Abstract
Background
Powdery mildew is a disease with one of the most substantial impacts on cucumber production globally. The most efficient approach for controlling powdery mildew is the development of genetic resistance; however, few genes associated with inherent variations in cucumber powdery mildew resistance have been identified as of yet.
Results
In this study, we re-sequence 299 cucumber accessions, which are divided into four geographical groups. A genome-wide association study identifies 50 sites significantly associated with natural variations in powdery mildew resistance. Linkage disequilibrium analysis further divides these 50 sites into 32 linkage disequilibrium blocks containing 41 putative genes. Virus-induced gene silencing and gene expression analysis implicate CsGy5G015960, which encodes a phosphate transporter, as the candidate gene regulating powdery mildew resistance. On the basis of the resequencing data, we generate five CsGy5G015960 haplotypes, identifying Hap.1 as the haplotype most likely associated with powdery mildew resistance. In addition, we determine that a 29-bp InDel in the 3′ untranslated region of CsGy5G015960 is responsible for mRNA stability. Overexpression of CsGy5G015960Hap.1 in the susceptible line enhances powdery mildew resistance and phosphorus accumulation. Further comparative RNA-seq analysis demonstrates that CsGy5G015960Hap.1 may regulate cucumber powdery mildew resistance by maintaining a higher H2O2 level through the depletion of multiple class III peroxidases.
Conclusions
Here we identify a candidate powdery mildew-resistant gene in cucumber using GWAS. The identified gene may be a promising target for molecular breeding and genetic engineering in cucumber to enhance powdery mildew resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-024-03402-8.
Keywords: Cucumber, Powdery mildew, GWAS, Phosphate transporter, H2O2
Background
Powdery mildew is a destructive foliar disease of greenhouse-grown cucumbers (Cucumis sativus L.) that is responsible for enormous economic losses worldwide [1]. Podosphaera xanthii (P. xanthii) is a causal organism that transmits powdery mildew, which is recognized by white or grayish powder-like spots on leaves and stems, which emerge initially on older leaves and shaded lower leaves. Fungicide treatment is one of the most common approaches to mitigate cucumber powdery mildew [2]. However, compared to other fungal diseases, P. xanthii has more easily acquired fungicide resistance [3]. Moreover, fungicide residues have detrimental effects on humans and the environment [4]. The amount of fungicide utilized in cucumber fields would be greatly reduced by comprehending the genetic basis of powdery mildew resistance and by developing powdery mildew-resistant cultivars.
The emergence of quantitative trait loci (QTL) in populations with diverse genetic backgrounds has helped us comprehend the genetic basis of powdery mildew resistance in cucumbers. According to the literature, at least 19 consensus powdery mildew resistance QTLs have been identified in various kinds of biparental populations and through linkage mapping [5]. For instance, Fukino et al. [6] mapped seven powdery mildew-resistant QTLs generated from PI197088 using 296 microsatellite markers. Other researchers, He et al. [7], used the study of simple sequence repeat markers from the powdery mildew-resistant inbred line WI2757 to locate six QTLs on chromosomes (Chr) 1, 3, 4, and 5. Recently, a study that combined whole-genome sequencing (WGS) and bulked segregant analysis (BSA) identified one powdery mildew-resistant QTL on Chr5 from the Cucumis hystrix introgression line IL52 [8]. Although it is straightforward to generate bi-parental segregation populations and they have been helpful for finding powdery mildew-resistant QTLs, QTL-based techniques have limitations (such as only a handful of genetic recombination events and parental alleles). To date, CsMLO1 (also known as CsaV3_5G036400 or Csa5G623470) is the only cucumber gene that has been cloned and functionally determined to be powdery mildew-resistant [9–11]. However, since CsMLO1 exhibits a recessive genetic pattern, programs for breeding may capitalize it less efficiently.
Phosphorus is an essential macronutrient that affects many biological processes, including photosynthesis, signal transduction, and plant metabolism, in addition to its contributions to development and growth [12]. Phosphate transporters (PHTs), which are principally responsible for soluble inorganic phosphate absorption, are the predominant means of phosphorus uptake and translocation in plants. Numerous PHTs have been identified in Arabidopsis and Oryza sativa to date. On the basis of their functional characteristics and subcellular distribution, these PHTs can be divided into four distinct subfamilies: PHT1, PHT2, PHT3, and PHT4. The PHT1 subfamily comprises an extensive number of members and is primarily located on the plasma membrane. Nevertheless, it should be noted that PHT2, PHT3, and PHT4 exhibit specific localization within cellular organelles, namely chloroplasts, mitochondria, and Golgi apparatus, respectively [13]. It is widely acknowledged that phosphorus is essential for plant growth; however, there is a paucity of information regarding its relationship with plant disease resistance. Phosphorus availability is critical for ensuring optimal crop growth and increasing plant resistance to diseases. In a previously documented field trial, the application of a 50 mM concentration of K2HPO4 has been shown to result in a significant increase in rice production, with a maximum improvement of 32%. Additionally, this treatment has been found to effectively mitigate the occurrence of neck blast, a disease caused by the pathogen Magnaporthe oryzae, with a maximum reduction rate of 42% [14]. In addition, it was discovered that the AtPHT4 mutants showed increased susceptibility to the pathogenic bacterium Pseudomonas syringae, a pathogen recognized to be associated with plant defense mechanisms mediated through salicylic acid. Another study has shown that the restricted movement of phosphate from the roots to the shoots causes the Arabidopsis pho1 mutant to exhibit symptoms associated with phosphate imbalance [15]. As a result, it is possible to conclude that PHTs play an important role in increasing plant resistance to pathogens.
Due to a higher mapping resolution and a larger amount of genetic variation than standard QTL mapping, GWAS employing LD is an effective and potentially valuable tool to comprehend the genetic architecture and maneuver complex features of interest in populations of unrelated lines [16]. The recent, rapid development of sequencing technologies has increased the appeal of GWAS to plant diseases. For instance, GWAS has been used to demonstrate the contributions of C2H2-type transcription factor (Bsr-d1) in the broad-spectrum blast resistance in rice, F-box protein (ZmFBL41) in the banded leaf and sheath blight resistance in maize, and non-specific lipid transfer protein (GhnsLTPsA10) in the resistance to Verticillium wilt in cotton [17–19]. In horticultural crops which include tomato [20], pepper [21], melon [22], and cucumber, GWAS has also been utilized effectively to rapidly locate the genes underlying particular features. Qi et al. [23] utilized WGS to construct a genetic variation map for cucumbers useful for a GWAS of agronomic parameters across 115 cucumber accessions from around the world. Wang et al. [24] examined 1234 USDA cucumber collections using a genotyping-by-sequencing (GBS) technique, identifying 24,319 single-nucleotide polymorphisms (SNPs). These SNPs were then used for detecting the genome regions significantly associated with 13 horticulturally important traits, including powdery mildew resistance, through a GWAS. Lee et al. [25] used GBS to identify 947,205 SNPs from a collection of 264 cucumber lines in a GWAS for powdery mildew resistance, spine color, and fruit stalk-end color. In this study, we describe the findings of a thorough analysis involving the resequencing of the 299 cucumber accessions. We examined the expression patterns, nucleotide diversity, and the effects of haplotypes on candidate genes by investigating the GWAS signals related with resistance to powdery mildew.
Results
Genotyping by whole-genome resequencing
The re-sequencing of 299 accessions yielded 27,843.8 million (M) paired-end reads with an average coverage of approximately 39 × . The reads were aligned to the Gy14 cucumber reference genome (version 2) using the Burrows–Wheeler Aligner program. The mapping rate ranged from 97.52% to 99.50% (Additional file 1: Table S1). Based on the mapped sequence, 5,394,682 SNPs and 1,292,277 insertions/deletions (InDels) were detected. After removing variable sites with a minor allele frequency (MAF) ≤ 5% and a missing data rate > 20%, 1,964,959 variable sites (1,625,341 SNPs and 339,618 InDels) were retained for the following analyses.
Marker density varied among chromosomes, with a minimum of 7393.6 markers per Mb on Chr 2 and a maximum of 10,029.6 markers per Mb on Chr 3 (Additional file 1: Table S2). Only two regions > 1 Mb, which were on chromosomes 2 and 6, lacked markers, and both regions were centromeric (Fig. 1A). The MAF for these markers ranged from 5 to 50%, with an average of 25.34% and a median of 24.08% (Fig. 1B), suggesting that all the retained markers had high imputation quality. Although the variable sites with a MAF between 5 and 10% were the most abundant, 1,525,064 variable sites (77.61%) had a MAF exceeding 10%, making them potentially useful for breeding-related applications (e.g., GWAS).
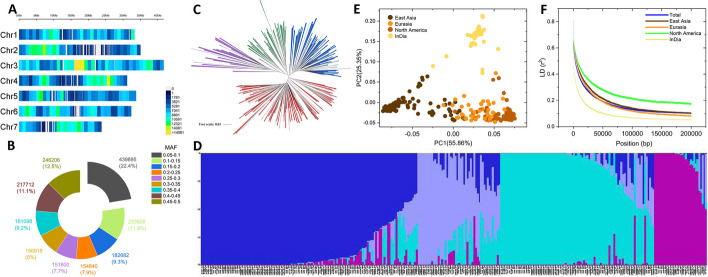
Fig. 1.
Population structure analysis of the cucumber accessions using high-quality SNPs and InDels. A SNP and InDel densities across the seven cucumber chromosomes. The number of SNPs and InDels in each 1-Mb non-overlapping window is provided. B Distribution of the minor allele frequency for the high-quality SNPs and InDels. C Phylogenetic tree for the resequenced accessions. The four groups, India (31 accessions), North America (49 accessions), Eurasia (91 accessions), and East Asia (128 accessions), are colored in violet, green, blue, and red, respectively. D Population structure plot for the 299 cucumber accessions with K = 4. Each color represents one subgroup, the Y axis quantifies subgroups membership, and the X axis shows the different cucumber accessions. E Principal component analysis of the resequenced accessions. Each plot indicated by a specific color represents an accession from the corresponding group. F Genome-wide average linkage disequilibrium (r2) decay for the accessions in the India, North America, Eurasia, and East Asia groups and for all accessions
Population structure and linkage disequilibrium
The phylogenetic tree generated using the neighbor-joining approach classified the 299 accessions into four distinct groupings (Fig. 1C). These groupings are identified by color and are connected to the accessions’ geographic distribution: East Asia (red, 128 accessions), North America (green, 49 accessions), Eurasia (blue, 91 accessions), and India (violet, 31 accessions) (Additional file 1: Table S1). The Admixture tool, which uses a maximum-likelihood approach to calculate individual ancestries and admixture proportions under the presumption that K populations exist, was used to analyze the population structure. The ideal K value was determined through Admixture clustering to be 4, since the cross-validation (CV) error quickly dropped until K = 4 and then stabilized. According to Fig. 1D, at K = 4, the 299 accessions could be split into four subpopulations, which is consistent with the findings of the phylogenetic analysis. In addition, principal component analysis (PCA) was performed to assess the genetic diversity of the panel. The top two PCs, PCA1 (55.86%) and PCA2 (25.35%), may account for more than 80% of the total genetic variance. In contrast to the other accessions, those from North America and Eurasia were clustered and easily distinguished from one another (Fig. 1E). These results indicated that the accessions were subdivided into subpopulations, which was a covariate in the GWAS model.
To determine the mapping resolution for the GWAS, we quantified the average extent of the LD using the PLINK software and evaluated the pairwise LD based on squared allele frequency correlations (r2). Using the 1,964,959 variable sites, the LD decay rate among all chromosomes in the whole population was calculated as approximately 11.25 kb, with r2 = 0.387 corresponding to half of the maximum value. The LD varied among the four groups (Fig. 1F), and the LD decay distances (to r2 = 0.387) for the India, Eurasia, East Asia, and North America groups were 2.5, 13.75, 15, and 22.5 kb, respectively. The higher LD value for cucumber accessions from North American is consistent with the earlier finding that this population has been subject to artificial selection [24].
We then evaluated the genetic diversity within groups. The average genome-wide nucleotide diversity (π) values for the India, North America, Eurasia, and East Asia groups were 0.371 × 10–3, 0.308 × 10–3, 0.248 × 10–3, and 0.216 × 10–3, respectively. The π value of the India group was the highest, consistent with the fact that India is the center of origin for cultivated cucumber. Therefore, we expect this region’s cucumber accessions to be more genetically diverse than those from other regions.
Phenotypic variations in response to powdery mildew
Based on the disease index (DI), 299 cucumber accessions were evaluated for powdery mildew resistance in a greenhouse at the adult plant stage in the autumn of 2022, winter of 2022, and spring of 2023. The overall distribution of DIs for the entire population and each of the four categories was determined. The resistance to powdery mildew differed between experiments. All accessions had DIs ranging from 0 to 32.04 (average of 15.67) in fall 2022, 0 to 51.2 (average of 20.52) in winter 2022, and 0 to 52.86 (average of 28.44) in spring 2023 (Fig. 2A–C). The mean DIs in the combined analysis ranged from 0 to 52.86, with an overall mean of 16.03. The Eurasia group had greater mean DIs and lower resistance levels than the other groups. Additionally, the DIs for the whole population over the three environments were significantly correlated, with Pearson’s correlation coefficients (r2) of 0.778 (fall–winter), 0.448 (fall–spring), and 0.458 (winter–spring), reflecting the high repeatability of the data as well as the effects of genetic factors on trait determination.
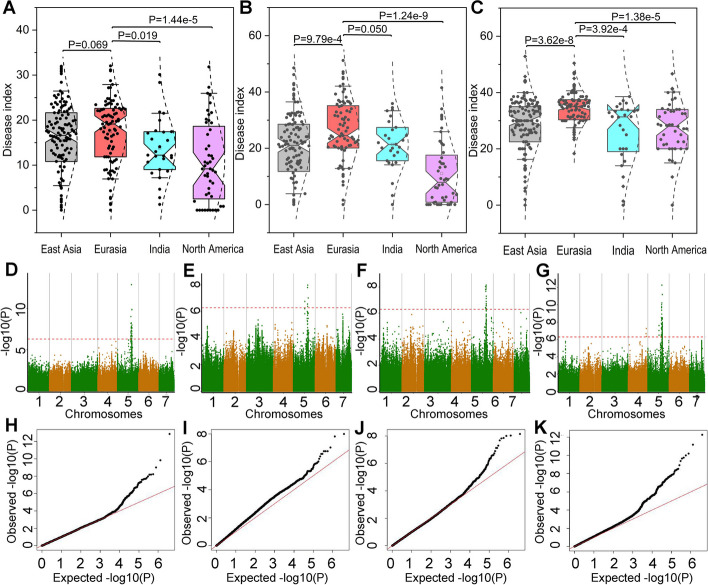
Fig. 2.
GWAS for cucumber powdery mildew resistance. Phenotypic variations in powdery mildew resistance based on the disease index among the India, North America, Eurasia, and East Asia groups over the following three cropping seasons: fall 2022 (A), winter 2022 (B), and spring 2023 (C). Each black dot represents the average disease index of a cucumber accession. Student’s t-test was used to calculate the P value. D Manhattan plot for the GWAS results using fall 2022 data. E Manhattan plot for the GWAS results using winter 2022 data. F Manhattan plot for the GWAS results using spring 2023 data. G Manhattan plot for the GWAS results using the BLUP (best linear unbiased prediction) values; H quantile–quantile (Q-Q) plot for the GWAS results using fall 2022 data. I Q-Q plot for the GWAS results using winter 2022 data. J Q-Q plot for the GWAS results using spring 2023 data. K Q-Q plot for the GWAS results using the BLUP (best linear unbiased prediction) values. Red dashed lines in the Manhattan plots indicate the significance threshold (− log10 P = 6.29). Black dots in the Q-Q plots represent the distribution of P values
Genome-wide association analysis for powdery mildew resistance in cucumber
The population structure (Q matrix) and kinship relationship (K matrix), which served as covariates, were taken into account in the association analyses based on the mixed linear model, together with the DIs of the 299 accessions for the three environments. The GWAS of this population revealed a strong signal for powdery mildew resistance on chromosome 5, which contained 68 significant variable sites (P < 5.16 × 10−7), including 40 from the fall 2022 data (Fig. 2D), 9 from the winter 2022 data (Fig. 2E), 19 from the spring 2023 data (Fig. 2F), and 105 from the BLUP model (Fig. 2G); these sites explained 9.95%–22.06% of the phenotypic variance (Additional file 1: Table S3). Two sites, chromosome 5_22246773 and chromosome 5_22936361, were detected in all three environments and by the BLUP method, with the most significant site being chromosome 5_22246773.
To obtain reliable results, we utilized only the significant sites that were identified at least twice. Of the 50 significant sites selected for further analysis, three (chromosome 5_17680102, chromosome 5_17704760, and chromosome 5_17713083) were located within the powdery mildew-resistant QTL Pm5.1, while the remaining sites were located within the powdery mildew-resistant QTL Pm5.2 [5], providing additional evidence that these sites are essential for powdery mildew resistance.
CsGy5G015960 was required for powdery mildew-resistance in cucumber
The LD analysis divided the 50 significant sites into 32 LD blocks, in which 41 candidate genes for powdery mildew resistance were predicted (Additional file 1: Table S4). We employed the cucumber green mottle mosaic virus vector (pV190) VIGS approach to knock down the expression of the 34 protein-coding candidate genes in the powdery mildew-resistant line EP6411 to discover the gene responsible for powdery mildew resistance [26]. We excluded seven genes (CsGy5G015240, CsGy5G015250, CsGy5G015730, CsGy5G015740, CsGy5G016440, CsGy5G016460, and CsGy5G016480) from further investigation because we cannot clone whole gene fragments. In fact, none of the 88 RNA-seq data sets included in the cucumber genome database showed the putative transcripts for these seven genes and were annotated with unknown functions (http://cucurbitgenomics.org/cgi-bin/RNA_project). Relative expression of the 34 candidate genes in plants infected by pV190 carrying genes fragment was significantly lower than those infected by pV190 empty vector (control) (Additional file 2: Fig. S1A). Cucumber phytoene desaturase (CsPDS encoded by CsGy4G002600) was used as a visual marker to monitor the silencing effect of the VIGS system. After 14 days of VIGS, the pV190-CsPDS-treated plants exhibited an albino phenotype, indicating that the VIGS system efficiently decreased the expression of the target gene in the powdery mildew-resistant cucumber line EP6411 (Additional file 2: Fig. S1B). After receiving the powdery mildew pathogen inoculation, the seedlings were checked 15 days after treatment. Only the pV190-CsGy5G015960-treated plants showed significantly higher DIs and more sporulation on the true leaves as compared to the pV190-treated plants (Additional file 2: Fig. S1B–D). Therefore, CsGy5G015960 (a phosphate transporter) was selected for future investigation because it may be a positive regulator of powdery mildew resistance in cucumbers. Tissue-specific expression analysis revealed that CsGy5G015960 was expressed in all eight examined tissues (i.e., mature leaves, young leaves, male flowers, female flowers, roots, stems, tendrils, and growth points). But in mature leaves, it was significantly expressed, as demonstrated by qRT-PCR (Additional file 2: Fig. S2).
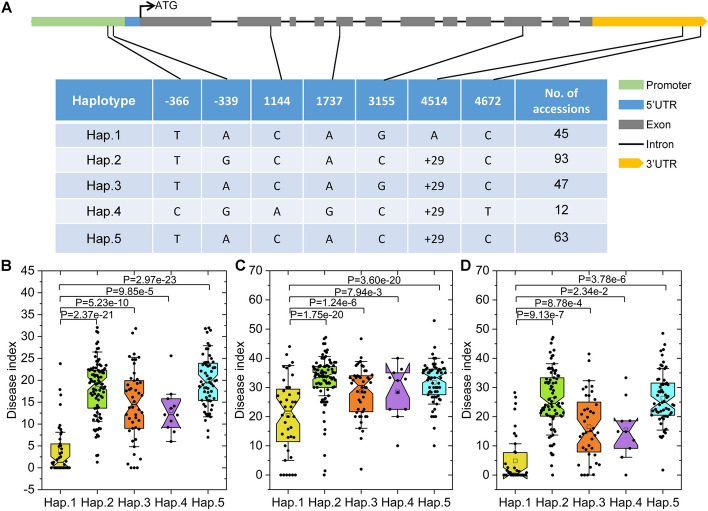
Using the re-sequencing data, the CsGy5G015960 haplotypes were built according to the homozygous SNPs and InDels detected in the promoter region (i.e., approximately 1 kb upstream of the transcription initiation site), the 5′ and 3′ UTRs, and the coding region. According to the translation start site of CsGy5G015960, we detected two SNPs in the promoter region (− 366 and − 339 bp), two synonymous SNPs (1141 and 1737 bp) and one non-synonymous SNP (3155 bp, Leu/Val) in the exons, one 29-bp InDel (4,514 bp, A/AAATGAAAATGAGAGTTTATTTTTAATGTT), and one SNP in the 3′ UTR (Fig. 3A). We concentrated on haplotypes shared by at least five accessions in the studied group. Among the 260 accessions, the haplotype analysis showed five haplotypes. The DIs were compared for the five haplotypes (Fig. 3B). The DI of Hap.1 was significantly lower than that of the other haplotypes (Fig. 3C). Notably, Hap.1 lacked the 29-bp InDel, implying that the InDel strongly influences powdery mildew resistance. These findings suggest that CsGy5G015960 could be a potential candidate gene for the GWAS locus.
Fig. 3.
Haplotypes of CsGy5G015960. A Graphical representation of the gene structure and details regarding the 5 haplotypes detected in 260 cucumber accessions. The SNP and InDel positions are provided in the first row. + 29, insertion of AATGAAAATGAGAGTTTATTTTTAATGTT. B Phenotypic effect of each haplotype for the fall 2022 data. C Phenotypic effect of each haplotype for the winter 2022 data. D Phenotypic effect of each haplotype for the spring 2023 data. Each black dot represents the average disease index of a cucumber accession. Student’s t-test was used to calculate the P value
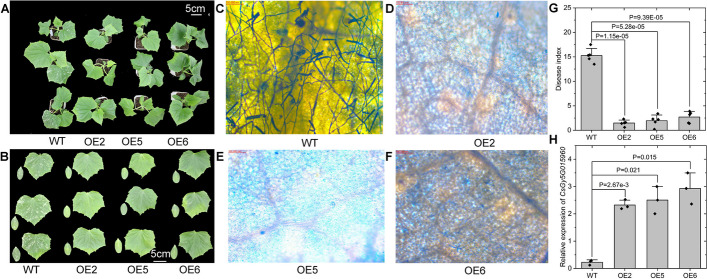
To further functionally characterize the potential impact of CsGy5G015960 on powdery mildew resistance, we overexpressed CsGy5G015960Hap.1 in the powdery mildew susceptible line CCMC. Three representative plants for each overexpression (OE) construct (OE2, OE5, and OE6) were self-pollinated to produce T2 transgenic lines that were further characterized. As shown in Fig. 4A–F, at 8 days after inoculation, the CsGy5G015960-OE lines exhibited only brownish lesions on the cotyledons and leaves, with barely noticeable mildew. In contrast, the infected leaves of wild-type (WT) plants showed clearly visible whitish mildew colonies. CsGy5G015960-OE lines treated with powdery mildew pathogens showed significantly lower DIs than the WT control under the same condition (Fig. 4G). The qRT-PCR result indicated that the expression levels of CsGy5G015960 in the OE2, OE5, and OE6 were 10.28-, 11.07-, and 12.96-fold higher than that of WT, respectively (Fig. 4H). Negligible powdery mildew symptoms were observed on the leaf surface of CsGy5G015960-OE lines and WT when inoculated with water (mock control, Additional file 2: Fig. S3). Therefore, our findings imply that CsGy5G015960 positively affects cucumber powdery mildew resistance.
Fig. 4.
Functional characterization of CsGy5G015960. A Representative whole-plant phenotypes of the CsGy5G015960 overexpressing lines (OE2, OE5, and OE6) and the wild-type (WT) control at 8 days post inoculation (dpi) with the powdery mildew pathogen. B Phenotype of the first true leaves and cotyledons of the CsGy5G015960 overexpressing lines and the WT control at 8 dpi of powdery mildew pathogen. Coomassie blue staining of WT (C), OE2 (D), OE5 (E), and OE6 (F) at 8 dpi of powdery mildew pathogens. G Disease indices of OE2, OE5, OE6, and the WT control at 8 dpi of powdery mildew pathogen. Each black dot represents one biological replicate that is an average of 15 plants. Bar = 100 nm. H Confirmation of CsGy5G015960 expression in the transgenic CsGy5G015960 overexpressing cucumber lines (OE2, OE5, and OE6) and the WT control by qRT-PCR. Each black dot represents one biological replicate. Student’s t-test was used to calculate the P value
The 29-bp indel in the 3′ UTR influences the mrna stability
We investigated CsGy5G015960 expression levels in 15 cucumber accessions from each of the five haplotypes. The qRT-PCR data demonstrated that powdery mildew significantly increased the expression of CsGy5G015960 in all 15 cucumber lines, but this was especially true in the three powdery mildew-resistant lines that belong to Hap.1 (YZ002A, YZU164A, and YZU038A). This was in contrast to the corresponding levels in Hap.2 (YZ014A, YZU166A, and YZU175A), Hap.3 (YZ047A, YZ038A, and YZ328), Hap.4 (YZU089A, YZU092A, and YZU099A), and Hap.5 (YZU007A, YZU055A, and YZU056A (Additional file 2: Fig. S4). Furthermore, we conducted the experiment to assess the expression of CsGy5G015960 in the roots. In contrast, there were no consistent trends in the expression of CsGy5G015960 among the 15 cucumber lines (Additional file 2: Fig. S5).
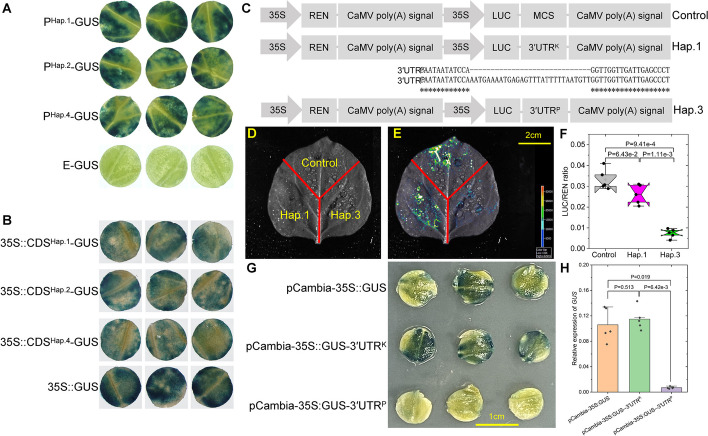
To address whether the altered expressions of CsGy5G015960 in Hap.2 to Hap.5 are caused by mutations in the promoters or exons, we cloned the promoters and coding sequence (CDS) from YZ002A (PHap.1 and CDSHap.1), YZ014A (PHap.2 and CDSHap.2), and YZU089A (PHap.4 and CDSHap.4) to produce promoter-GUS and 35S::CDS-GUS fusion constructs and detected their activities using an Agrobacterium-mediated β-glucuronidase (GUS) transient assay in cucumber leaves. Histochemical staining results showed that cucumber leaves with PHap.1-GUS, PHap.2-GUS, and PHap.4-GUS exhibited similar GUS expressions (Fig. 5A, Additional file 2: Fig. S6A). Similar results are also found among cucumber leaves with 35S::CDSHap.1-GUS, 35S::CDSHap.2-GUS, and 35S::CDSHap.3-GUS (Fig. 5B, Additional file 2: Fig. S6B). We then hypothesized that the CsGy5G015960 expression in powdery mildew-susceptible lines would be impaired by the 29-bp insertion in the 3′ UTR region. We cloned the 3′ UTR sequences from the powdery mildew-resistant YZ020A (KP2, Hap.1) and powdery mildew-susceptible YZ038A (PW, Hap.3) into the appropriate sites between the luciferase reporter gene and CaMV poly(A) signal to assess this (Fig. 5C). Sequence alignment proved that there was only one 29-bp InDel between the two lines, and transient transformation results showed that leaves expressing the 3′ UTR from YZ020A exhibited similar luminescence signals to the empty vector but were stronger than those expressing the 3′ UTR from YZ038A (Fig. 5C, D, E). Similarly to luciferase imaging, we discovered that leaves expressing the 3′ UTR from YZ038A had reduced LUC/REN ratio (Fig. 5F), indicating that the 29-bp insertion in the 3′ UTR region influenced the mRNA stability of CsGy5G015960. In order to further evaluate the impact of the 29-bp InDel on mRNA stability, we subsequently cloned the 3′ UTR sequences from the powdery mildew-resistant YZ020A (Hap.1) and powdery mildew-susceptible YZ038A (Hap.3) into the proper locations between the GUS reporter gene and NOS terminator. Cucumber leaves expressing the 3′ UTR from YZ020A displayed comparable GUS expression to the empty vector but were greater than those expressing the 3′ UTR from YZ038A, according to histochemical staining data (Fig. 5G). These results were in close agreement with cucumber leaf GUS mRNA expression levels (Fig. 5H).
Fig. 5.
Transient expression assays show that the 29-bp InDel in the 3′UTR region of CsGy5G015960 affects the mRNA stability. A Qualitative analyses of the activities of the CsGy5G015960 promoters cloned from YZ002A (PHap.1-GUS), YZ014A (PHap.2-GUS), and YZU089A (PHap.4-GUS) in cucumber cotyledons based on GUS histochemical staining. The empty vector (E-GUS) was used as a control. The diameter of each cucumber cotyledon sample is 1 cm. B Histochemical GUS staining of cucumber cotyledons inoculated with the coding sequence of CsGy5G015960 cloned from YZ002A (CDSHap.1-GUS), YZ014A (CDSHap.2-GUS), and YZU089A (CDSHap.4-GUS). The empty vector (35S::GUS) was used as a control. The diameter of each cucumber cotyledon sample is 0.6 cm. C Schematic diagram of the constructs used in the transient expression assay. The sequences alignment highlights the location of the mutation. MCS multiple cloning site, 35S CaMV 35S promoter, REN Renilla luciferase, LUC firefly luciferase, 3′UTRK 3′ UTR sequences cloned from powdery mildew-resistant YZ020A (KP2, Hap.1), 3′UTRP 3′ UTR sequences cloned from powdery mildew-susceptible YZ038A (PW, Hap.3). D A schematic representation of the different N. benthamiana leaf regions infiltrated with different constructs. E The representative luminescence image of one N. benthamiana leaf transiently expressing the three constructs. The signal intensity is indicated with different colors from low (blue) to high (red) as shown in the light spectrum bar. F Measurement of the LUC/REN ratio after transiently expression of the constructs in tobacco leaves. The black dots are given as the LUC/REN ratio of each replicate. G Histochemical staining of GUS in cucumber cotyledons transiently expressing the three constructs. H Quantitative analysis of the effects of the 29-bp InDel in the 3′ UTR region of CsGy5G015960 in cucumber leaves based on GUS gene expression. Each black dot represents one biological replicate. Student’s t-test was used to calculate the P value
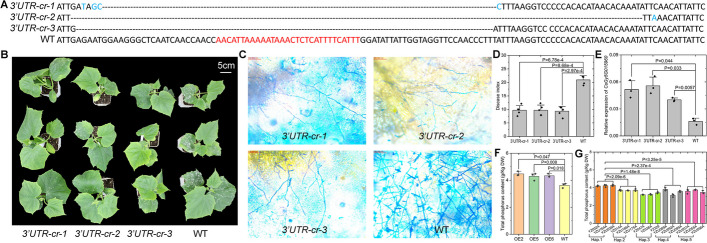
In order to evaluate the impact of the 29-bp InDel on powdery mildew resistance, knockout mutant lines lacking the 29-bp insertion in the powdery mildew-susceptible line YZU096A (Hap.3) were obtained by the CRISPR/Cas9-mediated genome editing. Three independent homozygous mutants (3’UTR-cr-1, 3’UTR-cr-2, and 3’UTR-cr-3) resulting in a complete deletion of the 29-bp insertion were selected for further characterizations (Fig. 6A). Compared to WT plants, all of the three mutants exhibited significantly lower DIs and less sporulation on the true leaves (Fig. 6B–D). We next studied the impact of 3’UTR deletion on CsGy5G015960 expression using RT-qPCR. The results showed that the relative expression of CsGy5G015960 in the leaves of the three mutants was significantly higher than that in WT (Fig. 6E). These results confirmed that the 29-bp InDel strongly influences cucumber powdery mildew resistance by altering CsGy5G015960 mRNA levels.
Fig. 6.
CRISPR/Cas9 induced mutations for the 29-bp insertion in the 3′ UTR of CsGy5G015960. A Comparison of 3′ UTR sequences among the three CRISPR/Cas9 induced mutants (3’UTR-cr-1, 3’UTR-cr-2, and 3’UTR-cr-3) and wild type (WT). Nucleotide deletions are indicated by dashes and nucleotide substitutions by blue color. The 29-bp insertion is indicated by red color. B Representative whole-plant phenotypes of the three CRISPR/Cas9 induced mutants and the WT control at 8 days post inoculation (dpi) of the powdery mildew pathogens. C Coomassie blue staining of powdery mildew fungal structures on leaves of the three CRISPR/Cas9-induced mutants (3’UTR-cr-1, 3’UTR-cr-2, and 3’UTR-cr-3) and WT at 8 dpi of powdery mildew pathogens. D Disease indices of 3’UTR-cr-1, 3’UTR-cr-2, 3’UTR-cr-3, and the WT control at 8 dpi of powdery mildew pathogen. Each black dot represents one biological replicate that is an average of 15 plants. E Confirmation of CsGy5G015960 expression in the three CRISPR/Cas9 induced mutants and the WT control by qRT-PCR. Each black dot represents one biological replicate. F Total phosphorus content in cucumber leaves of the transgenic CsGy5G015960 overexpressing cucumber lines (OE2, OE5, and OE6) and the WT control. G Total phosphorus content in cucumber leaves of the 15 selected cucumber inbred lines. Student’s t-test was used to calculate the P value
CsGy5G015960 mediates phosphorus accumulation
We assessed the internal phosphorus contents of CsGy5G015960-OE lines and WT leaves cultivated in fields with normal phosphorus concentrations in order to comprehend the role of CsGy5G015960 in phosphorus uptake. When cultivated with normal phosphorus application, transgenic plants with overexpression of CsGy5G015960 demonstrated enhanced phosphorus accumulation in leaves compared to WT plants (Fig. 6F). We also tested the phosphorus levels in the leaves of the five haplotypes. Notably, we discovered that the three Hap.1 lines had much higher phosphorous content than the Hap.2, Hap.3, Hap.4, and Hap.5 lines (Fig. 6G). This data suggests that CsGy5G015960 is involved in the accumulation of phosphorus in cucumbers.
CsGy5G015960 regulates powdery mildew-resistance by mediating H2O2 accumulation
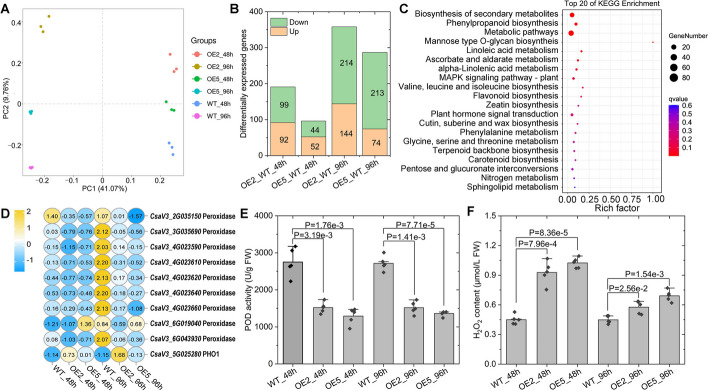
To discover potential downstream genes that CsGy5G015960 might govern, we used RNA-seq to compare the global expression profiles of powdery mildew-infected leaf samples from OE2 and OE5 at 48 and 96 h to leaves from WT. At least 40.2 M clean reads were obtained for each sequencing library, where PCA of the RNA-seq data showed that these samples were well separated along with the first two components. This explains 50.83% of the variability in the dataset (Fig. 7A). A total of 647 non-overlapping differentially expressed genes (DEGs) were identified using a |Log2 (fold change)|≥ 1 and FDR ≤ 0.05 as cutoffs. In summary, 182, 96, 358, and 287 DEGs were identified in the comparisons of OE2_vs_WT_48h, OE5_vs_WT_48h, OE2_vs_WT_96h, and OE5_vs_WT_96h, respectively (Fig. 7B). Additional file 1: Table S5 lists all the DEGs identified for each pairwise comparison. KEGG enrichment analysis of the 647 non-overlapping DEGs showed that “biosynthesis of secondary metabolites” (ko01110, 58 DEGs, p = 6.40E-06) and “phenylpropanoid biosynthesis” (ko00940, 18DEGs, p = 1.01E − 05) were the top two significantly enriched signaling pathways affected by overexpression of CsGy5G015960 (Fig. 7C). We found that CsGy5G015960 was significantly upregulated in both of OE2 and OE5 (Fig. 7D, details in Additional file 1:Table S5) than in the WT, indicating that 35S promoter is active in leaves and CsGy5G015960 is also overexpressed in the leaves of OE lines. Surprisingly, we observed that nine DEGs encoding peroxidase (Prx) were shared in the KEGG term “biosynthesis of secondary metabolites” and “phenylpropanoid biosynthesis,” which were all considerably higher in the WT than in the OE2 and OE5 upon powdery mildew inoculation (Fig. 7D). The nine Prxs were classified using PeroxiScan (https://peroxibase.toulouse.inra.fr/tools/peroxiscan) and the findings revealed that all nine DEGs belong to the class III Prxs, which are known for producing or detoxifying hydrogen peroxide (H2O2) in plant cells [27]. We further evaluated the expression of these nine Prxs by qRT-PCR analysis. As expected, the nine Prxs were all significantly downregulated in expression in both OE2 and OE5 at 48 h and 96 h after powdery mildew inoculation, consistent with the expression profiles derived from RNA-seq results, indicating the reliability of the RNA-seq results (Additional file 2: Fig. S7). In light of the reported transcriptome responses, we used spectrophotometry to assess the enzymatic activity of Prx and H2O2 concentrations in the leaves of OE2, OE5, and WT 48 and 96 h after powdery mildew pathogen inoculation. The total Prx activities were lower (Fig. 7E) and the H2O2 contents were higher (Fig. 7F) in the CsGy5G015960-OE lines in comparison to WT leaves. The results therefore demonstrated that the overexpression of CsGy5G015960 induced a high concentration of H2O2 via suppressing Prx activity, which enhanced resistance to powdery mildew infection.
Fig. 7.
Transcriptomic analysis of CsGy5G015960 overexpressed line (OE2 and OE5) and wild-type (WT). A Principal components (PC) analysis of the RNA-seq data. Each point represents the whole-gene profile of one biological replicate. Numbers in parentheses on the axes represent the percentage of total variance explained by each PC. B Number of differentially expressed genes (DEGs) in leaves of OE2 and OE5 compared to WT. C KEEG enrichment analysis of the 647 non-overlapping DEGs. D Heatmap showing the expression changes of CsGy5G015960 (CsaV3_5G025280) and the nine DEGs encoding peroxidase in leaves of OE2 and OE5 compared to WT upon 48 and 96 h of powdery mildew pathogen inoculation. The average FPKM value for each gene across three libraries was resized to row Z-score scale with blue for low expression and yellow for the high expression levels. E Comparison of peroxidase activities of WT, OE2, and OE5 leaves at 48 and 96 h of powdery mildew inoculation. Each dot denotes the mean activity of three replicates. F Comparison of H2O2 contents of WT, OE2, and OE5 leaves at 48 and 96 h of powdery mildew inoculation. Each dot denotes the mean content of three replicates. Student’s t-test was used to calculate the P value
Discussion
Marker density influences the ability of a GWAS to identify the loci associated with a particular trait [28]. Using high-density genotyping data obtained from the whole-genome resequencing of 299 cucumber accessions, the allelic variations responsible for powdery mildew resistance were identified in this study. The genotyping data for GWAS studies consisted of approximately 1.96 million high-quality variations, comprising 1,625,341 SNPs and 339,618 InDels. Given the substantial population divergence and the high-density genotyping data generated in this investigation, it seemed very likely that the GWAS would identify variants linked to powdery mildew resistance. When compared to earlier GBS-based GWAS analyses of cucumber, the resolution offered by the dense variations was at least 80 times higher [24, 25].
The 299 cucumber accessions were divided into the following four groups: India (31 accessions), North America (49 accessions), Eurasia (91 accessions), and East Asia (128 accessions) based on the population structure, PCA results, and phylogenetic relationships. Our study’s findings were consistent with previous studies and our current understanding of the genetic and geographic origins of cucumbers. The geographically related accessions of cucumber were classified together. This was owing to the relatively low genetic diversity among accessions from the same region compared to other regions [29]. However, our evaluation of 299 cucumber accessions uncovered inconsistencies. Specifically, 21 accessions (bolded in Additional file 1: Table S1) were not categorized by their geographic origin. For example, YZU121A was originally collected in Hungary but was included in the India group for the objectives of this investigation. This indicates that there may have been a germplasm exchange between regions. Regarding the phylogenetic relationships among cucumber accessions, there were some other differences between the findings of the current study and those of the earlier GBS-based analysis. For example, YZU025A was assigned to the North America group in the present study but to the East Asia group in the earlier study. This cultivar, which has Chinese ancestry, yields fruit excellent for pickling. In contrast to previous research, the new WGS-based method enabled a more sophisticated evaluation of the genetic relationships between cucumber accessions.
The thorough identification of genes regulating adult plant stress resistance will benefit the agricultural industry since these genes provide plants with long-term disease resistance [30]. As a result, we evaluated the powdery mildew resistance of cucumber accessions at the adult plant stage in greenhouse conditions, and the symptoms were undetectable on YZ002A, YZU111A, and YZU038A plants (DI = 0) in all 3 years. These accessions might serve as useful powdery mildew-resistant gene resources for cucumber breeding initiatives. In addition, Block and Reitsma [1] demonstrated that in the USA, YZU111A is highly resistant to powdery mildew. In the present study, the average DIs of certain accessions (e.g., YZ297, YZ332, and YZU074A) varied between years; this variation may be attributable to environmental factors such as humidity, water, and nutrient availability. In order to minimize environmental interference, we used a mixed linear model for the GWAS. The 50 significant locations found on Chr5 were within the powdery mildew-resistant QTLs (Pm5.1 and Pm5.2) that had previously been mapped, indicating that these loci include genes that give long-lasting powdery mildew resistance. It is still unknown, though, whether these regions contain the same genes or different ones. Because many generations can be progressed in a controlled environment in the time it takes to evaluate a single generation under field circumstances, the outcomes of this work could aid breeders in accelerating the development of powdery mildew-resistant cucumber varieties [31].
GWAS analysis identified 41 candidate genes on Chr5, but VIGS and cucumber transformation analysis suggest CsGy5G015960 positively influences cucumber powdery mildew resistance (Fig. 4, Additional file 2: Fig. S1). According to the current study, GWAS is a valid tool for discovering genes that are responsible for complex phenotypes. The 3′ UTR is critical to the regulation of phenotypic richness within a species because it governs mRNA stability/degradation, subcellular localization, and translation efficiency [32, 33]. The presence of the 3′ UTR of a soybean cytosolic glutamine synthetase gene exerts a silencing impact on transgene expression in transgenic alfalfa [34]. The del T mutation in the 3′ UTR of the CDK2-AP1 gene dramatically reduces CDK2-AP1 expression in human microsatellite unstable colorectal cancer cell lines [35]. In a recent study, the C-to-T transition in TaPHS1′ 3′ UTR has been associated with decreased TaPHS1 expression and higher pre-harvest sprouting susceptibility in wheat [36]. Therefore, we believe that the variation in CsGy5G015960 expression among the 15 selected cucumber lines inoculated with the powdery mildew pathogen is at least partially attributable to the 29-bp InDel in the 3' UTR. GUS and luciferase assays (Fig. 5) and CRISPR/Cas9-induced mutations in the 3' UTR (Fig. 6) provide additional support for this hypothesis.
The Arabidopsis pho1 (AT1G14040) mutant displays symptoms of phosphate deficit caused by restricted phosphate transport from the roots to the shoots [15]. However, xenotropic polytropic virus receptor 1 (XPR1), a PHO1 homolog that is present in both mice and humans, has been identified as a particular cell surface receptor for murine leukemia viruses [37, 38]. Mouse XPR1 heterologous expression in tobacco leaves results in selective phosphate efflux [39]. We hypothesize that the cucumber CsGy5G015960 gene is functionally similar based on its similarity to Arabidopsis PHO1 (66.13% sequence similarity). Moreover, total phosphorus content measurements provide additional support for this hypothesis (Fig. 6F, G). One of the primary constraints on plant growth is phosphate, yet little is currently known about the connection between phosphate export and powdery mildew resistance. Recent research has shown that the PHO1 homolog OsPT8’s controlled expression in rice affects plants’ resistance to the diseases Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae [14]. CsGy5G015960 may influence cucumber powdery mildew resistance by maintaining a high level of H2O2 by depleting numerous class III Prx genes, according to the results of our comparative RNA-seq study (Fig. 7, Additional file 2: Fig. S7). Complementary enzymatic activity experiments provide additional evidence that class III Prxs play a significant part in pathogen resistance. For example, the rice protein OsPrx30, which is primarily found in the endoplasmic reticulum, showed enhanced H2O2 accumulation and induced resistance to bacterial blight caused by Xanthomonas oryzae pv. oryzae when it was altered by the CRISPR/Cas9 method [40]. Similarly, knocking down TaPOD70 increased wheat resistance to powdery mildew caused by Blumeria graminis f. sp. tritici [41]. It is compelling to hypothesize that increased H2O2 might function as an antimicrobial substance that inhibits powdery mildew pathogen penetration into cucumber leaf epidermal cells or as an early event in the induction of hypersensitive host cell death which restricts powdery mildew pathogen growth, considering that powdery mildew pathogens are obligate biotrophs that can only complete their life cycles on living host cells [40, 42]. We hypothesized that the increased H2O2 content in CsGy5G015960 overexpression lines was due to physical interactions between CsGy5G015960 and Prx. As a result, we used CsGy5G015960 as a bait in yeast two-hybrid (Y2H) investigation and cloned the nine DEGs encoding Prx as a fusion to the activation domain. The Y2H data indicate that no direct interaction was observed between any of the nine Prx and CsGy5G015960. In addition to Prx, we found that two genes (CsaV3_1G002910 and CsaV3_1G038860) encoding respiratory burst oxidase and three genes (CsaV3_3G014270, CsaV3_7G022280, and CsaV3_7G028830) encoding glutaredoxin were also differentially expressed between CsGy5G015960-OE lines and WT. As both of respiratory burst oxidase and glutaredoxin were previously reported to regulate H2O2 accumulation, the precise roles of these DEGs in CsGy5G015960-regulated powdery mildew resistances require further investigation [43, 44]. According to earlier research, the rice phosphate transporter protein OsPT8 interacts directly with BWMK1, a mitogen-activated protein kinase (MAPK) that regulates disease resistance to pathogens by activating transcription factors such as OsEREBP1 and OsWRKY33 [14, 45]. In various studies, transcription factor activation confers altered Prx expression. Moreover, many genes encoding MAPK signaling pathways and transcription factors were found to be differently expressed between the CsGy5G015960-OE and WT lines (Fig. 7C, Additional file 1: Table S6). Therefore, it is possible to speculate that CsGy5G015960 affects Prx expressions and H2O2 levels by activating transcription factors post-translationally via physical interaction with MAPK. However, future research is required to elucidate the CsGy5G015960-mediated powdery mildew resistance signaling network in more depth.
Conclusions
This is, to the best of our knowledge, the first comprehensive GWAS of cucumber powdery mildew resistance utilizing WGS data for cucumber accessions with various geographic origins. This study provides valuable insight into the genes implicated in cucumber powdery mildew resistance and may be applicable to future efforts to develop novel cucumber cultivars with persistent powdery mildew resistance. Furthermore, our findings demonstrate that the phosphate transporter CsGy5G015960 enhances cucumber powdery mildew resistance by mediating H2O2 accumulation through the depletion of many class III Prx genes.
Methods
Whole-genome resequencing and variant calling
The GWAS panel consisted of 299 cucumber accessions, which were acquired as seeds from the USA national plant germplasm system collection, the Yangzhou University cucumber breeding lab, or from commercial sources (Additional file 1: Table S1). Collection and use of plant materials was conducted in accordance with institutional, national, and international guidelines and legislation. For each accession, total genomic DNA was extracted from the young leaves of individual healthy cucumber plants according to the traditional cetyltrimethylammonium bromide method [46]. Genomic DNA concentrations were determined using the Qubit 3.0 fluorometer (Life technologies, Bleiswijk, The Netherlands), whereas purity was assessed using a NanoDrop One spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA). Approximately 5 µg genomic DNA was used to construct paired-end sequencing libraries as described by the manufacturer of a commercial kit (Illumina, Foster City, CA, USA). The 299 libraries were subjected to paired-end WGS on the Illumina HiSeq XTen system. The libraries were constructed and sequenced by ANOROAD Technologies (Beijing, China) and Beijing Genomics Institute (Shenzhen, China).
After removing reads containing adapters and low-quality reads (i.e., > 30% of bases with a sequencing quality score < 20) with fastp (version 0.20.1) [47], the remaining high-quality reads were mapped onto the Gy14 cucumber reference genome (version 2.0) (http://cucurbitgenomics.org/) using the Burrows–Wheeler Aligner program (version 0.7.17-r1188) [48]. The Genome Analysis Toolkit (version 4.1.4.1) [49] was used for calling SNPs and InDels.
Phylogenetic, population structure, and LD analyses
The SNPs and InDels with MAF ≥ 5% and a missing data rate ≤ 20% were selected for further analyses. Evolutionary distances were calculated using MEGA-X [50] and a phylogenetic tree was constructed according to the neighbor-joining method (500 bootstraps) using an online toolkit (https://itol.embl.de/itol.cgi) [51]. The population structure of 299 cucumber accessions was analyzed using the model-based evolutionary clustering approaches implemented in Admixture (version 0.1.13) [52]. To optimize and accelerate the analysis, only the SNPs and InDels with LD > 0.2 were used. The number of groups was set as K = 1–13, and the CV error of each K was estimated to determine the optimal cluster and genetic background matrix (Q). The 299 accessions were classified into different groups according to their Q values. A PCA was performed using Plink (version 1.9) [53]. To estimate the LD level in each population, the squared allele-frequency correlation (r2) between pairs of SNPs was calculated using PopLDdecay (version 3.40) [54].
Phenotyping
Adult plant resistance to powdery mildew was evaluated over three cropping seasons (i.e., fall 2022, winter 2022, and spring 2023) in the same greenhouse at Yangzhou University (Yangzhou, China). For each accession, 10–15 plants were grown (50 × 30 cm spacing). Each plant was self-pollinated to generate one or two fruits. No fungicides were applied during the growth season. The resistance to powdery mildew was assessed when the highly susceptible line D8 was completely infected. The severity of the symptoms on leaves was determined using the following 0–5 scale: 0, no visible symptoms; 1, infection area < 30%; 2, infection area 30%–60%; 3, infection area 60%–80%; 4, infection area > 80%; and 5, powdery mildew spots covered the whole leaf, which was yellow. The DI was calculated using the following formula: DI = ∑(disease scale × number of leaves corresponding to that scale)/(total number of leaves examined × the highest disease grade) × 100 [55].
Data analysis
To minimize the adverse effects of the environment on the GWAS results, we calculated the breeding values according to the best linear unbiased predictors (BLUP) method using phenotypic data for two environments. The BLUP analysis was completed using the lme4 package of R (version 4.0.1) (https://github.com/lme4/lme4). The breeding values were treated as phenotypic values for the GWAS.
Genome-wide association analysis
The genome-wide association analysis was performed using the mixed linear model (MLM, Q + K) in the Tassel (version 5.0) software package [56]. To decrease the environmental error, BLUP values were estimated using the lme4 package of the R program (https://github.com/lme4/lme4/). The values for a single environment and the BLUP values were used for the association mapping. The kinship matrix (K) was calculated using GCTA (version 1.92–2.2) [57] and was considered as a random-effect factor. The significant P value threshold was calculated using R (version 4.0.1) according to the Bonferroni correction with a suggestive threshold (P < 1/n, n = number of markers) [58]. The GWAS results were visualized using Manhattan and quantile–quantile (Q-Q) plots that were produced by the R package “qqman” [59].
Gene prediction and qrt-PCR analysis
Linkage disequilibrium patterns surrounding the significant sites identified by the GWAS were constructed using the R package “LDBlockShow” [60]. Genes within the block region were predicted on the basis of the annotated Gy14 cucumber reference genome (version 2.0) (http://cucurbitgenomics.org/organism/16).
To analyze the relative expression levels of the candidate gene CsGy5G015960, leaves were collected from 15 cucumber lines at 48 h after they were inoculated with the powdery mildew pathogen (1 × 106 spores/mL solution containing 0.01% Tween-20). Mock control samples were sprayed with distilled water containing 0.01% Tween-20. Plants were inoculated at the three-leaf stage (three independent biological replicates) in a growth chamber. Conidia were collected from naturally infected D8 leaves in the greenhouse. Total RNA was isolated from each sample using RNAiso Plus (Takara, China). The qRT-PCR analysis was completed with cucumber β-actin (GenBank AB010922) and ubiquitin (CsaV3_5G031430) selected as the internal control. The qRT-PCR was performed using the ChamQ SYBR qPCR Master Mix (SYBR Green I) kit (Vazyme, Nanjing, China) and the iQ5 multicolor real-time PCR detection instrument (Bio-Rad, Hercules, CA). Relative gene expression levels were calculated according to the 2−ΔΔCt method. Primers were designed using Primer 3 (https://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by Sango Biotech Co., Ltd. (Shanghai, China). Details regarding the qRT-PCR primers, which were designed to avoid conserved regions, are provided in Additional file 1: Table S6.
Virus-induced gene silencing
Gene functions were studied using VIGS system based on the cucumber green mottle mosaic virus [26]. The recombinant virus pV190 vector was kindly provided by Dr. Qinsheng Gu from Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences. The candidate genes and CsGy4G002600 (CsPDS; + 484 to + 783 bp) coding sequences in YZ002A were amplified by PCR to produce ~ 300-bp fragments (Additional file 1: Table S6), which were then cloned into the BamHI site of pV190 vectors to generate constructs pV190-gene and pV190-CsPDS, respectively. After confirming the construct sequences were correct by Sanger sequencing, the recombinant constructs were transformed into Agrobacterium tumefaciens strain GV3101 cells. The VIGS inoculum was infiltrated into the cotyledons of powdery mildew-resistant cucumber EP6411 seedlings at the cotyledon stage (5 days after germination). To quantify the VIGS efficiency, the expressions of candidate genes in the Agro-infiltrated EP6411 leaves were determined by qRT-PCR using gene-specific primers. For qRT-PCR, the leaves samples were harvested at 2 weeks after the VIGS, when the true leaves of plants infiltrated with pV190-CsPDS exhibited an albino phenotype. Then, plants were inoculated with the powdery mildew pathogen (1 × 106 spores/mL solution containing 0.01% Tween-20) and then phenotyped at 15 days post inoculation. The analysis was completed using three biological replicates, with 25 seedlings infiltrated per replicate.
Cucumber transformation
For CsGy5G015960-OE analysis, the full-length coding sequence of CsGy5G015960Hap.1 was amplified from YZ002A and cloned into the SalI and BamHI sites of pCS5123aada vector carrying the aminoglycoside adenyl transferase gene (aadA) for selection to generate the 35S:CsGy5G015960Hap.1-aada construct. To generate mutations in the 3′UTR region of CsGy5G015960 using the CRISPR/Cas9 system, two specific single-guide RNA (sgRNA) target sites (sgRNA-1, TGTGGGGGACCTTAAATAAAGGG; sgRNA-2, GTTGGTTGTGAAATTGAGAATGG) were designed using the web-based tool CRISPR-P 2.0 (http://cbi.hzau.edu.cn/CRISPR2). sgRNA sequences were synthesized, amplified, and cloned into a CRISPR/Cas9 vector pCS4190aada (http://www.wimibio.com) carrying the aadA for selection. Agrobacterium-mediated cucumber transformation was conducted through a commercial service (http://www.wimibio.com/). Briefly, 2-day-old cotyledon explants from the North China-type inbred line CCMC (OE) or YZU096A (CRISPR/Cas9) were co-cultivated with Agrobacterium tumefaciens strain EHA105 carrying the recombinant constructs for 30 min at 28 °C. After 72 h of incubation in the dark condition at 23 °C and 7 days of incubation in the light condition at 25 °C, the explants were washed twice with sterile water and then transferred onto the selective medium containing 50 mg/L spectinomycin for the selection of spectinomycin-resistant transformation events. After 6 weeks of growth, the putative transgenic explants were transferred to an elongation medium, followed by a rooting medium with 50 mg/L spectinomycin. After confirming with PCR using primers flanking aadA (Additional file 1: Table S6), the putative transgenic seedlings were planted in the greenhouse, and their self-pollinated seeds were harvested separately. T2 homozygous seeds were grown in a growth chamber for further powdery mildew inoculation experiment and qRT-PCR analysis. To identify CRISPR/Cas9-mediated mutations, total DNA was extracted from T2 plants, and fragments containing the target sites were amplified and sequenced using target-specific primers (Additional file 1: Table S6).
Luciferase imaging and reporter assay
The 3′ UTR of CsGy5G015960 was amplified from YZ020A and YZ038A by PCR using primers (Additional file 1: Table S6). After confirmation by Sanger sequencing, the fragments were subcloned into the pGreenII0800-miRNA vector between the luciferase reporter gene and the Cauliflower mosaic virus (CaMV) poly(A) signal. Binary vectors were separately introduced into A. tumefaciens strain GV3101 (harboring pSoup-P19). The resulting strains were used to infiltrate the leaves of 5-week-old Nicotiana benthamiana. Leaves were harvested 48 h post infiltration, and the luciferase signals were captured using NightShade LB985 system (Berthold Technologies, Bad Wildbad, Germany) after spraying 1 mM D-Luciferin potassium salt (GoldBio, St. Louis, MO, USA). For luciferase reporter assay, the activities of firefly luciferase (LUC) and renilla luciferase (REN) were assayed following the protocol of dual-luciferase reporter assay system kit (Promega, Madison, WI, USA). The LUC activity was normalized with the REN activity (LUC/REN). The pGreenII0800-miRNA empty vector was used as a control. Five biological replicates were performed.
GUS histochemical analysis
To examine whether the mutations in the promoter of CsGy5G015960 affect the transcription, the 1-kb DNA fragments upstream of CsGy5G015960 were amplified using the genomic DNA of YZ002A, YZ014A, and YZU089A with corresponding primers that included the HindIII and PstI restriction sites (Additional file 1: Table S6). The fragments were subcloned into the pCAMBIA1391-GUS vector following confirmation by Sanger sequencing. To examine whether the point mutations in the exon region of CsGy5G015960 affect the gene stabilities, the full-length coding sequence of CsGy5G015960 (without stop codon) were amplified from YZ002A, YZ014A, and YZU089A with corresponding primers that included the NcoI and BglII restriction sites between CaMV 35S promoter and GUS (Additional file 1: Table S6). After confirmation by Sanger sequencing, the sequences were subcloned into the pCAMBIA1303-35S vector containing the GUS reporter gene. To determine whether the insertion affects transcription, the 3′ UTR of CsGy5G015960 was amplified from YZ020A and YZ38A using BstEII restriction site-containing primers (Additional file 1: Table S6). The fragments were subcloned into the pCAMBIA3301-GUS vector following confirmation by Sanger sequencing. Binary vectors were separately delivered into A. tumefaciens strain GV3101 and transiently transformed into 1-week-old cucumber cotyledons. The tissues were collected after 48 h of inoculation and immersed in X-Gluc solution (Solarbio, Beijing, China). After 12 h of incubation at 37 °C, the tissues were destained with 70% ethanol (v/v) until transparent. A digital camera was used to capture the GUS staining patterns. For GUS expression, the cucumber cotyledons were harvested at 48 h of A. tumefaciens inoculation for qRT-PCR analysis.
Coomassie blue staining
To visualize the powdery mildew fungal structures, leaf samples were cleared in destaining solution (alcohol:trichloromethane, 75:25, v/v) for 24 h and stained with Coomassie brillant blue R-250 (0.6%, w/v) for 10 min. Excess dye was rinsed off carefully with distilled water. The colonization of powdery mildew fungi was then observed under a microscope (NE610, Nexcope, Ningbo, China).
Measurement of total phosphorus concentration
To determine total phosphorus concentration, 300 mg of leaf tissue were crushed to a fine powder (SCIENTZ-48, Xinzhi, China) and digested with 10 mL of concentrated sulfuric acid at 100 °C for 10 min, 280 °C for 10 min, and 400 °C for 40 min. The digested solution was cooled to 28 °C and diluted by the addition of 1 mL of 30% H2O2. Following digestion (200 °C for 10 min, following 320 °C for 10 min, and finally 400 °C for 30 min), two further dilution steps with H2O2 were performed until the solution turned colorless. The total phosphorus concentration was then quantified using a microplate reader and molybdenum blue colorimetry at 700 nm (Ensight, PerkinElmer, Waltham, MA, USA).
RNA-seq analysis
For RNA sequencing, the leave samples were collected at 48 and 96 h after powdery mildew inoculation. The extracted RNA molecules containing poly (A) were purified using oligo (dT) magnetic beads. The library was prepared from purified RNA with a MGIEasy RNA Kit V3 (BGI, Shenzhen, China) following the manufacturer’s recommendation. After the quality check, 18 libraries were sequenced on an Illumina novaseq PE150 platform at Genepioneer biotechnologies Co. Ltd (Nanjing, China). After data filtering, clean reads were mapped to the ChineseLong_V3 genome with HISAT2 [61]. The expression value of each gene was calculated and normalized as FPKM (fragments per kilobase of transcript per million fragments mapped) by HTSeq v 0.6.0 [62]. DEGs were screened via the R package “DESeq2” v 1.20. 0 [63]. Pathway analysis was conducted to identify significant pathways of DEGs according to the KEGG database (http://www.genome.jp/kegg). To validate the RNA-seq results by qRT-PCR, an independent set of samples with three independent biological replicates was collected at 0, 48, and 96 h after powdery mildew inoculation (1 × 106 spores/mL solution containing 0.01% Tween-20). Cucumber plants were grown and inoculated in a same growth chamber as for RNA-seq.
Measurement of Prx activity and H2O2 content
The Prx activity of leave samples was assayed by a commercial assay kit (Comin Biotechnology Co., Ltd., Suzhou, China). The absorbance was recorded at 470 nm with a spectrophotometer (Spectrum SP-752, Shanghai, China). One unit (U) of Prx activity was defined as 0.01 change in absorbance per min under assay conditions. To determine the H2O2 content, weighted leave samples were ground in ice-cold acetone (100% v/v). The resulting homogenate was centrifuged at 8000 × g for 10 min at 4℃ for further use. Quantification was carried out according to the protocol supplied by the manufacturer (H2O2-2-Y, Comin), and the absorbance was measured at 415 nm with a spectrophotometer (Spectrum SP-752).
Supplementary Information
Additional file 1: Supplementary Tables S1–S6. This file contains the supplementary tables referenced in the main text. Table S1. Summary of the sampled cucumber accessions. Table S2. Chromosomal distribution of the high-quality markers detected in this study. Table S3. Significant markers associated with powdery mildew resistance detected at least twice. Table S4. Linkage disequilibrium (LD) blocks surrounding the 50 significant sites identified by the GWAS. Table S5. List of 647 non-overlapping differentially expressed genes identified by RNA-seq. Table S6. Primers used in this study.
Additional file 2: Supplementary Figs. S1–S7. This file contains the supplementary figures referenced in the main text. Fig. S1. Functional characterization of the candidate genes identified by the GWAS using VIGS. Fig. S2. Relative expression levels of CsGy5G015960 in different tissues of the PM-resistant line Jin5-508. Fig. S3. Representative whole-plant phenotypes of the CsGy5G015960 overexpression lines and the wild type (WT) control at 8 dpi with water. Fig. S4. Relative expression levels of CsGy5G015960 in 15 selected cucumber inbred lines at 48 h post powdery mildew inoculation. Fig. S5. Relative expression levels of CsGy5G015960 in roots of 15 selected cucumber inbred lines. Fig. S6. GUS expression analysis. Fig. S7. Relative expression levels of nine genes encoding peroxidase in leaves of OE2 and OE5 compared to WT upon 0, 48 and 96 h (h) of powdery mildew pathogen inoculation.
Acknowledgements
We thank Dr. Qinsheng Gu from Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences for kindly providing the pV190 vector.
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3.
Authors' contributions
X.C. and X.Y. conceived the original screening and research plans. X.X., Y.D., S.L., and M.T. conducted the experiments. M.T., Y.D., X.L., and X.Q. collected the data. X.X., H.S., and M.T. analyzed the data. X.X. and M.T. wrote the manuscript with input from all authors. All authors reviewed and approved this submission. X.C. and X.Y. agree to serve as the authors responsible for contact and ensures communication.
Funding
This research was supported by the National Natural Science Foundation of China (grant nos. 32030093, 32172570, and 31672176), the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS[2021]018), the Jiangsu Agricultural Innovation of New Cultivars (PZCZ201720), the Jiangsu Provincial Key R&D Programme-Modern Agriculture (BE2022339), and the Jiangsu Agricultural Science and Technology Independent Innovation Fund (CX(23)1009).
Availability of data and materials
The raw sequencing data generated in this study have been deposited in the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa/) of the National Genomics Data Center under the accession number CRA002417 [64]. The raw RNA-seq reads have been deposited in NCBI Sequence Read Archive (SRA) under accession PRJNA998480 [65]. The Gy14 cucumber reference genome (v2) was accessed from the Cucurbit Genomics Database (http://cucurbitgenomics.org/). The source codes used for data analysis in this study were deposited under GPL-3.0 license in Github (https://github.com/xiaodongy86/PM_GWAS_Cucumber) [66] and Zenodo (https://zenodo.org/records/13635209) [67]. No other scripts and software were used other than those mentioned in the “Methods” section.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaodong Yang, Email: yxd@yzu.edu.cn.
Xuehao Chen, Email: xhchen@yzu.edu.cn.
References
- 1.Block C, Reitsma K. Powdery mildew resistance in the U.S. national plant germplasm system cucumber collection. Hortscience. 2005;40:414–20. [Google Scholar]
- 2.Lebeda A, Mieslerová B. Taxonomy, distribution and biology of lettuce powdery mildew (Golovinomyces cichoracearum sensu stricto). Plant Pathol. 2011;60(3):400–15. [Google Scholar]
- 3.Keinath AP, Dubose VB. Controlling powdery mildew on cucurbit rootstock seedlings in the greenhouse with fungicides and biofungicides. Crop Prot. 2012;42:338–44. [Google Scholar]
- 4.Hafez YM, Attia KA, Kamel S, Alamery SF, Abdelaal K. Bacillus subtilis as a bio-agent combined with nano molecules can control powdery mildew disease through histochemical and physiobiochemical changes in cucumber plants. Physiol Mol Plant Pathol. 2020;11:101489. [Google Scholar]
- 5.Wang Y, Bo K, Gu X, Pan J, Li Y, Chen J, Wen C, Ren Z, Ren H, Chen X, Grumet R, Weng Y. Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic Res. 2020;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukino N, Yoshioka Y, Sugiyama M, Sakata Y, Matsumoto S. Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol Breed. 2013;32:267–77. [Google Scholar]
- 7.He X, Li Y, Pandey S, Yandell BS, Pathak M, Weng Y. QTL mapping of powdery mildew resistance in WI2757 cucumber (Cucumis sativus L.). Theor Appl Genet. 2013;126:2149–61. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K, Wang X, Zhu W, Qin X, Xu J, Cheng C, Lou Q, Li J, Chen J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor Appl Genet. 2018;131:2229–43. [DOI] [PubMed] [Google Scholar]
- 9.Berg JA, Appiano M, Santillán Martínez M, Hermans FW, Vriezen WH, Visser RG, Bai Y, Schouten HJ. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015;15:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie J, Wang Y, He H, Guo C, Zhu W, Pan J, Li D, Lian H, Pan J, Cai R. Loss-of-function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus L.). Front Plant Sci. 2015;6:1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badri Anarjan M, Bae I, Lee S. Marker-assisted evaluation of two powdery mildew resistance candidate genes in Korean cucumber inbred lines. Agronomy. 2021;11:2191. [Google Scholar]
- 12.Lu H, Wang F, Wang Y, Lin R, Wang Z, Mao C. Molecular mechanisms and genetic improvement of low-phosphorus tolerance in rice. Plant Cell Environ. 2023;46(4):1104–19. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Deng M, Xu J, Zhu X, Mao C. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin Cell Dev Biol. 2018;74:114–22. [DOI] [PubMed] [Google Scholar]
- 14.Dong Z, Li W, Liu J, Li L, Pan S, Liu S, Gao J, Liu L, Liu X, Wang GL, Dai L. The rice phosphate transporter protein OsPT8 regulates disease resistance and plant growth. Sci Rep. 2019;9:2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirier Y, Thoma S, Somerville C, Schiefelbein J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arruda MP, Brown P, Brown-Guedira G, Krill AM, Thurber C, Merrill KR, Foresman BJ, Kolb FL. Genome-wide association mapping of fusarium head blight resistance in wheat using genotyping-by-sequencing. Plant Genome. 2016;9:1–14. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, Zhou X, Zhu X, Chen Z, Wang J, Zhao W, Ma B, Qin P, Chen W, Wang Y, Liu J, Wang W, Wu X, Li P, Wang J, Zhu L, Li S, Chen X. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126.e15. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Lin B, Wang H, Li X, Yang F, Ding X, Yan J, Chu Z. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nature Genet. 2019;51:1540–8. [DOI] [PubMed] [Google Scholar]
- 19.Chen B, Zhang Y, Sun Z, Liu Z, Zhang D, Yang J, Wang G, Wu J, Ke H, Meng C, Wu L, Yan Y, Cui Y, Li Z, Wu L, Zhang G, Wang X, Ma Z. Tissue-specific expression of GhnsLTPs identified via GWAS sophisticatedly coordinates disease and insect resistance by regulating metabolic flux redirection in cotton. Plant J. 2021;107:831–46. [DOI] [PubMed] [Google Scholar]
- 20.Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, Huang Z, Li J, Zhang C, Wang T, Zhang Y, Wang A, Zhang Y, Lin K, Li C, Xiong G, Xue Y, Mazzucato A, Causse M, Fei Z, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li J, Ye Z, Du Y, Huang S. Genomic analyses provide insights into the history of tomato breeding. Nature Genet. 2014;46:1220–6. [DOI] [PubMed] [Google Scholar]
- 21.Han K, Lee HY, Ro NY, Hur OS, Lee JH, Kwon JK, Kang BC. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol J. 2018;16:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Gao P, Zhu Q, Zhu Z, Liu H, Wang X, Weng Y, Gao M, Luan F. Resequencing of 297 melon accessions reveals the genomic history of improvement and loci related to fruit traits in melon. Plant Biotechnol J. 2020;18:2545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi J, Liu X, Shen D, Miao H, Xie B, Li X, Zeng P, Wang S, Shang Y, Gu X, Du Y, Li Y, Lin T, Yuan J, Yang X, Chen J, Chen H, Xiong X, Huang K, Fei Z, Mao L, Tian L, Städler T, Renner SS, Kamoun S, Lucas WJ, Zhang Z, Huang S. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nature Genet. 2013;45:1510–5. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Bao K, Reddy UK, Bai Y, Hammar SA, Jiao C, Wehner TC, Ramírez-Madera AO, Weng Y, Grumet R, Fei Z. The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies, and core collection development. Horti Res. 2018;5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HY, Kim JG, Kang BC. Assessment of the genetic diversity of the breeding lines and a genome wide association study of three horticultural traits using worldwide cucumber (Cucumis spp.) germplasm collection. Agronomy. 2020;10:1736. [Google Scholar]
- 26.Liu M, Liang Z, Aranda MA, Hong N, Liu L, Kang B, Gu Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods. 2020;16:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Liu M, Hu Q, Yan W, Pan J, Yan Y, Chen X. A CsEIL3-CsARN6.1 module promotes waterlogging-triggered adventitious root formation in cucumber by activating the expression of CsPrx5. Plant J. 2023;114(4):824–35. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Huang H, Yi X, Zhang Y, Yang Q, Zhang C, Fan C, Zhou Y. Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study. Plant Biotechnol J. 2020;18:1472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Innark P, Khanobdee C, Samipak S, Jantasuriyarat C. Evaluation of genetic diversity in cucumber (Cucumis sativus L.) germplasm using agro-economic traits and microsatellite markers. Sci Hortic. 2013;162:278–84. [Google Scholar]
- 30.Mohler V, Stadlmeier M. Dynamic qtl for adult plant resistance to powdery mildew in common wheat (Triticum aestivum L.). J Appl Genet. 2019;60:291–300. [DOI] [PubMed] [Google Scholar]
- 31.Castro AJ, Chen X, Hayes PM, Knapp SJ, Vivar H. Coincident QTL which determine seedling and adult plant resistance to stripe rust in barley. Crop Sci. 2002;42:1701–8. [Google Scholar]
- 32.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–76. [DOI] [PubMed] [Google Scholar]
- 33.Mayr C. Regulation by 3’-untranslated regions. Annu Rev Genet. 2017;51:171–94. [DOI] [PubMed] [Google Scholar]
- 34.Ortega JL, Moguel-Esponda S, Potenza C, Conklin CF, Quintana A, Sengupta-Gopalan C. The 3’ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J. 2006;45:832–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin J, Yuan Z, Fordyce K, Sreeramoju P, Kent TS, Kim J, Wang V, Schneyer D, Weber TK. A del T poly T (8) mutation in the 3’ untranslated region (UTR) of the CDK2-AP1 gene is functionally significant causing decreased mRNA stability resulting in decreased CDK2-AP1 expression in human microsatellite unstable (MSI) colorectal cancer (CRC). Surgery. 2007;142:222–7. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, Wang D, Lin M, Sehgal SK, Dong L, Wu Y, Bai G. Artificial selection in breeding extensively enriched a functional allelic variation in taphs1 for pre-harvest sprouting resistance in wheat. Theor Appl Genet. 2020;134:339–50. [DOI] [PubMed] [Google Scholar]
- 37.Yang YL, Guo L, Xu SA, Holland CA, Kitamura T, Hunter K, Cunningham JM. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nature Genet. 1999;21:216–9. [DOI] [PubMed] [Google Scholar]
- 38.Battini JL, Rasko JEJ, Miller AD. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. P Natl Acad Sci USA. 1999;96:1385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wege S, Poirier Y. Expression of the mammalian xenotropic polytropic virus receptor 1 (xpr1) in tobacco leaves leads to phosphate export. FEBS Lett. 2014;588:482–9. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Dong S, Li M, Gu F, Yang G, Guo T, Chen Z, Wang J. The class III peroxidase gene OsPrx30, transcriptionally modulated by the AT-hook protein OsATH1, mediates rice bacterial blight-induced ROS accumulation. J Integr Plant Biol. 2021;63(2):393–408. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Zhang X, Zhao B, Song P, Zhang X, Wang B, Li Q. Wheat class III peroxidase TaPOD70 is a potential susceptibility factor negatively regulating wheat resistance to Blumeria graminis f. sp. tritici. Phytopathology. 2023;113(5):873–83. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Liu X, Yan Y, Wang W, Gebretsadik K, Qi X, Xu Q, Chen X. Comparative proteomic analysis of cucumber powdery mildew resistance between a single-segment substitution line and its recurrent parent. Hortic Res. 2019;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, Jiang F, Cen B, Wen J, Zhou Y, Wu Z. Respiratory burst oxidase homologue-dependent H2O2 and chloroplast H2O2 are essential for the maintenance of acquired thermotolerance during recovery after acclimation. Plant Cell Environ. 2018;41(10):2373–89. [DOI] [PubMed] [Google Scholar]
- 44.Ma B, Suo Y, Zhang J, Xing N, Gao Z, Lin X, Zheng L, Wang Y. Glutaredoxin like protein (RtGRL1) regulates H2O2 and Na+ accumulation by maintaining the glutathione pool during abiotic stress. Plant Physiol Biochem. 2021;159:135–47. [DOI] [PubMed] [Google Scholar]
- 45.Koo SC, Moon BC, Kim JK, Kim CY, Sung SJ, Kim MC, Cho MJ, Cheong YH. OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem Biophys Res Commun. 2009;387(2):365–70. [DOI] [PubMed] [Google Scholar]
- 46.Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander D, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C, Dong S, Xu J, He W, Yang T. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–8. [DOI] [PubMed] [Google Scholar]
- 55.Xu X, Yu T, Xu R, Shi Y, Lin X, Xu Q, Qi X, Weng Y, Chen X. Fine mapping of a dominantly inherited powdery mildew resistance major-effect QTL, Pm1.1, in cucumber identifies a 41.1 kb region containing two tandemly arrayed cysteine-rich receptor-like protein kinase genes. Theor Appl Genet. 2016;129:507–16. [DOI] [PubMed] [Google Scholar]
- 56.Bradbury P, Zhang Z, Kroon D, Casstevens T, Ramdoss Y, Buckler E. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–5. [DOI] [PubMed] [Google Scholar]
- 57.Yang J, Lee S, Goddard M, Visscher P. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang N, Lu Y, Yang X, Huang J, Zhou Y, Ali F, Wen W, Liu J, Li J, Yan J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014;10:e1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner S. Qqman: q-q and manhattan plots for GWAS data. J Open Source Softw. 2018;3:731. [Google Scholar]
- 60.Dong S, He W, Ji J, Zhang C, Guo Y, Yang T. Ldblockshow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform. 2020;30:bbaa227. [DOI] [PubMed] [Google Scholar]
- 61.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnol. 2019;37(8):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X, Du Y, Li S, Tan M, Sohail H, Liu X, Qi X, Yang X, Chen X. A genome-wide association study reveals molecular mechanism underlying powdery mildew resistance in cucumber. Datasets. Genome Sequence Archive. 2021. https://ngdc.cncb.ac.cn/gsa/browse/CRA002417.
- 65.Xu X, Du Y, Li S, Tan M, Sohail H, Liu X, Qi X, Yang X, Chen X. A genome-wide association study reveals the genetic basis of powdery mildew resistance in cucumber. Datasets. Sequence Read Archive. 2023. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA998480.
- 66.Xu X, Du Y, Li S, Tan M, Sohail H, Liu X, Qi X, Yang X, Chen X. A genome-wide association study reveals molecular mechanism underlying powdery mildew resistance in cucumber. Github. 2024. https://github.com/xiaodongy86/PM_GWAS_Cucumber.
- 67.Xu X, Du Y, Li S, Tan M, Sohail H, Liu X, Qi X, Yang X, Chen X. A genome-wide association study reveals molecular mechanism underlying powdery mildew resistance in cucumber. Zenodo. 2024. https://zenodo.org/records/13635209.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Tables S1–S6. This file contains the supplementary tables referenced in the main text. Table S1. Summary of the sampled cucumber accessions. Table S2. Chromosomal distribution of the high-quality markers detected in this study. Table S3. Significant markers associated with powdery mildew resistance detected at least twice. Table S4. Linkage disequilibrium (LD) blocks surrounding the 50 significant sites identified by the GWAS. Table S5. List of 647 non-overlapping differentially expressed genes identified by RNA-seq. Table S6. Primers used in this study.
Additional file 2: Supplementary Figs. S1–S7. This file contains the supplementary figures referenced in the main text. Fig. S1. Functional characterization of the candidate genes identified by the GWAS using VIGS. Fig. S2. Relative expression levels of CsGy5G015960 in different tissues of the PM-resistant line Jin5-508. Fig. S3. Representative whole-plant phenotypes of the CsGy5G015960 overexpression lines and the wild type (WT) control at 8 dpi with water. Fig. S4. Relative expression levels of CsGy5G015960 in 15 selected cucumber inbred lines at 48 h post powdery mildew inoculation. Fig. S5. Relative expression levels of CsGy5G015960 in roots of 15 selected cucumber inbred lines. Fig. S6. GUS expression analysis. Fig. S7. Relative expression levels of nine genes encoding peroxidase in leaves of OE2 and OE5 compared to WT upon 0, 48 and 96 h (h) of powdery mildew pathogen inoculation.
Data Availability Statement
The raw sequencing data generated in this study have been deposited in the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa/) of the National Genomics Data Center under the accession number CRA002417 [64]. The raw RNA-seq reads have been deposited in NCBI Sequence Read Archive (SRA) under accession PRJNA998480 [65]. The Gy14 cucumber reference genome (v2) was accessed from the Cucurbit Genomics Database (http://cucurbitgenomics.org/). The source codes used for data analysis in this study were deposited under GPL-3.0 license in Github (https://github.com/xiaodongy86/PM_GWAS_Cucumber) [66] and Zenodo (https://zenodo.org/records/13635209) [67]. No other scripts and software were used other than those mentioned in the “Methods” section.