Abstract
Brain tumor, a leading cause of uncontrolled cell growth in the central nervous system, presents substantial challenges in medical diagnosis and treatment. Early and accurate detection is essential for effective intervention. This study aims to enhance the detection and classification of brain tumors in Magnetic Resonance Imaging (MRI) scans using an innovative framework combining Vision Transformer (ViT) and Gated Recurrent Unit (GRU) models. We utilized primary MRI data from Bangabandhu Sheikh Mujib Medical College Hospital (BSMMCH) in Faridpur, Bangladesh. Our hybrid ViT-GRU model extracts essential features via ViT and identifies relationships between these features using GRU, addressing class imbalance and outperforming existing diagnostic methods. We extensively processed the dataset, and then trained the model using various optimizers (SGD, Adam, AdamW) and evaluated through rigorous 10-fold cross-validation. Additionally, we incorporated Explainable Artificial Intelligence (XAI) techniques-Attention Map, SHAP, and LIME-to enhance the interpretability of the model’s predictions. For the primary dataset BrTMHD-2023, the ViT-GRU model achieved precision, recall, and F1-score metrics of 97%. The highest accuracies obtained with SGD, Adam, and AdamW optimizers were 81.66%, 96.56%, and 98.97%, respectively. Our model outperformed existing Transfer Learning models by 1.26%, as validated through comparative analysis and cross-validation. The proposed model also shows excellent performances with another Brain Tumor Kaggle Dataset outperforming the existing research done on the same dataset with 96.08% accuracy. The proposed ViT-GRU framework significantly improves the detection and classification of brain tumors in MRI scans. The integration of XAI techniques enhances the model’s transparency and reliability, fostering trust among clinicians and facilitating clinical application. Future work will expand the dataset and apply findings to real-time diagnostic devices, advancing the field.
Keywords: Brain tumor, Deep learning, Pre-processing, ViT-GRU, Explainable Artificial Intelligence, Attention map
Subject terms: Medical research, Engineering
Introduction
The human brain consists of billions of neurons, synapses, and nerve cells that control many essential bodily functions. Like other bodily organs, some problems may arise in the brain, such as growths known as brain tumor. These tumors occur when cells grow abnormally in various regions of the brain and can lead to significant neurological complications and have significant consequences on a patient’s quality of life1. Brain tumors are widely acknowledged as among the most lethal diseases on a global scale, imposing considerable impact on mortality rates across all age groups. This condition brings challenges in terms of medical diagnosis and treatment2. These tumors manifest in various forms, including glioma, meningioma, pituitary, and cases with no tumor. With almost 120 varieties of tumors found to date, their diverse characteristics and dimensions make them difficult to detect, complicated by the intricate structure of the brain3. The identification and exact segmentation of tumorous areas, which include edema, necrotic centers, and tumorous tissues, is critical for accurate diagnosis and therapy planning4. The significance of the automatic detection and classification of medical images, particularly in brain tumor diagnosis, cannot be overstated. Early detection and categorization of brain tumors are critical for rapid treatment and improved patient outcomes. Traditional manual procedures for locating and classifying brain tumors in large medical image databases are time-consuming and resource-intensive5. Medical imaging modalities, including Magnetic Resonance Imaging (MRI), Computed Tomography (CT), and Positron Emission Tomography (PET) scans, are indispensable in the detection of brain tumors. MRI is the approach that is most frequently utilized in the investigation of brain structures. Therefore, MRI was selected as the preferred modality for our study due to its widespread utilization and efficacy in examining brain structures. Despite its ability to effectively model regions of interest, MRI has difficulties in accurately distinguishing tumors due to the volatility in intensity levels caused by varying machine settings. Multimodal MRI scans, including T1-weighted, T2-weighted, T5-weighted, and Fluid Attenuation Inversion Recovery (FLAIR) images, provide supplementary profiles for various glioma sub-regions, allowing for more accurate information extraction6,7. The manual segmentation of 3D MRI images conducted by health care professionals relies on the expertise as well as the experience of the physician. Even with the advancement in medical imaging approaches, automatically dividing brain tumors in multimodal MRI scans remained among the hardest issues in medical image processing. To prevent user-based difficulties, automatic classification techniques can be employed in medical images due to their measurable nature and ability to deliver dependable outcomes8. Brain tumor identification has been explored utilizing many machine-learning approaches and imaging types throughout the years9,10. In this context, Deep Learning (DL) approaches are up-and-coming brain tumor classification and segmentation tools. Their application in computer-aided systems for diagnosing brain tumors aims to provide accurate and reliable information regarding tumor presence, location, and type.
Existing brain tumor identification models face significant challenges, including performance issues, data handling inefficiencies, and limitations in model robustness and interpretability. Encouraged by these drawbacks, this study aims to develop an innovative approach for brain tumor identification using a combination of Vision Transformer (ViT) and Gated Recurrent Unit (GRU) models11. Our proposed ViT-GRU model addresses these issues by achieving higher accuracy, balancing data representation, and enhancing interpretability with Explainable Artificial Intelligence (XAI) techniques such as Shapley Additive Explanations (SHAP), Local Interpretable Model-Agnostic Explanations (LIME), and Attention mapping12. Additionally, it processes diverse data sources, which improves adaptability and relevance across various demographics and imaging methods. Thus, the hybrid ViT-GRU model addresses the limitations of previous models by utilizing the spatial attention mechanisms of ViTs and the temporal modeling capabilities of GRUs, providing a more robust and effective solution for brain tumor identification.
This study is motivated by the severe consequences of brain tumors, including death and costly, risky treatments. In Southern Bangladesh, particularly Faridpur, patients endure significant pain and adverse outcomes, highlighting the urgent need for improved detection and classification methods. To make a meaningful impact, the focus is on primary data collection, engaging directly with the community and healthcare providers to obtain specific information about brain tumor cases. The objective of this study is to implement a hybrid DL approach to enhance precision in detection and classification, addressing the challenges in brain tumor management. This comprehensive approach seeks to increase performance accuracy, facilitate early interventions, and improve the quality of life for individuals affected by brain tumors in Faridpur and similar regions.
In this research, the study tried to fulfill some research questions, including, How to increase the detection and classification accuracy of brain tumors?, How to enhance the overall performance of the proposed model?, How to reduce the training approach or model training time without compromising accuracy?, and How to make the model’s decision-making process more transparent and reliable?
In this system, the model’s structure incorporates multiple layers and adjustable parameters. Each MRI image goes through a series of ViT layers first, then through GRU layers for processing13,14. By integrating these components, we aim to improve feature representation and temporal modeling for robust decision-making in the final stage, making it a comprehensive and effective tool for accurate brain tumor detection and classification in medical imaging.
In this research, we achieved the following major contributions:
Our study uses hospital-based primary MRI data to analyze brain tumor patients and improve diagnosis.
We developed a hybrid ViT-GRU model that demonstrated superior results on our dataset, thereby enhancing the effectiveness of brain tumor diagnosis.
Additionally, we applied several preprocessing techniques on our primary dataset to optimize the performance of our proposed ViT-GRU model.
Finally, we employed three XAI techniques-Attention Map, SHAP, and LIME to offer interpretable and understandable explanations for the predictions made by the proposed model.
The structure of the rest of the paper is organized as follows: Section “Challenges of brain tumor disease detection with AI” describes the challenges associated with the identification of neurodegenerative diseases using AI. Section “Literature review” represents a thorough review of the most recent studies on identifying and classifying brain tumor disease. The comprehensive description of the materials and method employed in this study is presented in reference section “Materials and method”. Sections “Results” and “Discussion” provide an in-depth evaluation of the outcomes and findings in our study described as results and discussion. Section “Conclusion and future work” provides an extensive summary of the entire study.
Challenges of brain tumor disease detection with AI
The efficient integration of Artificial Intelligence (AI) into healthcare necessitates both transparency and efficacy, despite its immense potential. The utilization of primary MRI data in conjunction with Machine Learning (ML) and DL algorithms to detect brain tumors poses several challenges. Here are important considerations for brain tumor detection and classification associated with our whole research procedure.
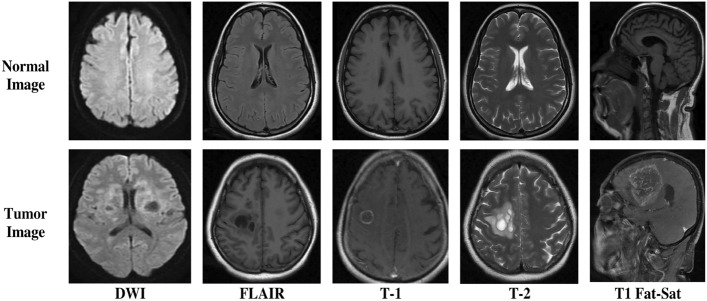
Complexity of Diverse Data: Multi-modal MRI datasets contain DWI, FLAIR, T1, T2, and T1 Fat-Sat images for brain information. Integrating these data types is complicated and requires study.
Data Preprocessing and Feature Extraction: In the case of preprocessing of primary MRI data, we face obstacles, including the need to standardize imaging protocols, mitigate artifacts, and reduce inter-subject variability. Feature extraction itself is challenging, requiring cautious selection to capture meaningful tumor characteristics.
Limited Availability of Annotated MRI data: Annotating primary MRI scans is a laborious and time-intensive technique. The lack of well-annotated data for training models may limit the development of robust algorithms for MRI datasets.
Class Imbalance and Tumor Rarity: The rarity of brain tumors in comparison to normal brain tissue within MRI datasets leads to class imbalance. This imbalance might result in biased models struggling to effectively detect the minority class (tumors), impacting overall performance.
Ethical and Legal Considerations in MRI Studies: The implementation of AI-based medical diagnostic systems employing MRI scans poses ethical and legal problems, including difficulties relating to patient privacy, permission, and potential biases in the algorithms. These concerns demand cautious attention and resolution.
Literature review
Early and precise identification of brain tumors is essential for optimal treatment. MRI scans play a crucial part in this procedure, however this might present challenges to interpret. DL has emerged as an efficient tool for brain tumor detection, and recent advances in the desire to automate and enhance the accuracy of brain tumor diagnosis are mentioned below.
An innovative Internet of Things (IoT) computational system in Ref.15 integrates a hybrid Convolutional Neural Network (CNN) and Long Short Term Memory (LSTM) approach for brain tumor detection and classification in MRI images which outperforms traditional CNN models, demonstrating superior accuracy in experiments conducted on a Kaggle dataset with 3264 MRI scans. Minimizing a lengthy training approach can increase the efficacy of this study. The authors16, introduce an innovative real-time intraoperative brain tumor diagnosis using stimulated Raman histology (SRH) and CNNs, obtaining a notable 94.6% accuracy, significantly reducing diagnosis time to under 150 seconds. The study suggests incorporating spectroscopic detection and investigating SRH’s role in molecular diagnosis. Researchers in Ref.17 proposed a cutting-edge, entirely automatic brain tumor segmentation and classification approach employing a multiscale Deep Convolutional Neural Network. Achieving a remarkable 97.3% accuracy on a dataset of 3064 MRI slices, which outperforms existing methods. However, dataset diversity, alternative architecture, and assessing the model may improve the results. According to the concept of Ref.18 this study represents a DL-based semantic segmentation method for automatic brain tumor segmentation on 3D BraTS datasets. Achieving a mean prediction ratio of 91.718% and excellent scores in IoU and BF metrics, the method exhibits accurate tumor prediction and 3D imaging. The authors in Ref.19 have comprehensively reviewed 147 recent studies on ML and DL approaches for detecting Alzheimer’s, brain lesions, epilepsy, and Parkinson’s. Analyzing 22 datasets, the study offers insights into effective methods of diagnosis, emphasizing future research directions and unresolved issues in brain disease diagnostics. This study highlights significant results, offering a foundational framework for future developments in the discipline. The author of the study20 explored glioma utilizing multimodal MRI scans, employing three 3D CNN architectures. This study obtained 5th place for segmentation and 2nd place for survival prediction in the 2018 MICCAI BraTS challenge among 60+ teams, obtaining a significant 61.0% accuracy in classifying survival categories. Diverse network architectures and training strategies may affect the further refinement of this study. The study21 employed a DL CNN model to automatically detect brain tumors using augmented MRI data from the Br35H dataset, achieving 98.99% accuracy. Future research could explore advanced architectures and integrate multimodal data to improve the interpretability of this study for more robust clinical applications in brain tumor diagnostics.
In Ref.22, the author of this study employs artificial intelligence (AI) techniques and pre-trained models (Xception, ResNet50, InceptionV3, VGG16, MobileNet) to enhance MRI-based brain tumor classification, accomplishing F1-scores ranging from 97.25% to 98.75%. Investigating biases, fine-tuning strategies, investigating hyperparameters, and expanding datasets may improve the efficiency of the study. The author of this paper23 proposed a CNN-based DL model for brain tumor classification using MRI images, achieving high accuracies of 96.13% and 98.7% across two studies. Improving model generalizability, benchmarking against existing methods, and enhancing interpretability for clinical applications may increase the efficacy of this study. Researchers introduce a strategy24 employing 3D U-Net Design and ResNet50 models for accurate brain tumor segmentation in MRI and CT scans, achieving high accuracies of 98.96% and 97.99%. Future work in this paper suggests that model enhancement, real-time training, and addressing class imbalance scenarios may yield better results. The paper1 presents a hybrid model for precise brain tumor classification in MRI images, integrating a novel CNN model with optimized ML algorithms. Achieving an exceptional mean accuracy of 97.15%, the proposed hybrid model outperforms advanced CNN models, exhibiting higher time efficiency. Exploring diverse ML algorithms and DL models and validating the hybrid model on larger and more diverse datasets may lead to better performance. In Ref.25, the author represented a modified U-Net structure for accurate brain tumor segmentation in MRI images, incorporating shuffling and sub-pixel convolution. Achieving notable accuracy of 93.40% and 92.20% on BraTS Challenge datasets, the proposed model outperforms existing approaches. Larger datasets and smooth computational assessment may assist in investigating the model’s generalizability and may become more efficient for diverse medical image segmentation tasks. The author of this paper26 utilized feature fusion and majority voting by introducing a class-weighted focal loss and working on imbalanced brain tumor classification datasets, yielding substantial improvements over conventional CNNs. Alternative fusion techniques and computational efficiency can overcome challenges in class weight determination and increase accuracy. In study7, the authors presented a robust brain tumor segmentation framework utilizing four MRI sequence images and an optimized CNN model. Demonstrating superior performance on the BRATS 2018 dataset with notable precision, recall, and Dice Score, the proposed method underscores its efficacy. Addressing additional pre-processing strategy exploration, scalability issues, and clinical challenges may contribute to an improvement in the model’s performance. The author of the study27 presented a DL fusion model for brain tumor classification, leveraging MRI images with a notable accuracy of 98.98%. The model incorporates features extracted from VGG16, ResNet50, and convolutional deep belief networks (CDBNs). However, enhancing data augmentation strategies and evaluating the model’s applicability across various clinical datasets and imaging techniques could further enhance its efficacy. In this paper28, the authors utilized the FPCIFHSS model to assess brain tumor susceptibility with complex intuitionistic fuzzy numbers (CIFNs), offering a versatile approach for managing diagnostic uncertainties, especially beneficial in resource-limited settings. Future studies should simplify the model for practical clinical application and validate its effectiveness through thorough comparative analyses with established diagnostic methods. The paper29 proposed two models: XGBoost achieved 98.620% accuracy in differentiating cancer stages using multi-omics data from the Cancer Genome Atlas, while a cascading Deep Forest ensemble accurately identified cancer subtypes from the METABRIC dataset (83.45% for five subtypes, 77.55% for ten). These advancements highlight the potential of AI in omics data analysis. However, future research should focus on refining AI algorithms, improving preprocessing techniques, validating models across diverse datasets, and conducting rigorous clinical trials for robust validation.
Previous models for identifying brain tumors have challenges such as low accuracy, class imbalance, large computing demands, poor interpretability, and limited generalizability. Our ViT-GRU model overcomes these difficulties, delivering greater accuracy and addressing class imbalance while enhancing interpretability using XAI techniques like LIME, SHAP, and Attention mapping. It also handles data from diverse sources, making it more adaptable and relevant across varied demographics and imaging methods.
Materials and method
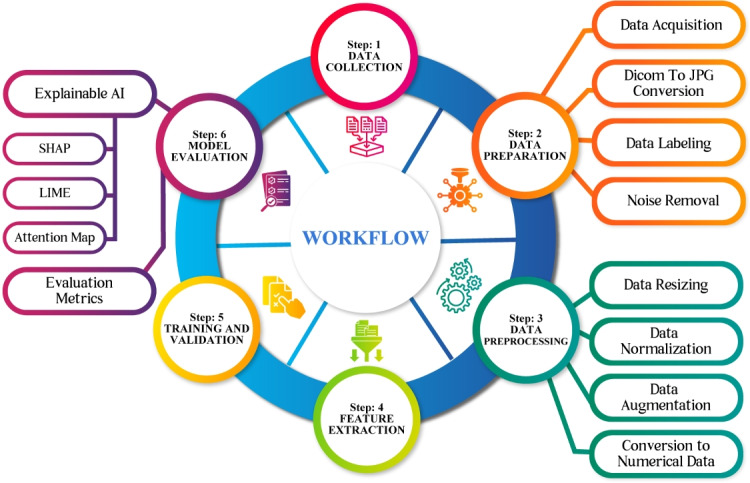
The methodology employed in this research consists of a meticulous and methodical process, which is depicted in Fig. 1, which provides a comprehensive description of the research methodology utilized in this study.
Fig. 1.
Flowchart of the proposed methodology illustrating the distinct phases involved in the research approach.
Dataset collection
In this research, we compiled a dataset named Brain Tumor MRI Hospital Data 2023 (BrTMHD-2023), consisting of 1166 MRI scans collected at Bangabandhu Sheikh Mujib Medical College Hospital (BSMMCH) in Faridpur, Bangladesh, over the period from January 1, 2023, to December 30, 2023. Our BrTMHD-2023 dataset contains different kinds of tumors and normal conditions, with images acquired from a Siemens Magnetom Skyra MR scanner for clarity and quality across Diffusion-Weighted Imaging (DWI), FLAIR, T1-weighted (T1), T2-weighted (T2), and T1 Fat-Sat categories. Legal regulations were followed, and informed consent was obtained from patients. Though the sharing of data is restricted for privacy, it is available upon request with ethical approval to assist in advancing brain tumor diagnosis and treatment. Furthermore, we confirm that all data collection and analysis methods adhered to the Declaration of Helsinki and Bangladesh Medical Research Council (BMRC) ethical guidelines.
A few samples of each category DWI, FLAIR, T1, T2- weighted, and T1 Fat-Sat from the dataset are shown in Fig. 2. This research also utilized an additional publicly available Brain Tumor Kaggle Dataset consisting 256 MRI images to evaluate the proposed model.
Fig. 2.
Sample images of the BrTMHD-2023 Dataset.
The dataset specifications are shown in Table 1.
Table 1.
Datasets of brain tumor disease utilized in this study.
| Dataset | Label of the dataset | Number of total collected patient | No of disease patient | No of MRI image | No of MRI image after augmentation | Male tumor patient | Female tumor patient | Total MRI image | Total MRI image after augmentation |
|---|---|---|---|---|---|---|---|---|---|
| BrTMHD-2023 | Normal patient | 232 | 163 | 945 | 1210 | 61 | 102 | 1166 | 2190 |
| Tumor patient | 69 | 221 | 980 | 45 | 24 | ||||
|
Brain tumor Kaggle dataset |
Normal | – | – | 156 | – | – | – | 256 | – |
| Tumor | – | 100 | – | – | – |
The dataset employs multimodal imaging protocol MRI scans, which ensures a consistent and standardized data acquisition approach. This makes the data in this research more relatable and trustworthy.
Data preparation
After acquiring MRI data from the medical facility, the samples were assembled using image reconstruction approaches and then optimized acquisition parameters for T1, T2-weighted, and other multimodal images30. We transformed DICOM files to JPG using MicroDicom for compatibility with image-processing tools and then performed enhancement and denoising31. Qualified radiologists classified the images into tumor and non-tumor types, ensuring high-quality data for ML training. Noise reduction techniques like filtering and advanced reconstruction were implemented to enhance the signal-to-noise ratio and image quality for accurate diagnosis32,33.
Data preprossessing
Preprocessing MRI images elevates their quality for DL analysis while retaining their integrity. We employed motion correction, data resizing, data normalization, data augmentation, and conversion to numerical data34. Motion artifacts were handled Utilizing retrospective and prospective motion correction approaches35. Dimensionality reduction enhances the computing efficiency and our model performance36. Pixel normalization between 0 and 1 improved feature extraction accuracy. To address the data imbalance caused by the unequal ratio of diseased to normal MRI images, we employed data augmentation techniques. This expanded our dataset to 2,190 MRI images, mitigating the scarcity and improving the learning and performance of our proposed model. Converting grayscale photos to RGB representation allowed us to apply pre-trained models, enhancing performance. These preprocessing methods assure the optimal results in the following image classification tasks37,38.
Proposed ViT-GRU model
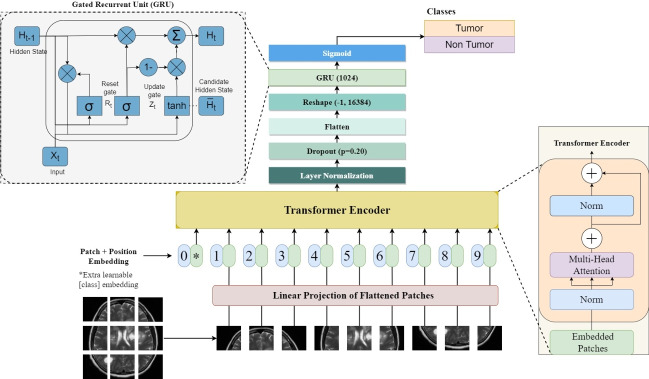
The proposed model called the hybrid ViT-GRU model, is for analyzing brain tumor detection and classification as illustrated in Fig. 3. Particularly in its utilization of ViT, this hybrid DL technique for classifying brain tumors exhibits an apparent connection to the original Transformer architecture39.
Fig. 3.
Architecture of the hybrid ViT-GRU model showing the interconnected components and their functional relationship in the proposed model.
ViT for feature extraction
This study builds upon the sophisticated ViT framework, which is renowned for its proficiency in acquiring visual data employing self-attention mechanisms; this architecture has been specifically fine-tuned to identify brain tumors from MRI scans. By exploiting the ViT backbone, our model excels at extracting hierarchical representations that show valuable patterns characteristic of brain tumors. Compared to traditional CNNs, the ViT utilizes self-attention mechanisms, which allow it to capture long-range dependencies and global context more effectively. To improve the ViT encoder for this purpose, we introduced strategic modifications. Firstly, we removed the Multi-Layer Perceptron (MLP) layer to increase computational efficiency and introduced layer normalization to accelerate convergence and stabilize training13. This adjustment contrasts with CNNs, where the dense layers are often a source of high computational load. Then, we added a dropout layer that reduces overfitting and enhances generalization. Finally, Incorporating a “flatten” layer improves the model’s ability to distinguish varied visual patterns in MRI data. These adjustments jointly increase the model’s reliability and effectiveness in brain tumor identification40,41.
GRU for temporal analysis
In this approach to the temporal analysis of brain MRI images, we incorporate a GRU, a specialized form of the neural network, after the extraction of critical features from the images. GRUs, which were initially proposed by Ref.42, can be likened to an enhanced iteration of conventional Recurrent Neural Networks (RNNs). They are capable of recognizing patterns and establishing relationships within data sequences. GRUs have these neat features called update and reset gates that help them manage information better. By using a GRU with 1024 units in our architecture, we take advantage of its ability to comprehend changes over time in brain MRI images. This allows us to identify better and classify brain tumors. The GRU was chosen over other RNNs like LSTM for its simpler architecture and faster training while maintaining comparable performance in capturing dependencies. LSTM represents a type of RNN architecture with two crucial components: an input gate and a forget gate. These components play a vital role in regulating the flow of information within the network. The input gate determines which input information is stored in the memory cell, while the forget gate decides which information should be discarded. GRUs, which have an essential function in a variety of Natural Language Processing (NLP) operations such as recognition of speech, machine translation, and language modeling, play an indispensable part in our methodology, which allows the model to distinguish and categorize brain tumors with improved precision and efficacy43.
Classification head
Once the feature extraction phase from the ViT and sequential data analysis of GRU architectures are finished, the final prediction is generated incorporating a classification head. The ViT-GRU-based process utilizes the final hidden state of the GRU unit as the input for a series of fully connected layers. The process of converting the hidden information into a probability distribution over the available categories is carried out by these layers through a sigmoid activation function. By utilizing the extracted features, the incorporation of this classification head substantially improves the model’s ability to generate precise predictions, which allows efficient classification within ViT-GRU frameworks.
Hyperparameters settings
In this study, we trained models with various configurations that enabled us to adapt how the model works in various manners. Our main focus was on hyperparameter tuning, a crucial phase for optimizing model performance. This involved finding the best hyperparameter values for our model, using a mix of usual settings, and trying out new approaches to make our model better at evaluating and predicting things. To compare the model parameters, we initially set them sequentially, adjusting one at a time while keeping others constant. After this systematic process, we identified the best-fitting parameters based on performance metrics. These optimized parameters were then used for our final analysis. Table 2 presents a comprehensive list of all the hyperparameters that were used in our ViT-GRU model. Pre-trained transfer learning model parameters were also selected using the same procedure.
Table 2.
Hyperparameters of our proposed ViT-GRU model with their values.
| Hyperparameters | Values |
|---|---|
| Epochs | 35 |
| Batch size | 64 |
| Image size | 224×224×3 |
| Learning rate | 0.0001 |
| Weight decay | 0.0001 |
| Optimizer | AdamW, Adam, SGD |
| Loss function | Categorical cross-entropy |
| Patch size | 8 |
| Number of patches | 256 |
| Projection dimension | 64 |
| Number of parallel self-attention heads | 4 |
| Number of transformer encoder layers | 8 |
The training was carried out on Google Colab by using a GPU with 12 GB of RAM.
Evaluation metrics
The proposed model’s performance is evaluated using various metrics, including accuracy, precision, recall, and F1-score. These measurements are calculated from the data provided in the confusion matrix, like True Positive, True Negative, False Positive, and False Negative, showing the model’s progress. We have used the following equations for measuring the performance of our proposed ViT-GRU model:
| 1 |
| 2 |
| 3 |
| 4 |
Explainable artificial intelligence
XAI is important for transparent and understandable AI systems and crucial in healthcare where AI judgements impact patient results44. As a pioneer in medical image categorization, XAI facilitates human interpretation of complex AI decisions. Particularly in brain tumor detection, techniques such as LIME, SHAP, and Attention Maps improve the interpretability of DL models13. In our study, we integrated XAI techniques into the proposed model to enhance its interpretability and provide valuable insights into its decision-making process. This integration allows healthcare practitioners to understand how the model arrives at its predictions, thereby building trust and facilitating better diagnostic and treatment decisions.
LIME as XAI
LIME interprets ML model choices, particularly advantageous for “black box” DL architectures45. Its fundamental function lies in providing perceptive heatmaps, emphasizing crucial locations affecting model predictions. The application of LIME resulted in intuitive heatmaps that highlighted key regions within MRI images critical for the model’s predictions, thereby enhancing the interpretability of our DL model. These heatmaps enabled us to visualize significant areas influencing the model’s predictions, providing valuable insights into its decision-making process. This integration facilitated a clearer understanding of the model’s decisions, thereby supporting more informed clinical judgments by healthcare professionals. By assigning relevance scores to pixels or image areas, LIME delivers a thorough comprehension of the key characteristics guiding categorization necessary for grasping intricate model behavior46. This process can be expressed by the following formula:
| 5 |
f(x) is the interpretable model’s projection for the image instance x.
N(x) shows the immediate vicinity of the image x.
L(.) is a loss function assessing the difference between the estimates of the interpretable model f(x) and the original model’s estimate in the surrounding area N(x).
is a normalization term.
SHAP as XAI
SHAP elucidates ML model outputs, specifically applying Shapley values to clarify feature contributions. Crucial for medical image categorization, SHAP indicates critical regions within images vital for model judgments47. We integrated SHAP into the model to calculate Shapley values for each pixel in the MRI images, identifying and visualizing the most critical regions influencing classification results. It graphically depicts essential feature values, which is helpful in predicting particular categories. Our application in brain tumor detection gave insights into critical brain areas, combining LIME explanations for a thorough comprehension of data patterns.
Attention map as XAI
Attention maps depict DL model focus areas in input pictures, boosting interpretability by illuminating key regions48. In medical image identification, they are essential for detecting tumor-impacted brain regions. We integrated attention maps by leveraging the multi-head attention layer in the proposed model, which highlights areas with high activation that contributed to the final classification. Leveraging the last multi-head attention layer to generate attention maps provided crucial insights into the model’s decision-making by highlighting key image regions and enhancing interpretability, thus emphasizing the need for further refinement to maximize patient care improvements49. Visualizing these maps considerably bolsters comprehension of DL model operations in sophisticated AI-driven assessments in healthcare.
Results
We conducted three experiments to assess our model’s performance. In experiment 1, We employed the primary BrTMHD-2023 dataset and applied a rigorous 10-fold cross-validation technique to verify the efficacy of our model during our evaluation. Experiment 2 involved a holdout analysis, on the BrTMHD-2023 dataset without k-fold validation. Finally, in experiment 3, we validated our model on an additional Brain Tumor Kaggle dataset to demonstrate its generalizability.
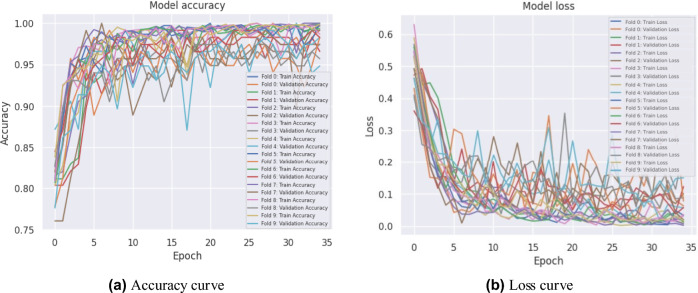
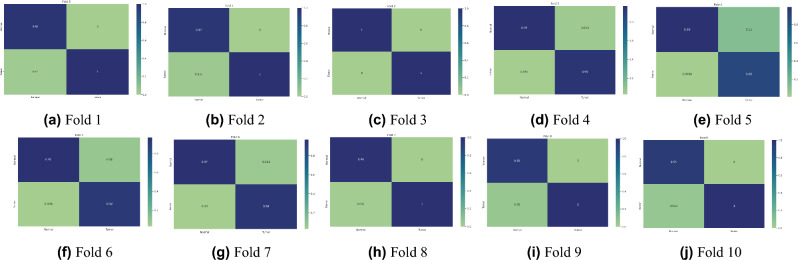
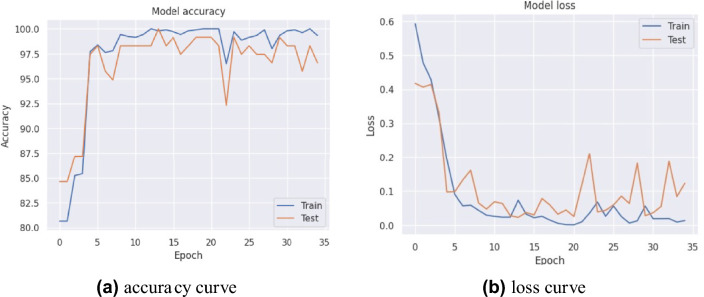
In experiment 1, our proposed model obtained the highest average accuracy of 98.97%, an average precision score of 97%, highlighting its accuracy in identifying positive instances. The model’s efficacy in accurately distinguishing true positives and minimizing false negatives was also demonstrated by its 97% average recall rate. The model’s dependable performance was affirmed by the F1-score, which achieved an exceptional average value of 97% by balancing precision and recall. Furthermore, we introduced various pre-trained transfer learning models into our study to investigate the training and performance capabilities of ML models. The results, summarized in Table 3, indicate that our model performed exceptionally well. Figure 4 illustrates the training accuracy and loss curves across various folds, highlighting the model’s learning trajectory. Figure 5 depicts the AUC-ROC contours for each fold, demonstrating the model’s class discrimination capability. Figure 6 presents the confusion matrices, providing a detailed analysis of each fold’s classification performance.
Table 3.
Results of 10-fold cross-validation in our proposed ViT-GRU model.
| Number of fold |
Precision (%) |
Recall (%) |
F1-score (%) |
Training loss (msec) |
Test loss (msec) |
Training acuracy (%) |
Test accuracy (%) |
|---|---|---|---|---|---|---|---|
| 1 | 98 | 98 | 98 | 0.0042 | 0.0107 | 100 | 100 |
| 2 | 98 | 97 | 97 | 0.0082 | 0.0307 | 99.9 | 99.15 |
| 3 | 100 | 100 | 100 | 0.0026 | 0.0688 | 100 | 97.44 |
| 4 | 97 | 97 | 97 | 0.0098 | 0.0378 | 99.81 | 99.15 |
| 5 | 98 | 97 | 97 | 0.0021 | 0.0153 | 100 | 100 |
| 6 | 96 | 96 | 96 | 0.0044 | 0.0478 | 100 | 98.29 |
| 7 | 96 | 96 | 96 | 0.0042 | 0.0374 | 99.81 | 99.14 |
| 8 | 99 | 99 | 99 | 0.0034 | 0.0097 | 99.9 | 100 |
| 9 | 96 | 96 | 95 | 0.0035 | 0.0562 | 100 | 98.28 |
| 10 | 96 | 96 | 96 | 0.0139 | 0.0591 | 99.9 | 98.28 |
| Average | 97 | 97 | 97 | 0.0056 | 0.0374 | 99.93 | 98.97 |
| STDEV | 0.014 | 0.013 | 0.015 | 0.004 | 0.021 | 0.079 | 0.884 |
Significant values are in bold.
Fig. 4.
Accuracy and loss curves for 10-fold cross-validation of the proposed ViT-GRU model.
Fig. 5.
AUC-ROC curves for 10-fold cross-validation of the proposed ViT-GRU model.
Fig. 6.
Confusion matrices for 10-fold cross-validation of the proposed ViT-GRU model.
In experiment 2, we adopted three different optimizers into our proposed ViT-GRU model, with the AdamW optimizer attaining the highest accuracy of 98.97%. This high mean test accuracy underscores the model’s precision in classifying data. Table 4 contrasts the results of multiple models with our proposed model using distinct optimizers. Additionally, Table 5 provides a comprehensive comparison of important approaches used for classifying brain tumors that focus on important factors such as the dataset used, number of classes, classification algorithm applied, image count, and performance measures.
Table 4.
Comparing the results of pre-trained transfer learning models with the proposed ViT-GRU on the AdamW, Adam, and SGD optimizer.
| Model | Optimizer | Precision (%) | Recall (%) | F1-score (%) | Training loss | Validation loss | Trainig time (s) |
Training accuracy (%) | Test accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|
| ResNet18 | AdamW | 91 | 91 | 90 | 0.0113 | 0.1577 | 5.83 | 99.88 | 94.29 |
| Adam | 94 | 93 | 93 | 0.0124 | 0.1193 | 6.07 | 99.88 | 96.86 | |
| SGD | 65 | 81 | 72 | 1.4124 | 1.4792 | 5.63 | 81.25 | 80.57 | |
| ResNet50 | AdamW | 91 | 91 | 91 | 0.0435 | 0.2218 | 12.94 | 98.65 | 91.43 |
| Adam | 92 | 92 | 92 | 0.0524 | 0.2454 | 13.03 | 98.65 | 92.86 | |
| SGD | 63 | 79 | 70 | 0.5341 | 0.5443 | 12.64 | 81.74 | 79.43 | |
| VGG16 | AdamW | 91 | 92 | 91 | 0.0588 | 0.1521 | 18.41 | 97.43 | 93.71 |
| Adam | 93 | 93 | 93 | 0.0547 | 0.1415 | 18.9 | 99.02 | 94 | |
| SGD | 72 | 81 | 75 | 0.5593 | 0.5347 | 17.73 | 78.55 | 81.43 | |
| VGG19 | AdamW | 92 | 91 | 91 | 0.0540 | 0.1755 | 20.92 | 98.28 | 94.29 |
| Adam | 91 | 91 | 91 | 0.0540 | 0.1755 | 20.92 | 98.9 | 94.29 | |
| SGD | 72 | 79 | 73 | 0.5951 | 0.5601 | 20.55 | 78.8 | 78.29 | |
| EfficientNet_V2_l | AdamW | 91 | 91 | 91 | 0.0723 | 0.1646 | 17.36 | 96.56 | 92.65 |
| Adam | 89 | 89 | 89 | 0.1131 | 0.2954 | 14.61 | 95.83 | 91.71 | |
| SGD | 68 | 76 | 72 | 6.0248 | 6.0263 | 14.57 | 50.86 | 51.14 | |
| MobileNet_V2 | AdamW | 85 | 86 | 85 | 0.2885 | 0.3368 | 6.43 | 90.2 | 85.14 |
| Adam | 80 | 83 | 80 | 0.2819 | 0.3643 | 6.54 | 88.97 | 83.71 | |
| SGD | 74 | 84 | 77 | 3.6244 | 2.5056 | 11.82 | 82.27 | 84.86 | |
| DenseNet121 | AdamW | 97 | 97 | 97 | 0.0182 | 0.0892 | 13.87 | 99.88 | 97.71 |
| Adam | 93 | 93 | 93 | 0.0311 | 0.1541 | 13.93 | 99.88 | 96.29 | |
| SGD | 72 | 83 | 73 | 2.7244 | 2.6056 | 13.82 | 80.27 | 82.86 | |
| AlexNet | AdamW | 86 | 87 | 86 | 0.1983 | 0.2656 | 4.57 | 92.89 | 89.43 |
| Adam | 89 | 89 | 88 | 0.1451 | 0.2399 | 4.05 | 96.08 | 91.14 | |
| SGD | 71 | 84 | 77 | 6.8541 | 6.8497 | 4.29 | 79.78 | 84 | |
| ViT-GRU | AdamW | 97 | 97 | 97 | 0.0056 | 0.0374 | 47.93 | 99.93 | 98.97 |
| Adam | 96 | 95 | 95 | 0.0089 | 0.1025 | 49.28 | 99.75 | 96.56 | |
| SGD | 66 | 81 | 73 | 0.4695 | 0.4606 | 48.3 | 81.02 | 81.66 |
Table 5.
Performance evaluation of existing state-of-the-art models and the proposed model.
| Study | Year | Model | Dataset | Image modalities | Number of classes | Number of images | Performance | XAI |
|---|---|---|---|---|---|---|---|---|
| Choudhury et al.50 | 2020 | CNN |
Brain tumor Kaggle dataset |
MRI | 2 | – |
Accuracy: 96.08% Precision: 94.87% Recall: 98.63% F1-score: 97.36% |
Not used |
| Saleh et al.22 | 2020 |
Pre-trained DL models |
Brain tumor Kaggle dataset |
MRI | 4 | 4480 |
Accuracy: 97.25-98.75% Precision: – Recall: – F1-score: 100% |
Not used |
| Bhanothu et al.51 | 2020 | Faster R-CNN |
Brain tumor Figshare dataset |
MRI | 3 | 2406 |
Accuracy: – Precision: 77.6% Recall: 80% F1-score: – |
Not used |
| Waghmare et al.52 | 2021 |
fine-tuned VGG-16 CNN |
Brain tumor Kaggle dataset |
MRI | 3 | 3664 |
Accuracy: 95.71% Precision: – Recall: 83.28% F1-score: – |
Not used |
| Díaz-Pernas et al.17 | 2021 | DCNN | BRATS 2013 | MRI | 3 | 3064 |
Accuracy: 97.3% Precision: – Recall: F1-score: |
Not used |
| Khairandish et al.53 | 2022 | CNN-SVM | BRATS 2015 | MRI | 2 | 230 |
Accuracy: 98.49% Precision: – Recall: – F1-score: – |
Not used |
| Mohan et al.54 | 2022 | DLBTDC-MRI |
BRATS2015 dataset |
MRI | 4 | 42,470 |
Accuracy: 98.145% Precision: 95.134% Recall: 97.348% F1-score: 96.25% |
Not used |
| Srinivas et al.55 | 2022 | VGG-16 |
Kaggle dataset |
MRI | 2 | 256 |
Accuracy: 96% Precision: 94% Recall: 100% F1-score: 98% |
Not used |
| Montaha et al.56 | 2022 | TD-CNN-LSTM |
BraTS 2018, BraTS 2019 and BraTS 2020 |
MRI | 2 | 978 |
Accuracy: 98.90% Precision: 98.95% Recall: 98.78% F1-score: 98.83% |
Not used |
| Hossain et al.24 | 2022 | ResNet50 |
TGCA-GBM dataset |
MRI and CT scans |
3 | 3064 |
Accuracy: 98.96% Precision: 98.67% Recall: 99.12% F1-score: 98.87% |
Not used |
| Vankdothu et. al.15 | 2022 | CNN-LSTM |
Brain tumor kaggle dataset |
MRI | 4 | 3264 |
Accuracy: 92% Precision: 96% Recall: 98.5% F1-score: – |
Not used |
| Rasheed et al.57 | 2023 | CNN |
Brain tumor Kaggle dataset |
MRI | 3 | 3064 |
Accuracy: 98.04% Precision: 98% Recall: 98% F1-score: 98% |
Not used |
| Abdusalomov et al.58 | 2023 | YOLOv7 |
Brain tumor Kaggle dataset |
MRI | 4 | 10,288 |
Accuracy: 99.5% Precision: 99.5% Recall: 99.3% F1-score: 99.4% |
Not used |
| Meena, Gaurav, et al.21 | 2024 | InceptionV3 | kaggle | – | 2 | 228 |
Accuracy: 98% Precision: – Recall: – F1-score: – |
Not used |
| BrTMHD-2023 | MRI | 2 | 1166 |
Accuracy: 98.97% Precision: 97% Recall: 97% F1-score: 97% |
Used | |||
| Proposed study | 2024 | ViT-GRU | Brain Tumor Kaggle Dataset | MRI | 2 | 256 |
Accuracy: 96.08% Precision: 97% Recall: 96% F1-score: 96% |
Not used |
Here, the model we used demonstrated consistently high performance, achieving a precision, recall, and F1-score of 97% each.
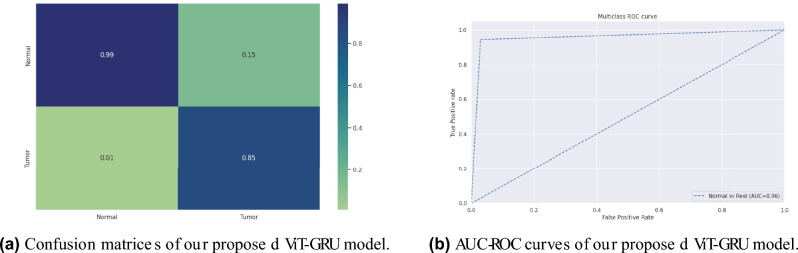
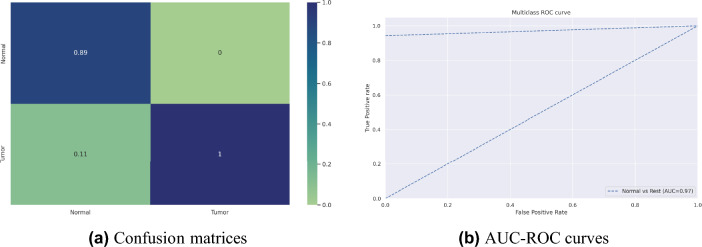
Our proposed model’s accuracy and loss curves are represented in Fig. 7. Visual analyses provided in Fig. 9a extend deeper into the confusion metrics details, and Fig. 9b displays the AUC-ROC curves. These visuals collectively emphasize the model’s exceptional performance across diverse testing circumstances.
Fig. 7.
Accuracy and loss curves analysis obtained from the proposed ViT-GRU model for BrTMHD-2023 dataset.
Fig. 9.
Confusion matrices and AUC-ROC curves of our proposed ViT-GRU model for BrTMHD-2023 dataset.
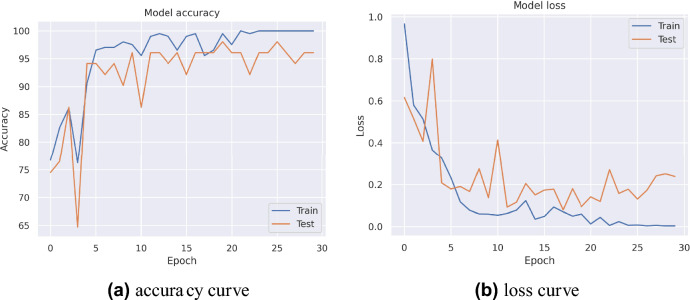
In experiment 3, the model demonstrated exceptional results, achieving an accuracy of 96.08%, a precision of 97%, a recall of 96%, and an F1 score of 96% on the Brain Tumor Kaggle dataset as presented in Table 5. This evaluation’s accuracy and loss curves are presented in Fig. 8. Also, the confusion matrices and the AUC-ROC curve are illustrated in Fig. 10.
Fig. 8.
Accuracy and loss curves analysis obtained from the proposed ViT-GRU model for Brain Tumor Kaggle Dataset.
Fig. 10.
Confusion matrices and AUC-ROC curves of our proposed ViT-GRU model for Brain Tumor Kaggle Dataset.
Discussion
This study provides a new framework combining a ViT with a GRU network for brain tumor assessment in MRI images. The hybrid ViT-GRU model employs ViT for regional feature extraction and GRU for temporal contextual assessment, increasing classification accuracy by collecting both spatial and temporal properties.
In our study, 10-fold cross-validation enhanced the model’s robustness and reliability, achieving an average 98.97% accuracy due to high-quality data, effective preprocessing, hyperparameter tuning, resulting in high precision, recall, and F1-scores of 97% respectively. In experiment 2, our proposed model showed excellent classification accuracy (98.97% with AdamW, 96.56% with Adam, and 81.66% with SGD). This study also evaluates our dataset using various pre-trained transfer learning models, including DenseNet121, ResNet18, and VGG19. After completing the necessary preprocessing techniques and parameter tuning, these models achieved the highest accuracies of 97.71%, 96.86%, and 94.29%, respectively. Our proposed model beat these pre-trained models in important parameters such as precision, recall, and F1-score and significantly shows 1.26% better accuracy than the highest performed TL models. We enhanced these models with hyperparameter adjustments and layer alterations. In addition, we presented and assessed our suggested model, which performed better than any other model in every metric. Table 4 presents the comparative results for AdamW, Adam, and SGD optimizers. The impact of each optimizer on the model’s performance is compared, and AdamW achieves higher accuracies by decoupling weight decay from gradient updates, enhancing model generalization, and allowing for better control over learning rates and regularization, resulting in improved overall metrics. Additionally, this study comprehensively compares existing technologies and previous work, which is shown in Table 5 with a detailed state-of-the-art comparison, highlighting how our hybrid model achieves better results across various metrics. The proposed model achieved high AUC-ROC scores of 96% on the BrTMHD-2023 dataset and 97% on the Brain Tumor Kaggle Dataset, demonstrating its robust effectiveness in brain tumor detection and classification, which is shown in Figs. 9b and 10b, respectively.
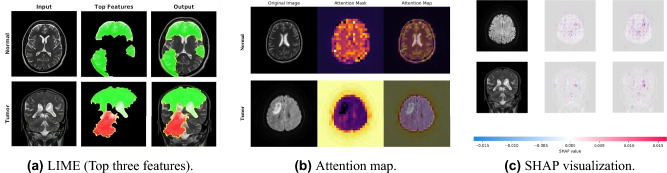
The innovativeness of this research lies in leveraging primary data with the development of the hybrid model, offering distinct advantages, including enhanced model interpretability through XAI techniques like LIME, SHAP, and Attention maps. This increased transparency supports clinicians in understanding the model’s judgments, thereby aligning our model with clinical expertise and improving its applicability in healthcare settings. Using XAI approaches like LIME, SHAP, and Attention maps, we increased model interpretability, emphasizing crucial aspects like white matter lesions and cortical atrophy, thereby supporting clinicians in comprehending the model’s judgments. The corresponding radiologist ensures and provides positive feedback about our model’s decision-making ability with the integration of XAI, allowing them to understand and trust the process that ensures the reliability and compatibility of the model. This transparency creates confidence and assures alignment with clinical expertise, boosting real-world application in healthcare. Our results, shown in numerous figures, support the model’s capacity to appropriately reflect brain tumor pathophysiology.
The results are shown in Fig. 11a, where each figure represents the LIME analytical assessment of our proposed model using sample images. Furthermore, SHAP analysis findings are shown in Fig. 11c, which again aligns with our suggested model and includes sample images. Lastly, sample images from the attention map analysis are used in Fig. 11b to demonstrate the results.
Fig. 11.
Visualization of XAI Techniques.
Despite the promising results, this study faces several limitations. The complexity of integrating multi-modal MRI data, challenges in data preprocessing and feature extraction, and the limited availability of well-annotated MRI scans impact the robustness of our model. Additionally, class imbalance due to the rarity of brain tumors and ethical and legal considerations related to AI implementation in medical diagnostics necessitate careful attention. Addressing these limitations in future research could further enhance the accuracy and applicability of AI-based brain tumor detection systems.
Conclusion and future work
In summary, this study introduces and examines the effectiveness of our newly developed ViT-GRU model for the detection and classification of brain tumors. We assessed our model’s performance by benchmarking it against several pre-trained models employing transfer learning using three distinct optimizers. Our ViT-GRU model achieved a remarkable accuracy of 98.97% using a 10-fold cross-validation technique. When tested on the BrTMHD-2023 dataset without cross-validation, it outperformed the top transfer learning model by 1.26% in accuracy. Additionally, in another Kaggle benchmark dataset, our model achieved an accuracy of 96.08%, exceeding previous results for this dataset. These results demonstrate that our model surpasses these existing models in accuracy, precision, recall, and F1-score metrics for MRI image analysis. The proposed model combines the regional feature extraction capabilities of ViT with the temporal contextual assessment strengths of GRU, leading to superior performance in comparison to several pre-trained models such as ResNet and VGG. Our model demonstrated exceptional accuracy, precision, recall, and F1-score metrics, particularly when optimized with the AdamW algorithm, achieving an accuracy of 98.97%. These theoretical advancements underscore the potential of hybrid models in enhancing diagnostic accuracy by integrating spatial and temporal data features. The practical implications of our research are further supported by extensive visual analyses, including accuracy and loss curves, confusion matrices, and AUC-ROC curves, which collectively highlight the model’s robust performance and reliability in diverse testing scenarios. Moreover, this study utilizes XAI techniques to increase the interpretability and transparency of the ViT-GRU model. We further illustrated the performance of our proposed model through accuracy and loss curves, confusion matrices, and AUC-ROC curves. The research incorporated primary MRI data collected from a hospital, which underwent preprocessing to prepare for analysis. Our approach combines the ViT with GRU and integrates the latest developments in Graph Neural Networks to effectively categorize brain tumor, deliver clinicians with essential information, and lead the way for efficient clinical implementations. The proposed study of this research demonstrates potential in facilitating a prompt response and enhancing the quality of life through early and precise diagnosis of brain tumor.
Future research should focus on expanding the dataset to improve model performance and generalizability across diverse patient populations and clinical settings. Additionally, integrating the ViT-GRU model into real-time diagnostic devices should be investigated to assess its practical utility in clinical environments, potentially transforming diagnostic workflows. Lastly, applying the model to other medical imaging tasks can validate its versatility and robustness, extending its utility beyond brain tumor classification.
Acknowledgements
We would like to acknowledge the support provided by the Bio-Imaging Research Lab, Department of Biomedical Engineering, Islamic University, Kushtia-7003, Bangladesh, in carrying out our research successfully.
Author contributions
Md. Mahfuz Ahmed: Conceptualization, Formal analysis, Methodology, Software, Data curation, Validation, Writing—Original Draft, Review & Editing. Md. Maruf Hossain: Methodology, Software, Validation, Writing - Review & Editing. Md Rakibul Islam: Data curation, Writing, Validation.Md. Shahin Ali: Methodology, Writing—Review & Editing. Abdullah Al Nomaan Nafi: Data curation, Writing, Validation, Visualization. Md Faisal Ahammed Data labeling, Validation. Kazi Mowdud Ahmed: Methodology, Writing—Review & Editing. Md Sipon Miah: Methodology, Writing—Review & Editing. Md Mahbubur Rahman: Methodology, Writing—Review & Editing, and Supervision. Mingbo Niu: Writing—Review & Editing, and Project Administration. Md Khairul Islam: Methodology, Formal analysis, Validation, Writing - Review & Editing, and Supervision.
Data availability
The BrTMHD-2023 dataset used to support the findings of this study is available from the corresponding author upon request. Additionally, the study utilized the Brain Tumor Kaggle Dataset, which is publicly accessible online.
Competing interests
The authors declare competing interests.
Ethical approval
The dataset utilized in this study was collected from BSMMCH, Faridpur, Bangladesh. We affirm that the dataset has received approval from the approval board of the hospital. The Ethical Approval Committee of Islamic University, Bangladesh, has also approved the use of this dataset in research, with the reference number EC/FBS/2024/06. Furthermore, we confirm that all data collection and statistical analysis methods were conducted in accordance with the relevant guidelines and regulations, including the Declaration of Helsinki and the BMRC ethical guidelines.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Celik, M. & Inik, O. Development of hybrid models based on deep learning and optimized machine learning algorithms for brain tumor multi-classification. Expert Syst. Appl.238, 122159 (2024). [Google Scholar]
- 2.Sharif, M., Amin, J., Raza, M., Yasmin, M. & Satapathy, S. C. An integrated design of particle swarm optimization (pso) with fusion of features for detection of brain tumor. Pattern Recogn. Lett.129, 150–157 (2020). [Google Scholar]
- 3.Mudda, M., Manjunath, R. & Krishnamurthy, N. Brain tumor classification using enhanced statistical texture features. IETE J. Res.68, 3695–3706 (2022). [Google Scholar]
- 4.Sajid, S., Hussain, S. & Sarwar, A. Brain tumor detection and segmentation in mr images using deep learning. Arab. J. Sci. Eng.44, 9249–9261 (2019). [Google Scholar]
- 5.Lan, Y.-L., Zou, S., Qin, B. & Zhu, X. Potential roles of transformers in brain tumor diagnosis and treatment. Brain-X1, e23 (2023). [Google Scholar]
- 6.Bhadra, S. & Kumar, C. J. An insight into diagnosis of depression using machine learning techniques: A systematic review. Curr. Med. Res. Opin.38, 749–771 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Ranjbarzadeh, R., Zarbakhsh, P., Caputo, A., Tirkolaee, E. B. & Bendechache, M. Brain tumor segmentation based on optimized convolutional neural network and improved chimp optimization algorithm. Comput. Biol. Med.168, 107723 (2024). [DOI] [PubMed] [Google Scholar]
- 8.Castiglioni, I. et al. Ai applications to medical images: From machine learning to deep learning. Physica Med.83, 9–24 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Gurusamy, R. & Subramaniam, V. A machine learning approach for MRI brain tumor classification. Comput. Mater. Continua53, 91–109 (2017). [Google Scholar]
- 10.Ali, M. S., Hossain, M. M., Kona, M. A., Nowrin, K. R. & Islam, M. K. An ensemble classification approach for cervical cancer prediction using behavioral risk factors. Healthc. Anal.5, 100324 (2024). [Google Scholar]
- 11.Hossain, S., Chakrabarty, A., Gadekallu, T. R., Alazab, M. & Piran, M. J. Vision transformers, ensemble model, and transfer learning leveraging explainable ai for brain tumor detection and classification. IEEE J. Biomed. Health Inform.28, 1261–1272 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Padmapriya, S. & Devi, M. G. Computer-aided diagnostic system for brain tumor classification using explainable ai. In 2024 IEEE International Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation (IATMSI) Vol. 2 (ed. Padmapriya, S.) 1–6 (IEEE, 2024). [Google Scholar]
- 13.Mahim, S. et al. Unlocking the potential of xai for improved alzheimer’s disease detection and classification using a vit-gru model. IEEE Access (2024).
- 14.Younis, A., Qiang, L., Khalid, M., Clemence, B. & Adamu, M. J. Deep learning techniques for the classification of brain tumor: A comprehensive survey. IEEE Access (2023).
- 15.Vankdothu, R., Hameed, M. A. & Fatima, H. A brain tumor identification and classification using deep learning based on cnn-lstm method. Comput. Electr. Eng.101, 107960 (2022). [Google Scholar]
- 16.Hollon, T. C. et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med.26, 52–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díaz-Pernas, F. J., Martínez-Zarzuela, M., Antón-Rodríguez, M. & González-Ortega, D. A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare9, 153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karayegen, G. & Aksahin, M. F. Brain tumor prediction on mr images with semantic segmentation by using deep learning network and 3d imaging of tumor region. Biomed. Signal Process. Control66, 102458 (2021). [Google Scholar]
- 19.Khan, P. et al. Machine learning and deep learning approaches for brain disease diagnosis: Principles and recent advances. IEEE Access9, 37622–37655 (2021). [Google Scholar]
- 20.Sun, L., Zhang, S., Chen, H. & Luo, L. Brain tumor segmentation and survival prediction using multimodal mri scans with deep learning. Front. Neurosci.13, 810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meena, G., Mohbey, K. K., Acharya, M. & Lokesh, K. Original research article an improved convolutional neural network-based model for detecting brain tumors from augmented mri images. J. Auton. Intell.6 (2023).
- 22.Saleh, A., Sukaik, R. & Abu-Naser, S. S. Brain tumor classification using deep learning. In 2020 International Conference on Assistive and Rehabilitation Technologies (iCareTech) (ed. Saleh, A.) 131–136 (IEEE, 2020). [Google Scholar]
- 23.Sultan, H. H., Salem, N. M. & Al-Atabany, W. Multi-classification of brain tumor images using deep neural network. IEEE Access7, 69215–69225 (2019). [Google Scholar]
- 24.Hossain, E. et al. Brain tumor auto-segmentation on multimodal imaging modalities using deep neural network. Comput. Mater. Continua72 (2022).
- 25.Pedada, K. R. et al. A novel approach for brain tumour detection using deep learning based technique. Biomed. Signal Process. Control82, 104549 (2023). [Google Scholar]
- 26.Deepak, S. & Ameer, P. Brain tumor categorization from imbalanced mri dataset using weighted loss and deep feature fusion. Neurocomputing520, 94–102 (2023). [Google Scholar]
- 27.Zebari, N. A. et al. A deep learning fusion model for accurate classification of brain tumours in magnetic resonance images. CAAI Trans. Intell. Technol. (2024).
- 28.Rahman, A. U. et al. A framework for susceptibility analysis of brain tumours based on uncertain analytical cum algorithmic modeling. Bioengineering10, 147 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali, A. M. & Mohammed, M. A. A comprehensive review of artificial intelligence approaches in omics data processing: Evaluating progress and challenges. Int. J. Math. Stat. Comput. Sci.2, 114–167 (2024). [Google Scholar]
- 30.Lin, D. J., Johnson, P. M., Knoll, F. & Lui, Y. W. Artificial intelligence for mr image reconstruction: An overview for clinicians. J. Magn. Reson. Imaging53, 1015–1028 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. et al. Advances in data preprocessing for biomedical data fusion: An overview of the methods, challenges, and prospects. Inf. Fusion76, 376–421 (2021). [Google Scholar]
- 32.Mohan, J., Krishnaveni, V. & Guo, Y. A survey on the magnetic resonance image denoising methods. Biomed. Signal Process. Control9, 56–69 (2014). [Google Scholar]
- 33.Bhadra, S. & Kumar, C. J. Enhancing the efficacy of depression detection system using optimal feature selection from ehr. Comput. Methods Biomech. Biomed. Engin.27, 222–236 (2024). [DOI] [PubMed] [Google Scholar]
- 34.Bernal, J. et al. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: A review. Artif. Intell. Med.95, 64–81 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Salvi, M., Acharya, U. R., Molinari, F. & Meiburger, K. M. The impact of pre-and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis. Comput. Biol. Med.128, 104129 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Spieker, V. et al. Deep learning for retrospective motion correction in MRI: A comprehensive review. IEEE Trans. Med. Imaging (2023). [DOI] [PubMed]
- 37.Ali, M. S. et al. Alzheimer’s disease detection using m-random forest algorithm with optimum features extraction. In 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA) (ed. Ali, M. S.) 1–6 (IEEE, 2021). [Google Scholar]
- 38.Islam, M. K. et al. Melanoma skin lesions classification using deep convolutional neural network with transfer learning. In 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA) (ed. Islam, M. K.) 48–53 (IEEE, 2021). [Google Scholar]
- 39.Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. Preprint at arXiv:2010.11929 (2020).
- 40.Islam, M. K., Rahman, M. M., Ali, M. S., Mahim, S. & Miah, M. S. Enhancing lung abnormalities diagnosis using hybrid dcnn-vit-gru model with explainable ai: A deep learning approach. Image Vis. Comput.142, 104918 (2024). [Google Scholar]
- 41.Lee, J.-h., Chae, J.-w. & Cho, H.-c. Improved classification of different brain tumors in mri scans using patterned-gridmask. IEEE Access (2024).
- 42.Chung, J., Gulcehre, C., Cho, K. & Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. Preprint at arXiv:1412.3555 (2014).
- 43.Yang, Y. et al. Early detection of brain tumors: Harnessing the power of gru networks and hybrid dwarf mongoose optimization algorithm. Biomed. Signal Process. Control91, 106093 (2024). [Google Scholar]
- 44.Hossain, M. M. et al. Cardiovascular disease identification using a hybrid cnn-lstm model with explainable AI. Inf. Med. Unlocked42, 101370 (2023). [Google Scholar]
- 45.Ahsan, M. M. et al. Enhancing monkeypox diagnosis and explanation through modified transfer learning, vision transformers, and federated learning. Inform. Med. Unlocked45, 101449 (2024). [Google Scholar]
- 46.Ribeiro, M. T., Singh, S. & Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In: Proc. 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 1135–1144 (2016).
- 47.Li, Z. Extracting spatial effects from machine learning model using local interpretation method: An example of shap and xgboost. Comput. Environ. Urban Syst.96, 101845 (2022). [Google Scholar]
- 48.Zhang, J., Jiang, Z., Dong, J., Hou, Y. & Liu, B. Attention gate resu-net for automatic mri brain tumor segmentation. IEEE Access8, 58533–58545 (2020). [Google Scholar]
- 49.Karimzadeh, R., Fatemizadeh, E. & Arabi, H. Attention-based deep learning segmentation: Application to brain tumor delineation. In 2021 28th National and 6th International Iranian Conference on Biomedical Engineering (ICBME) (ed. Karimzadeh, R.) 248–252 (IEEE, 2021). [Google Scholar]
- 50.Choudhury, C. L., Mahanty, C., Kumar, R. & Mishra, B. K. Brain tumor detection and classification using convolutional neural network and deep neural network. In 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA) (ed. Choudhury, C. L.) 1–4 (IEEE, 2020). [Google Scholar]
- 51.Bhanothu, Y., Kamalakannan, A. & Rajamanickam, G. Detection and classification of brain tumor in MRI images using deep convolutional network. In 2020 6th International Conference on Advanced Computing and Communication Systems (ICACCS) (ed. Bhanothu, Y.) 248–252 (IEEE, 2020). [Google Scholar]
- 52.Waghmare, V. K. & Kolekar, M. H. Brain tumor classification using deep learning. Internet of things for healthcare technologies 155–175 (2021).
- 53.Khairandish, M. O., Sharma, M., Jain, V., Chatterjee, J. M. & Jhanjhi, N. A hybrid cnn-svm threshold segmentation approach for tumor detection and classification of mri brain images. Irbm43, 290–299 (2022). [Google Scholar]
- 54.Mohan, P., Veerappampalayam Easwaramoorthy, S., Subramani, N., Subramanian, M. & Meckanzi, S. Handcrafted deep-feature-based brain tumor detection and classification using mri images. Electronics11, 4178 (2022). [Google Scholar]
- 55.Srinivas, C. et al. Deep transfer learning approaches in performance analysis of brain tumor classification using MRI images. J. Healthc. Eng.2022, 3264367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montaha, S. et al. Timedistributed-cnn-lstm: A hybrid approach combining cnn and lstm to classify brain tumor on 3d mri scans performing ablation study. IEEE Access10, 60039–60059 (2022). [Google Scholar]
- 57.Rasheed, Z. et al. Automated classification of brain tumors from magnetic resonance imaging using deep learning. Brain Sci.13, 602 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdusalomov, A. B., Mukhiddinov, M. & Whangbo, T. K. Brain tumor detection based on deep learning approaches and magnetic resonance imaging. Cancers15, 4172 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The BrTMHD-2023 dataset used to support the findings of this study is available from the corresponding author upon request. Additionally, the study utilized the Brain Tumor Kaggle Dataset, which is publicly accessible online.