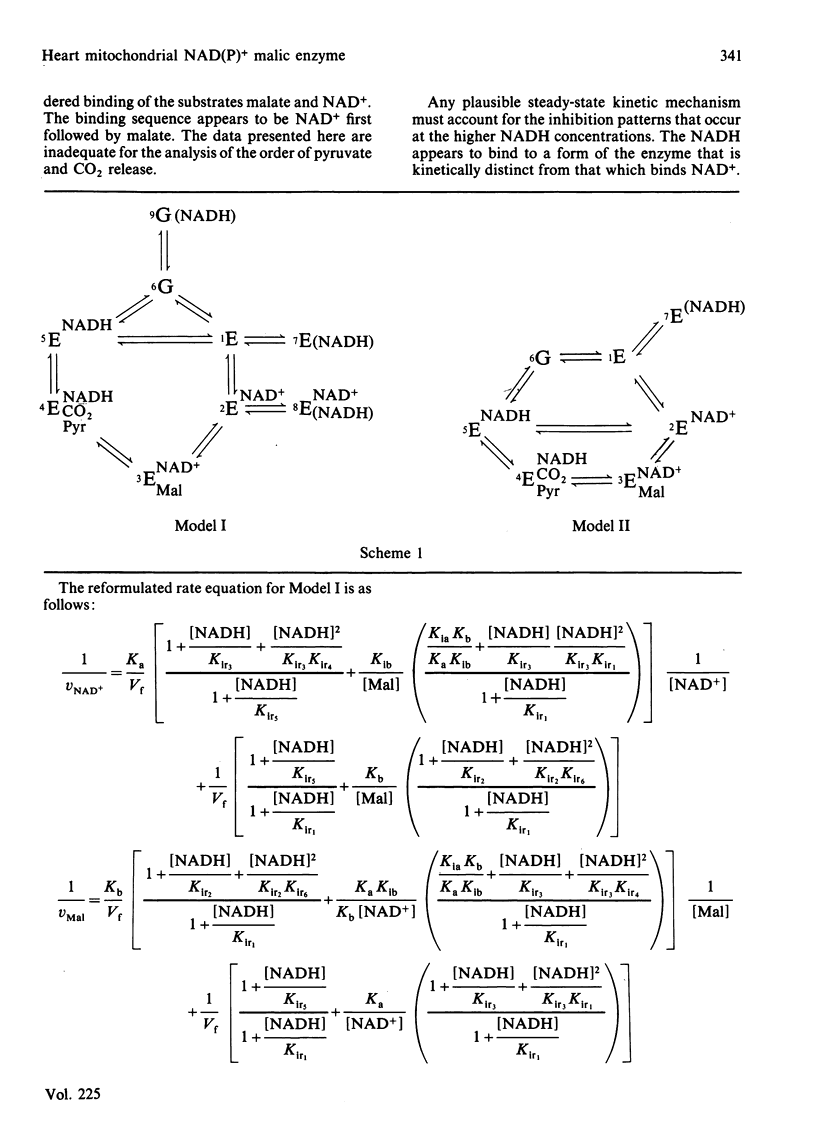
Abstract
The mitochondrial NAD(P)+ malic enzyme [EC 1.1.1.39, L-malate:NAD+ oxidoreductase (decarboxylating)] was purified from rabbit heart to a specific activity of 7 units (mumol/min)/mg at 23 degrees C. A study of the reductive carboxylation reaction indicates that this enzymic reaction is reversible. The rate of the reductive carboxylation reaction appears to be completely inhibited at an NADH concentration of 0.92 mM. A substrate saturation curve of this reaction with NADH as the varied substrate describes this inhibition. The apparent kinetic parameters for this reaction are Ka(NADH) = 239 microM and Vr = 1.1 mumol/min per mg at 23 degrees C. The steady-state product-inhibition patterns for pyruvate and NADH indicate a sequential binding of the substrates: NAD+ followed by L-malate. These data also indicate that NADH is the last product released. A steady-state kinetic model is proposed that incorporates NADH-enzyme dead-end complexes.
Full text
PDF

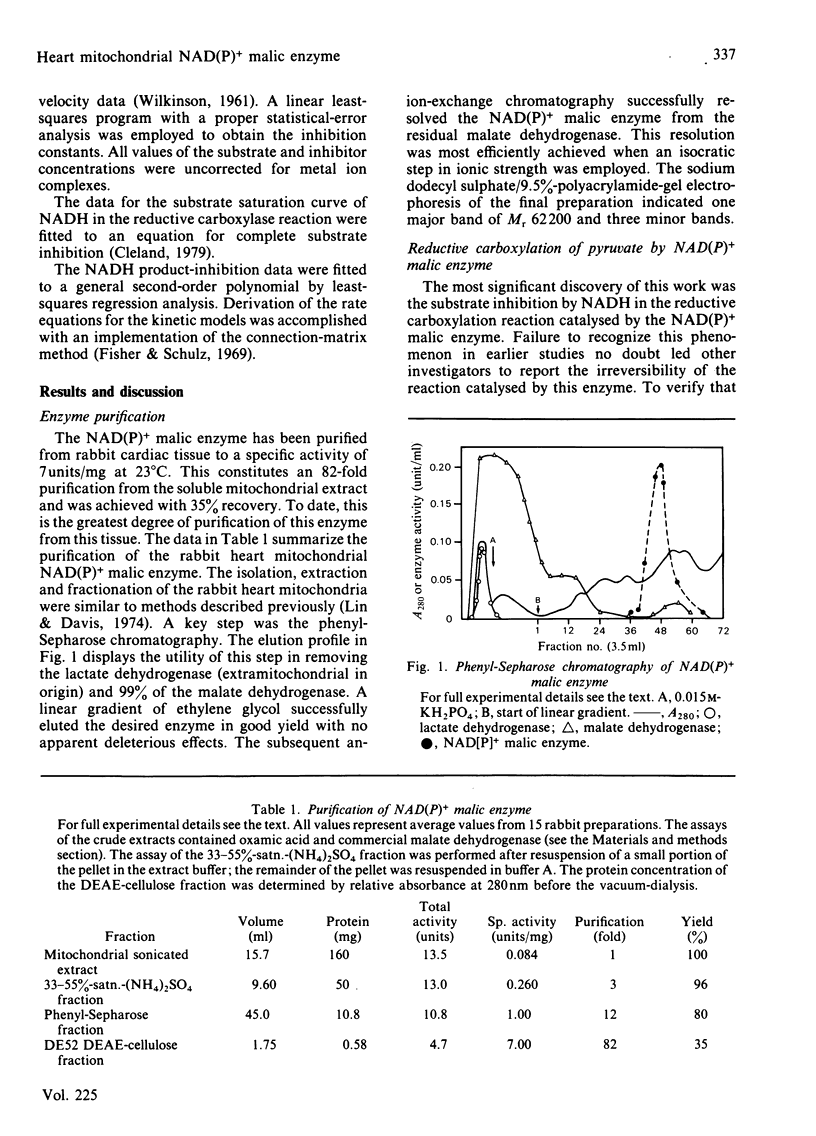
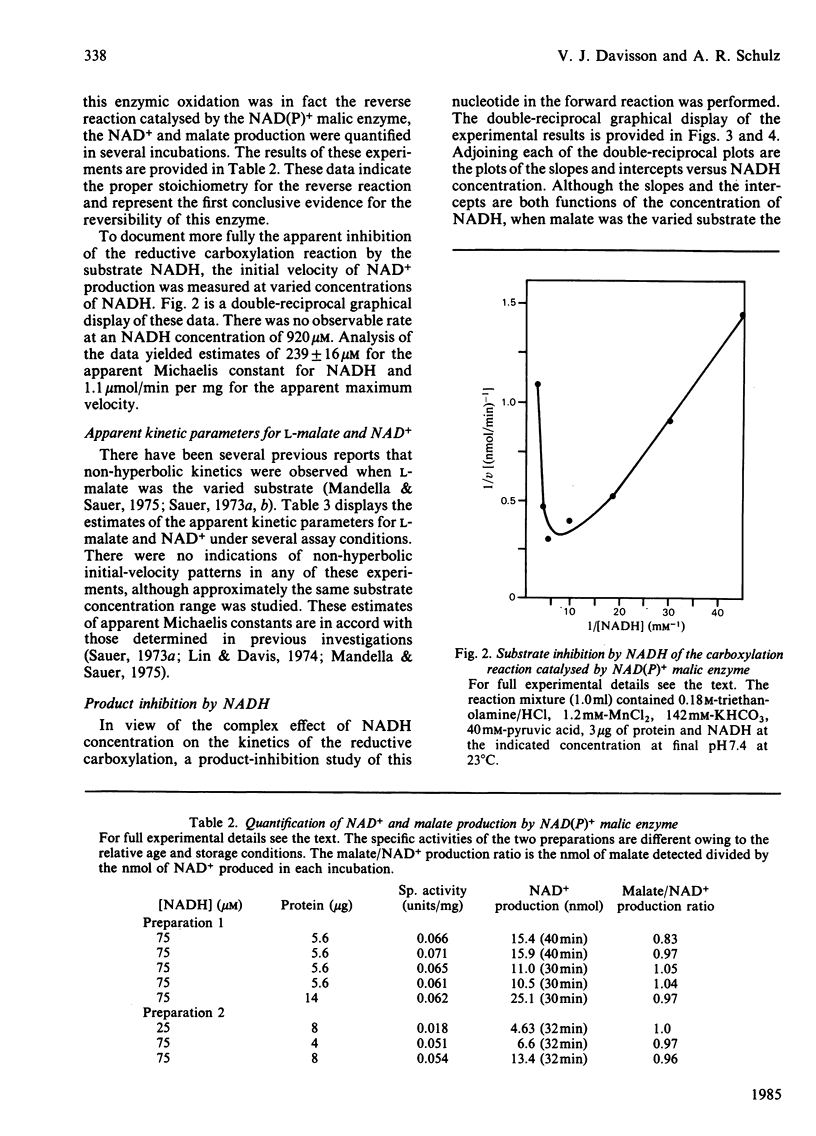
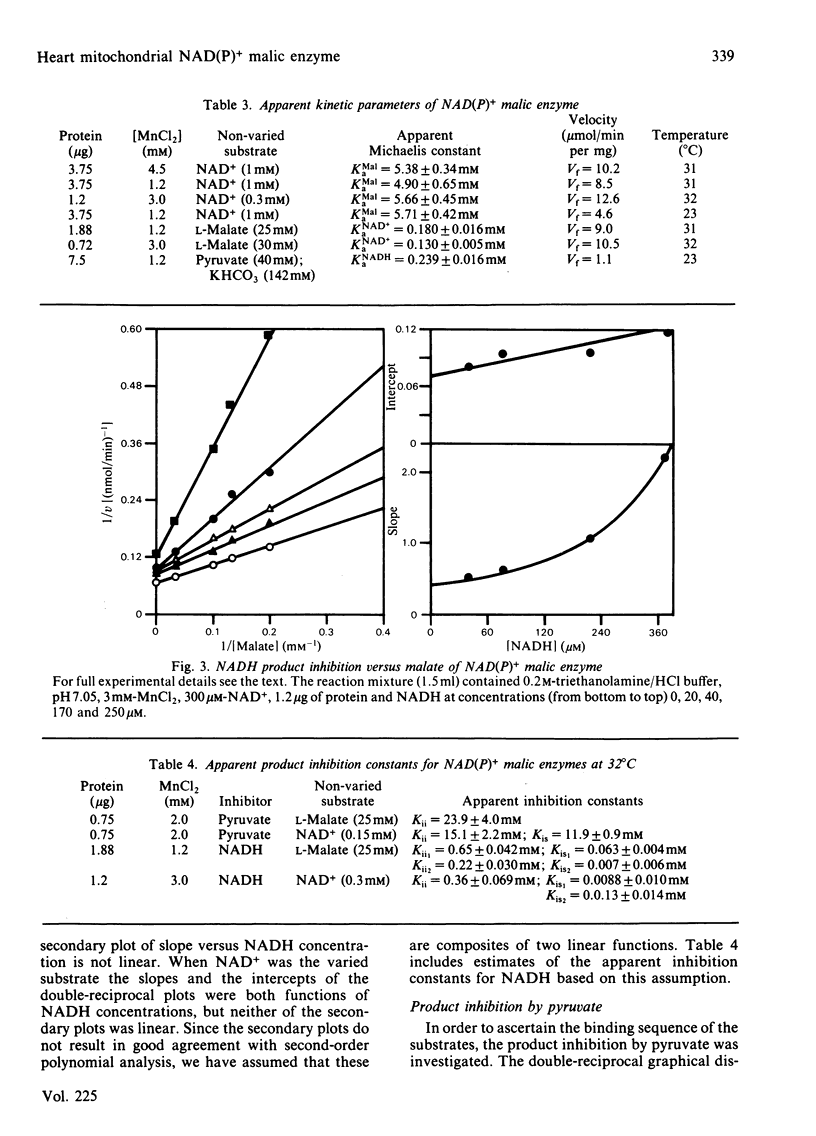
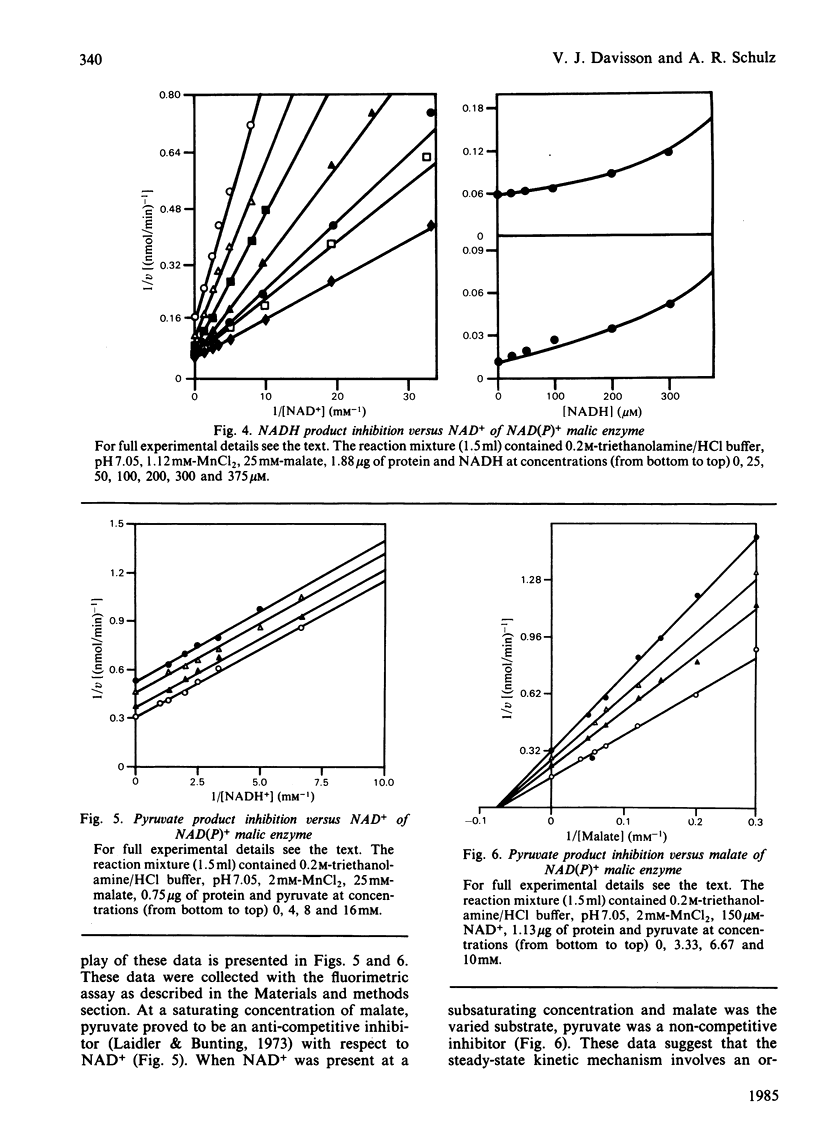

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Canellas P. F., Grissom C. B., Wedding R. T. Allosteric regulation of the NAD malic enzyme from cauliflower: activation by sulfate. Arch Biochem Biophys. 1983 Jan;220(1):116–132. doi: 10.1016/0003-9861(83)90393-4. [DOI] [PubMed] [Google Scholar]
- Canellas P. F., Wedding R. T. Substrate and metal ion interactions in the NAD+ malic enzyme from cauliflower. Arch Biochem Biophys. 1980 Jan;199(1):259–264. doi: 10.1016/0003-9861(80)90279-9. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Fodge D. W., Gracy R. W., Harris B. G. Studies on enzymes from parasitic helminths. I. Purification and physical properties of malic enzyme from the muscle tissue of Ascaris suum. Biochim Biophys Acta. 1972 May 12;268(2):271–284. doi: 10.1016/0005-2744(72)90322-1. [DOI] [PubMed] [Google Scholar]
- Frenkel R. Regulation and physiological functions of malic enzymes. Curr Top Cell Regul. 1975;9:157–181. doi: 10.1016/b978-0-12-152809-6.50012-3. [DOI] [PubMed] [Google Scholar]
- Grissom C. B., Canellas P. F., Wedding R. T. Allosteric regulation of the NAD malic enzyme from cauliflower: activation by fumarate and coenzyme A. Arch Biochem Biophys. 1983 Jan;220(1):133–144. doi: 10.1016/0003-9861(83)90394-6. [DOI] [PubMed] [Google Scholar]
- Grover S. D., Canellas P. F., Wedding R. T. Purification of NAD malic enzyme from potato and investigation of some physical and kinetic properties. Arch Biochem Biophys. 1981 Jul;209(2):396–407. doi: 10.1016/0003-9861(81)90297-6. [DOI] [PubMed] [Google Scholar]
- Hansford R. G., Lehninger A. L. Active oxidative decarboxylation of malate by mitochondria isolated from L-1210 ascites tumor cells. Biochem Biophys Res Commun. 1973 Mar 17;51(2):480–486. doi: 10.1016/0006-291x(73)91282-5. [DOI] [PubMed] [Google Scholar]
- Hsu R. Y., Lardy H. A., Cleland W. W. Pigeon liver malic enzyme. V. Kinetic studies. J Biol Chem. 1967 Nov 25;242(22):5315–5322. [PubMed] [Google Scholar]
- Landsperger W. J., Fodge D. W., Harris B. G. Kinetic and isotope partitioning studies on the NAD+-malic enzyme from Ascaris suum. J Biol Chem. 1978 Mar 25;253(6):1868–1873. [PubMed] [Google Scholar]
- Lin R. C., Davis E. J. Malic enzymes of rabbit heart mitochondria. Separation and comparison of some characteristics of a nicotinamide adenine dinucleotide-preferring and a nicotinamide adenine dinucleotide phosphate-specific enzyme. J Biol Chem. 1974 Jun 25;249(12):3867–3875. [PubMed] [Google Scholar]
- Macrae A. R. Isolation and properties of a 'malic' enzyme from cauliflower bud mitochondria. Biochem J. 1971 May;122(4):495–501. doi: 10.1042/bj1220495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandella R. D., Sauer L. A. The mitochondrial malic enzymes. I. Submitochondrial localization and purification and properties of the NAD(P)+-dependent enzyme from adrenal cortex. J Biol Chem. 1975 Aug 10;250(15):5877–5884. [PubMed] [Google Scholar]
- Nagel W. O., Sauer L. A. Mitochondrial malic enzymes. Purification and properties of the NAD(P)-dependent malic enzyme from canine small intestinal mucosa. J Biol Chem. 1982 Oct 25;257(20):12405–12411. [PubMed] [Google Scholar]
- Sauer L. A. An NAD- and NADP-dependent malic enzyme with regulatory properties in rat liver and adrenal cortex mitochondrial fractions. Biochem Biophys Res Commun. 1973 Jan 23;50(2):524–531. doi: 10.1016/0006-291x(73)90871-1. [DOI] [PubMed] [Google Scholar]
- Sauer L. A. Mitochondrial NAD-dependent malic enzyme: a new regulatory enzyme. FEBS Lett. 1973 Jul 1;33(2):251–255. doi: 10.1016/0014-5793(73)80205-4. [DOI] [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter C., Barrett M. J. The information content of enzyme kinetic data. 3. A comparison of four methods for fitting kinetic data to a power series. Enzymologia. 1970 Mar 31;38(3):147–160. [PubMed] [Google Scholar]


