Abstract
Despite novel therapeutic strategies, advanced-stage prostate cancer (PCa) remains highly lethal, pointing out the urgent need for effective therapeutic strategies. While dysregulation of the splicing process is considered a cancer hallmark, the role of certain splicing factors remains unknown in PCa. This study focuses on characterizing the levels and role of SRSF6 in this disease. Comprehensive analyses of SRSF6 alterations (copy number/mRNA/protein) were conducted across eight well-characterized PCa cohorts and the Hi-MYC transgenic model. SRSF6 was up-regulated in PCa samples, correlating with adverse clinical parameters. Functional assays, both in vitro (cell proliferation, migration, colony, and tumorsphere formation) and in vivo (xenograft tumors), demonstrated the impact of SRSF6 modulation on critical cancer hallmarks. Mechanistically, SRSF6 regulates the splicing pattern of the histone-chaperone HIRA, consequently affecting the activity of H3.3 in PCa and breast cancer cell models and disrupting pivotal oncogenic pathways (AR and E2F) in PCa cells. These findings underscore SRSF6 as a promising therapeutic target for PCa/advanced-stage PCa.
SRSF6 up-regulation in PCa cells controls malignant AR/E2F pathways by regulating HIRA splicing and histone H3.3 activity.
INTRODUCTION
Prostate cancer (PCa) is one of the most diagnosed cancer types worldwide and represents the fifth leading cause of cancer-related death worldwide (1). The main therapeutic approach to tackle PCa consists of targeting androgen receptor (AR) signaling, initially by using the so-called androgen deprivation therapy, followed by second-generation antiandrogens such as enzalutamide or abiraterone (2). Unfortunately, advanced PCa will eventually become resistant to AR blockade mainly due to aberrations driving AR-persistent signaling, such as AR amplification, AR-truncated variants, AR mutations, and/or dysregulation of AR coregulators (2, 3) resulting in castration-resistant prostate cancer (CRPC). Although many efforts have been put into blocking AR-persistent signaling, to date, there are not clinically available therapeutic strategies for that purpose. Therefore, discovering new molecular/therapeutic approaches that could be exploited to uncover novel strategies to target ongoing AR signaling in advanced PCa represents a clinical unmet need. In this scenario, it has been suggested that the dysregulation of the splicing process represents an intrinsic characteristic of PCa inasmuch as a broad number of splicing variants (SVs) are altered in any stage of the disease, which can act as drivers of PCa progression and/or modulate the expression, function, or activity of key oncogenes and tumor-suppressor genes (4). We and others have also recently shown that the components of the cellular machinery that catalyzes and regulates the splicing process [i.e., spliceosome components (SCs) and splicing factors (SFs)] are deeply dysregulated in PCa and can control pivotal PCa-related pathways, including AR activity (5–9).
In this sense, one important family of RNA processing factors is the serine/arginine (SR)–rich splicing factor family, which is characterized by containing one or two RNA recognition motifs at the N terminus, and a domain rich in arginine and serine dipeptide repeats at the C terminus that mediates protein-protein interactions (10, 11). Among the SR family members, SRSF6 stands up because of its key pathophysiological role in relevant diseases, including pleural fibrosis (12), Huntington’s disease (13), Alzheimer’s disease (14), diabetes (15), and systemic sclerosis (16) as well as different cancer types such as breast cancer (BCa) (17), melanoma (18), lung (19), and colorectal cancer (19, 20). Despite the connection between SRSF6 and the oncogenic transformation and tumor progression in the abovementioned cancer types, there are no reports addressing the potential pathophysiological role that SRSF6 might play in PCa. For that reason, this study aimed to characterize the levels of SRSF6 in PCa, its pathophysiological function, and molecular consequences of modulating (overexpression and silencing) its levels using both in vitro and in vivo PCa models, as well its potential role as diagnostic/prognostic biomarker and therapeutic target in this disease.
RESULTS
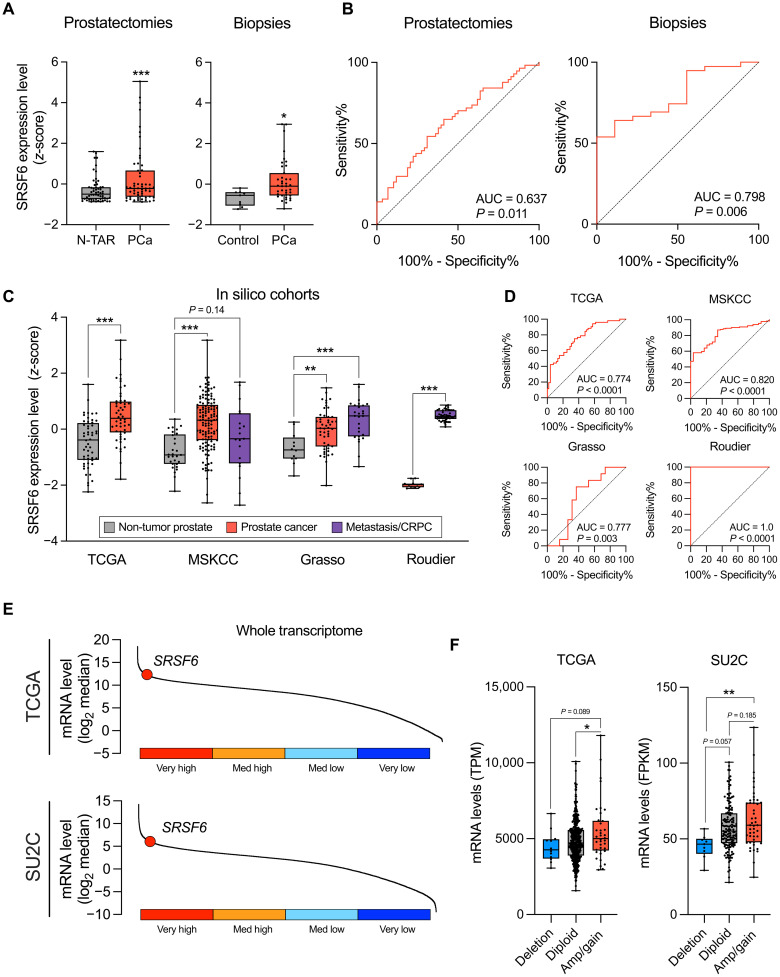
SRSF6 is overexpressed in PCa
We first interrogated the mRNA levels of SRSF6 in two independent human cohorts available in our laboratory [prostatectomies (cohort 1) and biopsies (cohort 2); tables S1 and S2, respectively], comprising PCa and non-tumor prostate tissue samples. SRSF6 mRNA levels were higher in PCa samples versus non-tumor prostate tissue in both cohorts (Fig. 1A). Moreover, SRSF6 expression significantly distinguished between PCa and non-tumor prostate samples in both cohorts (Fig. 1B). Consistently, we found that SRSF6 was also up-regulated in PCa and its levels distinguished between PCa and non-tumor prostate tissues in three additional human external cohorts [The Cancer Genome Atlas (TCGA) (21), Memorial Sloan Kettering Cancer Center (MSKCC) (22), and Grasso (23) datasets (Fig. 1, C and D)]. Notably, SRSF6 levels were especially elevated in CRPC samples in the Grasso cohort and were significantly higher in CRPC samples compared to those in primary PCa samples from the Roudier (24) cohort (Fig. 1C). SRSF6 expression levels perfectly discriminated between primary PCa and CRPC samples in the Roudier cohort [area under the curve (AUC) = 1.0, P < 0.001; Fig. 1D]. Of note, whole-transcriptome analyses from TCGA and Stand Up to Cancer/Prostate Cancer Foundation (SU2C) cohorts revealed that SRSF6 was among the top 25% of most expressed transcripts in both primary and CRPC tumors, respectively (Fig. 1E). Then, to investigate putative genomic alterations associated with SRSF6 overexpression in PCa, we explored SRSF6 genomic copy number alteration (CNA) and its association with SRSF6 mRNA levels in the TCGA [primary PCa; (21)] and SU2C [CRPC; (25)] cohorts. Specifically, SRSF6 was amplified/copy gained in 7.8% and deep deleted in 2.5% of the PCa tumors from TCGA cohort (fig. S1A). Furthermore, SRSF6 was amplified/copy gained in 22.6% and deep deleted in 4.3% of the PCa tumors from SU2C cohort (fig. S1B). In both TCGA and SU2C cohorts, SRSF6 CNA exhibited a significant association with SRSF6 mRNA levels (Fig. 1F). Besides that, SRSF6 mRNA levels did not consistently associate with any molecular subtype or common genomic aberration of PCa (fig. S1, C and D).
Fig. 1. mRNA levels and CNA of SRSF6 in patients’ prostate samples.
(A) Comparison of SRSF6 mRNA levels between non-tumor adjacent regions (N-TARs) versus PCa samples from the prostatectomy cohort (left; n = 84), and control (n = 9) versus PCa samples (n = 42) from the biopsy cohort (right). Data represent the min-to-max boxplot, with median of mRNA expression levels adjusted by normalization factor (calculated from ACTB and GAPDH expression levels) and standardized by z-score. (B) Receiver operating characteristic (ROC) curves of SRSF6 mRNA levels to distinguish between tumor and non-tumor samples from prostatectomy (left) and biopsy (right) cohorts. Area under the curve (AUC) and P value are depicted in the plots. (C) Comparison of SRSF6 mRNA levels between non-tumor prostate tissues, PCa, and/or metastatic/CRPC samples from The Cancer Genome Atlas (TCGA), Memorial Sloan Kettering Cancer Center (MSKCC), Grasso, and Roudier cohorts. Data represent the min-to-max boxplot, with median of SRSF6 expression levels standardized by z-score. (D) ROC curves of SRSF6 mRNA levels to distinguish between PCa and non-tumor prostate samples from the TCGA (top left), MSKCC (top right), and Grasso (bottom left) cohorts and between CRPC and primary PCa samples from Roudier cohort (bottom right). AUC and P value are depicted in the plots. (E) Ranked expression of SRSF6 across whole transcriptome in the TCGA (top) and SU2C (bottom) cohorts. (F) Association between SRSF6 mRNA levels (min-to-max boxplot, with median) and SRSF6 copy number alterations (CNAs) in the TCGA (left) and SU2C (right) cohorts. Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups. TPM, transcript per million; FPKM, fragments per kilobase million.
SRSF6 expression levels are associated with key clinical and molecular aggressiveness features in PCa
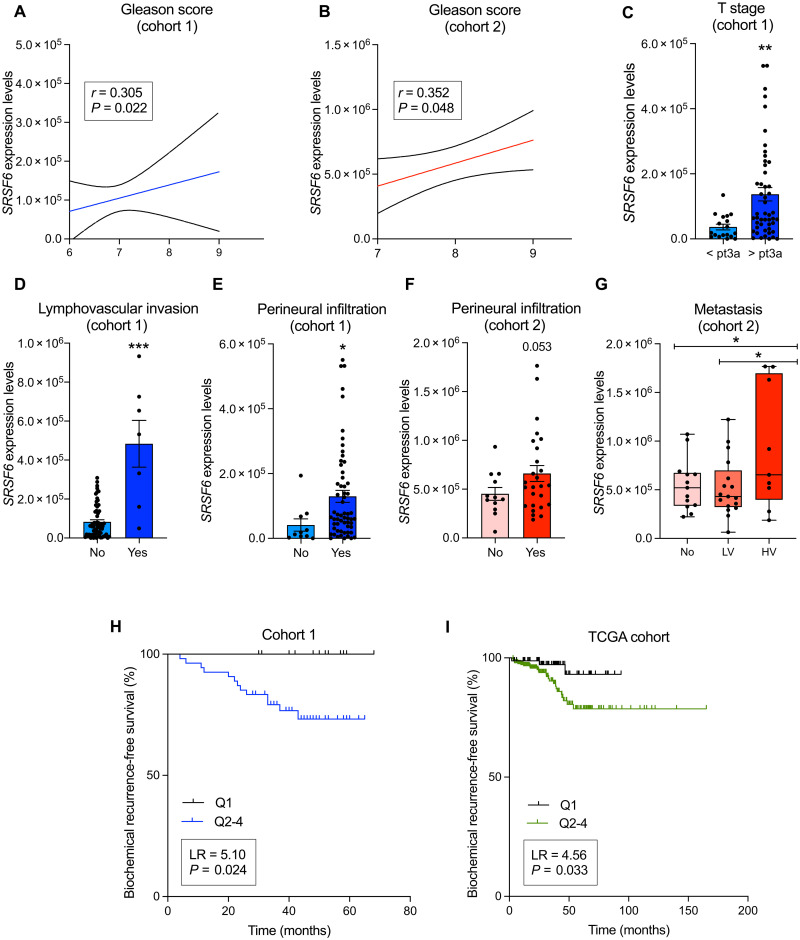
SRSF6 mRNA levels were positively correlated with Gleason score in our cohorts of PCa samples [prostatectomies (Fig. 2A) and biopsies (Fig. 2B)]. Consistently, PCa samples with high T stage (pT > 3a), lymphovascular invasion, or perineural infiltration were associated with high SRSF6 expression levels in our cohort of samples derived from prostatectomies (Fig. 2, C to E, respectively). On the same line, when analyzing a more aggressive cohort of PCa samples (cohort 2), we found that patients with high SRSF6 levels tended to be associated with tumors presenting perineural infiltration (Fig. 2F). Moreover, primary PCa tumors from patients presenting high volume metastases at diagnosis had higher SRSF6 expression levels compared to those who presented low volume metastases or those not presenting metastasis at diagnosis (Fig. 2G). Patients with low SRSF6 expression levels (quartile 1) showed significantly longer biochemical recurrence-free survival as compared to the rest of the patients in the prostatectomy samples (cohort 1) and in the TCGA cohort (Fig. 2, H and I, respectively). Aside that, SRSF6 levels did not associate with T stage or differ between different metastasis sites in TCGA and SU2C, respectively (fig. S2, A and B).
Fig. 2. Associations and correlations of SRSF6 expression levels with clinical parameters of PCa aggressiveness.
(A and B) Correlation of Gleason score with SRSF6 mRNA levels in the prostatectomy (A) and biopsy (B) cohorts. (C to E) Associations of SRSF6 mRNA levels and T stage (C), lymphovascular invasion (D), and perineural infiltration (E) in the prostatectomy cohort. (F and G) Associations of SRSF6 mRNA levels and perineural infiltration (F) and metastasis (LV, low volume; HV, high volume) (G) in the biopsy cohort. (H and I) Association between biochemical progression-free survival and SRSF6 mRNA levels [quartile 1 (Q1) versus Q2-4)] in the prostatectomy (H) and TCGA (I) cohorts. SRSF6 mRNA levels are adjusted by normalization factor (calculated from ACTB and GAPDH expression levels). Correlations data are represented by mean (connecting line) and error bands (pointed line). Data of associations represent the means ± SEM of mRNA expression levels. Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups. LR, log-rank.
Furthermore, we interrogated the cohort 2 to explore the association of SRSF6 expression levels with molecular parameters of PCa aggressiveness. Specifically, we observed that SRSF6 mRNA levels tended to be directly correlated with those of PCA3, SST5TMD4, In1-Ghrelin, and ESRP1 (fig. S2, C to E and H, respectively). Last, no correlation was found between SRSF6 and AR or AR-V7 mRNA levels (fig. S2, F and G, respectively).
SRSF6 protein levels are elevated in PCa
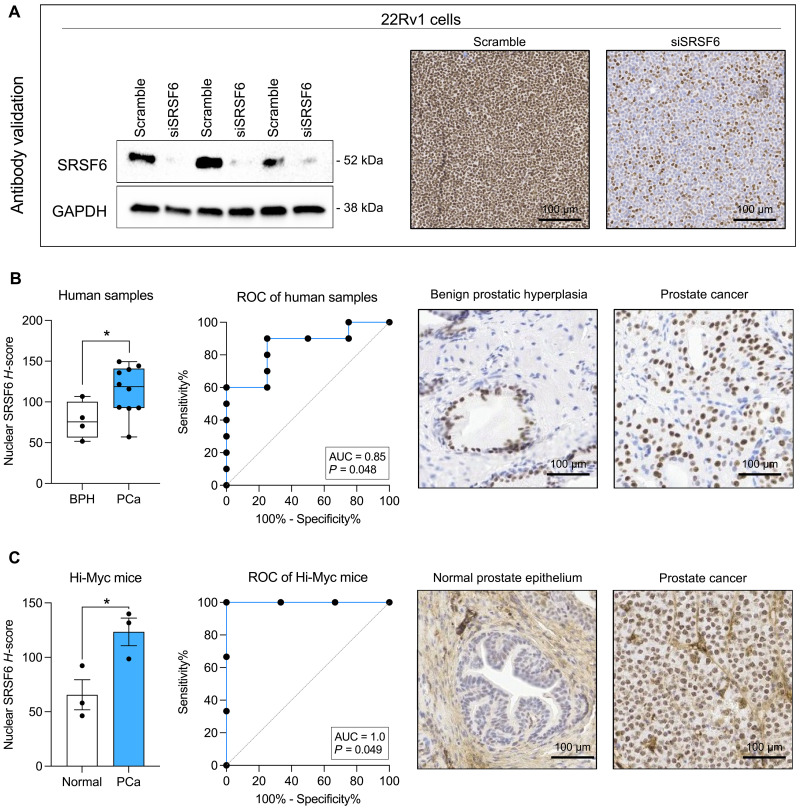
To study the protein levels of SRSF6 in PCa, we first validated the Abcam SRSF6 antibody (ab140623). SRSF6 signal and staining decreased in response to SRSF6–small interfering RNA (siRNA) by Western blot and immunohistochemistry (IHC), respectively, in 22Rv1 cells (Fig. 3A). Then, we analyzed a representative number of samples (cohort 3), including benign prostatic hyperplasia (BPH; n = 4) and PCa (n = 10) and observed that SRSF6 protein levels were significantly higher in PCa as compared to those in control BPH samples and that these levels distinguished between both groups (AUC = 0.85, P = 0.048; Fig. 3B). Furthermore, when analyzing samples derived from the Hi-Myc transgenic mouse model, we found that SRSF6 protein levels were also higher in PCa compared to those in non-tumor prostate tissue samples and perfectly discriminated between the two groups (AUC = 1.0, P = 0.049; Fig. 3C).
Fig. 3. Immunohistochemical analysis of SRSF6 in non-tumor prostate and PCa samples.
(A) Antibody validation by Western blot (left) and immunohistochemistry (IHC; right) in response to SRSF6 small interfering RNA (siRNA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as housekeeping for Western blot. (B) Comparison of SRSF6 protein levels by IHC between benign prostatic hyperplasia (BPH; n = 4) and PCa (n = 10) samples (left). Data are expressed as min-to-max boxplot, with median of nuclear SRSF6 H-score. ROC curve of SRSF6 protein levels to distinguish between BPH (n = 4) and PCa (n = 10) samples (center). AUC and P value are depicted in the plots. Representative images of BPH and PCa samples stained with SRSF6 antibody (right). (C) Comparison of SRSF6 protein levels between PCa (n = 3) and non-tumor prostate tissue (n = 3) samples from Hi-Myc mice (left). ROC curve of SRSF6 protein levels to distinguish between PCa (n = 3) and non-tumor prostate tissue (n = 3) from Hi-Myc mice (center). AUC and P value are depicted in the plots. Representative images of normal prostate epithelium and PCa samples from Hi-Myc mice stained with SRSF6 antibody (right). Asterisks (*P < 0.05) indicate statistically significant differences between groups.
SRSF6 mRNA levels are associated with MYC activity
On the basis of the previous data, we next interrogated the potential implication of the MYC pathway in the pathological regulation of SRSF6 levels. Specifically, we found that SRSF6 is one of the top-scoring potential target genes for MYC across different cell lines based on the ChIP-Atlas database (fig. S3A) (26). We observed that MYC overexpression increased SRSF6 mRNA levels in LNCaP and 22Rv1 cells (fig. S3, B and C). Furthermore, MYC expression and activity were positively correlated with SRSF6 mRNA levels in TCGA and SU2C cohorts (fig. S3, F to I), despite no significant associations were found for MYC gene amplification (fig. S3, D and E).
Modulation of SRSF6 expression alters aggressiveness of PCa cells in vitro
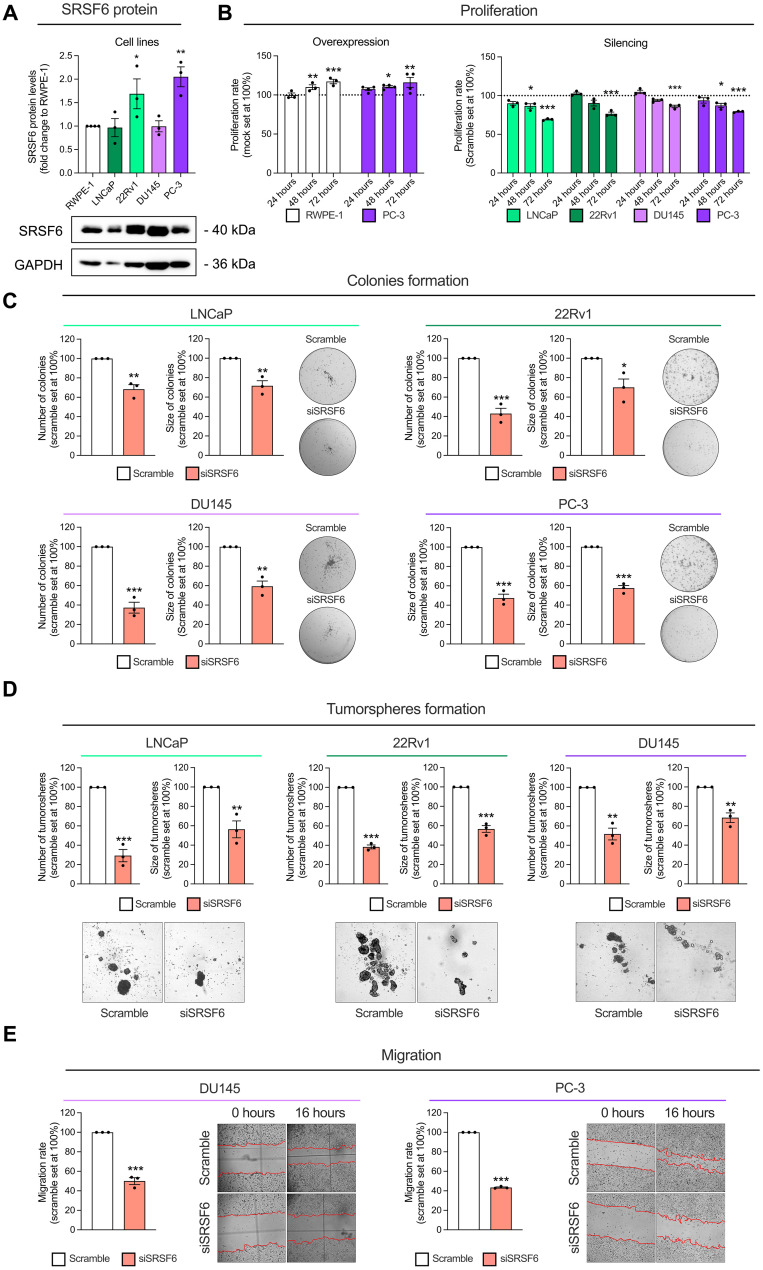
As observed in human tissues, we also found that SRSF6 mRNA levels are elevated in all the PCa cell lines analyzed herein (i.e., LNCaP, 22Rv1, DU145, and PC-3) compared to those in the non-tumor prostate-derived cell line RWPE-1 (fig. S4A). Furthermore, SRSF6 protein levels were consistently detectable in all the cell lines and significantly elevated in 22Rv1 and PC-3 compared to those in the RWPE-1 cells (Fig. 4A). On the basis of these data, we tested whether the modulation in the expression of SRSF6 could alter aggressiveness parameters in these prostate-derived cell models. In vitro SRSF6 overexpression in RWPE-1 and PC-3 cell lines [validation shown in fig. S4 (B and G)] increased proliferation rate at 48 and 72 hours (Fig. 4B). On the other hand, SRSF6 silencing in LNCaP, 22Rv1, DU145, and PC-3 cells [validation shown in fig. S4 (C to F and H to K)] decreased the proliferation rate at 48 and/or 72 hours (Fig. 4B). Moreover, SRSF6 silencing decreased the number and size of colonies formed by LNCaP, 22Rv1, DU145, and PC-3 (Fig. 4C) and reduced the number and size of tumorspheres formed by LNCaP, 22Rv1, and DU145 cells (Fig. 4D). In addition, SRSF6 silencing decreased the migration rate of DU145 and PC-3 after 16 hours (Fig. 4E).
Fig. 4. Functional consequences in response to SRSF6 expression modulation in prostate-derived cell lines.
(A) Comparison of SRSF6 protein levels between a non-tumor prostate cell line (RWPE-1) and PCa cell lines LNCaP, 22Rv1, DU145, and PC-3 (n = 3). SRSF6 protein levels were determined by Western blot and adjusted by GAPDH. Data are represented as fold change of RWPE-1 cells (means ± SEM). Representative images of Western blot are depicted on the bottom panels. (B) Proliferation rate in response to SRSF6 overexpression in RWPE-1 and PC-3 cell lines (left), and in response to SRSF6 silencing in LNCaP, 22Rv1, DU145, and PC-3 cell lines (right) at 24, 48, and 72 hours determined by Resazurin assay. Data are represented as percentage to control cells (means ± SEM). (C) Number (left) and size (right) of colonies in response to SRSF6 siRNA in LNCaP (top left), 22Rv1 (top right), DU145 (bottom left), and PC-3 (bottom right). Images of representative wells are depicted. Data are represented as percentage to scramble cells (means ± SEM). (D) Number (left) and size (right) of tumorspheres in response to SRSF6 siRNA in LNCaP (left), 22Rv1 (center), and DU145 (right). Images of representative areas of wells are depicted. Data are represented as percentage to scramble cells (means ± SEM). (E) Migration rate in response to SRSF6 silencing after 16 hours of incubation in DU145 (left) and PC-3 (right) cell lines determined by wound-healing assay. Representative images are depicted. Data are represented as percentage to scramble cells (means ± SEM). Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups.
Modulation of SRSF6 expression alters the aggressiveness of preclinical models of PCa
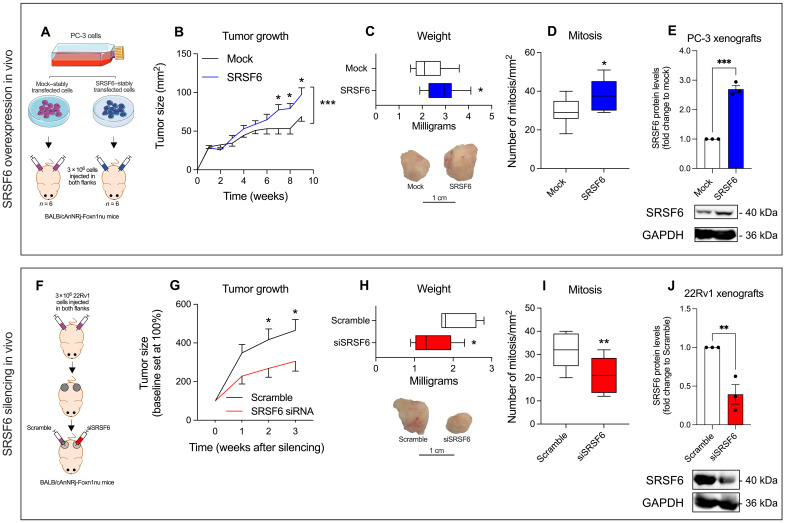
To study the pathophysiological role of SRSF6 in PCa in vivo, we stably overexpressed SRSF6 in PC-3 cells and inoculated them in immunosuppressed mice to follow up the tumor growth (Fig. 5A). SRSF6-overexpressing tumors grew faster, formed larger tumors, and exhibited higher number of mitotic cells compared to control tumors (Fig. 5, B to D, respectively). As a proof of concept, we explored the effect of SRSF6 silencing in vivo in already formed 22Rv1-derived tumors in immunosuppressed mice (Fig. 5F). Notably, the SRSF6 silencing resulted in a significant decrease of tumor growth and volume, as well as in a less proportion of mitosis as compared to control tumors (Fig. 5, G to I, respectively). Validation of SRSF6 overexpression and silencing is shown in Fig. 5 (E and J), respectively.
Fig. 5. In vivo tumor growth in response to SRSF6 expression modulation.
(A and F) Schematic representation of the in vivo tumor growth experiment in response to SRSF6 overexpression (A) and silencing (F). (B to E) Comparison between the growth over time (B), weight at the end of experiment (representative images of tumors are depicted in bottom) (C), number of mitosis (D), and SRSF6 protein levels (E) of xenograft tumors derived from mock-transfected cells or SRSF6-overexpressing PC-3 cells. SRSF6 protein levels were determined by Western blot and adjusted by GAPDH. Representative images of Western blot are depicted on bottom panels. (G to J) Comparison between the growth over time (G), weight at the end of experiment (representative images of tumors are depicted in bottom) (H), number of mitosis (I), and SRSF6 protein levels (E) of xenograft tumors derived from 22Rv1 cells treated in vivo with scramble or SRSF6 siRNA. SRSF6 protein levels were determined by Western blot and adjusted by GAPDH. Representative images of Western blot are depicted on the bottom panels. Data are represented as means ± SEM. Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups.
SRSF6 controls AR and E2F signaling pathways in PCa cells
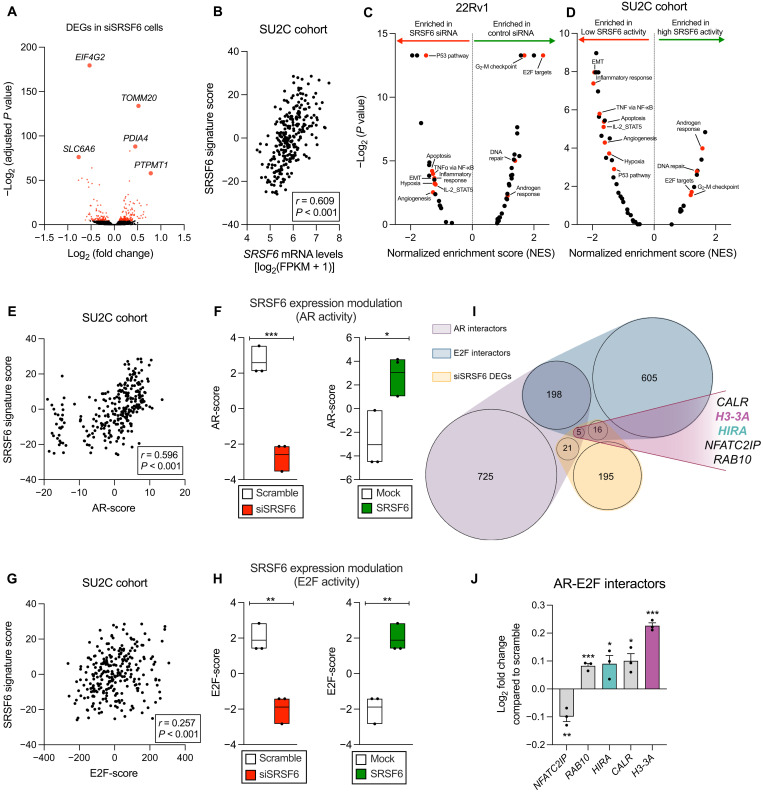
To study the potential molecular consequences of SRSF6 alteration, we performed an RNA sequencing (RNA-seq) in SRSF6-silenced 22Rv1 cells. Specifically, SRSF6 silencing significantly altered the expression of 239 genes, including those associated with tumor progression and drug resistance such as EIF4G2 (27, 28), PDIA4 (29), PTPMT1 (30), SLC6A6 (31), and TOMM20 (32) (Fig. 6A and data S1). To explore potential molecular redundancies, we checked the expression levels of the SR family members in response to SRSF6 silencing and found that none of them were significantly altered (fig. S5). As previously reported, the mere mRNA expression of a given RNA binding protein (RBP) may not perfectly reflect its protein activity. Specifically, given that RBPs commonly play a role in regulating gene expression (33), the mRNA levels of their potential downstream targets, rather than their own expression, are considered an optimal reflection of their activity, as shown by some authors (34, 35). For that reason, we defined a SRSF6 gene signature score to better correlate with its real activity, comprising the down-regulated genes in siSRSF6 versus scramble 22Rv1 cells (101 genes; P < 0.05; data S1) that were positively correlated with SRSF6 mRNA levels in the SU2C cohort. The SRSF6 signature was reduced in siSRSF6 22Rv1 cells (fig. S6A) and positively correlated with SRSF6 mRNA levels in SU2C (Fig. 6B) and TCGA (fig. S6B) cohorts. To further characterize the molecular implications of SRSF6 in PCa, a gene set enrichment analysis (GSEA) was conducted, revealing that certain hallmark gene sets were consistently associated with low levels of SRSF6 in 22Rv1 cells (Fig. 6C) as well as in patients from SU2C cohort (Fig. 6D), including genes related with apoptosis, P53, and epithelial-mesenchymal transition (EMT) pathways, while other gene sets were enriched in high levels of SRSF6, including AR and E2 promoter binding factor (E2F) pathways. Given the key role of AR and E2F pathways on PCa development and aggressiveness (2, 36, 37), we explored their potential functional relation with SRSF6 in PCa. Specifically, SRSF6 signature score and SRSF6 mRNA levels were positively correlated with AR downstream genes that represent markers of AR activity [AR-score; (38)] in SU2C and TCGA cohorts (Fig. 6E and fig. S6, C and D). In addition, SRSF6 silencing decreased AR-score in 22Rv1 and LNCaP cells, while SRSF6 overexpression increased it in 22Rv1 cells [Fig. 6F and fig. S7, C to F; validation of SRSF6 overexpression in 22Rv1 cells shown in fig. S7 (A and B)]. Moreover, we demonstrated that AR signaling inhibition increased, while AR stimulation decreased, the expression of SRSF6 in LNCaP cells (fig. S7M), suggesting a feedback mechanism that could potentially fuel AR activity in response to anti-androgens in a hormone-sensitive PCa setting. On the other hand, SRSF6 gene signature score, but not SRSF6 mRNA levels, positively correlated with E2F downstream genes that represent markers of E2F pathway activation (E2F-score), in SU2C and TCGA cohorts (Fig. 6G and fig. S6, E and F). Consistently, E2F-score was decreased in response to SRSF6 silencing in 22Rv1 and LNCaP cells, while SRSF6 overexpression increased it in 22Rv1 cells (Fig. 6H and fig. S7, G to J). In addition, consistent with the well-known role of AR and E2F pathways in stimulating PCa cell proliferation, we found that both SRSF6 expression and signature score were positively correlated with the Whitfield cell cycle signature (fig. S6, G and H). In line with this, SRSF6 silencing significantly reduced the Whitfield cell cycle signature score in 22Rv1 cells (fig. S6I), reinforcing the role of SRSF6 in promoting cell proliferation as previously observed in this study (Figs. 4B and 5). However, SRSF6 silencing did not alter the expression of AR, AR-V7, or E2F1-8 in 22Rv1 cells (fig. S7, K and L), suggesting that SRSF6 could be controlling the activity of AR and E2F through the modulation of co-regulators. We found that SRSF6 silencing altered the expression of five co-regulators of AR and E2F family members (Fig. 6, I and J). Specifically, NFATC2IP was down-regulated, while RAB10, HIRA, CALR, and H3-3A were up-regulated in response to SRSF6 silencing in 22Rv1 cells (Fig. 6J).
Fig. 6. Molecular consequences in response to the modulation of SRSF6 expression in PCa cells.
(A) Volcano plot showing differentially expressed genes (DEGs) in siSRSF6 versus scramble 22Rv1 cells. Top five significantly altered genes are depicted. Red dots represent statistically significant (P < 0.05), and black dots indicate nonsignificant DEGs. (B) Correlation between SRSF6 mRNA levels and SRSF6 activity in the SU2C cohort. (C and D) Volcano plot showing enriched hallmark gene sets defined by GSEA in 22Rv1 cells (siSRSF6 versus scramble) (C) and SU2C cohort (patients with low versus high SRSF6 activity) (D). TNFα, tumor necrosis factor–α; NF-κB, nuclear factor κB; IL-2, interleukin-2; STAT5, signal transducer and activator of transcription 5. (E and G) Correlation between SRSF6 activity and AR (AR-score; E) and E2F activity (E2F-score; G) in the SU2C cohort. (F and H) AR (F) and E2F (H) activity in response to silencing (siSRSF6 versus scramble) and overexpression (SRSF6 versus mock) of SRSF6 in 22Rv1 cells. (I and J) Venn diagram representing common AR and E2F protein interactors that are altered in response to SRSF6 silencing. (J) Expression profile by RNA-seq of AR and E2F-interactors in siSRSF6 versus scramble 22Rv1 cells. Data are represented as means ± SEM. Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups.
SRSF6 silencing alters HIRA splicing pattern and H3.3 activity
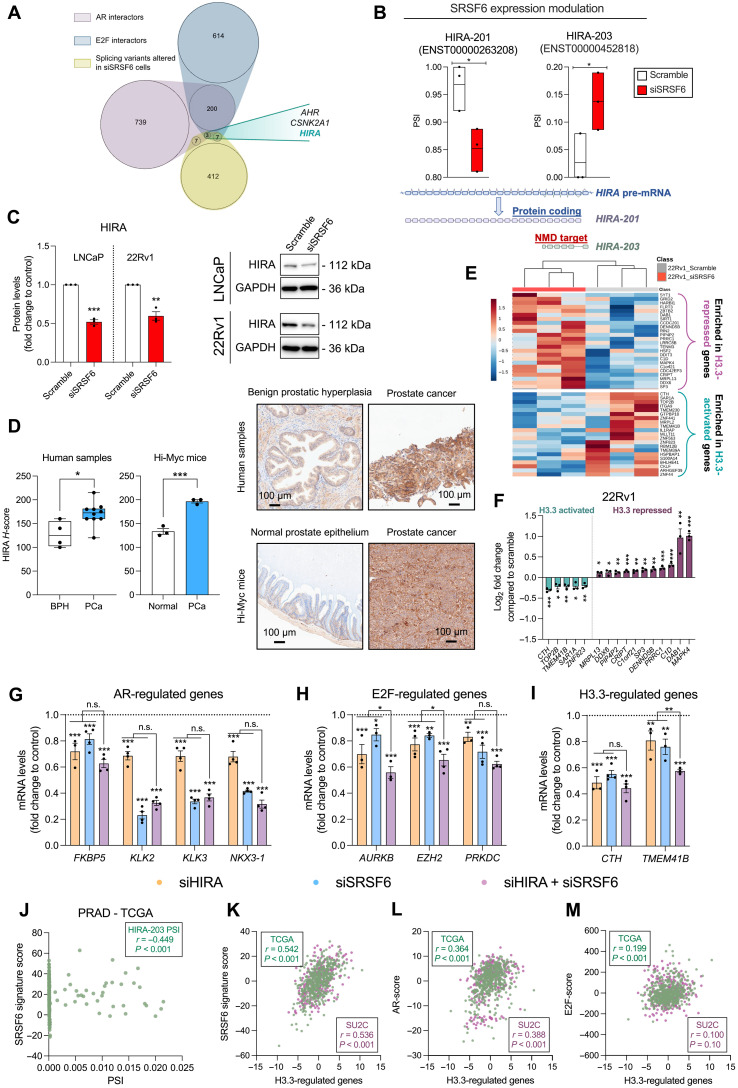
To address the molecular mechanism underlying the control of AR and E2F activity by SRSF6, and, given its function as an SF, we interrogated the splicing landscape of 22Rv1 cells in response to SRSF6 silencing. Specifically, 1323 transcripts were found to be significantly altered by SRSF6 siRNA (data S2). Among the different splicing events, alternative first exon and exon skipping were the most dysregulated ones (32.0 and 29.2%, respectively; fig. S8A). Notably, the splicing pattern of three common AR and E2F interactors (AHR, CSNK2A1, and HIRA) was significantly altered in response to SRSF6 silencing (Fig. 7, A and B, and fig. S8, B and C). Among them, we focused on HIRA because the silencing of SRSF6 induced the expression of the truncated variant HIRA-203, which is targeted and degraded by the nonsense-mediated mRNA decay (NMD) as defined by Ensembl (release 110) (Fig. 7B) (39), presumably resulting in a decrease of the levels of the canonical histone cell cycle regulator (HIRA) protein. In keeping with this, SRSF6 silencing reduced HIRA protein levels in LNCaP and 22Rv1 cells (Fig. 7C and fig. S9A), while no significant alteration was found in DU145 and PC-3 cells (fig. S9B). HIRA down-regulation was further validated in response to in vivo SRSF6 siRNA injection of 22Rv1 xenograft models (fig. S9C). Notably, we also found that SRSF6 overexpression consistently increased HIRA protein levels in 22Rv1 and PC-3 cells, as well as in the PC-3 xenograft model (fig. S9D). The expression levels of CABIN1, a key component of the HIRA complex (40), were also reduced by SRSF6 siRNA (fig. S9F). Similarly to SRSF6 protein levels, we found that HIRA protein (antibody validation in response to HIRA siRNA is shown in fig. S9E) was up-regulated in both human and mouse (Hi-Myc model) PCa samples compared to that in non-tumor samples (Fig. 7D).
Fig. 7. Splicing alteration in response to the modulation of SRSF6 expression in PCa cells.
(A) Venn diagram depicting common AR and E2F interactors whose splicing pattern has been altered in response to SRSF6 silencing (siSRSF6). (B) Effect of SRSF6 silencing on the splicing of HIRA pre-mRNA (top). Representation of HIRA splicing event [alternative first exon (AF)] altered in response to SRSF6 silencing (bottom). (C) HIRA protein levels in response to SRSF6 silencing in LNCaP and 22Rv1 cells. Representative images are depicted on the right panel. (D) Comparison of HIRA protein levels by IHC between BPH (n = 4) and PCa (n = 10) samples (left) and between PCa (n = 3) and non-tumor prostate tissue (n = 3) samples from Hi-Myc mice (right). Data are expressed as min-to-max boxplot, with median of HIRA H-score. Representative images of human and mouse samples stained with HIRA antibody are depicted. (E) Heatmap of the expression of H3.3-regulated genes determined by RNA-seq in response to SRSF6 silencing in 22Rv1 cells. (F) Significantly altered H3.3-activated and H3.3-repressed genes in response to SRSF6 silencing in 22Rv1 cells. (G to I) Expression levels by qPCR of dysregulated AR- (G), E2F- (H), and H3.3-regulated genes (I) in response to HIRA and SRSF6 silencing in LNCaP cells. Data are represented as means ± SEM. n.s., not significant. (J) Correlation between HIRA-203 percent spliced-in (PSI) with SRSF6 signature score in prostate adenocarcinoma (PRAD)–TCGA cohort. (K to M) Correlation between the expression of H3.3-regulated genes and the transcriptionally inferred activity of SRSF6 (K), AR (L), and E2F (M) in the TCGA (green colored) and SU2C (purple colored) cohorts. Asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001) indicate statistically significant differences between groups.
When analyzing the expression levels of the genes encoding for common AR, E2F, and HIRA interactors (fig. S9G), only H3-3A was dysregulated in response to SRSF6 silencing in 22Rv1 cells (fig. S9H), which was consistently altered in LNCaP cells (fig. S9I). H3-3A encodes for H3.3 histone variant, the main target of the HIRA complex, which has been reported as a modulator of AR and E2F activity (41–44). Because H3.3 deposition (orchestrated by HIRA) regulates the expression of certain genes (41, 44, 45), we explored the impact of SRSF6 on the H3.3-regulated transcriptome in 22Rv1 cells. To address this, we analyzed the expression levels of a set of genes found to be significantly up-regulated (H3.3-repressed genes) or down-regulated (H3.3-activated genes) in response to Hira knockout, as previously reported in the literature (42). Specifically, we found that SRSF6 silencing decreased the expression levels of H3.3-activated genes in 22Rv1 and LNCaP cells while increased those of H3.3-repressed genes in 22Rv1 cells (Fig. 7, E and F, and fig. S9, J to L), suggesting that SRSF6 controls H3.3 activity presumably, at least in part, through HIRA splicing dysregulation. HIRA silencing significantly reduced the expression of key AR-, E2F-, and H3.3-regulated genes and increased the expression of H3-3A (Fig. 7, G to I, and fig. S9, O to Q; validation of HIRA silencing shown in fig. S9, M and N), suggesting that SRSF6 could regulate these pathways through, at least in part, HIRA modulation. Most genes regulated by AR, E2F, and H3.3 and down-regulated by SRSF6 were not further reduced by simultaneous silencing of HIRA and SRSF6 in LNCaP cells (Fig. 7 G-I), suggesting a potential linear regulatory relationship between HIRA and SRSF6 in the control of these genes.
We then explored whether SRSF6 was associated in patient samples with the molecular changes described above. Specifically, SRSF6 mRNA and signature score were inversely correlated with HIRA-203 percent spliced-in (PSI) in prostate adenocarcinoma TCGA cohort, while no significant association was observed with HIRA-201 PSI (Fig. 7J and fig. S10, A and B). Consequently, SRSF6 mRNA and signature score were positively correlated with H3.3-regulated genes in TCGA and SU2C cohorts (Fig. 7K and fig. S10C). H3.3-score was directly correlated with AR-score and E2F-score in TCGA and SU2C cohorts (Fig. 7, L and M).
The PanCancer TCGA dataset analysis revealed ubiquitous expression of SRSF6 across all tumor types (fig. S11A). Notably, its highest expression was observed in PCa, closely followed by BCa [Breast Invasive Carcinoma (BRCA) dataset] (fig. S11A). This pattern aligns with the particularly elevated SRSF6 signature score in PCa and BCa tumor types (fig. S11B). SRSF6 mRNA and signature score exhibit a negative correlation with HIRA-203 PSI in the PanCancer TCGA dataset (fig. S11C), implying that the observed mechanism in PCa cells may extend across various tumor types. This observation is supported by the recapitulation of the molecular response observed in PCa cells, when SRSF6 was silenced in the BCa cell line BT-549, resulting in the up-regulation of H3-3A mRNA levels and the down-regulation of H3.3-regulated genes (fig. S11D).
DISCUSSION
Splicing process is an adaptative mechanism that allows the cells to increase the flexibility of their transcriptome, therefore playing a key role in tumor evolution and response to treatment (46, 47). Consequently, its dysregulation has emerged as a new key hallmark of cancer development and progression (48). Specifically, our group and others have proven that multiple alternative and aberrant SVs emerge in tumor conditions, including PCa (49–51). This altered SV landscape could be a consequence of a marked alteration of the expression levels of SCs and SFs in cancer, including PCa (5–8, 52). Given the relevance of certain SVs in the development, progression, and response to treatment of PCa, splicing regulators play a pivotal role in this pathology (5–9). In this context, the SR-rich proteins comprise one of the most relevant families of regulators of the splicing process (10, 11), with many members, such as SRSF1 and SRSF3, involved in the progression of several cancer types, including PCa (5–9). Among the SR family members, SRSF6 may represent a leading factor based on its involvement in the regulation of several biological processes other than splicing process, such as transcription and protein translation (11, 53, 54).
In this study, we analyzed several PCa cohorts to define the levels of SRSF6 and its potential implication in this disease. We demonstrated that both mRNA and protein levels of SRSF6 levels are higher in PCa tissues as compared to those in non-tumor prostate or BPH-derived samples, which is in keeping with previous studies showing that SRSF6 is up-regulated in certain tumor types, such as melanoma (18), colorectal (19, 20), lung cancer (19), and glioblastoma (55). We also found that SRSF6 is amplified in a relevant proportion of patients with mCRPC from the SU2C cohort and, at a lower proportion, in low-risk early disease from the TCGA cohort, suggesting that SRSF6 amplification/copy gain–driven up-regulation might be an early event that is enriched as PCa progresses. This may highlight a potential prognostic value for SRSF6 levels, which is reinforced by the fact that patients with higher SRSF6 expression levels showed shorter biochemical recurrence-free survival and were associated with relevant clinical parameter of tumor aggressiveness. In line with this hypothesis, the amplification of the SRSF6 locus, 20q13, has been proposed as an early event involved in cancer initiation (56). However, several patients with elevated SRSF6 expression levels did not show SRSF6 amplification. This indicates that there may be other regulatory mechanisms at play, contributing to the up-regulation of SRSF6 in PCa, which could involve transcriptional regulation and mRNA stability. In this regard, MYC hyperactivation has been reported to regulate the expression of certain SFs in cancer cells (57). Therefore, we orthogonally explored the association between MYC and SRSF6 by independent in vitro and in vivo approaches, together with human-derived data, revealing that SRSF6 up-regulation in PCa may be partly driven by MYC, as seen for other members of the SR protein family (57, 58).
Moreover, we also found that SRSF6 directly modulate PCa cells aggressiveness. Specifically, the overexpression of SRSF6 increased, while its silencing decreased, critical functional parameters of tumor aggressiveness in vitro and in vivo. Therefore, these data suggest that SRSF6 plays a relevant pathophysiological role in the progression of PCa, in a similar manner to other SR members, such as SRSF3 (5). Mechanistically, we also identified exon skipping as one of the most altered splicing event types in response to SRSF6 silencing in PCa cells, which is consistent with previously reported data in colorectal cells (20). These data may suggest that some of the mechanisms underlying SRSF6-mediated tumorigenesis and tumor progression might be shared across different tumor types, although further experimental evidence is largely required in this regard. An unbiased interrogation of whole-transcriptome data from (i) patients with CRPC and (ii) PCa cells upon SRSF6 depletion revealed AR and E2F as clinically relevant pathways potentially controlled by SRSF6 in this tumor type. In keeping with this, we found that SRSF6 silencing decreased, while its overexpression increased, the activity of AR and E2F in PCa cells. This might explain why AR-driven but also AR-negative PCa cell lines (i.e., DU145 and PC-3) were affected by SRSF6 silencing. Likewise, SRSF6 mRNA levels and activity positively correlated with AR and E2F activity in patients with CRPC. These data carry clinical relevance due to the fact that AR and E2F are hyperactivated and drive CRPC; the first one by AR-amplification, AR-truncated variants, AR mutations, and/or dysregulation of AR coregulators (2, 3), while the last one mainly by RB1-loss, which is a common genomic aberration in CRPC (59–61). Therefore, on the basis of our data, targeting SRSF6 would impair AR and E2F activity/function, potentially resulting in a therapeutic benefit for patients with CRPC. SRSF6 modulation did not result in a change in the expression levels of AR, AR-V7, or E2F1-8, thus suggesting that SRSF6 regulates AR and E2F activity through the modulation of AR/E2F co-regulators, pioneer factors, etc.
Precisely, HIRA was among the AR and E2F common interactors that were found to be dysregulated by SRSF6 silencing. Specifically, the alteration of SRSF6 activity induced changes in the splicing pattern of the HIRA pre-mRNA, leading to the up-regulation of an NMD-targeted variant (HIRA-203) likely contributing to the reduction in HIRA activity (62–64). In this context, SRSF6 silencing led to a reduction in the protein levels of the canonical and functional HIRA protein in LNCaP and 22Rv1 cells. This effect might be mediated by the dysregulation of its splicing pattern, as suggested by the observation that the full-length transcript was not down-regulated in response to SRSF6 silencing. The functional and direct association between SRSF6 and HIRA splicing was strengthened by the observed inverse correlation between SRSF6 levels/activity and HIRA-203 in patients’ samples. This correlation was not limited to patients with PCa but extended across various tumor types, indicating a pan-cancer association. Particularly, HIRA represents the hub of a protein complex (HIRA complex) whose main function involves the regulation of H3.3 and H4 histone variant deposition within the genome (40–43). Therefore, the loss or impairment of HIRA functionality can lead to a notable reduction of the deposition and activity of H3.3 as a gene expression regulator (41, 44, 45). In this sense, we found that SRSF6 silencing decreased H3.3 transcriptional activity (down-regulation of H3.3 positively regulated genes and up-regulation of H3.3 negatively regulated genes), thus demonstrating that HIRA function is impaired by SRSF6 silencing (presumably through HIRA splicing dysregulation). Several reports indicate that HIRA regulates AR and E2F activity in an H3.3-dependent manner (41, 44), which was reinforced by the down-regulation of AR-, E2F-, and H3.3-regulated genes in response to the silencing of HIRA in PCa cells and by the direct and positive correlations found between SRSF6 signature, H3.3 activity, and AR/E2F activity in patients’ samples. These data imply that SRSF6 regulates AR and E2F activity by impairing HIRA function through disbalancing its splicing pattern toward an NMD-targeted transcript, which was robustly corroborated by the redundant actions of HIRA and SRSF6 silencing on AR/E2F/H3.3 activity. Nonetheless, no significant changes regarding HIRA protein levels were found in response to SRSF6 silencing in DU145 and PC-3, suggesting that HIRA splicing–mediated SRSF6 actions may be occurring under certain cell contexts and/or genetic background in PCa cells (i.e., those found in LNCaP and 22Rv1), advocating a potential implication of AR and/or functional TP53 and further indicating a potential HIRA-independent mechanism in DU145 and PC-3 cells. Despite that, in vitro and in vivo SRSF6 overexpression consistently increased HIRA protein levels, revealing a potential direct association between SRSF6 and HIRA proteins in PCa, suggesting that SRSF6 oncogenic actions might be buffered by a cell context-dependent intricate molecular mechanism that may be worth to explore in PCa.
Together, the data herein presented provide solid and convincing evidence demonstrating that SRSF6 plays a relevant functional role in the pathophysiology of PCa. Furthermore, our study underscores the complex regulatory network orchestrated by SRSF6 in PCa cells and invites to suggest that SRSF6 may represent an innovative exploitable therapeutic target aimed at disrupting AR and E2F activity to improve advanced PCa outcome.
MATERIALS AND METHODS
Study approval
The present study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Reina Sofia University Hospital Ethics Committee (ref. 0170-N-23). The regional Biobank coordinated the collection, processing, management, and assignment of biological samples according to standard procedures. All patients provided a written informed consent. Experiments with mice were carried out according to the European Regulations for Animal Care under the approval of the university/regional government research ethics committees (ref. 19/04/2023/13). Our study exclusively examined male individuals and mice because PCa only affects male population.
Human samples
Our study analyzed three different cohorts of prostate samples:
Cohort 1: Consisting of formalin-fixed, paraffin-embedded (FFPE) PCa tissues (n = 84) and their corresponding non-tumor adjacent regions (used as control tissues; n = 84), collected from patients diagnosed with clinically localized PCa who underwent radical prostatectomies (table S1). The presence or absence of tumors was established by expert clinical pathologists histologically examining hematoxylin/eosin-stained tissue.
Cohort 2: Comprising fresh samples procured through (i) core needle biopsies from patients with significant PCa (n = 42; table S2) and (ii) cystoprostatectomies from patients without PCa (n = 9; used as control tissues; patients’ clinical data are summarized in table S2). The presence or absence of tumors was also histologically confirmed by expert uro-pathologists.
Cohort 3: Used for IHC analysis, consisting of tissue samples from BPH (n = 4) and PCa (n = 10; a representative set of samples from cohort 2), ensuring a similar distribution of patients with Gleason scores ranging from 7 to 9 [n = 4 (Gleason score 7), n = 3 (Gleason score 8), and n = 3 (Gleason score 9)].
Various clinical parameters were collected from each patient, including the Gleason score [evaluated by uro-pathologists according to the modified 2014 International Society of Urological Pathology (ISUP) criteria], T stage, perineural invasion, lymphovascular invasion, and the presence of metastases at the time of diagnosis (identified by computed tomography and bone scan).
In addition, transcriptomic and genomic data from TCGA (n = 545) (21), SU2C (n = 266) (25), MSKCC (n = 149) (22), Grasso (n = 88) (23), and Roudier (GSE74367; n = 56) (24) PCa cohorts were obtained from cBioPortal (65, 66) and Gene Expression Omnibus (67) repositories. PanCancer (n = 4915) gene and transcript expression levels were obtained from UCSC Xena repository (68). HIRA-201 and HIRA-203 PSI value was determined as ENST00000263208 (HIRA-201) and ENST00000452818 (HIRA-203) transcript levels normalized by HIRA (ENSG00000100084) mRNA levels.
Cell culture and reagents
Cell lines derived from normal prostate epithelium (RWPE-1), PCa (LNCaP, 22Rv1, DU145, and PC-3), and BCa (BT-549), were obtained from the American Type Culture Collection following the manufacturer’s recommendations. Specifically, LNCaP, 22Rv1, DU145, PC-3, and BT-549 were cultured with RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and glutamine (2 mM), while RWPE-1 cells were cultured with keratinocyte serum–free medium supplemented with bovine pituitary extract (0.05 mg/ml) and human recombinant epidermal growth factor (EGF; 5 ng/ml) and glutamine (2 mM). All cell lines were maintained in a humidified incubator with 5% CO2 at 37°C. Cell line identity was validated by short tandem repeat sequence analysis. All cell lines were tested for mycoplasma contamination by polymerase chain reaction (PCR), as previously reported (69). Dimethyl sulfoxide (DMSO; A3672, PanReac AppliChem), enzalutamide (S1250, Selleckchem), dihydrotestosterone (DHT; D-073, Sigma-Aldrich), and R1881 (R0908, Sigma-Aldrich) were used to perform molecular assays (see the “AR activity modulation” section).
Preclinical models of PCa
To evaluate in vivo tumor growth in response to SRSF6 overexpression, 10-week-old male athymic BALB/cAnNRj-Foxn1nu mice (Janvier Labs) were subcutaneously grafted in both flanks with 100 μl of basement membrane extract (Trevigen) and RPMI 1640 complemented with 10% FBS (F7524, Sigma-Aldrich) (1:1 ratio) containing 3 × 106 viable mock-transfected [empty pcDNA3.1 plasmid (GenScript); n = 5 mice; n = 10 tumors] or SRSF6–stably transfected PC-3 cells [SRSF6-pcDNA3.1 plasmid (OHu19224, GenScript); n = 5 mice; n = 10 tumors]. Tumor growth was monitored once per week for 2 months using a digital caliper.
To evaluate in vivo tumor growth in response to SRSF6 silencing, 10-week-old male athymic BALB/cAnNRj-Foxn1nu mice were subcutaneously grafted in both flanks with 3 × 106 viable naïve 22Rv1 cells (n = 5 mice; n = 10 tumors) that were resuspended in 100 μl of basement membrane extract and RPMI 1640 complemented with 10% FBS (1:1 ratio). Once the tumors reached 100 mm3, each flank was transfected with scramble-control (AM4611, Thermo Fisher Scientific) or SRSF6-targeting siRNA (siSRSF6; s12740, Thermo Fisher Scientific) by using AteloGene reagent (Koken), the following manufacturer’s recommendations.
In both cases, animals were euthanized, and each tumor was processed and divided in specular fragments for formalin fixation followed by paraffin inclusion and stored at −80°C for later RNA extraction using TRIzol reagent (Thermo Fisher Scientific).
In addition, we used the Hi-Myc (ARR2/Pbsn-Myc) mouse strain maintained in an FVB background (70), which was obtained originally from National Cancer Institute (NCI) and backcrossed to C57BL/6 for more than seven generations at Olmos’ laboratory to obtain pure genetic background. Hi-Myc mice were euthanized at 4 (no presence of tumor) and 12 to 15 months (predominately invasive carcinoma). Genotyping was performed in genomic DNA extracted from tail snip by PCR by using the primers recommended by NCI (sense: AAACATGATGACTACCAAGCTTGGC; antisense: ATGATAGCATCTTGTTCTT AGTCTTTTTCTTAATAGGG). For immunohistological analyses, a piece of the prostate tissues was processed for formalin fixation followed by paraffin inclusion.
RNA extraction, retrotranscription, and real-time qPCR
RNA was isolated from FFPE samples, fresh tissues, and cell lines using previously reported methods (5, 7). To isolate RNA from FFPE samples, the Maxwell 16 LEVRNA FFPE Kit (Promega) was used with the Maxwell MDx 16 Instrument (Promega). RNA was isolated from fresh tissues and PCa cell lines using the AllPrep DNA/RNA/Protein Mini Kit (QIAGEN) and TRIzol reagent, respectively. The RNA was treated with the RNase-Free DNase Kit (QIAGEN) to remove DNA. The Nanodrop One Spectrophotometer (Thermo Fisher Scientific) was used to determine the total RNA concentration and purity. cDNA was synthesized from total RNA using the cDNA First Strand Synthesis Kit (Thermo Fisher Scientific) and random hexamer primers. Real-time quantitative PCR (qPCR) was performed using the Stratagene Mx3000p device with the Brilliant III SYBR Green Master Mix (Stratagene), as previously described (71). Normalization was done using a normalization factor calculated with GeNorm 3.3 software (72) using ACTB and GAPDH expression levels, as previously reported (5). The primers used in this study can be found in table S3.
Overexpression and silencing on in vitro prostate cell models
To generate a stable overexpression of SRSF6, RWPE-1, 22Rv1, and PC-3 cells were transfected with empty pcDNA3.1 (mock) or SRSF6-pcDNA3.1 plasmid as previously reported (7). Similarly, to stimulate MYC expression, LNCaP and 22Rv1 cells were transfected with empty pCW57.1_blasti (mock) or pCW57.1 CMYC (MYC) plasmids. pCW57.1_blasti was a gift from RESOLUTE Consortium & Giulio Superti-Furga (Addgene, plasmid no. 194067; http://n2t.net/addgene:194067; RRID: Addgene_194067). pCW57.1 CMYC was a gift from R. Possemato (Addgene, plasmid no. 164145; http://n2t.net/addgene:164145; RRID: Addgene_164145). Experiments were performed with 1 μg of plasmid using Lipofectamine 2000 reagent (Thermo Fisher Scientific), and medium supplemented with 1% Geneticin (Thermo Fisher Scientific) was used to obtain stably transfected cells. To induce MYC expression, transfected cells were treated with doxycycline hyclate 2 μg/ml (Sigma-Aldrich) for at least 1 week. In addition, LNCaP, 22Rv1, DU145, PC-3, and BT-549 cells were transfected with scramble (AM4611, Thermo Fisher Scientific) or SRSF6 siRNA (siSRSF6; s12740, Thermo Fisher Scientific) at 100 nM using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) for 48 hours following the manufacturers’ indications. LNCaP cells were transfected with scramble or HIRA siRNA pool (siHIRA; L-013610-00-0005, Dharmacon) at 25 nM using Lipofectamine RNAiMAX reagent for 72 hours following the manufacturer’s indications.
Western blot
SRSF6 and HIRA protein levels were determined by Western blot. Briefly, proteins were isolated using radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) supplemented with Pierce Protease and Phosphatase inhibitors (Thermo Fisher Scientific). Then, proteins were sonicated and separated by SDS–polyacrylamide gel electrophoresis (any kD Mini-Protean TGX gels, Bio-Rad) and then transferred to Immun-Blot PVDF Membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk in tris-buffered saline/0.05% tween 20 (Sigma-Aldrich) and incubated overnight with the specific antibodies for SRSF6 (ab140623, Abcam), HIRA (12463, Cell Signaling Technology), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sc-32233, Santa Cruz Biotechnology) at 1:1000 and then with the secondary antibody horseradish peroxidase (HRP)–conjugated anti-rabbit immunoglobulin G (IgG; 7074, Cell Signaling Technology) or anti-mouse IgG (7076, Cell Signaling Technology) at 1:2000. Proteins were detected using an enhanced chemiluminescence detection system (Bio-Rad). A densitometry analysis of the bands obtained was carried out with ImageJ software. GAPDH protein levels were used to normalize SRSF6 and HIRA protein levels.
Immunohistochemistry
IHC analysis was performed on samples from cohort 3 and on samples from the Hi-Myc mice (control and PCa samples, n = 3 per group). Briefly, deparaffinized sections were incubated overnight (4°C) with anti-SRSF6 and anti-HIRA antibodies at 1:100 dilution, followed by incubation with an anti-rabbit HRP-conjugated secondary antibody. Last, sections were developed with 3,3-diaminobenzidine (EnVision system, Agilent) and contrasted with hematoxylin. 22Rv1 cell pellets (scramble, siSRSF6, and siHIRA) were used to evaluate SRSF6 and HIRA antibody specificity (Fig. 3A and fig. S9E, respectively). H-score was calculated as the sum of the percentage of stained nuclei with low, moderate, and high intensity following a blinded protocol as described elsewhere (8).
Cell proliferation and migration
Cell growth was examined by Resazurin assay (CA035, Canvax Biotech) following the manufacturer’s instructions, as previously described (5, 7). Cell migration was determined by wound healing assay as described elsewhere (5, 7). Briefly, cells were plated and, once the cell confluence was reached, serum starved overnight. Then, a scratch was made, wells were washed with phosphate-buffered saline, and cells were incubated with no serum medium. Images were taken immediately after scratching and after 16 hours of incubation. Migration was calculated as the area observed 16 hours after the wound was made versus the area observed just after wounding, using ImageJ software.
Colonies and tumorspheres formation
To evaluate clonogenic capacity, six-well plates were seeded with 2000 cells per well and incubated for 7 days. Colonies were then fixed and stained with a solution containing 6% glutaraldehyde and 1% crystal violet for 30 min, followed by air-drying. Tumorsphere formation was assessed using Corning Costar 24-well ultralow attachment plates (CLS3473, Sigma-Aldrich), with 2000 cells per well in Dulbecco’s modified Eagle’s medium–F12 medium supplemented with EGF (20 ng/ml; Sigma-Aldrich) and refreshed every 3 days. The number and size of colonies and tumorspheres were analyzed after 14 days using ImageJ software (Fiji plugins) (73).
RNA sequencing
An RNeasy Plus Mini kit (QIAGEN) was used to isolate high-quality RNA from siSRSF6 (n = 3) and scramble (n = 3) 22Rv1 cells. The integrity of total RNA was assessed using the 2100 Bioanalyzer (Agilent). RNA-seq was performed at the Genomics Core Unit of the National Centre for Cancer Research (CNIO, Madrid, Spain) as previously reported (7). Differential gene expression analysis was performed using DESeq2 (74) as described elsewhere (7). Alternative splicing analysis was performed using SUPPA2 (75). Briefly, transcript per million derived from Salmon tool was used to quantify alternative splicing events as PSI. The differential PSI and its associated P value were calculated for all the splicing events. Splicing events with P < 0.1 when comparing siSRSF6 versus scramble 22Rv1 samples were considered as statistically different.
Molecular gene signatures
SRSF6 gene signature score (SRSF6 signature score) was defined as the down-regulated genes in siSRSF6 versus scramble 22Rv1 cells (101 genes; P < 0.05) that were positively correlated with SRSF6 mRNA levels in the SU2C cohort. AR signaling activity (AR-score) was determined as a sum of the ranked expression levels of eight canonical AR-regulated genes (ACSL3, FKBP5, KLK2, KLK3, NKX3-1, PLPP1, RAB3B, and STEAP1) as previously described (38, 76). E2F signaling activity (E2F-score) was determined as a sum of the ranked expression levels of (i) six canonical E2F-regulated genes (used for inferring E2F activity from cell lines: AURKB, BIRC5, CCNE1, EZH2, PRKDC, and TP53) or (ii) E2F hallmark gene set obtained from GSEA database (77) (used for inferring E2F activity from SU2C and TCGA cohorts). H3.3 activity was calculated by the sum of the ranked expression levels of five genes that were positively regulated by H3.3 (CTH, SAR1A, TOP2B, TMEM41B, and ZNF823) (42) and were also significantly down-regulated in our study in response to SRSF6 silencing in 22Rv1 cells. Similarly, MYC gene sets and Whitfield cell cycle signature were used to infer MYC activity and proliferative status in patients with PCa, respectively. The gene sets used in this study are summarized in data S4.
AR activity modulation
To evaluate SRSF6 expression in response to AR modulation, LNCaP cells were cultured for 3 days in RPMI 1640 supplemented with 10% FBS or 10% charcoal-stripped serum (12676029, Thermo Fisher Scientific) and treated with DMSO (vehicle), enzalutamide (1 μM), DHT (10 nM), or R1881 (1 nM) for 24 hours. Then, TRIzol reagent was added to the wells, and RNA was extracted following the manufacturer’s instructions.
Bioinformatic and statistical data analysis
At least three independent experiments were performed for all analyses (n ≥ 3). Statistical differences between two groups were calculated using unpaired parametric t test and nonparametric Mann-Whitney U test, depending on normality, which was assessed by Kolmogorov-Smirnov test. For differences among three groups, one-way analysis of variance (ANOVA) analysis was used. For correlations, Spearman or Pearson coefficients were calculated on the basis of normality of variables. Statistical significance was considered when P < 0.05. A trend for significance was indicated when P values ranged between >0.05 and <0.1. All analyses were assessed using GraphPad Prism 8 (GraphPad software). Heatmaps were created by nVenn (78). AR, E2F, and HIRA interactors were obtained from the Biological General Repository for Interaction Datasets repository (data S3) (79). Continuous and categorical GSEAs were performed using hallmark gene sets in the SU2C cohort and 22Rv1 cells (siSRSF6 versus scramble), respectively, using GSEA 4.2.0 software (77, 80).
Acknowledgments
We deeply thank all the patients and the patients’ families for donating the samples and clinical data for research purposes. Special thanks to the staff of Biobank of the IMIBIC and the experimental animal service (SAE) of the UCO/IMIBIC.
Funding: This work was supported by the Spanish Ministry of Science, Innovation, and Universities, grants PID2022-1381850B-I00 (R.M.L.), PID2019-105564RB-I00 (R.M.L.), FPU16/05059 (A.C.F.-F), FPU17/00263 (P.S.-M.), FPU18/02485 (A.J.M.-H.), FPU18/06009 (J.M.P.-G.), FPU18/02275 (R.B.-E.), and PRE2020-094225 (F.P.-P.); Instituto de Salud Carlos III, co-funded by European Union (ERDF/ESF, “Investing in your future” grant DTS20/00050 (R.M.L.); Junta de Andalucia BIO-0139 (R.M.L.); Junta de Andalucia grant PI-0094-2020 (A.S.-C.); Prostate Cancer UK Travelling Prize Fellowship (J.M.J.-V.); Wellcome Trust Clinical Research Career Development Fellowship (A.S.); and CIBERobn (CIBER is an initiative of Instituto de Salud Carlos III, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain).
Author contributions: Conceptualization: A.J.M.-H., J.M.J.-V., P.S.-M., A.C.F.-F., J.C.-V., A.J.M.-F., J.P.C., M.D.-G., and R.M.L. Data curation: A.J.M.-H., J.M.J.-V., A.J.M.-F., and R.M.L. Formal analysis: A.J.M.-H., J.M.J.-V., P.S.-M., J.M.P.-G., R.B.-E., A.S.-C., A.J.M.-F., D.O., and R.M.L. Funding acquisition: J.P.C. and R.M.L. Investigation: A.J.M.-H., J.M.J.-V., E.G.-G., F.P.-P., P.S.-M., J.M.P.-G., A.C.F.-F., R.B.-E., E.C., P.J.L.-S., A.S.-C., A.J.M.-F., and R.M.L. Methodology: A.J.M.-H., J.M.J.-V., P.S.-M., A.C.F.-F., J.C.-V., A.J.M.-F., J.P.C., D.O., M.D.-G., and R.M.L. Project administration: J.M.J.-V., A.J.M.-F., D.O., M.D.-G., and R.M.L. Resources: A.J.M.-H., F.P.-P., J.M.P.-G., R.S.-S., T.G.-S., E.C., J.C.-V., A.J.M.-F., J.P.C., D.O., and R.M.L. Software: J.M.J.-V., R.B.E., J.C.-V., and A.J.M.-F. Supervision: J.M.J.-V., A.J.M.-F., A.S., M.D.-G., and R.M.L. Validation: A.J.M.-H., J.M.J.-V., P.S.-M., E.C., J.C.-V., A.J.M.-F., D.O., and R.M.L. Visualization: A.J.M.-H., J.M.J.-V., E.G.-G., R.B.-E., E.C., A.J.M.-F., and R.M.L. Writing—original draft: A.J.M.-H., J.M.J.-V., A.J.M.-F., E.E., and R.M.L. Writing—review and editing: A.J.M.-H., J.M.J.-V., P.S.-M., A.C.F.-F., R.B.-E., E.C., A.J.M.-F., E.E., J.P.C., A.S., and R.M.L.
Competing interests: A.S. is an employee of the ICR, which has a commercial interest in abiraterone, poly(adenosine 5′-diphosphate–ribose) polymerase inhibition in DNA repair defective cancers, and phosphatidylinositol 3-kinase/Akt pathway inhibitors (no personal income). A.S. has received travel support from Sanofi, Roche-Genentech, and Nurix and speaker honoraria from Astellas Pharma and Merck Sharp & Dohme. He has served as an advisor to DE Shaw Research, CHARM Therapeutics, Ellipses Pharma, and Droia Ventures. A.S. has been the CI/PI of industry-sponsored clinical trials. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S11
Tables S1 to S3
Legends for data S1 to S4
Other Supplementary Material for this manuscript includes the following:
Data S1 to S4
REFERENCES AND NOTES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Westaby D., Fenor de La Maza M. L. D., Paschalis A., Jimenez-Vacas J. M., Welti J., de Bono J., Sharp A., A new old target: Androgen receptor signaling and advanced prostate cancer. Annu. Rev. Pharmacol. Toxicol. 62, 131–153 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Watson P. A., Arora V. K., Sawyers C. L., Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschalis A., Sharp A., Welti J. C., Neeb A., Raj G. V., Luo J., Plymate S. R., de Bono J. S., Alternative splicing in prostate cancer. Nat. Rev. Clin. Oncol. 15, 663–675 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Jiménez-Vacas J. M., Herrero-Aguayo V., Montero-Hidalgo A. J., Gómez-Gómez E., Fuentes-Fayos A. C., León-González A. J., Sáez-Martínez P., Alors-Pérez E., Pedraza-Arévalo S., González-Serrano T., Reyes O., Martínez-López A., Sánchez-Sánchez R., Ventura S., Yubero-Serrano E. M., Requena-Tapia M. J., Castaño J. P., Gahete M. D., Luque R. M., Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer. EBioMedicine 51, 102547 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiménez-Vacas J. M., Herrero-Aguayo V., Gómez-Gómez E., León-González A. J., Sáez-Martínez P., Alors-Pérez E., Fuentes-Fayos A. C., Martínez-López A., Sánchez-Sánchez R., González-Serrano T., López-Ruiz D. J., Requena-Tapia M. J., Castaño J. P., Gahete M. D., Luque R. M., Spliceosome component SF3B1 as novel prognostic biomarker and therapeutic target for prostate cancer. Transl. Res. 212, 89–103 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Jiménez-Vacas J. M., Montero-Hidalgo A. J., Gómez-Gómez E., Sáez-Martínez P., Fuentes-Fayos A. C., Closa A., González-Serrano T., Martínez-López A., Sánchez-Sánchez R., López-Casas P. P., Sarmento-Cabral A., Olmos D., Eyras E., Castaño J. P., Gahete M. D., Luque R. M., Tumor suppressor role of RBM22 in prostate cancer acting as a dual-factor regulating alternative splicing and transcription of key oncogenic genes. Transl. Res. 253, 68–79 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Paschalis A., Welti J., Neeb A. J., Yuan W., Figueiredo I., Pereira R., Ferreira A., Riisnaes R., Rodrigues D. N., Jiménez-Vacas J. M., Kim S., Uo T., Micco P. D., Tumber A., Islam M. S., Moesser M. A., Abboud M., Kawamura A., Gurel B., Christova R., Gil V. S., Buroni L., Crespo M., Miranda S., Lambros M. B., Carreira S., Tunariu N., Alimonti A., Al-Lazikani B., Schofield C. J., Plymate S. R., Sharp A., de Bono J. S., JMJD6 is a druggable oxygenase that regulates AR-V7 expression in prostate cancer. Cancer Res. 81, 1087–1100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L. L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X., Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33, 3140–3150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher L., Ouzounis C. A., Enright A. J., Blencowe B. J., A genome-wide survey of RS domain proteins. RNA 7, 1693–1701 (2001). [PMC free article] [PubMed] [Google Scholar]
- 11.Howard J. M., Sanford J. R., The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L.-M., Xiong L., Cheng P.-P., Chen S.-J., Feng X., Zhou Y.-Y., Niu Q., Wang M., Chen Q., Song L.-J., Yu F., He X.-L., Xiang F., Wang X., Ye H., Ma W.-L., Splicing factor SRSF6 mediates pleural fibrosis. JCI Insight 6, e146197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neueder A., Dumas A. A., Benjamin A. C., Bates G. P., Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat. Commun. 9, 3955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mai H., Fan W., Wang Y., Cai Y., Li X., Chen F., Chen X., Yang J., Tang P., Chen H., Zou T., Hong T., Wan C., Zhao B., Cui L., Intranasal administration of miR-146a agomir rescued the pathological process and cognitive impairment in an AD mouse model. Mol. Ther. Nucleic Acids 18, 681–695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juan-Mateu J., Alvelos M. I., Turatsinze J.-V., Villate O., Lizarraga-Mollinedo E., Grieco F. A., Marroquí L., Bugliani M., Marchetti P., Eizirik D. L., SRp55 regulates a splicing network that controls human pancreatic β-cell function and survival. Diabetes 67, 423–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manetti M., Guiducci S., Romano E., Ceccarelli C., Bellando-Randone S., Conforti M. L., Ibba-Manneschi L., Matucci-Cerinic M., Overexpression of VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, leads to insufficient angiogenesis in patients with systemic sclerosis. Circ. Res. 109, e14–e26 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Park S., Brugiolo M., Akerman M., Das S., Urbanski L., Geier A., Kesarwani A. K., Fan M., Leclair N., Lin K. T., Hu L., Hua I., George J., Muthuswamy S. K., Krainer A. R., Anczuków O., Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Rep. 29, 2672–2688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen M. A., Wilkinson J. E., Krainer A. R., Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat. Struct. Mol. Biol. 21, 189–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen-Eliav M., Golan-Gerstl R., Siegfried Z., Andersen C. L., Thorsen K., Ørntoft T. F., Mu D., Karni R., The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J. Pathol. 229, 630–639 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Wan L., Yu W., Shen E., Sun W., Liu Y., Kong J., Wu Y., Han F., Zhang L., Yu T., Zhou Y., Xie S., Xu E., Zhang H., Lai M., SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut 68, 118–129 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Vega F., Mina M., Armenia J., Chatila W. K., Luna A., La K. C., Dimitriadoy S., Liu D. L., Kantheti H. S., Saghafinia S., Chakravarty D., Daian F., Gao Q., Bailey M. H., Liang W.-W., Foltz S. M., Shmulevich I., Ding L., Heins Z., Ochoa A., Gross B., Gao J., Zhang H., Kundra R., Kandoth C., Bahceci I., Dervishi L., Dogrusoz U., Zhou W., Shen H., Laird P. W., Way G. P., Greene C. S., Liang H., Xiao Y., Wang C., Iavarone A., Berger A. H., Bivona T. G., Lazar A. J., Hammer G. D., Giordano T., Kwong L. N., McArthur G., Huang C., Tward A. D., Frederick M. J., McCormick F., Meyerson M.; The Cancer Genome Atlas Research Network, van Allen E. M., Cherniack A. D., Ciriello G., Sander C., Schultz N., Oncogenic signaling pathways in the cancer genome atlas. Cell 173, 321–337.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., Gerald W. L., Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grasso C. S., Wu Y. M., Robinson D. R., Cao X., Dhanasekaran S. M., Khan A. P., Quist M. J., Jing X., Lonigro R. J., Brenner J. C., Asangani I. A., Ateeq B., Chun S. Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J. R., Palanisamy N., Ryslik G. A., Vandin F., Raphael B. J., Kunju L. P., Rhodes D. R., Pienta K. J., Chinnaiyan A. M., Tomlins S. A., The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roudier M. P., Winters B. R., Coleman I., Lam H. M., Zhang X., Coleman R., Chéry L., True L. D., Higano C. S., Montgomery B., Lange P. H., Snyder L. A., Srivastava S., Corey E., Vessella R. L., Nelson P. S., Üren A., Morrissey C., Characterizing the molecular features of ERG-positive tumors in primary and castration resistant prostate cancer. Prostate 76, 810–822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I., Cieslik M., Benelli M., Robinson D., van Allen E. M., Sboner A., Fedrizzi T., Mosquera J. M., Robinson B. D., de Sarkar N., Kunju L. P., Tomlins S., Wu Y. M., Nava Rodrigues D., Loda M., Gopalan A., Reuter V. E., Pritchard C. C., Mateo J., Bianchini D., Miranda S., Carreira S., Rescigno P., Filipenko J., Vinson J., Montgomery R. B., Beltran H., Heath E. I., Scher H. I., Kantoff P. W., Taplin M.-E., Schultz N., deBono J. S., Demichelis F., Nelson P. S., Rubin M. A., Chinnaiyan A. M., Sawyers C. L., Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C., ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S., Shao J., Lou G., Wu C., Liu Y., Zheng M., MiR-144-3p-mediated dysregulation of EIF4G2 contributes to the development of hepatocellular carcinoma through the ERK pathway. J. Exp. Clin. Cancer Res. 40, 53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S., Wagner G., Deep computational analysis details dysregulation of eukaryotic translation initiation complex eIF4F in human cancers. Cell Syst. 12, 907–923.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang L., Wang D., Shen T., Liu X., Dai B., Zhou D., Shen H., Gong J., Li G., Hu Y., Wang P., Mi X., Zhang Y., Tan X., PDIA4 confers resistance to ferroptosis via induction of ATF4/SLC7A11 in renal cell carcinoma. Cell Death Dis. 14, 193 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao M. H.-R., Yang C., A. P.-W. Tse, Wei L., Lee D., Zhang M. S., Goh C. C., Chiu D. K.-C., Yuen V. W.-H., Law C.-T., Chin W.-C., Chui N. N.-Q., Wong B. P.-Y., Chan C. Y.-K., Ng I. O.-L., Chung C. Y.-S., Wong C.-M., Wong C. C.-L., Genome-wide CRISPR-Cas9 knockout library screening identified PTPMT1 in cardiolipin synthesis is crucial to survival in hypoxia in liver cancer. Cell Rep. 34, 108676 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Yasunaga M., Matsumura Y., Role of SLC6A6 in promoting the survival and multidrug resistance of colorectal cancer. Sci. Rep. 4, 4852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B., Zhang J.-Y., Liu X.-S., Chen H.-Z., Ai Y. L., Cheng K., Sun R.-Y., Zhou D., Han J., Wu Q., Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res. 28, 1171–1185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerstberger S., Hafner M., Tuschl T., A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider-Lunitz V., Ruiz-Orera J., Hubner N., van Heesch S., Multifunctional RNA-binding proteins influence mRNA abundance and translational efficiency of distinct sets of target genes. PLOS Comput. Biol. 17, e1009658 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Q., Saghafinia S., Chryplewicz A., Fournier N., Christe L., Xie Y.-Q., Guillot J., Yucel S., Li P., Galván J. A., Karamitopoulou E., Zlobec I., Ataca D., Gallean F., Zhang P., Rodriguez-Calero J. A., Rubin M., Tichet M., Homicsko K., Hanahan D., Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science 378, eabl7207 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Mandigo A. C., Shafi A. A., McCann J. J., Yuan W., Laufer T. S., Bogdan D., Gallagher L., Dylgjeri E., Semenova G., Vasilevskaya I. A., Schiewer M. J., McNair C. M., de Bono J. S., Knudsen K. E., Novel oncogenic transcription factor cooperation in RB-deficient cancer. Cancer Res. 82, 221–234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandigo A. C., Yuan W., Xu K., Gallagher P., Pang A., Guan Y. F., Shafi A. A., Thangavel C., Sheehan B., Bogdan D., Paschalis A., McCann J. J., Laufer T. S., Gordon N., Vasilevskaya I. A., Dylgjeri E., Chand S. N., Schiewer M. J., Domingo-Domenech J., Den R. B., Holst J., McCue P. A., de Bono J. S., McNair C., Knudsen K. E., RB/E2F1 as a master regulator of cancer cell metabolism in advanced disease. Cancer Discov. 11, 2334–2353 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt D. E., Alshalalfa M., Fishbane N., Weiner A. B., Mehra R., Mahal B. A., Lehrer J., Liu Y., Zhao S. G., Speers C., Morgan T. M., Dicker A. P., Freedland S. J., Karnes R. J., Weinmann S., Davicioni E., Ross A. E., Den R. B., Nguyen P. L., Feng F. Y., Lotan T. L., Chinnaiyan A. M., Schaeffer E. M., Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment naïve primary prostate cancer. Clin. Cancer Res. 25, 6721–6730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin F. J., Amode M. R., Aneja A., Austine-Orimoloye O., Azov A. G., Barnes I., Becker A., Bennett R., Berry A., Bhai J., Bhurji S. K., Bignell A., Boddu S., Branco Lins P. R., Brooks L., Ramaraju S. B., Charkhchi M., Cockburn A., da Rin Fiorretto L., Davidson C., Dodiya K., Donaldson S., El Houdaigui B., El Naboulsi T., Fatima R., Giron C. G., Genez T., Ghattaoraya G. S., Martinez J. G., Guijarro C., Hardy M., Hollis Z., Hourlier T., Hunt T., Kay M., Kaykala V., Le T., Lemos D., Marques-Coelho D., Marugán J. C., Merino G. A., Mirabueno L. P., Mushtaq A., Hossain S. N., Ogeh D. N., Sakthivel M. P., Parker A., Perry M., Piližota I., Prosovetskaia I., Pérez-Silva J. G., Salam A. I. A., Saraiva-Agostinho N., Schuilenburg H., Sheppard D., Sinha S., Sipos B., Stark W., Steed E., Sukumaran R., Sumathipala D., Suner M. M., Surapaneni L., Sutinen K., Szpak M., Tricomi F. F., Urbina-Gómez D., Veidenberg A., Walsh T. A., Walts B., Wass E., Willhoft N., Allen J., Alvarez-Jarreta J., Chakiachvili M., Flint B., Giorgetti S., Haggerty L., Ilsley G. R., Loveland J. E., Moore B., Mudge J. M., Tate J., Thybert D., Trevanion S. J., Winterbottom A., Frankish A., Hunt S. E., Ruffier M., Cunningham F., Dyer S., Finn R. D., Howe K. L., Harrison P. W., Yates A. D., Flicek P., Ensembl 2023. Nucleic Acids Res. 51, D933–D941 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray-Gallet D., Ricketts M. D., Sato Y., Gupta K., Boyarchuk E., Senda T., Marmorstein R., Almouzni G., Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat. Commun. 9, 3103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morozov V. M., Riva A., Sarwar S., Kim W., Li J., Zhou L., Licht J. D., Daaka Y., Ishov A. M., HIRA-mediated loading of histone variant H3.3 controls androgen-induced transcription by regulation of AR/BRD4 complex assembly at enhancers. Nucleic Acids Res. 51, 10194–10217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nashun B., Hill P. W. S., Smallwood S. A., Dharmalingam G., Amouroux R., Clark S. J., Sharma V., Ndjetehe E., Pelczar P., Festenstein R. J., Kelsey G., Hajkova P., Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell 60, 611–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rai T. S., Cole J. J., Nelson D. M., Dikovskaya D., Faller W. J., Vizioli M. G., Hewitt R. N., Anannya O., McBryan T., Manoharan I., van Tuyn J., Morrice N., Pchelintsev N. A., Ivanov A., Brock C., Drotar M. E., Nixon C., Clark W., Sansom O. J., Anderson K. I., King A., Blyth K., Adams P. D., HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 28, 2712–2725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daury L., Chailleux C., Bonvallet J., Trouche D., Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 7, 66–71 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tafessu A., O'Hara R., Martire S., Dube A. L., Saha P., Gant V. U., Banaszynski L. A., H3.3 contributes to chromatin accessibility and transcription factor binding at promoter-proximal regulatory elements in embryonic stem cells. Genome Biol. 24, 25 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matera A. G., Wang Z., A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scotti M. M., Swanson M. S., RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oltean S., Bates D. O., Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Jiménez-Vacas J. M., Montero-Hidalgo A. J., Gómez-Gómez E., Fuentes-Fayos A. C., Ruiz-Pino F., Guler I., Camargo A., Anglada F. J., Carrasco-Valiente J., Tena-Sempere M., Sarmento-Cabral A., Castaño J. P., Gahete M. D., Luque R. M., In1-ghrelin splicing variant as a key element in the pathophysiological association between obesity and prostate cancer. J. Clin. Endocrinol. Metab. 106, e4956–e4968 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Hormaechea-Agulla D., Gahete M. D., Jiménez-Vacas J. M., Gómez-Gómez E., Ibáñez-Costa A., López F. L., Rivero-Cortés E., Sarmento-Cabral A., Valero-Rosa J., Carrasco-Valiente J., Sánchez-Sánchez R., Ortega-Salas R., Moreno M. M., Tsomaia N., Swanson S. M., Culler M. D., Requena M. J., Castaño J. P., Luque R. M., The oncogenic role of the In1-ghrelin splicing variant in prostate cancer aggressiveness. Mol. Cancer 16, 146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hormaechea-Agulla D., Jiménez-Vacas J. M., Gómez-Gómez E., López F. L., Carrasco-Valiente J., Valero-Rosa J., Moreno M. M., Sánchez-Sánchez R., Ortega-Salas R., Gracia-Navarro F., Culler M. D., Ibáñez-Costa A., Gahete M. D., Requena M. J., Castaño J. P., Luque R. M., The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J. 31, 4682–4696 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zhang D., Hu Q., Liu X., Ji Y., Chao H.-P., Liu Y., Tracz A., Kirk J., Buonamici S., Zhu P., Wang J., Liu S., Tang D. G., Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 11, 2089 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanford J. R., Gray N. K., Beckmann K., Cáceres J. F., A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson C. M., Sherer N. M., Malim M. H., SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J. Virol. 84, 6748–6759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuentes-Fayos A. C., Vázquez-Borrego M. C., Jiménez-Vacas J. M., Bejarano L., Pedraza-Arévalo S., López F. L., Blanco-Acevedo C., Sánchez-Sánchez R., Reyes O., Ventura S., Solivera J., Breunig J. J., Blasco M. A., Gahete M. D., Castaño J. P., Luque R. M., Splicing machinery dysregulation drives glioblastoma development/aggressiveness: Oncogenic role of SRSF3. Brain 143, 3273–3293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tabach Y., Kogan-Sakin I., Buganim Y., Solomon H., Goldfinger N., Hovland R., Ke X.-S., Oyan A. M., Kalland K.-H., Rotter V., Domany E., Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLOS ONE 6, e14632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urbanski L., Brugiolo M., Park S., Angarola B. L., Leclair N. K., Yurieva M., Palmer P., Sahu S. K., Anczuków O., MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. 41, 111704 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das S., Anczuków O., Akerman M., Krainer A. R., Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma A., Yeow W.-S., Ertel A., Coleman I., Clegg N., Thangavel C., Morrissey C., Zhang X., Comstock C. E., Witkiewicz A. K., Gomella L., Knudsen E. S., Nelson P. S., Knudsen K. E., The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J. Clin. Invest. 120, 4478–4492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dick F. A., Rubin S. M., Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 14, 297–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nyquist M. D., Corella A., Coleman I., de Sarkar N., Kaipainen A., Ha G., Gulati R., Ang L., Chatterjee P., Lucas J., Pritchard C., Risbridger G., Isaacs J., Montgomery B., Morrissey C., Corey E., Nelson P. S., Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep. 31, 107669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karousis E. D., Mühlemann O., Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb. Perspect. Biol. 11, a032862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kervestin S., Jacobson A., NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13, 700–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rivas M. A., Pirinen M., Conrad D. F., Lek M., Tsang E. K., Karczewski K. J., Maller J. B., Kukurba K. R., DeLuca D. S., Fromer M., Ferreira P. G., Smith K. S., Zhang R., Zhao F., Banks E., Poplin R., Ruderfer D. M., Purcell S. M., Tukiainen T., Minikel E. V., Stenson P. D., Cooper D. N., Huang K. H., Sullivan T. J., Nedzel J.; GTEx Consortium; Geuvadis Consortium, Bustamante C. D., Li J. B., Daly M. J., Guigo R., Donnelly P., Ardlie K., Sammeth M., Dermitzakis E. T., McCarthy M. I., Montgomery S. B., Lappalainen T., MacArthur D. G., Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348, 666–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldman M. J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A. N., Zhu J., Haussler D., Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uphoff C. C., Drexler H. G., Detection of mycoplasma contaminations. Methods Mol. Biol. 290, 13–23 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Ellwood-Yen K., Graeber T. G., Wongvipat J., Iruela-Arispe M. L., Zhang J., Matusik R., Thomas G. V., Sawyers C. L., Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Hormaechea-Agulla D., Gómez-Gómez E., Ibáñez-Costa A., Carrasco-Valiente J., Rivero-Cortés E., López F. L., Pedraza-Arevalo S., Valero-Rosa J., Sánchez-Sánchez R., Ortega-Salas R., Moreno M. M., Gahete M. D., López-Miranda J., Requena M. J., Castaño J. P., Luque R. M., Ghrelin O-acyltransferase (GOAT) enzyme is overexpressed in prostate cancer, and its levels are associated with patient's metabolic status: Potential value as a non-invasive biomarker. Cancer Lett. 383, 125–134 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., de Paepe A., Speleman F., Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, Research0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trincado J. L., Entizne J. C., Hysenaj G., Singh B., Skalic M., Elliott D. J., Eyras E., SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 19, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faisal F. A., Sundi D., Tosoian J. J., Choeurng V., Alshalalfa M., Ross A. E., Klein E., Den R., Dicker A., Erho N., Davicioni E., Lotan T. L., Schaeffer E. M., Racial variations in prostate cancer molecular subtypes and androgen receptor signaling reflect anatomic tumor location. Eur. Urol. 70, 14–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pérez-Silva J. G., Araujo-Voces M., Quesada V., nVenn: Generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics 34, 2322–2324 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., Tyers M., BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S3
Legends for data S1 to S4
Data S1 to S4