Abstract
The majority of viruses classified as pandemic threats are enveloped viruses which enter the cell through receptor-mediated endocytosis and take advantage of endosomal acidification to activate their fusion machinery. Here we report that the endosomal fusion of low pH-requiring viruses is highly dependent on TRPM7, a widely expressed TRP channel that is located on the plasma membrane and in intracellular vesicles. Using several viral infection systems expressing the envelope glycoproteins of various viruses, we find that loss of TRPM7 protects cells from infection by Lassa, LCMV, Ebola, Influenza, MERS, SARS-CoV-1, and SARS-CoV-2. TRPM7 ion channel activity is intrinsically necessary to acidify virus-laden endosomes but is expendable for several other endosomal acidification pathways. We propose a model wherein TRPM7 ion channel activity provides a countercurrent of cations from endosomal lumen to cytosol necessary to sustain the pumping of protons into these virus-laden endosomes. This study demonstrates the possibility of developing a broad-spectrum, TRPM7-targeting antiviral drug to subvert the endosomal fusion of low pH-dependent enveloped viruses.
Subject terms: Cellular imaging, Ion transport, Virology
The TRPM7 channel enables endosomal acidification, which is vital for the entry of many enveloped viruses. Here, the authors show that loss of TRPM7 protects cells from various pandemic-threat viruses, pointing to a new strategy for broad-spectrum antiviral drugs.
Introduction
The vast majority of zoonotic viruses that are prioritized by the National Institute of Allergy and Infectious Diseases as emerging or re-emerging threats (coronaviruses, orthomyxoviruses, flaviviruses, filoviruses, etc.) are enveloped viruses. Commonly, these viruses are internalized into host cells through different types of endocytic mechanisms, and the subsequent fusion of the viral envelope to the endosomal membrane releases the virus into the cytoplasm, often in close proximity to the nucleus1–3. Vaccines provide the ideal defense against viral infections and pathogenesis, but their development is not fast enough to prevent the rapid spread of a newly emergent virus. Similarly, many available antiviral drugs have been developed against distinct viral targets and their effectiveness is limited to a narrow spectrum of related viruses. It might be possible to develop broad-spectrum antivirals against enveloped viruses by identifying and targeting host cell factors that are required for infection and pathogenesis4–6. Such broad-spectrum antivirals can be an important component of an immediate and early response to blunt the severity of disease in the infected patients and as a prophylactic measure to prevent new infections in high-risk populations.
After a virus is internalized into a cell, the virus-containing endosomes go through a maturation process. As the endosomes mature from early to late endosomes, they undergo proteomic reconfiguration, and the pH of the lumen decreases progressively. Typically, during a successful infection, the enveloped viruses fuse to the endosomal membrane and escape into the cytosol. Many enveloped viruses take advantage of endosomal acidification to drive fusion to the endosome1,3,7. These pH-sensitive mechanisms may entail protonation-triggered conformational changes of crucial envelope-resident glycoproteins8,9 or activation of acid-optimized cathepsins, which can then activate key components of the viral fusion machinery through proteolytic cleavage10–12. Endosomes are acidified by vacuolar (H+)-ATPases (V-ATPases), which are assembled on the endosomal membrane as large multi-subunit proton pumps13. V-ATPases use cytosolic ATP to pump protons across the endosomal membrane, but as protons accumulate inside the endosomal lumen, the endosomal membrane is hyperpolarized (positive charge accumulates on the luminal side of the endosomes). If the increase of positive charge is not electrically balanced by a parallel flux of counter-ions, the energetic cost for the H+-pumping activity of V-ATPase continues to rise, and endosomal acidification cannot be sustained14,15. Theoretically, this countercurrent can be achieved by an anionic flux into the endosomal lumen (movement of Cl- from the cytosol to endosomal lumen) or a cationic flux from the lumen (movement of Na+, K+, Ca2+ from the lumen of the endosomes to the cytosol), but as yet, the ion channels that mediate such countercurrents and thus drive the acidification of virus-laden endosomes and viral membrane fusion are not well defined.
Intracellular membranes, including endosomes, contain a variety of ion channels16,17. The Transient Receptor Potential (TRP) channels constitute a large family of cation-selective ion channels18–20 and many of its members, most notably the lysosomal TRPML1-3, have important functions in the intracellular membranes21,22. The chanzyme TRPM7, a TRP channel that also contains a functional kinase domain23,24, resides in the plasma membrane and in intracellular vesicles25. In 2005, a study reported an unbiased genome-wide screen of the human kinome to identify the kinases regulating different forms of endocytosis26. Investigators used infection by vesicular stomatitis virus (VSV) as a readout for clathrin-mediated endocytosis and identified TRPM7 among 128 other putative hits as a significant effector of VSV infection. However further investigation as to the breadth of viruses affected and the mechanism of TRPM7 inhibition was never explored.
TRPM7 is nonselective for cations and is permeable to Na+, K+, Ca2+, Mg2+, Zn2+, and other divalent cations27. It conducts only a few pA of inward current at physiological pH but is greatly potentiated by low pH28. TRPM7 is expressed at very high levels in mouse embryos, but its expression diminishes greatly as embryogenesis is completed29. When deleted globally, Trpm7−/− embryos do not survive past day 7 of embryogenesis29,30 but its deletion after organ development is well tolerated30. In adult mice, TRPM7 currents are readily detected in most cell types by electrophysiology31–33 but the protein levels are low, and therefore enrichment of the membrane fractions or immunoprecipitation is required to detect the protein by immunoblotting34,35. Cell biological investigations of TRPM7 have suggested a role for TRPM7 in membrane trafficking. For instance, TRPM7 has been implicated in receptor internalization that is required for Fas-signaling in T cells34 and TLR4 signaling in macrophages32. Recently, it was also shown that TRPM7 affects acidification of macrophage efferophagosomes (phagosomes containing apoptotic cell corpses)36. However, the precise molecular function of TRPM7 remains elusive. Identifying the molecular mechanisms underpinning cellular and physiological phenotypes associated with TRPM7 is necessary to advance TRPM7 as a rational and feasible drug target.
In this study, we tested the central hypothesis that TRPM7 ion channel activity regulates enveloped virus infection. We found that TRPM7 is required for infection by a wide range of enveloped viruses that use low endosomal pH for membrane fusion. We show that TRPM7 is required to acidify the virus-containing endosomes, and through other mechanistic inquiries, our evidence leads to a model wherein TRPM7 mediates the cationic countercurrent necessary for the V-ATPase-mediated acidification of viral endosomes.
Results
Validation of TRPM7 knockdown in SVG-A cells
TRPM7 was depleted in SVG-A cells, a human fetal astroglial cell line, using siRNA. Compared to control (Ctl) cells which were treated with non-targeting siRNA, the knockdown (M7KD) SVG-A cells were depleted of TRPM7 mRNA to ~20% of levels in control cells (Fig. 1a). Whole-cell patch clamp electrophysiology showed that siRNA-mediated depletion of TRPM7 greatly reduced the typical outwardly rectifying TRPM7 currents (IM7) that are readily detected in control cells. The current-voltage (IV) relationships seen in Ctl and M7KD cells are shown overlaid (Fig. 1b, left panel). Quantification of peak current densities observed at +100 mV is shown (Fig. 1b, right panel). These analyses confirm the efficient depletion of TRPM7 in M7KD cells.
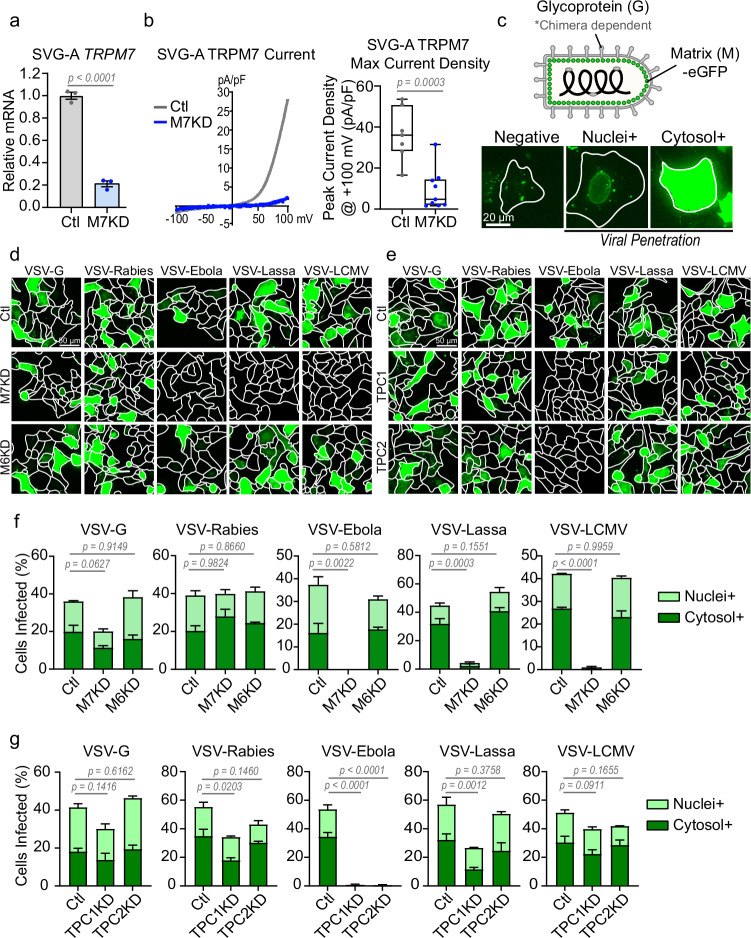
Fig. 1. Loss of TRPM7 broadly inhibits infection by various VSV-chimeras.
a qRT-PCR of TRPM7 in control non-targeting (gray, Ctl) and TRPM7 (blue, M7KD) siRNA treated SVG-A cells from independent transfections. P values determined by two-tailed Student’s t test. Mean, and SEM plotted, n = 3. b (Left) Representative whole-cell patch-clamp recordings shown as current-voltage (IV) relationship. The IVs of Control (gray) and TRPM7 KD (blue) SVG-A cells are overlayed. (Right) A quantitative summary of peak current densities from individual cells at +100 mV is shown (Ctl n = 7, M7KD n = 9 cells). P values determined by two-tailed Student’s t test. Median, 25th, and 75th quartiles are shown with maximum and minimum. c (Top) Schematic of VSV-chimeras used for viral infection assays. Chimeras express the glycoprotein of the indicated viruses. All viruses express MeGFP fusion protein. (Bottom) Viruses that have penetrated the cytosol release MeGFP which subsequently binds to nuclear pores (nuclei+). As viral genomes are translated, MeGFP accumulates in the cytosol (cytosol+). Cytosol+ and nuclei+ are both representative of successful viral escape from the endosomes into the cytosol. Representative images from two independent experiments are shown. d Representative maximum projection fluorescence microscopy images of viral infection (eGFP) in control (Ctl), TRPM7 knockdown (M7KD), and TRPM6 knockdown (M6KD) SVG-A cells. e Representative maximum projection fluorescence microscopy images of viral infection (eGFP) in control (Ctl), TPC1 knockdown (TPC1KD), and TPC2 knockdown (TPC2KD) SVG-A cells. f Quantification of viral penetration in control, M7KD, and M6KD cells by various VSV-chimeras at 6 hours post-infection. Percentage of nuclei+ (light green) and cytosol+ (dark green) cells together represent total virus-infected cells. Mean and SEM plotted of average infection across three independent infections. P values (one-way ANOVA with Tukey’s multiple comparison test) are respective to total viral infection values. g Quantification of virus penetration in control, TPC1KD, and TPC2KD cells by indicated VSV-chimeras at 6 hours post-infection. Percentage of nuclei+ (light green) and cytosol+ (dark green) cells represent total virus-infected cells. Mean and SEM plotted of average infection across three independent infections. P values (one-way ANOVA with Tukey’s multiple comparison test) are respective to total viral infection values.
Assessment of infection by eGFP-expressing VSV particles
VSV particles engineered to express eGFP-tagged Matrix (M) protein (Fig. 1c, top panel schematic) were used to infect the SVG-A cells. Three stages of infection can be observed using this approach, as shown in the bottom panel of Fig. 1c and schematized in Supplementary Fig. 1a. In the first stage, the internalization of the viral particles yields endosomes that contain the fluorescent virus, and these can be visualized as internalized puncta. In this stage, the virus has not yet escaped from the endosomes, and from the standpoint of productive infection, these cells are still counted as “negative” for infection. In the second stage, successful fusion of these viral particles enables the virus to escape from the endosome and into the cytosol. Once the virus penetrates the cytoplasm, the M-protein dissociates and coats the nuclear membrane of the host cell, and this is visualized as nuclear rings37. These cells are counted as “Nuclei+”. Finally, in the third stage of the infection, the virus replicates, and the amplified MeGFP protein fills the cytosol (Cytosol+). Thus, the latter two stages, Nuclei+ and Cytosol+, represent successful penetration of the virus across the endosomal membrane which is indicative of infection. The outlines of the cells are determined based on staining with fluorescently labeled Wheat Germ Agglutinin (WGA) – representative images are shown (Supplementary Fig. 1b).
TRPM7-depleted SVG-A cells are resistant to VSV-chimeras of Ebola, Lassa, and LCMV viruses
The VSV genome can be further manipulated to generate VSV chimeras in which the VSV glycoprotein is substituted by the molecular analogs belonging to other enveloped viruses. The glycoprotein is responsible for binding and fusion of enveloped viral proteins to the cell, and thus chimeric viruses are an excellent tool to study virus entry into the cell37. We infected SVG-A cells with VSV chimeras expressing the glycoproteins specific to VSV (indicated as VSV-G), Rabies, Ebola, Lassa, and LCMV. Cells were imaged before budding of viruses could occur, ensuring that cells were only infected by the initial one-hour virus incubation. As shown in Fig. 1d, TRPM7-depleted SVG-A cells (M7KD) were protected to varying extent against infections by most VSV chimeras. We quantified these infections in terms of the infection stages described earlier (Fig. 1f). The TRPM7-depleted cells were only mildly resistant to VSV-G and had no resistance to VSV-Rabies infection but were significantly more resistant to VSV-Ebola, VSV-Lassa and VSV-LCMV infections. This inhibition was seen across two multiplicities of infection (MOIs) of the virus (Supplementary Fig. 2). Since these VSV chimeras differ only in the molecular components needed for membrane fusion, these effects are not due to differences in the replication and translation of the virus genomes, which would present as cells only being nuclei+. Loss of TRPM6, a close homolog of TRPM7, had no effect on virus infection in SVG-A cells (Fig. 1d, f).
TPC channels, host factors for Ebola infection, do not regulate infection by several other VSV chimeras
TPC1 and 2 are endosomal cation channels known to be host factors for Ebola, Middle East Respiratory Syndrome coronavirus (MERS), and SARS-CoV-2 infection38–40. To assess the selectivity of the TRPM7 effect, we tested whether TPC channels also regulated enveloped virus infections in a manner similar to TRPM7. As expected, the loss of TPC1 or TPC2 completely abrogated the infection by VSV-Ebola (Fig. 1e, g). However, the loss of TPC1 only had a modest effect on infection by other VSV-chimeras tested, and the loss of TPC2 had no significant effect on LCMV and only a modest effect on Lassa. These differences suggest that TRPM7 and TPC channels execute different functions during enveloped virus infection.
Loss of TRPM7 decreases plaque formation
To supplement the imaging-based infection assays, we performed virus plaque assays (Supplementary Fig. 1c). Unlike the imaging assay, which utilizes a higher multiplicity of infection (MOI) of virus and assesses the first round of viral infection, plaque assays integrate infections resulting from many rounds of virus replication. Control (Ctl) and TRPM7-knockdown (M7KD) SVG-A cells were infected with VSV-Lassa (MOI = 0.01) and incubated for 48 hours prior to harvesting for plaque assay. Plaque assays of VSV-G, VSV-Lassa, and VSV-LCMV in Vero cells show that depletion of TRPM7 by siRNA significantly decreases the infection by these three viruses. Note that due to the exponential expansion of the virus during the 48 h infection period, smaller differences, like the ~20% decrease seen in the imaging-based infection assay of VSV-G, are amplified further in the plaque assay.
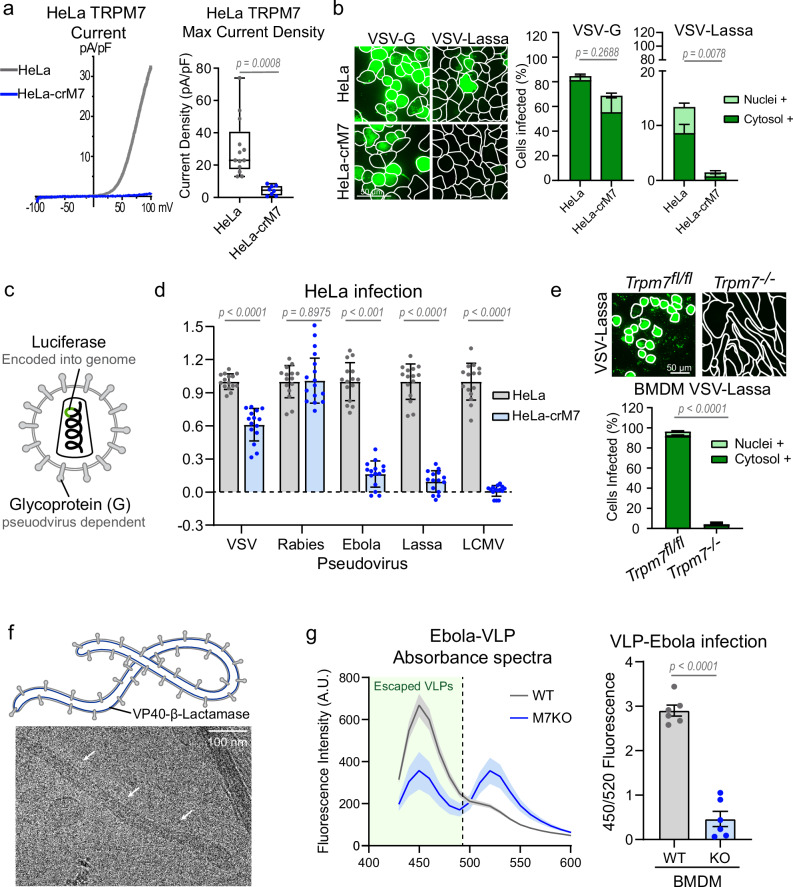
TRPM7 knockout in HeLa cells prevents infection from VSV-Lassa and various pseudoviruses
Next, we used HeLa-crM7 cells in which TRPM7 is deleted by CRISPR-Cas9 technique. As expected, the HeLa-crM7 cells do not display IM7 (Fig. 2a). The HeLa-crM7 cells are strongly resistant to VSV-Lassa (Fig. 2b), demonstrating a similar response to the SVG-A knockdown. To ensure that these observations were not an artifact of the VSV-chimera system, we infected HeLa cells with pseudoviruses produced using a lentivirus system41. These constructs are lentivirus particles, which express the glycoprotein of various enveloped viruses (Fig. 2c). These particles also encode luciferase in the viral genome, which is expressed in the host cell following successful infection. Control HeLa cells were readily infected with all pseudoviruses. HeLa-crM7 cells were infected with VSV and rabies pseudotyped viruses, but were resistant to infection by Ebola, Lassa, or LCMV constructs (Fig. 2d). These results are consistent with those obtained with VSV chimeras.
Fig. 2. Loss of TRPM7 protects a variety of cell types from various enveloped viral infections.
a Representative whole cell patch clamp recordings shown as current-voltage (IV) relationship of HeLa (gray) and HeLa-crM7 (blue) wherein Trpm7 was deleted by Crispr-cas9. Quantitative summaries of peak current densities at +100 mV are shown to the right (HeLa n = 13, HeLa-crM7 n = 8). P values determined by two-tailed Student’s t test. Median, 25th, and 75th quartiles are shown, as well as maximum and minimum. b (Left) Representative 20 μm maximum intensity projection of HeLa and HeLa-crM7 cells infected with VSV-G and VSV-Lassa. (Right) Quantification of nuclei+ and cytosol+ infected cells. P values determined by two-tailed Student’s t test of total positive cells. Each experiment is plotted as the average percent infected across multiple fields of view, n = 3. Mean and SEM are plotted. c Schematic of lenti-pseudovirus expressing various enveloped virus glycoproteins. Each pseudovirus genome encodes luciferase. d Relative luciferase luminescence of lenti-pseudovirus infected HeLa cells. Graph shows data points from three independent experiments with five well replicates of HeLa (gray) and HeLa-crM7 (blue). Data points were normalized to average control HeLa luciferase intensity per experiment. P values determined by two-tailed Student’s t test, n = 15. Mean and standard deviation are plotted. e (Left) Representative images of infection Trpm7fl/fl and Trpm7−/− BMDMs with VSV-Lassa. WGA outlines are shown. (Right) Quantification of average percent infected from n = 3 independent experiments. P values determined by two-tailed Student’s t test of total positive cells. Mean and SEM are plotted. f (Top) Schematic of ebola virus-like particle (VLP) expressing membrane-bound VP40 tagged with β-lactamase. (Bottom) Cryo-EM of ebola VLP. White arrows indicate one VLP. The cryo-EM image shown is representative of multiple VLPs observed in two independent sample preparations. g (Left) Spectrum of β-lactamase intensity of WT (gray) and TRPM7 KO (blue) BMDMs at 400 nm excitation. Mean and SEM are plotted. β-lactamase tagged VP40 from cytosolic-penetrated VLPs can cleave CCF-2 AM dye, decreasing FRET 520 nm signal and increasing 450 nm signal. (Right) Average ratio of 450 nm to 520 nm emission is plotted. Two-tailed Student’s t test was used to determine p values. Mean and standard errors are shown. n = 6.
Trpm7−/− primary mouse macrophages are resistant to VSV-Lassa and Ebola virus-like particles
We then sought to extend these results to primary cells. Since Lassa and Ebola initially infect human macrophages during the course of their systemic infection42, we tested whether TRPM7 was necessary for such infection in murine bone marrow-derived macrophages (BMDMs). We found that VSV-Ebola does not infect murine BMDMs, but VSV-Lassa infects BMDMs with high efficiency. Trpm7−/− BMDMs, derived from Trpm7fl/fl LysM-Cre mice, were highly resistant to infection by VSV-Lassa (Fig. 2e). Lastly, we used Ebola virus-like particles (VLPs) which retain the shape and membrane composition of ebola in a non-replicative vector43. This particular construct expresses β-Lactamase tagged to VP40, a protein lining the inner leaflet of the ebola membrane (Fig. 2f). Viruses that successfully escape the endosome release VP-40-β-lactamase into the cytosol, which can then cleave CCF2-AM dye. CCF2 consists of 2 fluorophores connected by a β-lactam ring that FRET pair when connected but are separated by β-lactamase. Therefore, the ratio of CCF2 that emits at 450/520 reflects the efficiency of virus escape into the cytosol. Bone marrow-derived macrophages were incubated with ebola VLPs for 3 hours and then loaded with CCF2-AM dye at room temperature before detecting emission. Compared to WT BMDMs, M7KO BMDMs exhibited a significant decrease in 450/520 nm emission, indicating a decrease in cytosolic viral penetration.
Overall, the results obtained using multiple approaches (siRNA, genetic deletion, imaging-based virus infection assays and plaque assays), different viral platforms (VSV chimeras, Lentivirus-based pseduviruses, and ebola VLPs), and different cell types (SVG-A, HeLa, primary BMDMs) reveal that TRPM7 is expendable for VSV and Rabies infection but has a crucial role in infection by other clinically relevant enveloped viruses (i.e. Lassa, LCMV, and Ebola).
TRPM7 promotes, but is not necessary for, the endocytosis of VSV chimeras
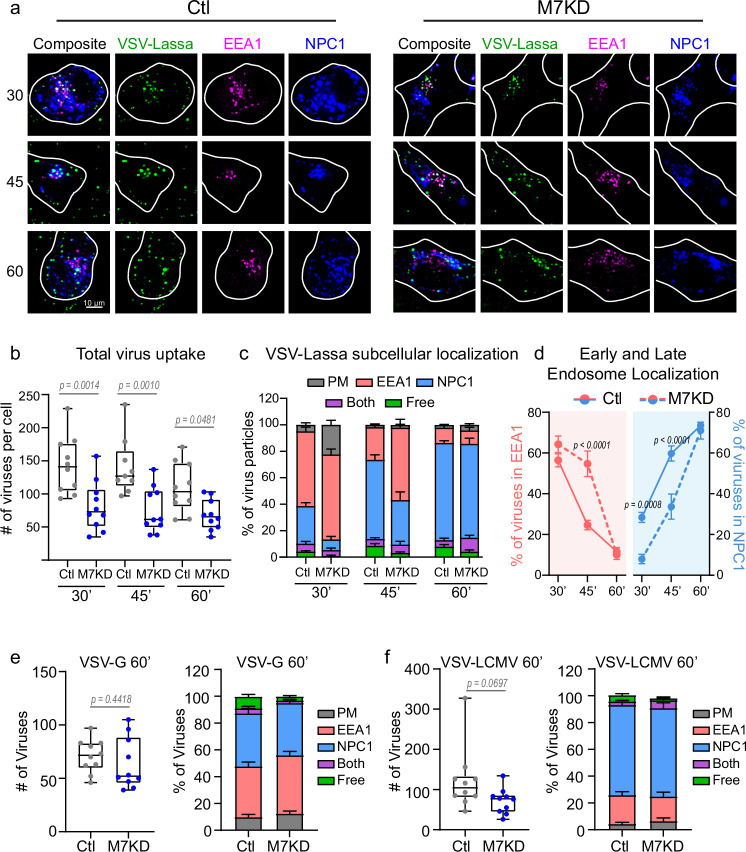
We first tested the hypothesis that TRPM7 regulates the endocytosis of the enveloped viruses. To test this, we used SVG-A cells that were gene-edited to incorporate fluorescent tags in endogenous alleles of proteins that mark early and late endosomes37. In these cells, mScarlet was fused to the N-terminus of the early endosome marker EEA1 (mScarlet-EEA1), and Halo was fused to the C-terminus of the late endosome marker NPC1 (NPC1-Halo). Halo is a modified haloalkane dehalogenase that can be covalently conjugated with synthetic, cell-permeable fluorescent dyes. Halo was fluorescently labeled with JFX647 for 30 minutes before exposure to virus. We infected these gene-edited SVG-A cells with VSV-Lassa-MeGFP. The cells were imaged at 30-, 45- and 60-minutes post-infection and individual viruses were determined to localize to the plasma membrane, early endosome, late endosome, dual labeled compartments, or free in the cytosol. The number of endocytosed viral particles was estimated based on the average fluorescent intensity of a single viral particle and the point spread function of fluorescent spots in the cells (Supplementary Fig. 3a–d). In total, 3,791 viruses in control cells and 2,258 viruses in knockdown cells were analyzed for the VSV-Lassa-treated cells. The endocytosed viral particles co-localized with early endosomes (EEA1) and late endosome/lysosomes (NPC1) in both control and TRPM7-depleted cells (M7KD) (Fig. 3a). We found that compared to control cells, the cells depleted of TRPM7 contained a reduced number of VSV-Lassa particles (Fig. 3b), but these particles progressed from early to late endosomes in both cell populations (Fig. 3c and Supplementary Fig. 3e). While the rate of progression was slowed in cells depleted of TRPM7, by 60 minutes the same proportion of viruses had progressed to NPC1+ stage (Fig. 3d). We also evaluated VSV-G and VSV-LCMV colocalization at the 60-minute time point. Of the viruses endocytosed, a similar proportion of viruses localized to early and late endosomal compartments (Fig. 3e, f, Supplementary Fig. 3f). VSV-LCMV also trended towards a decrease in the number of viruses endocytosed. These results indicate that although TRPM7 promotes the endocytosis of VSV chimeras, it is not essential for this process, and the striking resistance to VSV-Lassa and VSV-LCMV infection is not adequately explained by this aspect of TRPM7 function.
Fig. 3. Viruses traffic through the endosomal maturation pathway in TRPM7 depleted cells.
a Representative images of VSV-Lassa-MeGFP infected Control (Ctl) and TRPM7 knockdown (M7KD) SVG-A cells. Early endosomes are tagged with EEA1-mScarlet (magenta), late endosomes/lysosomes with NPC1-Halo-JFX647 (blue). Cells were incubated with VSV-Lassa-MeGFP for 20 minutes, washed, and imaged at 30-, 45-, and 60-minutes post-infection. Maximum projection of 1.08 μm is shown. b Box and whisker plot of total number of virus particles detected per cell at each time point in Control (gray) and M7KD (blue) cells at 30-, 45-, and 60-minutes post-infection. Median, quartiles, maximum, and minimum are shown. P values determined by two-way ANOVA with Sidak multiple comparisons test. For control-treated cells at 30, 45, and 60 minutes, N = 10, 9, 10. For all M7KD timepoints, N = 10. Same cells are used for analysis in (c, d). c Average percent of viruses localizing to the cell surface (gray), early endosome (red, EEA1), late endosome (blue, NPC1), or free in cytosol (green) per cell at 30-, 45-, and 60-minutes post-infection. Some viruses localized to endosomes that were EEA1+ and NPC1+ (both, purple). Mean and SEM plotted. d Average percent of viruses per cell in early (red) and late (blue) endosomes. Control cells are represented by solid lines, while M7KD cells are represented with dashed lines. Mean and SEM are shown. P values determined by two-way ANOVA with Sidak multiple comparisons test. e (Left) Box and whisker plot of VSV-G particle uptake at 60 minutes post-infection in Control (gray) and M7KD (blue) cells. Median, quartiles, maximum, and minimum are shown. P values determined by two-tailed Student’s t test. (Right) Subcellular localization in Control and M7KD SVG-A cells at 60 minutes post-infection. n = 10 cells per condition. Mean and SEM plotted. f (Left) Box and whisker plot of total VSV-LCMV viral uptake at 60 minutes post-infection in Control (gray) and M7KD (blue) cells. Median, quartiles, maximum, and minimum are shown. P values determined by two-tailed Student’s t test. (Right) Subcellular localization in Control and M7KD SVG-A cells at 60 minutes post-infection. n = 10 cells per condition. Mean and SEM plotted.
TRPM7 is recruited to dense virus-laden endosomes
To determine if TRPM7 localizes to virus-containing endosomes, we used density gradient centrifugation to fractionate the intracellular vesicular compartments (Supplementary Fig. 3h). In this experiment, the MeGFP expressed by the VSV-Lassa-MeGFP enables the identification of fractions that contain the virus-laden endosomes. In uninfected cells, Rab5, a marker of early endosomes is found predominantly in the lighter fractions (fraction 3 and 4). However, in infected cells, the early endosomes that contain the viruses are dense and hence Rab5 is detected in the denser fractions 8 and 9. In uninfected SVGA cells, TRPM7 is seen in vesicular fractions of various densities. However, in VSV-Lassa-MeGFP infected cells, TRPM7 is found predominantly in the dense fractions (fractions 8 and 9) and depleted from less dense fractions. These data suggest that TRPM7 is a component of virus-laden endosomes.
TRPM7-deficient cells are particularly resistant to viruses that depend on low pH for endosomal fusion
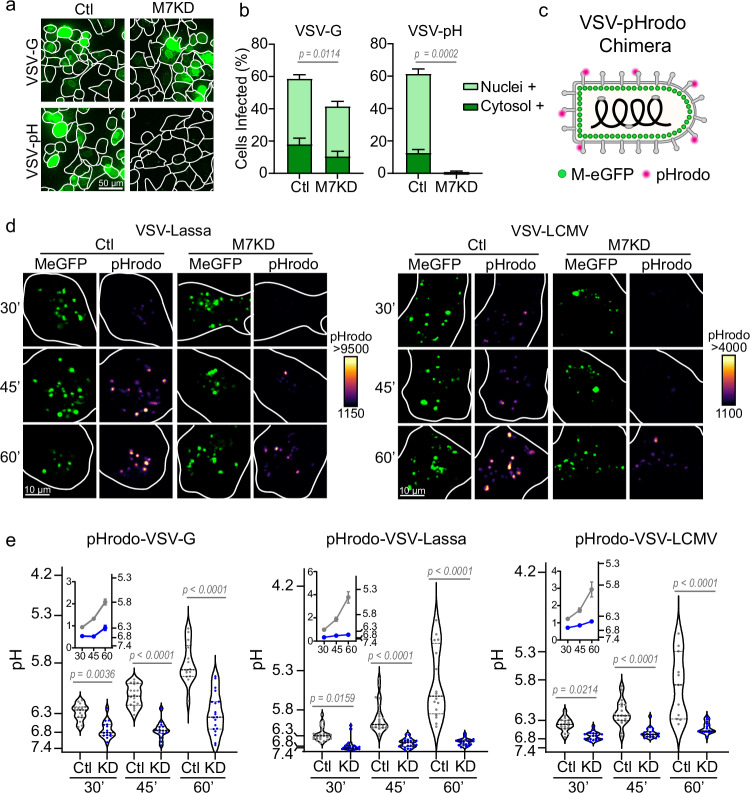
The viral glycoproteins we tested undergo pH-dependent conformational changes to trigger membrane fusion. The envelope glycoproteins of VSV and Rabies are estimated to undergo a conformational change at a pH of ~6.2 and ~6.15, respectively44,45. The Lassa and LCMV glycoproteins require a pH lower than 5.5 for viral escape46–48. The Ebola glycoprotein requires proteolytic processing by acid-activated host cell cathepsin in the endolysosomal network11,47,49. Since cathepsins are optimized for the low pH found in the lysosome, Ebola escapes in the later stages of the endolysosomal pathway50. We noted that TRPM7-depleted cells exhibited significantly higher resistance to viruses that require low pH for membrane fusion. To test the hypothesis TRPM7 regulates the fusion of viruses that require a low endosomal pH, we focused on VSV-G. While VSV binds to its receptor at the plasma membrane and is endocytosed like other viruses, VSV does not need the receptor once internalized and only requires a modest decrease in endosomal pH to trigger fusion3. It has been shown that a substitution of a single amino acid (V269H) in the VSV glycoprotein allows the virus to get internalized into the host cell normally but requires a lower pH (<5.8) for virus fusion37. We here refer to this VSV variant as VSV-pH. We tested VSV-G and VSV-pH for infection in control and TRPM7-depleted SVG-A cells. As seen previously, depletion of TRPM7 conferred a modest resistance to VSV-G, but the infection by VSV-pH was completely inhibited (Fig. 4a, b). Since VSV-G infects HeLa cells readily, we also tested VSV-pH infection in HeLa-crM7 cells wherein endogenous TRPM7 is deleted. Again, infection by VSV-pH was robustly inhibited in the absence of TRPM7 (Supplementary Fig. 4a). These results substantiated our hypothesis that TRPM7 activity is required for infection by enveloped viruses that depend on low endosomal pH for membrane fusion. This also indicates that changes in receptor availability or endocytosis are not the main factors of infection resistance in TRPM7-depleted cells. Loss of other endosomal ion channels, TPC1 and TPC2, did not significantly inhibit VSV-pH infection, nor did the loss of TRPM6 (Supplementary Fig. 4b), which is only expressed at very low levels.
Fig. 4. TRPM7 regulates viral endosome acidification.
a Representative maximum projections of Control (Ctl) and TRPM7 knockdown (M7KD) SVG-A cells infected by parental VSV-G or the VSV pH-sensitive mutant, VSV-V269H. Cells were fixed 6 hours post-infection. b Quantification of VSV-G and VSV-pH sensitive mutant infection. Percent of Nuclei+ cells are shown in light green and Cytosol+ cells in dark green. P values determined from two-tailed Student’s t test of total positive cells from three independent infection assays. Mean and SEM are plotted. c Schematic of pHrodo-tagged VSV-chimera viruses expressing MeGFP. d Representative images of MeGFP and pHrodo signal time course in Ctl and M7KD SVG-A cells. VSV-Lassa-MeGFP or VSV-LCMV-MeGFP is shown in green. pHrodo signal is shown pseudo-colored based on the intensity spectrum, with black/purple being the lower intensity and yellow being the highest intensity. Images are 2.43 μm maximum projections. e Single particle quantification of the viral endosome pH at 30-, 45-, and 60 minutes following virus incubation. Control (Ctl) cells are shown in gray and TRPM7 knockdown (KD) cells in blue. Each data point represents the average pH of all virus particles detected across a single cell. Violin plots display median (thick dotted line) as well as 25th and 75th quartiles (thin dotted lines). P values are determined by two-way ANOVA with Sidak multiple comparisons test. Inset graphs show mean pH per cell over time with SEM. At 30, 45, and 60 minutes VSV N = 15, 18, 14; Lassa N = 25, 22; 16; LCMV N = 17, 19, 11 for control cells and VSV N = 15, 15, 19; Lassa N = 24, 23, 20; LCMV N = 19, 20, 18 for M7KD cells.
TRPM7 is required for the acidification of virus-laden endosomes
To assess the acidification of virus-containing endosomes, we conjugated the envelope proteins of VSV-MeGFP with the pH-sensitive dye pHrodo, which reports increases in fluorescence intensity as pH decreases (Fig. 4c). While the pHrodo tag was exposed to the endosomal lumen, the eGFP-labeled M-protein located inside the envelope of the virus is protected from acid-induced quenching by endosomal acidification. This configuration enables ratiometric assessment of the change in endosomal pH. A decrease in endosomal pH is reported by an increase in the ratio of pHrodo:eGFP. We calibrated these measurements to absolute pH by imaging the virus particles alone in varying pH buffers (Supplementary Fig. 4c). Across time points and treatments, 4633 VSV-G, 9430 VSV-Lassa, and 1885 VSV-LCMV particles were analyzed, and the average particle pH was reported for each cell. As expected, the control SVG-A cells infected with pHrodo-labeled VSV-Lassa-MeGFP display a progressive decrease in pH over 60 minutes. In contrast, the TRPM7-depleted SVG-A cells (M7KD) display reduced acidification (Fig. 4d, left). A similar difference is also seen in cells infected with VSV-LCMV (Fig. 4d, right). A quantitative summary of these data shows that depletion of TRPM7 results in a substantial and significant defect in acidification of the virus-loaded endosomes, allowing a pH range of 6.8–6.5 at one hour while controls exhibit a pH range of 5.8–5.4 (Fig. 4e). Infection of HeLa and HeLa-crM7 cells showed a very similar inhibition of acidification (Supplementary Fig. 4d). Addition of 50 nM Bafilomycin, a potent inhibitor of the V-ATPase, completely abrogated endosomal acidification. We conclude from these data that TRPM7 is necessary for the acidification of virus-loaded endosomes.
TRPM7 does not regulate the acidification of all endosomes
If TRPM7 regulates the pH of all endosomal compartments, we would expect drastic effects on cell proliferation and survival, as nutrient uptake and autophagy are highly dependent on endosomal acidification. However, HeLa-crM7 cells grow and proliferate readily (Supplementary Fig. 5a). To test whether loss of TRPM7 induced global changes to the pH of highly acidified compartments, cells were loaded with Lysosensor, a dye that accumulates in acidic compartments and increases in fluorescence as pH decreases (Supplementary Fig. 5c). Compared to parental HeLa cells, there were no robust changes of fluorescence in HeLa-crM7 cells. Bafilomycin treatment led to a robust loss of Lysosensor fluorescence, indicating that intracellular compartments in HeLa-crM7 were substantially acidified. Lastly, we tested whether TRPM7 regulates the degradation of the epidermal growth factor receptor (EGFR). EGF binding to EGFR triggers the complex to be endocytosed and trafficked to lysosomes for pH-mediated degradation that can be inhibited by Bafilomycin51. Immunoblot of cell lysates treated with EGF demonstrates that loss of TRPM7 in SVG-A or HeLa cells does not impede EGFR degradation (Supplementary Fig. 5b). These results indicate that TRPM7 regulates the acidification of a subset of endosomes (including virus-containing endosomes), but not all endosomes.
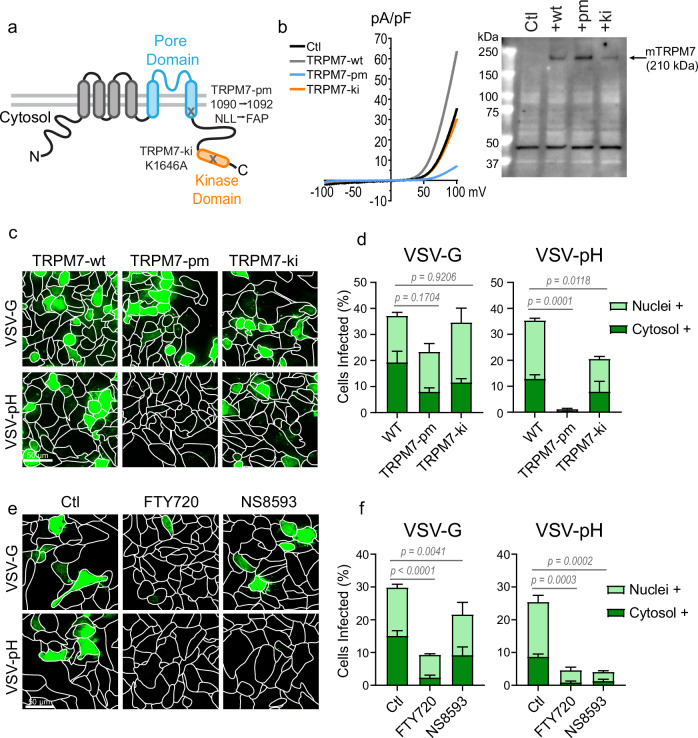
The ion channel activity of TRPM7 is necessary for infection by pH-dependent VSV-pH
TRPM7 contains both a functional ion channel and a kinase domain. To determine which of these activities was salient for endosomal pH regulation, SVG-A cells were transfected with mouse TRPM7 variants wherein either the ion channel pore or the kinase activity is inactivated by site-directed mutagenesis (Fig. 5). The unmodified wild-type TRPM7 (TRPM7-wt), the pore mutant (TRPM7-pm) or the kinase inactivated variant (TRPM7-ki) were reported previously34. Tetramerization of TRPM7 monomer is required to form a functional channel and thus, incorporation of TRPM7-pm monomers into the tetrameric complex renders the entire ion channel pore dysfunctional. Each of these variants was ectopically expressed in SVG-A cells and evaluated for infection by VSV-G and VSV-pH. The ectopic expression of TRPM7-pm in SVG-A cells had the expected dominant-negative effect on endogenous TRPM7 currents (IM7) (Fig. 5b). Immunoblotting of membrane fractions for mouse TRPM7 demonstrated successful expression of all three variants. TRPM7-wt and TRPM7-ki had lower expression, most likely because high expression of the functional ion channel can lead to cell death and therefore favoring the survival of cells with moderate expression34. Compared to cells transfected with TRPM7-wt, the TRPM7-pm transfected cells were modestly deficient in infection by VSV-G. However, TRPM7-pm expression resulted in complete abrogation of infection by the more pH-dependent VSV-pH virus (Fig. 5c, d) and inhibited the acidification of the virus-laden endosomes (Supplementary Fig. 6a). These results established that the ion channel activity of TRPM7 is required for endosomal acidification. In contrast, the cells ectopically expressing TRPM7-ki displayed only a modest resistance to infection by VSV-pH (Fig. 5c, d). The role of TRPM7 kinase activity cannot be ruled out due to differences in expression levels, but the available evidence indicates that it plays a modest role in the regulation of viral endosome acidification. Since TRPM7 channel activity is required for VSV-pH infection, we tested the effect of two structurally distinct small molecule TRPM7 channel inhibitors on VSV-pH infection. Based on previous reports, we evaluated FTY720 at 5 μM52 and NS8593 at 30 μM53,54. At these concentrations, both compounds inhibited the native IM7 in SVG-A cells (Supplementary Fig. 6b). Cells treated with either compound exhibit more resistance to VSV-pH than to VSV-G (Fig. 5e, f). Infections of TRPM7 knockdown cells in the presence of TRPM7 inhibitors reveal that FTY720 further inhibits the infection while NS8593 does not (Supplementary Fig. 6c). These data argue that NS8593 is a more selective inhibitor of TRPM7 and that other molecular targets of NS8593 are not factors in virus infection of SVG-A cells. These findings establish a salient function for TRPM7 ion channel activity in the acidification of virus-laden endosomes and in the infection by pH-dependent viruses.
Fig. 5. Ion channel activity of TRPM7 is required for pH-sensitive virus infection.
a Diagram of TRPM7 domains with annotations of pore mutant (pm) and kinase-inactive (ki) mutations. b (Left) Whole-cell patch-clamp recordings shown as current-voltage (IV) relationship of mouse TRPM7 constructs expressing SVG-A cells. The IV of non-transfected (black), wild-type (TRPM7-wt, dark gray), pore mutant (TRPM7-pm, blue), and kinase-inactive (TRPM7-ki, orange) transfected SVG-A cells are overlayed. (Right) Immunoblot of membrane fraction of non-transfected SVG-A cells (ctl) and cells transfected with mouse TRPM7-wt, -pm, and -ki constructs. Immunoblotting is done with an anti-mouse TRPM7 antibody which does not detect human TRPM7. A non-specific band seen at 45 kDa is used as a loading control. The immunoblot results were reproduced in two independent experiments. c Representative maximum projection (20 μm) images of VSV-G and VSV-pH infections (green) in SVG-A cells ectopically expressing the TRPM7 variants: wild-type (TRPM7-wt), pore mutant (TRPM7-pm), or kinase-inactive (TRPM7-ki). d Quantification of infection in SVG-A cells ectopically expressing TRPM7-wt, TRPM7-pm, and TRPM7-ki variants. Cells were infected with either parental VSV-G or VSV-pH. Percent cytosolic+ (dark green) and nuclei+ (light green) cells are shown. Averages are derived from three independent infections. P values were determined from total cells infected by one-way ANOVA with Tukey’s multiple comparison test. Mean and SEM are plotted. e Representative maximum projection (20 μm) images of VSV-G and VSV-pH infections (green) in SVG-A cells treated with TRPM7 inhibitors FTY720 and NS8593 at 5 µM or 30 µM respectively. The inhibitors were added to the cells 15 minutes prior to infection and are present throughout infection. f Quantification of VSV-G and VSV-pH infection of SVG-A cells in the presence of TRPM7 inhibitors FTY720 (5 μM) and NS8593 (30 μM). Percent cytosolic+ (dark green) and nuclei+ (light green) are shown. Averages are derived from three independent infections. P values were determined from total cells infected by one-way ANOVA with Tukey’s multiple comparison test. Mean and SEM are plotted.
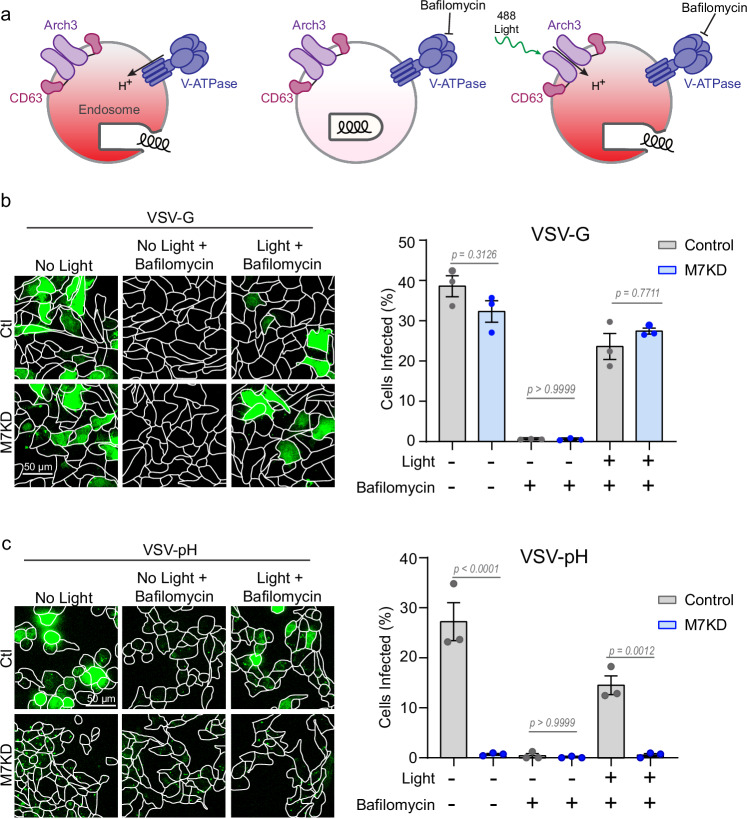
Endosomal proton-pumping cannot be sustained without TRPM7
A substantial pool of TRPM7 is located intracellularly in vesicular compartments25. We reasoned that intracellular TRPM7 may have an important role in early and late endosomes as an ion channel mediating the cationic countercurrent to the proton-pumping activity of V-ATPases. To test this hypothesis, we used an optogenetic tool that can manipulate the intraluminal pH on command. Arch3-CD63-Halo (A3CH), a light-driven proton pump generated by fusing Archaerhodopsin 3 (Arch3) with the late endosome/lysosome protein CD63 (known as lysophoenix)55, was modified to contain a Halo-tag and was stably expressed in SVG-A cells. A3CH co-localized to endosomes containing VSV-pH (Supplementary Fig. 7b). When V-ATPase is inhibited by Bafilomycin, endosomes fail to acidify, and the cells are resistant to infection by viruses that require an acidified endosome for membrane fusion and infection. However, Arch3 is not sensitive to Bafilomycin. Therefore, exposure of cells to light activates the Arch3 pump to acidify the endosome and allows viral fusion even in the presence of Bafilomycin (Fig. 6a). We demonstrate that blue light-mediated activation of A3CH can compensate for Bafilomycin-mediated inhibition of V-ATPase in control cells, thereby enabling infection by VSV-G, which requires mild acidification to a pH of approximately 6.244 (Fig. 6b). Similarly, activation of A3CH in Bafilomycin treated TRPM7 knockdown cells is also sufficient to restore VSV-G infection. A quantitative summary of these data is shown (Fig. 6b, right panel). However, infection by VSV-pH requires a significantly lower pH (<5.837). As expected, VSV-pH infection is abrogated by Bafilomycin, and it can be restored by activation of A3CH in control cells (Fig. 6c). However, VSV-pH infection cannot be restored by optogenetic acidification in TRPM7-depleted cells at the same dose of light stimulation, arguing that TRPM7 is intrinsically necessary to reach the optimal endosomal pH for VSV-pH infection. A calibration curve of light exposure to VSV-G and VSV-pH treated control and M7KD cells shows that sustained light stimulation in M7KD cells can restore a small amount of virus infection in VSV-pH treated cells (Supplementary Fig. 7c). This is likely because the photonic energy provided by prolonged stimulation is sufficient to overcome the energy barrier of the unfavorable electrical gradient that is created by the accumulation of endosomal protons. Light stimulation of cells expressing the Arch3 pump inactive mutation (D95N) does not restore VSV-pH infection demonstrating that this effect requires proton-pumping activity and is not a result of membrane damage inflicted by photons (Supplementary Fig. 7d).
Fig. 6. TRPM7 modulates proton pumping into the lumen of the viral endosome.
a Diagram showing the use of the optogenetic tool Arch3, a light-activated proton pump that can be used to acidify endosomes in the presence of Bafilomycin. Arch3 fused to CD63 is targeted to late endosome/lysosomes. Normally, the endogenous V-ATPase pumps protons to acidify the endosomal lumen and this enables the virus to escape the endosome (left). When cells are treated with V-ATPase inhibitor Bafilomycin (50 nM), the endosome fails to acidify, and the virus stays in the endosomes (middle). But even in the presence of Bafilomycin, light-mediated activation of Arch3 acidifies the endosome and enables the virus to enter the cytosol (right). b Maximum projections of Arch3-CD63 expressing, Ctl (gray) and TRPM7 knockdown (M7KD, blue) SVG-A cells infected with VSV-G. Cells were treated with Bafilomycin (50 nM) and/or exposed to 15 µWatt 488 nm laser (60 seconds). Quantification of cells positive for infection (green) from three independent infections is shown in the right panel. P values determined by one-way ANOVA with Tukey’s multiple comparison analysis. Mean and SEM is plotted. c Maximum projections of Arch3-CD63 expressing Ctl (gray) and M7KD (blue) SVG-A cells infected with VSV-pH mutant with or without Bafilomycin (50 nM) or light stimulation (15 µWatt 488 nm, 60 seconds). Quantification of total cells positive for infection (green) from three independent infections is shown in the right panel. P values determined by one-way ANOVA with Tukey’s multiple comparison analysis. Mean and SEM is plotted.
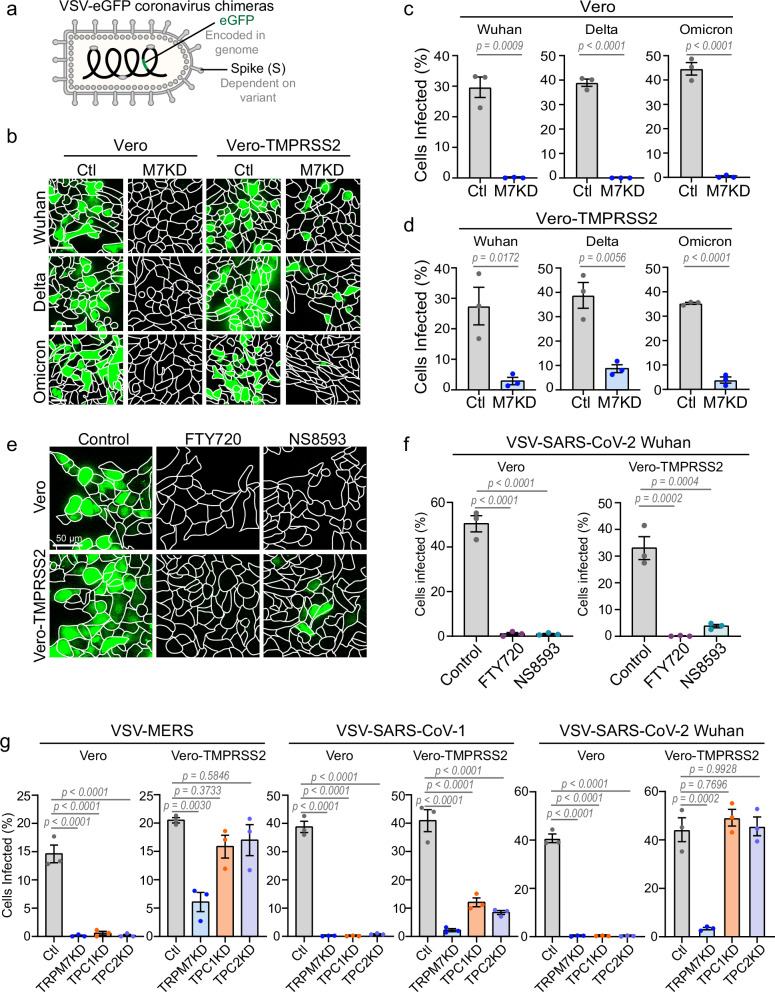
TRPM7 ion channel activity is necessary for infection by SARS-CoV-2 and other coronaviruses
Next, we investigated whether TRPM7 regulates infection by VSV-SARS-CoV-2 variants expressing the SARS-CoV-2 Spike glycoprotein from the Wuhan, Delta, and Omicron strains of SARS-CoV-2. All chimeras also include a genetically encoded eGFP that is transcribed upon infection (Fig. 7a). Previous studies have shown that SARS-CoV-2 enters the cells via endocytosis and requires proteolytic processing in acidified endosomes for escape into the cytosol56–59. The Spike glycoprotein of SARS-CoV-2 requires cleavage by low pH-activated cathepsins to reach its fusion competent conformation, except in TMPRSS2-expressing cells. The protease TMPRSS2 cleaves the spike protein earlier in the endocytic pathway and permits virus fusion at a less acidic pH than that necessary for cathepsin activation56,58,60. Vero cells which express endogenous ACE2 were readily infected by VSV-SARS-CoV-2 variants with or without the ectopic expression of TMPRSS2. Depletion of Trpm7 by siRNA inhibited VSV-SARS-CoV-2 infection by all variants in both parental Vero cells as well as Vero cells over-expressing TMPRSS2 (Fig. 7b–d). Expression of TMPRSS2 did slightly increase the number of cells infected by the Wuhan, Delta, and Omicron variants in the absence of TRPM7, likely due to the change in pH dependence of virus fusion through TMPRSS2-cleavage of the spike protein. To check if VSV-SARS-CoV-2 is endocytosed, we used a construct of VSV-SARS-CoV-2 (Wuhan spike) with an eGFP tag on the P protein of VSV and imaged them in SVG-A cells expressing endogenously tagged EEA1 and NPC1 and ectopically expressing ACE2. The fluorescent viral particles were found co-localized with EEA1 and NPC1 in these cells with or without TRPM7 (Supplementary Fig. 8d). These data indicates that, like VSV-Lassa in Fig. 3c, loss of TRPM7 permits endocytosis of VSV-SARS-CoV-2 but still suppresses infection, likely by preventing the acidification of the virus-laden endosomes. This model is supported by the observation that pharmacologic inhibition of TRPM7 ion channel activity is sufficient to prevent infection by VSV-SARS-CoV-2. We subjected Vero and Vero-TMPRSS2 cells to VSV-SARS-CoV-2 in the presence of TRPM7 inhibitors FTY720 and NS8593. Both inhibitors blocked infection (Figs. 7e, f). Like TRPM7-depleted cells, expression of TMPRSS2 enabled a moderate rescue of the infection in cells treated with NS8593. FTY720, a sphingosine analog, can be phosphorylated by some cells to yield FTY720-phosphate which is a potent agonist of S1P receptor, with EC50 less than 10 nM61,62. The ability to block SARS-CoV-2 infection however requires considerably higher concentration of FTY720 ( ~ 1.5 μM) (Supplementary Fig. 8e).
Fig. 7. Loss or inhibition of TRPM7 is protective against SARS-CoV-2 and other coronavirus infection.
a Schematic of VSV-coronavirus chimera construct with genetically encoded eGFP that is transcribed upon infection. Different variant-specific spike proteins were expressed as the envelope glycoprotein. b Maximum projection of cells infected with VSV-SARS-CoV-2-eGFP variants (green). Vero cells +/- TMPRSS2 were transfected with Control (Ctl) and TRPM7 siRNA (M7KD). Cells were then infected with VSV-SARS-CoV-2-eGFP expressing the Spike protein of the Wuhan, Delta, or Omicron variants. Cells were incubated with virus for 8 hours before fixation and imaging. The scale bar at the bottom left of the images is representative of 50 µm for all images across each row. c Quantification VSV-SARS-CoV-2-eGFP variant infections in control (gray) and M7KD (blue) Vero cells. Cells were infected with VSV pseudotyped with Wuhan, Delta, and Omicron spike variants. P values determined by two-tailed Student’s t test. n = 3 independent experiments. Mean and SEM plotted. d Quantification of infection in Vero cells stably overexpressing TMPRSS2 with VSV-SARS-CoV-2-eGFP variants in control (gray) and M7KD (blue) cells. P values determined by two-tailed Student’s t test. n = 3 independent experiments. Mean and SEM is plotted. e Maximum projection (20 μm) of Vero and Vero-TMPRSS2 cells infected with VSV-SARS-CoV-2-eGFP Wuhan strain in the presence of 5 μM FTY720 or 30 μM NS8593. Cells were pretreated with FTY720 and NS8593 for 15 minutes before exposure to virus and throughout the infection assay. f Quantification of VSV-SARS-CoV-2-eGFP infection in Vero and Vero-TMPRSS2 cells in the presence of 5 μM FTY720 (purple) or 30 μM NS8593 (teal) or without drug (gray). P values were determined by one-way ANOVA with Tukey’s multiple comparison test. n = 3 independent experiments. Mean and SEM plotted. g Quantification of infection by VSV-MERS, VSV-SARS-CoV-1, and VSV-SARS-CoV-2 Wuhan variant in Control (Ctl), TRPM7 (M7KD), TPC1 (TPC1KD), and TPC2 (TPC2KD) knockdown Vero cells. Infection assays were performed in parental Vero cells and Vero cells stably overexpressing TMPRSS2. P values determined by one-way ANOVA with Tukey’s multiple comparison test. n = 3 independent experiments. Mean and SEM plotted.
The endosomal ion channel TPC2 has previously been shown to inhibit infection by several coronaviruses: the MERS virus and SARS-CoV-239,40. We directly compared the effect of depleting TRPM7, TPC1, and TPC2 on VSV-eGFP chimeras of MERS, SARS-CoV-1, and SARS-CoV-2 infection in Vero cells, with or without the ectopic expression of TMPRSS2 (Fig. 7g). Loss of TRPM7, TPC1, or TPC2 led to a complete inhibition of infection in parental Vero cells infected with either VSV-MERS, VSV-SARS-CoV-1, or VSV-SARS-CoV-2. However, in Vero-TMPRSS2 cells, we observed differences. In TPC1 and TPC2 depleted cells, presence of TMPRSS2 led to full recovery of VSV-MERS and VSV-SARS-CoV-2 infection and a mild recovery of VSV-SARS-CoV-1 infection. Conversely, presence of TMPRSS2 in TRPM7-depleted cells only led to a slight recovery of infection across VSV-MERS, -SARS-CoV-1, and -SARS-CoV-2 (<10% recovery of infection). Thus, while TRPM7 and TPC channels are clearly involved in infection by enveloped viruses, the salience of TPC channels is confined to fewer types of virus infections.
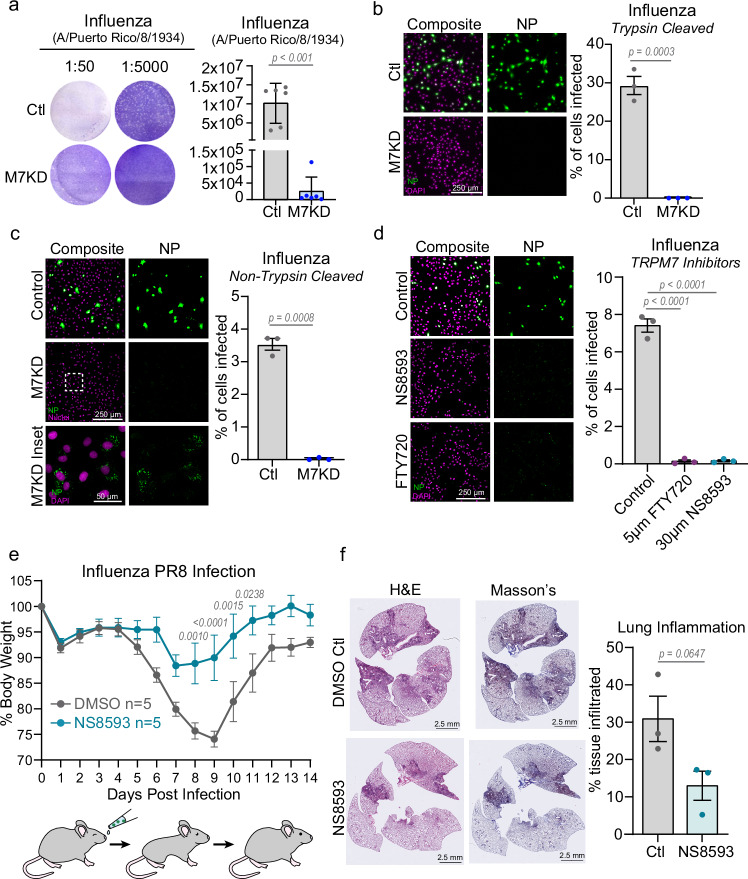
Targeting the ion channel activity of TRPM7 inhibits influenza infection
To extend our findings to a live, unmodified enveloped virus, we assessed the role of TRPM7 in infection by the Influenza strain A/Puerto Rico/8/1934. The fusion of Influenza to host membranes requires a pH of ~5.8–5.0 for viral entry, depending on the isoform of Hemagglutinin, the influenza fusion protein. A/Puerto Rico/8/1934 fuses at a pH of ~5.163. In plaque assays of influenza infection, we see a robust inhibition of plaque formation in M7KD Vero cells (Fig. 8a). We then assessed Influenza entry in control (Ctl) and M7-depleted (M7KD) Vero cells by staining the cells for the accumulation of Influenza A Nucleoprotein (NP), a viral structural protein that is highly expressed in infected cells64. The M7KD Vero cells were highly resistant to infection by both trypsin-cleaved and non-trypsin cleaved influenza (Fig. 8b, c). As trypsin cleavage increases the efficiency of influenza entry, less virus is needed to achieve infection. Notably in cells treated with influenza that was not treated with trypsin, virus particles can be seen accumulating in M7KD intracellular compartments, consistent with the observation that TRPM7 is necessary for efficient viral egress from the endosome. FTY720 and NS8593 also inhibited Influenza infection of Vero cells (Supplementary Fig. 9). The virus particles can also be seen in the intracellular compartments of Vero cells treated with these inhibitors during infection. Finally, we tested the effect of NS8593 in a mouse model of Influenza A infection. Mice were inoculated intranasally with PBS (40 µL) containing 250 µM NS8593 or DMSO along with 150 PFUs of Influenza (A/Puerto Rico/8/1934). After infection, the mice were monitored and weighed for the following 14 days as weight loss is representative of the severity of influenza infection in mice65. NS8593 treatment prevented significant weight loss (Fig. 8e). The onset of weight loss was also delayed in NS8593 treated mice, suggesting that the drug inhibited the early stages of infection. To assess the overall pathology in the lung tissue, we checked the degree of immune infiltration. Three mice from each cohort were used for histological analysis of the lungs by hematoxylin and eosin stain as well as the Masson’s Trichrome strain. Lungs of the NS8593 treated mice showed a trend towards less immune infiltration, correlating with the decreased illness exhibited by NS8593 treated mice (Fig. 8f).
Fig. 8. Inhibition of TRPM7 ion channel activity protects from Influenza infection in vitro and in vivo.
a Plaque assay of Influenza A/Puerto Rico/8/1934 at two different dilutions. Viruses were propagated in Control (Ctl, gray) or TRPM7 knockdown (M7KD, blue) cells, and plaque assays were performed with parental Vero cells. P values determined by two-tailed Student’s t test. n = 6 independent plaque assays. Mean and standard deviation are plotted. b (Left) First round trypsin-cleaved influenza infection in control and M7KD Vero cells stained for influenza nucleoprotein (NP, green). Individual cells are identified by DAPI staining (Magenta). (Right) Quantification of percent cells infected in Ctl and M7KD vero cells. P values determined by two-tailed Student’s t test. n = 3 independent experiments. Mean and SEM plotted. c (Left) Maximum projection of first round non-trypsin-cleaved influenza infection in control and M7KD Vero cells stained for influenza nucleoprotein. Inset shows viral nucleoprotein retained in intracellular compartments after 8-hour incubation. (Right) Quantification of infection in Ctl and M7KD Vero cells with p values determined by two-tailed Student’s t test. n = 3 independent experiments. Mean and SEM plotted. d (Left) Representative maximum projection images of non-trypsin cleaved influenza infection in Vero cells treated with 5 µM FTY720 or 30 µM NS8593. (Right) Quantification of infection in Vero cells treated with FTY720 or NS8593. P values determined by one-way ANOVA with Tukey’s multiple comparison test. n = 3 independent experiments. Mean and SEM plotted. e Weight of mice treated with 150 Plaque Forming Units (PFU) of Influenza A/Puerto Rico/8/1934. Control (DMSO) mice are shown in gray, and NS8593 treated mice in teal. Virus was nasally administered in 40 µL of PBS containing DMSO or 250 µM NS8593. P values determined by Repeated Measures two-way ANOVA with Sidak multiple comparisons test. f (Left) Histology of lungs from mice treated with Influenza +/− NS8593. Hematoxylin and eosin (H&E) stain as well as Mason’s trichrome stain are shown. (Right) Quantification of tissue area with immune infiltration from H&E staining in Control and NS8593 murine lungs. P values determined by two-tailed Student’s t test. n = 3 lungs from individual mice. Mean and standard deviation are plotted.
Discussion
In this study we show that the endosomal fusion of a broad range of pH-dependent enveloped viruses, including SARS-CoV-2 and influenza, is highly dependent on the ion channel activity of TRPM7. Infection by viruses that require lower pH, such as Ebola, LCMV, Lassa, Influenza, MERS, SARS-CoV-1, and SARS-CoV-2, is robustly inhibited by loss or inhibition of TRPM7. Preventing virus entry into the cytosol has been a successful pharmacological therapy for viruses such as HIV and Herpes66 but there is a need for broad-spectrum antivirals that can serve as a first line of defense against emerging viruses. This work advances TRPM7 as a novel pharmacological target for the prevention and treatment of enveloped virus infections. We show that FTY720 and NS8593, known small molecule inhibitors of TRPM7 ion channel activity, abrogate infection by pH-dependent enveloped viruses. In SVG-A and Vero cells, NS8593 most closely replicates the phenotype of TRPM7 protein depletion during viral infection. In mice, nasally administered NS8593 reduces illness from influenza, further supporting TRPM7 as a potential molecular target for a broad-spectrum anti-viral drug. Previously, we showed that TRPM7 is highly expressed in developing embryos and is required for mouse embryogenesis29,30. However, Trpm7 has been deleted selectively in many different organs after organogenesis without any overt pathology indicating that tissues can tolerate the loss of TRPM730. This observation argues that pharmacological targeting of TRPM7 could be tolerated during an acute therapeutic window. While counter-intuitive, proteins that are essential during embryogenesis are often good drug targets. Prominent examples include mTOR and Calcineurin, targets of widely used FDA-approved drugs rapamycin (sirolimus) and FK506 (tacrolimus).
Our previous work has shown that TRPM7 regulates endocytosis of the Fas receptor34 and TLR432. Consistently, we found that in the absence of TRPM7, cells tend to endocytose less virus. However, the resistance to infection is especially pronounced for the viruses that depend on low endosomal pH for infection. This is underscored by the observation that infection by a VSV variant that requires a lower pH for infection is highly dependent on TRPM7. We show that virus-laden endosomes cofractionate with TRPM7, and in the absence of TRPM7, these endosomes do not acidify normally. We propose a model wherein TRPM7 is recruited to virus-laden endosomes. The initial acidification of the virus-laden endosomes by the V-ATPase can be initiated without the presence of TRPM7, up to a pH of ~6.3. The ion channel activity of TRPM7, which is pH-sensitive28,36, is then activated as acidification progresses. The ionic composition of the early endosomes resembles the extracellular environment with high [Na+] (~145 mM). Given the low [Na+] in the cytosol (<7 mM), the high driving force for Na+ flux from the endosomal lumen into the cytosol is well suited for permeation through TRPM7 in the membrane of the virus-laden endosomes. The TRPM7-mediated cationic counter current then plays the critical role of dissipating the build-up of positive charge on the luminal side of the endosomal membrane. The TRPM7-mediated countercurrent thereby sustains progressive endosomal acidification by the V-ATPase. Without TRPM7, the steady accumulation of positive charge on the luminal side of the endosomal membrane increases the energy barrier for V-ATPase-mediated proton-pumping, slowing the acidification of the endosomes.
We show that TRPM7 promotes the acidification of only a subset of endo/lysosomal compartments. EGFR degradation, a process dependent on endocytosis and pH-dependent lysosomal degradation, is normal in TRPM7-deficient cells. TRPM7−/− cells also maintain acidity in their lysosomes. Long-term inhibition of V-ATPase by Bafilomycin treatment is highly toxic to cells67. In contrast, TRPM7−/− cells can grow and proliferate. In this study we show this for Trpm7−/− HeLa cells, and we have previously demonstrated that primary Trpm7−/− T cells are normal in their growth and proliferation33. Trpm7−/− bone marrow progenitors also differentiate normally into macrophages32,36. These observations show that TRPM7 is necessary for robust acidification of a subset of endosomal compartments, including virus-laden endosomes. The mechanisms underlying the role of TRPM7 in only a subset of endosomes remain unknown at this juncture. Possibilities include differential recruitment or activation of TRPM7 in endosomes, depending on the size of the compartments or the identity of receptors that triggered endocytosis. Larger compartments may require more sustained V-ATPase activity for acidification and would have greater need for a TRPM7-mediated counter current. This is consistent with our previous observation that TRPM7 is critical for the acidification of efferophagosomes, which result from the phagocytosis of apoptotic cell corpses36.
Although the V-ATPase is the central controller of endosomal acidification, ion channels and transporters appear to play important ancillary roles and are more suited for subtle pharmacological manipulation of this process. Chloride transporters and channels such as chloride-conducting channels/exchangers (CLCs), the cystic fibrosis transmembrane conductance regulator (CFTR), and the proton activated chloride channel (PAC) have been proposed to regulate Cl− flux across the endosomal membrane to adjust endosomal pH15,68. It is conceivable that a variety of ion channels differentially regulate endosomal acidification at various stages of maturation and in different endosome populations. TPC1 and TPC2 have previously been implicated in several viral infection processes38–40. We found that while loss of TPC channels does inhibit the entry of several enveloped viruses, they do not show the same pattern of inhibition in a battery of virus infection assays as TRPM7 and likely mediate resistance to virus infection through mechanisms distinct from TRPM7.
An early model of SARS-CoV-2 infection assumed that in cells expressing the protease TMPRSS2 the virus can fuse at the plasma membrane, while in cells lacking TMPRSS2, the virus is endocytosed and fuses to the endosomal membrane56,69. However, recent work demonstrates that SARS-CoV-2 particles are endocytosed and TMPRSS2 mediated cleavage enables the virus to escape from early endosomal compartments compared to cells lacking TMPRSS258. In agreement, we found that loss of TRPM7 inhibits VSV-SARS-CoV-2 infection in Vero cells, with or without the expression of TMPRSS2. The slight recovery of VSV-SARS-CoV-2 Wuhan and Delta infection in TRPM7 depleted cells overexpressing TMPRSS2 is most likely due to the escape of some viruses from earlier, less acidic endosomal compartments. This may precede a stage where TRPM7 is necessary for further acidification.
Overall, our study establishes a critical role for TRPM7 in the membrane fusion of pH-dependent enveloped viruses. Our findings are consistent with a model wherein TRPM7 channels resident in the virus-laden endosomes mediate a cationic counter-current necessary to sustain the acidification of these endosomes by V-ATPases. This aspect of TRPM7 function is likely to regulate other endosomes but the underlying mechanisms of such specialization are not understood at this point. Although the biophysical characteristics of TRPM7 ion channel activity have been defined, it has long remained a mysterious channel from the standpoint of its core cell biological function. Our discovery that TRPM7 is recruited to a subset of endosomes and regulates their acidification is an advance that will serve as an important groundwork for all subsequent inquiries into the cell biological studies of TRPM7.
Methods
The research reported here research complies with all relevant ethical regulations. Mouse protocols were approved by the University of Virginia IACUC (protocol 3916).
Cell culture
HeLa cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Vero E6 cells were cultured in DMEM with 25 mM HEPES, 10% FBS, and 1% penicillin-streptomycin. SVG-A cells were grown in MEM with 10% FBS, 10 mM HEPES, and 1% penicillin-streptomycin. HEK293T cells for lentivirus production were grown in DMEM with 10% FBS. All cells were grown at 37 °C and 5% CO2 and passaged using 0.25% trypsin-EDTA.
siRNA knockdown and mouse TRPM7 mutant plasmid transfections
The siRNA treatments were performed as follows. Cells were plated in six-well TC treated dishes (day 0) and transfected at ~90% confluency with siRNA using RNAimax lipofectamine (day 1). On day 2, cells were split and re-plated into six-well dish and then on day 3, they were re-transfected with a second hit of siRNA. The cells were plated for experiments on day 4 and typically used for experiments on day 5. Transfections of mouse TRPM7 wild-type, pore mutant, and kinase inactive plasmids were performed with lipofectamine 2000 or 3000 using 2.5 μg of each plasmid for each well of a six-well plate and utilized for testing two days after transfection.
Generation of stable cell lines
Knockout of TRPM7 from HeLa cells was achieved as follows. Lenti-CRISPR-Cas9 plasmid with sgRNA targeting human TRPM7 was purchased from Sigma (5’-AAGCAACTGCTTGATTCGTGGGG-3’). WT HeLa (ATCC CCL-2) cell line was transfected with 1–5 μg of the Lenti-CRISPR-Cas9 plasmid DNA and on day 3 put under puromycin selection for 7 days. Cells were then single-cell cloned by dilution, amplified, and subsequently tested for Cas9 expression and TRPM7 deletion. Arch3-CD63-Halo lentiviral transfer plasmids were generated from pCMV-lyso-pHoenix (addgene #70112) by substituting mKate for Halo. Pump mutant Arch3 was generated with the substitution D95N of Arch3. Stable SVG-A ACE2 expressing cells and Vero-TMPRSS2 cells were generated using lentivirus transduction as previously described70.
Generation of Trpm7−/− BMDMs
Mice at age 12 weeks were used for BMDM production. Trpm7 fl/fl(LysM cre) mice were generated as described previously to express Cre in myeloid cells32. Mice were housed and bred in accordance with policies and procedures of the University of Virginia Institutional Animal Care and Use Committee (IACUC). BMDMs were isolated as previously described36. Briefly, Tibia, Fibula, and Femurs were isolated from Trpm7 fl/fl and Trpm7 fl/fl (LysM-cre) mice. Bone marrow was removed by centrifugation and lysed with ACK Lysis buffer. Cells were differentiated for 7 days in BMDM media (RPMI + 10%FBS + 20% L929-conditioned media + PenStrep). On day 7 cells were scraped with a cell lifter and plated for experimentation the following day. L929 media was generated by growing L929 cells in DMEM + 10%FBS for 14 days. Media was harvested and replenished on day 7.
Patch clamp electrophysiology
TRPM7 currents (ITRPM7) were measured in whole cell configuration as described previously32. Briefly, the external cell solution contained (in mM): 135 Na-methanesulfonate, 5 Cs-gluconate, 2.5 CaCl2, 10 HEPES, pH 7.3 (adjusted with NaOH), and an osmolality of 280–290 mOsm/Kg. The pipette solution contained (in mM): 110 Cs-gluconate, 0.5 NaCl, 0.75 CaCl2, 10 HEPES, 10 HEDTA, 1.8 Cs4-BAPTA, 2 Na2ATP, pH 7.3 (adjusted with CsOH), and an osmolality of 273 mOsm/Kg. Free [Ca2+] was approximately 100 nM and was calculated via the Maxchelator algorithm (http://maxchelator.stanford.edu/webmaxc/webmaxcS.htmhttps://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/webmaxc/webmaxcS.htm). The recordings were performed with 400 ms ramps from −100 mV to +100 mV and a holding potential of 0 mV. Signals were low-pass filtered at 5 kHz and sampled at 10 kHz. All electrophysiology experiments were conducted at RT (~23 °C) using an Axopatch 200B amplifier (Molecular Devices). ITRPM7 was inhibited by flowing on FTY720- or NS8593-containing external solution at concentration of 5 μM or 30 μM respectively.
RNA isolation and qRT-PCR
RNA was isolated using Qiagen RNAeasy mini plus isolation kit. RNA was converted to cDNA using Promega GoScript Reverse Transcriptase kit. Primers for human β2M (housekeeping gene) and human TRPM7 were used for amplification. SYBR was used to detect amplification on CFX Biorad real-time PCR System.
VSV chimera viruses
VSV chimeras described in this paper were propagated as previously described37,70,71. Briefly, the propagation of recombinant VSV (Indiana) and chimeras expressing MeGFP (VSV-G, VSV-Rabies, VSV-Ebola, VSV-Lassa, VSV-LCMV) were grown by infection of BSR-T7/5 cells. VSV-MERS, VSV-SARS-CoV-1, VSV-SARS-CoV-2 variants (Wuhan, Delta, Omicron) and VSV-SARS-CoV-2-PeGFP were propagated by infection of MA104 cells. Cells were grown in 15 cm dishes at a multiplicity of infection of 0.01. After 48 hours post-infection, media was collected and pelleted at 30,000 × g for 2 hours at 4 °C using a Ti45 fixed angle rotor. The pellet was resuspended in NTE buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 7.4) and layered on a 15% sucrose-NTE cushion and spun at 110,000 × g for 2 hours at 4 °C in a SW55 rotor. Buffer and sucrose were removed, and the pellet was resuspended in NTE and incubated overnight at 4 °C. The virus was then loaded on a discontinuous sucrose gradient of 15–45% sucrose-NTE and centrifuged at 150,000 × g for 1.5 hours at 4 °C. Virus was resuspended in NTE in a volume of 200–500 μL and stored at 4 °C for use. To generate VSV-MERS and VSV-SARS-CoV-1, the S gene of MERS (HCoV-EMC/2012 isolate; Genbank accession: YP_009047204.1; codon optimized) or SARS-CoV-1 (Urbani strain; Genbank accession: AAP13441.1), with the C-terminal 21 amino acids removed, were inserted into an infectious molecular clone of vesicular stomatitis virus72 in place of the native glycoprotein in a manner analogous to the method previously described for VSV-SARS-CoV-273. To recover virus from cDNA, BSRT7/5 cells were infected with a recombinant vaccinia virus that expresses T7 RNA polymerase74 and transfected with T7-based expression plasmids encoding VSV N, P, L, and G, and an antigenomic copy of the viral genome72,73. Cell culture supernatants were collected at 72 hours post-transfection, clarified by centrifugation (5 min at 1000 × g), and passed through a 0.22 μm filter. Virus was plaque-purified on Vero CCL81 cells in the presence of 25 μg ml-1 of cytosine arabinoside, and viral stocks were amplified on Vero CCL81 cells at an MOI of 0.01 in DMEM containing 2% FBS and 20 mM HEPES pH 7.7 at 34 °C. VSV chimeras of coronavirus strains are patent pending by Washington University in St. Louis.
Influenza virus strains
Influenza (A/Puerto Rico/9/1934) used for plaque assay was a generous gift from Daniel Maurer and Dr. Aaron Schmidt (Ragon Institute MGH, MIT, Harvard) and generated by reverse genetics in a co-culture of HEK293 and MDCK cells and then passaged twice on MDCK cells. For all other influenza infection assays, Influenza (A/Puerto Rico/9/1934) was purchased from ATCC (VR-1469) and propagated in MDCK cells with or without TPCK trypsin.
VSV chimera infection assays
All VSV-MeGFP chimera infections were performed as follows: Glass coverslips were cleaned by sonicating in isopropanol followed by sonication in 0.5 M KOH. Polydimethylsiloxane (PDMS) was prepared by thoroughly mixing at a 1–10 dilution with the curing reagent. 5 grams of the mixture was poured into a 10 cm plate and incubated at 90 °C for ~4 hours. Dishes were removed and stored at room temperature. 3-millimeter or 5-millimeter diameter holes were punched out of the PDMS. Holes were cut out and placed in a PDC-001 plasma cleaner along with cleaned glass slides at 750 mtorr at 30 W for 2 minutes. PDMS was then firmly pressed against the glass and placed in 60 °C oven for 20 minutes. Glass coverslips with attached PDMS were placed in 70% ethanol for 20 minutes and dried under UV light for sterilization. Appropriate cells were trypsinized, pelleted, resuspended in media, and plated into the wells of PDMS to achieve a density of 50–80% for the following day. 10 μL of cells were plated into each 3-mm well and 20 μL into each 5-mm well. Cells were allowed to adhere for 1 hour with media surrounding the PDMS. Wells were then completely submerged in media overnight. The next day, for cells with Halo tags, JFX-647 was incubated at a concentration of 200 nM for 30 minutes at 37 °C. Cells were washed 3× with media. Cells were then incubated with virus for 1 hour. Cells were briefly washed with DMEM and incubated at 37 °C with DMEM for five hours for all conditions except cells incubated VSV-SARS-CoV-2 variants which were incubated for seven hours and infection of Bone Marrow-Derived Macrophages with VSV-Lassa which required incubation for 18 hours before infection was achieved. If being treated with pharmacological inhibitors, cells were pretreated for 15 minutes with 5 μM FTY720 and 30 μM NS8593 or 1 hour with 50 nM Bafilomycin A1 and appropriate dilutions were included in the virus-containing media and the incubation media. Arch3 activation was achieved by stimulating cells with 15µWatts of 488 nm illumination for the indicated time length. After the experiment, media was removed from cells and cells were incubated at room temperature with Wheat Germ Agglutinin for 30 seconds, washed with PBS, and fixed in 4% paraformaldehyde. All infection assays were imaged using the same laser power and exposure time for accurate comparison between Control and Knockdown infections. Cells were deemed nuclei+ if a visible GFP+ ring was seen in the cytosol of the cell. Cells were deemed cytosolic+ if the fluorescent MeGFP signal in the cytosol was greater than 1300 A.U. All virus infection graphs demonstrate the mean and SEM of the average proportion of cells with nuclei+ or cytosolic+ GFP staining from a minimum of five separate planes of view with three independent infections performed per condition. Influenza and VSV-Lassa-eGFP infection assays were performed in eight-well glass bottom chamber plates. Vero cells were plated and the next day treated with virus. Influenza was incubated with cells for one hour, then washed and incubated for a subsequent 8 hours before fixation with paraformaldehyde for 15 minutes. Cells were permeabilized for 15 minutes with 0.25% triton-x-100 in PBS, blocked in 5% goat serum in PBST, and probed with a 1:1000 dilution of Nucleoprotein antibody in blocking solution at 4 °C overnight. The following day cells were washed, stained with secondary antibody and DAPI, and imaged on Leica Thunder microscope.
Plaque assays
Vero cells were treated with Control or TRPM7 siRNA as described above. Cells were then plated into the wells of a six-well plate. Cells were treated with VSV-chimeras or Influenza (A/Puerto Rico/8/1934) at an MOI of approximately 0.01. The virus was allowed to propagate in cells for 48 hours. Supernatant was harvested. Untreated Vero cells were then plated in six-well plates and treated for one hour with 150 μL of viral supernatant. The media was then removed and DMEM with 25 mM HEPES, 1× pen/strep and 0.5% agarose was gently layered on top of the cells. Agarose media was allowed to congeal at room temperature for 15 minutes and then cells were subsequently incubated at 10% CO2 and 37 °C for 36 hours. Cells were fixed with 3:1 Methanol:Acetic Acid for 30 minutes. Agarose media was then removed by washing with DI H2O. Plaques were stained with 150 μL of Crystal Violet (1%) for 30 minutes. Wells were washed of stain and plaques were counted. Plaque forming units (PFUs)/mL were determined as follows: plaque counts × dilution × 6.67.
Live cell single-viral particle imaging
SVGA cells were plated on 5-mm PDMS chambers as described above. The following day, cells were incubated with the appropriate virus for 20 minutes at 37 °C/5% CO2. For endosomal colocalization, wells were washed and placed in fluorobrite media with 20 mM HEPES before imaging at 37 °C/5% CO2. For MeGFP with pHrodo imaging, cells were washed, incubated with WGA-647 for 30 seconds, washed, and then imaged in fluorobrite at 37 °C/5% CO2 at 30-, 45- and 60-minutes post-infection. Images were taken at 0.27 μm z-stacks encompassing the entire cell volume. At least 9 cells were imaged at each time point. After imaging one-time point, new cells were imaged for the subsequent time point to avoid photobleaching and excessive radiation exposure to the cells that could impact endosomal dynamics.
Analysis of single viral particle imaging
Diffraction-limited spots from the fluorescence signals elicited by eGFP tagged to the M protein (or P protein) of VSV were detected through a least-squares minimization numerical fitting with a model of the microscope’s PSF approximated by a 3D Gaussian function. Estimated fluorescence intensities associated with each spot were calculated from the corresponding amplitudes of the fitted 3D Gaussian function75,76. To calibrate the intensities of single virus, we allowed virus to bind to a glass cover slip and imaged a 20-plane volume (0.27 µm space, the same as in each experiment) and determined the intensity of every virus in multiple fields of view and fit the intensity distribution of each particle. To analyze co-localization with endosomal markers, an image of 21 × 21 pixels (x vs y) with a projection of 2 pixels in z was extracted for the coordinates of every detected spot. The co-localization of each spot was then visually examined using a custom-built software written in LabView (National Instruments) previously described77. To analyze virus labeled with pHrodo-Red, an image of 11 × 11 pixels (x vs y) with a projection of 2 pixels in z was extracted for the coordinates of every detected spot. The intensity of the projected image of each spot was extracted in the LabView written software and the ratio of pHrodo/GFP was determined. Data was analyzed on unaltered images; representative images were smoothened once in image j for illustration.
Lenti pseudovirus generation and infection assays
Lenti pseudovirus were made by transfecting the packaging plasmids Gag/Pol (HDM-Hgpm2 Addgene: 204152 Dadonaite et al.41 Cell. A gift from Jesse Bloom) HIV-1 Rev (pRC-CMV_Rev1b Addgene 204153 Dadonaite et al. Cell 2023 a gift from Jesse Bloom) and Tat (HDM-tat1b Addgene 204154 Dadonaite et al. Cell 2023 a gift from Jesse Bloom) the transfer plasmid co-expressing luciferase and ZsGreen (pHAGE-CMV-Luc2-IRES-ZsGreen-W Addgene 164432 a gift from Alejandro Balazs) and the indicated glycoprotein41. A 15-cm plate of HEK293T cells were transfected with 5.9 μg Luc/ZsGreen, 1.3 μg, Gag/Pol, 1.3 μg Rev, 1.3 μg tat1b, and 2 μg of indicated glycoprotein using PEI transfection reagent. Cells were incubated for 24 hrs then the supernatant was collected and replaced. After an additional 24 hrs, supernatant was collected. Cellular debris was removed by centrifugation at 1000 × g for 10 min. Supernantant was removed from pellet and passed through a 0.45 μm filter. Supernatant was then flash-frozen and stored at −80 °C until use. Infection assay was performed in a 96 well plate. Cells were plated at 70,000 cells per well, and infection began the following day. Media was removed, and pseudovirus media was added after mixing 1:1 with DMEM to the respective well. After 24 hours media was removed, cells were lysed and firefly luciferase activity was determined using a luciferase assay system (Promega E4550).
Ebola virus-like particle generation and infection assays
Virus-like particles were made as described in Simmons et al.43 by co-expressing VP40 and the GP of Ebola Zaire strain. A fluorescent tag of mCherry was attached to a VP40 as well as a β-Lactamase attached to VP40 (All plasmids were a gift from Dr. Judith White). A 15 cm plate of HEK293T cells was transfected with 2 μg of VP40, 4.5 μg β-Lactamase-VP40, and 4.5 μg of mCherry-VP40, and 3 μg of Ebola GP. After 24 hours the supernatant was collected and replaced. After an additional 24 hours supernatant was collected and pooled. Cell debris was pelleted at 1000 × g for 10 min. Media was layered on top of 20% sucrose and pelleted using an SW40 rotor (110,000 × g for 2 hours). Pellets were placed in resuspension buffer (20 mM HEPES, 20 mM MES, 130 mM NaCl, pH 7.4) and then layered on top of a 60%, 20% sucrose step gradient in a SW55 rotor and centrifuged at 194,000 × g for 5 hrs. VLPs were then collected from the interface of the 60% and 20% sucrose solutions. Sucrose was diluted from VLPs by adding 3 mL of resuspension buffer. VLPs were then pelleted through a 20% sucrose layer in an SW55 by centrifugation at 110,00 × g for 1 hr. Pellet was resuspended in resuspension buffer with 10% sucrose. Aliquots were then frozen and thawed as needed for assay. For infection assays, BMDMs were plated (80,000 cells per well) in a 96 well plate. On day of experiment media was replaced. 1 μg of VLP was added to each well of the indicated condition. VLPs were allowed to incubate with cells for 3 hours at 37 °C. Cells were then moved to room temperature and loaded with substrate CCF2-AM and using a Beta-lactamase loading solutions assay (Invitrogen K1085). After one hour plate spectrum was taking on plate reader with excitation of 400 nm, a 425 nm cutoff filter with 10 nm scans from 430–600 nm. The peak fluorescence at 450 nm and that at 520 nm was used to determine the relative amount of VLP entry.
pHrodo conjugation to VSV-chimeras
0.25 μg pHrodo red-succinimidyl ester was aliquoted and dried under vacuum. pHrodo dye was resuspended in 200 μL aliquots of virus (250 μg/mL of viral RNA) with 0.1 M sodium bicarbonate and incubated at room temperature for 1 hour covered from light. Free dye was removed by passaging the mixture through 40 kDa ZEBA spin desalting columns twice.
Density gradient centrifugation and intracellular membrane preparations
For VSV-Lassa-MeGFP treated membrane density centrifugation, five 15 cm plates of SVG-A cells were treated with or without the virus for one hour. Cells were washed and scraped in ice-cold PBS. For whole-cell intracellular membrane preparations, a 10 cm plate of transfected cells was washed once with PBS, scraped, and resuspended in ice-cold PBS. Cells were then pelleted at 500 × g for 5 minutes, and the pellet was subsequently resuspended in 5 mL of homogenization media (0.26 M sucrose, 10 mM MOPS, 0.2 mM EDTA, pH = 7.2). Cells were pelleted at 500 × g for 5 minutes and resuspended in homogenization media containing protease inhibitors. Cells were gently ruptured using a ball-bearing homogenizer (isobiotec). Cell lysates were first pelleted at 6800 × g for 10 minutes at 4 °C to pellet nuclei. The supernatant was collected and pelleted at 11,300 × g for 15 minutes at 4 °C to pellet mitochondria. For density gradient centrifugation, post mitochondrial pellet supernatant was gently layered on top of an iodixanol gradient of 0.5 mL 30% iodixanol and 3.3 mL 14.5% iodixanol in an Ultra-Clear SW55 centrifuge tube. PNS were subjected to centrifugation in a SW55 rotor at 150,000 × g for 15 hours at 4 °C. Fractions were then removed in 0.5 mL increments from the top of the gradient. Fractions from density gradient or post-mitochondrial supernatant for whole cell membrane preparations were then pelleted at 130,000 × g for 30 minutes at 4 °C using a TLA-120.2 rotor in a Beckman tabletop ultracentrifuge. The membrane pellet was resuspended in 30 μL of PBS and 10 μL of 4× Laemlli buffer was added to the samples.
Immunoblotting
Samples were boiled for 7 minutes, centrifuged briefly on a tabletop centrifuge, and loaded onto a 4–20% Tris-glycine SDS page gel. The resolved gels were transferred to a PVDF membrane using the BioRad Trans-blot Turbo Transfer device, dried, and rehydrated in methanol. Blots were blocked with 5% milk in TBST for 1 hour. Primary and secondary antibodies were suspended in Signal Boost Immunoreaction buffers at 1:1000 and 1:10,000 dilution, respectively. Membranes were incubated with primary antibody overnight at 4 °C, washed 3× with TBST, incubated with secondary antibody (HRP-conjugated) for one hour at room temperature, washed 3×, then developed Pico-Plus Supersignal chemiluminescent buffers and visualized with FujiFilm LAS-3000. Rabbit anti-mouse TRPM7 antibody was generated by LifeProtein. All other antibodies were obtained commercially: Rabbit anti-human TRPM7: ab245408; Rabbit anti-rab5: CST C8B1; Rabbit anti-GFP: abcam ab6556; Rabbit anti-EGFR: CST D38B1; Rabbit anti-β-actin: CST D6A8.
EGFR degradation
500,000 cells were plated into each well of a 12-well dish and incubated overnight. The following day, cells were serum starved for 2.5 hours. Then 50 ng/mL of EGF was added to the appropriate wells. Cells were lysed in ice-cold RIPA buffer containing protease inhibitors. BCA assay was performed on cell lysates, and an equal protein amount was loaded into a 4–20% Tris-glycine SDS page gel.
Lysosensor imaging
HeLa cells were loaded with 1 μM lysosensor DND-189 dye in serum-free media for 1 hour at 37 °C and 5% CO2. Bafilomycin condition cells were incubated with 50 nM Bafilomycin for 30 minutes before dye loading and throughout the experiment. Cells were washed with DMEM without phenol red and imaged using Zeiss LSM 880 with a ×60 objective at 0.27 μm intervals.
Cell growth assay
200,000 HeLa cells were plated into the well of a 12-well dish in complete media (DMEM + 10% FBS + PenStrep) or serum-free media (DMEM + PenStrep). 24 hours later, cells were trypsinized and counted using BioRad TC20 cell counter.
Murine influenza infection
8-week-old female C57BL/6 mice (Jackson) were anesthetized with ketamine/dexmedetomidine and nasally administered 150 PFU of Influenza A (A/Puerto Rico/8/1934) simultaneously with 250 µM of NS8593 or DMSO control resuspended in 40 µL of PBS. Mice were then weighed daily for 14 days as a readout of severity of illness. Mice were sacrificed by CO2 asphyxiation, perfused with ice-cold PBS, and lungs were inflated with 10% neutral buffered formalin. Lungs were isolated and fixed in 10% neutral buffered formalin and processed for histology by the UVA histology core. The protocol was approved by the University of Virginia IACUC.
Statistical analyses
All statistical analyses were performed using GraphPad Prism (version 10.3.0, GraphPad Software, San Diego, CA). Data are presented as mean ± standard error of the mean (SEM) unless otherwise specified in the figure legend. For comparisons between two groups, two-tailed Student’s t tests were used. For comparisons involving more than two groups, one-way ANOVA followed by Tukey’s multiple comparisons test was employed. Two-way ANOVA with Sidak’s multiple comparisons test was used to analyze data with two independent variables in some experiments. Concentration-response curves were fitted using a four-parameter logistic equation. For pH calibration, four-parameter logistic equations were used to fit pHrodo:GFP ratios to pH values. Electrophysiological data were analyzed by comparing peak current densities between conditions using appropriate t tests or ANOVA. For all analyses, p < 0.05 was considered statistically significant. The specific statistical test used and the number of experiments performed for each analysis are indicated in the corresponding figure legend. Normal distribution was assumed for all analyses, and no data points were excluded. All percent infection assays are representative of three independent experiments with a minimum of five fields of view imaged per experiment to determine the average percent infected. Data from single particle imaging experiments are represented as values (either counts or average intensity) on a per cell basis. Single particle regions of interest (ROIs) were identified with MATLAB and assessed using LabView (GitHub links included in software availability). Initial TRPM7 KD SVG-A and HeLa infection assays (Supplementary Figs. 2, and 4a and Figs. 4a and 2b) were performed blinded to genotype and VSV-chimera treatment. Vero influenza (Fig. 8c), Vero VSV-Lassa (Supplementary Fig. 8f), and histology analysis (Fig. 8f) experiments were also performed blinded to genotype. For the in vivo influenza experiments, repeated measures two-way ANOVA with Sidak’s multiple comparisons test was used to analyze mouse weight data over time.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the members of the Desai and Tamm labs for their helpful discussions. We also thank Dr. Judith White (UVA), Dr. Michael Schappe (HMS), Dr. Tom Kirchhausen (HMS), and Dr. Stephen C. Harrison (HMS) for their scientific insights and feedback. We thank Drs. Kevin Lynch (UVA), Doug Bayliss (UVA), Bob Nakamoto (UVA), Norbert Leitinger (UVA), and James Casanova (UVA) for helpful and timely advice as Thesis Committee advisors to C.A.D. We thank Dr. Mark Kreutzberger (UVA) and Dr. Ed Egelman (UVA) for the cryo-em image of Ebola-VLPs. We thank Jessica Sobeck and Susan Landes of the UVA Center for Comparative Medicine for their assistance with the influenza mouse model. We thank the following core facilities at UVA: Advanced Microscopy Facility, Research Histology Core, and the Biorepository and Tissue Research Facility for assistance with histology. The work was predominantly funded by following NIH research grants: AI155808 (B.N.D.), GM108989 (B.N.D.), AI030557 (L.K.T.), AI163019 (SPJW), AI147394 (J.S.), AG069264 (J.S.), AI112844 (J.S.), HL170961 (J.S.), AI154598 (J.S.), Harvard Virology Program T32 AI007245 (A.J.B.K.), UVA MSTP Training Grant T32 GM007267 (C.A.D.), and Pharmacological Sciences Training Grant T32 GM007055 (C.A.D.). Additional funding was provided by the Owens Family Foundation (B.N.D.).
Author contributions
Conceptualization and research design: C.A.D., A.J.B.K., B.N.D., investigation: C.A.D., G.W.B., A.J.B.K., W.H.I., C.R., H.D., M.C.M., E.J.S., M.A., S.N., J.K., data analyses: C.A.D., V.K., C.R., H.D., W.H.I., G.W.B., software development: V.K., resource generation: B.N.D., C.A.D., M.E.S., A.J.B.K., W.W., S.P.J.W., L.M.B., C.E.T., P.W.R., funding acquisition: S.P.J.W., J.S., L.K.T., B.N.D., figures: C.A.D., writing–draft and editing: C.A.D., B.N.D., project administration: B.N.D.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided in this paper.
Materials availability
Reagents generated in this study are available from the B.N.D. with a completed Materials Transfer Agreement. Requests for VSV-chimeras should be made to S.P.J.W. at spjwhelan@wustl.edu.
Code availability
Software for single particle ROI identification is available on GitHub: (https://github.com/VolkerKirchheim/TrackBrowser_Matlab.git) (https://github.com/VolkerKirchheim/TrackBrowser_LabView.git).
Competing interests
S.P.J.W. is a consultant for Thylacine Biosciences and receives support from Vir Biotechnology to study the properties of VSV-coronavirus chimeras unrelated to this project. J.S. receives support for research unrelated to this project from Icosavax. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52773-w.
References
- 1.Mercer, J., Schelhaas, M. & Helenius, A. Virus entry by endocytosis. Annu. Rev. Biochem.79, 803–833 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Yamauchi, Y. & Helenius, A. Virus entry at a glance. J. Cell Sci.126, 1289–1295 (2013). [DOI] [PubMed] [Google Scholar]
- 3.White, J. M. & Whittaker, G. R. Fusion of enveloped viruses in endosomes. Traffic17, 593–614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigant, F., Santos, N. C. & Lee, B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol.13, 426–437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wisskirchen, K., Lucifora, J., Michler, T. & Protzer, U. New pharmacological strategies to fight enveloped viruses. Trends Pharm. Sci.35, 470–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White, J. M. et al. Drug combinations as a first line of defense against coronaviruses and other emerging viruses. mBio12, e0334721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White, J., Kartenbeck, J. & Helenius, A. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J. Cell Biol.87, 264–272 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz, R., Stiasny, K. & Heinz, F. X. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol.183, 353–361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, J., Gregory, S. M., Nelson, E. A., White, J. M. & Tamm, L. K. The roles of histidines and charged residues as potential triggers of a conformational change in the fusion loop of ebola virus glycoprotein. PLoS One11, e0152527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brecher, M. et al. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol.86, 364–372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schornberg, K. et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol.80, 4174–4178 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons, G. et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA102, 11876–11881 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, L., Wu, D., Robinson, C. V., Wu, H. & Fu, T. M. Structures of a complete human V-ATPase reveal mechanisms of its assembly. Mol. Cell80, 501–511.e503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casey, J. R., Grinstein, S. & Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol.11, 50–61 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Chadwick, S. R., Grinstein, S. & Freeman, S. A. From the inside out: Ion fluxes at the centre of endocytic traffic. Curr. Opin. Cell Biol.71, 77–86 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Abe, K. & Puertollano, R. Role of TRP channels in the regulation of the endosomal pathway. Physiol. (Bethesda)26, 14–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, H. & Ren, D. Lysosomal physiology. Annu. Rev. Physiol.77, 57–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clapham, D. E. TRP channels as cellular sensors. Nature426, 517–524 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Clapham, D. E. SnapShot: mammalian TRP channels. Cell129, 220 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Venkatachalam, K. & Montell, C. TRP channels. Annu. Rev. Biochem.76, 387–417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong, X. P. et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature455, 992–996 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, P., Gu, M. & Xu, H. Lysosomal ion channels as decoders of cellular signals. Trends Biochem. Sci.44, 110–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadler, M. J. et al. LTRPC7 is a Mg.ATP-regulated divalent cation channel required for cell viability. Nature411, 590–595 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Runnels, L. W., Yue, L. & Clapham, D. E. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science291, 1043–1047 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Abiria, S. A. et al. TRPM7 senses oxidative stress to release Zn2+ from unique intracellular vesicles. Proc. Natl. Acad. Sci. USA114, E6079–E6088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature436, 78–86 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Monteilh-Zoller, M. K. et al. TRPM7 provides an ion channel mechanism for cellular entry of trace metal ions. J. Gen. Physiol.121, 49–60 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, J., Li, M. & Yue, L. Potentiation of TRPM7 inward currents by protons. J. Gen. Physiol.126, 137–150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, J. et al. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science322, 756–760 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, J. et al. The channel kinase, TRPM7, is required for early embryonic development. Proc. Natl. Acad. Sci. USA109, E225–E233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sah, R. et al. Timing of myocardial trpm7 deletion during cardiogenesis variably disrupts adult ventricular function, conduction, and repolarization. Circulation128, 101–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schappe, M. S. et al. Chanzyme TRPM7 mediates the Ca(2+) influx essential for lipopolysaccharide-induced toll-like receptor 4 endocytosis and macrophage activation. Immunity48, 59–74.e55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendu, S. K. et al. Targeting the ion channel TRPM7 promotes the thymic development of regulatory T cells by promoting IL-2 signaling. Sci. Signal.13, eabb0619 (2020) [DOI] [PMC free article] [PubMed]
- 34.Desai, B. N. et al. Cleavage of TRPM7 releases the kinase domain from the ion channel and regulates its participation in Fas-induced apoptosis. Dev. Cell22, 1149–1162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krapivinsky, G., Krapivinsky, L., Manasian, Y. & Clapham, D. E. The TRPM7 chanzyme is cleaved to release a chromatin-modifying kinase. Cell157, 1061–1072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schappe, M. S. et al. Efferocytosis requires periphagosomal Ca(2+)-signaling and TRPM7-mediated electrical activity. Nat. Commun.13, 3230 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang, Y. L. et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. USA117, 20803–20813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai, Y. et al. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science347, 995–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunaratne, G. S., Yang, Y., Li, F., Walseth, T. F. & Marchant, J. S. NAADP-dependent Ca(2+) signaling regulates Middle East respiratory syndrome-coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium75, 30–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou, X. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun.11, 1620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dadonaite, B. et al. A pseudovirus system enables deep mutational scanning of the full SARS-CoV-2 spike. Cell186, 1263–1278.e1220 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prescott, J. B. et al. Immunobiology of Ebola and Lassa virus infections. Nat. Rev. Immunol.17, 195–207 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Simmons, J. A. et al. Ebolavirus glycoprotein directs fusion through NPC1+ endolysosomes. J. Virol.90, 605–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, I. S. et al. Mechanism of membrane fusion induced by vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA114, E28–E36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roche, S. & Gaudin, Y. Evidence that rabies virus forms different kinds of fusion machines with different pH thresholds for fusion. J. Virol.78, 8746–8752 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, S. et al. Acidic pH-induced conformations and LAMP1 binding of the lassa virus glycoprotein spike. PLoS Pathog.12, e1005418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, J. et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. USA114, E7987–E7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Simone, C. & Buchmeier, M. J. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology209, 3–9 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Lee, J. et al. Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry. Nat. Struct. Mol. Biol.28, 181–189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turk, B. et al. Acidic pH as a physiological regulator of human cathepsin L activity. Eur. J. Biochem.259, 926–932 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Alwan, H. A., van Zoelen, E. J. & van Leeuwen, J. E. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J. Biol. Chem.278, 35781–35790 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Qin, X. et al. Sphingosine and FTY720 are potent inhibitors of the transient receptor potential melastatin 7 (TRPM7) channels. Br. J. Pharm.168, 1294–1312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chubanov, V. & Gudermann, T. Mapping TRPM7 function by NS8593. Int. J. Mol. Sci21, 7017 (2020) [DOI] [PMC free article] [PubMed]
- 54.Chubanov, V. et al. Natural and synthetic modulators of SK (K(ca)2) potassium channels inhibit magnesium-dependent activity of the kinase-coupled cation channel TRPM7. Br. J. Pharm.166, 1357–1376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rost, B. R. et al. Optogenetic acidification of synaptic vesicles and lysosomes. Nat. Neurosci.18, 1845–1852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell181, 271–280.e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bayati, A., Kumar, R., Francis, V. & McPherson, P. S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem.296, 100306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koch, J. et al. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J.40, e107821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Birtles, D., Oh, A. E. & Lee, J. Exploring the pH dependence of the SARS-CoV-2 complete fusion domain and the role of its unique structural features. Protein Sci.31, e4390 (2022)
- 60.Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol.5. 10.1126/sciimmunol.abc3582 (2020). [DOI] [PMC free article] [PubMed]
- 61.Graler, M. H. & Goetzl, E. J. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G-protein-coupled receptors. FASEB J.18, 551–553 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Brinkmann, V. et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem.277, 21453–21457 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Galloway, S. E., Reed, M. L., Russell, C. J. & Steinhauer, D. A. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog.9, e1003151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu, Y. M., Sneyd, H., Dekant, R. & Wang, J. Influenza A virus nucleoprotein: a highly conserved multi-functional viral protein as a hot antiviral drug target. Curr. Top. Med. Chem.17, 2271–2285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouvier, N. M. & Lowen, A. C. Animal models for influenza virus pathogenesis and transmission. Viruses2, 1530–1563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Clercq, E. & Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol Rev.29, 695–747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drose, S. & Altendorf, K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol.200, 1–8 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Osei-Owusu, J. et al. Proton-activated chloride channel PAC regulates endosomal acidification and transferrin receptor-mediated endocytosis. Cell Rep.34, 108683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol.23, 3–20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kreutzberger, A. J. B. et al. Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. J. Virol.95, e0097521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Case, J. B. et al. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe28, 465–474.e464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whelan, S. P., Ball, L. A., Barr, J. N. & Wertz, G. T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA92, 8388–8392 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Case, J. B. et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe28, 475–485.e475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuerst, T. R., Niles, E. G., Studier, F. W. & Moss, B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA83, 8122–8126 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aguet, F. et al. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol. Biol. Cell27, 3418–3435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou, Y. Y. et al. Inherited nuclear pore substructures template post-mitotic pore assembly. Dev. Cell56, 1786–1803.e1789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cabot, M., Kiessling, V., White, J. M. & Tamm, L. K. Endosomes supporting fusion mediated by vesicular stomatitis virus glycoprotein have distinctive motion and acidification. Traffic23, 221–234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided in this paper.
Reagents generated in this study are available from the B.N.D. with a completed Materials Transfer Agreement. Requests for VSV-chimeras should be made to S.P.J.W. at spjwhelan@wustl.edu.
Software for single particle ROI identification is available on GitHub: (https://github.com/VolkerKirchheim/TrackBrowser_Matlab.git) (https://github.com/VolkerKirchheim/TrackBrowser_LabView.git).