Abstract
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) induces apoptosis through two receptors, TRAIL-R1 (also known as death receptor 4) and TRAIL-R2 (also known as death receptor 5), that are members of the TNF receptor superfamily of death domain-containing receptors. We show that human adenovirus type 5 encodes three proteins, named RID (previously named E3-10.4K/14.5K), E3-14.7K, and E1B-19K, that independently inhibit TRAIL-induced apoptosis of infected human cells. This conclusion was derived from studies using wild-type adenovirus, adenovirus replication-competent mutants that lack one or more of the RID, E3-14.7K, and E1B-19K genes, and adenovirus E1-minus replication-defective vectors that express all E3 genes, RID plus E3-14.7K only, RID only, or E3-14.7K only. RID inhibits TRAIL-induced apoptosis when cells are sensitized to TRAIL either by adenovirus infection or treatment with cycloheximide. RID induces the internalization of TRAIL-R1 from the cell surface, as shown by flow cytometry and indirect immunofluorescence for TRAIL-R1. TRAIL-R1 was internalized in distinct vesicles which are very likely to be endosomes and lysosomes. TRAIL-R1 is degraded, as indicated by the disappearance of the TRAIL-R1 immunofluorescence signal. Degradation was inhibited by bafilomycin A1, a drug that prevents acidification of vesicles and the sorting of receptors from late endosomes to lysosomes, implying that degradation occurs in lysosomes. RID was also shown previously to internalize and degrade another death domain receptor, Fas, and to prevent apoptosis through Fas and the TNF receptor. RID was shown previously to force the internalization and degradation of the epidermal growth factor receptor. E1B-19K was shown previously to block apoptosis through Fas, and both E1B-19K and E3-14.7K were found to prevent apoptosis through the TNF receptor. These findings suggest that the receptors for TRAIL, Fas ligand, and TNF play a role in limiting virus infections. The ability of adenovirus to inhibit killing through these receptors may prolong acute and persistent infections.
Adenovirus (Ad) has been widely studied as a model for virus replication, gene regulation, oncogenic cell transformation, and immune evasion. Ad infection in cell culture proceeds in well-regulated phases. The immediate-early E1A proteins, derived from the E1A transcription unit, induce transcription of delayed-early genes in the E1B, E2, E3, and E4 transcription units. Viral DNA begins to replicate at about 7 h postinfection (p.i.), and then late, primarily structural genes are expressed. Virions begin to assemble in the cell nucleus at about 1 day p.i. The cells begin to lyse at 2 to 3 days p.i. and release virus particles.
It is important that the infected cell remain intact during this extended period of infection. Indeed, Ads have evolved proteins that protect infected cells against apoptosis induced by cells and agents of the immune system (reviewed in references 14, 49, 69, 83, 87, 89, and 90). Most of these Ad proteins are encoded by the E3 and E1B transcription units. One such protein, named E3-gp19K, is a membrane glycoprotein localized in the endoplasmic reticulum. E3-gp19K forms a complex with major histocompatibility complex class I antigens, blocks their transport to the cell surface, and prevents killing of infected cells by cytotoxic T lymphocytes (CTL). Three Ad proteins inhibit apoptosis induced by tumor necrosis factor alpha (TNF-α) and Fas ligand (FasL; also known as CD95L). These ligands are expressed on activated leukocytes and are also shed in functional form; interact with their cognate receptors, TNF receptor 1 (TNFR1) and Fas (also known as CD95 and ApoI), respectively; and induce apoptosis by activation of caspases. The E3 protein named RID (for receptor internalization and degradation), a complex of the RIDα and RIDβ proteins (formerly known as E3-10.4K and E3-14.5K), is an integral membrane protein localized primarily on the cell surface (34, 67, 74, 75). RID inhibits apoptosis through the Fas pathway (19, 65, 72) by stimulating the internalization of cell surface Fas into endosomes, which are transported to lysosomes, where Fas is degraded (72). RID also inhibits TNF-induced apoptosis (23, 42). Another E3 protein, a nonmembrane protein named E3-14.7K (78), independently inhibits TNF-induced apoptosis (22, 24, 42). E3-14.7K is also reported to inhibit apoptosis induced through the Fas pathway (13). Finally, the protein named E1B-19K inhibits apoptosis induced through the TNF and Fas pathways (21, 31, 56, 72, 84).
TNF and FasL are members of the TNF superfamily. TNFR1 and Fas are members of the TNFR superfamily and contain “death domains” (reviewed in references 28, 53, 61, and 62). Death domains are conserved protein domains that participate in protein-protein interactions leading to activation of caspases that mediate apoptosis. TNF-related apoptosis-inducing ligand (TRAIL [also known as Apo2L]) is another member of the TNF superfamily that induces apoptosis (51, 58, 85), and two of the TRAIL receptors, TRAIL-R1 (also known as death receptor 4) and TRAIL-R2 (also known as death receptor 5), contain death domains (12, 54, 55, 64, 81). TRAIL and its receptors are expressed on many cell types (25).
TRAIL and the TRAIL receptors have been shown to play a role in a number of viral infections. T cells from human immunodeficiency virus-infected patients are killed by TRAIL (35, 38). Human cytomegalovirus (CMV) infection of primary human fibroblasts increased cell surface expression of TRAIL-R1 and TRAIL-R2, and TRAIL displayed potent antiviral activity in vitro on human CMV-infected fibroblasts (63). TRAIL and TRAIL receptors contribute to the apoptosis and pathology associated with reovirus infection (16) and are suggested to be involved in immunosuppression observed with measles infection (80).
TRAIL is emerging as another molecule used by cells of the immune system to kill virus-infected and tumor cells. Reports indicate that activated T and B cells express TRAIL (50, 52) and that TRAIL mediates killing by CD4+ CTL (39, 70). Several groups have found that human natural killer (NK) cells express TRAIL (37) and show TRAIL-dependent cytotoxicity (35, 37). TRAIL and TRAIL receptor expression could also be induced in a number of cells by interferon (IFN) treatment. IFN-γ and TNF induced TRAIL expression in primary human fibroblasts (63). Type I IFNs induced TRAIL expression on both CD4+ and CD8+ peripheral blood T cells (40). After IFN-γ or IFN-α treatment, human monocytes (27) and dendritic cells (20) expressed TRAIL and were able to kill tumor cells. Following treatment of monocytes with Type I IFNs, monocytes developed into TRAIL-expressing dendritic cells, which showed antiviral and antitumor effects (62). Thus, it might be expected that the TRAIL pathway would be targeted for inactivation by adenoviruses.
In this study we show that the Ad RID, E3-14.7K, and E1B-19K proteins independently inhibit TRAIL-induced apoptosis. As is the case with Fas, RID stimulates the internalization and degradation of TRAIL-R1.
MATERIALS AND METHODS
Cell lines.
Human A549 lung carcinoma cells (American Type Culture Collection [ATCC]), human 293 cells, and human HeLa cervical carcinoma cells were grown in Dulbecco's modified essential medium containing 10% fetal bovine serum. HT29.14S cells (10) (received from Jeff Browning, Biogen, Inc., Cambridge, Mass.; a clone derived from the HT29 colon carcinoma cell line [ATCC]) were grown in McCoy's medium with 10% fetal bovine serum. The E3-14.7K-expressing lines 4-6-8F and 37-2-1-2B were derived from A549 cells stably cotransfected with plasmids pSV2neo and either pMT2-14.7K or pMT2-14.7K(D37A) and selected in G418. The cell lines were isolated following subcloning of individual colonies. Clone 4-6-8F expresses the wild-type (WT) Ad5 E3-14.7K protein; clone 37-2-1-2B expresses an E3-14.7K protein in which a point mutation (D37 changed to A37) has been built into E3-14.7K (this has been shown not to alter E3-14.7K function).
Viruses.
Viruses used in these studies include Ad type 5 (Ad5), dl309 (Ad5 derivative; RID− 14,700-molecular-weight protein [14.7K]−) (36), dl111 (Ad5 derivative; RID− 14.7K− E1B-19K−) (3), lp5 (Ad2 derivative with a point mutation in E1B-19K) (68), dl250 (Ad2 derivative; E1B-19K−) (68), rec700 (the WT parental virus for viruses with 700 numbers; an Ad5-Ad2-Ad5 recombinant) (88), dl758 (14.7K−) (8), dl7000 (Ad5 derivative expressing only 14.7K from E3) (59), dl701 (6.7K−) (6), dl754 (deletion and modification of E3-6.7K C terminus and deletion of E3-gp19K) (22), dl704 (E3-gp19K−) (6), dl731 (E3-12.5K−) (8), pm734.1 (double point mutations eliminate the first two methionine codons of the Ad death protein [ADP]; ADP is not expressed) (76), dl762 (14.7K−) (8), dl764 (RIDβ−) (74), dl799 (RID−) (23), and pm760 (increased expression of RID; decreased expression of 14.7K) (9). Table 1 provides additional information. Virus stocks were grown in KB suspension cultures, and virus titers were determined by plaque assay on A549 cells as described previously(73).
TABLE 1.
Viruses used in this studya
| Virusb | RIDα | RIDβ | E3-14.7K | E1B-19K |
|---|---|---|---|---|
| rec700 | + | + | + | + |
| Ad5 | + | + | + | + |
| dl111 | − | − | − | − |
| dl309 | − | − | − | + |
| lp5 | + | + | + | − |
| dl250 | + | + | + | − |
| dl764 | + | − | + | + |
| dl799 | − | − | + | + |
| dl758 | + | + | − | + |
| dl762 | + | + | − | + |
| pm760 | ++ | ++ | ± | + |
| dl7000 | − | − | + | + |
| dl701 | + | + | + | + |
| dl704 | + | + | + | + |
| dl731 | + | + | + | + |
| pm734.1 | + | + | + | + |
| Ad/E3 | + | + | + | − |
| Ad/RID/14.7K | + | + | + | − |
| Ad/RID | + | + | − | − |
| Ad/14.7K | − | − | + | − |
All mutants except Ad/E3, Ad/RID/14.7K, Ad/RID, and Ad/14.7K are fully competent for replication in cultured cells. Ad/E3 is an E1− replication-defective Ad vector expressing all E3 Ad proteins except ADP from the CMV promoter. E3 c-encoded proteins are named RIDα, RIDβ, 14.7K, gp19K, 6.7K, 12.5K, and ADP. RIDα and RIDβ function together as the protein complex named RID. Ad/RID/14.7K is an E1− vector expressing only RID and 14.7K. Ad/RID is an E1− vector expressing only RID. Ad/14.7K is an E1− vector expressing only 14.7K. +, expressed; −, not expressed; ++, overexpressed; ±, underexpressed.
rec700 is an Ad5-Ad2-Ad5 recombinant and is the WT parent for mutants with 700 or 7000 numbers. Ad5 is the WT parent for dl111 and dl309. lp5 and dl250 are derived from Ad2, which is very closely related to Ad5 and rec700. dl701 lacks the gene for the E3-6.7K protein. dl704 lacks the gene for the E3-gp19K protein. dl731 lacks the gene for the E3-12.5K protein. pm734.1 lacks the gene for ADP (previously named E3-11.6K).
Ad vectors.
Ad/E3, Ad/RID/14.7K, Ad/14.7K, Ad/RID, and Ad/null are replication-deficient Ad vectors and were constructed according to the method described by Bett et al. (5); the construction will be described in detail elsewhere. Briefly, the E3 transcription unit of pm734.1 (76) was cloned into pcDNA3.1Zeo(+) (Invitrogen, Carlsbad, Calif.). Ad pm734.1 contains point mutations in the Met1 and Met41 codons of the adp gene and therefore does not express functional ADP. The whole expression cassette (CMV promoter, intron, and E3 genes) was excised and cloned into pdlE1sp1A (Microbix, Toronto, Canada), resulting in plasmid p231. p231 (the precursor plasmid for Ad/E3) expresses all E3 proteins except ADP, as shown by immunoblotting and immunofluorescence. pOD1 is very similar to p231, except it has the genes for the E3-12.5K, E3-6.7K, and E3-gp19K proteins deleted; this plasmid was used for construction of Ad/RID/14.7K. pOD2 and pOD3 express only the 14.7K or the RID protein, respectively, and were used to construct Ad/14.7K and Ad/RID, respectively. p371 has all the E3 genes deleted but retains the CMV promoter; it was used to produce the control Ad/null (empty) vector. These plasmids were sequenced to verify the inserts and then were cotransfected with pBHG10 (Microbix) into 293 cells. The viruses that resulted from recombinations were plaque purified three times on 293 cells, analyzed by DNA digestion using HindIII, and tested for the absence or presence of specific E3 proteins by immunoblotting, indirect immunofluorescence, and immunoprecipitation (data not shown). The viruses were grown in 293 suspension cultures and purified by CsCl banding. Titers were determined by plaque assay on 293 cells.
Antibodies.
A mouse monoclonal antibody (MAb) specific for TRAIL-R1 (M271) (26) was obtained from Immunex Corp. (Seattle, Wash.). Antibodies to transferrin receptor (TfnR) (OKT9) and Fas (M38) were from hybridoma cell lines obtained from ATCC. Antibody to the epidermal growth factor receptor (EGFR) (528) was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Rabbit polyclonal anti-peptide antibody to the Ad DNA binding protein (DBP) was a gift from Maurice Green (48). Rabbit polyclonal anti-peptide antibody to RIDβ has been described previously (74). Fluorochrome-conjugated, affinity-purified secondary antibodies were purchased from Cappel/ICN (Costa Mesa, Calif.).
Phase microscopy.
A549 cells were infected with 100 PFU (or 10 PFU/cell for dl111) of the indicated viruses/cell. At 14 h p.i., the cells were treated with TRAIL (200 ng/ml) in medium containing 25 μg of cycloheximide (CHX)/ml. After 7 h of TRAIL treatment, the cells were photographed on Tmax 400 film on a Nikon TMS inverted microscope.
Indirect immunofluorescence.
A549 or HeLa cells were plated on glass coverslips. The cells were infected with 50 to 400 PFU of virus/cell (as indicated in the figure legends). Some cells were treated with bafilomycin A1 (Baf) (0.1 μM) to inhibit acidification of lysosomes and to disrupt lysosomal degradation of internalized proteins (79, 91). The cells were fixed at the times indicated in the figure legends. For EGFR and RIDβ staining, cells were fixed in methanol (−20°C) containing 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. For TRAIL-R1 or DBP staining, cells were fixed in 3.7% paraformaldehyde for 10 min at room temperature and then permeabilized with methanol (−20°C) containing DAPI for 6 min. The cells were rehydrated with three washes of phosphate-buffered saline and then were stained with antiserum to TRAIL-R1 (M271) at a concentration of 10 μg/ml. EGFR MAb (528) was diluted to 1 μg/ml; DBP and RIDβ antisera were used at 1:400 and 1:250 dilutions, respectively. All antibodies were diluted in phosphate-buffered saline containing 1% bovine serum albumin and 0.1% sodium azide. The secondary antibodies were affinity-purified goat anti-mouse immunoglobulin G (IgG)-fluorescein isothiocyanate (FITC) conjugate for the MAb and goat anti-rabbit IgG-FITC conjugate for the rabbit polyclonal antibodies (Cappel/ICN). The mounting medium contained p-phenylenediamine as an antifading agent. The cells were photographed on Tmax 400 film on a Nikon Optiphot microscope equipped with epifluorescence. The film was developed in Diafine developer (Accufine) and fixed in Kodak fixer.
Apoptosis assays.
Apoptosis assays were conducted at 1 to 2 days p.i., a period well before these assays detect virus-induced cytotoxicity. Cells were infected with 100 PFU of the E1B-19K-positive replication-competent viruses/cell. For the E1B-19K-negative viruses dl111, dl250, and lp5, 5 to 25 PFU/cell was used (to reduce the cytolytic phenotype of these mutants). To confirm that cells were well infected, an immunofluorescence assay of DBP expression was quantitated at approximately 24 h p.i. For the E1-minus replication-defective vectors, 5 to 20 PFU/cell was used. For all viruses and vectors, cells at 4 to 5 h p.i. were trypsinized, diluted, and plated into 96-well plates. At approximately 24 h p.i., the cells were treated with serial dilutions of TRAIL (0.5 to 50 ng of leucine zipper TRAIL/ml; received from Immunex) (82) in medium containing 25 μg of CHX/ml. After approximately 24 h of TRAIL treatment, the supernatants were removed from the wells and assayed for lactate dehydrogenase (LDH) release using the CytoTox96 assay (Promega, Madison, Wis.). The following equation was used: percent specific lysis = (absorbance with TRAIL − absorbance with CHX)/(maximum absorbance − absorbance with CHX) × 100. The E3-14.7K stably transfected cell lines and their parental A549 cells were treated with 1 ng of TRAIL/ml for 25 h, and then trypan blue exclusion was used to determine the percentage of viable cells.
In some experiments, cell viability was assayed in parallel with the CellTiter 96 Aqueous One Solution cell proliferation assay (Promega). MTS (Owen's reagent) is bioreduced to a colored formazan product, which is soluble in the tissue culture medium and is measured at 490 nm in the original tissue culture plate.
Flow cytometry.
Cells were infected at 100 to 150 PFU of replication-competent viruses/cell or 5 to 20 PFU of E1-minus replication-defective viruses/cell, and staining was begun at 23 to 24 h p.i. as indicated in the figure legends. Live cells were incubated on ice with mouse monoclonal primary antibodies in fluorescence-activated cell sorter buffer at the following concentrations: TRAIL-R1 (M271), 5 μg/ml; Fas MAb (M38), 1:4 dilution of culture supernatant; EGFR MAb (Santa Cruz 528), 1 μg/ml; TfnR MAb (OKT9), 1:4 dilution of culture supernatant. The secondary antibody was affinity-purified goat anti-mouse IgG-FITC conjugate (whole molecule; Cappel/ICN). The cells were analyzed on a FACScaliber flow cytometer using Cell Quest software. The figures are presented as three-dimensional overlays of the flow cytometry data. Each curve represents data from 10,000 gated events.
RESULTS
RID, E3-14.7K, or E1B-19K protein is required to inhibit TRAIL-induced apoptosis in Ad-infected cells.
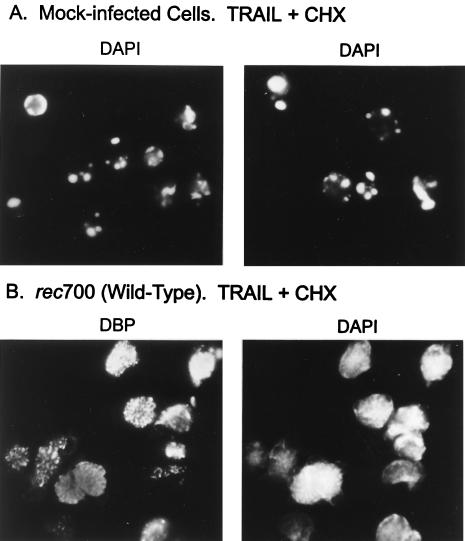
Human A549 cells were mock infected or infected with WT Ad (named rec700). At 19 h p.i., the cells were treated for 26 h with TRAIL (20 ng/ml) plus CHX (25 μg/ml). CHX was used because it increases the sensitivity of cells to apoptosis induced by TRAIL (44) and TNF (41, 44, 53). The cells were fixed, permeabilized with methanol containing DAPI (to stain nuclei), and then immunostained for the Ad-coded DBP. With mock-infected cells, nuclei were apoptotic, i.e., they were shrunken, the DNA was condensed, and apoptotic bodies were apparent (Fig. 1A). Cells infected with WT Ad, indicated by the speckled staining pattern for DBP in the cell nucleus, had nonapoptotic nuclei (Fig. 1B). We conclude that TRAIL induces apoptosis in A549 cells and that this apoptosis is inhibited by Ad infection.
FIG. 1.
Ad infection inhibits TRAIL-induced apoptosis as assessed by nuclear morphology. A549 cells were mock infected or infected with WT Ad (rec700) (100 PFU/cell). At 19 h p.i., the cells were treated with leucine zipper TRAIL (20 ng/ml) plus CHX (25 μg/ml). After 26 h, the cells were fixed in paraformaldehyde and then permeabilized with methanol containing DAPI. The cells were immunostained for the Ad-encoded DBP (72). (A) Two fields of mock-infected DAPI-stained nuclei. All the nuclei shown are apoptotic. (B) The same field of WT Ad-infected cells immunostained for DBP (left) and stained with DAPI (right). All the infected cells shown, as indicated by the speckled staining for DBP in the nucleus, had nonapoptotic nuclei.
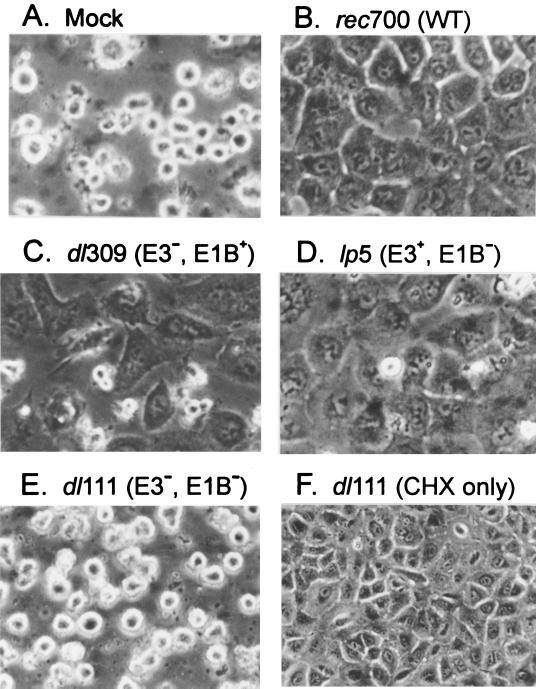
Virus mutants lacking combinations of the RID, E3-14.7K, and E1B-19K proteins (Table 1), previously shown to inhibit TNF- and Fas agonist-induced apoptosis, were tested to determine whether these viral proteins also inhibit TRAIL-induced apoptosis. A549 cells were infected, treated with TRAIL plus CHX, and then examined by phase-contrast microscopy. Mock-infected cells were apoptotic, but WT Ad-infected cells remained flat, attached, and viable (Fig. 2A and B). With a mutant lacking RID and E3-14.7K but expressing E1B-19K (i.e., RID− 14.7K− E1B+) (dl309) or a mutant expressing RID and E3-14.7K but not E1B-19K (RID+ 14.7K+ E1B-19K−) (lp5), most of the cells remained viable (Fig. 2C and D). However, with a RID− 14.7K− E1B-19K− mutant (dl111), the cells were killed by TRAIL plus CHX (Fig. 2E) but not by CHX alone (Fig. 2F). These results indicate that Ad has at least two independent functions that inhibit TRAIL-induced apoptosis, one being E1B-19K and the other being RID, E3-14.7K, or both E3 proteins.
FIG. 2.
Ad E3 and E1B-19K proteins inhibit TRAIL-induced apoptosis as indicated by cell morphology. A549 cells were infected with Ad mutants (100 PFU/cell), treated with leucine zipper TRAIL (200 ng/ml) plus CHX (25 μg/ml) at 14 h p.i., and then photographed after 7 h under phase-contrast microscopy. The lens magnifications were 100× (A to E) and 50× (F). See Table 1 for a description of the mutant genotypes. Mock-infected and dl111-infected (dl111 lacks E1B-19K, RID, and 14.7K) cells are apoptotic; viruses expressing E1B-19K or E3 proteins are protected. dl111-infected cells treated only with CHX are not apoptotic.
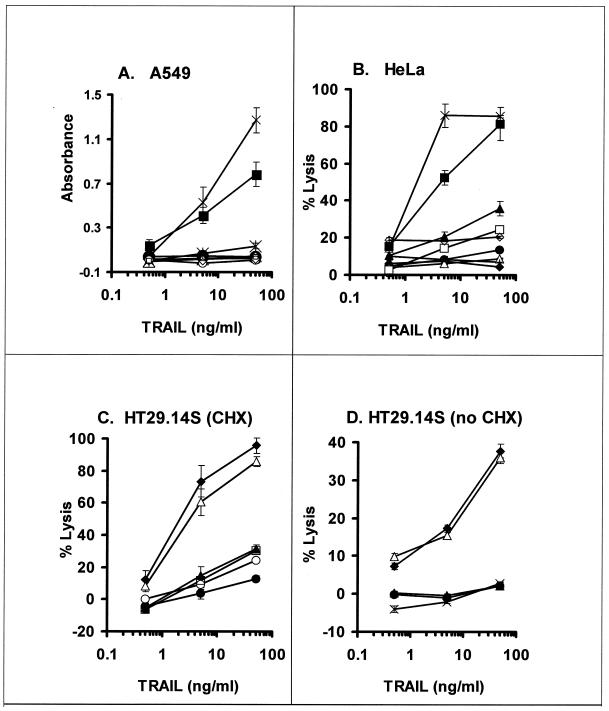
TRAIL-induced apoptosis was further examined by release of LDH from A549 cells, and additional virus mutants were tested (Table 1). TRAIL plus CHX induced apoptosis in mock-infected cells, and this was inhibited by WT Ad (rec700 or Ad5) but not by a RID− 14.7K− E1B-19K− mutant (dl111), as indicated by three independent assays: LDH release (Fig. 3A), trypan blue exclusion (data not shown), and the MTS assay for mitochondrial activity (data not shown). Similar LDH release results were obtained with HeLa cells (Fig. 3B). Parallel immunofluorescence studies indicated that nearly all the dl111-infected cells were expressing DBP (i.e., the cells were well infected) and that the DBP-positive cells had apoptotic nuclei (data not shown). TRAIL-induced killing was prevented by any mutant that expresses at least one of the E1B-19K, RID, or 14.7K proteins; the data are represented by the overlapping curves near the bottom of Fig. 3A and B. For example, TRAIL-induced apoptosis was blocked by mutants with the following genotypes: RID+ 14.7K+ E1B-19K− (lp5 and dl250), RID− 14.7K− E1B-19K+ (dl309), RID− 14.7K+ E1B-19K+ (dl764 and dl7000), RID+ 14.7K− Ε1B-19K+ (dl758), and RID++ 14.7K± E1B-19K+ (i.e., RID overexpressed and 14.7K underexpressed) (pm760). These data are consistent with those in Fig. 2 and indicate that E1B-19K and one or both of the RID and E3-14.7K proteins inhibit TRAIL-induced apoptosis of Ad-infected A549 or HeLa cells. When examined by immunofluorescence, both mock- and dl111-infected cells showed loss of cytochrome c from mitochondria and activation of caspase 3, indicating that the cells were undergoing apoptosis (data not shown).
FIG. 3.
The RID, E3-14.7K, and E1B-19K proteins inhibit TRAIL-induced apoptosis of Ad-infected cells. (A) Human A549 cells were infected with Ad mutants (100 PFU/cell). At 22 h p.i., the cells were treated with leucine zipper TRAIL (0.5, 5.0, and 50 ng/ml) plus CHX (25 μg/ml). After 28 h, apoptosis was determined colorimetrically based on release of LDH. The viruses used were mock, rec700, dl111, dl309, lp5, dl250, pm760, dl758, dl764, and dl7000. (B) Human HeLa cells infected with Ad mutants (100 PFU/cell) were treated with TRAIL and CHX at 21 h p.i. and assayed for LDH release after 24 h of treatment. The viruses used were mock, rec700, dl111, dl309, lp5, dl764, dl758, dl7000, and Ad5. (C) Human HT29.14S cells were infected with Ad mutants (100 PFU/cell). At 22 h p.i., the cells were treated with TRAIL and CHX. An LDH assay was done after 26 h of treatment. The viruses used were rec700, dl309, lp5, dl764, dl758, and pm760. (D) Human HT29.14S cells were infected with Ad mutants (100 PFU/cell). The cells were treated with TRAIL (no CHX) at 23 h p.i. and assayed for LDH release after 23 h of TRAIL treatment. The viruses used were mock, rec700, dl309, dl764, and lp5. ×, mock; ▪, dl111 (RID− 14.7K− E1B−); ●, rec700 (WT); ⧫, dl309 (RID− 14.7K−); ▴, lp5 (E1B−); ∗, dl250 (E1B−); ▵, dl764 (RIDβ−); □, dl758 (14.7K−); ○, pm760 (RID+ increased; 14.7K+ decreased); ◊, dl7000 (RID−); +, Ad5 (WT). The error bars indicate standard deviations.
The mutants shown in Fig. 3A and B do not distinguish between RID and E3-14.7K because every mutant that lacks one of these proteins also expresses E1B-19K. To examine RID specifically, an experiment was conducted in human HT29.14S cells, a clone of HT29 cells selected for sensitivity to TNF, Fas agonist MAb, and lymphotoxin α1 and β2 (10). In these cells, neither the E1B-19K nor E3-14.7K protein prevents Fas agonist-induced apoptosis (65), and this proves also to be true for TRAIL. TRAIL plus CHX lysed cells infected with a RID− 14.7K− E1B+ mutant (dl309) or a RID− 14.7K+ E1B+ mutant (dl764) (Fig. 3C); since dl309 expresses E1B-19K and dl764 expresses E1B-19K and E3-14.7K, the data indicate that E3-14.7K and E1B-19K do not inhibit TRAIL-induced apoptosis in these cells. TRAIL plus CHX did not kill cells infected with three mutants that express RID, dl758 (14.7K−), pm760 (RID++ 14.7K±), and lp5 (E1B-19K−) (Fig. 3C). These results establish that RID, but not E3-14.7K or E1B-19K, is required to inhibit TRAIL-induced apoptosis in Ad-infected HT29.14S cells.
Ad infection sensitizes cells to TRAIL-induced apoptosis, and RID is required to inhibit TRAIL-induced apoptosis in HT29.14S cells.
In the results shown so far, cells were treated with TRAIL in the presence of CHX. Ad-infected HT29.14S cells were also examined for apoptosis induced by TRAIL in the absence of CHX. The cells were mock infected or infected with various mutants and treated with 0.5, 5.0, or 50 ng of TRAIL/ml at 23 h p.i., and then cell lysis was determined at 46 h p.i. by release of LDH. Mock-infected cells were not killed by TRAIL, nor were cells infected with WT Ad (rec700) or an E3+ E1B-19K− mutant (lp5) (Fig. 3D). In contrast, cells were lysed by TRAIL when infected with a mutant (dl764) that lacks only RID and a mutant (dl309) that lacks RID and 14.7K (Fig. 3D).
These data support three conclusions. First, uninfected HT29.14S cells are not sensitive to TRAIL in the absence of CHX. Second, Ad infection sensitizes the cells to TRAIL; this property is revealed in the infections with the mutants that lack RID, i.e., Ad infection rendered the cells susceptible to TRAIL. Third, RID, but not E3-14.7K or E1B-19K, is required to inhibit TRAIL-induced apoptosis of Ad-infected HT29.14S cells in the absence of CHX.
RID stimulates the internalization of cell surface TRAIL-R1 into putative lysosomes.
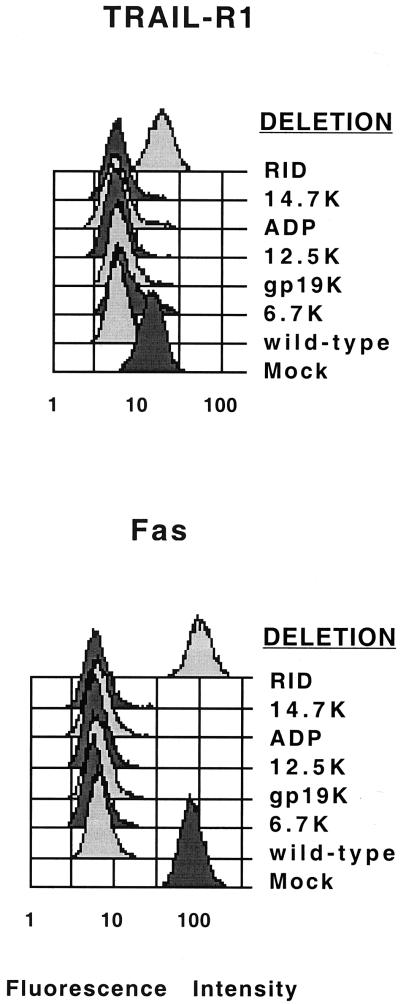
We have shown that RID causes the internalization of cell surface Fas into putative endosomes, which are transported to lysosomes where Fas is degraded (72). As a result, RID inhibits Fas agonist MAb-induced apoptosis. We have also shown that RID forces the internalization of EGFR into endosomes and lysosomes, where EGFR is degraded (11, 77). We have now addressed whether this scenario also applies to TRAIL-R1. HeLa cells were mock infected, infected with WT Ad (rec700), or infected with a series of mutants that have individual E3 genes deleted. These mutants were rec700 (WT), dl701 (E3-6.7K−), dl704 (E3-gp19K−), dl731 (E3-12.5K−), pm734.1 (ADP−), dl762 (14.7K−), and dl799 (RID−). At 23 h p.i., unfixed cells were stained and examined by flow cytometry for cell surface TRAIL-R1 and Fas. Both receptors were removed from the cell surface by WT Ad and any mutant that expresses RID (Fig. 4). In contrast, these receptors were not cleared by the mutant (dl799) that lacks only RID. Similar results were obtained in A549 cells (data not shown). Thus, RID is necessary to remove TRAIL-R1 and Fas from the cell surface.
FIG. 4.
RID is required for internalization of TRAIL-R1 and Fas from the surface of Ad-infected HeLa cells. Cells were mock-infected or infected (150 PFU/cell) with WT Ad (rec700) or with E3 deletion mutants in which the genes for the individual E3 proteins have been deleted. At 23 h p.i., unfixed cells were stained with MAbs against TRAIL-R1 or Fas, incubated with goat anti-mouse FITC-conjugated secondary antibody, and analyzed by flow cytometry using a FACScaliber flow cytometer and Cell Quest software (72). The mutants are as follows: rec700 (WT), dl701 (E3-6.7K−), dl704 (E3-gp19K−), dl731 (E3-12.5K−), pm734.1 (ADP−), dl762 (E3-14.7K−), and dl799 (RID−).
An additional point to note is that the RID− mutants express both E3-14.7K and E1B-19K. Therefore, these proteins apparently do not play a role in down-regulating these receptors.
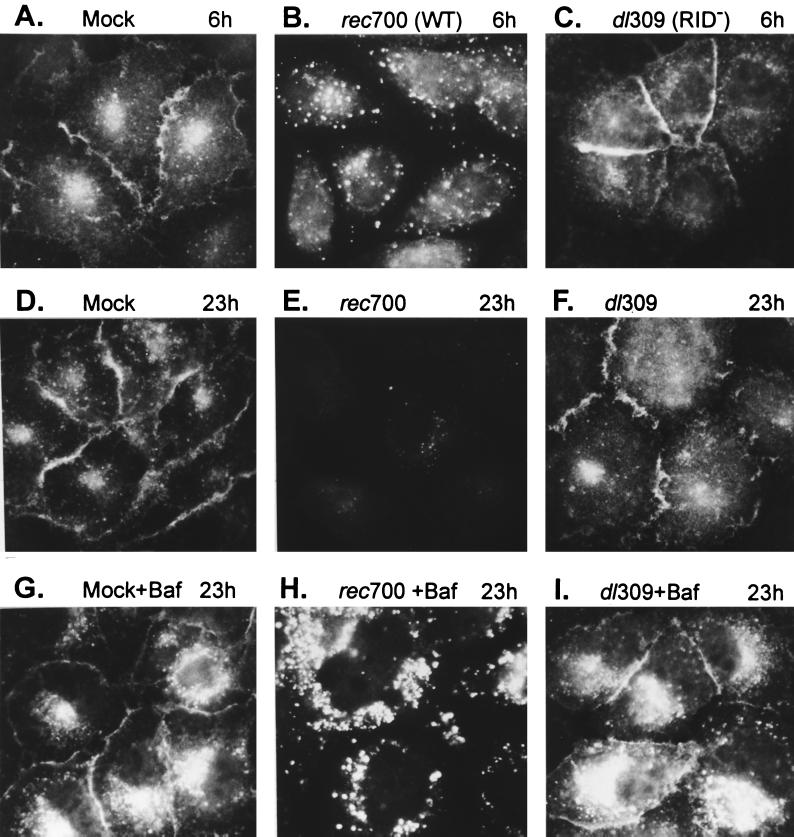
To address whether RID causes TRAIL-R1 to internalize into endosomes and lysosomes, A549 cells were mock infected or infected with WT Ad (rec700) or a RID− mutant (dl309) and then examined for TRAIL-R1 by indirect immunofluorescence at 6 and 23 h p.i. At 6 h p.i., the Ad infection is in the early phase (prior to Ad DNA replication), when RID is present in quite small amounts. At 23 h, the infection is in late stages, and RID has been in the cell for about 20 h and has accumulated in larger amounts. With mock-infected cells, there was uniform TRAIL-R1 staining on the cell surface and also some internal Golgi-like punctate staining (Fig. 5A and D). With WT Ad at 6 h, TRAIL-R1 had been cleared from the surfaces of about half the cells, and many vesicles were apparent (Fig. 5B). At 23 h, there was very little staining of TRAIL-R1, and only a few cells had vesicles (the field shown in Fig. 5E was selected to show these few cells). With the RID− mutant (dl309), TRAIL-R1 was not removed from the cell surface (Fig. 5C and F). Thus, RID causes internalization of cell surface TRAIL-R1 into putative endosomes and lysosomes, resulting in disappearance, presumably degradation, of TRAIL-R1. Attempts to show by immunoblotting that RID causes degradation of TRAIL-R1 were unsuccessful because TRAIL-R1 could not be detected.
FIG. 5.
RID mediates internalization of cell surface TRAIL-R1 into putative endosomes and lysosomes, where TRAIL-R1 is degraded. (A to F) Cells were mock infected or infected with WT Ad (rec700) or a RID− 14.7K− mutant (dl309). At 6 and 23 h p.i., the cells were fixed in paraformaldehyde and then permeabilized with methanol. The cells were immunostained for TRAIL-R1 using the M271 MAb (Immunex) and FITC-conjugated goat anti-mouse IgG. For the 6- and 23-h time points, 400 and 50 PFU of virus/cell, respectively, were used. (G to I) Same as for panels D to F, except the cells were treated with Baf (0.1 μM) beginning at 6 h p.i.
If TRAIL-R1 is degraded in lysosomes, then its degradation should be inhibited by Baf. Baf, a specific inhibitor of the vacuolar-type H+ ATPase, prevents acidification of vesicles and the sorting of receptors from late endosomes to lysosomes (79, 91). Baf inhibits the RID-mediated degradation of Fas (72) and EGFR (data not shown). In the present study, Baf had only a marginal effect on mock- or dl309-infected A549 cells, causing a modest accumulation of TRAIL-R1-containing vesicles, probably by blocking a low level of constitutive degradation of TRAIL-R1 (Fig. 5G and I). With WT Ad infection (rec700), Baf caused TRAIL-R1 to accumulate in large vesicles rather than being degraded (Fig. 5H). Baf did not affect clearance of TRAIL-R1 from the cell surface. These results strongly support the conclusion that RID mediates the degradation of TRAIL-R1 in lysosomes.
RID and E3-14.7K are sufficient to inhibit TRAIL-induced apoptosis.
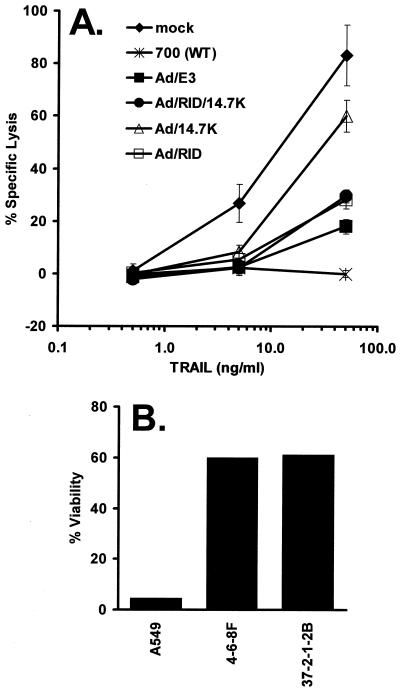
The results shown above address, in the context of Ad infection, whether RID, E3-14.7K, and E1B-19K are required to inhibit TRAIL-induced apoptosis and force TRAIL-R1 from the cell surface into putative endosomes and lysosomes. As a means to examine whether the E3 proteins are sufficient for these TRAIL-related effects, we employed replication-defective Ad vectors that express E3 proteins (K. Toth, M. Kuppuswamy, K. Doronin, O. A. Doronina, A. E. Tollefson, and W. S. M. Wold, unpublished data). One vector, named Ad/E3, contains an expression cassette with the entire E3 transcription unit driven by the CMV promoter-enhancer. Six E3 proteins are expressed from this vector, namely, RIDα, RIDβ, E3-14.7K, E3-gp19K, E3-6.7K, and E3-12.5K (data not shown). Other Ad proteins are not synthesized. A second vector, named Ad/RID/14.7K, expresses only RID and E3-14.7K from the CMV promoter. A third vector, named Ad/RID, expresses only RID from the CMV promoter. A fourth vector, named Ad/14.7K, expresses only E3-14.7K from the CMV promoter. The control for the Ad vectors was infection with an empty Ad vector (Ad/null). A549 cells were mock infected or infected with WT Ad (rec700) or the Ad vectors. The cells were treated with TRAIL plus CHX at 24.5 h p.i., and apoptosis was measured at 52.5 h p.i. by release of LDH. Mock-infected cells were lysed efficiently by TRAIL, and this lysis was strongly inhibited by WT Ad (Fig. 6A). The RID-expressing vectors also provided strong protection against TRAIL; the protection was slightly less than that of WT Ad, probably because the vectors lack E1B-19K (Fig 6A). Ad/14.7K gave partial but significant protection, especially when TRAIL was added at 5 ng/ml (Fig. 6A). These results show that RID and, to a lesser extent, E3-14.7K are sufficient, in the context of these Ad vectors, to inhibit TRAIL-induced apoptosis. The RID-expressing vectors also efficiently inhibited apoptosis mediated through the Fas pathway (data not shown).
FIG. 6.
RID protein is sufficient to inhibit TRAIL-induced apoptosis; 14.7K protects cells from TRAIL-induced apoptosis in stable transfectants. (A) A549 cells were mock infected or infected with 5 to 20 PFU of WT Ad (rec700), Ad/E3 (expressing all E3 proteins), Ad/RID/14.7K, Ad/RID, or Ad/14.7K per cell and then treated with TRAIL (plus CHX at 25 μg/ml) at 24.5 h p.i. as indicated. After 28 h, apoptosis was determined by release of LDH. The error bars indicate standard deviations. (B) A549 cells as well as two independent stably transfected A549 clonal cell lines expressing the E3-14.7K protein, named 4-6-8F (WT 14.7K expression) and 37-2-1-2B (14.7K with point mutation of D37A, not affecting function), were treated with TRAIL (1.0 ng/ml) plus CHX (25 μg/ml). After 25 h of treatment, the percentage of viable cells was determined by trypan blue exclusion.
To address further whether E3-14.7K can function alone to block TRAIL-induced apoptosis, two clonal lines of stably transfected A549 cells expressing E3-14.7K were examined. These lines express good levels of E3-14.7K that are readily detected by immunoblotting or immunofluorescence (data not shown). Apoptosis was determined by a trypan blue exclusion assay. Most parental A549 cells were killed by TRAIL plus CHX, whereas about 60% of both cell lines remained viable (Fig. 6B). A number of additional E3-14.7K-expressing clones had similar phenotypes (data not shown). Thus, E3-14.7K expression is sufficient to inhibit TRAIL-induced apoptosis in these cell lines.
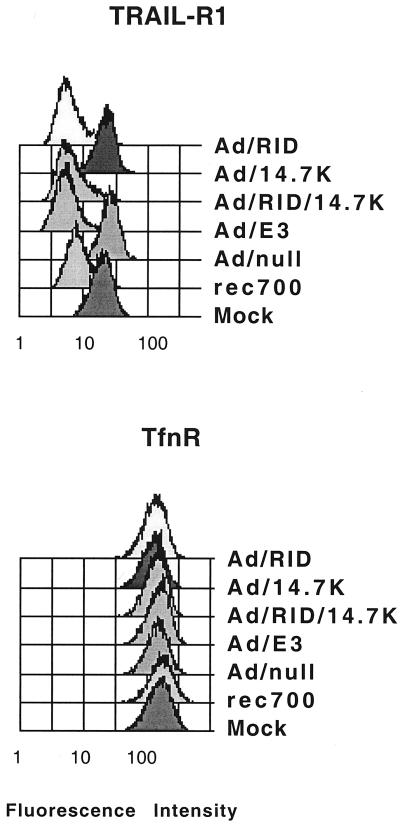
The Ad vectors were also used to address whether RID is sufficient to clear TRAIL-R1 from the cell surface. HeLa cells were mock infected, infected with rec700, or infected with the Ad vectors. At 23 h p.i., the cells were stained for TRAIL-R1 or TfnR and then analyzed by flow cytometry. TfnR, the negative control, was not affected (Fig. 7). TRAIL-R1 was cleared by rec700 and all vectors that express RID (Fig. 7). TRAIL-R1 was not cleared by Ad/14.7K or the empty vector.
FIG. 7.
Infection with replication-defective E1-minus Ad vectors expressing RID removes TRAIL-R1 but not TfnR from the cell surface. HeLa cells were mock infected or infected with 5 to 20 PFU of rec700 or Ad vectors/cell. At 23 h p.i., unfixed cells were stained with MAbs for TRAIL-R1 or TfnR and assayed by flow cytometry. Ad/E3 encodes and expresses all E3 proteins except ADP (E3-12.5K, E3-6.7K, E3-gp19K, RIDα, RIDβ, and E3-14.7K). Ad/E3, Ad/RID/14.7K, Ad/RID, and Ad/14.7K express only the E3 proteins indicated.
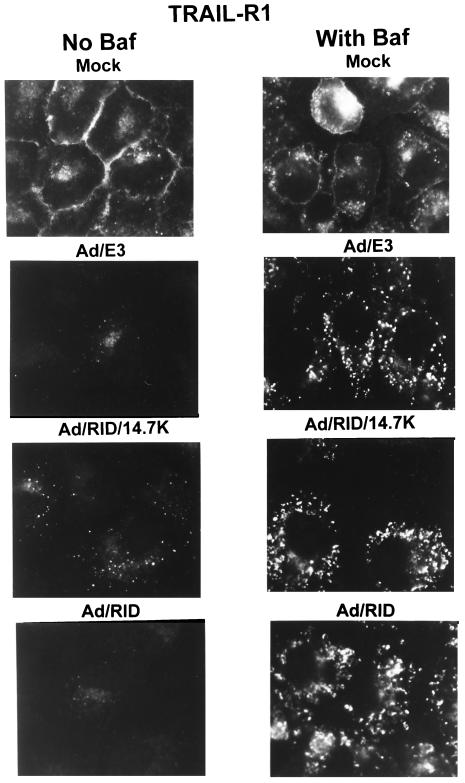
The Ad vectors were also used to examine the internalization of TRAIL-R1 into vesicles in the absence and presence of Baf. A549 cells were mock-infected or infected with the vectors, treated with Baf (0.1 μM) at 3 h p.i., and then fixed and stained for TRAIL-R1 at 25 h p.i. With mock-infected cells not treated with Baf, there was strong staining for TRAIL-R1 on the cell surface (Fig. 8). Baf had little effect on mock-infected cells. With the Ad/E3, Ad/RID/14.7K, and Ad/RID infections at 25 h p.i., most cells were no longer stained for TRAIL-R1, and with those that did stain, TRAIL-R1 was in vesicles rather than on the cell surface (Fig. 8). Similar results were obtained in a parallel experiment in which the cells were immunostained for EGFR (data not shown). When cells infected with RID-expressing vectors were treated with Baf, TRAIL-R1 was cleared from the cell surface and it accumulated in brightly staining vesicles (Fig. 8). These results are similar to those observed with rec700 (Fig. 5). The Ad/14.7K vector, which express E3-14.7K but not RID, did not affect TRAIL-R1 localization (data not shown). We conclude that RID expressed from the vectors is sufficient to force TRAIL-R1 from the cell surface into vesicles and to cause degradation (disappearance) of TRAIL-R1. The degradation of TRAIL-R1 very likely occurs in lysosomes, because it was inhibited by Baf.
FIG. 8.
Expression of RID by Ad vectors mediates down-regulation and degradation of TRAIL-R1, presumably by a lysosomal degradation pathway. A549 cells were mock infected or infected with 50 to 200 PFU of Ad vectors/cell. Baf was added at 3 h p.i. The cells were fixed and immunostained for TRAIL-R1 at 25 h p.i.
DISCUSSION
We have shown that Ad has three independent proteins that inhibit TRAIL-induced apoptosis, RID, E3-14.7K, and E1B-19K. RID stimulates the internalization of TRAIL-R1 from the cell surface, which probably explains why RID inhibits killing by TRAIL. TRAIL-R1 is internalized into vesicles, putative endosomes, and lysosomes and is degraded as indicated by the severe reduction in immunostaining for TRAIL-R1. (An alternative explanation for the lack of TRAIL-R1 immunostaining is that the epitope for TRAIL-R1 becomes masked.) The inhibition of TRAIL-R1 degradation by Baf argues strongly that TRAIL-R1 is degraded in lysosomes.
There are six known members of the TNFR superfamily that have death domains, namely, TNFR1, Fas, death receptor 3, TRAIL-R1, TRAIL-R2, and death receptor 6. Ligand-induced activation of TNFR1, Fas, TRAIL-R1, and TRAIL-R2 causes a series of protein-protein interactions involving the death domains that leads to activation of caspases and apoptosis (reviewed in references 1, 2, 17, 53, and 82). RID, E1B-19K, and E3-14.7K inhibit apoptosis induced by three of these ligands, TNF, FasL, and TRAIL (references 19, 21, 22, 24, 65, and 72 and this study). The Ad proteins block death ligand-induced apoptosis at several levels. RID gets rid of TRAIL-R1 and Fas by forcing them from the cell surface into lysosomes, where they are degraded. While this report was in preparation, Benedict et al. (4) reported that in addition to RID (referred to as E3-10.4K/14.5K), an E3 protein named E3-6.7K is required to clear TRAIL from the cell surface. Their studies were conducted with retroviruses expressing the RID and/or E3-6.7K proteins. We clearly have not observed a requirement for E3-6.7K in RID-mediated down-regulation of TRAIL-R1. Perhaps these differences in results are due to different experimental systems. Benedict et al. (4) also reported that both RID and E3-6.7K are required to down-regulate TRAIL-R2. We have similar findings under our experimental conditions (A. E. Tollefson, D. L. Lichtenstein, M. Kuppuswamy, K. Toth, K. Doronin, O. A. Doronina, C. A. Smith, and W. S. M. Wold, unpublished data).
RID also inhibits TNF-induced translocation of the 85-kDa cytosolic phospholipase A2 (cPLA2) from the cytosol to membranes (18). There is evidence that TNF-induced activation of cPLA2 is important in TNF-induced apoptosis (32, 42, 71, 86). We do not know if cPLA2 is activated through the Fas and TRAIL pathways.
E3-14.7K apparently inhibits apoptosis at more than one level. Yeast two-hybrid studies indicate that E3-14.7K binds to three cellular proteins named FIP-1, FIP-2, and FIP-3 (45–47). FIP-2 and FIP-3 may be part of the apoptotic signaling pathway that leads from TNFR1 and Fas through RIP; E3-14.7K could potentially inhibit this pathway. E3-14.7K is also reported to inhibit apoptosis induced by an Ad vector expressing FasL or by transient transfection of procaspase 8 (13). E3-14.7K forms a complex with procaspase 8 (13). E3-14.7K also inhibits TNF-induced activation of cPLA2 (42, 71, 92). It is not known if any of these functions of E3-14.7K account for its ability to inhibit TRAIL-induced apoptosis.
E1B-19K is a functional homolog of BCL-2 (7). It interacts with and inhibits the activity of proapoptotic members of the BCL-2 family (14, 83, 87). These proteins appear to promote apoptosis in part by displacing adapters from antiapoptotic BCL-2 family members, allowing caspases to become activated. E1B-19K was recently reported to interact with a conformationally altered form of Bax in mitochondria; this interaction inhibits cytochrome c release and caspase-9 activation (57). E1B-19K did not prevent activation of caspase-8. It seems likely that these functions of E1B-19K explain why E1B-19K inhibits TRAIL-induced apoptosis.
Routes et al. (60) recently reported that the Ad E1A proteins sensitize human A2058 melanoma and H4 fibrosarcoma cells to TRAIL-dependent killing. Such a function of E1A would explain the results of our experiment (Fig. 3D) showing that Ad infection sensitizes HT29.14S cells to TRAIL-induced apoptosis. Routes et al. (60) also reported an experiment suggesting that unspecified E3 proteins, and to a lesser extent E1B-19K, are required to inhibit TRAIL killing of Ad-infected A2058 cells.
Apoptosis would be deleterious to virus replication if it should occur before replication is complete (15). Thus, it makes sense that the virus should target receptors that mediate apoptosis. Apoptosis is a major mechanism by which the immune system eliminates unwanted cells (53). CTL kill through two main systems, the perforin-granzymes and Fas. They also kill through the TNF pathway, as indicated by long-term cell culture cytolysis assays. It is likely that the TRAIL system is involved in CTL killing of virus-infected cells, considering that T cells express TRAIL. Ad is well equipped to prevent killing of infected cells by CTL. The Ad-encoded gp19K prevents recognition of infected cells by the T-cell receptor. The RID, E3-14.7K, and E1B-19K proteins block apoptosis induced through Fas, TNFR1, and TRAIL-R1. Activated NK cells kill through the perforin-granzyme and perhaps the Fas and TRAIL systems, and activated macrophages synthesize TNF. Thus, it is possible that these Ad proteins inhibit CTL killing not only at the adaptive stage of the immune response but also at the innate stage.
Other signal transduction pathways, including NF-κB, stress-induced kinases, and neutral and acidic sphingomyelinases, are also activated through the TNFR1, Fas, and TRAIL receptors (17, 25, 53). Perhaps activation of one or more of these signaling pathways is deleterious to Ad replication, and accordingly, the receptors are eliminated by RID. The idea would be consistent with the ability of RID to cause internalization and degradation of EGFR, insulin receptor, and insulinlike growth factor-1 (IGF-1) receptor (11, 43, 77). For example, cPLA2 can be activated not only by TNF, but also by growth factors through the Ras–mitogen-activated protein kinase pathway. Arachidonic acid is the precursor to the proinflammatory eicosinoids, and by inhibiting arachidonic acid synthesis, the Ad proteins could inhibit inflammation. Indeed, the RID and E3-14.7K proteins inhibit inflammation in mouse models (29, 30, 66).
Considering that RID down-regulates very distinct receptors in the TNFR and protein tyrosine kinase families, the question arises as to the specificity of RID. There are some receptors that are not affected by RID, namely, TfnR (Fig. 7) (72, 77), major histocompatibility complex class I antigens (33), platelet-derived growth factor receptor, HER2 (43), CD46 (19), and CD44 (unpublished results). Thus, there is considerable specificity to RID.
As discussed above, RID causes the receptors for TRAIL, FasL, TNF, EGF, insulin, and IGF-1 to be internalized from the cell surface and degraded in lysosomes (11, 43, 72, 77; T. Dimitrov, C. F. Colle, A. E. Tollefson, D. L. Lichtenstein, and W. S. M. Wold, unpublished data). EGF, insulin, and IGF-1 are well known to stimulate internalization and degradation of their receptors. Surprisingly, little is known about this property for the death receptor ligands. Growth factor-induced degradation of receptors is believed to attenuate the growth factor signal, and this could also be true for the death receptor ligands. Attenuation may not be necessary if the cell has already been triggered to die. However, many cells are normally resistant to these death ligands and must become sensitized in order for the cell to undergo apoptosis; for nonsensitized cells, ligand-induced clearance of the death receptors would preclude cell death if the cells should subsequently receive a sensitizing signal. In any case, RID appears to serve as a surrogate ligand to cause internalization and degradation of these receptors. (There is no evidence that RID performs the other response to these ligands, i.e., induction of signal transduction.) The molecular mechanism of action of RID must be very interesting, and knowledge of it will increase our understanding of receptor signaling and sorting.
ACKNOWLEDGMENTS
K.T., K.D., and M.K. contributed equally to this work.
We thank Chris Wells for technical assistance, Todd Ranheim for dl7000, Abraham Scaria for pm734.1, G. Chinnadurai for lp5 and dl250, Tom Shenk for dl309, Harold Ginsberg and Lee Babiss for dl111, Maurice Green for antiserum to DBP, Jeff Browning for HT29.14S cells, Lynn Dustin for assistance with flow cytometry, Jayma Mikes for preparation of the figures, and Dawn Schwartz for preparation of the manuscript.
This work was supported by grants CA58538 and CA24710 from the National Institutes of Health.
REFERENCES
- 1.Ashkenazi A, Dixit V M. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 3.Babiss L E, Fisher P B, Ginsberg H S. Effect on transformation of mutations in the early region 1b-encoded 21- and 55-kilodalton proteins of adenovirus 5. J Virol. 1984;52:389–395. doi: 10.1128/jvi.52.2.389-395.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict C, Norris P, Prigozy T, Bodmer J L, Mahr J A, Garnett C, Martinon F, Tschopp J, Gooding L R, Ware C F. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J Biol Chem. 2001;276:3270–3278. doi: 10.1074/jbc.M008218200. [DOI] [PubMed] [Google Scholar]
- 5.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat B M, Wold W S M. A small deletion distant from a splice or polyadenylation site dramatically alters pre-mRNA processing in region E3 of adenovirus. J Virol. 1987;61:3938–3945. doi: 10.1128/jvi.61.12.3938-3945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 8.Brady H A, Scaria A, Wold W S M. Map of cis-acting sequences that determine alternative pre-mRNA processing in the E3 complex transcription unit of adenovirus. J Virol. 1992;66:5914–5923. doi: 10.1128/jvi.66.10.5914-5923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady H A, Wold W S M. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol Cell Biol. 1988;8:3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning J L, Miatkowski K, Sizing I, Griffiths D, Zafari M, Benjamin C D, Meier W, Mackay F. Signaling through the lymphotoxin beta receptor induces the death of some adenocarcinoma tumor lines. J Exp Med. 1996;183:867–878. doi: 10.1084/jem.183.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlin C R, Tollefson A E, Brady H A, Hoffman B L, Wold W S M. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell. 1989;57:135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary P M, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kB pathway. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Tian J, Kovesdi I, Bruder J B. Interaction of the adenovirus 14.7K protein with FLICE inhibits Fas ligand-induced apoptosis. J Biol Chem. 1998;273:5815–5820. doi: 10.1074/jbc.273.10.5815. [DOI] [PubMed] [Google Scholar]
- 14.Chinnadurai G. Control of apoptosis by human adenovirus genes. Semin Virol. 1998;8:399–408. [Google Scholar]
- 15.Chiou S K, White E. Inhibition of ICE-like proteases inhibits apoptosis and increases virus production during adenovirus infection. Virology. 1998;244:108–118. doi: 10.1006/viro.1998.9077. [DOI] [PubMed] [Google Scholar]
- 16.Clarke P, Meintzer S, Gibson S, Widmann C, Garrington T, Johnson G, Tyler K. Reovirus-induced apoptosis is mediated by TRAIL. J Virol. 2000;74:8135–8139. doi: 10.1128/jvi.74.17.8135-8139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnay B G, Aggarwal B B. Early events in TNF signaling: a story of associations and dissociations. J Leukoc Biol. 1997;61:559–566. doi: 10.1002/jlb.61.5.559. [DOI] [PubMed] [Google Scholar]
- 18.Dimitrov T, Krajcsi P, Hermiston T W, Tollefson A E, Hannink M, Wold W S M. Adenovirus E3-10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J Virol. 1997;71:2830–2837. doi: 10.1128/jvi.71.4.2830-2837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elsing A, Burgert H-G. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc Natl Acad Sci USA. 1998;95:10072–10077. doi: 10.1073/pnas.95.17.10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanger N, Maliszewski C, Schooley K, Griffith T S. Human dendritic cells mediate cellular apoptosis via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) J Exp Med. 1999;190:1155–1164. doi: 10.1084/jem.190.8.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gooding L R, Aquino L, Duerksen-Hughes P J, Day D, Horton T M, Yei S P, Wold W S M. The E1B 19,000-molecular-weight protein of group C adenoviruses prevents tumor necrosis factor cytolysis of human cells but not of mouse cells. J Virol. 1991;65:3083–3094. doi: 10.1128/jvi.65.6.3083-3094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gooding L R, Elmore L W, Tollefson A E, Brady H A, Wold W S M. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell. 1988;53:341–346. doi: 10.1016/0092-8674(88)90154-7. [DOI] [PubMed] [Google Scholar]
- 23.Gooding L R, Ranheim T S, Tollefson A E, Aquino L, Duerksen-Hughes P, Horton T M, Wold W S M. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J Virol. 1991;65:4114–4123. doi: 10.1128/jvi.65.8.4114-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gooding L R, Sofola I O, Tollefson A E, Duerksen-Hughes P, Wold W S M. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J Immunol. 1990;145:3080–3086. [PubMed] [Google Scholar]
- 25.Griffith T, Lynch D H. TRAIL: a molecule with multiple receptors and control mechanisms. Curr Opin Immunol. 1998;10:559–563. doi: 10.1016/s0952-7915(98)80224-0. [DOI] [PubMed] [Google Scholar]
- 26.Griffith T S, Rauch C T, Smolak P J, Waugh J Y, Boiani N, Lynch D H, Smith C A, Goodwin R G, Kubin M Z. Functional analysis of TRAIL receptors using monoclonal antibodies. J Immunol. 1999;162:2597–2605. [PubMed] [Google Scholar]
- 27.Griffith T S, Wiley S R, Kubin M, Sedger L, Maliszewski C, Fanger N. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J Exp Med. 1999;189:1343–1353. doi: 10.1084/jem.189.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross A, McDonnell J, Korsmeyer S. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 29.Harrod K S, Hermiston T W, Trapnell B C, Wold W S M, Whitsett J A. Lung-specific expression of E3-14.7K in transgenic mice attenuates adenoviral vector-mediated lung inflammation and enhances transgene expression. Hum Gene Ther. 1998;9:1885–1898. doi: 10.1089/hum.1998.9.13-1885. [DOI] [PubMed] [Google Scholar]
- 30.Harrod K S, Mounday A D W J A. Adenoviral E3-14.7K protein in LPS-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2000;278:631–639. doi: 10.1152/ajplung.2000.278.4.L631. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto S, Ishii A, Yonehara S. The E1b oncogene of adenovirus confers cellular resistance to cytotoxicity of tumor necrosis factor and monoclonal anti-Fas antibody. Int Immunol. 1991;3:343–351. doi: 10.1093/intimm/3.4.343. [DOI] [PubMed] [Google Scholar]
- 32.Hayakawa M, Ishida N, Takeuchi K, Shibamoto S, Hori T, Oku N, Ito F, Tsujimoto M. Arachidonic acid-selective cytosolic phospholipase A2 is crucial in the cytotoxic action of tumor necrosis factor. J Biol Chem. 1993;268:11290–11295. [PubMed] [Google Scholar]
- 33.Hermiston T W, Tripp R A, Sparer T, Gooding L R, Wold W S M. Deletion mutation analysis of the adenovirus type 2 E3-gp19K protein: identification of sequences within the endoplasmic reticulum lumenal domain that are required for class I antigen binding and protection from adenovirus-specific cytotoxic T lymphocytes. J Virol. 1993;67:5289–5298. doi: 10.1128/jvi.67.9.5289-5298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman P, Yaffe M B, Hoffman B L, Yei S, Wold W S M, Carlin C. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. Evidence for intermolecular disulfide bonding and plasma membrane localization. J Biol Chem. 1992;267:13480–13487. [PubMed] [Google Scholar]
- 35.Jeremias I, Herr I, Boehler T, Debatin K-M. TRAIL/Apo-2-ligand-induced apoptosis in human T cells. Eur J Immunol. 1998;28:143–152. doi: 10.1002/(SICI)1521-4141(199801)28:01<143::AID-IMMU143>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Jones N, Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 37.Kashii Y, Giorda R, Herberman R, Whiteside T, Vujanovic N. Constitutive expression and role of the TNF family ligands in apoptotic killing of tumor cells by human NK cells. J Immunol. 1999;163:5358–5366. [PubMed] [Google Scholar]
- 38.Katsikis P D, Garcia-Ojeda M E, Torres-Roca J F, Tijoe I M, Smith C A, Herzenberg L A, Herzenberg L A. Interleukin-1 beta converting enzyme-like protease involvement in Fas-induced and activation-induced peripheral blood T cell apoptosis in HIV infection. TNF-related apoptosis-inducing ligand can mediate activation-induced T cell death in HIV infection. J Exp Med. 1997;186:1365–1372. doi: 10.1084/jem.186.8.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayagaki N, Yamaguchi N, Nakayama M, Kawasaki A, Akiba H, Okumura K, Yagita H. Involvement of TNF-related apoptosis-inducing ligand in human CD4+ T cell-mediated cytotoxicity. J Immunol. 1999;162:2639–2647. [PubMed] [Google Scholar]
- 40.Kayakaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: a novel mechanism for the antitumor effects of type I IFNs. J Exp Med. 1999;189:1451–1460. doi: 10.1084/jem.189.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirstein M, Baglioni C. Tumor necrosis factor induces synthesis of two proteins in human fibroblasts. J Biol Chem. 1986;261:9565–9567. [PubMed] [Google Scholar]
- 42.Krajcsi P, Dimitrov T, Hermiston T W, Tollefson A E, Ranheim T S, Vande Pol S B, Stephenson A H, Wold W S M. The adenovirus E3-14.7K protein and the E3-10.4K/14.5K complex of proteins, which independently inhibit tumor necrosis factor (TNF)-induced apoptosis, also independently inhibit TNF-induced release of arachidonic acid. J Virol. 1996;70:4904–4913. doi: 10.1128/jvi.70.8.4904-4913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuivinen E, Hoffman B L, Hoffman P A, Carlin C R. Structurally related class I and class II receptor protein tyrosine kinases are down-regulated by the same E3 protein coded for by human group C adenoviruses. J Cell Biol. 1993;120:1271–1279. doi: 10.1083/jcb.120.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leverkus M, Neumann M, Mengling T, Rauch C, Brocker E-B, Krammer P H, Walczak H. Regulation of tumor necrosis factor-related apoptosis-inducing ligand sensitivity in primary and transformed human keratinocytes. Cancer Res. 2000;60:553–559. [PubMed] [Google Scholar]
- 45.Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, Wallach D, Horwitz M S. Identification of a cell protein (FIP-3) as a modulator of NF-κB activity and as a target of an adenovirus inhibitor of tumor necrosis factor α-induced apoptosis. Proc Natl Acad Sci USA. 1999;96:1042–1047. doi: 10.1073/pnas.96.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Kang J, Horwitz M S. Interaction of an adenovirus 14.7-kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J Virol. 1997;71:1576–1582. doi: 10.1128/jvi.71.2.1576-1582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Kang J, Horwitz M S. Interaction of an adenovirus E3 14.7-kilodalton protein with a novel tumor necrosis factor alpha-inducible cellular protein containing leucine zipper domains. Mol Cell Biol. 1998;18:1601–1610. doi: 10.1128/mcb.18.3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lillie J W, Loewenstein P M, Green M R, Green M. Functional domains of adenovirus type 5 E1a proteins. Cell. 1987;50:1091–1100. doi: 10.1016/0092-8674(87)90175-9. [DOI] [PubMed] [Google Scholar]
- 49.Mahr J A, Gooding L R. Immune evasion by adenoviruses. Immunol Rev. 1999;168:121–130. doi: 10.1111/j.1600-065x.1999.tb01287.x. [DOI] [PubMed] [Google Scholar]
- 50.Mariani S M, Krammer P H. Surface expression of TRAIL/Apo-2 ligand in activated mouse T and B cells. Eur J Immunol. 1998;28:1492–1498. doi: 10.1002/(SICI)1521-4141(199805)28:05<1492::AID-IMMU1492>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 51.Mariani S M, Matiba B, Armandola E A, Krammer P H. Interleukin 1β-converting enzyme related proteases/caspases are involved in TRAIL-induced apoptosis of myeloma and leukemia cells. J Cell Biol. 1997;137:221–229. doi: 10.1083/jcb.137.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Lorenzo M J, Alava M A, Gamen S, Kim K J, Chuntharapai A, Pineiro A, Naval J, Anel A. Involvement of APO2 ligand/TRAIL in activation-induced death of Jurkat and human peripheral blood T cells. Eur J Immunol. 1998;28:2714–2725. doi: 10.1002/(SICI)1521-4141(199809)28:09<2714::AID-IMMU2714>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 54.Pan G, Ni J, Wei Y F, Yu G, Gentz R, Dixit V M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 55.Pan G, O'Rourke K, Chinnaiyan A M, Gentz R, Ebner R, Ni J, Dixit V M. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 56.Perez D, White E. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J Cell Biol. 1998;141:1255–1266. doi: 10.1083/jcb.141.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez D, White E. TNFα signals apoptosis through a Bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol Cell. 2000;6:53–63. [PubMed] [Google Scholar]
- 58.Pitti R M, Marsters S A, Ruppert S, Donahue C J, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 59.Ranheim T S, Shisler J, Horton T M, Wold L J, Gooding L R, Wold W S M. Characterization of mutants within the gene for the adenovirus E3 14.7-kilodalton protein which prevents cytolysis by tumor necrosis factor. J Virol. 1993;67:2159–2167. doi: 10.1128/jvi.67.4.2159-2167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Routes J M, Ryan S, Clase A, Miura T, Kuhl A, Potter T A, Cook J L. Adenovirus E1A oncogene expression in tumor cells enhances killing by TNF-related apoptosis-inducing ligand. J Immunol. 2000;165:4522–4527. doi: 10.4049/jimmunol.165.8.4522. [DOI] [PubMed] [Google Scholar]
- 61.Sabelko-Downes K, Russell J H. The role of fas ligand in vivo as a cause and regulator of pathogenesis. Curr Opin Immunol. 2000;12:330–335. doi: 10.1016/s0952-7915(00)00095-9. [DOI] [PubMed] [Google Scholar]
- 62.Screaton G, Xu X-N. T cell life and death signalling via TNF-receptor family members. Curr Opin Immunol. 2000;12:316–322. doi: 10.1016/s0952-7915(00)00093-5. [DOI] [PubMed] [Google Scholar]
- 63.Sedger L, Shows D, Blanton R, Peschon J, Goodwin R, Cosman D, Wiley S. IFN-γ mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol. 1999;163:920–926. [PubMed] [Google Scholar]
- 64.Sheridan J P, Marsters S A, Pitti R M, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, Goddard A D, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 65.Shisler J, Yang C, Walter B, Ware C F, Gooding L R. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sparer T, Tripp R A, Dillehay D L, Hermiston T W, Wold W S M, Gooding L R. The role of adenovirus early region 3 proteins (gp19K, 10.4K, 14.5K, and 14.7K) in a murine pneumonia model. J Virol. 1996;70:2431–2439. doi: 10.1128/jvi.70.4.2431-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart A R, Tollefson A E, Krajcsi P, Yei S P, Wold W S M. The adenovirus E3 10.4K and 14.5K proteins, which function to prevent cytolysis by tumor necrosis factor and to down-regulate the epidermal growth factor receptor, are localized in the plasma membrane. J Virol. 1995;69:172–181. doi: 10.1128/jvi.69.1.172-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian T, Kuppuswamy M, Gysbers J, Mak S, Chinnadurai G. 19-kDa tumor antigen coded by early region E1b of adenovirus 2 is required for efficient synthesis and for protection of viral DNA. J Biol Chem. 1984;259:11777–11783. [PubMed] [Google Scholar]
- 69.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas W D, Hersey P. TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. J Immunol. 1998;161:2195–2200. [PubMed] [Google Scholar]
- 71.Thorne T E, Voelkel-Johnson C, Casey W M, Parks L W, Laster S M. The activity of cytosolic phospholipase A2 is required for the lysis of adenovirus-infected cells by tumor necrosis factor. J Virol. 1996;70:8502–8507. doi: 10.1128/jvi.70.12.8502-8507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tollefson A E, Hermiston T W, Lichtenstein D L, Colle C F, Tripp R A, Dimitrov T, Toth K, Wells C E, Doherty P C, Wold W S M. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature. 1998;392:726–730. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]
- 73.Tollefson A E, Hermiston T W, Wold W S M. Preparation and titration of CsCl-banded adenovirus stocks. In: Wold W S M, editor. Adenovirus methods and protocols. Totowa, N.J: The Humana Press; 1998. pp. 1–9. [Google Scholar]
- 74.Tollefson A E, Krajcsi P, Pursley M H, Gooding L R, Wold W S M. A 14,500 MW protein is coded by region E3 of group C human adenoviruses. Virology. 1990;175:19–29. doi: 10.1016/0042-6822(90)90182-q. [DOI] [PubMed] [Google Scholar]
- 75.Tollefson A E, Krajcsi P, Yei S P, Carlin C R, Wold W S M. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J Virol. 1990;64:794–801. doi: 10.1128/jvi.64.2.794-801.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tollefson A E, Stewart A R, Yei S P, Saha S K, Wold W S M. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J Virol. 1991;65:3095–3105. doi: 10.1128/jvi.65.6.3095-3105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tollefson A E, Wold W S M. Identification and gene mapping of a 14,700-molecular-weight protein encoded by region E3 of group C adenoviruses. J Virol. 1988;62:33–39. doi: 10.1128/jvi.62.1.33-39.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Weert A W M, Dunn K W, Geuze H J, Maxfield F R, Stoorvogel W. Transport from late endosomes to lysosomes, but not sorting of integral membrane proteins in endosomes, depends on the vacuolar proton pump. J Cell Biol. 1995;130:821–834. doi: 10.1083/jcb.130.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidalain P, Azocar O, Lamouille B, Astier A, Rabourdin-Combe C, Servet-Delprat C. Measles virus induces functional TRAIL production by human dendritic cells. J Virol. 2000;74:556–559. doi: 10.1128/jvi.74.1.556-559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walczak H, Degli-Esposti M A, Johnson R S, Smolak P J, Waugh J Y, Boiani N, Timour M S, Gerhart M J, Schooley K A, Smith C A, Goodwin R G, Rauch C T. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walczak H, Krammer P H. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis system. Exp Cell Res. 2000;256:58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 83.White E. Regulation of apoptosis by adenovirus E1A and E1B oncogenes. Semin Virol. 1998;8:505–513. [Google Scholar]
- 84.White E, Sabbatini P, Debbas M, Wold W S M, Kusher D I, Gooding L R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor α. Mol Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiley S R, Schooley K, Smolak P J, Din W S, Huang C-P, Nicholl J K, Sutherland G R, Smith T D, Rauch C, Smith C A, Goodwin R G. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 86.Wissing D, Mouritzen H, Egeblad M, Poirier G G, Jaattela M. Involvement of caspase-dependent activation of cytosolic phospholipase A2 in tumor necrosis factor-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:5073–5077. doi: 10.1073/pnas.94.10.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wold W S M, Chinnadurai G. Adenovirus proteins that regulate apoptosis. In: Cann A J, editor. DNA virus replication: frontiers in molecular biology. Oxford, United Kingdom: Oxford University Press; 2000. pp. 200–232. [Google Scholar]
- 88.Wold W S M, Deutscher S L, Takemori N, Bhat B M, Magie S C. Evidence that AGUAUAUGA and CCAAGAUGA initiate translation in the same mRNA region E3 of adenovirus. Virology. 1986;148:168–180. doi: 10.1016/0042-6822(86)90412-5. [DOI] [PubMed] [Google Scholar]
- 89.Wold W S M, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 90.Wold W S M, Tollefson A E. Adenovirus E3 proteins: 14.7K, RID, and gp19K inhibit immune-induced cell death; adenovirus death protein promotes cell death. Semin Virol. 1998;8:515–523. [Google Scholar]
- 91.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 92.Zilli D, Voelkel-Johnson C, Skinner T, Laster S M. The adenovirus E3 region 14.7 kDa protein, heat and sodium arsenite inhibit the TNF-induced release of arachidonic acid. Biochem Biophys Res Commun. 1992;188:177–183. doi: 10.1016/0006-291x(92)92366-6. [DOI] [PubMed] [Google Scholar]