Abstract
How eukaryotic ribosomes traverse messenger RNA (mRNA) leader sequences to search for protein-synthesis start sites remains one of the most mysterious aspects of translation and its regulation. While the search process is conventionally described by a linear “scanning” model, its exquisitely dynamic nature has restricted detailed mechanistic study. Here, we observed single Saccharomyces cerevisiae ribosomal scanning complexes in real time, finding that they scan diverse mRNA leaders at a rate of 10 to 20 nt s−1. We show that specific binding of a protein to its mRNA leader sequence substantially arrests scanning. Conversely, impairing scanning-complex guanosine 5′-triphosphate hydrolysis results in native start-site bypass. Our results illustrate an mRNA-centric, kinetically controlled regulatory model where the ribosomal pre-initiation complex amplifies a nuanced energetic landscape to regulate scanning and start-site selection fidelity.
Real-time observation of ribosomal scanning reveals cis- and trans-regulatory mechanisms.
INTRODUCTION
During eukaryotic translation initiation, a 43S pre-initiation complex (PIC) of the small (40S) ribosomal subunit, the methionylated initiator tRNA (Met-tRNAiMet) and protein initiation factors, loads at the mRNA 5′ end (1). The resulting 48S PIC must then navigate the mRNA leader to accurately locate an AUG start codon tens to hundreds of nucleotides (nt) distant. A linear, unidirectional “scanning” mechanism for PIC motion through the leader was proposed by Kozak nearly a half-century ago (2). The requirement to continuously sample a 3-nt codon register differentiates scanning mechanistically from other search processes on single-stranded nucleic acids (3, 4). However, even basic physical properties of the scanning mechanism remain poorly understood. While past indirect bulk measurements inferred a scanning rate of ~10 nt s−1 (5, 6), a rate of significantly over 100 nt s−1 was reported in a recent single-molecule fluorescence resonance energy transfer (FRET) study (7). Precisely quantifying scanning dynamics is important, as they underpin control mechanisms central to health and disease. For instance, interception of scanning PICs lies at the core of translational control by mRNA upstream open reading frames (uORFs), found in up to 50% of mRNAs across eukaryotes (8, 9). Moreover, trinucleotide repeat expansions in specific mRNA leaders have been proposed to slow scanning, facilitating initiation of the repeat-associated non-AUG translation central to amyotrophic lateral sclerosis, and fragile X-associated tremor/ataxia syndrome (10, 11).
Genome-scale snapshots captured by translation-complex profiling (TCP-seq) have also underscored the trans-regulatory potential of RNA binding proteins during scanning, raising the question of where, how, and to what extent they may impact initiation (12). Recent data also highlight unexpectedly prevalent upstream initiation at near-cognate start codons (13); whether scanning dynamics contribute to their utilization remains unknown. At a deeper mechanistic level, the contributions of individual PIC components to scanning regulation also remain poorly understood. In particular, there are contrasting models for how and when a key guanosine 5′-triphosphate (GTP) hydrolysis step within the PIC ensures start-site selection fidelity (14, 15). To gain a targeted view of scanning dynamics, we deployed a real-time single-molecule fluorescence assay to directly visualize a key biochemical event that marks the end of the process, ejection of eukaryotic initiation factor 1 (eIF1) from the PIC. With this assay, we directly observed scanning on single mRNAs with reconstituted Saccharomyces cerevisiae PICs. The assay leverages zero-mode waveguides (ZMWs) (16) to simultaneously observe scanning on hundreds of mRNA molecules at physiological PIC and factor concentrations with millisecond time resolution.
RESULTS
Tracking eIF1 quantifies real-time, single-mRNA scanning dynamics
In our assay, eIF1 provides a direct readout on events occurring at the core of the PIC during scanning and start-site recognition. PIC arrival at the start codon triggers rapid eIF1 ejection, which gates scanning arrest through irreversible release of the phosphate product of GTP hydrolysis by eIF2, the GTPase that delivers Met-tRNAiMet to the 48S P site (14). To visualize scanning, capped mRNAs are surface immobilized by poly(A)-tail capture, then pre-assembled 43S PICs are delivered to mRNA at 20°C, forming 48S PICs. The PICs contain a fluorescently labeled 40S subunit and eIF1, in addition to the complement of native PIC components: eIF1A, eIF2, eIF3, and eIF5, and Met-tRNAiMet (Fig. 1A and fig. S1, A to C). PIC delivery occurs in the presence of the eIF4F cap-recognition complex (eIF4E•eIF4G1•eIF4A), the eIF4F cofactor eIF4B, and adenosine 5′-triphosphate (ATP). 40S ribosomal subunits are labeled with Cy3 (17), eIF1 is Cy5 labeled at its C terminus (18), and mRNAs are Cy5.5 labeled via the immobilization oligonucleotide.
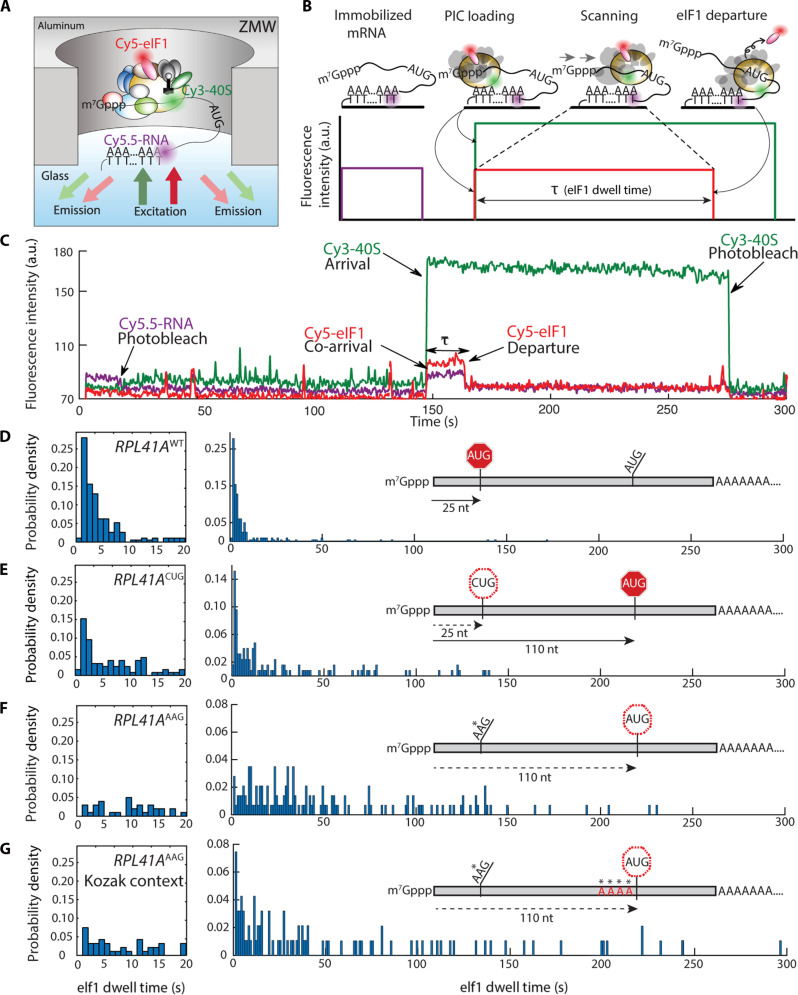
Fig. 1. Direct observation of scanning in real time.
(A) Experimental setup for single-molecule fluorescence scanning assays in zero-mode waveguides. (B) Idealized single-molecule fluorescence trace for scanning signal, indicating eIF1 dwell time, τ. (C) Representative trace with scanning signal observed on the wild-type RPL41A mRNA. (D to G) Representative eIF1 dwell-time distributions on wild-type RPL41A mRNA (n = 193 molecules), RPL41AAAG (n = 143), RPL41AAAG/Kozak (n = 94), and RPL41ACUG (n = 254) mRNAs, respectively. The distribution in the 0 to 20 s time domain is expanded to the left of the full-timescale distribution. Distributions from replicate experiments were statistically indistinguishable. Inset schematics indicate distances (in nucleotides) between the +1 nt and the initiation codon.
At the single-molecule level, PIC-mRNA recruitment results in simultaneous appearance of Cy3-40S and Cy5-eIF1 fluorescence (Fig. 1B). Both signals persist as the PIC scans, but the Cy5-eIF1 signal is lost with eIF1 ejection upon start-codon recognition. Thus, the eIF1 dwell time quantifies the total time the PIC spends in preparation for scanning, the scanning process, start-codon recognition, and eIF1 ejection.
To validate that our system recapitulates on-pathway, cap-dependent translation initiation, we confirmed that labeled eIF1 is incorporated into PICs (fig. S2, A and B). eIF1 fluorescent labeling, via a fluorophore-conjugated C-terminal dipeptide extension, has been applied in bulk assays to quantify the eIF1 ejection rate upon start-codon recognition (19). A similar C-terminal extension of eIF1 is phenotypically silent in vivo (18). In our assay, efficient stable PIC-mRNA recruitment also depended on eIF4F (fig. S2, C and D) (20, 21).
Scanning occurs at a net forward rate of ~20 nt s−1
We applied our assay to characterize scanning on the yeast RPL41A mRNA (22–24). We observed the expected Cy3-40S/Cy5-eIF1 co-arrival to immobilized RPL41A and subsequent loss of Cy5-eIF1 (Fig. 1C). The mean eIF1 dwell time was 8.8 ± 1.9 s (mean ± SD); the median time was 3.3 ± 0.1 s (table S1 and Fig. 1D). Dwell-time distributions were highly reproducible between replicates (fig. S3, A and B) and between different eIF3 preparations (fig. S3C). We expected dwell times to be short on this 25-nt leader because its length is within the 19- to 45-nt 48S PIC footprint (12).
The dwell-time distribution rose to a short-time peak at ~1 to 2 s, followed by a quasi-exponential decay. This dominant exponential component implies that a single kinetic step largely limits the time interval from PIC recruitment to eIF1 ejection. Exponential fitting of the decay phase (2 to 10 s) yielded an eIF1 dissociation rate of ~0.36 ± 0.04 s−1, approaching 0.6 s−1 measured in bulk (fig. S3D) (14). The distribution was also well modeled by the probability density function of an irreversible 25-step process, with 24 steps occurring at a rate constant of 20 s−1 and a 25th occurring at 0.35 s−1 (fig. S3E) (25).
To quantify how a near-cognate triplet affected scanning, we prepared an RPL41ACUG variant, substituting CUG for the native (+25) start codon. Among near-cognate codons, CUG initiation efficiency is closest to AUG (26–28). Mean and median eIF1 dwell times for RPL41ACUG (28.9 ± 2.5 s and 9.2 ± 2 s, respectively) lengthened with respect to RPL41Awt. Notably, the dwell-time distribution for RPL41ACUG was biphasic (Fig. 1E and fig. S3F), corresponding to two populations of scanning PICs. The short-time component of the RPL41ACUG distribution resembled the entire RPL41Awt distribution (fig. S3F). Thus, CUG-for-AUG substitution did not affect the scanning rate but induced bypass of the +25 triplet for a proportion of PICs.
Substituting the native start codon with another near-cognate codon, AAG (RPL41AAAG) fully eliminated the short-time component common to the RPL41Awt and RPL41ACUG distributions (Fig. 1F). The mean dwell time rose to 59.6 ± 1.8 s, an almost sevenfold increase over RPL41Awt; the median time increased over 11-fold, to 38 ± 4 s. Although near-cognate to AUG, fungal PICs commit to AAG initiation sites at <0.5% efficiency relative to AUG (27, 28). Thus, elimination of the short dwell-time component is consistent with full bypass of the +25 triplet and continued downstream scanning.
Since the 5′-proximal AUG codon of RPL41AAAG lies at position +110, a mean dwell time of ~60 s could result either from recognition of this triplet after scanning at a rate of ~1.8 nt s−1 or from additional bypass of the +110 AUG triplet. Substituting the +110 AUG triplet with AAG did not alter the eIF1 dwell-time distribution relative to RPL41AAG, indicating the +110 site is very inefficiently recognized by the PIC (fig. S3, G and H). Furthermore, changing the nucleotide context flanking the +110 AUG (“Kozak sequence”), which tunes the efficiency of start-codon recognition (26, 29, 30), from the relatively poor native sequence (GATTAUG) (31) to an optimal sequence (AAAAAUG), led only to a small increase in the number of short eIF1 dwells (Fig. 1G). Evidently, even with an optimal Kozak sequence, the +110 AUG triplet is not effective in translation initiation, possibly because it is in the native 3′ untranslated region of the RPL41A mRNA.
Single-molecule dwell-time distributions for multistep processes encode in their shapes the information on the number and the relative rates of the steps in the underlying process (fig. S4) (32). Together, the distributions for these RPL41A mRNAs consistently indicate that scanning occurs at a rate of ~20 nt s−1, regardless of commitment to a particular initiation site.
The scanning rate is maintained on longer leaders
The dynamics of scanning on longer mRNA leaders (over 250 nt) remain unknown, raising the question of whether the more structurally complex landscape of these mRNAs might alter scanning. We next measured the eIF1 dwell-time distribution on the 574-nt GCN4 leader, in a construct containing the entire leader sequence and the first 30 ORF nucleotides. This leader contains four uORFs, which play a paradigmatic role in translational control (33). We serially substituted the uORF AUG triplets with AAG, producing five RNAs with increasing cap-to-5′ proximal AUG spacing (Fig. 2A).
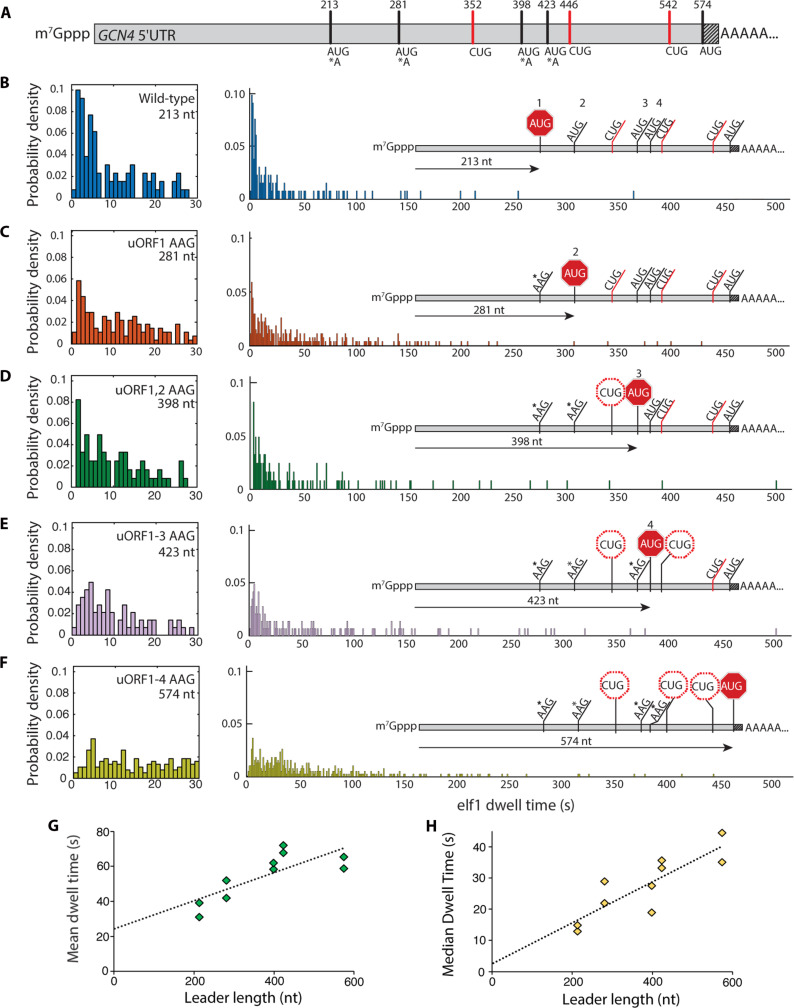
Fig. 2. Scanning dynamics on the GCN4 leader.
(A) Schematic of the GCN4 leader indicating positions of canonical upstream ORF AUG codons, near-cognate CUG triplets, and the GCN4 main-ORF AUG codon. The shaded portion indicates 30 nt of the GCN4 coding sequence included in the transcript. (B to F) eIF1 dwell-time distributions for the wild-type GCN4 leader sequence and the four sequential upstream ORF AUG-to-AAG variants. Left panels expand the short-time component of the distributions. Insets indicate relative linear distance traversed by the PIC for each leader sequence. For (B) to (F), the number of molecules in each distribution was 136, 115, 165, 98, and 160, respectively. (G and H) Mean and median eIF1 dwell times plotted against the cap to AUG leader length in nucleotides. Trend lines are from linear regression to all datapoints.
The eIF1 dwell-time distribution for the wild-type GCN4 leader included a short-time peak at ~2 s, followed by a quasi-exponential decay (Fig. 2B) (mean and median dwell times: 35.1 ± 4.1 s and 13.9 ± 1.0 s). Since GCN4wt spans 213 nt upstream of the 5′-proximal AUG, the distribution is consistent with scanning at ~10 nt s−1. eIF1 mean and median dwell times increased with increasing cap-to-AUG spacing in the variant leaders (table S1 and Fig. 2, C to F). As the spacing increased, the exponential component of the dwell-time distribution was subsumed into a more symmetric distribution, corresponding to PIC passage times becoming dominated by multistep transit through the leader, rather than by a single kinetic step (fig. S4). Linear regression indicated the eIF1 dwell time increased by ~0.08 s nt−1 (95% confidence intervals: 0.028 to 0.13 s nt−1) and ~0.065 s nt−1 (0.032 to 0.099 s nt−1) based on the mean and median values, respectively (Fig. 2, G and H). This corresponds to scanning rates of ~12 and ~15 nt s−1, similar to RPL41A. These values were also well supported by global analysis of eIF1 dwell times for RPL41A and GCN4 sequences, in addition to the PAB1 and SSA1 mRNAs (fig. S5). The linear dependence on leader length observed over two orders of eIF1 dwell-time magnitude also indicates that premature Cy5 photobleaching does not detectably influence our scanning rate measurement. Together, our data indicate that scanning rates of ~10 to 20 nt s−1 are maintained regardless of leader length, sequence, or structural features.
Efficient start-codon recognition requires GTP hydrolysis
Past studies that probed the mechanistic role of GTP hydrolysis kinetics in scanning were carried out on model, uncapped RNA (14, 15). To delineate the contributions of GTP hydrolysis to cap-/eIF4F-dependent scanning on full-length mRNAs, we measured eIF1 dwell-time distributions in the presence of the nonhydrolyzable GTP analog, 5′-guanylyl imidodiphosphate (GDPNP). We compared distributions for RPL41Awt and the GCN41–4 AAG quadruple AUG mutant, which have 5′-proximal AUG sites at +25 and +574 nt from the cap (Fig. 3, A and B).
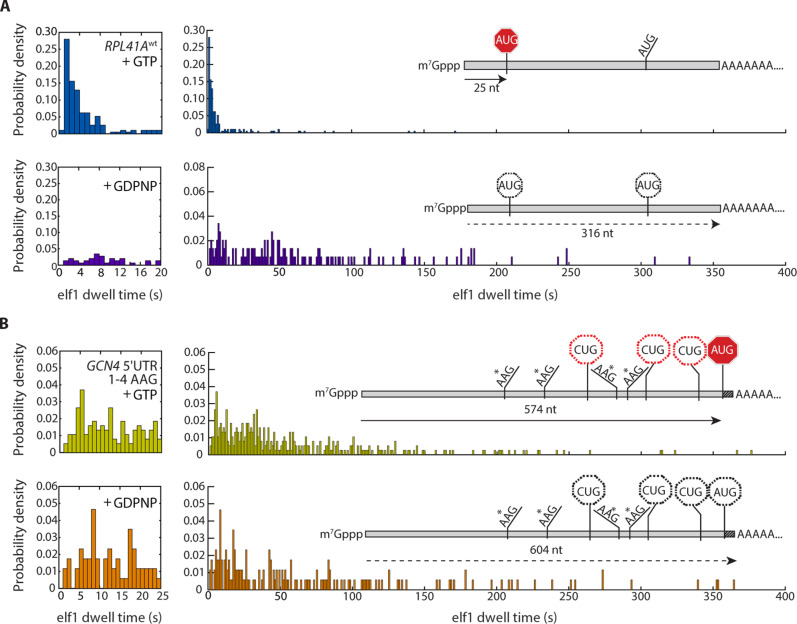
Fig. 3. Regulation of scanning by GTP hydrolysis.
(A and B) eIF1 dwell-time distributions for scanning on (A) RPL41Awt (n = 193 in the presence of GTP; n = 146 in the presence of GDPNP) and (B) the GCN4 quadruple uORF AUG-to-AAG variant (n = 378 in the presence of GTP; n = 172 in the presence of GDPNP), contrasted between PICs formed with GTP versus GDPNP in the eIF2 TC (top versus bottom panel, respectively).
GDPNP-for-GTP substitution drastically altered the RPL41Awt eIF1 dwell-time distribution (Fig. 3, A and B). The mean dwell time increased about eightfold, to 74 ± 5 s; the median increased ~15-fold, to 48 ± 1 s. The peak position increased from 1.4 to 5.8 s. As for RPL41ACUG, RPL41AAAG, and GCN4, these changes indicate an increased number of scanning steps before loss of eIF1 fluorescence. Thus, GDPNP caused PIC bypass of the +25 AUG start codon.
If the PIC bypassed all start codons, scanning to the RNA 3′ end, we hypothesized that GDPNP would have a lesser impact on the eIF1 dwell-time distribution for GCN41–4 AAG since this RNA includes only ~30 unhybridized nucleotides downstream of its main AUG and significant downstream PIC motion would be impossible. Mean and median eIF1 dwell times lengthened for GCN41–4 AAG by only ~1.3-fold with GDPNP (table S1 and Fig. 3, A and B). Similarly, omission of eIF5, the GTPase-activating protein for eIF2, also shifted eIF1 dwell-time distribution parameters to longer times for GCN4wt and PAB1AAG, consistent with bypassing the AUG site when GTP hydrolysis is impaired (fig. S6, A and B).
GDPNP is conventionally regarded as a nonhydrolyzable GTP analog. However, a direct comparison for the GTPase H-Ras revealed only ~110-fold slower GDPNP-versus-GTP hydrolysis at 37°C (34). When GDPNP-based ternary complexes (TCs) were incubated for 1, 2, or 3 hours at 30°C, then used to assemble PICs, the median eIF1 dwell time for scanning on the RPL41A mRNA progressively shortened toward a value of ~6 s with increased TC pre-incubation, close to the value of ~3 s observed with GTP; meanwhile, on the same timescale, phosphate accumulated in the isolated TC samples at around the estimated TC concentration (fig. S6C). Put otherwise, the GDPNP-based scanning behavior transitioned to the GTP-based behavior as GDPNP-based TCs aged. These data provide an avenue to reconcile our single-molecule results with past work where PICs committed to start sites when the TC included GDPNP.
A specific protein-mRNA interaction arrests scanning
TCP-seq analysis for the PAB1 mRNA, which encodes the poly(A)-binding protein Pab1p, revealed 48S PIC accumulation at an 11-mer oligo(A) leader sequence beginning at position +91 of the transcript. These and other data (12, 35, 36) have led to proposals that Pab1p might negatively trans-regulate scanning on its own mRNA.
The eIF1 dwell-time distribution on PAB1 mRNA was biphasic, with a major component (peak at ~1 s) corresponding to rapid eIF1 ejection similar to RPL41Awt (Fig. 4A). The rapid-ejection population might suggest scanning is intrinsically faster on PAB1 than RPL41A or GCN4. However, the PAB1 leader includes two upstream near-cognate (CUG) start codons, at positions +47 and +67. When these CUG triplets were substituted by AAG, the eIF1 dwell-time distribution prominently peaked at ~6 s (Fig. 4B), consistent with introduction of additional scanning steps between the leader 5′ end and eventual start-codon recognition site. Since the PAB1 cap-to-AUG spacing is 137 nt, the data again indicate a scanning rate of ~14 nt s−1. Together, these data indicate eIF1 ejection at the upstream PAB1 CUG sites is efficient, similar to RPL41ACUG.
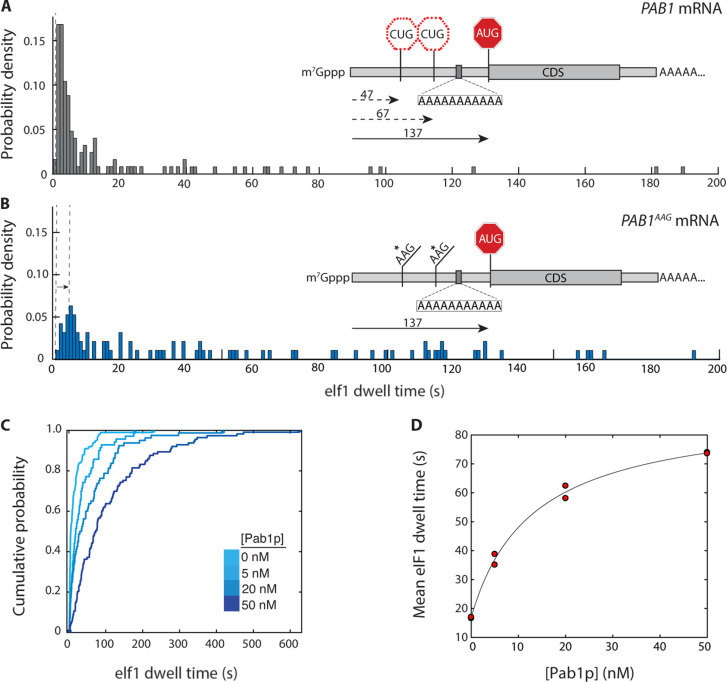
Fig. 4. Regulation of scanning by S. cerevisiae poly(A)-binding protein.
(A) eIF1 dwell-time distribution for the PAB1 mRNA (n = 125 molecules). Inset: Relative locations of upstream CUG triplets and PAB1 main-ORF AUG codon, and location of internal oligo(A) 11-mer. (B) eIF1 dwell-time distribution for the PAB1AAG mRNA, in which both upstream CUG triplets are substituted by AAG (n = 95). (C) eIF1 dwell-time cumulative distribution functions for scanning on the PAB1 mRNA in the presence of varying concentrations of the poly(A)-binding protein, Pab1p (5 nM, n = 110; 20 nM, n = 103; 50 nM, n = 112). (D) Dependence of mean eIF1 dwell time on PAB1 mRNA on the concentration of Pab1p, with hyperbolic fit (Kd 13 nM; 95% confidence intervals 1.1, 25.6 nM). Replicate data points at 0 and 50 nM Pab1p overlap. The time added to scanning at saturating Pab1p concentration is 72 s (95% confidence intervals: 64 to 79 s).
To quantify the extent that Pab1p might impede PIC access to the PAB1 start codon, we measured eIF1 dwell-time distributions on the PAB1wt mRNA in the presence of Pab1p. Addition of increasing Pab1p concentrations shifted the distribution to longer times (Fig. 4C and table S1). The mean eIF1 dwell time rose saturably from 16.8 ± 0.2 s in the absence of Pab1p to 73.8 ± 0.2 s at 50 nM Pab1p. Dwell times were also lengthened for the PAB1 leader where both upstream CUGs were mutated to AAG (table S1 and fig. S7, A and B), confirming the increased scanning time is specific to the AUG codon. In contrast, addition of 50 nM Pab1p to the RPL41AAAG mRNA, which lacks an internal poly(A) tract, slightly shortened eIF1 dwell times (fig. S7, C and D). The data point to the PAB1 internal oligo(A) sequence, rather than the poly(A) tail or other PIC components, as the mediator of Pab1p-induced scanning arrest.
Hyperbolic fitting indicated ~13 nM Pab1p half-maximally prolonged eIF1 dwell times on PAB1 mRNA (Fig. 4D). This approaches the equilibrium dissociation constant of S. cerevisiae Pab1p from a 12-mer oligo(A), ~30 nM (37) and the Pab1p apparent equilibrium dissociation constant for nonpolyadenylated PAB1 (~25 nM; Fig. S7F). Hill analysis was consistent with Pab1p titrating a single mRNA site (nH = 0.98; fig. S7E). Together, these data indicate an mRNA-specific equilibrium-binding interaction of Pab1p with a single site in its own mRNA leader creates a steric block that drastically lengthens scanning.
DISCUSSION
Codon-anticodon base pairing controls scanning rates
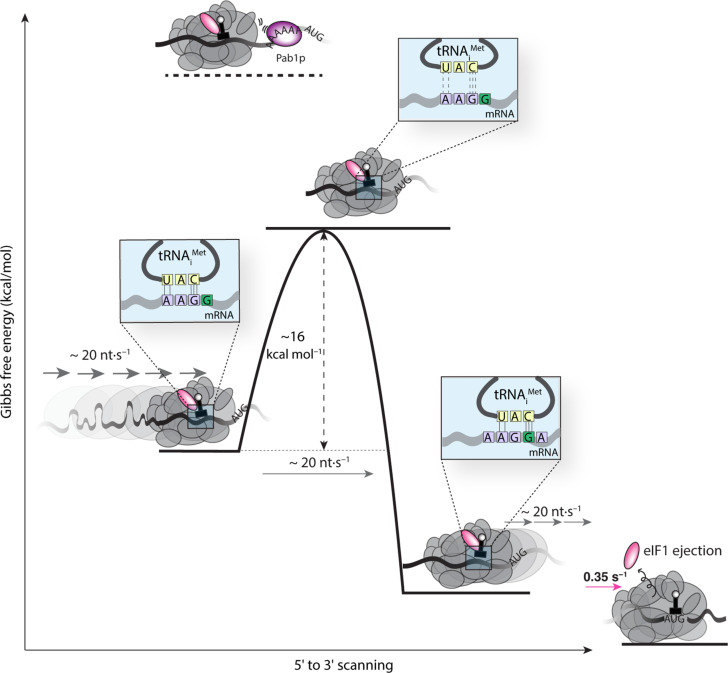
A scanning rate of ~10 to 20 nt s−1 across widely diverse leaders points to a common process limiting PIC motion regardless of mRNA identity. Eyring analysis of this rate sets an upper limit of ~16 kcal mol−1 on the average energy barrier for PIC movement through the leader, consistent with breakage of a few hydrogen bonds at each step (Fig. 5). Since the tRNAiMet anticodon (3′ UAC 5′) includes three of the four RNA nucleotides, it can sample at least one base pair, or between two and five hydrogen bonds, at all non-AUG mRNA leader triplets except CCC. The stability of isolated tRNA duplexes with tri- or tetraribonucleotides lies in the region of −3 kcal mol−1 for the Gibbs free energy (38) and sets a lower limit on the activation barrier. This includes an enthalpic contribution of around −15 kcal mol−1, which, in the spatially constrained environment of the PIC, is expected to dominate the barrier to tRNA-mRNA disengagement (38). While secondary PIC-mRNA interactions, such as with the Kozak sequence, may fine-tune scanning rates, our findings point to mRNA-tRNAiMet interactions playing a dominant role in controlling scanning progress. From an evolutionary perspective, the scanning rate is thus a direct consequence of the requirement to sample the leader sequence in a three-base register one nucleotide at a time.
Fig. 5. Energetics of scanning.
The measured scanning rate of ~20 nt s−1 implies an energy barrier of ~16 kcal mol−1, on average, for PIC movement to the next nucleotide at each scanning step. The magnitude of this barrier corresponds to breakage of the hydrogen bonds in base-paired tRNA and mRNA; this base pairing by itself has an overall stability in the range of −3 kcal mol−1, with an enthalpic component of around −15 kcal mol−1. On the left of the schematic, one possible interaction during scanning includes five hydrogen bonds formed between a near-cognate mRNA triplet with a noncognate tRNA. Thermal fluctuation that disengages the base pair is expected to traverse an activation barrier equal to or exceeding the enthalpy of the hydrogen-bond interactions, as measured by our assay. Steric hindrance from Pablp binding to an oligo(A) site abutting the PIC increases this activation barrier, resulting in blocked scanning. eIF1 ejection stabilizes the PIC to the point where no further forward motion is possible.
While our measured scanning rates agree with past bulk estimates, they are approximately an order of magnitude slower than recently measured by a FRET-based single-molecule fluorescence assay (7). This difference likely results in part from the lower temperature used in our assay (20°C versus 30°C). In addition, the previous single-molecule study inferred PIC progression along the mRNA from a FRET signal between the 40S subunit and an mRNA position downstream of the start codon. A potential limitation of the FRET signal is that it reports on the distance between fluorophores embedded within the intricate three-dimensional structure of the PIC. Thus, in addition to scanning, the FRET signal may report on structural rearrangement of the PIC that occurs on a faster timescale than scanning. In the same single-molecule study, the time for 60S-subunit recruitment after the proposed end of scanning was determined. For three of four mRNAs tested, the 60S-subunit recruitment time showed an apparent linear increase with leader length. This delay in 60S-subunit recruitment is comparable to our measured scanning time. Further studies will be required to fully reconcile these differences.
Scanning at 10 to 20 nt s−1 enables kinetic control of initiation
Our data point to a kinetic-control model where the scanning speed and eIF1 ejection rate are balanced to ensure start-site selection fidelity. At each leader position, the PIC decides whether to continue scanning or eject eIF1. If scanning occurred at a faster rate, PIC forward progress would outcompete eIF1 ejection, increasing the likelihood of start-codon bypass. Conversely, scanning at a slower rate would enhance spurious eIF1 ejection at incorrect start sites. Moreover, since the scanning times lie within the same timescale as translation overall (39), a constant scanning rate on varying mRNA leaders offers the potential to directly connect leader length to flux through initiation. Thus, our findings rationalize the observation that leader length anticorrelates with initiation probability (40). More broadly, our results evoke an mRNA-centric regulatory model with layered control of scanning, balanced between global and local leader features, including RNA binding protein sites that facilitate trans-regulatory activities.
Our data underscore that while the scanning motion does not intrinsically require PIC GTP hydrolysis, start-codon recognition does. Because our study involves full-length, capped mRNAs and the full complement of initiation factors, we suggest that regulation of GTP hydrolysis is a function of the entire machinery within the PIC, rather than purely of local activities of factors interacting with the GTPase eIF2. Put otherwise, events taking place at the core of the PIC are dynamically connected to events occurring at distant PIC and mRNA elements.
Our data directly calibrate the energetics of the scanning motion. The hyperbolic dependence of scanning times on Pab1p concentration indicates reversible Pab1p•PAB1-mRNA binding limits PIC passage through the leader. This is consistent with a sequential mechanism where the PIC arrives at the blockage site and is obliged to wait for protein dissociation before further scanning (fig. S8). The maximum delay at saturating Pab1p concentration (72 s) corresponds to Pab1p dissociation at ~0.014 s−1, with a barrier height of ~21 kcal mol−1. Evidently, the PIC does not intrinsically have sufficient energy to move past a block of this magnitude.
Because the scanning rate is exponentially related to the barrier height for PIC progress, subtle energy-landscape changes in scanning may be transduced into large effects on start-site commitment. Thus, while formation of the full complement of tRNA-mRNA hydrogen bonds upon P-site AUG arrival imposes only a modest increase on the barrier to PIC forward motion, relative to a near-cognate codon, the kinetic consequences are sufficient to significantly slow the PIC for eIF1 ejection and the structural changes that conclude scanning (41). Our findings underscore how even nuanced modulation of barriers to scanning, such as by features of disease-associated mRNA leaders, is sufficient to halt scanning whilst maintaining the potential for efficient non–AUG-initiated translation.
Experimental timescale affects GDPNP effects on start-site recognition efficiency
Our finding that the presence of GDPNP in the TC impairs efficient start-site selection contrasts with past bulk toeprinting assays, which indicate that yeast PICs are competent to scan and commit to the translation start site in the presence of GDPNP (23). A significant difference between the bulk and single-molecule assays is that on the timescale of the single-molecule assay, each mRNA is scanned only once, whereas on the timescale of bulk assays, mRNAs can be scanned multiple times. Our data (Fig. 3, A and B) suggest that, when GDPNP is present, PICs in the single-molecule assay bypass start sites and continue scanning the mRNA, but stall at the mRNA 3′ end, likely due to the immobilization oligo and potentially the NeutrAvidin (Fig. 1A). In contrast, under the conditions of the bulk assays, PICs that bypass the start codon are in a position to continue scanning and then run off the mRNA 3′ end.
We considered two possible explanations, which are not mutually exclusive, to reconcile the bulk and single-molecule results. The first possibility is that GDPNP-bound PICs can commit to start sites, but less efficiently than GTP-bound complexes, and that stable PIC-mRNA recruitment in bulk assays with GDPNP accrues over multiple cycles of scanning. This model is compatible with PIC-mRNA recruitment rates of 0.25 to 0.74 min−1 (corresponding to mean recruitment times of 1.3 to 4 min) determined for the 314-nt RPL41Awt mRNA by bulk electrophoretic mobility shift assays (21, 23). At a scanning rate of ~20 nt s−1, PICs bypassing the start codon would take 15 to 20 s to scan the full length of the RLP41Awt mRNA. Each mRNA molecule could be scanned at least 5 to 16 times on the measured timescale of stable recruitment. Under these conditions, a start-site commitment probability with GDPNP of ~20%, or even as low as 6%, would reconcile our results with the bulk data. A recent translatome-wide implementation of the reconstituted bulk assay system indicated that start-codon bypass (“leaky scanning”) is possible in the system with GDPNP-based TCs: Around 3% of start-site recognition events inherently occurred at AUG triplets internal to annotated ORFs with GDPNP, and this rose to 7.8% when the translational helicase Ded1p was added, which is thought to enhance scanning processivity (42).
The second, related possibility is that under the conditions of the bulk assays some fraction of the GDPNP-based eIF2 TCs undergoes GDPNP hydrolysis and that these complexes mediate start-site commitment. The PIC stimulates TC GTP hydrolysis by six orders of magnitude even in the absence of mRNA [kGTP,PIC ~ 9 s−1; (14)]. On the basis of our data for isolated TCs, PIC-stimulated GDPNP hydrolysis could therefore occur in the bulk experiments, which involve pre-incubation of TCs with other PIC components for tens of minutes or up to hours.
These mechanisms could operate on their own or in parallel to enable the start-site recognition previously observed with GDPNP-based TCs. Their relative prevalence notwithstanding, while our data do not rule out that start-site commitment is possible with GDPNP, they strongly suggest a model where GTP hydrolysis is an important requirement for it to occur efficiently during a single round of scanning. Further detailed studies that bridge the timescales of the single-molecule and bulk assays will provide additional insights to fully parse these differences.
MATERIALS AND METHODS
Experimental design
Our objective was to robustly quantify the scanning rate on different mRNA sequences through single-molecule fluorescence analysis with reconstituted ribosomal PICs. To this end, we focused on an event that marks the biochemical endpoint of scanning, eIF1 ejection from the PIC. Single-molecule fluorescence assays with purified and biochemically validated components were designed to measure eIF1 dwell times in scanning PICs with appropriate temporal resolution on sufficient numbers of single mRNA molecules to reproducibly determine dwell-time distributions between technical replicates of the experiments.
Purification of protein factors
Published constructs and strains were used to purify yeast eIF1 (20), eIF1A (20), eIF5 (20), eIF2 (43), eIF3, eIF4G (44), eIF4E (45), eIF4A (46), Pab1p (45), 40S ribosomal subunit (20), tRNA (20), and the yeast methionyl-tRNA synthetase (MetRS) (20). All proteins were stored in 50 mM Hepes-KOH (pH 7.5), 100 mM KOAc, 3 mM MgOAc2, 2.5 mM TCEP, 10% glycerol except MetRS [40 mM tris, 10 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol (DTT)].
Purification of eIF1, eIF1A, and eIF5
Yeast eIF1, eIF1A, and eIF5 were purified as previously described (20). A pTYB2 vector containing the gene of interest, fused to a C-terminal intein-chitin–binding domain, was transformed into BL21 Codon Plus RIL cells (Agilent). The proteins were overexpressed with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and the cells were lysed by sonication in lysis buffer [20 mM Hepes-KOH (pH 7.4), 0.5 M KCl, 0.1% Triton X-100, 1 mM EDTA, and 1× protease inhibitor cocktail]. The lysates were clarified and loaded on 1-ml chitin resin [New England Biolabs (NEB)] and washed with 1 M KCl. The protein-bound resin was incubated overnight with cleavage buffer [20 mM Hepes-KOH (pH 8.0), 500 mM KCl, and 75 mM DTT] to induce intein cleavage, then eluted the next day. For eIF1 C-terminal labeling, the protein was incubated with the cleavage buffer containing 200 mM sodium 2-mercaptoethanesulfonate (MESNA) instead of DTT and Cys-Lys(ε-Cy5) overnight. The eluted proteins were further purified on a 1-ml heparin-HP column (Cytiva), followed by size-exclusion chromatography on Superdex 75. The purified proteins were flash-frozen with liquid nitrogen and stored at −80°C.
Purification of eIF2
The yeast strain, GP3511, which expresses N-terminally hexahistidine-tagged eIF2γ, in addition to eIF2a and eIF2b, was a gift from A. G. Hinnebusch. The cells were grown in yeast extract/peptone/adenine sulfate/dextrose (YPAD) media [to a final optical density at 600 nm (OD600) of 4 to 6] and were harvested, washed, and resuspended [~2 g of cells/1 ml of lysis buffer; 30 mM Hepes-KOH (pH 7.4), 800 mM KCl, 10 mM imidazole, 0.1 mM MgCl2, 5 mM β-mercaptoethanol, 10% glycerol, and 1× protease inhibitor cocktail] and were flash-frozen with liquid nitrogen. The cells were lysed using a cryogenic mill (Spex Sample Prep, model 6875), for 15 repetitions of a 2-min run at 15 cycles per second, with 2-min cool time between repetitions. The debris was removed by filtration as previously described (43). The lysate was further clarified by centrifugation and then loaded onto a 5-ml Ni–nitrilotriacetic acid (NTA) resin by gravity flow. The protein was eluted with lysis buffer containing 500 mM imidazole and was further purified using a heparin column followed by a Q column. The protein was then dialyzed into storage buffer. The purified protein was flash-frozen with liquid nitrogen and stored at −80°C.
Purification of eIF3
A yeast strain (YOR361C-TAP) bearing a genomically encoded, C-terminally TAP-tagged eIF3b subunit, was purchased from Dharmacon. The growth condition was the same as for eIF2. The cells were harvested and washed with lysis buffer [10 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.1% NP-40, and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. The washed cells were then resuspended in lysis buffer and flash-frozen with liquid nitrogen for storage until purification. The cells were lysed with a bead beater (45-s grinding followed by 2-min, 15-s rest, for five cycles). The clarified and filtered lysate was loaded on 1 ml of immunoglobulin G (IgG) Sepharose fast flow resin (NEB). The column was washed with lysis buffer and TEV cleavage buffer (lysis buffer with 75 mM NaCl, 0.5 mM EDTA, and without DTT). The IgG-bound sample was cleaved with TEV protease overnight and further purified by size-exclusion chromatography on Superdex 200. Residual TEV protease was removed by incubating the eluted sample with Ni-NTA (500 μl) for 10 min at 4°C. eIF3 was collected, aliquoted and was flash-frozen with liquid nitrogen and stored at −80°C.
Purification of eIF4G
Yeast eIF4G was purified as described previously (44). A pTYB2 vector encoding a C-terminal fusion of eIF4G with an intein and chitin-binding domain was transformed into BL21 CodonPlus RIL cells (Agilent). Protein expression was induced with 0.5 mM IPTG for overnight at 16°C. The cells were lysed by sonication in 50 mM Hepes-KOH (pH 7.4), 500 mM KCl, 1 mM EDTA, and 1× protease inhibitor cocktail. The lysate was clarified before loading onto the chitin resin (NEB). The sample was treated with 3 U/μL of micrococcal nuclease (NEB) in 50 mM Hepes-KOH (pH 7.4), 100 mM KCl, and 2 mM CaCl2 and was incubated with cleavage buffer containing 50 mM Hepes-KOH (pH 7.4), 250 mM KCl, 1 mM EDTA, and 50 mM DTT overnight. Eluted eIF4G was purified by anion-exchange chromatography on a 1-ml Q-HP column (Cytiva) and dialyzed into the storage buffer, then flash-frozen with liquid nitrogen and stored at −80°C.
Purification of eIF4A and eIF4E
Yeast eIF4A and eIF4E were purified as described previously (45, 46). A pET28a(+) vector encoding N-terminally his-tagged eIF4A or eIF4E was transformed into BL21 Codon Plus RIL cells. Protein overexpression was induced with 1 mM IPTG overnight at 16°C. Cells were lysed by sonication, and the lysate was clarified by centrifugation before loading on the Ni-NTA column. For eIF4A, the protein eluted from the Ni-NTA step was further purified on a 1-ml Q-HP column followed by size-exclusion chromatography (Superdex 75; Cytiva) in storage buffer. eIF4A was flash-frozen with liquid nitrogen and stored at −80°C. For eIF4E, the Ni-NTA–purified protein was buffer exchanged into the storage buffer over a 10-DG column (Bio-Rad) and was stored at 4°C. Purified eIF4E preparations were used for no more than 2 weeks.
Purification of eIF4B
A pET-22b(+) vector encoding hexahistidine-tagged eIF4B (TIF3) was transformed into BL21 CodonPlus RIL cells. The protein was purified as described previously (47). Protein overexpression was induced with 0.2 mM IPTG for 6 hours at 30°C. The cells were resuspended in lysis buffer [50 mM tris-HCl (pH 7.5), 500 mM KCl, and 20 mM imidazole] and lysed by sonication. The clarified lysate was loaded on 1 ml of Ni-NTA resin and was washed with lysis buffer containing 50 mM imidazole. The protein was eluted with lysis buffer containing 250 mM imidazole. The protein eluate was diluted into a low-salt buffer [50 mM tris-HCl (pH 7.5), 100 mM KCl, and 2.5 mM TCEP] and loaded on a pre-equilibrated 1-ml Q-HP column. The protein was further purified by size-exclusion chromatography (Superdex 200) in storage buffer. The purified protein was flash-frozen with liquid nitrogen and stored at −80°C.
Purification of Pab1p
A pET-28a(+) plasmid, encoding the PAB1 gene fused to an N-terminal hexahistidine tag (45), was transformed into BL21-Codon Plus RIL cells. The cells were grown and lysed under the same condition as for eIF4E/4A, except the lysis buffer was composed of 50 mM Hepes-KOH (pH 7.5), 300 mM KCl, 2.5 mM TCEP, 1× protease inhibitor cocktail, and 1× PMSF. The protein was loaded on 5-ml Ni-NTA resin and was washed with lysis buffer containing 10 mM imidazole. The resin-bound protein was treated with micrococcal nuclease, as for eIF4G. His6-Pab1p was then eluted with lysis buffer containing 250 mM imidazole. The protein was further purified on a 1-ml Q-HP column and buffer exchanged into the storage buffer on a 10-DG column. The purified proteins were flash-frozen with liquid nitrogen and stored at −80°C.
40S ribosomal subunit purification
Yeast 40S purification and labeling were carried out as described previously, with a few modifications (17, 20). The yeast strain encodes a 23-nt extension to helix 39 of the rRNA, which enables fluorescent labeling through hybridization of a fluorophore-conjugated complementary oligonucleotide. Approximately 2 liters of yeast culture was grown and harvested at OD600 of ~1 to 2. The cells were washed with lysis buffer [30 mM Hepes-KOH (pH 7.5), 100 mM KCl, 20 mM MgCl2, 2 mM DTT, heparin (2 mg/ml), and RNAse inhibitor (20 U/ml)] and then flash-frozen. The cells were lysed using a bead beater and loaded on a 3-ml 1 M sucrose cushion. The sample was centrifuged in a 70Ti rotor at 55,000 rpm for 3.5 hours at 4°C. The pellet was washed and resuspended with separation buffer [50 mM Hepes-KOH (pH 7.5), 500 mM KCl, 2 mM MgCl2, and 2 mM DTT]. The resuspended sample was further clarified using a table-top centrifuge by centrifugation at 16,000g for 15 min. Puromycin (1 mM) was added to the clarified sample, which was then incubated on ice for 15 min, followed by incubation at 37°C for 10 min. The resulting sample was loaded onto a 5 to 20% sucrose gradient and then centrifuged for 10 hours at 22,000 rpm (SW-32 rotor). The gradient was pumped off with a peristaltic pump while monitoring the 254 nm absorbance and collecting 1-ml fractions. The fractions containing pure 40S subunits were pooled and buffer exchanged into storage buffer, then flash-frozen with liquid nitrogen and stored at −80°C.
In vitro transcription of mRNAs and purification
DNA templates for transcription of full-length mRNAs of RPL41A and PAB1, the GCN4 leader, and a model AUG mRNA sequence (20) were inserted into the pUC119 vector. A 5′-GpG dinucleotide RNA sequence was added to each template sequence, to maximize transcription efficiency. A suitable restriction site was added to each template sequence 3′ end, to allow linearization for runoff transcription. Plasmids were transformed into DH5α (NEB). The transformed cells were grown in 300 ml of LB media at 37°C overnight. Plasmid DNA was extracted using a maxi-prep kit (Takara Bio) and restriction-digested (RPL41A and GCN4-UTR templates with Bam HI, PAB1 template with Eco RI), to linearize the template at its 3′ end. The template was then transcribed with T7 polymerase (NEB), in transcription buffer [100 mM tris-HCl, (pH 8.0), 10 mM spermidine, 1% Triton X-100, 16 mM nucleotide triphosphates, 3% dimethyl sulfoxide, 15 mM DTT, 25 mM MgCl2, and ribonuclease inhibitor (NEB)]. Reactions were incubated at 37°C for 2 hours, followed by deoxyribonuclease treatment for 30 min. RNAs were extracted using phenol-chloroform-isoamyl alcohol (pH 4.3, 125:25:1) and ethanol precipitated. The SSA1 DNA transcription template was prepared by polymerase chain reaction amplification from yeast genomic DNA using a forward primer, 5′-TAATACGACTCACTATAGGGCACATCAAAAGAAAAGTAATC-3′, and a reverse primer, 5′-TCTATAGCGTATAGAATATATGTTACATGTATATATATATATAAAG TAAAAACGTTCGG-3′. SSA1 transcription reactions were carried out as described above, but at a MgCl2 concentration of 6 mM and for 1 hour. The RNAs were capped with the Vaccinia capping enzyme (NEB) and poly(A)-tailed with Escherichia coli poly(A) polymerase (NEB), following the manufacturer’s protocols. RNAs were freshly prepared before each single-molecule assay.
Hybridization of mRNAs with Cy5.5-oligo
Capped and poly(A)-tailed mRNAs (1 μM) were hybridized to biotin-5′-d(T)100–3′-Cy5.5 (100 nM) in 100 mM Hepes-KOH (pH 7.5) and 300 mM KCl, by heating to 98°C for 3 min using a thermocycler with slow cooling to 4°C at 0.1°C s−1. The annealed product was used directly for single-molecule experiments.
In vitro transcription and purification of initiator tRNA
A pUC19 construct containing the yeast tRNAiMet template sequence fused to a 3′ hammerhead ribozyme sequence was transformed into DH5⍺ cells (20). tRNAiMet was transcribed as for mRNAs, except on a larger (5 ml) scale for 4 hours, in 12 mM MgCl2. Ribozyme self-cleavage was initiated by increasing MgCl2 concentration to 30 mM and incubating at 60°C for an additional hour. The tRNA was further purified by gel electrophoresis and gel extraction. The tRNA was buffer exchanged into 10 mM bis-tris-HCl (pH 7.0) and 10 mM NaCl using a 5 kDa centrifugal filter (Milipore) and was stored at −80°C.
Purification of S. cerevisiae methionyl-tRNA synthetase (yMetRS)
A yeast strain harboring a plasmid that encodes GST-tagged MetRS gene was grown in 3 liters of YPAD media to ~2 OD600. The cells were harvested and washed with 1× phosphate-buffered saline (PBS), then processed as for purification of eIF3. The clarified lysate was loaded on a 1-ml GSTrap column (Cytiva), which was then washed with 1× PBS. MetRS was eluted with 10 mM reduced glutathione in 50 mM tris-HCl (pH 8.0) buffer. The eluted sample was buffer exchanged into storage buffer [40 mM tris-HCl (pH 7.4), 10 mM MgCl2, 10% glycerol, and 2 mM DTT] on a 10-DG column (Bio-Rad). The purified protein was flash-frozen with liquid nitrogen and stored at −80°C.
Methionylation of Met-tRNAiMet and acid-gel analysis
Yeast Met-tRNAiMet was methionylated as previously described (48). The tRNA was refolded by heating at 70°C for 10 min with 10 mM MgCl2 and slowly cooling to room temperature. The refolded tRNA was incubated with MetRS in acylation buffer containing 100 mM Hepes-KOH (pH 7.6), 100 μM methionine, 10 mM ATP, 1 mM DTT, 10 mM KCl, and 20 mM MgCl2. The reaction was incubated at 37°C for 20 min and extracted with acidic phenol, followed by chloroform-isoamyl alcohol (24:1). The methionylated tRNA was purified on Sephadex G-25. The acylated tRNA was analyzed by acid urea–polyacrylamide gel electrophoresis [PAGE; acrylamide:bisacrylamide ratio of 19:1, 15%, in 100 mM NaOAc (pH 4.3)] for verification and quantitation of the aminoacylation efficiency.
Native PAGE analysis of PIC formation
PIC formation was verified by 4% native PAGE as previously described (20). TC was formed by incubating eIF2 (800 nM) with GDPNP (2 mM) for 10 min at room temperature, followed by addition of Met-tRNAiMet to 2 μM and incubation for another 5 min. For native-PAGE analysis, a 43-mer model mRNA (AUG) was used (20). The RNA was ligated to a 5′-phosphorylated Cy3-oligo at the 3′ end of the RNA by T4 RNA ligase (NEB) by following the manufacturer’s protocol. The Cy3-ligated AUG model mRNA was heated at 90°C for 3 min and snap-cooled on ice for 5 min. The RNA was mixed with the TC, eIF1 (1 μM), eIF1A (1 μM), and 40S (100 nM) in the binding buffer [30 mM Hepes-KOH (pH 7.5), 2 mM Mg(OAc)2, 100 mM KOAc, and 2 mM DTT]. The mixture was incubated for an hour at room temperature and 10% (w/v) sucrose was added before loading on the gel. The gel was electrophoresed for 1 hour 30 min at 4°C and visualized using a Typhoon imager. This protocol was also used for analysis of PIC formation with Cy3-labeled 40S subunits.
Pab1p electrophoretic mobility shift analysis
To quantify Pab1p binding to PAB1 mRNA, the mRNA (50 nM) was incubated with varying concentrations of yeast Pab1p (200 to 1 nM) in 1× binding buffer [30 mM Hepes-KOH pH 7.4, 20 mM KCl, 1 mM MgCl2]. The final salt and glycerol concentration of each sample was adjusted to 100 mM KCl and 10% glycerol. The reaction was incubated for 30 min at room temperature, and then samples were loaded on a 0.8% tris-borate–EDTA buffer (0.5×) agarose gel, prestained with ethidium bromide. The gel was electrophoresed at 80 V for 1.5 hours at 4°C and then visualized with a ChemiDoc imaging system (Bio-Rad).
Measurement of phosphate concentration in TC samples
To quantitate the concentration of phosphate in incubated the TC samples, we used the commercially available EnzCheck Phosphate Assay Kit (Thermo Fisher Scientific). TC was formed initially by incubating eIF2 with 5′-GDPNP for 10 min and immediately with Met-tRNAi for 5 min at room temperature. TC was then incubated with 1× reaction buffer [50 mM tris-HCl and 1 mM MgCl2 (pH 7.5)], 2-Amino-6-mercapto-7-methylpurine riboside (200 μM), and 1 U of purine nucleoside phosphorylase for 30 min at room temperature. The absorbance of the resulting reaction mixture was measured at 360 nm. The phosphate concentration in the sample was calculated based on a standard curve prepared with inorganic phosphate (0 to 10 μM). TC samples incubated at 30°C for 1, 2, and 3 hours, and control-incubation samples containing only GDPNP, were processed using the same procedure.
Single-molecule fluorescence assay
The scanning assay was performed in a customized PacBio RSII instrument (16). The general procedure described previously (46) was modified as follows. For 40S subunit labeling, Cy3-oligo was incubated with 40S in ~1.1:1 ratio (2.5 μM 40S:2.25 μM Cy3-oligo), and the sample was heated at 42°C for 2 min, then slowly cooled at 37°C for 15 min and 30°C for another 15 min. The TC was formed by incubating eIF2 (2 μM) with GTP (3.5 mM) for 10 min and then with initiator tRNA (2 μM) for a further 5 min, at room temperature. Initially, a SMRT cell (Pacific Biosciences) containing 150,000 ZMWs was reacted with 16 μM NeutrAvidin (Invitrogen) for 5 min at room temperature. The surface was then washed with assay buffer three times [30 mM Hepes-KOH (pH 7.5) and 3 mM Mg(OAc)2, and 100 mM KOAc]. The biotin-5′-d(T)100–3′-Cy5.5 annealed mRNA sample (10 nM) was immobilized for 5 min. The surface was again washed with assay buffer and then blocked with unlabeled eIF1 (1 μM), a mixture of Biolipidure 203 and 206 [5% (v/v) each, NOF America], and bovine serum albumin (1 mg/ml; BSA), to abrogate nonspecific surface binding of Cy5-eIF1 and Cy3-40S subunits. Excess block-mixture components were removed by one wash with assay buffer supplemented with 2 mM protocatechuic acid, 2 mM triplet-state quencher (Pacific Biosciences), and 1× protocatechate-3,4-dioxygenase oxygen scavenging system (Pacific Biosciences). Twenty microliters of imaging buffer containing the oxygen scavenging system and BSA (1 mg/ml) was then added to the SMRT cell. During cell preparation, PICs were reconstituted by combining 50 nM Cy5-eIF1, 500 nM eIF1A, 50 nM Cy3-40S, 300 nM TC, 500 nM eIF3, 200 nM eIF4E, 80 nM eIF4G, 1 μM eIF4A, 500 nM eIF4B [varying concentrations (10 to 100 nM) of Pab1p were added to experiments containing Pab1p for PAB1 mRNA], 2 mM ATP, 200 nM eIF5, oxygen scavenging system, and 1× casein, in the order given. The final concentration of all components is halved with respect to the values above, upon dilution of the sample into the 20 μl of imaging buffer previously added to the cell. The reconstituted PIC (20 μl) was then robotically delivered at the beginning of the experiment and Cy3, Cy5, and Cy5.5 fluorophores were excited by illuminating with green (532 nm) and red (642 nm) laser at 0.7 and 0.07 μW μm−2, respectively. Movies were recorded at 10 frames per second for 15 min. For the experiments with GDPNP, the TC was formed with GDPNP instead of GTP.
Single-molecule data analysis
The movies were analyzed using MATLAB scripts reported previously (16). Fluorescence traces were first extracted from raw movie data with automated selection of ZMWs containing a Cy3 signal. The resulting traces were then manually curated to ensure that they each contained the following: (i) a Cy5.5-mRNA signal at the start of the movie and (ii) an event with co-arrival of Cy3-40S and Cy5-eIF1. Put otherwise, traces were only included in the dataset if they demonstrably reported on a surface-immobilized RNA being recognized by a complex of eIF1 with the 40S subunit at the ZMW surface. The durations of the Cy5-eIF1 events that followed co-arrival with a Cy3-40S signal were manually assigned and tabulated. Data were binned in 1-s increments to produce the normalized-density eIF1 dwell-time histograms depicted in the figures.
Statistical analysis
All statistical analysis and fitting was performed in MATLAB. Wilcoxon rank-sum tests to compare dwell-time distribution pairs were performed at a significance level of 0.05. For empirical exponential fitting of eIF1 dwell-time distributions, the tabulated dwell times were converted to an empirical cumulative distribution function, then this function was fit by nonlinear regression using the Trust-Region algorithm with bisquares weighting. Goodness of fit was evaluated by inspection of root mean square error values, which were typically in the range of probability −0.01 to −0.09. An identical nonlinear regression approach was taken for hyperbolic and Hill fitting of the scanning time dependence on Pab1p concentration. For linear regression of mean or median eIF1 dwell times versus GCN4 leader lengths, the data were fit using the Trust-Region algorithm. Goodness of fit was evaluated based on the R2 value, which ranged from 0.7 to 0.8. Ninety-five percent confidence intervals for estimation of the slopes are reported in the text.
Acknowledgments
We thank A. G. Hinnebusch (National Institute of Child Health and Human Development) for providing yeast strain GP3511, and A. Petrov (Auburn University) for providing the strain for production of the fluorescently labelable yeast 40S subunits. pUC19-HHtRNA, for preparation of the initiator tRNA, was a gift from J. Lorsch (Addgene plasmid no. 37222). Plasmid pSW149, for eIF4G1 overexpression, was a gift from S. Walker (University at Buffalo).
Funding: This work was supported by NIH grants R01GM138939 and R00GM111898 (S.E.O.).
Author contributions: Conceptualization: S.E.O. and G.B. Methodology: S.E.O., H.J.H., A.L.Z., and G.B. Investigation: H.J.H., A.L.Z., and A.B.C. Visualization: H.J.H. and S.E.O.. Validation: H.J.H., A.L.Z., A.B.C., and S.E.O. Formal analysis: S.E.O. and H.J.H.. Funding acquisition: S.E.O. Project administration: S.E.O., A.L.Z., and H.J.H. Resources: H.J.H. and G.B. Supervision: S.E.O. Writing–original draft: H.J.H. and S.E.O.. Writing–review and editing: H.J.H., and S.E.O.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Materials are available on written request to the corresponding author. Custom MATLAB code (16) is available at: https://drive.google.com/drive/folders/1k1ZvZqb-TMgjpysfUVnEEwbiFJ7s0rsr?usp=share_link.
Supplementary Materials
The PDF file includes:
Figs. S1 to S8
Table S1
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Aitken C. E., Lorsch J. R., A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Kozak M., How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 15, 1109–1123 (1978). [DOI] [PubMed] [Google Scholar]
- 3.Peters J. M., Vangeloff A. D., Landick R., Bacterial transcription terminators: The RNA 3′-end chronicles. J. Mol. Biol. 412, 793–813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson J. P., Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. G., Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51, 987–1001 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Vassilenko K. S., Alekhina O. M., Dmitriev S. E., Shatsky I. N., Spirin A. S., Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 39, 5555–5567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Shin B.-S., Alvarado C., Kim J.-R., Bohlen J., Dever T. E., Puglisi J. D., Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation. Cell 185, 4474–4487.e17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearse M. G., Green K. M., Krans A., Rodriguez C. M., Linsalata A. E., Goldstrohm A. C., Todd P. K., CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell 62, 314–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McManus C. J., May G. E., Spealman P., Shteyman A., Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 24, 422–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary J. D., Pattamatta A., Ranum L. P. W., Repeat-associated non-ATG (RAN) translation. J. Biol. Chem. 293, 16127–16141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGillivray P., Ault R., Pawashe M., Kitchen R., Balasubramanian S., Gerstein M., A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res. 46, 3326–3338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer S. K., Shirokikh N. E., Beilharz T. H., Preiss T., Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg A. R., Higdon A. L., Hollerer I., Fields A. P., Jungreis I., Diamond P. D., Kellis M., Jovanovic M., Brar G. A., G. A., Translation initiation site profiling reveals widespread synthesis of non-AUG-initiated protein isoforms in yeast. Cell Syst. 11, 145–160.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Algire M. A., Maag D., Lorsch J. R., Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20, 251–262 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G., Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21, 1217–1230 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Dalal R. V., Petrov A. N., Tsai A., O’Leary S. E., Chapin K., Cheng J., Ewan M., Hsiung P.-L., Lundquist P., Turner S. W., Hsu D. R., Puglisi J. D., High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 111, 664–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrov A. N., Puglisi J. D., Site-specific labeling of Saccharomyces cerevisiae ribosomes for single-molecule manipulations. Nucleic Acids Res. 38, e143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maag D., Lorsch J. R., Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330, 917–924 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Maag D., Fekete C. A., Gryczynksi Z., Lorsch J. R., A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17, 265–275 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R., Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Yourik P., Aitken C. E., Zhou F., Gupta N., Hinnebusch A. G., Lorsch J. R., Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. eLife 6, e31476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jivotovskaya A. V., Valasek L., Hinnebusch A. G., Nielsen K. H., Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 26, 1355–1372 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell S. F., Walker S. E., Algire M. A., Park E.-H., Hinnebusch A. G., Lorsch J. R., The 5′-7-methylguanosine cap on eukaryotic mRNAs serves to both stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell 39, 950–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker S. E., Zhou F., Mitchell S. F., Larson V. S., Valasek L., Hinnebusch A. G., Lorsch J. R., Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 19, 191–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floyd D. L., Harrison S. C., van Oijen A. M., Analysis of kinetic intermediates in single-particle dwell-time distributions. Biophys. J. 99, 360–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch A. G., Ivanov I. P., Sonenberg N., Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clements J. M., Laz T. M., Sherman F., Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4533–4536 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei J., Zhang Y., Ivanov I. P., Sachs M. S., The stringency of start codon selection in the filamentous fungus Neurospora crassa. J. Biol. Chem. 288, 9549–9562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinnebusch A. G., Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M., Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 87, 8301–8305 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L., Liu P., Dai Z., Fan F., Zhang X., Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants. Microb. Cell Fact. 20, 148 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaevitz J. W., Block S. M., Schnitzer M. J., Statistical kinetics of macromolecular dynamics. Biophys. J. 89, 2277–2285 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinnebusch A. G., Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Stumber M., Herrmann C., Wohlgemuth S., Kalbitzer H. R., Jahn W., Geyer M., Synthesis, characterization and application of two nucleoside triphosphate analogues, GTPγNH2 and GTPγF. Eur. J. Biochem. 269, 3270–3278 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Gilbert W. V., Zhou K., Butler T. K., Doudna J. A., Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317, 1224–1227 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Melo E. O., Dhalia R., Sa C. M., Standart N., de Melo Neto O. P., Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: Role in cooperative binding to poly (A) and efficient cap distal translational repression. J. Biol. Chem. 278, 46357–46368 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Sachs A. B., Davis R. W., Kornberg R. D., A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and viability. Mol. Cell. Biol. 7, 3268–3276 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisinger J., Feuer B., Yamane T., Codon-anticodon binding in tRNAPhe. Nat. New Biol. 231, 126–128 (1971). [DOI] [PubMed] [Google Scholar]
- 39.Palmiter R. D., Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell 4, 189–197 (1975). [DOI] [PubMed] [Google Scholar]
- 40.Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J. B., Rate-limiting steps in yeast protein translation. Cell 153, 1589–1601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomakin I. B., Steitz T. A., The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500, 307–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou F., Bocetti J. M., Hou M., Qin D., Hinnebusch A. G., Lorsch J. R., Transcriptome-wide analysis of the function of Ded1 in translation preinitiation complex assembly in a reconstituted system. eLife 13, RP93255 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jennings M. D., Pavitt G. D., Quantifying the binding of fluorescently labeled guanine nucleotides and initiator tRNA to eukaryotic translation initiation factor 2. Methods Mol. Biol. 2428, 89–99 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Schuessler P. J., Sahoo A., Walker S. E., Reconstitution and analyses of RNA interactions with eukaryotic translation initiation factors and ribosomal preinitiation complexes. Methods 162–163, 42–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Leary S. E., Petrov A., Chen J., Puglisi J. D., Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure 21, 2197–2207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Çetin B., O'Leary S. E., mRNA- and factor-driven dynamic variability controls eIF4F-cap recognition for translation initiation. Nucleic Acids Res. 50, 8240–8261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreou A. Z., Klostermeier D., eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 426, 51–61 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Walker S. E., Fredrick K., Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1
Legend for data S1
Data S1