Abstract
Herpesvirus envelopment is a two-step process which includes acquisition of a primary envelope resulting from budding of intranuclear capsids through the inner nuclear membrane. Fusion with the outer leaflet of the nuclear membrane releases nucleocapsids into the cytoplasm, which then gain their final envelope by budding into trans-Golgi vesicles. It has been shown that the UL34 gene product is required for primary envelopment of the alphaherpesvirus pseudorabies virus (PrV) (B. G. Klupp, H. Granzow, and T. C. Mettenleiter, J. Virol. 74:10063–10073, 2000). For secondary envelopment, several virus-encoded PrV proteins are necessary, including glycoproteins E, I, and M (A. R. Brack, J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 73:5364–5372, 1999). We show here that the product of the UL37 gene of PrV, which is a constituent of mature virions, is involved in secondary envelopment. Replication of a UL37 deletion mutant, PrV-ΔUL37, was impaired in normal cells; this defect could be complemented on cells stably expressing UL37. Ultrastructural analysis demonstrated that intranuclear capsid maturation and budding of capsids into and release from the perinuclear space were unimpaired. However, secondary envelopment was drastically reduced. Instead, apparently DNA-filled capsids accumulated in the cytoplasm in large aggregates similar to those observed in the absence of glycoproteins E/I and M but lacking the surrounding electron-dense tegument material. Although displaying an ordered structure, capsids did not contact each other directly. We postulate that the UL37 protein is necessary for correct addition of other tegument proteins, which are required for secondary envelopment. In the absence of the UL37 protein, capsids interact with each other through unknown components but do not acquire the electron-dense tegument which is normally found around wild-type capsids during and after secondary envelopment. Thus, apposition of the UL37 protein to cytoplasmic capsids may be crucial for the addition of other tegument proteins, which in turn are able to interact with viral glycoproteins to mediate secondary envelopment.
Herpesvirus particles are characterized by the presence of four morphologically differentiable components: the inner nucleoprotein core with double-stranded genomic DNA; the icosahedral capsid shell; an amorphous material of protein called the tegument; and an envelope of host cell-derived lipids containing virus-encoded (glyco)proteins (26). Assembly of herpesvirus capsids occurs in the nuclei of infected cells. Intranuclear capsids acquire a primary envelope by budding through the inner nuclear membrane, resulting in perinuclear enveloped virions (13, 14, 26). These virions have been shown to be biochemically and ultrastructurally different from mature extracellular virus particles. In particular, perinuclear pseudorabies virus (PrV) virions contain the UL34 protein, which has been shown to be required for primary envelopment of herpes simplex virus type 1 (HSV-1) (27) as well as PrV (19). This protein is absent from intracytoplasmic or extracellular mature PrV virions. In contrast, the UL49 tegument protein is present in mature virions but absent from perinuclear virus particles (19). Therefore, the protein compositions of perinuclear immature and of intracytoplasmic and extracellular mature virus particles are different, a fact which is easily explained by a two-step envelopment process. This includes deenvelopment by fusion of the primary envelope with the outer leaflet of the nuclear membrane and final envelopment by budding of intracytoplasmic capsids into trans-Golgi vesicles (5, 10, 13, 14, 15, 17, 24, 34, 35, 39).
So far, the molecular basis for either of the envelopment processes is largely unknown. Recently, the importance of so-called “nonessential” glycoproteins for secondary envelopment was demonstrated. In the absence of the glycoprotein E-I complex (gE/I) as well as glycoprotein M (gM), secondary envelopment was blocked and large intracytoplasmic accumulations containing capsids surrounded by electron-dense tegument material were observed (2, 3). Thus, in the absence of these glycoproteins, capsids were apparently still able to assemble tegument but did not gain access to the budding machinery for secondary envelopment. This situation resulted in the formation of intracytoplasmic inclusion bodies containing large numbers of capsids associated with tegument. In contrast, in the absence of the PrV UL3.5 protein, only naked nucleocapsids without visible tegument were observed scattered throughout the cytoplasm (8), a result which suggested that the apposition of tegument is required for the observed interactions resulting in inclusions. Since these data indicated that tegument proteins were responsible for the formation of aberrant particle-to-particle contacts leading to inclusions, we set out to analyze the individual contributions of tegument proteins to virion maturation.
From X-ray diffraction studies, the tegument is hypothesized to be largely unstructured, apart from the inner portions, which make contact with the icosahedral capsid (6, 38). For HSV-1, many proteins have been demonstrated or hypothesized to be part of the tegument (31). These include factors modulating the host cell after virus infection, such as the UL41-encoded virion-host cell shutoff protein. They also include viral regulatory proteins, such as UL48-encoded VP16, a transinducing transcriptional activator, or the immediate-early proteins ICP0 and ICP4 (36, 37). Moreover, virus-encoded enzymes, such as the UL13 protein kinase, are constituents of the tegument. Besides these well-characterized components, the major portions of the tegument are made up of functionally rather poorly characterized proteins. They include, among others, the products of the UL36, UL37, UL46, UL47, and UL49 genes (26, 31).
Most of these proteins are not required for productive viral replication in cultured cells. However, it has recently been reported that the largest protein encoded by alphaherpesviruses, the UL36 gene product, is required for intracytoplasmic maturation of virus particles. In the absence of the UL36 protein, capsids traverse the nuclear membrane but do not acquire a secondary envelope. Instead, they are found clustered in the cytoplasm without any detectable tegument structure (7).
The UL37 gene product is one of the few alphaherpesvirus proteins whose function has not been revealed so far. Although initially described as a nonstructural phosphoprotein (1, 29, 30), it has subsequently been detected as a component of virions (22) and, more specifically, the tegument (20, 28). To study the function of the UL37 homolog in PrV, we produced an antiserum against the PrV UL37 protein, isolated a UL37 deletion mutant, and characterized the mutant virus in cell cultures.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants are derived from the wild-type laboratory strain Kaplan (PrV-Ka) (18). Viruses were grown on rabbit kidney (RK13) or porcine kidney (PSEK) cells in Eagle's minimum essential medium supplemented with 10% fetal calf serum.
Sequencing.
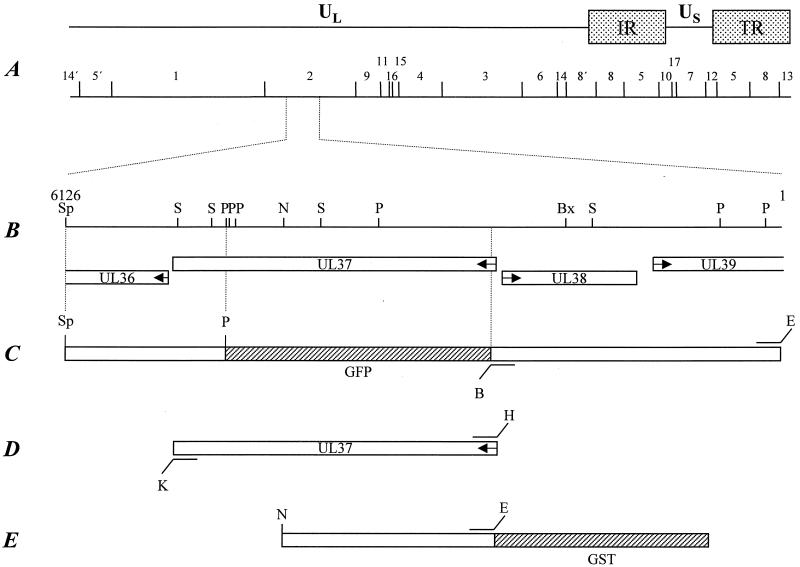
The nucleotide sequence comprising 750 bp of the UL37 open reading frame (ORF) has been published recently (GenBank accession no. X80797) (4). To complete the UL37 sequence, a 4.3-kb BstXI/SphI fragment of BamHI fragment 2 (Fig. 1) was cloned into SmaI- and SphI-cleaved pUC19. For further subcloning, plasmid pUC-BstX/Sph4.3 was digested with PstI and SphI. The resulting 1.3-kb SphI/PstI and 1.2-kb PstI fragments were ligated into appropriately cleaved pUC19, and the remaining 1.8-kb PstI/BstXI fragment with the vector sequences was religated after blunt ending. All subclones were subjected to nested deletion reactions, and selected clones were sequenced. The 920-bp SalI subfragment of pUC-BstX/Sph4.3 overlapping the junction between the 1.3-kb SphI/PstI and 1.2-kb PstI fragments was also cloned and sequenced. To verify the junction of the 1.2-kb PstI and 1.8-kb PstI/BstXI subfragments, sequencing was done using specific primers with pUC-BstX/Sph4.3. Sequencing was performed manually with double-stranded plasmid DNA using T7 and 7-deaza T7 sequencing kits (Amersham Pharmacia Biotech, Freiburg, Germany). The sequence was assembled and arranged using Wisconsin Package Version 10.1 (Genetics Computer Group, Madison, Wis.).
FIG. 1.
Construction of a UL37-negative PrV mutant and UL37-expressing cells. (A) Diagram of the PrV genome shown above a BamHI restriction fragment map. The PrV genome is divided into unique long (UL) and unique short (US) regions by internal and terminal repeats (IR and TR, respectively). (B) Enlarged view of the relevant portion of the genome. The locations of the ORFs are shown, and transcriptional orientation is indicated by arrows. (C) Construction of a transfer plasmid to create PrV-ΔUL37. (D) Construct used to establish cell line RK13-UL37. (E) Construct used for the expression of UL37 as a GST-UL37 fusion protein. Relevant cleavage sites are indicated: B, BamHI; Bx, BstXI; E, EcoRI; H, HindIII; K, KpnI; N, NotI; P, PstI; S, SalI; and Sp, SphI. Sequences encoding GFP or GST are not drawn to scale.
Preparation of monospecific anti-UL37 serum.
For the generation of UL37-specific antiserum, the UL37 ORF was PCR amplified using Platinum pfx DNA polymerase (Life Technologies, Karlsruhe, Germany), primers UL37FOR2 (5′-CACAGAATTCCGCGCGGACCCTCTTATAAT-3′; nucleotides [nt] 2439 to 2460; GenBank accession no. AJ318065) and UL37REV (5′-CACAGGTACCGCTGAAATAACACACGCGCG-3′; nt 5254 to 5235; GenBank accession no. AJ318065), and cloned BamHI fragment 2 as a template; EcoRI and KpnI sites introduced for convenient cloning are indicated by italics. The resulting 2.8-kb PCR product was cleaved with NotI using an internal NotI site (Fig. 1), and a 1.8-kb EcoRI/NotI fragment was inserted into EcoRI- and SmaI-cleaved pGEX-4T-1. A ca. 93-kDa glutathione S-transferase (GST)-UL37 fusion protein was used for immunization of a rabbit after separation on a sodium dodecyl sulfate-polyacrylamide gel and electroelution. Immunization was done as described recently (19). Serum obtained after the third immunization was used in this study.
Isolation of UL37-expressing cells.
For construction of complementing cells, the UL37 ORF was amplified as described above using primers UL37FOR (5′-CACAAAGCTTCGCGCGGACCCTCTTATAAT-3′; nt 2439 to 2460; GenBank accession no. AJ318065) and UL37REV, which contained HindIII and KpnI sites for convenient cloning (indicated by italics). The 2.8-kb PCR product was cloned into appropriately cleaved pcDNA3 (InVitrogen, Groningen, The Netherlands), resulting in plasmid pcDNA3-UL37. This plasmid was transfected into RK13 cells, and G418-resistant cell clones were picked and tested for UL37 expression by indirect immunofluorescence using the UL37-specific antiserum. One cell clone, RK13-UL37, was selected and used in this study.
Isolation of a UL37-negative PrV mutant.
To construct a recombination plasmid with UL37 sequences deleted, 5′-flanking sequences comprising the UL38 and part of the UL39 genes were PCR amplified using Platinum pfx DNA polymerase with primers UL38For (5′-CACAGGATCCACGGCGTGGTCGGCCTCCTC-3′; nt 2502 to 2483 [GenBank accession no. AJ318065]) and UL38REV (5′-CACAGAATTCAGCACGGCGCGCACGTCC-3′; nt 1 to 19; GenBank accession no. AJ318065). The 2.5-kb PCR product was cloned as an EcoRI/BamHI fragment in front of the 1.3-kb SphI/PstI fragment. For easier selection of recombinant virus, a green fluorescent protein (GFP) marker cassette was inserted into the BamHI and PstI sites separating the UL37 5′- and 3′-flanking regions (for locations of the fragments used, see Fig. 1). The transfer plasmid was cotransfected with wild-type PrV-Ka DNA into RK13 cells (12), and green fluorescent plaques were picked and purified. One single-plaque isolate, PrV-ΔUL37, was further characterized.
Virus purification, plaque assay, and one-step growth analysis.
Virus purification, plaque assay, and one-step growth analysis were performed as previously described (19). For one-step growth analysis, cells were infected at an multiplicity of infection (MOI) of 10 with PrV-ΔUL37 which had been grown on RK13-UL37 cells or with PrV-Ka.
Western blotting, EM, and immunolabeling.
Western blotting, electron microscopy (EM), and immunolabeling were performed as described recently (19).
Nucleotide sequence accession number.
The sequence obtained has been deposited in GenBank under accession no. AJ318065.
RESULTS
Sequence and expression of PrV UL37.
The sequence of the UL38 gene and part of the UL37 gene has been published recently (4). The UL37 sequence comprised 750 bp of the UL37 ORF. To complete the UL37 sequence, the adjacent region was cloned and sequenced. The UL37 ORF comprises 2,757 bp coding for 919 amino acids. A “TATA” box (nt 2319 to 2323) was found 140 bp upstream of the start codon (nt 2459 to 2461), as already described (4). A poly(A) signal (AATAAA; nt 5214 to 5219) overlaps the stop codon at nt 5216 to 5218. The size of the UL37 transcript was estimated to be 3.5 to 4 kb (4), in the expected size range. The calculated molecular mass for the UL37 protein is 98 kDa. The deduced amino acid sequence shows 33% identity with the homolog of HSV-1, 34% with that of varicella-zoster virus, and 43% with that of equine herpesvirus 1 (data not shown).
Identification and kinetics of expression of the PrV UL37 protein.
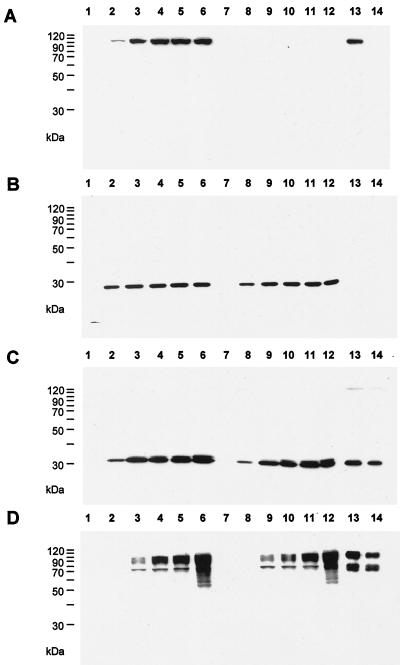
To identify the PrV UL37 protein, a monospecific polyclonal rabbit antiserum was generated against a GST-UL37 fusion protein (Fig. 1). In a Western blot of wild-type PrV-infected cells, this antiserum recognized a ca. 100-kDa protein (Fig. 2A) which was already detectable at 3 h after infection. Expression kinetics were similar to those of the UL34 (Fig. 2B) and UL49 (Fig. 2C) proteins, whereas the late gene product glycoprotein C (gC) (Fig. 2D) was not detectable before 5 h after infection.
FIG. 2.
Identification and kinetics of expression of the PrV UL37 protein. RK13 cells were infected at an MOI of 10 with PrV-Ka (lanes 1 to 6) or PrV-ΔUL37 (lanes 7 to 12) and harvested at 1 (lanes 1 and 7), 3 (lanes 2 and 8), 5 (lanes 3 and 9), 7 (lanes 4 and 10), 9 (lanes 5 and 11), or 24 (lanes 6 and 12) h after infection. Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and, after Western blotting, incubated with polyclonal sera against the UL37 protein (A), the UL34 protein (B), and the UL49 protein (C) or a monoclonal antibody against gC (D). Lanes 13, purified PrV-Ka virions. Lanes 14, purified PrV-ΔUL37 virions.
The PrV UL37 protein is a component of extracellular virions.
For HSV-1, the UL37 protein has been described as a structural component of virions (22). To analyze whether this is also true for the PrV UL37 homolog, sucrose gradient-purified virions were analyzed by Western blotting using the PrV UL37 protein-specific rabbit antiserum. As shown in Fig. 2A, lane 13, the antiserum detected a ca. 100-kDa protein in purified virions, as it did in infected cells. As a negative control, the UL34 protein, which had previously been found to be absent from mature virions (19), was not detectable in the purified virion fraction (Fig. 2B); however, the UL49 protein (Fig. 2C) and gC (Fig. 2D) were present. Thus, the PrV UL37 protein is a component of extracellular virions.
Absence of the UL37 protein results in restricted replication.
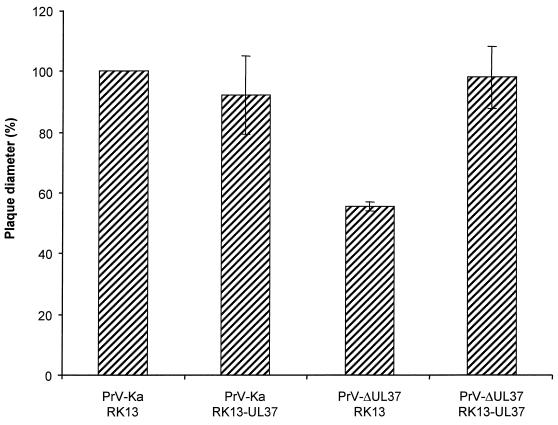
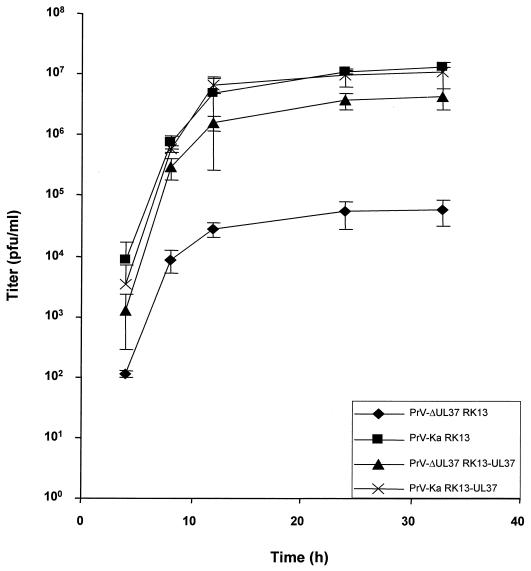
After cotransfection into normal RK13 cells of wild-type PrV DNA and a plasmid encompassing a partial deletion of the UL37 ORF with a GFP expression cassette insertion, we were able to isolate a recombinant virus, PrV-ΔUL37, which expressed GFP. Southern blot analyses indicated correct deletion of the UL37 sequences (data not shown), and Western blotting confirmed the absence of the UL37 protein in mutant virus-infected cells (Fig. 2A, lanes 7 to 12) and mutant virions (Fig. 2A, lane 14). These data indicated that the UL37 protein was not strictly required for PrV replication. However, the mutant virus plaques were significantly smaller, by about 50%, than those produced by wild-type PrV, reflecting a growth deficiency of the UL37-negative PrV mutant. This reduction in plaque size was not observed on UL37-expressing cells, demonstrating that the defect was indeed due to the absence of the UL37 protein (Fig. 3). In one-step growth analyses (Fig. 4), PrV-ΔUL37 exhibited a clear reduction in replication on normal RK13 cells, with final titers of ca. 5 × 104 PFU/ml. In contrast, wild-type PrV-Ka easily reached titers of as high as 107 PFU/ml. The replication defect was rescued on UL37-expressing cells, highlighting the importance of UL37 for the observed impairment in replication.
FIG. 3.
Plaque size of PrV-ΔUL37. RK13 or RK13-UL37 cells were infected under plaque assay conditions with PrV-Ka or PrV-ΔUL37. Two days after infection, plaque diameters were measured microscopically and compared with the average diameter of plaques induced by parental PrV-Ka, which was set at 100%. Average values and standard deviations after measurement of at least 50 plaques in three independent experiments each are indicated.
FIG. 4.
One-step growth analysis. RK13 or RK13-UL37 cells were infected as described in Materials and Methods with wild-type PrV-Ka or PrV-ΔUL37. At the indicated times after infection, supernatants and cells were harvested and titers on RK13 cells were determined and added. Average values and standard deviations of data from three independent experiments are shown.
The PrV UL37 protein is involved in secondary envelopment.
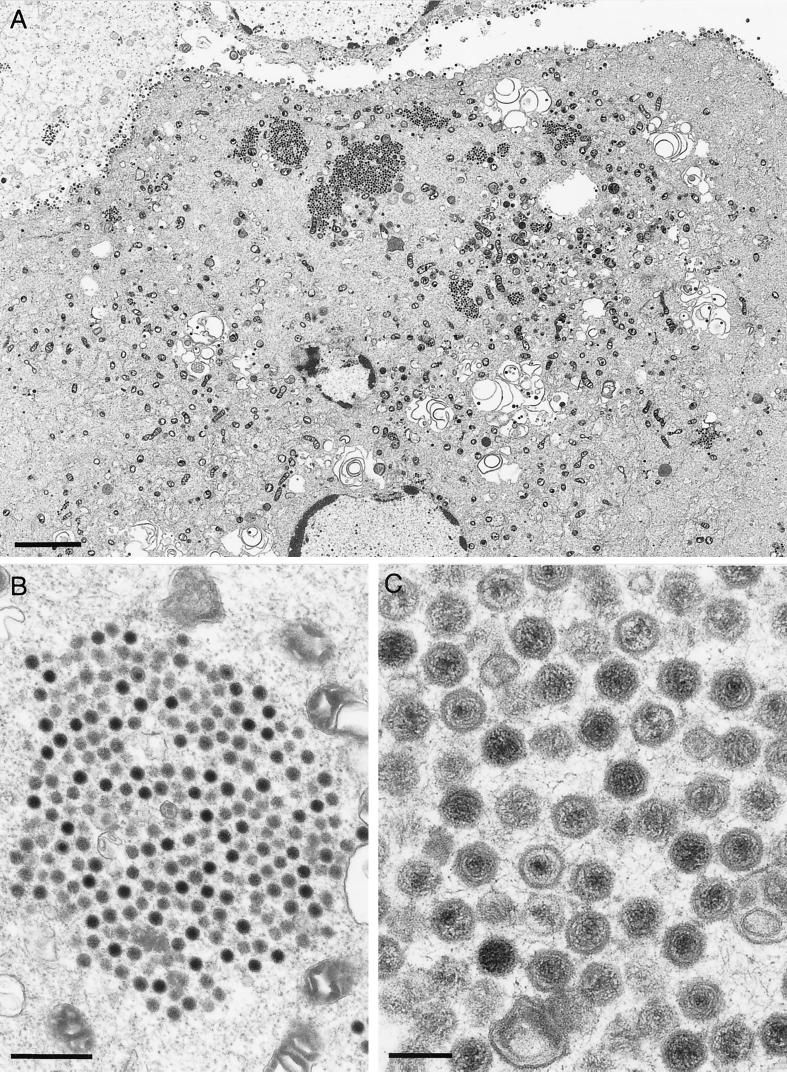
To examine the replication defect of PrV-ΔUL37 in more detail, EM was performed on PrV-ΔUL37-infected RK13 cells 16 h after infection. As shown in Fig. 5A, there was a striking phenotype, with large accumulations of intracytoplasmic capsids. In contrast to observed in the absence of gE/I and gM (2), in the absence of UL37 no apparent tegument material surrounded the capsids (Fig. 5B). However, they appeared to contact each other indirectly (Fig. 5C), resulting in an orderly arrangement. Thus, it appears as if viral maturation is blocked before the addition of tegument and before secondary envelopment. It is interesting that another PrV mutant which exhibits a block prior to the addition of tegument and secondary envelopment, PrV-UL3.5−, did not show any aggregation of capsids. Rather, dispersed capsids in the cytoplasm were observed in PrV-UL3.5−-infected cells (8).
FIG. 5.
EM of PrV-ΔUL37-infected RK13 cells. RK13 cells were infected at an MOI of 1 and analyzed 16 h after infection. (A) Overview of an infected cell. (B and C) Enlargements of aggregates of cytoplasmic capsids. Bars in panels A, B, and C are 3.5 μm, 500 nm, and 150 nm, respectively.
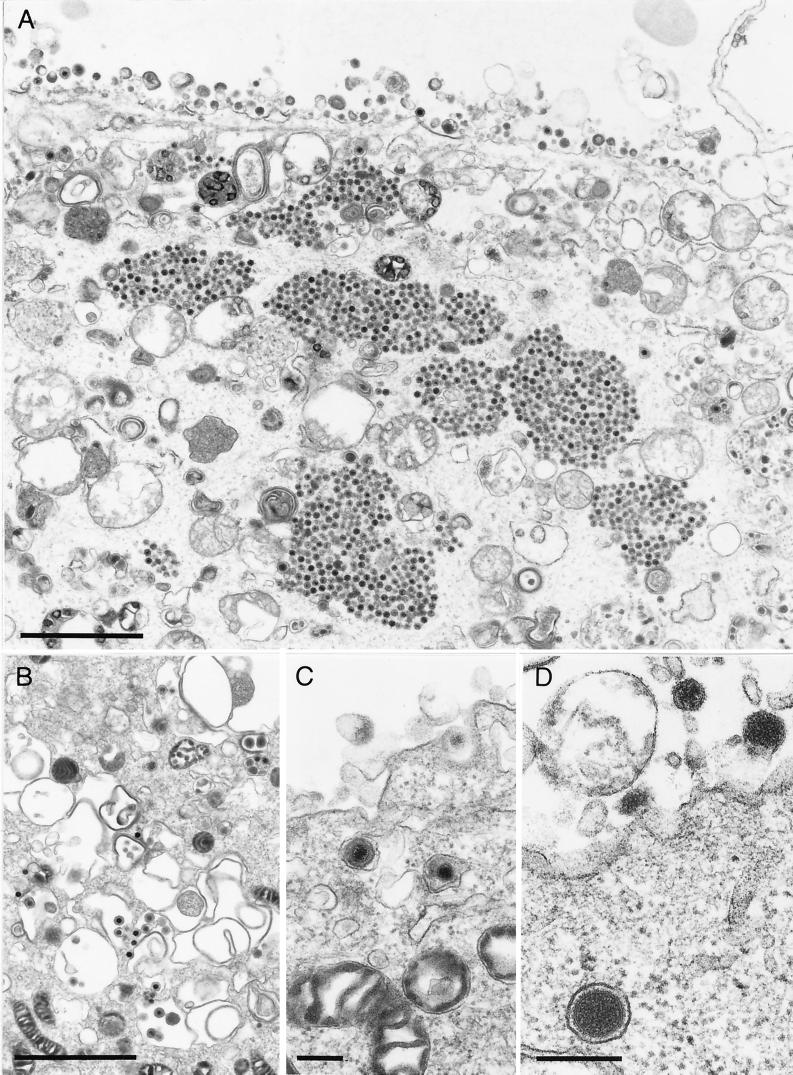
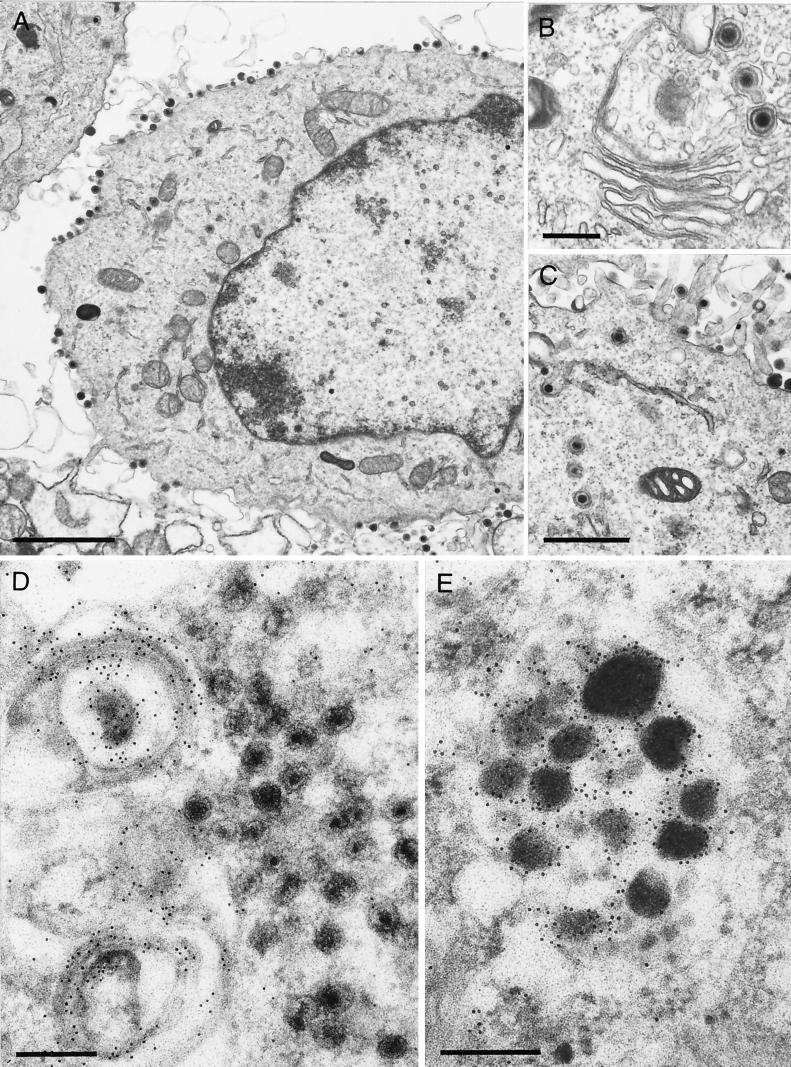
However, the block in PrV-ΔUL37-infected cells is not absolute. As shown in an overview in Fig. 6A, besides the accumulations of capsids, normal maturation of virions was also observed, including secondary envelopment by budding into trans-Golgi vesicles (Fig. 6B) and the presence of enveloped virions in vesicles (Fig. 6C). However, most strikingly, L-particle formation occurred efficiently (Fig. 6D), and numerous L- particles lined the surface of PrV-ΔUL37-infected cells (Fig. 6A). To correlate the observed phenotype with the absence of the UL37 protein, EM was also performed on RK13-UL37 cells infected with PrV-ΔUL37. As shown in Fig. 7A, no accumulations of capsids were observed and all stages of virion maturation could be visualized easily, including secondary envelopment (Fig. 7B), enveloped virions in vesicles presumably during transport to the cell surface, and the presence of numerous extracellular virus particles (Fig. 7A and C). Thus, the observed defect was indeed due to the absence of the UL37 protein.
FIG. 6.
Production of L-particles in PrV-ΔUL37-infected RK13 cells. RK13 cells were infected as described in the legend to Fig. 5. (A) Overview demonstrating aggregation of intracytoplasmic capsids as well as numerous extracellular L particles. (B and C) Normal secondary envelopment (B) as well as enveloped virions in vesicles presumably during transport to the surface (C) were also observed, although rarely. (D) Particularly striking was the production of L particles (see also panel B). Bars in panels A and B and panels C and D are 2 μm and 250 nm, respectively.
FIG. 7.
EM and immuno-EM of PrV-ΔUL37-infected RK13-UL37 and RK13 cells. (A to C) After infection of complementing RK13-UL37 cells with PrV-ΔUL37, all stages of normal virion maturation were observed (A), including unimpeded secondary envelopment (B) as well as transport to and accumulation at the plasma membrane (C). (D) To detect the major tegument protein UL49, PrV-ΔUL37-infected RK13 cells were labeled with a monospecific polyclonal anti-UL49 serum followed by incubation with gold-tagged secondary antibodies. Label was primarily detected in conjunction with vesicular membranes in the trans-Golgi area, the site of secondary envelopment. No label was detected in the accumulated capsids. (E) In contrast, L particles were heavily labeled by the anti-UL49 serum. Bars in panels A, B, C, and D and E are 2 μm, 0.5 μm, 1 μm, and 250 nm, respectively.
In the absence of the UL37 protein, the UL49 protein is not added to capsids.
Given the above results, we postulated that the addition of the UL37 protein to maturing capsids is required for the apposition of other tegument proteins prior to secondary envelopment. To assay for the presence of one of the major tegument proteins, the UL49 gene product, in maturing virions, RK13 cells were infected with PrV-ΔUL37 and analyzed by immuno-EM using a monospecific anti-UL49 serum and immunogold-labeled secondary antibodies (3). As shown in Fig. 7D, label was detected in conjunction with vesicular membranes in the trans-Golgi area, whereas the accumulations of capsids were free of label. In contrast, L-particles, which were formed in PrV-ΔUL37-infected cells, were strongly labeled by the anti-UL49 serum (Fig. 7E), indicating that the expression of UL49 and the interaction of tegument proteins with the secondary envelope were not drastically disturbed. Thus, the absence of the UL37 protein appeared to specifically interfere with a step in virion morphogenesis which precedes the addition of other tegument proteins, including UL49, as a requirement for secondary envelopment.
DISCUSSION
Our results show that the UL37 protein of PrV is involved in virion formation and that in its absence, secondary envelopment in the cytoplasm is impaired, resulting in aggregation of capsids in the cytoplasm. Thus, this is another in a series of mutants described by this laboratory with defects in virion morphogenesis resulting in the accumulation of capsids in the cytoplasm. However, the phenotypes of these mutants clearly differ. In the absence of the UL3.5 protein, secondary envelopment is also blocked, but capsids are found dispersed in the cytoplasm, with no sign of aggregation. In contrast, inhibition of secondary envelopment in the absence of gE/I and gM results in huge intracytoplasmic aggregates of capsids associated with protein material which includes the UL49 tegument protein. Therefore, these two phenotypes represent different stages in intracytoplasmic virion morphogenesis. In the absence of the UL37 protein, we found another distinct phenotype: capsids accumulate in ordered structures in the cytoplasm, but they are not surrounded by electron-dense tegument material. In addition, these capsids are not labeled by the anti-UL49 serum. However, besides the observation that these capsids accumulate without contacting each other directly, higher-magnification electron micrographs (Fig. 5C) indicate that indirect contacts between the capsids do indeed result in the observed orderly aggregates. At present, it is unclear which protein mediates these interactions. The largest tegument protein, the product of the UL36 gene, is one prime candidate. In the absence of the UL36 protein in HSV-1, capsids lacking tegument have been observed to accumulate in the cytoplasm (7). However, they do not regularly form the ordered aggregates which we observed in the absence of the UL37 protein. Therefore, the UL36 protein may mediate the interactions resulting in capsid aggregation. In this scenario, UL36 may be one of the first tegument proteins added to maturing capsids, a hypothesis which is supported by electron cryomicroscopy of HSV-1 and human cytomegalovirus cytoplasmic capsids (6, 38). UL36 may then, in turn, direct the deposition of the UL37 protein.
It is interesting that, except perhaps for the UL36 gene product in HSV-1 (7), none of the identified tegument proteins has been found to be indispensable for virus maturation. This finding also holds true for PrV UL37 which, despite a striking maturation defect, is able to replicate productively on noncomplementing cells, although only to a limited extent. This finding may indicate that, as has been observed for so-called nonessential envelope glycoproteins (2, 3), there may in fact be redundancy in the function of the individual tegument proteins inasmuch as in the absence of one or even more species, the remaining others are sufficient to promote secondary envelopment. We are currently isolating PrV mutants with multiple tegument protein deletions to shed light on this peculiar phenomenon.
Currently, it is unclear whether the reduction in the titers of mutant virus progeny produced on noncomplementing cells is solely due to the impairment in the formation of mature virions or whether there is also a decrease in the specific infectivity of released virions. However, our EM data clearly show a severe maturation defect, and only a few mature virions were detectable in mutant virus-infected noncomplementing cells, in contrast to numerous L particles (Fig. 6). A ca. 100-fold increase in titers was observed on complementing RK13-UL37 cells; however, the final titers still were ca. 10-fold lower than those obtained with the wild-type virus. The reason for this result is unclear at present but either may involve other gene products (e.g., UL36 or UL38) or may reflect a certain inefficiency of the complementing cell line. This result notwithstanding, the fact that the mutant virus titers increased by ca. 2 log10 units after propagation on RK13-UL37 cells and the observation of apparently normal virion morphogenesis after infection of complementing RK13-UL37 cells by PrV-ΔUL37 clearly demonstrate the role of UL37 in virion formation.
In the absence of the UL36 (7) or UL37 (this study) protein, capsids accumulate in the cytoplasm. This fact demonstrates that neither of these tegument proteins is required for capsids which are assembled in the nucleus to traverse the nuclear membrane. So far, it is still debated whether a certain amount or a certain species of tegument protein is added to capsids in the nucleus and is required for driving the budding process during primary envelopment. Electron micrographs indicate that enveloped capsids in the perinuclear space indeed contain immediately beneath the envelope an electron-dense material (13, 14) whose composition is unclear. So far, we have not been able to isolate a mutant virus lacking a tegument protein(s) which is blocked in primary envelopment. This phenotype has been observed only in the absence of the UL34 membrane protein, which localizes to both leaflets of the nuclear membrane (19).
Capsid formation is not required for secondary envelopment, since L-particles, i.e., tegument proteins surrounded by an envelope (16, 32), were readily observed in mutants inhibited in intranuclear capsid formation (21, 25). In contrast, in the absence of envelope gE/I and M, not only the production of mature virions but also the formation of L particles was blocked. Thus, an interaction between tegument proteins and envelope glycoproteins, including gE/I and M, is required and sufficient for budding during secondary envelopment. Since L-particle formation still proceeds in the absence of the UL37 protein, UL37 is not a component of the tegument essential for making contact with the future envelope.
We hypothesize that the UL37 protein is added as one of the first tegument proteins to maturing capsids, perhaps immediately after the addition of the UL36 protein. A direct interaction between HSV-1 UL36 and the capsid has been proposed (23), as has been an interaction between the homologs of UL36 and UL37 in human cytomegalovirus (11). The presence of the UL37 protein may then be required for the efficient addition of other tegument proteins, including UL49, which in turn make contact with viral envelope glycoproteins in trans-Golgi vesicles. This interaction then promotes budding and acquisition of the secondary (and final) envelope.
In this scenario, tegument proteins would fulfill the role of the matrix proteins in RNA viruses, bridging the nucleocapsid and the envelope (9, 33). However, in herpesviruses, the complexity, abundance, and flexibility of the tegument may, in addition, allow the incorporation into virus particles of “cargo” without disruption of the virion architecture. This cargo may include viral proteins required for modulating cell metabolism in favor of the virus prior to the onset of viral gene expression. Examples may be transcriptional activators such as ICP0 and IPC4 or the UL41 host cell shutoff protein.
ACKNOWLEDGMENTS
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/5-1).
We thank Uta Hartwig, Petra Meyer, and Nadine Müller for expert technical assistance.
REFERENCES
- 1.Albright A, Jenkins F. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67:4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brack A R, Klupp B G, Granzow H, Tirabassi R, Enquist L W, Mettenleiter T C. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J Virol. 2000;74:4004–4016. doi: 10.1128/jvi.74.9.4004-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun A, Kaliman A, Boldogköi Z, Aszodi A, Fodor I. Sequence and expression analyses of the UL37 and UL38 genes of Aujeszky's disease virus. Acta Vet Hung. 2000;48:125–136. doi: 10.1556/AVet.48.2000.1.14. [DOI] [PubMed] [Google Scholar]
- 5.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D H, Jiang H, Lee M, Liu F, Zhou Z H. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 7.Desai P J. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs W, Klupp B G, Granzow H, Rziha H-J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galinski M S, Wechsler S L. The molecular biology of the paramyxovirus genus. In: Kingsbury D, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 41–82. [Google Scholar]
- 10.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 12.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 13.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granzow H, Klupp B G, Fuchs W, Veits J, Osterrieder N, Mettenleiter T C. Egress of alphaherpesviruses: a comparative ultrastructural study. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harson R, Grose C. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J Virol. 1995;69:4994–5010. doi: 10.1128/jvi.69.8.4994-5010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 17.Jones F, Grose C. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J Virol. 1988;62:2701–2711. doi: 10.1128/jvi.62.8.2701-2711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan A S, Vatter A. A comparison of herpes simplex and pseudorabies virus. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 19.Klupp B G, Granzow H, Mettenleiter T C. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol. 2000;74:10063–10073. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLauchlan J. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J Gen Virol. 1997;78:189–194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- 21.McLauchlan J, Rixon F J. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 22.McLauchlan J, Liefkens K, Stow N D. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J Gen Virol. 1994;75:2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- 23.McNabb D, Courtney R J. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 1992;190:221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- 24.Mettenleiter T C. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 25.Rixon F J, Addison C, McLauchlan J. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J Gen Virol. 1992;73:277–284. doi: 10.1099/0022-1317-73-2-277. [DOI] [PubMed] [Google Scholar]
- 26.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2231. [Google Scholar]
- 27.Roller R, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz J B, Albright A, Kinchington P, Jenkins F. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206:1055–1066. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 29.Shelton L G, Albright A, Ruyechan W, Jenkins F. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shelton L G, Pensiero M, Jenkins F. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J Virol. 1990;64:6101–6109. doi: 10.1128/jvi.64.12.6101-6109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens A, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 32.Szilagyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 33.Weldon R A, Hunter E. Molecular requirements for retrovirus assembly. In: Chiu W, Burnett R, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 381–410. [Google Scholar]
- 34.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao F, Courtney R J. A major transcriptional regulatory protein (ICP4) of herpes simplex virus type 1 is associated with purified virions. J Virol. 1989;63:3338–3344. doi: 10.1128/jvi.63.8.3338-3344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao F, Courtney R J. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66:2709–2716. doi: 10.1128/jvi.66.5.2709-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]