Abstract
The seventh cholera pandemic started in 1961 in Indonesia and spread across the world in three waves in the decades that followed. Here, we utilised genomic evidence to detail the first wave of the seventh pandemic. Genomes of 22 seventh pandemic Vibrio cholerae isolates from 1961 to 1979 were completely sequenced. Together with 152 publicly available genomes from the same period, they fell into seven phylogenetic clusters (CL1–CL7). By multilevel genome typing (MGT), all were assigned to MGT2 ST1 (Wave 1) except three isolates in CL7 which were typed as MGT2 ST2 (Wave 2). The Wave 1 seventh pandemic expanded in two stages, with Stage 1 (CL1–CL5) spread across Asia and Stage 2 (CL6 and CL7) spread to the Middle East and Africa. Three non-synonymous mutations, one each, in three regulatory genes, csrD (global regulator), acfB (chemotaxis), and luxO (quorum sensing) may have critically contributed to its pandemicity. The three MGT2 ST2 isolates in CL7 were the progenitors of Wave 2 and evolved from within Wave 1 with acquisition of a novel IncA/C plasmid. Our findings provide new insight into the evolution and transmission of the early seventh pandemic, which may aid future cholera prevention and control.
Subject terms: Pathogens, Molecular evolution, Epidemiology
The seventh cholera pandemic spread across the globe in three waves from 1961. Here, the authors sequence 22 genomes from 1961 to 1979 and show that the first wave of the pandemic occurred in two distinct stages with different geographic and genomic characteristics.
Introduction
Cholera is an acute and rapidly progressing diarrhoeal disease, causing an estimated 1.3 to 4.0 million cases and 21,000 to 143,000 deaths worldwide each year1. The causative agent of cholera is a comma-shaped Gram-negative bacterium, Vibrio cholerae2. There are more than 200 serogroups in V. cholerae but only O1 and O139 serogroups have been recorded to cause epidemic and pandemic-level disease3.
V. cholerae has caused seven cholera pandemics since the first pandemic spread from India in 18174,5. The current ongoing seventh pandemic was caused by V. cholerae of O1 serogroup and El Tor biotype, originating in Sulawesi, Indonesia in 19616. The biotype shift from Classical to El Tor coincided with the first large-scale transmission wave in 1961–1966 and spread to most of Asia. The El Tor biotype was dominant in India by then, but the Classical biotype was still predominant in Bangladesh during this period. There was a second sharp increase in cholera cases in 1971 which spread out of Asia to Africa and Europe7. There was a lull of cholera in the 1980s during which cholera was confined to Asia and Africa and then upsurges and repeated spread across continents in the following decades until today2.
Many genomics studies have examined the origins and evolution of the seventh pandemic8–16. The seventh pandemic clone arose from its precursor in Makassar (Sulawesi), Indonesia in 1960 through mostly mutational changes to gain its pandemic spread capability8 and spread to other countries in 1961, marking the start of the pandemic7. Wide global spread was followed and an endemic environmental niche in Bangladesh and India was also established17–20. The pandemic spread can be divided into three waves9. Wave 1 (1961–1999) spread globally from its origin, Indonesia, to the Bay of Bengal, India and further to East Africa and South-West Americas. Wave 2 (1978–1984) spread to East Asia and Africa from the Indian subcontinent9,10,21. Wave 3 (1991 onwards) spread to Africa again and South America9,10,14,21,22.
The transmission of the seventh pandemic from 1970s onwards in Europe, Africa and the Americas has been well studied by genomic analysis10–14. However, the transmission of the seventh pandemic in its earlier years from 1961 to 1979 (before entering the low cholera period in the 1980s) remained less well studied due to the lack of genomic data. In this study, we generated 22 complete V. cholerae genomes from isolates sampled between 1961 and 1979 and used phylogenetic analysis to compare them with 152 publicly available genomes from strains collected before 1980, to provide high-resolution evidence of global transmissions in the early stages of the current seventh pandemic and to achieve a clearer understanding of the evolution of the cholera pandemic.
Results
Complete genome sequences of 22 V. cholerae isolates and single nucleotide polymorphisms (SNP) comparison
A total of 22 isolates from 1961 to 1979 were completely sequenced using a combination of Oxford nanopore and Illumina sequencing (Table 1). The isolates sequenced in this study were distributed in 16 countries but limited to three continents, Asia (15), Africa (6) and Europe (1). Among the 22 newly sequenced isolates, four were from Bangladesh, three from India, two from Vietnam and the remaining 13 from 13 different countries. Six publicly available complete genomes from Asia including five early seventh pandemic isolates (1961–1978) and one pre-seventh pandemic strain (C5) from 1957 were retrieved from the GenBank and included in this study. C5 was the closest pre-seventh pandemic isolate8 and was used as an outgroup and for comparison to identify seventh pandemic-specific changes. The isolates were distributed over 14 discontinuous years. All isolates were identified as sequence type (ST) ST69 and serogroup O1.
Table 1.
Complete genomes of V. cholerae O1 used in this study
| Strain | Year | Location | Sourcea | Original laboratory identification | Genome sequence reference | genome completeness |
|---|---|---|---|---|---|---|
| C5 | 1957 | Indonesia | GCF_001887395b | 8 | closed | |
| E9120 | 1961 | Indonesia | GCF_001887655b | 8 | closed | |
| M803 | 1961 | Hong Kong | Institut Pasteur | HK1 | this study | closed |
| E1162 | 1962 | China | GCF_001887495b | 8 | closed | |
| M805 | 1963 | Cambodia | Institut Pasteur | 930059 | this study | closed |
| M808 | 1969 | Vietnam | Institut Pasteur | 1536 | this study | closed |
| M807 | 1969 | Vietnam | Institut Pasteur | 601 | this study | closed |
| M686 | 1968 | Thailand | AFRIMS | SP-EV-29-1 | this study | closed |
| M820 | 1978 | Malaysia | Institut Pasteur | EB 251/1MR | this study | closed |
| M811 | 1971 | Burma | Institut Pasteur | 930029 | this study | not closed |
| M806 | 1964 | India | Institut Pasteur | CRC1106c | this study | closed |
| M804 | 1962 | India | Institut Pasteur | 930030 | this study | closed |
| CRC711 | 1964 | India | GCF_001887435b | 8 | closed | |
| M812 | 1971 | Chad | Institut Pasteur | 930046 | this study | closed |
| M815 | 1973 | Philippines | Institut Pasteur | 430035 | this study | not closed |
| M813 | 1972 | Senegal | Institut Pasteur | 9292 | this study | closed |
| M819 | 1975 | Germany | Institut Pasteur | 232 | this study | closed |
| M814 | 1972 | Morocco | Institut Pasteur | 113 | this study | closed |
| M818 | 1975 | Comoros Islands | Institut Pasteur | 102 | this study | closed |
| M809 | 1970 | Sierra Leone | Institut Pasteur | 930037 | this study | closed |
| M810 | 1970 | Ethiopia | Institut Pasteur | 930038 | this study | closed |
| M647 | 1970 | Bangladesh | CCUG | 13119 | this study | closed |
| N16961 | 1975 | Bangladesh | GCF_900205735b | closed | ||
| M795 | 1976 | Bangladesh | University of Maryland | 30167 | this study | closed |
| P27459 | 1976 | Bangladesh | GCF_013085125b | 70 | closed | |
| M714 | 1979 | Bangladesh | AFRIMS | 96A/CO | this study | not closed |
| M646 | 1979 | Bangladesh | CCUG | 9193 | this study | closed |
| M650 | 1976 | India | Institut Pasteur | 762/76 | this study | not closed |
| E7946 | 1978 | Bahrain | GCF_013085165b | 70 | closed |
aInstitut Pasteur, Paris, France; AFRIMS, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand; CCUG, Culture Collection of the University of Goteborg, Goteborg, Sweden; University of Maryland, Centre for Vaccine Development, University of Maryland, Baltimore.
bNCBI RefSeq sequence.
cThis strain was sequenced previously using PacBio (accession number: GCF_001887455.1)8 but re-sequenced in this study.
Comparison of the 28 isolates identified 491 SNPs, of which 377 SNPs were on chromosome 1 and 114 SNPs on chromosome 2 (Supplementary Data 1), and 432 and 59 SNPs were located on genes and intergenic regions respectively. These SNP-carrying genes can be categorised into at least one functional category and were allocated into 23 functional categories. The signal transduction mechanisms category was the largest with 42 genes, followed by the amino acid transport and metabolism category which had 34 genes (Supplementary Fig. S1, Supplementary Data 2). Most of the genes (207) had single SNPs, while 26 genes had two or more SNPs, among which 17 genes had two SNPs each, four genes had three SNPs each, two genes had eight SNPs, and one gene each had five and 12 SNPs (Supplementary Data 2). Notably, the majority of the SNPs (372/491) were non-synonymous SNPs (Supplementary Data 1). By functional categories, 8 categories contained genes carrying mostly (>80%) non-synonymous SNPs (Supplementary Fig. S1).
Phylogenetic analysis of complete genomes and phylogenetic distribution of SNPs
All SNPs identified were used for phylogenetic analysis. The pre-pandemic strain C5 isolated in 1957 was used as an outgroup (Fig. 1) and is known as the closest to the seventh pandemic clone8. To assess support for the tree, SNPs were mapped onto the tree, with 473 SNPs uniquely mapped to one of the 54 tree branches (Supplementary Fig. S2), and 18 SNPs mapped to multiple branches which were excluded.
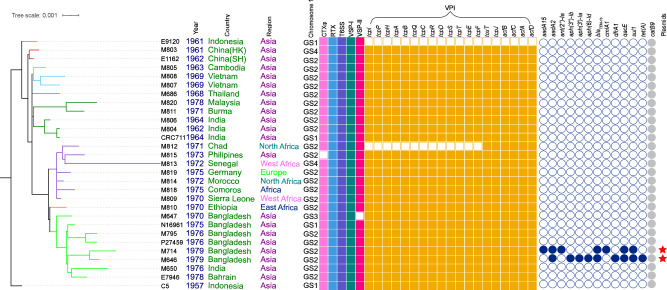
Fig. 1. Maximum-likelihood phylogenetic tree of the complete genomes of the early seventh pandemic (1961–1979).
The tree was rooted using V. cholerae strain C5 as an outgroup. The metadata is noted on the right side of the tree. Genome rearrangement structures of chromosome 1 are marked as GS1-GS4 (see Supplementary Fig S5 for details). Filled and empty squares represent gene presence and absence of virulence factors or genes shown respectively: the cholera toxin (CTXφ), the Vibrio pathogenicity island (VPI), Vibrio seventh pandemic islands (VSP-I and VSP-II), type VI secretion system (T6SS) and the repeats-in-toxin (MARTX) toxin (RTX). Filled circles represent the presence of an antibiotic resistance gene as listed. catB9 is annotated in grey since it has not been associated with phenotypic resistance. Red stars denote the two new IncA/C plasmids. The colours of the branches correspond to the clusters identified in Fig. 2 (see Fig. 2 for details). Other colours in different columns were for visual separation purposes.
The mapping of SNPs to branches allowed us to examine the distribution of multiple SNPs from the same gene or intergenic region along the phylogenetic tree (Supplementary Fig. S2). Co-occurrence of multiple SNPs from the same gene/intergenic region on the same branch may indicate recombination while singular appearance of SNPs from the same gene/intergenic region on different branches suggests independent mutational events. For the 41 genes with more than one SNP, these SNPs were distributed on 37 tree branches, of which 12 were internal branches and 25 were terminal branches leading to a single genome. Most of the SNPs from the same gene were distributed in different branches (Supplementary Data 3). However, six genes had two or more SNPs located on the same branch. They were located on seven branches, of which six were terminal branches. luxO had two SNPs each located on branches 28 and 34 with inter-SNP distance of 635 bp and 1047 bp, respectively. acp had two SNPs 423 bp apart on branch 36. The different copies of the hcpA gene on chromosome 1 and chromosome 2 had three and five SNPs on branch 51 respectively, with SNPs distances varied from 9 bp to 75 bp. A gene encoding a hypothetical protein (VC_A0432) had three SNPs, 65 bp and 103 bp apart, on branch 17 (an internal branch). One intergenic region had 11 SNPs on branch 23 with SNPs distance from 2 bp to 96 bp (39 bp on average).
We also compared the seventh pandemic isolates with the precursor strain C5 for any unique SNPs to the early pandemic. Twelve mutations were previously identified as seventh pandemic specific8. These unique SNPs were confirmed in this study. We further identified three nonsynonymous mutations, one each, on the csrD, acfB and luxO genes that were specific to the early seventh pandemic isolates of the complete genomes (Supplementary Data 4). These SNPs were not identified as seventh pandemic specific by Hu et al.8. We interrogated 7574 seventh pandemic isolates from all three waves on these SNPs. The SNP on csrD reversed back to the allele on the C5 strain in three seventh pandemic isolates (ERR576981 (Wave 1), ERR4175611 (Wave 2), ERR9716121 (Wave 3)) and thus was seventh pandemic specific. The SNPs on acfB and luxO were found to be present in all the seventh pandemic isolates examined. Thus, we can ascertain that the three SNPs observed were unique to the seventh pandemic with the exceptions described above.
Phylogenetic analysis of all genomes from the early seventh pandemic period
The newly sequenced complete genomes were compared with 152 Illumina sequenced genomes from the early seventh pandemic period (1961–1979) by phylogenetic analysis (Fig. 2, Supplementary Fig. S3). To better describe key spatiotemporal and evolutionary events, we have labelled seven clades as clusters (CL1–CL7). Each of the clusters contained at least one complete genome. The clusters were well supported by bootstrap values with all being >74%. It should be noted that along the phylogenetic tree and within the clusters, there were many isolates showing a star phylogeny. Such branching patterns are typical of rapid population expansion of a pandemic organism.
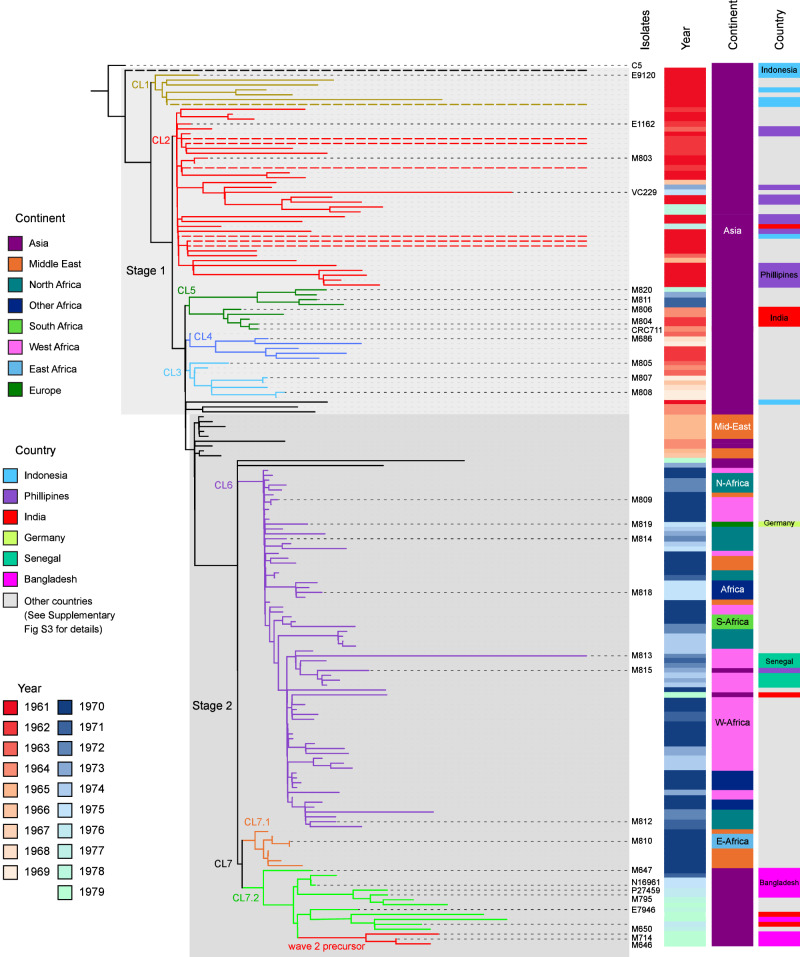
Fig. 2. Maximum-likelihood phylogenetic tree of the early seventh pandemic (1961–1979).
The tree was constructed using 29 complete genomes and 152 Illumina sequenced genomes from the seventh pandemic isolates between 1961 to 1979 and rooted using strain C5 as an outgroup. Stage 1 and Stage 2 were demarcated with light and dark grey shading. Cluster 1 (CL1) to Cluster 7 (CL7) are demarcated with different colour branches and labelled at each node. Dashed branches were shortened for clarity. Year of isolation is shown using a heatmap with reds for 1960s and blues for 1970s. Continent/Subcontinent of origin for each isolate is displayed with a colour bar and country of origin is displayed when that country was mentioned specifically in the text. All other countries were marked in grey colour with country details in Supplementary Fig S3. Only complete genomes are named for clarity. See Supplementary Fig. S3 for full details of isolate names and complete metadata.
Based on phylogenetic clustering and metadata, two clusters (CL1, CL2) contained at least one isolate from Indonesia in 1961. In addition, two isolates (SRR6027720 and ERR579063) from Indonesia isolated in 1961 were well separated from each other and from the two clusters. These two clusters and the two singleton isolates clearly diverged in Indonesia before they spread to other Asian countries. Therefore, we assigned this initial spread as Stage 1 of the seventh pandemic spread, which originated from Indonesia. Within Stage 1, there were three other clusters (CL3–CL5). Earliest isolate in CL3–CL5 were from Cambodia (1963), Japan (1962), and India (1962) respectively. Although these clusters did not contain any Indonesian isolates, they were clearly early spread from Indonesia directly or indirectly.
The majority of the remaining isolates on the tree were grouped as a single clade which includes Iran isolates from 1965 as well as CL6 and CL7 which contain isolates from the Middle East and Africa. We assigned this spread of the seventh pandemic as Stage 2. The Stage 2 node was supported by two SNPs from two genes (VC1482 and VC2487, Supplementary Data 1). Thus, early spread of the seventh pandemic from Indonesia is divided into two stages.
The seventh pandemic was divided into waves9 and the three waves can be distinguished using multilevel genome typing (MGT)23. By MGT typing, 173 isolates/genomes were identified as MGT2 ST1 which belongs to Wave 1. Three isolates/genomes, M646 and M714 and one Illumina sequenced genome (ERR025383) were typed as MGT2 ST2, which belongs to Wave 2, suggesting that Wave 2 was derived from a precursor in C7.2. Further phylogenetic analysis of these three isolates with other Wave 2 isolates found that the three isolates diverged the earliest (Supplementary Fig S4). Another three Illumina sequenced genomes were typed as MGT2 ST16, MGT2 ST19 and MGT2 ST829, interspersed among MGT2 ST1 isolates on the phylogeny and clearly belonged to Wave 1.
Virulence elements in the complete genomes
We identified the genes on each known virulence element including the cholera toxin phage (CTXφ), the Vibrio pathogenicity island (VPI), Vibrio seventh pandemic islands (VSP-I and VSP-II), the type III secretion system (T3SS) and the type VI secretion system (T6SS). All ctxB genes were typed to ctxB genotype 3. All the tcpA genes of the complete genomes from this study and the strain C5 were identical. All complete genomes carried the 11 VSP-I genes, the 19-gene T6SS gene cluster and the four genes of the multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin gene cluster (rtxA, rtxB, rtxC and rtxD) (Fig. 1). Nearly all isolates carried VSP-II, CTXφ and the VPI, with the following exceptions. M647 lacked VSP-II, M815 lacked CTXφ, E9120) lacked VPI as reported previously24, and M812 contained only six (toxT, tcpJ, acfA, acfB, acfC and acfD,) of the 19 VPI genes.
Plasmids and resistance genes in the complete genomes
Two isolates (M646 and M714) carried an IncA/C family plasmid. Both isolates were in CL7.2 and isolated from Bangladesh in 1979 which was the latest isolation year in this study (Fig. 1). None of the other isolates from the early pandemic period carried any plasmids and no isolates carried antimicrobial resistance (AMR) genes or mutations.
The plasmid in M646 was named as pM646 and was assembled as circular DNA of 170,552 bp in length. G + C content was 52.59% which was higher than its host genome (47.51%). A total of 207 coding sequences (CDSs) were predicted. pM646 was found to carry the AMR genes aadA2, aph(3”)-Ib, aph(3’)-Ia, aph(6)-Id, blaTEM-1b, dfrA1, qacE, sul1 and tet(A).
The plasmid in M714 was named as pM714 and was assembled as circular DNA of 148,732 bp in length. G + C content was 51.58% and 180 CDSs were predicted. pM714 carried AMR genes, aadA15, aadA2, ant(2”)-Ia, blaTEM-1b, cmlA1, qacE and sul1.
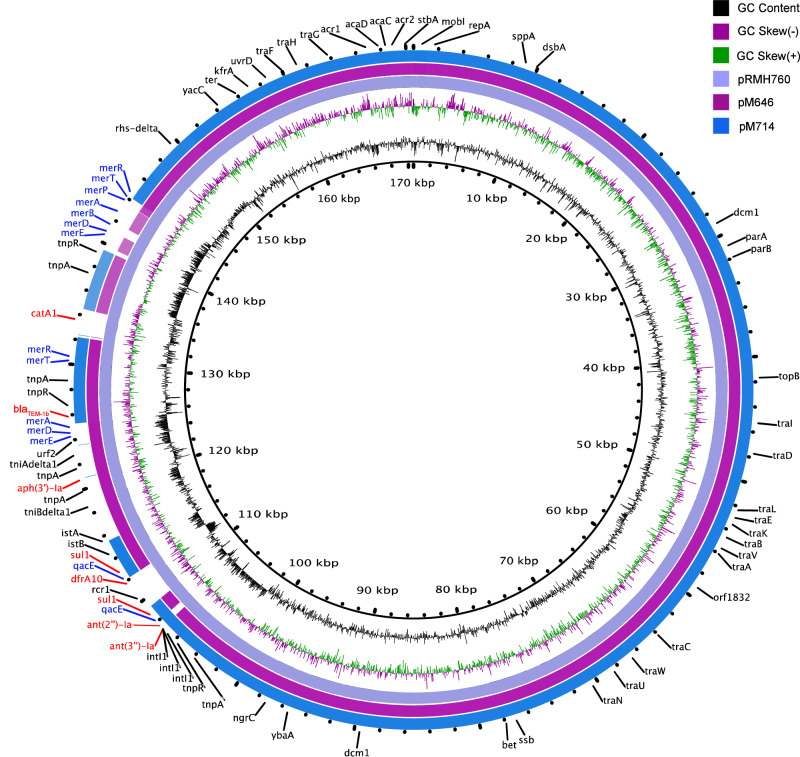
The two plasmids were closely related to each other and to a plasmid found in a Klebsiella pneumoniae strain isolated in Australia in 1997 (pRMH760,170,613 bp), carrying aadB, sul1, dfrA10, aphA1, blaTEM-1, and catA1genes25 (Fig. 3). Compared to pRMH760 (NCBI Reference Sequence: NC_023898.1), the coverages of pM646 and pM714 were 95% and 88% respectively, and the nucleotide sequence identities were both above 99.9%. Both pM646 and pM714 lacked two fragments on pRMH760 containing catA1 and dfrA10 genes. The coverage and identity of pM714 to pM646 were 87% and 99.91%. pM714 lacked two fragments on pM646 that carried several mercury resistance genes (merA, merB, merD, merE and merP) and an aminoglycoside resistance gene aph.
Fig. 3. Two new IncA/C plasmids compared with the most closely related plasmid pRMH760 of Klebsiella pneumoniae.
From the inner circle to the outer circle are the three plasmids pRMH760 (Klebsiella pneumoniae), pM646 (V. cholerae M646) and pM714 (V. cholerae M714). The genes are annotated based on pRMH760 (NCBI Reference Sequence: NC_023898.1). Antibiotic resistance genes are highlighted in red while antiseptic and metal resistance genes are marked in blue. The inner rings are size scale, G + C content and G + C skews per colour legend.
Gene duplications in the complete genomes
We also screened the complete genomes for gene duplications (Supplementary Data 5). Eight genes with known functions and six genes encoding hypothetical proteins were found to have been duplicated variably in different genomes with two to three copies. In comparison to the pandemic precursor strain C5, there were no genes that were uniquely duplicated in the seventh pandemic isolates. No duplicated C5 genes were deduplicated to single copy in the early seventh pandemic isolates. Interestingly, one gene (locus_tag=VC_A0301) encoding an oxidoreductase that was duplicated in all except two Stage 1 genomes was deduplicated to single copy in all Stage 2 genomes.
Superintegrons (SIs) in the complete genomes
SIs were extracted from chromosome 2 of the 25 genomes and aligned to the SI from the outgroup strain C5 as a reference. A total of 32 functional genes and up to 257 hypothetical proteins were annotated in an SI (Supplementary Data 6). catB9 was identified in all SIs.
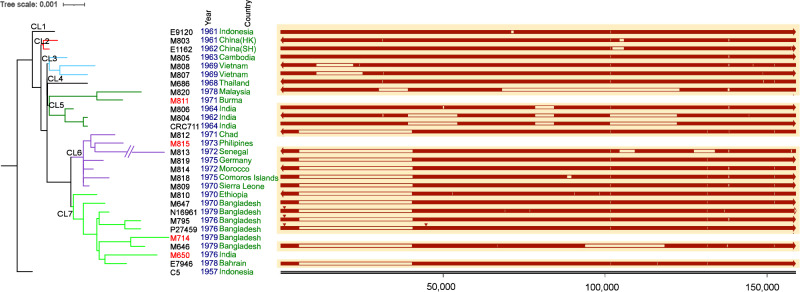
There was one large deletion (34,872 bp) found in both CL7.1 and CL7.2 SIs (Fig. 4). Three small deletions were found in genomes from India in CL5. The SI size and number of open reading frames (ORFs) varied among lineages (Supplementary Data 7, Fig. 4). The largest deletion of 54,651 bp was observed in M820 (in CL5), a 1978 isolate from Malaysia.
Fig. 4. Alignments of the superintegrons from complete genomes.
Superintegrons (right panel) were extracted from chromosome 2 of each complete genome and aligned against strain C5. The number under the tick marks of the integron size is in base pairs (bp). The empty line segments represent deletions. The red triangles on the top of the segments represent large insertions. The genome sequences of the chromosome 2 of the strains marked in red had not been closed and thus the superintegrons were not presented. The arrows denote the orientation of each aligned sequence. The white arrow denotes the missing bases. Colours for year and country are for visual separation purpose. The tree was the same as that of Fig. 1.
Two insertion events were found (Fig. 4). An identical insertion (Insertion 1) of 1376 bp was found in three genomes (N16961, P27459 and M795) at position 1192 on the SI. The insertion region was a duplication of a region present in the SI of each genome. It was identical to regions on both chromosome 1 and chromosome 2 of strain O395 encoding for hypothetical proteins. The other insertion (Insertion 2) of 1270 bp was identified only in P27459 at position 44,867 on the SI. Insertion 2 was an insertion sequence, ISVch4, inserted in the SI. ISVch4 is also present on chromosome 1 of the 26 complete genomes encoding transposase OrfA and OrfB (Supplementary Data 8).
Genome rearrangement in complete genomes
Four genomic structures (GS) of chromosome 1 were identified with rearrangements of large sections of the genomes (GS1-GS4) (Supplementary Fig. S5). The majority of the isolates (22/29) belonged to GS2. Four belonged to GS1 (strain C5, E9120, CRC711 and N16961), two to GS4 (M803 and M813) and one to GS3 (M647). We also aligned chromosome 2 from all isolates and found no rearrangements.
Discussion
Stages of cholera transmission during the early seventh pandemic
The first of the three waves of the seventh cholera pandemic (Wave 1) occurred from 1961 to 1999 and spread from Indonesia to neighbouring countries as well as East Africa and South-West America7,9. Further studies revealed numerous transmission events within Africa (T1–T13)11, Latin America (LAT1–LAT3)10 and Europe (EUR1–EUR8)14. In this study, we examined the evolution of the early seventh pandemic (from 1961 to 1979) using complete genomes as well as a large set of draft genomes from the same period. We found that there were two distinct transmission stages in this early pandemic period. Stage 1 represented the initial spread from Indonesia, comprised of cluster CL1-CL5 isolates from Asian countries. We found that in Stage 1, V. cholerae had already diverged in Indonesia in 1961 and these diverged strains spread in parallel to other Asian countries in the early 1960s at the same time. Some of the clusters persisted until the late 1970s. Since we only included isolates before 1980 in this study, it was likely that these clusters were circulated widely in Asia before wave 1 was replaced by other waves9.
It is perhaps not surprising that the seventh pandemic has diverged before it spread out of Indonesia. The precursor of the seventh pandemic clone was known to have caused outbreaks between 1937 and 1957 in the island of Sulawesi, Indonesia7. Then in 1960, an outbreak of cholera occurred in Makassar (Sulawesi) and spread to other Asian countries in 1961 which was recognised as the year of the start of the seventh pandemic26,27. Therefore, there were at least three years from 1957 for the seventh pandemic clone to diverge in Indonesia before it spread to other regions.
Stage 2 was marked by the spread to the Middle East as the last leg of cholera spread within Asia and crossing to Africa in the 1970s11. At this stage, the seventh pandemic spread out of Asia to Africa as clusters CL6 and CL7. CL6 corresponded to Africa introduction event T111, which spread from the Middle Eastern countries to South, and North African countries in the 1970s. There were two subclusters identified in CL7. CL7.1 isolates circulated in the Middle East and East Africa in 1970s which corresponded to Africa introduction event T311. CL7.2 isolates were found mainly in Bangladesh and other Asian countries in the late 1970s. An isolate (M647) in Bangladesh in 1970 shared the most recent common ancestor (MRCA) with other isolates in CL7.2. We assumed that both CL7.1 and CL7.2 diverged from a common ancestor in 1970 with CL7.1 spreading to East Africa through the Middle East11 while CL7.2 remained in Asia, and was mostly sampled in Bangladesh.
CL7.2 may hold an important insight into the further development of the seventh pandemic. Although Bangladesh was affected by the initial seventh pandemic spread in the 1960s, the Classical biotype predominated until 1972 when it was replaced by the El Tor biotype7. It was likely that CL7.2 was the cause of that replacement and subsequently gave rise to wave 2.
Three isolates belonged to MGT2 ST2 (two complete genomes from this study and one public draft genome) and thus belonged to seventh pandemic wave 223. These isolates were all obtained in Bangladesh in 1979 and each carried an IncA/C plasmid. Previous studies describe the acquisition of the SXT/R391 integrative and conjugative element (ICE) as the beginning of the transition from Wave 1 to Wave 29. Since these three isolates diverged the earliest among the Wave 2 isolates, they were likely the earliest progenitors of Wave 2 and evolved from Wave 1 by acquiring an IncA/C plasmid carrying AMR genes rather than an ICE. The subsequent acquisition of the SXT/R391 with multidrug resistance must have conferred an advantage to the SXT/R391 positive strains facilitating the spread of Wave 29.
Interestingly, in cluster CL6, a strain from Germany (M819) isolated in 1975 shared MRCA with African strains. The transmission from Africa to European countries was probably due to Portuguese troops traveling frequently between South/West Africa and Portugal in the early 1970s28. However, there were many other possible transmission routes through trade and human travel between Africa and Europe. Moreover, another CL6 isolate (M815) from the Philippines in 1973, where 2075 cholera cases were reported in that year7, shared an MRCA with a strain from Senegal (M813). The finding suggests that the West African lineage might have transmitted back and forth between South Asian and African countries.
Mutations unique to early seventh pandemic that may have contributed to its pandemic transition
A previous study found that the seventh pandemic has gained only 12 mutations in comparison to its precursor C5 to gain the high capacity of spread8. These mutations were confirmed to be seventh pandemic-specific in this study with more isolates from the start of the pandemic. However, there is no apparent explanation how these genes or their SNP changes contributed to their pandemic transition8. In this study, we found three additional nonsynonymous mutations on the csrD, acfB and luxO genes that have been acquired by the early seventh pandemic. These genes are all involved in gene regulation and cell signalling. CsrA is an RNA-binding global regulator which regulates the major virulence gene regulator ToxR and quorum sensing regulon29. AcfB is a methyl-accepting chemotaxis protein and LuxO is the quorum sensing regulator protein30,31. We confirmed these mutations were present in all or nearly all Wave 2 and Wave 3 isolates and thus maintained in the seventh pandemic clone. Our findings suggest that the accumulations of nonsynonymous mutations in the three genes that play key roles in adaptation and virulence may have enabled the pandemicity of the seventh pandemic clone in the initial development in Indonesia after it diverged from its precursor.
Low level of recombination and elevated mutation rate of adaptive genes in the early seventh pandemic
To determine whether recombination contributed to the evolution of the early seventh pandemic, we examined genes with more than one SNP. Since the mutation rate was about three SNPs per genome per year9, the probability of two SNPs occurring on the same gene is low. We further used phylogenetic information to determine whether the SNPs were occurring at the same time, i.e. located on the same branch. We found that the majority of the SNPs from the same gene occurred on different branches on the phylogenetic tree, suggesting that these SNPs arose through mutations independently. However, there were 41 occasions where two or more SNPs from the same gene or intergenic region were located on the same branch, indicating that these may have arisen through recombination. Thus, occasional recombination events were likely to have happened during the early seventh pandemic. As hypothesised by Hu et al.32, the low recombination rate in the seventh pandemic clone may be due to that they spend less time in the environment during pandemic and outbreak periods, and they may also enter a viable but nonculturable form with less opportunity for recombination when they are in the environment.
Since the multiple SNPs on the same gene were mostly derived independently through mutations and many of these SNPs were non-synonymous SNPs, it was likely that some of these changes were adaptive. The luxO gene stood out most, with 12 SNPs, all being non-synonymous, among the completely sequenced genomes. Compared to the estimated mutation rate for the seventh pandemic V. cholerae of 3.3 SNPs per genome per year9, luxO clearly had elevated mutations. LuxO and HapR/LuxR encoded by luxO and hapR respectively are two critical regulatory proteins of the quorum sensing (QS) system which is a top regulator for virulence gene expression by monitoring population density30,33. QS upregulates biofilm formation and virulence gene expression to colonise the intestine during human infection and downregulates these processes before exiting human hosts to promote transmission34,35. This high frequency of mutations indicates positive selection on QS to facilitate increased transmission during the early seventh pandemic. The elevated mutation rate in luxO and hapR was maintained throughout the three-seventh pandemic waves (Supplementary Fig. S6).
Since luxO and hapR had elevated mutation rates, we examined all 43 genes with mutations in the signal transduction category in the early seventh pandemic isolates for elevated mutation rates and found an additional four genes (arcA, dctB, dctR and VC0694) with a persistently elevated mutation rate in the seventh pandemic across different waves (Supplementary Fig. S6). ArcA is a global regulator of the ArcB/A two component system (TCS) and known to regulate virulence gene expression and biofilm formation36,37. DctB (VC1925) and DctR (VC1926) pair as a TCS that sense C4-dicarboxylates38,39, but little is known of regulatory targets or functions. VC0694 encodes an uncharacterised TCS histidine kinase with VC0693 encoding its cognate response regulator and plays a role in intestinal colonization in infant mice40 and biofilm formation41. It seems that mutations in a subset of the TCSs may have played an adaptive role in the development of the seventh pandemic, which has not been recognised previously.
It is noteworthy that a previous study of Chinese isolates by Didelot et al.42 found that nine of the 17 mutator strains among 260 seventh pandemic isolates studied were from the early 1960s. Mutators that carry mutations in DNA mismatch repair genes have increased mutation rates and may facilitate adaptation43. However, our complete genomes did not contain any mutator mutations with the mismatch repair genes examined (mutS, mutH, mutL and uvrD).
Evolution of AMR and plasmids in the early seventh pandemic V. cholerae
Two IncA/C plasmids, pM646 and pM714, were found in this study. The plasmids were in two strains from Bangladesh in 1979. A multidrug-resistant plasmid was reported from isolates from an outbreak in Bangladesh in the same year44, these isolates were resistant to tetracycline, ampicillin, kanamycin, streptomycin, gentamicin and trimethoprim. Both pM646 and pM714 carried genes conferring resistance to streptomycin, ampicillin, and sulfamethoxazole. Additionally, pM646 carried trimethoprim and tetracycline resistance genes while pM714 carried gentamicin and chloramphenicol resistance genes. Another V. cholerae incomplete genome (ERR025383) from Bangladesh isolated in 1971 also carried the same plasmid as pM646 with identical AMR genes.
AMR genes such as aadA, dfrA, qacE and cmlA were often found on integrons45. However, in this study, these genes were found on two IncA/C plasmids. The acquisition of these plasmids by the seventh pandemic clone is interesting. It is known that very few plasmids were present in the pandemic clone, possibly due to two plasmid defence systems encoded by VPI-2 and VSP-II46. This may also explain why later seventh pandemic strains acquired the SXT ICEs carrying AMR genes rather than AMR plasmids for its selective advantage. Nevertheless, IncA/C plasmids play an important role in AMR acquisition in V. cholerae47.
Both plasmids carry qacE, a disinfectant resistance gene that is rare in V. cholerae48. It was first reported in clinical and aquatic environmental V. cholerae non-O1/non-O139 isolates49 and later found in two V. cholerae plasmids isolated from an environmental non-toxigenic O1 strain50 and a clinical O139 strain51 in China. In our previous studies of isolates from China, the qacE gene was not present in any of the IncA/C plasmids of V. cholerae O139 isolates16 or environmental non-O1/non-O139 isolates52.
The superintegrons are marked with few genetic events in the early pandemic
There were relatively few insertion and deletion events found on the SIs of the genomes sequenced in this study, which contrasts with the diverse SIs in environmental V. cholerae53,54. We found that the SI structure varied among the phylogenetic clusters. Our findings suggest that the SI in the early seventh pandemic was quite stable with occasional losses but no gain of new cassettes. In contrast, studies showed that the SI is dynamic and involved in the acquisition of gene cassettes from other species for adaptation to local environments55. Environmental adaptation may not have played a large selection pressure in the seventh pandemic clone as it was primarily transmitted from human to human in large outbreaks with little time in the local environment in the earlier years of its spread.
Impact of genome rearrangement in the early seventh pandemic
Genome rearrangements can affect both gene expression and growth rate56 and consequently may impact on virulence and transmissibility. We identified four GSs of chromosome 1 in the early seventh pandemic V. cholerae. GS1 was the older structure found in pre-seventh pandemic strain C5 and the first-seventh pandemic strain E9120. However, GS2 was the dominant structure in 76% of the complete chromosome 1 sequences in this study. Occasional inversions in GS3, GS4 and back to GS1 were identified in both Stage 1 and Stage 2 isolates. Chromosome 2 was stable with no genomic rearrangement except for variations within the SI. More complete genomes and further phenotypical studies are needed to determine the effect of genome rearrangement, in particular, GS2 in comparison to GS1, on gene expression and fitness.
In conclusion, analysis of complete genomes of isolates from the start of the seventh pandemic to the late 1970s, supplemented with draft genomes, allowed us to dissect the first wave of the seventh pandemic. The early seventh pandemic expanded in two stages. In Stage 1, the seventh pandemic clone diverged into at least two clusters and two singletons within Indonesia and spread in parallel to other Asian countries. In Stage 2, the seventh pandemic spread to the Middle East and further spread to African countries. The Wave 1 seventh pandemic evolved into Wave 2 marked by the initial acquisition of an IncA/C plasmid carrying multiple AMR genes. Three non-synonymous mutations in regulatory genes, csrD, acfB, luxO, involved in global gene regulation, chemotaxis and quorum sensing respectively, were uniquely acquired by the early seventh pandemic in comparison to the pre-seventh pandemic strain C5 and may have critically contributed to its pandemicity. Further, adaptive mutations in multiple TCS regulatory genes, especially luxO and hapR, were elevated, contributing to the divergence and adaptation of the seventh pandemic. This study offered a high-resolution dissection and an enhanced understanding of the current cholera pandemic in its early stages of spread and may help design strategies for the prevention and control of cholera.
Methods
Genomes
A total of 22 V. cholerae isolates were sequenced in this study (Table 1). These isolates were collected by other laboratories from different countries, with source laboratories listed in Table 1. These strains have used in our previous studies15 and were historically archived strains. Note that one isolate (M806) was the same strain as CRC1106 sequenced by Hu et al. using PacBio8. Comparison of the two genome sequences showed 70 base differences and three genomic structure differences (Supplementary Data 9). Therefore, we used our sequence to represent the strain for consistency. Seven publicly available complete V. cholerae genomes from the seventh pandemic were obtained from NCBI (on 26/05/2022) and included for comparison. V. cholerae strain N16961 (NCBI accession: GCF_900205735.1) was used as the reference genome and the pre-seventh pandemic strain C5 (GCF_001887395) was used as an outgroup for phylogenetic analysis (Table 1). All genomes were identified as V. cholerae O1 serogroup ST69 by nucleotide BLAST (version 2.9.0+) and in silico MLST (https://github.com/tseemann/mlst) using default settings. Additionally, raw sequence data of 152 V. cholerae isolated between 1961 and 1979 from African and Asian countries with genome sequence quality that passed MGT typing filters were downloaded from NCBI for comparison (Supplementary Data 10).
WGS with nanopore technology
DNA of 22 isolates was extracted using the phenol-chloroform extraction method and sequenced using both nanopore technology and Illumina platform. Canu (version 2.2)57 and Unicycler (version 0.4.7)58 were used to assemble the genomes. All genome sequences in this study have been submitted as raw reads under BioProject accession number PRJNA970070 in the NCBI SRA database.
SNPs calling and phylogenetic analysis
The SNPs of 28 complete genomes were called using SaRTree pipeline59 against chromosome 1 and chromosome 2 of V. cholerae O1 biovar El Tor strain N16961 (GCF_900205735.1) separately. Proportion threshold 100 was set for calling the SNPs and all recombinant SNPs were identified. The SNPs of publicly available genomes and the complete genomes in this study were called by SaRTree pipeline using proportion threshold 20 and all recombinant SNPs were removed. We used IQ-Tree (version 2.0.4)60 with default parameters and 1000 ultrafast bootstrap replicates61 to construct the maximum likelihood (ML) tree using the SNPs from both two chromosomes (Best-fit model: TVMe+ASC [complete genome tree] and TVMe+ASC + R2 [NCBI genome tree]). The strain C5 was used as an outgroup for both trees. Tree files were annotated and visualised in iTOL (version 6.5.2)62. SNPs were mapped onto branches of the tree by the SaRTree pipeline.
The functional categories used were defined by Database of Clusters of Orthologous Genes (COGs) on NCBI (data updated: March 2022). Each gene of the genomes from this study was aligned against the reference genome and locus tags were used to link the functional categories. Note that to relate the gene locus tags of GCF_900205735.1 with VC numbers used in old annotations and many publications including this study, we have provided a list of correspondence of the two annotations for genes mentioned in this study in Supplementary Data 11.
Note that we attempted BEAST analysis on the complete genome dataset and the total dataset. Unfortunately for the complete genome dataset, TempEst (version1.5.3) analysis found that there was not enough temporal signal to perform BEAST analysis. For the total dataset of Illumina draft genomes and the complete genomes together, there was good temporal signal based on TempEst analysis. However, there were eight genomes with very long terminal branch lengths which led to violation of all the evolutionary models tested in BEAST analysis. One of the eight genomes was a strain from Indonesia isolated in 1961 which diverged the earliest, so we attempted BEAST analysis by 1) removing the eight genomes; 2) keeping the Indonesian one but removing the other seven genomes. In both cases, forcing strain C5 (the pre-pandemic strain) as the outgroup, the date estimate for the MCRA of the seventh pandemic was 1940s. In particular, in the latter, the effective sample size (ESS) for clock rate and overall priors were very low.
Multilevel genome typing
All genomes in this study including complete genomes and Illumina sequenced genomes were processed and submitted to the V. cholerae MGT database23. MGT STs were assigned automatically by the MGTdb server (https://mgtdb.unsw.edu.au/vibrio/). MGT2 ST1, ST2 and ST3 correspond to the seventh pandemic wave 1, wave 2 and wave 3 respectively23.
Genetic elements analysis
ABRicate (https://github.com/tseemann/abricate) (version 0.9.8) was used to predict the AMR genes and plasmids in the databases of ResFinder (on 23/08/2021)63 and PlasmidFinder64, respectively. We used AMRFinderPlus version 3.12.8 with database version 2024-05-02.2 to identify point mutations in the assemblies65. The presence of 67 virulence genes from CTXφ, VPI, VSP-I and VSP-II, T6SS, T3SS and RTX were also screened using ABRicate. Duplications were identified using nucleotide BLAST (version 2.9.0+) with V. cholerae seventh pandemic strain N16961 coding sequences (RefSeq accession numbers NC_002505.1 and NC_002506.1). The identity of 99% was used as the cut-off for the duplication screening.
Superintegron analysis
intI466 and 115 bp VCR sequences were used to identify the SIs. SIs were extracted from 25 closed chromosomes. SnapGene software (version 4.2.4) and progressiveMauve (version 2.4.0)67 were used to align and analyse the insertions and deletions on the SIs. Prokka (version 1.14.6)68 was used to annotate genes on the SIs.
Genome rearrangement and alignment
The two chromosomes of complete genomes were reordered separately using Circlator (version 1.5.5)69. In chromosome 1, dnaA was used as a start position to reorder the sequences and in chromosome 2, intI4 was used to reorder the sequences. The reordered chromosomes were aligned in progressiveMauve (version 2.4.0)67. Four chromosome 2 sequences that were not completely assembled were excluded.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
Yun Luo was a PhD student supported by Australian Government Research Training Program Scholarship. This research was supported in part by a grant from the National Health and Medical Council of Australia (2011806, RL). The authors acknowledge Liam Cheney for helping with the sequencing.
Author contributions
R.L. conceived the study. Y.L. performed experiments and data analysis and wrote the draft of the manuscript. M.P., S.D., S.O. and R.L. verified the underlying data. M.P., S.K., S.O. and R.L. reviewed and edited the manuscript. All authors reviewed and approved the final version of the manuscript. All authors had full access to all the data in this study. R.L. had final responsibility for the decision to submit for publication.
Peer review
Peer review information
Nature Communications thanks Ola Brynildsrud and Thandavarayan Ramamurthy, who co-reviewed with Agila Pragasam, for their contribution to the peer review of this work. A peer review file is available.
Data availability
All 22 genomes sequenced in this study have been submitted as raw reads under BioProject accession number PRJNA970070 in the NCBI SRA database. The accession numbers for all other publicly available sequences used in this study are listed in Supplementary Data 10.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52800-w.
References
- 1.Ali, M., Nelson, A. R., Lopez, A. L. & Sack, D. A. Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis.9, e0003832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanungo, S., Azman, A. S., Ramamurthy, T., Deen, J. & Dutta, S. Cholera. Lancet399, 1429–1440 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Kaper, J. B., Morris, J. G. Jr & Levine, M. M. Cholera. Clin. Microbiol Rev.8, 48–86 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert, S. Reports on the epidemic cholera which has raged throughout Hindostan and the peninsula of India, since August 1817. Published under the Authority Of Government. Med. Chir. J.2, 547–557 (1820).
- 5.Weil, A. A. & Ryan, E. T. Cholera: recent updates. Curr. Opin. Infect. Dis.31, 455–461 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Sack, D. A., Sack, R. B., Nair, G. B. & Siddique, A. K. Cholera. Lancet363, 223–233 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Barua, D. in History of Cholera. (eds Barua, D., Greenough, W. B.). Cholera. 1–36 (Boston, MA, Springer US, 1992).
- 8.Hu, D. et al. Origins of the current seventh cholera pandemic. Proc. Natl Acad. Sci. USA113, E7730–E7739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature477, 462–465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domman, D. et al. Integrated view of Vibrio cholerae in the Americas. Science358, 789–793 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Weill, F. X. et al. Genomic history of the seventh pandemic of cholera in Africa. Science358, 785–789 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Bwire, G. et al. Molecular characterization of Vibrio cholerae responsible for cholera epidemics in Uganda by PCR, MLVA and WGS. PLoS Negl. Trop. Dis.12, e0006492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weill, F. X. et al. Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature565, 230–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oprea, M. et al. The seventh pandemic of cholera in Europe revisited by microbial genomics. Nat. Commun.11, 5347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam, C., Octavia, S., Reeves, P., Wang, L. & Lan, R. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg. Infect. Dis.16, 1130–1132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, Y. et al. Genomic epidemiology of Vibrio cholerae O139, Zhejiang province, China, 1994-2018. Emerg. Infect. Dis.28, 2253–2260 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John, T. J. & Jesudason, M. V. The spread of Vibrio cholerae O139 in India. J. Infect. Dis.171, 759–760 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Islam, M. S., Alam, M. J. & Khan, S. I. Occurrence and distribution of culturable Vibrio cholerae O1 in aquatic environments of Bangladesh. Int J. Evol. Stud.47, 217–223 (1995). [Google Scholar]
- 19.Alam, M. et al. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ. Microbiol72, 4096–4104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam, M. et al. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ. Microbiol72, 2849–2855 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramamurthy, T. et al. Revisiting the global epidemiology of cholera in conjunction with the genomics of Vibrio cholerae. Front. Public Health7, 203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monir, M. M. et al. Genomic attributes of Vibrio cholerae O1 responsible for 2022 massive cholera outbreak in Bangladesh. Nat. Commun.14, 1154 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheney, L., Payne, M., Kaur, S. & Lan, R. Multilevel genome typing describes short- and long-term Vibrio cholerae molecular epidemiology. mSystems6, e0013421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaolis, D. K. et al. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl Acad. Sci. USA95, 3134–3139 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmer, C. J. & Hall, R. M. pRMH760, a precursor of A/C(2) plasmids carrying blaCMY and blaNDM genes. Micro. Drug Resist20, 416–423 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Mukerjee, S. Problems of cholera (EL TOR). Am. J. Trop. Med. Hyg.12, 388–392 (1963). [DOI] [PubMed] [Google Scholar]
- 27.Felsenfeld, O. Some observations on the cholera (E1 Tor) epidemic in 1961-62. Bull. World Health Organ28, 289–296 (1963). [PMC free article] [PubMed] [Google Scholar]
- 28.Blake, P. A. et al. Cholera in Portugal, 1974.I. modes of transmission. Am. J. Epidemiol.105, 337–343 (1977). [DOI] [PubMed] [Google Scholar]
- 29.Mey, A. R., Butz, H. A. & Payne, S. M. Vibrio cholerae CsrA regulates ToxR levels in response to amino acids and is essential for virulence. mBio6, e01064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milton, D. L. Quorum sensing in vibrios: complexity for diversification. Int J. Med Microbiol296, 61–71 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Chaparro, A. P., Ali, S. K. & Klose, K. E. The ToxT-dependent methyl-accepting chemoreceptors AcfB and TcpI contribute to Vibrio cholerae intestinal colonization. FEMS Microbiol Lett.302, 99–105 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Conner, J. G., Teschler, J. K., Jones, C. J., Yildiz, F. H. Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol. Spectr. 4, 10 (2016). [DOI] [PMC free article] [PubMed]
- 33.Jobling, M. G. & Holmes, R. K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol.26, 1023–1034 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Rothenbacher, F. P. & Zhu, J. Efficient responses to host and bacterial signals during Vibrio cholerae colonization. Gut Microbes5, 120–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiao, A. & Zhu, J. Pathogenicity and virulence regulation of Vibrio cholerae at the interface of host-gut microbiome interactions. Virulence11, 1582–1599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta, N., Paul, K. & Chowdhury, R. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect. Immun.71, 5583–5589 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi, D. et al. The response regulator ArcA enhances biofilm formation in the vpsT manner under the anaerobic condition in Vibrio cholerae. Micro. Pathog.144, 104197 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Cheung, J. & Hendrickson, W. A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem.283, 30256–30265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, G. H. & Boyd, E. F. On sialic acid transport and utilization by Vibrio cholerae. Microbiology157, 3253–3254 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Cheng, A. T., Ottemann, K. M. & Yildiz, F. H. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog.11, e1004933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitts, G. et al. The Rvv two-component regulatory system regulates biofilm formation and colonization in Vibrio cholerae. PLoS Pathog.19, e1011415 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Didelot, X. et al. The role of China in the global spread of the current cholera pandemic. PLoS Genet. 11, e1005072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyce, K. J. Mutators enhance adaptive micro-evolution in pathogenic microbes. Microorganisms. 10, 442 (2022). [DOI] [PMC free article] [PubMed]
- 44.Glass, R. I. et al. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1, biotype El Tor: evidence for a point-source outbreak in Bangladesh. J. Infect. Dis.147, 204–209 (1983). [DOI] [PubMed] [Google Scholar]
- 45.Dalsgaard, A., Forslund, A., Serichantalergs, O. & Sandvang, D. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother.44, 1315–1321 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaskolska, M., Adams, D. W. & Blokesch, M. Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature604, 323–329 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carraro, N., Rivard, N., Ceccarelli, D., Colwell, R. R., Burrus, V. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. mBio. 7, e00509 (2016). [DOI] [PMC free article] [PubMed]
- 48.Carraro, N., Matteau, D., Luo, P., Rodrigue, S. & Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet.10, e1004714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazama, H., Hamashima, H., Sasatsu, M. & Arai, T. Characterization of the antiseptic-resistance gene qacE delta 1 isolated from clinical and environmental isolates of Vibrio parahaemolyticus and Vibrio cholerae non-O1. FEMS Microbiol. Lett.174, 379–384 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Wang, R., Liu, H., Zhao, X., Li, J. & Wan, K. IncA/C plasmids conferring high azithromycin resistance in vibrio cholerae. Int J. Antimicrob. Agents51, 140–144 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. et al. IncA/C plasmids harboured in serious multidrug-resistant Vibrio cholerae serogroup O139 strains in China. Int J. Antimicrob. Agents45, 249–254 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Luo, Y. et al. Population structure and multidrug resistance of non-O1/non-O139 Vibrio cholerae in freshwater rivers in Zhejiang, China. Micro. Ecol.82, 319–333 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Orata, F. D. et al. The dynamics of genetic interactions between Vibrio metoecus and Vibrio cholerae, two close relatives co-occurring in the environment. Genome Biol. Evol.7, 2941–2954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao, Y. et al. Structural variation of the superintegron in the toxigenic Vibrio cholerae O1 EI Tor. Biomed. Environ. Sci.24, 579–592 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Boucher, Y. et al. Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. mBio2, e00335–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waters, E. V., Tucker, L. A., Ahmed, J. K., Wain, J. & Langridge, G. C. Impact of Salmonella genome rearrangement on gene expression. Evol. Lett.6, 426–437 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res.27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol.13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu, D., Liu, B., Wang, L. & Reeves, P. R. Living trees: high-quality reproducible and reusable construction of bacterial phylogenetic trees. Mol. Biol. Evol.37, 563–575 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol.35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother.67, 2640–2644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother.58, 3895–3903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feldgarden, M. et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep.11, 12728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, C. et al. The purifying trend in the chromosomal integron in Vibrio cholerae strains during the seventh pandemic. Infect. Genet Evol.26, 241–249 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res.14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Hunt, M. et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol.16, 294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stutzmann, S. & Blokesch, M. Comparison of chitin-induced natural transformation in pandemic. Vibrio cholerae O1 El Tor strains. Environ. Microb.22, 4149–4166 (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All 22 genomes sequenced in this study have been submitted as raw reads under BioProject accession number PRJNA970070 in the NCBI SRA database. The accession numbers for all other publicly available sequences used in this study are listed in Supplementary Data 10.