SUMMARY
Transcranial magnetic stimulation (TMS) is used to treat multiple psychiatric and neurological conditions by manipulating activity in particular brain networks and circuits, but individual responses are highly variable. In clinical settings, TMS coil placement is typically based on either group-average functional maps or scalp heuristics. Here, we found that this approach can inadvertently target different functional networks in depressed patients due to variability in their functional brain organization. More precise TMS targeting should be feasible by accounting for each patient’s unique functional neuroanatomy. To this end, we developed a targeting approach, termed “TANS” (Targeted Functional Network Stimulation). The TANS approach improved stimulation specificity in silico in 8 highly sampled patients with depression and 6 healthy individuals, and in vivo when targeting somatomotor functional networks representing the upper and lower limbs. Code for implementing TANS and an example dataset are provided as a resource.
Keywords: Precision functional mapping, transcranial magnetic stimulation, electric field modeling, functional brain networks
eTOC
Transcranial magnetic stimulation (TMS) is used to treat multiple psychiatric and neurological conditions by manipulating activity in particular brain networks and circuits. Lynch et al. demonstrate that more precise TMS targeting is possible by accounting for each patient’s unique functional neuroanatomy and cortical folding patterns using their method called Targeted Functional Network Stimulation (“TANS”).
INTRODUCTION
Repetitive transcranial magnetic stimulation (TMS) is a neuromodulatory technique used to treat multiple psychiatric and neurological conditions (Baeken et al., 2014; Carpenter et al., 2012; Connolly et al., 2012; Hanlon et al., 2015; Lefaucheur et al., 2020), including major depression, by manipulating activity in particular brain circuits and networks. Clinical response to TMS is variable, however, with only 30 to 40% of patients with treatment-resistant depression achieving remission in response to conventional protocols (Avery et al., 2008; Gaynes et al., 2014; Luborzewski et al., 2007; McGirr et al., 2021). The lack of response in some patients may be due in part to a failure to engage the desired functional networks, since individual human brains exhibit an idiosyncratic functional organization (Gordon and Nelson, 2021b; Gordon et al., 2017a; Mueller et al., 2013; Wang and Liu, 2014) and spatial targeting in TMS protocols is not usually individualized (Cash et al., 2020; Cocchi and Zalesky, 2018; Klooster et al., 2021). In this report, we examine functional neuroanatomy in two datasets comprising 8 highly sampled individuals with major depression and 6 individuals with no reported history of depression. In both samples, we show how individual variability in their functional brain organization interacts with the ability of TMS to selectively target specific brain circuits. We go on to develop a new method for more precise individualized targeting and validate it in vivo.
Precision functional mapping — the practice of delineating functional brain organization at the individual level using resting-state functional magnetic resonance imaging (fMRI) — has accelerated rapidly in recent years (Braga and Buckner, 2017; Braga et al., 2019; Gordon et al., 2017a; Gratton et al., 2019; Hacker et al., 2013; Lynch et al., 2020a; Poldrack et al., 2015; Wang et al., 2015; Zheng et al.). Multiple studies of highly sampled individuals have revealed features of an individual’s functional brain organization that deviate from central tendencies in large groups (Kraus et al., 2021; Seitzman et al., 2019) and have also shown how different functional networks can be represented in the same cortical area (Gordon and Nelson, 2021a; Gordon et al., 2017a; Laumann et al., 2015) or subcortical zone (Greene et al., 2019; Marek et al., 2018; Sylvester et al., 2020; Xue et al., 2021; Zheng et al.) across individuals. Studies have also reported that the functional connectivity (Cash et al., 2019, 2021; Fox et al., 2012; Hopman et al., 2021; Siddiqi et al., 2021; Weigand et al., 2018) or network properties (Siddiqi et al., 2019) of the TMS stimulation site affect treatment outcomes. These findings have created an expectation that targeting the same anatomical area with TMS could inadvertently engage different functional networks in different patients (Gratton et al., 2019; Lynch and Liston, 2020), which might in turn contribute to variability in treatment responses.
Although there is increasing interest in using functional connectivity to inform target site selection in basic research settings (Eldaief et al., 2011; Halko et al., 2014; Lynch et al., 2018; Wang et al., 2014) and for TMS interventions (Cash et al., 2019; Cole et al., 2022; Fox et al., 2012; Klooster et al., 2021; Siddiqi et al., 2022), most conventional TMS protocols are not guided by the individual patient’s functional brain organization. Instead, generic coil placements based on scalp heuristics (Beam et al., 2009; Mir-Moghtadaei et al., 2022) or stereotaxic coordinates derived from group-average functional maps (Blumberger et al., 2018; Weigand et al., 2018) have been used. One reason for these generic approaches is that reliable mapping of functional networks at the individual level can require large quantities of data per subject when using traditional single-echo fMRI methods (Gordon et al., 2017a; Laumann et al., 2015; Lynch et al., 2020a), which is a significant obstacle in clinical settings. We recently found that less data per subject is needed for precision functional mapping when using multi-echo fMRI methods (Kundu et al., 2017; Lynch et al., 2020a, 2021), which should increase the feasibility of individualized TMS targeting in clinical settings (Gratton et al., 2019).
This paper consists of three parts. In the first section, we present maps of functional networks in 8 depressed patients and 6 non-depressed individuals and use E-field modeling to demonstrate how a standard TMS targeting approach would likely engage multiple different functional networks in each individual, exactly the kind of variability that from first principles might not always produce the desired clinical outcome. Next, we propose a new technique, which we call “TANS” (Targeted Functional Network Stimulation), that leverages knowledge of each individual’s unique functional topology and cortical folding patterns to find a coil placement on the scalp and that will stimulate a given target functional network with as little off-target stimulation as is possible. The level of stimulation specificity that can be obtained using TANS is evaluated in silico and compared to alternative targeting approaches. Finally, we validate the TANS approach in vivo by using it to selectively engage somatomotor functional networks corresponding to the upper and lower limbs in three healthy individuals. The targeting workflow and code complete with an example dataset has been made available online as a public resource (Lynch, 2022).
RESULTS
Targeting the same brain area may engage different functional networks across patients
We first examined the extent to which delivering TMS in a manner consistent with current clinical practice (placing the stimulating coil directly above the same anatomical area) is likely to stimulate different functional networks in different patients. The primary goal of this first experiment was to evaluate the need for a targeting approach that is guided by how functional networks are arranged spatially in each patient’s brain.
Functional networks were mapped in 8 patients (N=3 females, mean age = 28.97 ± 7.06 years) with a mood disorder diagnosis and 6 individuals with no reported psychiatric history (N=6 males, mean age = 34.6 ± 9.30 years) using InfoMap (Rosvall and Bergstrom, 2008), a widely-used community detection algorithm, and the precision functional mapping routine used in (Gordon et al., 2020, 2021). Consistent with other studies of highly sampled individuals (Gordon et al., 2017a, 2017b; Laumann et al., 2015), we observed variability in the topology (size, shape, spatial arrangement) of functional networks in patients (see Figure 1A). Functional networks were mapped using each individual’s entire resting-state fMRI dataset (9.42 ± 9.94 hours of data per-subject, range: 0.72 to 29.86 hours). However, highly similar maps could be obtained with as little as 30-minutes of data from the first study visit (see Figure S1A-B), indicating that smaller clinically-tractable quantities of multi-echo fMRI data are sufficient for delineating patient-specific functional neuroanatomy for individualized TMS targeting.
Figure 1:
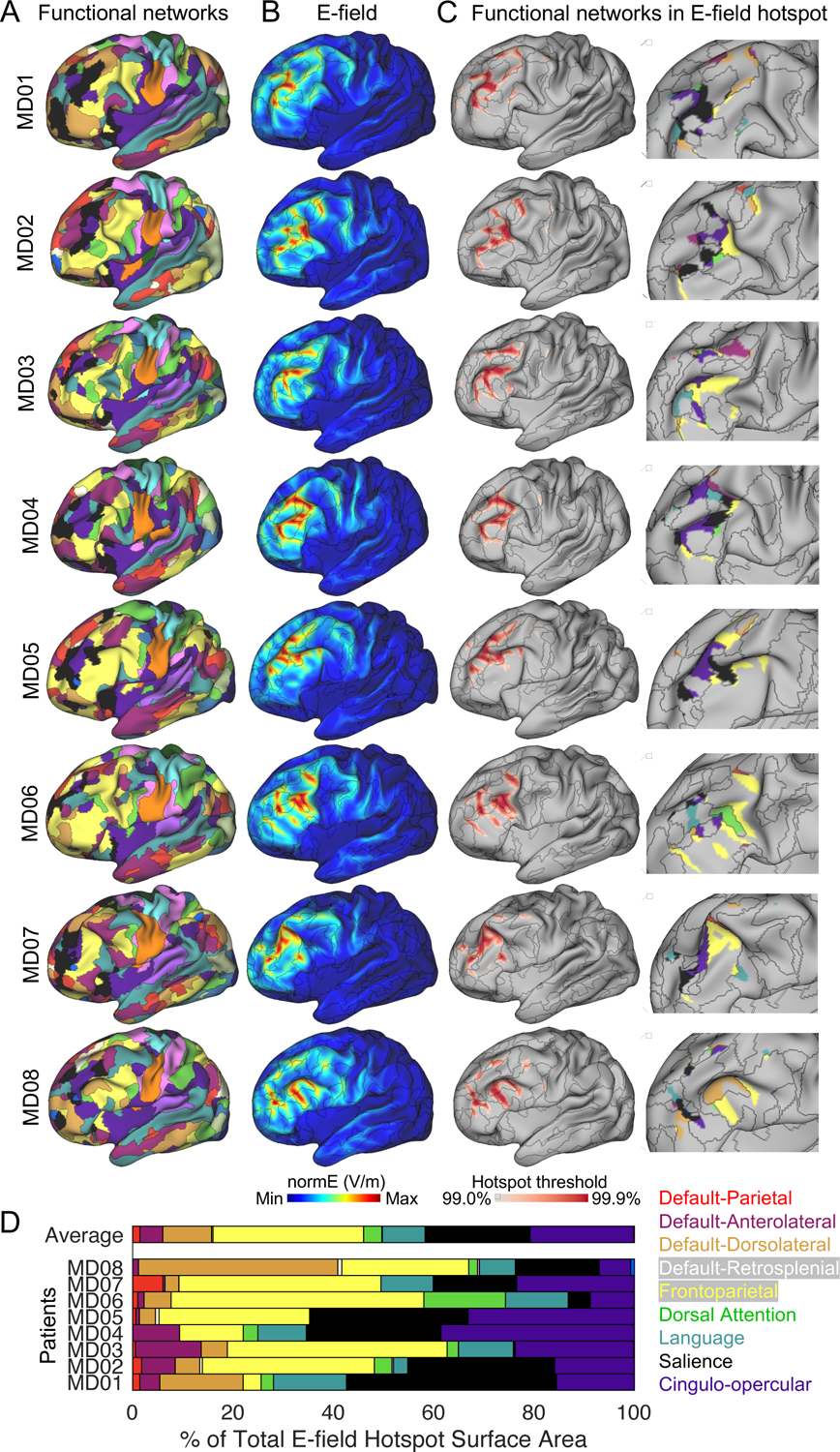
Targeting the same area of the left dorsolateral prefrontal cortex likely stimulates different functional brain networks in different patients. (A) Functional brain networks were identified in each patient using a precision functional mapping approach. (B) The electric field (E-field) generated by the TMS coil when it is set directly over MNI coordinates X=−42, Y=44, Z=30 in each patient. (C) The E-field hotspot defined using percentile based thresholds (99% to 99.9%, in 0.1% steps). A representative middle (99.5%) threshold is selected for visualizing functional networks inside the E-field hotspot in the insets (D). Variability in functional network stimulation across patients summarized in a horizontal stacked bar graph.
Next, we modeled how a generic targeting approach might engage functional networks in each individual using E-field modeling (Thielscher et al., 2015), the primary method by which the spatial distribution of TMS effects can be estimated. The MNI coordinates X=−42, Y=44, Z=30 — a stimulation target used in multiple recent TMS studies (Cash et al., 2019; Fox et al., 2012; Weigand et al., 2018) — were transformed into each individual’s native image space. These coordinates correspond to peak subgenual cingulate anticorrelation in group-average resting-state fMRI data. E-field modeling was performed using SimNIBS (Thielscher et al., 2015). To mimic how TMS is typically administered, the coil center was positioned directly above the target with the handle oriented posteriorly. The E-field generated by this coil placement is displayed on each patient’s inflated cortical surface (Figure 1B). The E-field “hotspot” (where the E-field is maximal) was defined using a range of percentile based thresholds (99% to 99.9%, in 0.1% steps). Multiple functional networks were observed in the E-field hotspot in each patient (Figure 1C). For example, in patient MD02, the E-field hotspot was primarily on the frontoparietal network (35% of the E-field hotspot), but the salience and cingulo-opercular networks (28% and 29% of the E-field hotspot, respectively) also received relatively high levels of stimulation. In other patients, such as MD01 and MD04, the salience network received more stimulation than the frontoparietal network (42% vs. 4% and 27% vs. 12% of the E-field hotspot in MD01 and MD04, respectively). The stacked horizontal bar plot summarizes the percentage of the total E-field hotspot surface area that was occupied by each functional brain network in all 8 patients (Figure 1D). A similar pattern was observed in the 6 non-depressed individuals (Figure S1C-F).
In summary, this first analysis revealed two findings that motivated the targeting approach introduced in this paper. First, highly reliable patient-specific functional maps could be obtained with as little as 30-minutes of multi-echo fMRI data per patient (see Figure S1A), indicating that individualized network mapping for TMS targeting should be feasible in clinical settings. Second, different functional networks were observed inside the E-field hotspot across individuals when using a generic targeting approach, despite the same anatomical area being specified as the stimulation target, and frequently multiple functional networks were simultaneously targeted within an individual. Because the efficacy of TMS is thought to depend on engaging specific functional networks underlying a patient’s symptoms (Cash et al., 2020; Siddiqi et al., 2019; Williams, 2016), the findings presented in Figure 1 suggest that a generic targeting approach may not be optimal for selectively engaging any specific functional network in patients. Thus, we sought to develop an individualized targeting approach that leverages information about how functional networks are arranged spatially in each patient’s brain in order to more selectively stimulate a given target network. In the following section, we describe this approach, termed “TANS” (Targeted Functional Network Stimulation).
The targeted functional network stimulation (“TANS”) approach
A graphical representation of the four main steps in the Targeted Functional Network Stimulation (“TANS”) approach is provided in Figure 2. The first step is to find the largest piece of the target functional network that is located on a gyral crown. Attempting to stimulate pieces of the target network in a sulcus would inevitably result in greater stimulation of whichever functional networks are represented on the nearest gyrus because the E-field is strongest on the gyral crown (Bungert et al., 2017; Siebner et al., 2022; Thielscher et al., 2011) (this effect is demonstrated in Figure S2A-B). An assumption of this first step is that it is important to stimulate within a particular functional network, and not any particular point within the network. In the second step, a search grid of coil placements is created on the scalp surface above the centroid of the target network cluster identified in Step 1. Subsampling is used to reduce the search grid density and reduce runtime. In the third step, E-field modeling is performed at each point in the search grid (multiple orientations are considered at each site at an angular resolution specified by the user) and the strength of the E-field generated by each simulation is mapped to the cortical surface. In the final step, the percentage of the E-field hotspot surface area occupied by the target network (referred to as the on-target value) is calculated for each threshold separately and then averaged. An avoidance region can be specified in order to avoid stimulation of a specific non-target region or network. If no avoidance region is provided, stimulation of any non-target network is penalized equally. The coil placement that maximizes the on-target value on average across E-field hotspot thresholds is considered the optimal choice. TANS is able to identify the optimal coil placement without the user specifying a specific stimulation intensity because the strength of the E-field varies linearly with dl/dt (the speed of variation of the current throughout the coil) and, for this reason, it has no effect on its spatial distribution (including where it is maximal, see Figure S2C). However, once the optimal coil placement has been identified, a range of possible stimulation intensities can be optionally modeled and the portion of the suprathreshold E-field that is on-target at each intensity level (given some absolute threshold representing the neural activation threshold, inferred from motor evoked potentials or some other means) can be estimated.
Figure 2:
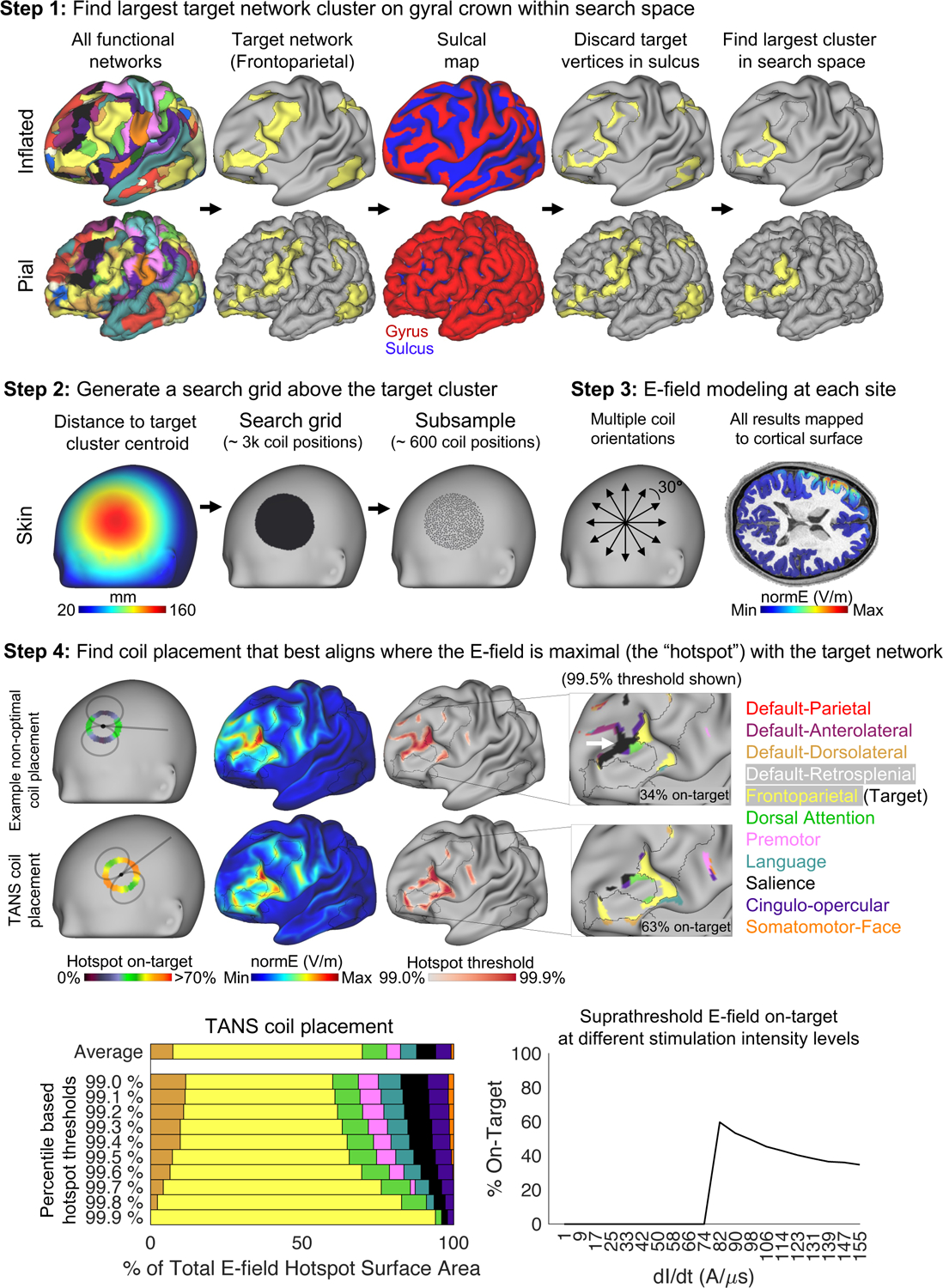
The four main steps of the Targeted Functional Network Stimulation (TANS) approach. Step 1 involves discarding target network vertices located in a sulcus in order to identify the largest piece of the target network on a gyral crown within the search space of interest (in this case, lateral prefrontal cortex). Step 2 involves creating a search grid directly above the target cluster centroid identified in Step 1. E-field modeling is performed at each point in the search grid during Step 3. Step 4 involves identifying the coil placement that best aligns where the E-field is maximal (the “hotspot”, defined for the purposes of the optimization using percentile-based thresholds) with the target functional network. The white arrow highlights off-target stimulation associated with an example non-optimal coil placement. After the optimal coil placement is found, a range of stimulation intensity levels can be modeled and the portion of the suprathreshold E-field that is on-target at each intensity level (given a specified neural activation threshold) is plotted. dl/dt = the speed of variation of the current throughout the coil.
The TANS approach incorporates elements of other E-field based TMS targeting approaches (Gomez et al., 2021; Weise et al.), including use of the SimNIBS E-field modeling software (Thielscher et al., 2015) and a grid-search procedure (evaluating many different coil positions and orientations on the scalp above the target). The TANS approach is distinguished from other targeting frameworks in two important ways. First, existing approaches attempt to maximize the E-field strength in a single spherical ROI defined in the volume. A sphere is generally ill-suited for describing the complex topology of functional networks in cortex defined at the individual level (Braga and Buckner, 2017; Gordon and Nelson, 2021b). Furthermore, because functional networks are multifocal and span multiple cortical zones (Power et al., 2011; Yeo et al., 2011), it is not known a priori where this ROI should be set for best results. Second, existing approaches do not penalize for off-target stimulation. In other words, they are designed to find a coil placement that maximizes the E-field strength at a single target location, even if that means another non-target region or network inadvertently receives equivalent or even greater stimulation.
Evaluating stimulation specificity achieved by TANS in silico
Next, we evaluated in silico how selectively TANS could stimulate one of the functional networks most often targeted in depression (based on the results shown in Figure 1D) — the frontoparietal network. Figure 3A shows the optimal coil placement found using TANS for stimulating the frontoparietal network (in either hemisphere) in each patient. Circular heatmaps describe how the on-target value changes with the coil handle orientation (the coil handle orientation displayed in Figure 3A is the one that maximized the on-target value). The E-field associated with the optimal coil placement and boundaries of the target functional network (black borders) in each patient is shown in Figure 3B. On average, 71.2% ± 12.4% of the E-field hotspot associated with the optimal coil placement was on-target. All functional networks in the E-field hotspot are shown in Figure 3C (the 99.5% threshold is used for visualization purposes). A stacked horizontal bar plot (Figure 3C) summarizes the percentage of the total E-field hotspot surface area that is occupied by each functional brain network. The same pattern was observed in the 6 non-depressed individuals (Figure S3).
Figure 3:
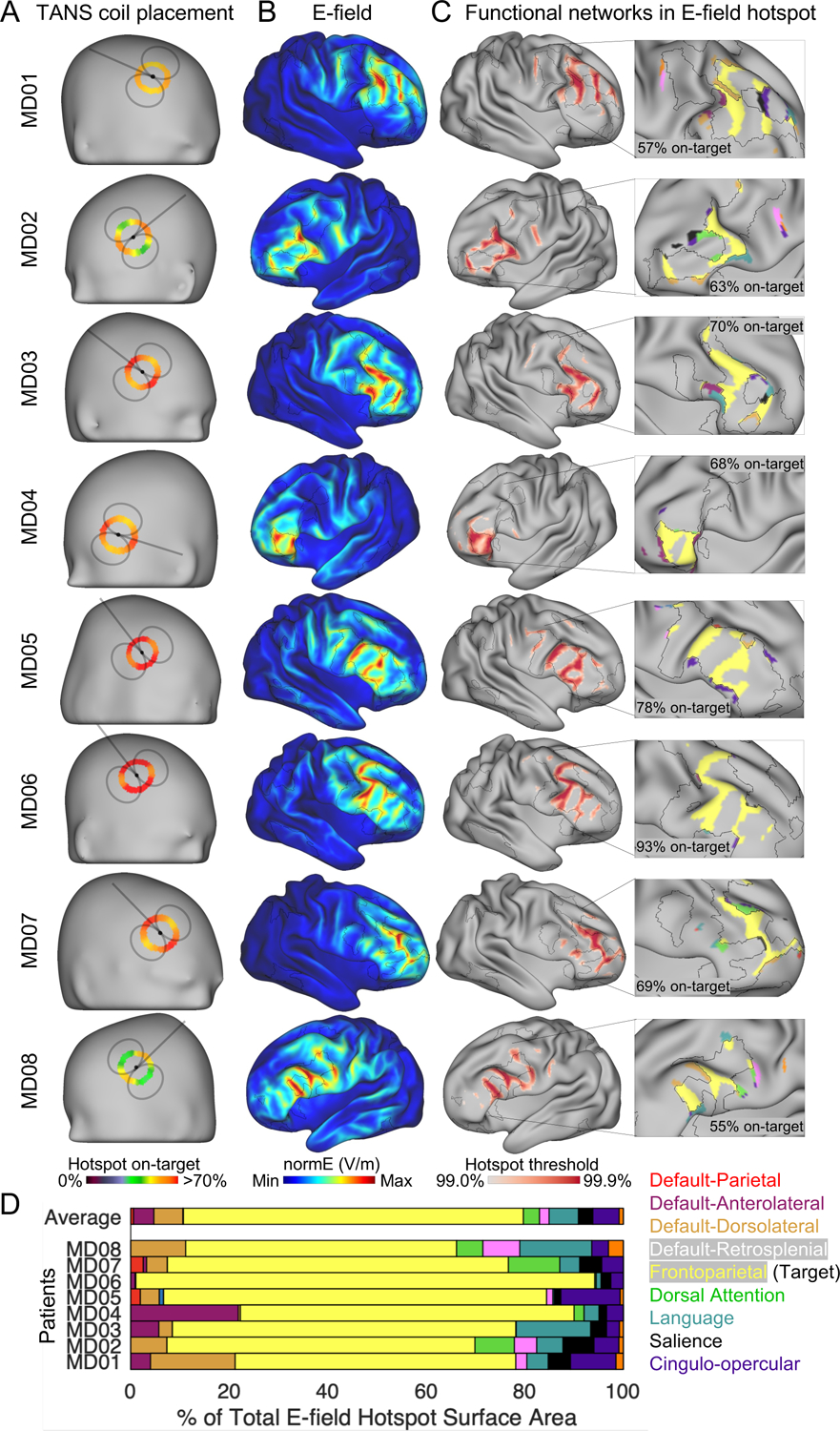
Selective stimulation of the frontoparietal network is achieved using TANS in silico in data from highly sampled patients with depression. (A) The coil placement that maximized the on-target value in each patient. The black foci represents the coil center. Circular heat maps show how the on-target value changes with coil orientation. (B) The electric field (E-field) generated by the optimal TMS coil placement. (C) The E-field hotspot defined using percentile based thresholds (99% to 99.9%, in 0.1% steps). The middle (99.5%) threshold is selected for visualizing functional networks inside the E-field hotspot in the inset. (D) Variability in functional network stimulation across patients summarized in a horizontal stacked bar graph. TANS = targeted functional network stimulation.
It is notable that the TANS approach achieved higher on-target values in some individuals than others. For example, only 57% and 55% of the hotspot was on-target in MD01 and MD08 versus 78% and 93% in MD05 and MD06. Further analysis revealed that this was largely explained by differences in the target network size across the patients (larger target network associated with better on-target values). The correlation between total surface area occupied by the target frontoparietal network cluster and on-target value was r = 0.75 (see Figure S3E), indicating that off-target stimulation occurs when the size of the E-field hotspot exceeds the largest piece of the target functional network. With this caveat in mind, the findings presented in Figure 3 suggest that TANS can be used to find a coil placement that can selectively stimulate a target functional network.
Comparison to other TMS targeting approaches
To contextualize these findings, we compared the performance of TANS to another E-field based targeting algorithm, the fast computational auxiliary dipole method (ADM; (Gomez et al., 2021). ADM is designed to quickly find a coil placement that maximizes the E-field strength in a single spherical ROI defined in the volume. ADM does not evaluate the E-field outside of this ROI, and functional networks other than the target could potentially receive equivalent or even greater levels of stimulation than the target network. These properties lead to an expectation that ADM would likely deliver more energy to the target location inside the target network, but that TANS would probably deliver more energy overall to a particular target network, and also less energy to non-target brain regions and networks.
In each individual, we first calculated the percentage of the E-field hotspot that is occupied by each functional network when using the generic (Figure 4A), ADM (Figure 4B), and TANS coil placements (Figure 4C). The relative improvement within each patient is shown in Figure 4D. TANS increased the on-target value by 51% ± 21% when compared to the generic approach (paired t-test, [t(13) = 9.61, p < 0.001]) and 31% ± 16% compared to ADM (paired t-test, [t(13) = 5.76, p < 0.001]). The same pattern was observed in the non-depressed individuals (Figure S4). The strength of E-field inside the spherical ROI (set at the target network cluster centroid) was on average 28% ± 23% greater when using ADM (paired t-test, [t(13) = 4.09, p = 0.001], see Figure 5A), indicating that the ADM approach was working as expected. We note that in some patients (for example, MD04) that difference in E-field strength at the target centroid can be attributed to the fact that the TANS coil placement delivered stimulation to a part of the functional network that was distant from its centroid.
Figure 4:
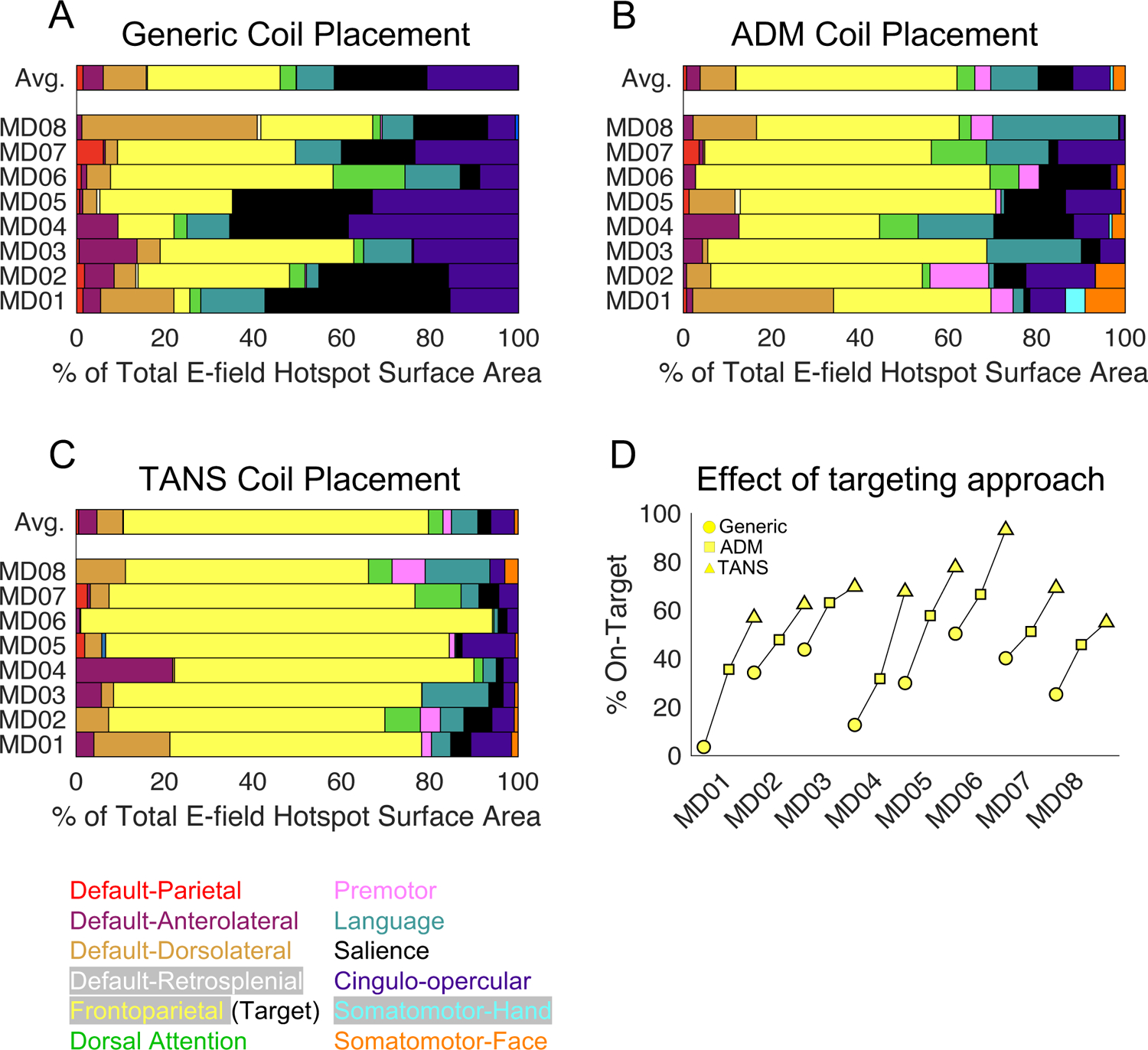
Comparing the performance of TANS in silico to two other targeting approaches in depressed patients. Variability in functional brain network stimulation when using a generic (A), ADM (B), and TANS (C) coil placement. The relative improvement in the on-target value (the proportion of the E-field hotspot aligned with the frontoparietal network) within each patient (D). TANS = targeted functional network stimulation, ADM = auxiliary dipole method. Circle = Generic, Square = ADM, Triangle = TANS.
Figure 5:
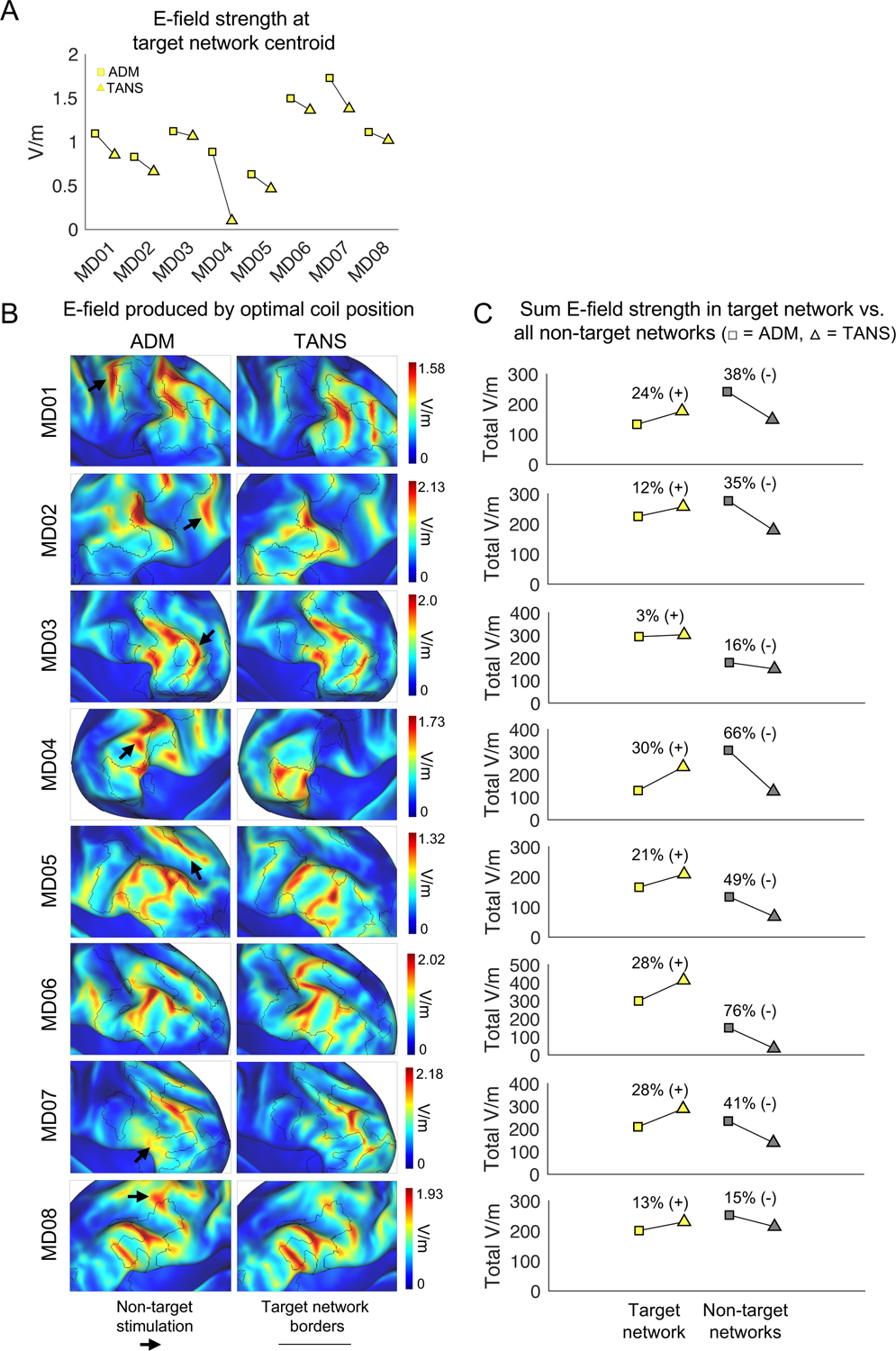
The TANS approach increases the total amount of stimulation in the target functional brain network (A) The average E-field strength inside a 5mm ROI sphere set at the centroid of the target network cluster when using ADM and TANS. Square = ADM, Triangle = TANS. (B) E-field associated with the optimal coil placements identified by ADM (left) and TANS (right). Black borders represent the boundaries of the target functional network. Black arrows highlight stimulation of non-target regions. (C) Total on-target and off-target stimulation (the sum V/m for all target and non-target network vertices inside the E-field hotspot) is shown for each patient. All simulation were performed with stimulation intensity set to dl/dt = 1 A/μ. ADM = auxiliary dipole method, TANS = targeted functional network stimulation.
Next, we calculated the total E-field strength summed across the vertices of each functional network within the E-field hotspot for each individual when using the ADM and TANS approaches. The E-fields associated with the TANS and ADM coil placements are shown on each patient’s inflated cortical surface in Figure 5B. Total on-target and off-target stimulation (the sum V/m for all target and non-target network vertices inside the E-field hotspot when dl/dt = 1 A/μs) is shown for each patient in Figure 5C. The amount of on-target stimulation increased by 24% ± 16% on average when compared to the ADM approach (paired t-test, [t(13) = 5.36, p < 0.001]). TANS also decreased off-target stimulation by 45% ± 23% on average when compared to the ADM approach (paired t-test, [t(13) = 6.26, p < 0.001]). In five of the eight patients (MD01, MD02, MD04, MD07 and MD08) and five of the six non-depressed individuals (ME01, ME03, ME04, ME05, and ME06), non-target functional networks actually received a comparable or greater level of stimulation than the target network. Data associated with the non-depressed individuals is shown in Figure S5. These findings collectively suggest that TANS and ADM can be used to address different goals. Specifically, TANS aligns where the E-field is maximal (the “hotspot”) with the target functional network. In contrast, ADM maximizes the E-field strength at a single location inside the network, which in some cases leads to simultaneous stimulation of adjacent non-target regions and networks.
Finally, we repeated our main analyses using an absolute threshold to define the suprathreshold portion of the E-field. This allowed us to evaluate the stimulation specificity that would be achieved by TANS relative to other targeting approaches given a particular stimulation intensity level and assumed neural activation threshold (Figure 6). In all 14 study participants, we calculated the percentage of the E-field hotspot (when the neural activation threshold is set to 100 V/m) that is occupied by the frontoparietal network when using the generic (Figure 6A), ADM (Figure 6B), and TANS coil placements (Figure 6C) over a range of stimulation intensity levels (from dl/dt = 1 A/μs to 155 A/μs, which corresponds approximately to the possible range of realized dl/dt on our MagPro X100 machine when using the B70 coil). The relative improvement in frontoparietal network stimulation specificity within each patient is shown in Figure 6D. TANS increased the on-target value by 50% ± 19% when compared to the generic approach (paired t-test, [t(13) = 9.27, p < 0.001]) and 32% ± 16% compared to ADM (paired t-test, [t(13) = 6.06, p < 0.001]). A similar pattern was observed in the 6 non-depressed individuals (Figure S6). We repeated this analysis using a lower absolute threshold (see right column of Figure 6) and observed a similar pattern of results, indicating that our evaluation of TANS relative to the other targeting approaches is robust to the choice of absolute threshold.
Figure 6:

Evaluating the effect of stimulation intensity on stimulation specificity achieved in silico when using TANS and two other targeting approaches in depressed patients. Two hypothetical neural activation thresholds are used, 100 V/m (left column) and 50 V/m (right column), for the generic (A), ADM (B), and TANS (C) coil placements. A range of stimulation intensities are considered (from dl/dt = 1 A/μs to 155 A/μs). (D) The relative improvement in the on-target value (the maximum proportion of the suprathreshold E-field hotspot that is aligned with the frontoparietal network) for each patient. The unique colors represent different study participants. TANS = targeted functional network stimulation, ADM = auxiliary dipole method. Circle = Generic, Square = ADM, Triangle = TANS.
Validation of the TANS approach in vivo
In our final analysis, we tested in vivo the prediction that the TANS approach can improve stimulation specificity. Single pulses of TMS administered to motor cortex (M1) can elicit a contraction in contralateral muscles, referred to as a motor evoked potential (MEP) (Bestmann and Krakauer, 2015; Wassermann et al., 1992). There are functional networks represented in M1 that can be delineated using resting-state fMRI at the individual level (Gordon et al., 2017a) that correspond to the foot, hand, and mouth regions (Barch et al., 2013; Buckner et al., 2011). We reasoned that MEPs could be used as a readout of both on-target effects (movement in target limb) and off-target effects (movement in non-target limb), and in this way test the prediction that the TANS approach can be used to more selectively engage a particular functional brain network.
TMS coil placements for targeting the somatomotor-hand and somatomotor-foot networks in three healthy individuals (see Figure 7A for ME01, study author CJL, and Figure S7A for ME02 and ME06, study authors JDP and IE) were generated using both ADM and TANS. Consistent with the modeling results reported earlier (Figure 5), E-field simulations predicted better stimulation specificity when using TANS (see white arrows in Figure 7B and the difference in total non-target stimulation summarized in Figure 7C). For this reason, we expected that TANS would be associated with less off-target stimulation than ADM – particularly when targeting the somatomotor-foot network (Figure 7C, top panel). We tested this prediction by administering TMS single pulses (intensity ranging from 35% to 80% of maximum stimulator output, 5 pulses per intensity) to the right somatomotor-hand (cyan) and somatomotor-foot (dark green) functional networks using optimal coil placements prescribed by ADM and TANS for each individual. Both the subject and the individual administering the TMS were blinded to target identity and the method used to generate each coil placement. The TMS administrator monitored visually for movements in the contralateral upper or lower limbs, and muscle contractions were confirmed using surface electrodes placed on the left first dorsal interosseus muscle and tibialis anterior muscles, which are connected to a 2-channel electromyography (EMG) device incorporated into the BrainSight neuronavigation system.
Figure 7:
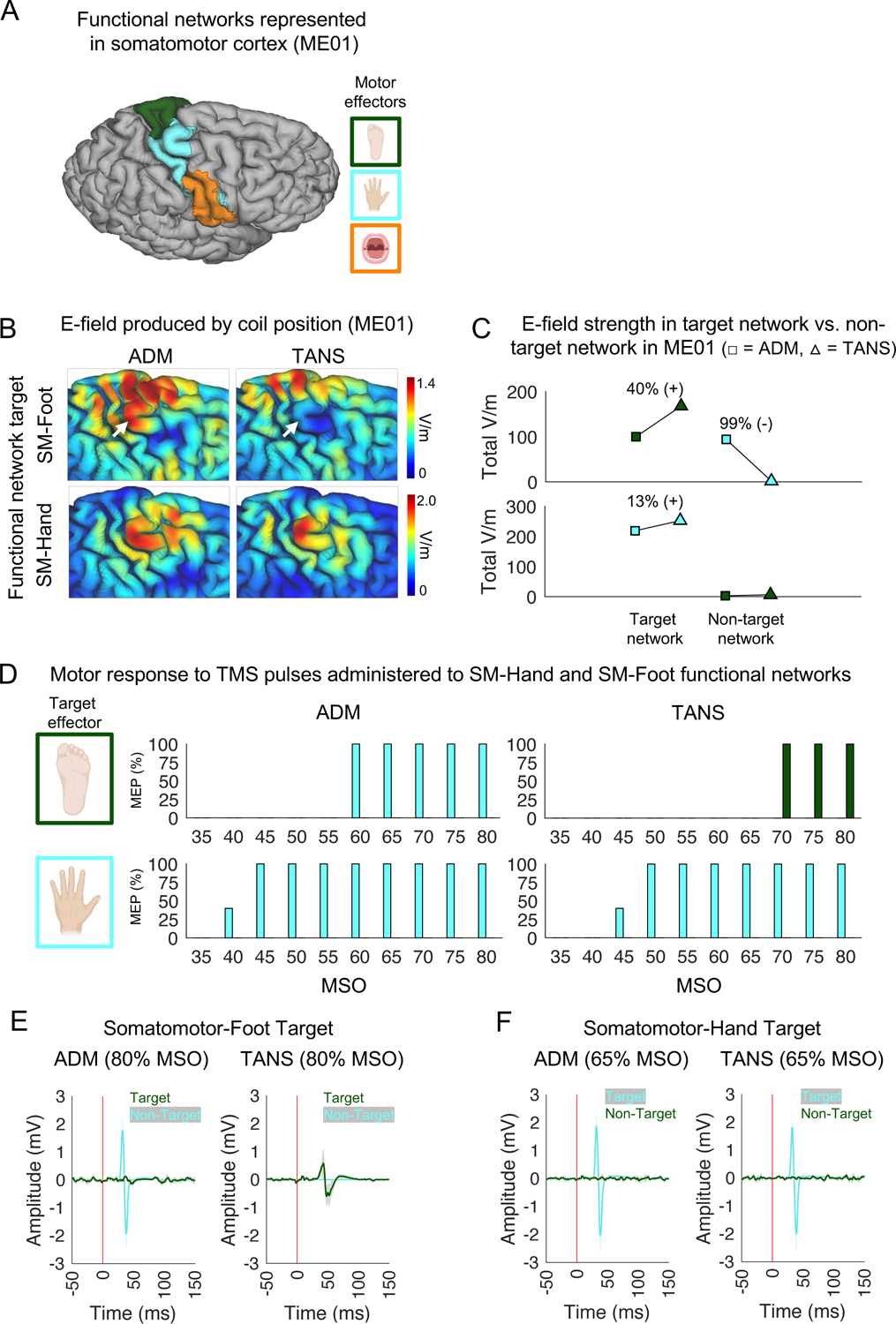
In vivo validation of the TANS approach. Adjacent somatomotor functional networks in a healthy individual (ME01) were targeted to confirm TANS can be used to selectively stimulate functional brain networks. (A) The functional networks represented in the right somatomotor cortex of ME01 include the somatomotor-foot (dark green), somatomotor-hand (cyan), and somatomotor-face (tan) networks. (B) The E-fields associated with the ADM and TANS coil placements for both the somatomotor-foot (top row) and somatomotor-hand (bottom row) network targets. For ADM, the target sphere was centered on the centroid of the target network cluster identified by TANS. White arrows highlight inadvertent stimulation of somatomotor-hand when using ADM to target somatomotor-foot. (C) The total amount of on-target and off-target stimulation (the sum V/m for all target and non-target network vertices inside the E-field hotspot) is increased and decreased, respectively, in ME01 when using TANS when compared to ADM. These simulations were performed with stimulation intensity set to dl/dt = 1 A/μs. (D) The percentage of single TMS pulses (stimulation intensity ranging from 35% to 80% of MSO, in 5% steps) delivered to somatomotor-foot and somatomotor-hand networks that produced a muscle contraction in either the contralateral lower (dark green) or upper limb (cyan). No movements in the upper limb were observed at any stimulation intensity when targeting the somatomotor-foot using TANS, hence the absence of cyan bars in the upper right panel. Similarly, movements in the lower limb were not observed at any stimulation intensity during the other three experiments, hence the absence of green bars in the other three panels. EMG recordings of the left tibialis anterior muscle and first dorsal interosseous muscle in response to single TMS pulses delivered to the somatomotor-foot (E) and somatomotor-hand (F) networks target when using the ADM and TANS coil placements. The vertical red line represents the onset of the TMS pulse. ADM = auxiliary dipole method, TANS = targeted functional network stimulation, SM = somatomotor, MSO = maximum stimulator output, EMG = Electromyography.
The results of this experiment were consistent with our predictions. In all three subjects, TANS produced a MEP in the target limb, and no movement in a non-target limb was observed at any stimulation intensity level. In contrast, ADM produced a MEP in a non-target limb in two of the three subjects, consistent with the E-field modeling results predicting non-target network stimulation in those instances. We describe the results observed in the first subject (ME01) in more detail below (see Figure 7D–F). The results for the two replication subjects (ME02 and ME06) are reported in Figure S7.
When targeting the somatomotor-foot network in ME01 (see the top row of Figure 7D and Figure 7E), only the TANS coil placement elicited MEPs in the contralateral foot, whereas the ADM coil placement elicited MEPs in the contralateral hand, as expected given the high level of off-target stimulation predicted in the top panel Figure 7C. No hand movement was observed at any intensity level when using the TANS coil placement to target the somatomotor-foot network (see top right panel of Figure 7D), suggesting that there was no off-target stimulation. In contrast, when targeting the somatomotor-hand network (Figure 7F), the ADM and TANS coil placements both elicited MEPs in the contralateral hand (see the bottom row of Figure 7D and Figure 7F) and neither elicited MEPs in the contralateral foot (at any intensity level), indicating target engagement for both methods with no off-target effects in this case. ADM actually elicited hand MEPs at a slightly lower intensity than TANS in ME01 (40% vs. 45% MSO), consistent with earlier simulations indicating that ADM can sometimes maximize stimulation intensity at a single location (Figure 5A), but at the cost of off-target stimulation (Figure 5C). A similar set of results was observed in ME02 and ME06 (see Figure S7). Collectively, these results confirm the prediction that the TANS approach improves stimulation specificity, whereas ADM is prone to also stimulating non-target brain regions or networks.
DISCUSSION
In the first part of this paper, we mapped functional brain networks precisely in 14 highly-sampled individuals (8 patients with depression and 6 healthy individuals) and found that administering TMS in a manner consistent with current clinical practice (positioning the stimulating coil directly above the same anatomical area) would likely engage different functional networks in different individuals due to variability in their functional brain organization (Figure 1). Although individual differences in functional network topology have now been characterized extensively in healthy populations (Braga and Buckner, 2017; Gordon et al., 2017a; Laumann et al., 2015), they have not been studied extensively in a clinical population, and they were critical to establishing how TMS would likely engage functional networks in patients for whom treatment with TMS is indicated. This finding led us to develop a new TMS targeting approach, referred to here as “TANS” (Targeted Functional Network Stimulation), that leverages individual-specific functional network maps and cortical folding patterns to find the coil placement that will maximize stimulation specificity (Figure 2). In simulations, TANS achieved significantly more selective stimulation of the frontoparietal network in 8 patients with depression (Figures 3–6) and 6 non-depressed individuals (Figures S3–6). In a prospective proof-of-principle demonstration, we confirmed that TANS performs as expected in vivo when targeting somatomotor functional networks corresponding to the upper and lower limbs in three healthy individuals (Figure 7 and Figure S7). Collectively, these findings demonstrate the feasibility of delineating and selectively targeting functional brain networks at the level of individual patients using TANS.
TMS is thought to alleviate symptoms of psychiatric illnesses, including major depressive disorder, by manipulating activity in the particular brain circuits responsible for them (Cash et al., 2020; Lynch and Liston, 2020; Siddiqi et al., 2019). Consistent with this possibility, (Siddiqi et al., 2019) recently found that patients with symptoms of sadness and anhedonia responded best to TMS when a functional circuit resembling the cingulo-opercular or dorsal attention network was stimulated, whereas the frontoparietal and default mode networks were more effective stimulation targets in patients with anxiety and somatic symptoms. The results of our experiment summarized in Figure 1 reveal how there are likely multiple and distinct sets of functional networks stimulated in different patients — even when coordinates corresponding to the same anatomical location are targeted — because of individual differences in how functional networks are arranged spatially in the region targeted by TMS. In general, stimulating different functional networks across patients or multiple functionally distinct networks simultaneously within a patient is not desirable to the extent it contributes to variability in the treatment response to TMS. At the same time, it is unclear at this time whether the same functional network will be the best TMS target for treating depression in all patients. Individuals with depression present with varying clinical symptom profiles, which may relate to distinct pathophysiological mechanisms (Feighner, 1981; Lynch et al., 2020b), and it might instead be beneficial to stimulate different functional networks or specific combinations of functional networks. The TANS approach should be useful independent of which functional network or networks need to be targeted in each patient, and would be especially impactful if different subtypes of depression turn out to benefit from targeting different functional networks.
A major outstanding conceptual question in this paper is whether TMS neuromodulation should prioritize energy to a fixed site or prioritize selective stimulation of a site or functional network. In certain contexts, stimulation of a fixed site would seem sensible, such as when eliciting a motor response (Bestmann and Krakauer, 2015) or interfering with processing in the visual word form area (Pattamadilok et al., 2019). If energy to a fixed site is the priority, algorithms optimizing E-field strength at a single location will serve that purpose well. However, in other contexts the specific site may be less important than the kind of tissue being targeted. The human brain is organized into large-scale, distributed networks that can be delineated in resting-state fMRI, networks that are recapitulated via coordinated coactivation in a variety of task settings (Cole et al., 2014; Power et al., 2011; Smith et al., 2009), and which are implicated in psychiatric and neurological disease. If the goal is to selectively modulate activity in a distributed multi-nodal network, our work indicates that targeting a single stereotaxic coordinate based on group-average maps or applying the ADM approach to an individualized target are prone to producing substantial off-target effects. The main contribution of the present work, from this perspective, is the development of the network-centric TANS approach.
There has been significant interest in leveraging patient-specific functional neuroanatomy to select targets for stimulation interventions (Cash et al., 2020; Gratton et al., 2019; Greene et al., 2019; Klooster et al., 2021; Lynch and Liston, 2020; Siddiqi et al., 2022). Only recently have multiple lines of research advanced sufficiently to set the stage for the TMS targeting framework introduced in this paper. Early efforts using individual differences in functional connectivity to guide stimulation site selection were challenging due in part to the poor signal characteristics and limited reliability associated with smaller quantities of fMRI data (Fox et al., 2013). Acquiring larger amounts of fMRI data from single individuals as opposed to smaller quantities of data from many individuals has enabled reliably delineating functional networks at the individual level for the first time (Gordon et al., 2017a; Laumann et al., 2015; Poldrack et al., 2015). When combined with a multi-echo acquisition and denoising framework (Kundu et al., 2017; Lynch et al., 2020a, 2021), reliable individual-specific functional network maps may be obtained in cortex from clinically feasible quantities of data per-patient (approximately 30-minutes, see Figure S1A-B) and used to guide stimulation interventions (Gratton et al., 2019). A sufficient amount of high-quality fMRI data per-patient is a prerequisite for precision functional mapping. In parallel, the development of easy-to-use tools for computational modeling of the E-field created in the brain by TMS (Thielscher et al., 2015) allows prospectively determining which brain regions will likely be engaged by a particular coil placement. These advancements have made personalizing the administration of TMS to patient-specific functional neuroanatomy more feasible.
The TANS approach has the potential to be useful in basic research settings as well. For example, TMS is commonly used in cognitive neuroscience to study the cognitive or behavioral effects of modulating activity within particular brain networks or regions (Sandrini et al., 2011). These experiments often control for the non-specific effects of TMS with an active control that can produce comparable scalp and auditory sensations, but different transcranial brain stimulation (Bergmann et al., 2021). In principle, the ideal active control should be a functionally distinct area that is close in proximity to the target of interest. However, the closer the active control is to the target of interest, the more difficult it becomes to selectively stimulate the two sites. A useful feature of the TANS approach is the option to specify avoidance regions, which will help ensure that the two brain areas are selectively stimulated. The results of the validation experiment (see Figure 7 and Figure S7) — where two somatomotor functional networks were selectively engaged using TANS — are consistent with this possibility. By enabling more specific stimulation of networks or regions of interest, while avoiding other regions or networks, TANS has the potential to facilitate the design of more sophisticated TMS experiments.
Several aspects of this work warrant additional consideration. First, the modeling experiments performed in this investigation indicate that selective stimulation of a functional network with TMS requires that the network has sufficient size and representation on a gyral crown (Figure S3E). Smaller networks or those more often represented in a fissure or on the medial surface of the brain, such as the cingulo-opercular network, may be less amenable to stimulation than larger networks with prominent representations on the lateral surface of the brain, such as the frontoparietal network. A related point is that the geometry of the coil (here we used the figure-of-eight MagVenture B70 coil) will determine in part how precisely TMS can be used to stimulate a given functional network (Deng et al., 2014; Hanlon, 2017). For example, coils with a double-cone geometry will create a stronger but also less focal E-field when compared to a figure-of-eight style coil (Deng et al., 2013; Schecklmann et al., 2020). Second, the TANS approach involves aligning spatial characteristics of the E-field (specifically, where it is maximal) with a specified target functional network, which alone may not be sufficient to predict a neuronal response to TMS (Aberra et al., 2019; Casula et al., 2018; Hannah and Rothwell, 2017; Peterchev et al., 2012). Other temporal parameters (such as the stimulation waveform, intensity, and frequency) and biological factors (neural activation thresholds) must also be considered after the optimal coil placement has been identified to achieve the desired effect on the targeted brain circuit. Finally, the majority of the analyses reported here are simulations (with the exception of the TMS experiments summarized in Figure 7 and Figure S7), and no prospective application of TANS was performed therapeutically in patients in order to evaluate whether the TANS approach would actually improve treatment outcomes. Future studies involving prospective targeting of functional networks using tools such as TANS will be an important next step towards validating the use of E-field modeling combined with precision mapping as a way to personalize the administration of TMS to better account for patient-specific functional neuroanatomy.
There is an unmet need for mental health treatments that are personalized to biological characteristics of individual patients (Insel and Cuthbert, 2015; Williams, 2016). Precise delineation and targeting of functional networks at the individual patient level has the potential to enhance treatment response in the growing number of clinical populations where TMS is indicated. Here, we demonstrated that patient-specific functional networks can be more selectively targeted in silico and in vivo using a new TMS targeting approach, termed TANS. As a part of this NeuroResource article, Matlab code for implementing TANS and example data from the highly sampled non-depressed individuals are provided as a public resource (Lynch, 2022).
STAR Methods
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Conor Liston (col2004@med.cornell.edu)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Matlab code for implementing the TANS algorithm has been deposited online (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation; DOI: 10.5281/zenodo.6958500). Code for preprocessing multi-echo fMRI data is maintained in a separate online repository (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline; DOI: 10.5281/zenodo.6958611).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Participants
The study sample consisted of five healthy individuals (N=6 males, mean age = 34.6 ± 9.3 years), four of whom were previously a part of separate investigation (Lynch et al., 2020a), and 8 patients (N=3 females, mean age = 34.6 ± 7.06) with a diagnosis of a mood disorder (N=6 with major depression disorder and N=2 bipolar disorder II). These diagnoses were based on the DSM-IV-TR criteria and confirmed by the Mini-International Neuropsychiatric Interview administered by a trained clinician. Seven of the eight patients were stably medicated and taking an antidepressant medication (SSRI, SNRI, or Bupropion). Two patients were additionally taking an augmentation agent (Lithium or Lamotrigine). Four patients were taking Benzodiazepines. One MDD patient received Lamotrigine monotherapy. One depressed patient was not taking medications. These eight patients were scanned longitudinally (for up to 1.5 years), with fluctuating mood symptoms during this time period (moderate to severe symptoms defined as a HAMD6 score greater than or equal to 5 present during 77% of the total study visits), and received treatment with their psychiatrists outside the context of the present study.
METHOD DETAILS
MRI image acquisition
Data were acquired on a Siemens Magnetom Prisma 3T scanner at the Citigroup Biomedical Imaging Center of Weill Cornell’s medical campus using a Siemens 32-channel head coil. Multi-echo, multi-band resting-state fMRI scans were collected using a T2*-weighted echo-planar sequence covering the full brain (TR: 1355 ms; TE1: 13.40 ms, TE2: 31.11 ms, TE3: 48.82 ms, TE4: 66.53 ms, and TE5: 84.24 ms; FOV: 216 mm; flip angle: 68° (the Ernst angle for gray matter assuming a T1 value of 1400 ms); 2.4 mm isotropic voxels; 72 slices; AP phase encoding direction; in-plane acceleration factor: 2; and multi-band acceleration factor: 6) with 640 volumes acquired per scan for a total acquisition time of 14 minutes and 27 seconds. This sequence was generously provided by the Center for Magnetic Resonance Research (CMRR) at the University of Minnesota. Spin echo EPI images with opposite phase encoding directions (AP and PA) but identical geometrical parameters and echo spacing were acquired before each resting-state scan. Multi-echo T1-weighted (TR/TI: 2500/1000 ms; TE1: 1.7 ms, TE2: 3.6 ms, TE3: 5.5 ms, TE4: 7.4 ms; FOV: 256; flip angle: 8°, and 208 sagittal slices with a 0.8 mm slice thickness) and T2-weighted anatomical images (TR: 3200 ms; TE: 563 ms; FOV: 256; flip angle: 8°, and 208 sagittal slices with a 0.8 mm slice thickness) were acquired.
Anatomical preprocessing and cortical surface generation
Anatomical data were preprocessed and cortical surfaces generated using the Human Connectome Project (HCP) PreFreeSurfer, FreeSurfer, and PostFreeSurfer pipelines (version 4.3).
Multi-echo fMRI preprocessing
Preprocessing of multi-echo data minimized spatial interpolation and volumetric smoothing while preserving the alignment of echoes. The single-band reference (SBR) images (five total; one per echo) for each scan were averaged. The resultant average SBR images were aligned, averaged, co-registered to the ACPC aligned T1-weighted anatomical image, and simultaneously corrected for spatial distortions using FSL’s topup and epi_reg programs. Freesurfer’s bbregister algorithm (Greve and Fischl, 2009) was used to refine this co-registration. For each scan, echoes were combined at each timepoint and a unique 6 DOF registration (one per volume) to the average SBR image was estimated using FSL’s MCFLIRT tool (Jenkinson et al., 2002), using a 4-stage (sinc) optimization. All of these steps (co-registration to the average SBR image, ACPC alignment, and correcting for spatial distortions) were concatenated using FSL’s convertwarp tool and applied as a single spline warp to individual volumes of each echo after correcting for slice time differences using FSL’s slicetimer program. The functional images underwent a brain extraction using the co-registered brain extracted T1-weighted anatomical image as a mask and corrected for signal intensity inhomogeneities using ANT’s N4BiasFieldCorrection tool. All denoising was performed on preprocessed, ACPC-aligned images.
Multi-echo fMRI denoising
Multi-echo ICA (ME-ICA; (Kundu et al., 2013)) denoising designed to isolate spatially structured T2*- (neurobiological; “BOLD-like”) and S0-dependent (non-neurobiological; “not BOLD-like”) signals was performed using the “tedana.py” workflow (DuPre et al., 2020). In short, the preprocessed, ACPC-aligned echoes were first combined according to the average rate of T2* decay at each voxel across all time points by fitting the monoexponential decay, S(t) = S0e −t / T2*. From these T2* values, an optimally-combined multi-echo (OC-ME) time-series was obtained by combining echoes using a weighted average (WTE = TE * e −TE/ T2*), as in (Posse et al., 1999). The covariance structure of all voxel time-courses was used to identify major signals in the OC-ME time-series using principal component and independent component analysis. Components were classified as either T2*-dependent (and retained) or S0-dependent (and discarded), primarily according to their decay properties across echoes. All component classifications were manually reviewed by author CJL and revised when necessary following the criteria described in (Griffanti et al., 2017). Mean gray matter time-series regression was performed to remove spatially diffuse noise. Temporal masks were generated for censoring high motion time-points using a framewise displacement (FD; (Power et al., 2012)) threshold of 0.3 mm and a backward difference of two TRs (2 * 1.355 = 2.77 seconds), for an effective sampling rate comparable to historical FD measurements (approximately 2 to 4 seconds). Prior to the FD calculation, head realignment parameters were filtered using a stopband Butterworth filter (0.2 – 0.35 Hz) to attenuate the influence of respiration (Power et al., 2019) on motion parameters.
Surface processing and CIFTI generation of fMRI data
The denoised fMRI time-series was mapped to the midthickness surfaces (“wb_command -volume-to-surface-mapping”, using the “-ribbon-constrained” method), combined into the Connectivity Informatics Technology Initiative (CIFTI) format, and spatially smoothed with geodesic (for surface data) and Euclidean (for volumetric data) Gaussian kernels (σ = 2.55 mm) using Connectome Workbench command line utilities (Smith et al., 2013). This yielded time courses representative of the entire cortical surface, subcortex (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, brainstem), and cerebellum, but excluding non-gray matter tissue. Spurious coupling between subcortical voxels and adjacent cortical tissue was mitigated by regressing the average time-series of cortical tissue < 20 mm in Euclidean space from a subcortical voxel.
Precision mapping of functional brain networks
A functional connectivity matrix summarizing the correlation between the time-courses of all cortical vertices and subcortical voxels across all study visits was constructed. Correlations between nodes less than 10 mm apart (geodesic and Euclidean space used for cortico-cortical and subcortical-cortical distance, respectively) were set to zero. Correlations between voxels belonging to subcortical structures were set to zero. Functional connectivity matrices were thresholded in such a way that they retained at least the strongest X% correlations (0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, and 5%) to each vertex and voxel and were used as inputs for the InfoMap community detection algorithm (Rosvall and Bergstrom, 2008). The optimal scale for further analysis was defined as the graph threshold producing the best size-weighted average homogeneity relative to the median of the size-weighted average homogeneity calculated from randomly rotated networks, as done in (Gordon et al., 2020). Size-weighted average homogeneity was found to be maximized relative to randomly rotated communities at the 0.1% graph density. The communities at this threshold were manually reviewed and functional network identities were assigned by study author CJL.
Evaluating functional networks stimulated when using a generic target
Electric field modeling was performed using SimNIBS (version 3.2). High-resolution tetrahedral head meshes (0.5 vertices per mm) were created from co-registered T1-weighted and T2-weighted anatomical images using headreco (Nielsen et al., 2018). The coil center was positioned directly above the stimulation site. The first stimulation site tested was obtained by converting the MNI152 coordinates X=−42, Y=44, Z=30 used in (Weigand et al., 2018) to each individual’s native volume space using FSL’s img2imgcoord tool. These coordinates correspond to peak subgenual cingulate anticorrelation in independent group-averaged data.The stimulation intensity and coil-to-scalp distance was set to the default values of dl/dt = 1 A/μs and 4 mm, respectively. The prolongation of the coil handle was oriented posteriorly in the direction of the CP1 landmark from the 10–20 EEG system. The norm of the electric field was mapped to the midthickness surfaces (“wb_command -volume-to-surface-mapping”, using the “-ribbon-constrained” method) and combined into the CIFTI format. The E-field hotspot was defined using a range of percentile thresholds (99% to 99.9%, in 0.1% steps).
Head model construction
A high-resolution tetrahedral headmesh (0.5 vertices per mm) was created from the co-registered T1-weighted and T2-weighted anatomical images using headreco (Nielsen et al., 2018). The skin mesh was converted from STL to GIFTI format using FreeSurfer’s mris_convert program. The skin mesh was smoothed for visualization purposes (“wb_command -surface-smoothing” with smoothing strength set to 0.25 and 500 total iterations). A matrix summarizing the distance in geodesic space between all skin vertices was constructed using Connectome Workbench command line utilities (“wb_command -surface-geodesic-distance”). These steps are all performed by the “tans_headmodels.m” function.
Region-of-interest selection and search grid construction
All elements of the target network amenable to stimulation (operationalized as vertices on a gyral crown) are identified. Medial wall vertices are removed from consideration. Optionally, a search space (e.g., a group of gryal labels from the Desikan-Killiany Atlas (Desikan et al., 2006)) can be specified in addition to avoid considering locations that are not of interest. The largest cluster of target network vertices retained after sulcus masking within this search space was identified and is hereafter referred to simply as the target network region-of-interest (ROI). These steps are performed by the “tans_roi.m” function. Next, all skin mesh vertices within a specified distance in Euclidean space (default is 40 mm) of the ROI centroid vertex were identified. The three-dimensional coordinates associated with these vertices formed the search grid. Search grid density is decreased to 2 mm to reduce the total runtime. These steps are performed by the “tans_searchgrid.m” function.
E-field modeling
A TMS simulation is performed at each point in the search grid using SimNibs (Thielscher et al., 2015). Each simulation involves a number of coil orientations determined by an angle (in degrees) specified by the user. The stimulation intensity and coil-to-scalp distance was set to the default values of dl/dt = 1 A/μs and 4 mm, respectively. We note that a fixed stimulation intensity is used during this stage of TANS because the strength of the E-field varies linearly with dl/dt (the speed of variation of the current throughout the coil) and, for this reason, has no effect on its spatial distribution (including where it is maximal relative to the target network). The coil model used in the simulation is also specified by the user (the MagVenture B70 coil model was used for all analyses reported here). These steps are performed by the “tans_simnibs.m” function.
Evaluating the quality of coil positions in the search grid
The quality of each coil placement in the search grid was evaluated by taking the total surface area of all target network vertices inside the hotspot in relation to the total surface area of the hotspot. This measure is referred to as the percentage of the hotspot that is on-target. After the optimal coil placement is identified, a range of stimulation intensities are modeled and the portion of the suprathreshold E-field (≥ 1000 mm2) that is on-target at each intensity level (given some absolute threshold representing a neural activation threshold, default value is set to 100 V/m, following (Tendler et al., 2016)) is calculated. These steps are performed by the “tans_optimize.m” function.
Comparison with other E-field targeting approaches
Performance of the targeted functional network stimulation (TANS) algorithm was compared to the fast computational auxiliary dipole method (ADM; (Gomez et al., 2021)). Default parameters for ADM were used. The 5mm ROI sphere was centered on the centroid of the target network cluster identified in step 1 of TANS.
Targeting somatomotor functional networks in vivo using TANS
The somatomotor-hand and somatomotor-foot functional networks in three healthy male adults (ME01: study author CJL, ME02: study author JDP, ME06: study author IE) were targeted with single TMS pulses using coil placements prescribed by the ADM and TANS approaches. For TANS, the search space was set to the right primary motor cortex and the non-target somatomotor network was set as an avoidance region. For ADM, the target location was set to the centroid of each somatomotor network cluster. The Brainsight 2 Frameless stereotactic system for image guided TMS research (Rogue Research). This system uses infrared reflectors attached to a headband worn by the subject to coregister the T1w image with the participant’s head. Single pulses of TMS were applied using a MagPro x100 device (MagVenture, Inc.) and an actively cooled B70 coil. Stimulation was delivered using the MAGnetic stimulator Interface Controller (MAGIC) toolbox (Saatlou et al., 2018). The stimulation intensity (ranging from 30% to 80% of maximum stimulator output, in 5% steps) and inter-pulse interval (between 4 and 8 seconds) was randomized prior to each run. There were 5 pulses total per intensity. Muscle contractions in the contralateral upper and lower limb were monitored for visually and confirmed using surface electrodes (on the left first dorsal interosseous and tibialis anterior muscle, respectively) connected to a 2-channel electromyography (EMG) device incorporated into the BrainSight system. Both the subject and the individuals administering TMS (either study author CJL or IE) were blinded to target identity and the method used to generate each coil placement.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and Algorithms | ||
| MATLAB | MathWorks | https://www.mathworks.com/ |
| Connectome Workbench | (Marcus et al., 2013) | https://www.humanconnectome.org/software/connectome-workbench |
| Tedana | (DuPre et al., 2020) | https://tedana.readthedocs.io/en/latest/ |
| FreeSurfer | (Fischl, 2012) | https://surfer.nmr.mgh.harvard.edu/ |
| Infomap | (Rosvall and Bergstrom, 2008) | http://www.mapequation.org/ |
| Targeted Functional Network Stimulation (TANS) | This manuscript | https://github.com/cjl2007/Targeted-Functional-Network-Stimulation; DOI: 10.5281/zenodo.6958500 |
Highlights.
Current TMS targeting approaches stimulate different functional networks across patients.
A method for selective stimulation of patient-specific functional networks is introduced.
Targeted Functional Network Stimulation (“TANS”) improves stimulation specificity.
Code for implementing TANS and an example dataset is provided as a public resource.
Acknowledgements:
We thank the staff at the Citigroup Biomedical Imaging Center for assistance with data collection. Dr. Zhi Deng provided comments on E-field modeling procedures and shared certain TMS coil files. This work was supported by grants to C.L. from the National Institute of Mental Health, the National Institute on Drug Addiction, the Hope for Depression Research Foundation, and the Foundation for OCD Research. C.J.L. was supported by an NIMH F32 National Research Service Award (F32MH120989).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest: C.L. is listed as an inventor for Cornell University patent applications on neuroimaging biomarkers for depression that are pending or in preparation. C.L. has served as a scientific advisor or consultant to Compass Pathways PLC, Delix Therapeutics, Magnus Medical, and Brainify.AI. The authors declare no other competing interests.
Inclusion and Diversity. We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. One or more authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
REFERENCES
- Aberra A, Grill W, and Peterchev A (2019). Simulation of controllable pulse parameter transcranial magnetic stimulation in realistic head model with morphologically-accurate cortical neurons. Brain Stimulation 12, 589. 10.1016/j.brs.2018.12.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery DH, Isenberg KE, Sampson SM, Janicak PG, Lisanby SH, Maixner DF, Loo C, Thase ME, Demitrack MA, and George MS (2008). Transcranial magnetic stimulation in the acute treatment of major depressive disorder: clinical response in an open-label extension trial. J. Clin. Psychiatry 69, 441–451. [DOI] [PubMed] [Google Scholar]
- Baeken C, Marinazzo D, Wu G-R, Van Schuerbeek P, De Mey J, Marchetti I, Vanderhasselt M-A, Remue J, Luypaert R, and De Raedt R (2014). Accelerated HF-rTMS in treatment-resistant unipolar depression: Insights from subgenual anterior cingulate functional connectivity. World J. Biol. Psychiatry 15, 286–297. [DOI] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, et al. (2013). Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam W, Borckardt JJ, Reeves ST, and George MS (2009). An efficient and accurate new method for locating the F3 position for prefrontal TMS applications. Brain Stimul. 2, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann TO, Varatheeswaran R, Hanlon CA, Madsen KH, Thielscher A, and Siebner HR (2021). Concurrent TMS-fMRI for causal network perturbation and proof of target engagement. Neuroimage 118093. [DOI] [PubMed] [Google Scholar]
- Bestmann S, and Krakauer JW (2015). The uses and interpretations of the motor-evoked potential for understanding behaviour. Exp. Brain Res 233, 679–689. [DOI] [PubMed] [Google Scholar]
- Blumberger DM, Vila-Rodriguez F, Thorpe KE, Feffer K, Noda Y, Giacobbe P, Knyahnytska Y, Kennedy SH, Lam RW, Daskalakis ZJ, et al. (2018). Effectiveness of theta burst versus high-frequency repetitive transcranial magnetic stimulation in patients with depression (THREE-D): a randomised non-inferiority trial. Lancet 391, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Braga RM, and Buckner RL (2017). Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron 95, 457–471.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga RM, Van Dijk KRA, Polimeni JR, Eldaief MC, and Buckner RL (2019). Parallel distributed networks resolved at high resolution reveal close juxtaposition of distinct regions. Journal of Neurophysiology 121, 1513–1534. 10.1152/jn.00808.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, and Yeo BTT (2011). The organization of the human cerebellum estimated by intrinsic functional connectivity. J. Neurophysiol 106, 2322–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungert A, Antunes A, Espenhahn S, and Thielscher A (2017). Where does TMS Stimulate the Motor Cortex? Combining Electrophysiological Measurements and Realistic Field Estimates to Reveal the Affected Cortex Position. Cereb. Cortex 27, 5083–5094. [DOI] [PubMed] [Google Scholar]
- Carpenter LL, Janicak PG, Aaronson ST, Boyadjis T, Brock DG, Cook IA, Dunner DL, Lanocha K, Brent Solvason H, and Demitrack MA (2012). Transcranial magnetic stimulation (TMS) for major depression: A multisite, naturalistic, observational study of acute treatment outcomes in clinical practice. Depression and Anxiety 29, 587–596. 10.1002/da.21969. [DOI] [PubMed] [Google Scholar]
- Cash RFH, Zalesky A, Thomson RH, Tian Y, Cocchi L, and Fitzgerald PB (2019). Subgenual functional connectivity predicts antidepressant treatment response to transcranial magnetic stimulation: independent validation and evaluation of personalization. Biol. Psychiatry 86, e5–e7. [DOI] [PubMed] [Google Scholar]
- Cash RFH, Weigand A, Zalesky A, Siddiqi SH, Downar J, Fitzgerald PB, and Fox MD (2020). Using brain imaging to improve spatial targeting of TMS for depression. Biol. Psychiatry [DOI] [PubMed] [Google Scholar]
- Cash RFH, Weigand A, Zalesky A, Siddiqi SH, Downar J, Fitzgerald PB, and Fox MD (2021). Using Brain Imaging to Improve Spatial Targeting of Transcranial Magnetic Stimulation for Depression. Biol. Psychiatry 90, 689–700. [DOI] [PubMed] [Google Scholar]
- Casula EP, Rocchi L, Hannah R, and Rothwell JC (2018). Effects of pulse width, waveform and current direction in the cortex: A combined cTMS-EEG study. Brain Stimul. 11, 1063–1070. [DOI] [PubMed] [Google Scholar]
- Cocchi L, and Zalesky A (2018). Personalized Transcranial Magnetic Stimulation in Psychiatry. Biol Psychiatry Cogn Neurosci Neuroimaging 3, 731–741. [DOI] [PubMed] [Google Scholar]
- Cole EJ, Phillips AL, Bentzley BS, Stimpson KH, Nejad R, Barmak F, Veerapal C, Khan N, Cherian K, Felber E, et al. (2022). Stanford Neuromodulation Therapy (SNT): A Double-Blind Randomized Controlled Trial. Am. J. Psychiatry 179, 132–141. [DOI] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, and Petersen SE (2014). Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly KR, Helmer A, Cristancho MA, Cristancho P, John PO, and Others (2012). Effectiveness of transcranial magnetic stimulation in clinical practice post-FDA approval in the United States: results observed with the first 100 consecutive cases of depression at an academic medical center. J. Clin. Psychiatry 73, 5611. [DOI] [PubMed] [Google Scholar]
- Deng Z-D, Lisanby SH, and Peterchev AV (2013). Electric field depth–focality tradeoff in transcranial magnetic stimulation: Simulation comparison of 50 coil designs. Brain Stimul. 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z-D, Lisanby SH, and Peterchev AV (2014). Coil design considerations for deep transcranial magnetic stimulation. Clin. Neurophysiol 125, 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. [DOI] [PubMed] [Google Scholar]
- DuPre E, Salo T, Markello R, Kundu P, Whitaker K, and Handwerker D (2020). ME-ICA/tedana: 0.0. 9a. Zenodo. [Google Scholar]
- Eldaief MC, Halko MA, Buckner RL, and Pascual-Leone A (2011). Transcranial magnetic stimulation modulates the brain’s intrinsic activity in a frequency-dependent manner. Proc. Natl. Acad. Sci. U. S. A 108, 21229–21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feighner JP (1981). Nosology of primary affective disorders and application to clinical research. Acta Psychiatr. Scand. Suppl 290, 29–41. [DOI] [PubMed] [Google Scholar]
- Fischl B (2012). FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Buckner RL, White MP, Greicius MD, and Pascual-Leone A (2012). Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol. Psychiatry 72, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Liu H, and Pascual-Leone A (2013). Identification of reproducible individualized targets for treatment of depression with TMS based on intrinsic connectivity. Neuroimage 66, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynes BN, Lloyd SW, Lux L, Gartlehner G, Hansen RA, Brode S, Jonas DE, Swinson Evans T, Viswanathan M, and Lohr KN (2014). Repetitive transcranial magnetic stimulation for treatment-resistant depression: a systematic review and meta-analysis. J. Clin. Psychiatry 75, 477–489; quiz 489. [DOI] [PubMed] [Google Scholar]
- Gomez LJ, Dannhauer M, and Peterchev AV (2021). Fast computational optimization of TMS coil placement for individualized electric field targeting. Neuroimage 228, 117696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, and Nelson SM (2021a). Three types of individual variation in brain networks revealed by single-subject functional connectivity analyses. Current Opinion in Behavioral Sciences 40, 79–86. [Google Scholar]
- Gordon E, and Nelson S (2021b). Three types of individual variation in brain networks revealed by single-subject functional connectivity analyses. Current Opinion in Behavioral Sciences. [Google Scholar]
- Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt-Drazen C, Gratton C, Sun H, et al. (2017a). Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NUF, and Petersen SE (2017b). Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage 146, 918–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Marek S, Raut RV, Gratton C, Newbold DJ, Greene DJ, Coalson RS, Snyder AZ, Schlaggar BL, et al. (2020). Default-mode network streams for coupling to language and control systems. Proc. Natl. Acad. Sci. U. S. A 117, 17308–17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Marek S, and Newbold DJ (2021). Human Fronto-Striatal Connectivity is Organized into Discrete Functional Subnetworks. bioRxiv. [Google Scholar]
- Gratton C, Kraus BT, Greene DJ, Gordon EM, Laumann TO, Nelson SM, Dosenbach NUF, and Petersen SE (2019). Defining Individual-Specific Functional Neuroanatomy for Precision Psychiatry. Biological Psychiatry 10.1016/j.biopsych.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DJ, Marek S, Gordon EM, Siegel JS, Gratton C, Laumann TO, Gilmore AW, Berg JJ, Nguyen AL, Dierker D, et al. (2019). Integrative and Network-Specific Connectivity of the Basal Ganglia and Thalamus Defined in Individuals. Neuron 10.1016/j.neuron.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, and Fischl B (2009). Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Douaud G, Bijsterbosch J, Evangelisti S, Alfaro-Almagro F, Glasser MF, Duff EP, Fitzgibbon S, Westphal R, Carone D, et al. (2017). Hand classification of fMRI ICA noise components. Neuroimage 154, 188–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, and Corbetta M (2013). Resting state network estimation in individual subjects. Neuroimage 82, 616–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halko MA, Farzan F, Eldaief MC, Schmahmann JD, and Pascual-Leone A (2014). Intermittent theta-burst stimulation of the lateral cerebellum increases functional connectivity of the default network. J. Neurosci 34, 12049–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon C (2017). Blunt or precise? A note about the relative precision of figure-of-eight rTMS coils. Brain Stimul. 10, 338–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon CA, Dowdle LT, Austelle CW, DeVries W, Mithoefer O, Badran BW, and George MS (2015). What goes up, can come down: Novel brain stimulation paradigms may attenuate craving and craving-related neural circuitry in substance dependent individuals. Brain Res. 1628, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah R, and Rothwell JC (2017). Pulse Duration as Well as Current Direction Determines the Specificity of Transcranial Magnetic Stimulation of Motor Cortex during Contraction. Brain Stimul. 10, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman HJ, Chan SMS, Chu WCW, Lu H, Tse C-Y, Chau SWH, Lam LCW, Mak ADP, and Neggers SFW (2021). Personalized prediction of transcranial magnetic stimulation clinical response in patients with treatment-refractory depression using neuroimaging biomarkers and machine learning. J. Affect. Disord 290, 261–271. [DOI] [PubMed] [Google Scholar]
- Insel TR, and Cuthbert BN (2015). Medicine. Brain disorders? Precisely. Science 348, 499–500. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, and Smith S (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841. [DOI] [PubMed] [Google Scholar]
- Klooster DCW, Ferguson MA, Boon PAJM, and Baeken C (2021). Personalizing Repetitive Transcranial Magnetic Stimulation Parameters for Depression Treatment Using Multimodal Neuroimaging. Biol Psychiatry Cogn Neurosci Neuroimaging 10.1016/j.bpsc.2021.11.004. [DOI] [PubMed] [Google Scholar]
- Kraus BT, Perez D, Ladwig Z, Seitzman BA, Dworetsky A, Petersen SE, and Gratton C (2021). Network variants are similar between task and rest states. Neuroimage 229, 117743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Brenowitz ND, Voon V, Worbe Y, Vértes PE, Inati SJ, Saad ZS, Bandettini PA, and Bullmore ET (2013). Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc. Natl. Acad. Sci. U. S. A 110, 16187–16192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Voon V, Balchandani P, Lombardo MV, Poser BA, and Bandettini PA (2017). Multi-echo fMRI: A review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage 154, 59–80. [DOI] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen M-Y, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NUF, et al. (2015). Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefaucheur J-P, Aleman A, Baeken C, Benninger DH, Brunelin J, Di Lazzaro V, Filipović SR, Grefkes C, Hasan A, Hummel FC, et al. (2020). Evidence-based guidelines on the therapeutic use of repetitive transcranial magnetic stimulation (rTMS): An update (2014–2018). Clin. Neurophysiol 131, 474–528. [DOI] [PubMed] [Google Scholar]
- Luborzewski A, Schubert F, Seifert F, Danker-Hopfe H, Brakemeier E-L, Schlattmann P, Anghelescu I, Colla M, and Bajbouj M (2007). Metabolic alterations in the dorsolateral prefrontal cortex after treatment with high-frequency repetitive transcranial magnetic stimulation in patients with unipolar major depression. Journal of Psychiatric Research 41, 606–615. 10.1016/j.jpsychires.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lynch CJ (2022). Targeted Functional Network Stimulation. 10.5281/zenodo.6958500 [DOI] [Google Scholar]
- Lynch CJ, and Liston C (2020). Precision Functional Mapping of Corticostriatal and Corticothalamic Circuits: Parallel Processing Reconsidered. Neuron 105, 595–597. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Breeden AL, Gordon EM, Cherry JBC, Turkeltaub PE, and Vaidya CJ (2018). Precision Inhibitory Stimulation of Individual-Specific Cortical Hubs Disrupts Information Processing in Humans. Cereb. Cortex 10.1093/cercor/bhy270. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Power JD, Scult MA, Dubin M, Gunning FM, and Liston C (2020a). Rapid Precision Functional Mapping of Individuals Using Multi-Echo fMRI. Cell Rep. 33, 108540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, Gunning FM, and Liston C (2020b). Causes and Consequences of Diagnostic Heterogeneity in Depression: Paths to Discovering Novel Biological Depression Subtypes. Biol. Psychiatry 10.1016/j.biopsych.2020.01.012. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Elbau I, and Liston C (2021). Improving precision functional mapping routines with multi-echo fMRI. Curr Opin Behav Sci 40, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, et al. (2013). Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage 80, 202–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S, Siegel JS, Gordon EM, Raut RV, Gratton C, Newbold DJ, Ortega M, Laumann TO, Adeyemo B, Miller DB, et al. (2018). Spatial and Temporal Organization of the Individual Human Cerebellum. Neuron 100, 977–993.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGirr A, Vila-Rodriguez F, Cole J, Torres IJ, Arumugham SS, Keramatian K, Saraf G, Lam RW, Chakrabarty T, and Yatham LN (2021). Efficacy of Active vs Sham Intermittent Theta Burst Transcranial Magnetic Stimulation for Patients With Bipolar Depression: A Randomized Clinical Trial. JAMA Netw Open 4, e210963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir-Moghtadaei A, Siddiqi SH, Mir-Moghtadaei K, Blumberger DM, Vila-Rodriguez F, Daskalakis ZJ, Fox MD, and Downar J (2022). Updated scalp heuristics for localizing the dorsolateral prefrontal cortex based on convergent evidence of lesion and brain stimulation studies in depression. Brain Stimul. 10.1016/j.brs.2022.01.013. [DOI] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, and Liu H (2013). Individual variability in functional connectivity architecture of the human brain. Neuron 77, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JD, Madsen KH, Puonti O, Siebner HR, Bauer C, Madsen CG, Saturnino GB, and Thielscher A (2018). Automatic skull segmentation from MR images for realistic volume conductor models of the head: Assessment of the state-of-the-art. Neuroimage 174, 587–598. [DOI] [PubMed] [Google Scholar]
- Pattamadilok C, Planton S, and Bonnard M (2019). Spoken language coding neurons in the Visual Word Form Area: Evidence from a TMS adaptation paradigm. Neuroimage 186, 278–285. [DOI] [PubMed] [Google Scholar]
- Peterchev AV, Wagner TA, Miranda PC, Nitsche MA, Paulus W, Lisanby SH, Pascual-Leone A, and Bikson M (2012). Fundamentals of transcranial electric and magnetic stimulation dose: definition, selection, and reporting practices. Brain Stimul. 5, 435–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen M-Y, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, et al. (2015). Long-term neural and physiological phenotyping of a single human. Nat. Commun 6, 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse S, Wiese S, Gembris D, Mathiak K, Kessler C, Grosse-Ruyken M-L, Elghahwagi B, Richards T, Dager SR, and Kiselev VG (1999). Enhancement of BOLD-contrast sensitivity by single-shot multi-echo functional MR imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 42, 87–97. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, et al. (2011). Functional network organization of the human brain. Neuron 72, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, and Petersen SE (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Lynch CJ, Silver BM, Dubin MJ, Martin A, and Jones RM (2019). Distinctions among real and apparent respiratory motions in human fMRI data. Neuroimage 201, 116041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, and Bergstrom CT (2008). Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. U. S. A 105, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatlou FH, Rogasch NC, McNair NA, Biabani M, Pillen SD, Marshall TR, and Bergmann TO (2018). MAGIC: An open-source MATLAB toolbox for external control of transcranial magnetic stimulation devices. Brain Stimulation: Basic, Translational, and Clinical Research in Neuromodulation 11, 1189–1191. [DOI] [PubMed] [Google Scholar]
- Sandrini M, Umiltà C, and Rusconi E (2011). The use of transcranial magnetic stimulation in cognitive neuroscience: a new synthesis of methodological issues. Neurosci. Biobehav. Rev 35, 516–536. [DOI] [PubMed] [Google Scholar]
- Schecklmann M, Schmaußer M, Klinger F, Kreuzer PM, Krenkel L, and Langguth B (2020). Resting motor threshold and magnetic field output of the figure-of-8 and the double-cone coil. Sci. Rep 10, 1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzman BA, Gratton C, Laumann TO, Gordon EM, Adeyemo B, Dworetsky A, Kraus BT, Gilmore AW, Berg JJ, Ortega M, et al. (2019). Trait-like variants in human functional brain networks. Proc. Natl. Acad. Sci. U. S. A 116, 22851–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi SH, Taylor S, Cooke D, Pascual-Leone A, George MS, and Fox MD (2019). Distinct symptom-specific treatment targets for circuit-based neuromodulation. Brain Stimulation 12, e138. 10.1016/j.brs.2019.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi SH, Weigand A, Pascual-Leone A, and Fox MD (2021). Identification of Personalized Transcranial Magnetic Stimulation Targets Based on Subgenual Cingulate Connectivity: An Independent Replication. Biol. Psychiatry 90, e55–e56. [DOI] [PubMed] [Google Scholar]
- Siddiqi SH, Taylor JJ, Horn A, and Fox MD (2022). Bringing Human Brain Connectomics to Clinical Practice in Psychiatry. Biol. Psychiatry 10.1016/j.biopsych.2022.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebner HR, Funke K, Aberra AS, Antal A, Bestmann S, Chen R, Classen J, Davare M, Di Lazzaro V, Fox PT, et al. (2022). Transcranial magnetic stimulation of the brain: What is stimulated? – a consensus and critical position paper. Clinical Neurophysiology 10.1016/j.clinph.2022.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. (2009). Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U. S. A 106, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Vidaurre D, Beckmann CF, Glasser MF, Jenkinson M, Miller KL, Nichols TE, Robinson EC, Salimi-Khorshidi G, Woolrich MW, et al. (2013). Functional connectomics from resting-state fMRI. Trends Cogn. Sci 17, 666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester CM, Yu Q, Srivastava AB, Marek S, Zheng A, Alexopoulos D, Smyser CD, Shimony JS, Ortega M, Dierker DL, et al. (2020). Individual-specific functional connectivity of the amygdala: A substrate for precision psychiatry. Proc. Natl. Acad. Sci. U. S. A 117, 3808–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendler A, Ygael NB, Roth Y, and Zangen A (2016). Deep transcranial magnetic stimulation (dTMS) – beyond depression. Expert Review of Medical Devices 13, 987–1000. 10.1080/17434440.2016.1233812. [DOI] [PubMed] [Google Scholar]
- Thielscher A, Opitz A, and Windhoff M (2011). Impact of the gyral geometry on the electric field induced by transcranial magnetic stimulation. Neuroimage 54, 234–243. [DOI] [PubMed] [Google Scholar]
- Thielscher A, Antunes A, and Saturnino GB (2015). Field modeling for transcranial magnetic stimulation: A useful tool to understand the physiological effects of TMS? Conf. Proc. IEEE Eng. Med. Biol. Soc 2015, 222–225. [DOI] [PubMed] [Google Scholar]
- Wang D, and Liu H (2014). Functional connectivity architecture of the human brain: not all the same. Neuroscientist 20, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, et al. (2015). Parcellating cortical functional networks in individuals. Nat. Neurosci 18, 1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Rogers LM, Gross EZ, Ryals AJ, Dokucu ME, Brandstatt KL, Hermiller MS, and Voss JL (2014). Targeted enhancement of cortical-hippocampal brain networks and associative memory. Science 345, 1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann EM, McShane LM, Hallett M, and Cohen LG (1992). Noninvasive mapping of muscle representations in human motor cortex. Electroencephalogr. Clin. Neurophysiol 85, 1–8. [DOI] [PubMed] [Google Scholar]
- Weigand A, Horn A, Caballero R, Cooke D, Stern AP, Taylor SF, Press D, Pascual-Leone A, and Fox MD (2018). Prospective Validation That Subgenual Connectivity Predicts Antidepressant Efficacy of Transcranial Magnetic Stimulation Sites. Biol. Psychiatry 84, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise K, Numssen O, Thielscher A, Hartwigsen G, and Knösche TR A novel approach to localize cortical TMS effects. 10.1101/595603. [DOI] [PubMed] [Google Scholar]
- Williams LM (2016). Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry 3, 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue A, Kong R, Yang Q, Eldaief MC, Angeli PA, DiNicola LM, Braga RM, Buckner RL, and Yeo BTT (2021). The detailed organization of the human cerebellum estimated by intrinsic functional connectivity within the individual. J. Neurophysiol 125, 358–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, et al. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology 106, 1125–1165. 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Montez DF, Marek S, Gilmore AW, Newbold DJ, Laumann TO, Kay BP, Seider NA, Van AN, Hampton JM, et al. Parallel Hippocampal-Parietal Circuits for Self- and Goal-oriented Processing. 10.1101/2020.12.01.395210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole EJ, Phillips AL, Bentzley BS, Stimpson KH, et al. (2021). Stanford Neuromodulation Therapy (SNT): A Double-Blind Randomized Controlled Trial. Am J. Psych 179, 132–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Matlab code for implementing the TANS algorithm has been deposited online (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation; DOI: 10.5281/zenodo.6958500). Code for preprocessing multi-echo fMRI data is maintained in a separate online repository (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline; DOI: 10.5281/zenodo.6958611).


