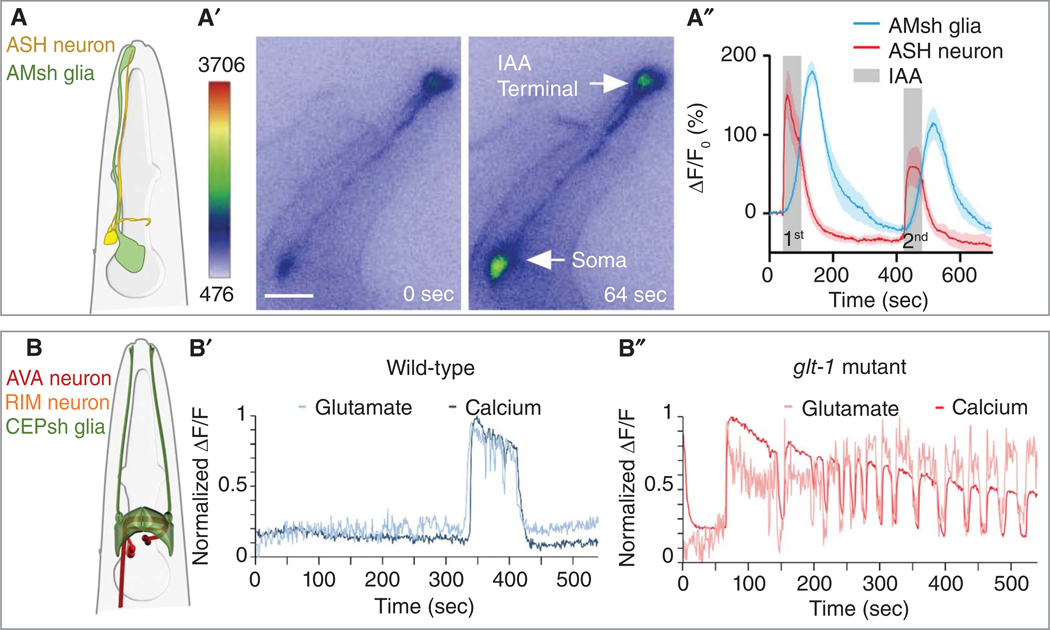
AMsh glia can detect sensory cues independently of associated neurons. They exhibit Ca2+ transients upon isoamyl alcohol or octanol exposure using glia-specific G-protein-coupled receptors, which triggers olfactory adaptation by glia-driven GABA signaling to sensory neurons (Duan et al. 2020). AMsh glia Ca2+ transients upon tactile nose-touch stimulation also modulates behavioral adaptation through chloride channels CLH-1 and GABA signaling (Fig. 4A; Ding et al. 2015; Fernandez-Abascal et al. 2022). Glial Ca2+ transients require EGL-19/L-type Ca2+ channel α1 subunit activity, and loss of egl-19 causes defects in olfactory adaptation (Chen et al. 2022). They also respond to environmental stress by inducing expression of VER-1/VEGFR to drive glia-neuron structural remodeling (reviewed above) (Procko et al. 2011; Duan et al. 2020; Fernandez-Abascal et al. 2022).
Figure 4.
Caenorhabditis elegans glial functions in animal behavior. (A–A′′) Schematic (A), representative micrograph (A′) (scale bar, 20 μm), and Ca2+ transient quantification (A′′) in AMsh glia and ASH neuron upon two pulses of isoamylalcohol (IAA) stimulation. (B–B′′) Schematic (B) depicting the region where CEPsh posterior processes contact AVA and RIM neurons. Representative traces of spontaneous glutamate (light, measured by iGluSnFR) and calcium (darkline, measured by GCaMP) dynamics near the AVA neuron in wild-type (B′) and glt-1 mutant (B′′) animals. (Images in A are reprinted with permission from Duan et al. 2020. Images in B are reprinted from Katz et al. 2019 under a Creative Commons Attribution 4.0 International License.)
Glial Regulation of Sensory-Neuron Activity
AMsh Glia
Besides modulating NRE shape to effect neuron functional changes, AMsh glia also use signaling pathways to influence neuron activity. AMsh glia FIG-1/thrombospondin domain protein is required for octanol detection and amphid neuron dye filling (Bacaj et al. 2008; Wallace et al. 2016). Astrocyte thrombospondins regulate vertebrate synapse assembly and function (Christopherson et al. 2005), perhaps suggesting similar activities. The Na+-selective AMsh glia DEG/EnaC subunit ACD-1 regulates AWC chemosensory neuron responses to specific odors (Wang et al. 2008, 2012), relying on glial acidification by the CLH-1/ClC chloride channel (Grant et al. 2015). CLH-1 also promotes glia-dependent GABA activation and cAMP signaling in ASH neurons (Fig. 4B; Grant et al. 2015; Park et al. 2021; Fernandez-Abascal et al. 2022). Human Clc2, expressed in glia, also regulates ion homeostasis and GABA signaling (Sík et al. 2000; Depienne et al. 2013).
OLQso/ILso Glia
These glia express DEG/EnaC channel subunits DELM-1/2 and NaK+-ATPase α-subunits EAT-6 and CATP-1, which regulate nose-touch sensitivity and foraging behaviors, likely through modulating neuron excitability (Fig. 4B; Han et al. 2013; Johnson et al. 2020; Ray and Singhvi 2021).
Glial Regulation of Synaptic Activity
Sleep Regulation
Postembryonic ablation of CEPsh glia does not alter nerve-ring morphology. Nonetheless, animals move at half speed, along circular trajectories, and lapse into ectopic sleep bouts (Katz et al. 2018, 2019). In vivo, Ca2+ imaging during sleep reveals that while most neurons are silent, ALA neurons exhibit calcium transients (Nichols et al. 2017). These neurons form inhibitory synapses onto AVE locomotion interneurons, and these synapses are inactivated by CEPsh glia that wrap around them (Katz et al. 2018). AVE activity precedes backward movement; however, CEPsh glia ablation results in prolonged AVE Ca2+ signals uncoupled from movement. Astrocyte regulation of sleep is conserved in Drosophila and in mice (Frank 2013; Poskanzer and Yuste 2016; Artiushin and Sehgal 2020; Blum et al. 2021).
Repetitive Behavior
Animals lacking CEPsh glia or GLT-1, a conserved glutamate transporter expressed in CEPsh glia and vertebrate astrocytes, exhibit repeated backward movement initiations (Mano et al. 2007; Katz et al. 2019). Dual-color imaging of extracellular glutamate and intracellular Ca2+ signals in AVA (a backward-locomotion interneuron) in glt-1 mutants, reveals oscillations of glutamate release near AVA and of AVA firing (Fig. 4B). These studies suggest that in the absence of glial GLT-1, glutamate diffuses away from AVA postsynaptic sites and engages an extrasynaptic glutamate receptor, MGL-2/mGluR5, on presynaptic neurons. This leads to unevoked release of glutamate, mediated by presynaptic EGL-30/Gαq, driving anautocrine feedforward loop that causes AVA to fire repeatedly (Katz et al. 2019). Conditional knockout of GLT1 in mouse astrocytes results in repetitive grooming behavior (Aida et al. 2015), and murine mGluR5 inhibition prevents repetitive grooming and head tics in mouse models of autism spectrum and other repetitive behavior disorders (Silverman et al. 2010). Thus, it is possible that mammalian repetitive behavior (Tingand Feng 2008) also originates from synaptic glutamate control defects.
Locomotion and Salt Resistance
let-381 mutants, which block postembryonic maintenance of GLR fate, as well as animals in which GLR glia are genetically ablated after nervous system development is largely complete, exhibit specific defects in locomotory behavior resembling those seen in CEPsh ablated animals (Katz et al. 2018, 2019). Among other defects, GLR glia-defective animals have reduced locomotion speed and increased reversal probability. In addition, animals with disrupted GLR glia are hypersensitive to salt, arresting locomotion for longer than wild-type animals, and recovering more slowly once normo-osmotic conditions are restored, suggesting important roles for these glia in coordinating neuronal activity.
GLIAL ROLES IN STRESS AND AGING
Immunity
C. elegans encounters microorganisms in its environment, adapting physiology and behavior accordingly. Transcriptome studies reveal that co-culture with Penicillium brevicompactum, an ecologically relevant mold, up-regulates stress-response genes, including xenobiotic metabolizing enzymes (XMEs), in the intestine and AMsh glia. The nuclear-hormone receptors NHR-45 and NHR-156 are induction regulators, and mutants that cannot induce XMEs in the intestine when exposed to P. brevicompactum experience mitochondrial stress and exhibit developmental defects. Wild isolates of C. elegans harbor sequence polymorphisms in nhr-156, resulting in phenotypic diversity in AMsh glia responses to microbes. Thus, as in flies and mammals, C. elegans glia may also mediate immunity (Wallace et al. 2021).
Longevity
C. elegans glia may also regulate aging. Expression in CEPsh glia of constitutively active XBP-1, a transcription factor mediating responses to endoplasmic reticulum stress, extends life span and has ameliorating effects on distal tissues (Apfeld and Kenyon 1999; Arey and Murphy 2017; Frakes et al. 2020). This response may be mediated in part through changes in neurotransmitters (Wang and Bianchi 2021). Loss of RGBA-1, a neuropeptide-like protein expressed in glia, or of the neuropeptide receptor NPR-28 influences age-related decline in worm mating behavior (Fig. 4D; Yin et al. 2017). However, since glia lack the canonical dense core vesicle release factor UNC-31/CAPS and EGL-3/convertase (Purice et al. 2023), how they secrete neuropeptides like RGBA-1 remains to be determined.
GLIAL FUNCTIONS OF EPITHELIAL CELLS
A C. elegans Model for Radial Glia-to-Motoneuron Differentiation
Canonical radial glia stem cells are absent in C. elegans; nonetheless, the cell division-independent transformation of the epithelial tube lining Y cell into the PDA motoneuron has been informative in understanding motoneuron generation (Jarriault et al. 2008; Zuryn et al. 2014; Rashid et al. 2022). This trans-differentiation event requires LIN-12/Notch acting through NGN-1/Ngn and its regulator HLH-16/Olig. lin-12 loss blocks transformation, while lin-12 (gf) promotes precocious PDA formation. Early basal expression of ngn-1/Ngn and hlh-16/Olig depends on sem-4/Sall and egl-5/Hox. Later, co-incident with Y-cell morphological changes, ngn-1/Ngn expression is up-regulated in a sem-4/Sall and egl-5/Hox-dependent but hlh-16/Olig-independent manner. Control of histone methylation by JMJD-3.1, an H3K27me3/me2 demethylase, and the SET-2/Set1 H3K4 methylation complex ensures robustness of this trans-differentiation. Homologous proteins regulate motoneuron generation from radial glia in the vertebrate spinal cord (Jessell 2000; Dasen and Jessell 2009), suggesting that C. elegans genetics can help identify additional genes and interactions mediating these events.
Epithelia-Mediated Neurite Morphogenesis and Maintenance
In addition to maintaining glia–neuron architecture with age (Chong et al. 2021; Coraggio et al. 2023; Martin et al. 2024), epithelia assume glial-like roles and provide substrates for peripheral neurites and regulate their form and integrity. Epithelial EGL-15/FGFR guides the outgrowth of specific axons (PVP, PVQ, PVT, DA/DB) along the anteroposterior and dorsoventral axes in a kinase-independent manner via LET-60/Ras GTPase and adaptors SOC-1/2 (Bülow et al. 2004). Epithelia-expressed DRAG-1 regulates axon branching via UNC-40/DCC in hermaphrodite-specific neurons (HSNs) (Tsutsui et al. 2021). COL-99/ColA1, and DPY-18/P4HA2 affect longitudinal axons and male tail ray morphology. They are also expressed by epithelia but their tissue requirement is unclear (Baird and Emmons 1990; Hill et al. 2000; Soete et al. 2007; Taylor et al. 2018). Epithelia also guide dendrite morphogenesis. Epithelial adhesion molecules MNR-1/Fam151 and SAX-7/L1CAM form a coligand complex, bind the LRR transmembrane receptor DMA-1 on PVD neurons and instruct PVD dendritic branching (Liu and Shen 2012; Dong et al. 2013; Salzberg et al. 2013).
Epithelia Protect Axons
Epithelial cells can also guide synaptogenesis and synaptic maintenance in the PNS. Hemidesmosome attachments couple PLM neuron axons to the epidermis (Emtage et al. 2004). Hemidesmosome components (LET-805/Myotactin, VAB-10/Plakin) localize periodically for this attachment, are required to protect axons from damage, and are disrupted with age (Coakley et al. 2020; Bonacossa-Pereira et al. 2022). Epidermal UNC-70/β-Spectrin, TBC-10/GAP, and RAB-35/GTPase synergize to preserve hemidesmosomes, axon-epidermal attachments, and axon integrity against breakage (Coakley et al. 2020). Thus, the adhesion of peripheral axons to epithelia ensures their mechanical resilience.
Epithelia Regulate Synapse Assembly
Two immunoglobulin-fibronectin-domain adhesion proteins, SYG-1 and SYG-2, expressed in HSNs and vulval epithelia, respectively, interact to specify synapses (Shen and Bargmann 2003; Shen et al. 2004). SYG-2 instructs SYG-1 localization at presynaptic sites. The Ig-domain transmembrane protein ZIG-10 controls synapse maintenance. ZIG-10, localized by MAGU-2 near neuromuscular junctions, is required in the epidermis and motor neurons for synapse maintenance (Cherra and Jin 2016; Cherra et al. 2020). Epithelial ZIG-10 regulates CED-1-mediated phagocytosis to constrain the cholinergic synaptic apparatus. This is reminiscent of MGEF10/CED-1-mediated synaptic pruning by astrocytes, microglia, and Drosophila glia (Stevens et al. 2007; Fuentes-Medel et al. 2009; Awasaki and Lee 2011; Chung et al. 2013; Raiders et al. 2021b).
LOOKING AHEAD
Studies of C. elegans already reveal extensive conservation across species in glia development and function, even at the molecular level. The invariant lineage and contacts each glial cell makes with neurons and nonneural tissue enable an understanding of the roles of these conserved molecules at single cell and single contact resolution. With such a powerful molecular-genetic toolkit, C. elegans is poised to shed light on many unresolved and exciting aspects of glia biology. For example, the molecular basis of specificity in glia–neuron interactions is generally unexplored. Findings on AMsh, CEPsh, and ILso glia interactions with their neuron partners provide excellent foundations to interrogate in vivo mechanisms and logic behind interaction specificity. The roles of Ca2+ transients in vertebrate glia remain highly debated. With a completely mapped connectome and molecular tools for selectively labeling glia, C. elegans may finally reveal whether glia Ca2+ serves roles in information processing.
The invariance in C. elegans glial cell numbers and anatomy provides a powerful opportunity to understand glia heterogeneity, across sex, age, position, circuit activity, stress, and other variables. Further, the complete molecular atlas of glia across both sexes in C. elegans has been compiled by snRNA-seq, the first such map for glia of a multicellular nervous system (Purice et al. 2023). Coupled with the animal’s invariant glia–neuron development and connectome, this now provides unparalleled resolution to dissect glia biology molecularly. Further work extending such analyses to how different glia are tuned to different variables should emerge over the next few years.
Caenorhabditis elegans also provides an appropriate setting to understand how glia acquire their fates, maintain their elaborate morphologies, and organize in tiled configuration, questions that still remain unanswered for most vertebrate glia.
The discovery of neuron-receptive-ending pruning by AMsh glia also places C. elegans at the forefront of understanding this disease-relevant but molecularly enigmatic ability. Likewise, the observation that CEPsh astrocytes contribute to sleep, and control repetitive behavior, using machinery conserved with vertebrates, suggests that insights into glial control of behavior are likely to emerge from future studies of the worm.
The last two decades of research on C. elegans glia have borne fruit to early promise by rapidly delivering surprising and novel insights into glia biology. This, however, is just the beginning. Building on this exciting momentum, the coming years are likely to reveal fundamental insights into how glia govern nearly every aspect of the nervous system.
ACKNOWLEDGMENTS
The authors apologize to those whose work was not cited due to unintentional oversight or space concerns. The authors thank members of the Singhvi, Shaham, and Rapti groups for discussions. G.R. thanks colleagues at the FKNE-Kavli and Interdisciplinary Center for Neuroscience for scientific discussions. A.S. sincerely thanks all the generous philanthropic support to her laboratory including from Stephanus, Brown, and Van Sloun Foundations. This work was funded by Simons Foundation/SFARI grant (488574), Esther A. and Joseph Klingenstein Fund and the Simons Foundation Award in Neuroscience (227823), Brain Research Foundation Seed grant (BRFSG-2023–10) and NIH/NINDS funding (NS114222) to A.S., and NIH grant R35NS105094 to S.S. A.S. thanks the Glenn Foundation for Medical Research and AFAR Junior Faculty Grant for support. G.R. was supported by the European Molecular Biology Laboratory (EMBL).
REFERENCES
- Aida T, Yoshida J, Nomura M, Tanimura A, Iino Y, Soma M, Bai N, Ito Y, Cui W, Aizawa H, et al. 2015. Astroglial glutamate transporter deficiency increases synaptic excitability and leads to pathological repetitive behaviors in mice. Neuropsychopharmacology 40: 1569–1579. doi: 10.1038/npp.2015.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert PS, Riddle DL. 1983. Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. J Comp Neurol 219: 461–481. doi: 10.1002/cne.902190407 [DOI] [PubMed] [Google Scholar]
- Allen NJ, Eroglu C. 2017. Cell biology of astrocyte-synapse interactions. Neuron 96: 697–708. doi: 10.1016/j.neuron.2017.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. 2012. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 486: 410–414. doi: 10.1038/nature11059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun ZF, Chen B, Wang ZW, Hall DH. 2009. High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn 238: 1936–1950. doi: 10.1002/dvdy.22025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809. doi: 10.1038/45544 [DOI] [PubMed] [Google Scholar]
- Arey RN, Murphy CT. 2017. Conserved regulators of cognitive aging: from worms to humans. Behav Brain Res 322: 299–310. doi: 10.1016/j.bbr.2016.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G, Sehgal A. 2020. The glial perspective on sleep and circadian rhythms. Annu Rev Neurosci 43: 119–140. doi: 10.1146/annurev-neuro-091819-094557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Lee T. 2011. New tools for the analysis of glial cell biology in Drosophila. Glia 59: 1377–1386. doi: 10.1002/glia.21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. 2008. Glia are essential for sensory organ function in C. elegans. Science 322: 744–747. doi: 10.1126/science.1163074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SE, Emmons SW. 1990. Properties of a class of genes required for ray morphogenesis in Caenorhabditis elegans. Genetics 126: 335–344. doi: 10.1093/genetics/126.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. 1993. Genetic and cellular analysis of behavior in C. elegans. Annu Rev Neurosci 16: 47–71. doi: 10.1146/annurev.ne.16.030193.000403 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Marder E. 2013. From the connectome to brain function. Nat Methods 10: 483–490. doi: 10.1038/nmeth.2451 [DOI] [PubMed] [Google Scholar]
- Barres BA. 2008. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 60: 430–440. doi: 10.1016/j.neuron.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Bénard CY, Boyanov A, Hall DH, Hobert O. 2006. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development 133: 3329–3340. doi: 10.1242/dev.02507 [DOI] [PubMed] [Google Scholar]
- Blum ID, Keleş MF, Baz ES, Han E, Park K, Luu S, Issa H, Brown M, Ho MCW, Tabuchi M, et al. 2021. Astroglial calcium signaling encodes sleep need in Drosophila. Curr Biol 31: 150–162.e7. doi: 10.1016/j.cub.2020.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacossa-Pereira I, Coakley S, Hilliard MA. 2022. Neuron-epidermal attachment protects hyper-fragile axons from mechanical strain. Cell Rep 38: 110501. doi: 10.1016/j.celrep.2022.110501 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. doi: 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow HE, Boulin T, Hobert O. 2004. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron 42: 367–374. doi: 10.1016/s0896-6273(04)00246-6 [DOI] [PubMed] [Google Scholar]
- Bunk EC, Ertaylan G, Ortega F, Pavlou MA, Gonzalez Cano L, Stergiopoulos A, Safaiyan S, Völs S, van Cann M, Politis PK, et al. 2016. Prox1 is required for oligodendrocyte cell identity in adult neural stem cells of the subventricular zone. Stem Cells 34: 2115–2129. doi: 10.1002/stem.2374 [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. 2002. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 22: 183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. 2017. Comprehensive single-cell transcriptional profiling of a multi-cellular organism. Science 357: 661–667. doi: 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada RC, Russell RL. 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46: 326–342. doi: 10.1016/0012-1606(75)90109-8 [DOI] [PubMed] [Google Scholar]
- Cebul ER, McLachlan IG, Heiman MG. 2020. Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development 147: dev180448. doi: 10.1242/dev.180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Cheng H, Liu S, Al-Sheikh U, Fan Y, Duan D, Zou W, Zhu L, Kang L. 2022. The voltage-gated calcium channel EGL-19 acts on glia to drive olfactory adaptation. Front Mol Neurosci 15: 907064. doi: 10.3389/fnmol.2022.907064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ III, Jin Y. 2016. A two-immunoglobulin-domain transmembrane protein mediates an epidermal-neuronal interaction to maintain synapse density. Neuron 89: 325–336. doi: 10.1016/j.neuron.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ III, Goncharov A, Boassa D, Ellisman M, Jin Y. 2020. C. elegans MAGU-2/Mpp5 homolog regulates epidermal phagocytosis and synapse density. J Neurogenet 34: 298–306. doi: 10.1080/01677063.2020.1726915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiareli RA, Carvalho GA, Marques BL, Mota LS, Oliveira-Lima OC, Gomes RM, Birbrair A, Gomez RS, Simão F, Klempin F, et al. 2021. The role of astrocytes in the neuro-repair process. Front Cell Dev Biol 9: 665795. doi: 10.3389/fcell.2021.665795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Hsiao TI. 2012. The Caenorhabditis elegans epidermis as a model skin. I: Development, patterning, and growth. Wiley Interdiscip Rev Dev Biol 1: 861–878. doi: 10.1002/wdev.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MK, Cebul ER, Mizeracka K, Heiman MG. 2021. Loss of the extracellular matrix protein DIG-1 causes glial fragmentation, dendrite breakage, and dendrite extension defects. J Dev Biol 9: 42. doi: 10.3390/jdb9040042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. 2005. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120: 421–433. doi: 10.1016/j.cell.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. 2013. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504: 394–400. doi: 10.1038/nature12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Verghese PB, Chakraborty C, Joung J, Hyman BT, Ulrich JD, Holtzman DM, Barres BA. 2016. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci 113: 10186–10191. doi: 10.1073/pnas.1609896113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Barres BA. 2013. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci 14: 311–321. doi: 10.1038/nrn3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley S, Ritchie FK, Galbraith KM, Hilliard MA. 2020. Epidermal control of axonal attachment via β-spectrin and the GTPase-activating protein TBC-10 prevents axonal degeneration. Nat Commun 11: 133. doi: 10.1038/s41467-019-13795-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Ramos DA, Margeta MA, Shen K. 2007. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science 318: 103–106. doi: 10.1126/science.1143762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LT, Bayer EA, Duerr JS, et al. 2019. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571: 63–71. doi: 10.1038/s41586-019-1352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coraggio F, Bhushan M, Roumeliotis S, Caroti F, Bevilacqua C, Prevedel R, Rapti G. 2023. An interplay of HSP-proteostasis, biomechanics and ECM-cell junctions ensures C. elegans astroglial architecture. bioRxiv doi: 10.1101/2023.10.28.564505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadras J, Martin G, Czternasty G, Bruner J. 1985. Gap-like junctions between neuron cell bodies and glial cells of crayfish. Brain Res 326: 149–151. doi: 10.1016/0006-8993(85)91394-0 [DOI] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. 2009. Chapter six hox networks and the origins of motor neuron diversity. Curr Top Dev Biol 88: 169–200. doi: 10.1016/S0070-2153(09)88006-X [DOI] [PubMed] [Google Scholar]
- Dawes JHP, Kelsh RN. 2021. Cell fate decisions in the neural crest, from pigment cell to neural development. Int J Mol Sci 22: 13531. doi: 10.3390/ijms222413531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Bugiani M, Dupuits C, Galanaud D, Touitou V, Postma N, van Berkel C, Polder E, Tollard E, Darios F, et al. 2013. Brain white matter oedema due to ClC-2 chloride channel deficiency: an observational analytical study. Lancet Neurol 12: 659–668. doi: 10.1016/S1474-4422(13)70053-X [DOI] [PubMed] [Google Scholar]
- Derouiche A, Pannicke T, Haseleu J, Blaess S, Grosche J, Reichenbach A. 2012. Beyond polarity: functional membrane domains in astrocytes and Müller cells. Neurochem Res 37: 2513–2523. doi: 10.1007/s11064-012-0824-z [DOI] [PubMed] [Google Scholar]
- Ding G, Zou W, Zhang H, Xue Y, Cai Y, Huang G, Chen L, Duan S, Kang L. 2015. In vivo tactile stimulation-evoked responses in Caenorhabditis elegans amphid sheath glia. PLoS ONE 10: e0117114. doi: 10.1371/journal.pone.0117114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici C, Moreno-Bravo JA, Puiggros SR, Rappeneau Q, Rama N, Vieugue P, Bernet A, Mehlen P, Chédotal A. 2017. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature 545: 350–354. doi: 10.1038/nature22331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, Shen K. 2013. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell 155: 296–307. doi: 10.1016/j.cell.2013.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Jin S, Shao Z. 2020. Glia promote synaptogenesis through an IQGAP PES-7 in C. elegans. Cell Rep 30: 2614–2626.e2. doi: 10.1016/j.celrep.2020.01.102 [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. eLife 3: e01948. doi: 10.7554/eLife.01948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Zhang H, Yue X, Fan Y, Xue Y, Shao J, Ding G, Chen D, Li S, Cheng H, et al. 2020. Sensory glia detect repulsive odorants and drive olfactory adaptation. Neuron 108: 707–721.e8. doi: 10.1016/j.neuron.2020.08.026 [DOI] [PubMed] [Google Scholar]
- Emmons SW. 2005. Male development. WormBook 1–22. doi: 10.1895/wormbook.1.33.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons SW. 2018. Neural circuits of sexual behavior in Caenorhabditis elegans. Annu Rev Neurosci 41: 349–369. doi: 10.1146/annurev-neuro-070815-014056 [DOI] [PubMed] [Google Scholar]
- Emtage L, Gu G, Hartwieg E, Chalfie M. 2004. Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron 44: 795–807. doi: 10.1016/j.neuron.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW, O’Rourke NA, Park CY, Özkan E, Chakraborty C, Mulinyawe SB, Annis DS, Huberman AD, et al. 2009. Gabapentin receptor α2δ−1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139: 380–392. doi: 10.1016/j.cell.2009.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ji T, Wang K, Huang J, Wang M, Manning L, Dong X, Shi Y, Zhang X, Shao Z, et al. 2020. A muscle-epidermis-glia signaling axis sustains synaptic specificity during allometric growth in Caenorhabditis elegans. eLife 9: e55890. doi: 10.7554/eLife.55890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy-Tselnicker I, Allen NJ. 2018. Astrocytes, neurons, synapses: a tripartite view on cortical circuit development. Neural Dev 13: 7. doi: 10.1186/s13064-018-0104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favuzzi E, Huang S, Saldi GA, Binan L, Ibrahim LA, Fernández-Otero M, Cao Y, Zeine A, Sefah A, Zheng K, et al. 2021. GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell 184: 4048–4063.e32. doi: 10.1016/j.cell.2021.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326: 259–267. doi: 10.1038/326259a0 [DOI] [PubMed] [Google Scholar]
- Fernandez-Abascal J, Johnson CK, Graziano B, Wang L, Encalada N, Bianchi L. 2022. A glial ClCCl− channel mediates nose touch responses in C. elegans. Neuron 110: 470–485. e7. doi: 10.1016/j.neuron.2021.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes AE, Metcalf MG, Tronnes SU, Bar-Ziv R, Durieux J, Gildea HK, Kandahari N, Monshietehadi S, Dillin A. 2020. Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science 367: 436–440. doi: 10.1126/science.aaz6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MG. 2013. Astroglial regulation of sleep homeostasis. Curr Opin Neurobiol 23: 812–818. doi: 10.1016/j.conb.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, Freeman MR. 2009. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol 7: e1000184. doi: 10.1371/journal.pbio.1000184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung W, Wexler L, Heiman MG. 2020. Cell-type-specific promoters for C. elegans glia. J Neurogenet 34: 335–346. doi: 10.1080/01677063.2020.1781851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung W, Tan TM, Kolotuev I, Heiman MG. 2023. A sex-specific switch in a single glial cell patterns the apical extracellular matrix. Curr Biol 33: 4174–4186 e7. doi: 10.1016/j.cub.2023.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Atlas EG, Hobert O. 2016. A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife 5: e17686. doi: 10.7554/eLife.17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CL, Balbona JT, Niedzwiecki A, Rodriguez P, Nguyen KCQ, Hall DH, Blakely RD. 2018. Glial loss of the metallo β-lactamase domain containing protein, SWIP-10, induces age- and glutamate-signaling dependent, dopamine neuron degeneration. PLoS Genet 14: e1007269. doi: 10.1371/journal.pgen.1007269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. 2016. Sydney Brenner on the genetics of Caenorhabditis elegans. Genetics 204: 1–2. doi: 10.1534/genetics.116.194084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Sengupta P. 2019. How Caenorhabditis elegans senses mechanical stress, temperature, and other physical stimuli. Genetics 212: 25–51. doi: 10.1534/genetics.118.300241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. 1997. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol 137: 1171–1183. doi: 10.1083/jcb.137.5.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J, Matthewman C, Bianchi L. 2015. A novel mechanism of pH buffering in C. elegans glia: bicarbonate transport via the voltage-gated ClC Cl− channel CLH-1. J Neurosci 35: 16377–16397. doi: 10.1523/JNEUROSCI.3237-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH. 1977. The posterior nervous system of the nematode Caenorhabditis elegans. California Institute of Technology, Pasadena, CA. [Google Scholar]
- Han L, Wang Y, Sangaletti R, D’Urso G, Lu Y, Shaham S, Bianchi L. 2013. Two novel DEG/ENaC channel subunits expressed in glia are needed for nose-touch sensitivity in Caenorhabditis elegans. J Neurosci 33: 936–949. doi: 10.1523/JNEUROSCI.2749-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Johnsen R, Lauter G, Baillie D, Bürglin TR. 2006. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics 7: 280. doi: 10.1186/1471-2164-7-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway JA, Sturgeon SM, Snarrenberg CL, Li Z, Xu XZ, Bermingham DP, Odiase P, Spencer WC, Miller DM, Carvelli L, et al. 2015. Glial expression of the Caenorhabditis elegans gene swip-10 supports glutamate dependent control of extrasynaptic dopamine signaling. J Neurosci 35: 9409–9423. doi: 10.1523/JNEUROSCI.0800-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, Shaham S. 2007. Ancestral roles of glia suggested by the nervous system of Caenorhabditis elegans. Neuron Glia Biol 3: 55–61. doi: 10.1017/S1740925X07000609 [DOI] [PubMed] [Google Scholar]
- Heiman MG, Shaham S. 2009. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137: 344–355. doi: 10.1016/j.cell.2009.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Harfe BD, Dobbins CA, L’Hernault SW. 2000. dpy-18 encodes an α-subunit of prolyl-4-hydroxylase in Caenorhabditis elegans. Genetics 155: 1139–1148. doi: 10.1093/genetics/155.3.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilu-Dadia R, Kurant E. 2020. Glial phagocytosis in developing and mature Drosophila CNS: tight regulation for a healthy brain. Curr Opin Immunol 62: 62–68. doi: 10.1016/j.coi.2019.11.010 [DOI] [PubMed] [Google Scholar]
- Hong H, Chen H, Zhang Y, Wu Z, Zhang Y, Zhang Y, Hu Z, Zhang JV, Ling K, Hu J, et al. 2021. DYF-4 regulates patched-related/DAF-6-mediated sensory compartment formation in C. elegans. PLoS Genet 17: e1009618. doi: 10.1371/journal.pgen.1009618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. 1995. Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82: 1025–1036. doi: 10.1016/0092-8674(95)90281-3 [DOI] [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. 2012. The connectome of a decision-making neural network. Science 337: 437–444. doi: 10.1126/science.1221762 [DOI] [PubMed] [Google Scholar]
- Jarriault S, Schwab Y, Greenwald I. 2008. A Caenorhabditis elegans model for epithelial–neuronal transdifferentiation. Proc Natl Acad Sci 105: 3790–3795. doi: 10.1073/pnas.0712159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM. 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1: 20–29. doi: 10.1038/35049541 [DOI] [PubMed] [Google Scholar]
- Johnson CK, Fernandez-Abascal J, Wang Y, Wang L, Bianchi L. 2020. The Na+-K+-ATPase is needed in glia of touch receptors for responses to touch in C. elegans. J Neurophysiol 123: 2064–2074. doi: 10.1152/jn.00636.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. 1995. Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82: 1013–1023. doi: 10.1016/0092-8674(95)90280-5 [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E, Ohta A, Ujisawa T, Sun S, Nishikawa Y, Kuhara A, Mitani S. 2016. Caenorhabditis elegans homologue of Prox1/Prospero is expressed in the glia and is required for sensory behavior and cold tolerance. Genes Cells 21: 936–948. doi: 10.1111/gtc.12394 [DOI] [PubMed] [Google Scholar]
- Kastriti ME, Adameyko I. 2017. Specification, plasticity and evolutionary origin of peripheral glial cells. Curr Opin Neurobiol 47: 196–202. doi: 10.1016/j.conb.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Katz M, Corson F, Iwanir S, Biron D, Shaham S. 2018. Glia modulate a neuronal circuit for locomotion suppression during sleep in C. elegans. Cell Rep 22: 2575–2583. doi: 10.1016/j.celrep.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Corson F, Keil W, Singhal A, Bae A, Lu Y, Liang Y, Shaham S. 2019. Glutamate spillover in C. elegans triggers repetitive behavior through presynaptic activation of MGL-2/mGluR5. Nat Commun 10: 1882. doi: 10.1038/s41467-019-09581-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS. 2019. Astrocyte–neuron interactions in the striatum: insights on identity, form, and function. Trends Neurosci 42: 617–630. doi: 10.1016/j.tins.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Harrison SW, Xu SQ, Chen L, Fire A. 1994. Elements regulating cell- and stage-specific expression of the C. elegans MyoDfamilyhomologhlh-1. Dev Biol 166: 133–148. doi: 10.1006/dbio.1994.1302 [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. 1996. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development 122: 2579–2588. doi: 10.1242/dev.122.9.2579 [DOI] [PubMed] [Google Scholar]
- Lamkin ER, Heiman MG. 2017. Coordinated morphogenesis of neurons and glia. Curr Opin Neurobiol 47: 58–64. doi: 10.1016/j.conb.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F, Quintin S, Labouesse M. 2004. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev Biol 265: 478–490. doi: 10.1016/j.ydbio.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Lee IH, Procko C, Lu Y, Shaham S. 2021a. Stress-induced neural plasticity mediated by glial GPCR REMO-1 promotes C. elegans adaptive behavior. Cell Rep 34: 108607. doi: 10.1016/j.celrep.2020.108607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Magescas J, Fetter RD, Feldman JL, Shen K. 2021b. Inherited apicobasal polarity defines the key features of axon-dendrite polarity in a sensory neuron. Curr Biol 31: 3768–3783.e3. doi: 10.1016/j.cub.2021.06.039 [DOI] [PubMed] [Google Scholar]
- Liu OW, Shen K. 2012. The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat Neurosci 15: 57–63. doi: 10.1038/nn.2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low IIC, Williams CR, Chong MK, McLachlan IG, Wierbowski BM, Kolotuev I, Heiman MG. 2019. Morphogenesis of neurons and glia within an epithelium. Development 146: dev171124. doi: 10.1242/dev.171124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano I, Straud S, Driscoll M. 2007. Caenorhabditis elegans glutamate transporters influence synaptic function and behavior at sites distant from the synapse. J Biol Chem 282: 34412–34419. doi: 10.1074/jbc.M704134200 [DOI] [PubMed] [Google Scholar]
- Martin CG, Bent JS, Hill T, Topalidou I, Singhvi A. 2024. Epithelia delimits glial apical polarity against mechanical shear to maintain glia-neuron—architecture. Devel Cell (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahira N, Takebayashi H, Ono K, Watanabe K, Ding L, Furusho M, Ogawa Y, Nabeshima Y-I, Alvarez-Buylla A, Shimizu K, et al. 2006. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev Biol 293: 358–369. doi: 10.1016/j.ydbio.2006.02.029 [DOI] [PubMed] [Google Scholar]
- McMiller TL, Johnson CM. 2005. Molecular characterization of HLH-17, a C. elegans bHLH protein required for normal larval development. Gene 356: 1–10. doi: 10.1016/j.gene.2005.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijlink F, Beverdam A, Bruower A, Oosterveen TC, Berge DT. 1999. Vertebrate aristaless-related genes. Int J Dev Biol 43: 651–663. [PubMed] [Google Scholar]
- Melkman T, Sengupta P. 2005. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development 132: 1935–1949. doi: 10.1242/dev.01788 [DOI] [PubMed] [Google Scholar]
- Meng L, Zhang A, Jin Y, Yan D. 2016. Regulation of neuronal axon specification by glia-neuron gap junctions in C. elegans. eLife 5: e19510. doi: 10.7554/eLife.19510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G, Gansmuller A, Hindelang C, Labouesse M. 2000. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Curr Biol 10: 1098–1107. doi: 10.1016/s0960-9822(00)00695-3 [DOI] [PubMed] [Google Scholar]
- Miller RH. 2005. Dorsally derived oligodendrocytes come of age. Neuron 45: 1–3. doi: 10.1016/j.neuron.2004.12.032 [DOI] [PubMed] [Google Scholar]
- Miller RM, Portman DS. 2011. The Wnt/β-catenin asymmetry pathway patterns the atonal ortholog lin-32 to diversify cell fate in a Caenorhabditis elegans sensory lineage. J Neurosci 31: 13281–13291. doi: 10.1523/JNEUROSCI.6504-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K, Heiman MG. 2015. The many glia of a tiny nematode: studying glial diversity using Caenorhabditis elegans. Wiley Interdiscip Rev Dev Biol 4: 151–160. doi: 10.1002/wdev.171 [DOI] [PubMed] [Google Scholar]
- Mizeracka K, Rogers JM, Rumley JD, Shaham S, Bulyk ML, Murray JI, Heiman MG. 2021. Lineage-specific control of convergent differentiation by a Forkhead repressor. Development 148: dev199493. doi: 10.1242/dev.199493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-García L, Lloret-Fernández C, Cook SJ, Kim B, Bonnington RC, Sammut M, O’Shea JM, Gilbert SP, Elliott DJ, Hall DH, et al. 2020. Direct glia-to-neuron transdifferentiation gives rise to a pair of male-specific neurons that ensure nimble male mating. eLife 9: e48361. doi: 10.7554/eLife.48361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Yu X, Papouin T, Cheong E, Freeman MR, Monk KR, Hastings MH, Haydon PG, Rowitch D, Shaham S, et al. 2021. Behaviorally consequential astrocytic regulation of neural circuits. Neuron 109: 576–596. doi: 10.1016/j.neuron.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM III, Blakely RD. 2002. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci 99: 3264–3269. doi: 10.1073/pnas.042497999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard M. 1994. Direct signaling from astrocytesto neurons in cultures of mammalian brain cells. Science 263: 1768–1771. doi: 10.1126/science.8134839 [DOI] [PubMed] [Google Scholar]
- Nichols ALA, Eichler T, Latham R, Zimmer M. 2017. A global brain state underlies C. elegans sleep behavior. Science 356: eaam6851. doi: 10.1126/science.aam6851 [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Kriegstein AR. 2008. Distinct behaviors of neural stem and progenitor cell sunderlie cortical neurogenesis. J Comp Neurol 508: 28–44. doi: 10.1002/cne.21669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura K, Bürglin TR. 2011. Dye-filling of the amphid sheath glia: implications for the functional relationship between sensory neurons and glia in Caenorhabditis elegans. Biochem Biophys Res Commun 406: 188–193. doi: 10.1016/j.bbrc.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Oikonomou G, Shaham S. 2011. The glia of Caenorhabditis elegans. Glia 59: 1253–1263. doi: 10.1002/glia.21084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Watanabe S, Jorgensen EM, Shaham S. 2011. Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS Biol 9: e1001121. doi: 10.1371/journal.pbio.1001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Shaham S. 2012. Some, but not all, retromer components promote morphogenesis of C. elegans sensory compartments. Dev Biol 362: 42–49. doi: 10.1016/j.ydbio.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, et al. 2019. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365: eaax1971. doi: 10.1126/science.aax1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Sakurai Y, Sato H, Kanda S, Iino Y, Kunitomo H. 2021. Roles of the ClC chloride channel CLH-1 in food-associated salt chemotaxis behavior of C. elegans. eLife 10: e55701. doi: 10.7554/eLife.55701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peco E, Davla S, Camp D, Stacey SM, Landgraf M, van Meyel DJ. 2016. Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development 143: 1170–1181. doi: 10.1242/dev.133165 [DOI] [PubMed] [Google Scholar]
- Perens EA, Shaham S. 2005. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev Cell 8: 893–906. doi: 10.1016/j.devcel.2005.03.009 [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117: 456–487. doi: 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Portman DS, Emmons SW. 2000. The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development 127: 5415–5426. doi: 10.1242/dev.127.24.5415 [DOI] [PubMed] [Google Scholar]
- Poskanzer KE, Yuste R. 2016. Astrocytes regulate cortical state switching in vivo. Proc Natl Acad Sci 113: E2675–E2684. doi: 10.1073/pnas.1520759113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S. 2011. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development 138: 1371–1381. doi: 10.1242/dev.058305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S. 2012. Sensory organ remodeling in Caenorhabditis elegans requires the zinc-finger protein ZTF-16. Genetics 190: 1405–1415. doi: 10.1534/genetics.111.137786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purice M, Quitevis E, Manning R, Severs L, Tran N, Sorrentino V, Setty M, Singhvi A. 2023. Molecular atlas of C. elegans glia across sexes reveals heterogeneity, variable sex-dimorphism, and glial properties. bioRxiv doi: 10.1101/2023.03.21.533668 [DOI] [Google Scholar]
- Qu Z, Zhang A, Yan D. 2020. Robo functions as an attractive cue for glial migration through SYG-1/Neph. eLife 9: e57921. doi: 10.7554/eLife.57921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani P, Rogalski T, Moerman DG. 2015. The C. elegans UNC-23 protein, a member of the BCL-2-associated athanogene (BAG) family of chaperone regulators, interacts with HSP-1 to regulate cell attachment and maintain hypodermal integrity. Worm 4: e1023496. doi: 10.1080/21624054.2015.1023496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders S, Black EC, Bae A, MacFarlane S, Klein M, Shaham S, Singhvi A. 2021a. Glia actively sculpt sensory neurons by controlled phagocytosis to tune animal behavior. eLife 10: e63532. doi: 10.7554/eLife.63532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders S, Han T, Scott-Hewitt N, Kucenas S, Lew D, Logan MA, Singhvi A. 2021b. Engulfed by glia: glial pruning in development, function, and injury across species. J Neurosci 41: 823–833. doi: 10.1523/JNEUROSCI.1660-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti G. 2021. Open frontiers in neural cell type investigations; lessons from Caenorhabditis elegans and beyond, toward a multimodal integration. Front Neurosci 15: 787753. doi: 10.3389/fnins.2021.787753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti G, Li C, Shan A, Lu Y, Shaham S. 2017. Glia initiate brain assembly through noncanonical Chimaerin-Furin axon guidance in C. elegans. Nat Neurosci 20: 1350–1360. doi: 10.1038/nn.4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid A, Tevlin M, Lu Y, Shaham S. 2022. A developmental pathway for epithelial-to-motoneuron transformation in C. elegans. Cell Rep 40: 111414. doi: 10.1016/j.celrep.2022.111414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Singhvi A. 2021. Charging up the periphery: glial ionic regulation in sensory perception. Front Cell Dev Biol 9: 687732. doi: 10.3389/fcell.2021.687732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Gurung P, Manning RS, Kravchuk AA, Singhvi A. 2024. Neuron cilia restrain glial KCC-3 to a microdomain to regulate multi-sensory processing. Cell Rep 43: 113844. doi: 10.1016/j.celrep.2024.113844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzauti A, Laurent P. 2021. Ectocytosis prevents accumulation of ciliary cargo in C. elegans sensory neurons. eLife 10: e67670. doi: 10.7554/eLife.67670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousse I, Robitaille R. 2006. Calcium signaling in Schwann cells at synaptic and extra-synaptic sites: active glial modulation of neuronal activity. Glia 54: 691–699. doi: 10.1002/glia.20388 [DOI] [PubMed] [Google Scholar]
- Rowitch DH. 2004. Glial specification in the vertebrate neural tube. Nat Rev Neurosci 5: 409–419. doi: 10.1038/nrn1389 [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Díaz-Balzac CA, Ramirez-Suarez NJ, Attreed M, Tecle E, Desbois M, Kaprielian Z, Bülow HE. 2013. Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell 155: 308–320. doi: 10.1016/j.cell.2013.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JL. 2003. Polarized domains of myelinated axons. Neuron 40: 297–318. doi: 10.1016/s0896-6273(03)00628-7 [DOI] [PubMed] [Google Scholar]
- Sammut M, Cook SJ, Nguyen KCQ, Felton T, Hall DH, Emmons SW, Poole RJ, Barrios A. 2015. Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526: 385–390. doi: 10.1038/nature15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR. 2018. Theworm connectome: back to the future. Trends Neurosci 41: 763–765. doi: 10.1016/j.tins.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Rakic P. 1979. A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat Embryol (Berl) 156: 115–152. doi: 10.1007/BF00300010 [DOI] [PubMed] [Google Scholar]
- Sengupta P, Samuel AD. 2009. Caenorhabditis elegans: a model system for systems neuroscience. Curr Opin Neurobiol 19: 637–643. doi: 10.1016/j.conb.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PK, Santella A, Jacobo A, Siletti K, Hudspeth AJ, Bao Z. 2017. An in toto approach to dissecting cellular interactions in complex tissues. Dev Cell 43: 530–540.e4. doi: 10.1016/j.devcel.2017.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. 2005. Glia–neuron interactions in nervous system function and development. Curr Top Dev Biol 69: 39–66. doi: 10.1016/S0070-2153(05)69003-5 [DOI] [PubMed] [Google Scholar]
- Shaham S. 2010. Chemosensory organs as models of neuronal synapses. Nat Rev Neurosci 11: 212–217. doi: 10.1038/nrn2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. 2015. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb Perspect Biol 7: a020578. doi: 10.1101/cshperspect.a020578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Watanabe S, Christensen R, Jorgensen EM, Colón-Ramos DA. 2013. Synapse location during growth depends on glia location. Cell 154: 337–350. doi: 10.1016/j.cell.2013.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Bargmann CI. 2003. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112: 619–630. doi: 10.1016/s0092-8674(03)00113-2 [DOI] [PubMed] [Google Scholar]
- Shen K, Fetter RD, Bargmann CI. 2004. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell 116: 869–881. doi: 10.1016/S0092-8674(04)00251-X [DOI] [PubMed] [Google Scholar]
- Shigetomi E, Patel S, Khakh BS. 2016. Probing the complexities of astrocyte calcium signaling. Trends Cell Biol 26: 300–312. doi: 10.1016/j.tcb.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sík A, Smith RL, Freund TF. 2000. Distribution of chloride channel-2-immunoreactive neuronal and astrocytic processes in the hippocampus. Neuroscience 101: 51–65. doi: 10.1016/S0306-4522(00)00360-2 [DOI] [PubMed] [Google Scholar]
- Silverman JL, Tolu SS, Barkan CL, Crawley JN. 2010. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology 35: 976–989. doi: 10.1038/npp.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, Shaham S. 2017. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat Commun 8: 14100. doi: 10.1038/ncomms14100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, Shaham S. 2019. Glia-neuron interactions in Caenorhabditis elegans. Annu Rev Neurosci 42: 149–168. doi: 10.1146/annurev-neuro-070918-050314 [DOI] [PubMed] [Google Scholar]
- Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, Huang XY, Shaham S. 2016. A glial K/Cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell 165: 936–948. doi: 10.1016/j.cell.2016.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soete G, Betist MC, Korswagen HC. 2007. Regulation of Caenorhabditis elegans body size and male tail development by the novel gene lon-8. BMC Dev Biol 7: 20. doi: 10.1186/1471-213X-7-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanakis N, Jiang J, Liang Y, Shaham S. 2024. LET-381/FoxF and UNC-30/Pitx2 control the development of C. elegans mesodermal glia that regulate motor behavior. EMBO J (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131: 1164–1178. doi: 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156. doi: 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. 1980. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol 78: 542–576. doi: 10.1016/0012-1606(80)90352-8 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119. doi: 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Isonishi A, Yamasaki M, Kawabe Y, Morita-Takemura S, Nakahara K, Terada Y, Shinjo T, Okuda H, Tanaka T, et al. 2018. Olig2-Lineage astrocytes: a distinct subtype of astrocytes that differs from GFAPastrocytes. Front Neuroanat 12: 8. doi: 10.3389/fnana.2018.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Unsoeld T, Hutter H. 2018. The transmembrane collagen COL-99 guides longitudinally extending axons in C. elegans. Mol Cell Neurosci 89: 9–19. doi: 10.1016/j.mcn.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, et al. 2021. Molecular topography of an entire nervous system. Cell 184: 4329–4347.e23. doi: 10.1016/j.cell.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Feng G. 2008. Glutamatergic synaptic dysfunction and obsessive-compulsive disorder. Curr Chem Genomics 2: 62–75. doi: 10.2174/1875397300802010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui K, Kim HS, Yoshikata C, Kimura K, Kubota Y, Shibata Y, Tian C, Liu J, Nishiwaki K. 2021. Repulsive guidance molecule acts in axon branching in Caenorhabditis elegans. Sci Rep 11: 22370. doi: 10.1038/s41598-021-01853-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Sieber M, Morphew M, Han M. 2005. The Caenorhabditis elegans aristaless orthologue, alr-1, is required for maintaining the functional and structural integrity of the amphid sensory organs. Mol Biol Cell 16: 4695–4704. doi: 10.1091/mbc.e05-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. 2001. Control of synapse number by glia. Science 291: 657–661. doi: 10.1126/science.291.5504.657 [DOI] [PubMed] [Google Scholar]
- Varadarajan SG, Kong JH, Phan KD, Kao TJ, Panaitof SC, Cardin J, Eltzschig H, Kania A, Novitch BG, Butler SJ. 2017. Netrin1 produced by neural progenitors, not floor plate cells, is required for axon guidance in the spinal cord. Neuron 94: 790–799.e3. doi: 10.1016/j.neuron.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. 2011. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol 7: e1001066. doi: 10.1371/journal.pcbi.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. 1996. Glide directs glial fate commitment and cell fate switch between neurons and glia. Development 122: 131–139. doi: 10.1242/dev.122.1.131 [DOI] [PubMed] [Google Scholar]
- Wagner B, Natarajan A, Grünaug S, Kroismayr R, Wagner EF, Sibilia M. 2006. Neuronal survival depends on EGFR signaling in cortical but not midbrain astrocytes. EMBO J 25: 752–762. doi: 10.1038/sj.emboj.7600988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SW, Singhvi A, Liang Y, Lu Y, Shaham S. 2016. PROS-1/Prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell Rep 15: 550–562. doi: 10.1016/j.celrep.2016.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SW, Lizzappi MC, Magemizoğlu E, Hur H, Liang Y, Shaham S. 2021. Nuclear hormone receptors promote gut and glia detoxifying enzyme induction and protect C. elegans from the mold P. brevicompactum. Cell Rep 37: 110166. doi: 10.1016/j.celrep.2021.110166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bianchi L. 2021. Maintenance of protein homeostasis in glia extends lifespan in C. elegans. Exp Neurol 339: 113648. doi: 10.1016/j.expneurol.2021.113648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Apicella A, Lee SK, Ezcurra M, Slone RD, Goldmit M, et al. 2008. A glial DEG/ENaC channel functions with neuronal channel DEG-1 to mediate specific sensory functions in C. elegans. EMBO J 27: 2388–2399. doi: 10.1038/emboj.2008.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, D’Urso G, Bianch L. 2012. iKnockout of glial channel ACD-1 exacerbates sensory deficits in a C. elegans mutant by regulating calcium levels of sensory neurons. J Neurophysiol 107: 148–158. doi: 10.1152/jn.00299.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, et al. 2015. Cell-specific transcriptional profiling of ciliated sensory neurons reveals regulators of behavior and extracellular vesicle biogenesis. Curr Biol 25: 3232–3238. doi: 10.1016/j.cub.2015.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Perens EA, Oikonomou G, Wallace SW, Lu Y, Shaham S. 2017. IGDB-2, an Ig/FNIII protein, binds the ion channel LGC-34 and controls sensory compartment morphogenesis in C. elegans. Dev Biol 430: 105–112. doi: 10.1016/j.ydbio.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. 1975. Electron microscopical reconstruction of the anterior sensoryanatomy of thenematode Caenorhabditis elegans. JComp Neurol 160: 313–337. doi: 10.1002/cne.901600305 [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. 1986. The structure of the nervous system of the nematode Caenorhabditiselegans. Philos Trans RSoc Lond BBiol Sci 314: 1–340. doi: 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Wilton DK, Dissing-Olesen L, Stevens B. 2019. Neuron-glia signaling in synapse elimination. Annu Rev Neurosci 42: 107–127. doi: 10.1146/annurev-neuro-070918-050306 [DOI] [PubMed] [Google Scholar]
- Witvliet D, Mulcahy B, Mitchell JK, Meirovitch Y, Berger DR, Wu Y, Liu Y, Koh WX, Parvathala R, Holmyard D, et al. 2021. Connectomes across development reveal principles of brain maturation. Nature 596: 257–261. doi: 10.1038/s41586-021-03778-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JA, Gao G, Liu XJ, Hao ZQ, Li K, Kang XL, Li H, Shan YH, Hu WL, Li HP, et al. 2017. Genetic variation in glia–neuron signalling modulates ageing rate. Nature 551: 198–203. doi: 10.1038/nature24463 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Nakano S, Suzuki T, Ihara K, Higashiyama T, Mori I. 2016. A glial K(+)/Cl(−) cotransporter modifies temperature-evoked dynamics in Caenorhabditis elegans sensory neurons. Genes Brain Behav 15: 429–440. doi: 10.1111/gbb.12260 [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. 2008. mls-2 and vab-3 control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135: 2263–2275. doi: 10.1242/dev.019547 [DOI] [PubMed] [Google Scholar]
- Yu RY, Nguyen CQ, Hall DH, Chow KL. 2000. Expression of ram-5 in the structural cell is required for sensory ray morphogenesis in Caenorhabditis elegans male tail. EMBO J 19: 3542–3555. doi: 10.1093/emboj/19.14.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Emmons SW. 1995. Specification of sense-organ identity by a Caenorhabditis elegans Pax-6 homologue. Nature 377: 55–59. doi: 10.1038/377055a0 [DOI] [PubMed] [Google Scholar]
- Zhang A, Ackley BD, Yan D. 2020a. Vitamin B12 regulates glial migration and synapse formation through isoform-specific control of PTP-3/LAR PRTP expression. Cell Rep 30: 3981–3988.e3. doi: 10.1016/j.celrep.2020.02.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Guan Z, Ockerman K, Dong P, Guo J, Wang Z, Yan D. 2020b. Regulation of glial size by eicosapentaenoic acid through a novel Golgi apparatus mechanism. PLoS Biol 18: e3001051. doi: 10.1371/journal.pbio.3001051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Noma K, Yan D. 2020c. Regulation of gliogenesis by lin-32/Atoh1 in Caenorhabditis elegans. G3 10: 3271–3278. doi: 10.1534/g3.120.401547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S, Ahier A, Portoso M, White ER, Morin MC, Margueron R, Jarriault S. 2014. Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science 345: 826–829. doi: 10.1126/science.1255885 [DOI] [PubMed] [Google Scholar]