Abstract
The efficacy of immunotherapy targeting the PD-1/PD-L1 pathway in hepatocellular carcinoma (HCC) is limited. NOD-like receptors (NLRs) comprise a highly evolutionarily conserved family of cytosolic bacterial sensors, yet their impact on antitumor immunity against HCC remains unclear. In this study, we uncovered that NOD1, a well-studied member of NLR family, exhibits predominant expression in tumor-associated macrophages (TAMs) and correlates positively with improved prognosis and responses to anti–PD-1 treatments in patients with HCC. Activation of NOD1 in vivo augments antitumor immunity and enhances the effectiveness of anti–PD-1 therapy. Mechanistically, NOD1 activation resulted in diminished expression of perilipin 5, thereby hindering fatty acid oxidation and inducing free fatty acid accumulation in TAMs. This metabolic alteration promoted membrane localization of the costimulatory molecule OX40L in a lipid modification–dependent manner, thereby activating CD8+ T cells. These findings unveil a previously unrecognized role for NOD1 in fortifying antitumor T cell immunity in HCC, potentially advancing cancer immunotherapy.
Targeting NOD1 provides a promising strategy for combination therapy aimed at improving the management of immunotherapy in HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC) is a highly prevalent malignancy and the third leading cause of cancer-related death worldwide (1). A considerable number of patients with HCC are diagnosed at an advanced stage, rendering them ineligible for curative surgery (2). Immune checkpoint blockade (ICB) therapy, particularly the use of antibodies targeting the programmed cell death (PD-1/PD-L1) signaling pathway, has revolutionized cancer treatment. Nevertheless, clinical trials, CheckMate 040 (3) and CheckMate 459 (4), revealed that the anti–PD-1 inhibitors achieved a modest objective response rate of only 14% and disease control rate of 56% in patients with advanced HCC. Therefore, there is an urgent need to identify a combined therapeutic strategy to improve the efficacy of ICB therapy for HCC.
The limited efficacy of ICB may be attributed to the ability of HCC to evade immune responses through various mechanisms, including the generation of a tumor microenvironment (TME) rich in tumor-associated macrophages (TAMs) (constitute more than 50% of immune cells) with formidable immunosuppressive properties (5, 6). Two primary strategies have emerged for targeting TAMs in treatment. The first involves the depletion of TAMs by disrupting their recruitment and viability, such as by inhibiting TAM recruitment via the C-C Motif Chemokine Ligand 2 (CCL2)-C-C Motif Chemokine Receptor 2 (CCR2) axis (7) or inducing TAM apoptosis through blockade of the Colony Stimulating Factor 1 (CSF1)-Colony Stimulating Factor 1 Receptor (CSF1R) axis (8). However, this approach compromises the intrinsic immunostimulatory functions of macrophages as key phagocytes and proficient antigen-presenting cells (APCs) within solid tumors (9). The second strategy entails reprogramming or repolarizing immunosuppressive TAMs into immunostimulatory phenotypes, which has exhibited greater promise in numerous preclinical investigations involving the blocking of Triggering Receptor Expressed on Myeloid Cells 2 (TREM2) (10), CD24-Siglec10 (11), and CD47-SIRPα (12) signaling pathways. These approaches aim to alter the cellular immunophenotype by targeting macrophage membrane receptors. Nevertheless, limited attention has been given to exploring the role of cytosolic receptors within TAMs in modulating tumor immunity.
NOD-like receptors (NLRs) constitute a highly evolutionarily conserved family of cytosolic bacterial sensors, primarily expressed in APCs including macrophages and dendrite cells. Because of their capability to detect distinct bacterial molecular signatures and trigger inflammatory responses, NLRs are generally recognized as key receptors for pathogen identification within the innate immune system (13). It is suggested that certain microorganisms exhibit evasion mechanisms, such as perforating the phagosome membrane and entering the cytoplasm, which necessitates the recognition of cytosolic NLRs (14). In addition, nonpathogenic bacteria could elicit responses in host cells via NLRs even in the absence of physical contact (15). Of note, Griffin et al. highlighted that Enterococcus can produce NOD2-active muropeptides, thereby enhancing the anti–PD-L1 antitumor efficacy in melanoma, indicating the potential of NLRs in advancing tumor immunotherapy (16). Considering the compromised intestinal barrier in patients with HCC, gut microbiota and their components may infiltrate the TME via enterohepatic circulation (17). Recent studies (18–20) and our previous work (21) have underscored the critical role of NLRs in the development and progression of HCC. However, existing researches primarily focus on the roles of NLRs in the malignant behavior of tumor cells, leaving a noticeable research gap in understanding the regulatory role of NLRs in the immune microenvironment of HCC.
In this study, among the NLR family, we identified that NOD1, primarily recognizing the conserved bacterial peptidoglycan molecular pattern, γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP) (13), was predominantly expressed in TAMs and associated with enhanced antitumor immunity and survival outcomes in patients with HCC. NOD1 activation induces TAMs to acquire an immunostimulatory phenotype and delay HCC progression. Mechanistically, NOD1 activation disrupts fatty acid oxidation (FAO) in TAMs through perilipin 5 (PLIN5), subsequently leading to the macrophage membrane enrichment of the costimulatory molecule OX40 ligand (OX40L), thereby supporting CD8+ T cell response. Furthermore, NOD1 serves as a potent adjunctive target for augmenting the effectiveness of ICB therapy.
RESULTS
NOD1 is predominantly expressed in TAMs and associated with enhanced antitumor immunity and survival outcomes in patients with HCC
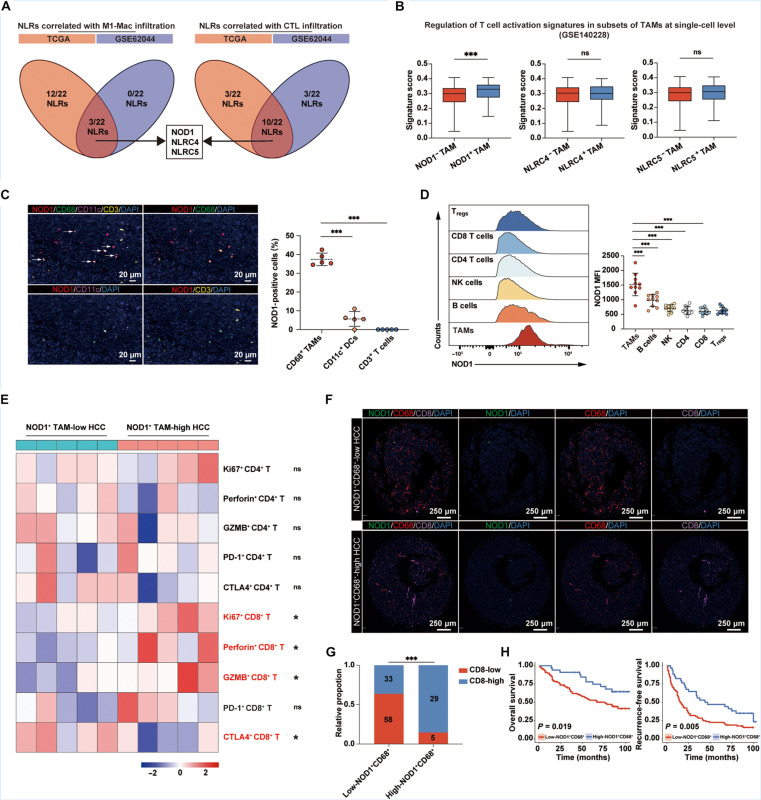
Although the role of NLRs in macrophages for pathogen identification within the innate immune response is well-established, how NLRs on TAMs mediate the microbial influence on antitumor immunity remains unclear. Numerous studies have elucidated that TAMs play a pivotal role in promoting the progression and immunotherapy resistance of HCC by suppressing T cell chemotaxis and activation (22–24). To identify essential regulatory molecules within the NLR family related to TAM phenotype and T cell activation, we analyzed two sets of bulk RNA sequencing (RNA-seq) data from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) and the Gene Expression Omnibus (GEO) (GSE62044). The abundance of M1-type macrophages and cytotoxic lymphocytes (CTLs) in tumor samples was quantified using the Cell Immune Biopsy Expression and Receptor Signature of Tumors (CIBERSORT) algorithm (25, 26) and the Microenvironment Cell Populations-counter (MCP-counter) (27), respectively. Correlations among NLRs, M1-type TAMs, and CTLs were delineated in fig. S1 (A and B). As anticipated, the abundance of M1-type TAMs was significantly associated with CTL infiltrations. Subsequently, we performed an intersection of statistically significant NLR members correlated with M1-type TAMs and CTLs, identifying three key genes: NOD1, NLRC4, and NLRC5 (Fig. 1A). Validation at the single-cell level (GSE140228) showed that NOD1+ TAMs exhibited a significantly enhanced regulatory ability for T cell activation compared to NOD1− TAMs within the HCC TME, a phenomenon not observed in NLRC4+ TAMs and NLRC5+ TAMs (Fig. 1B and fig. S1, C and D). Consequently, NOD1 was prioritized for our subsequent analysis. Furthermore, we demonstrated positive correlations between NOD1 levels and the infiltrations of M1-type macrophages and CTLs across various tumors using the CIBERSORT and the Tumor Immune Dysfunction and Exclusion (TIDE) (28) tool, respectively (fig. S1E), suggesting that NOD1 may be crucial in fostering T cell immunity.
Fig. 1. The infiltration of NOD1+ TAM correlates with CD8+ T cell activation and improved survival in patients with HCC.
(A) Venn diagrams showing the overlap of statistically significant NLR members associated with M1-type macrophages and CTL infiltrations in both TCGA-LIHC and GSE62044 cohorts. (B) Single-cell analysis comparing expression-based scores for the regulation of T cell activation across different TAM subsets. (C) mIF analysis for NOD1, CD68, CD11c, and CD3 markers in human HCC tissues (n = 5). White arrows indicate NOD1+ TAMs. Scale bars, 20 μm. (D and E) Flow cytometric analysis of human HCC samples (n = 10). NOD1 expression across different immune cell populations within the HCC TME (D). Heatmap displaying relative expressions of functional molecules in CD4+ and CD8+ T cells in the low- and high-NOD1+ TAM groups (E). (F to H) mIF analysis in a cohort of patients with HCC with survival data (n = 125). Patients were categorized into low-NOD1+ CD68+ (n = 91) and high-NOD1+ CD68+ (n = 34) groups according to the optimal cutoff value determined by X-title software. Representative mIF images of NOD1, CD68, and CD8 were presented (F). Scale bars, 250 μm. The levels of CD8+ T cells among patients with HCC with low- or high-NOD1+ CD68+ cells (G). Kaplan-Meier analysis comparing the OS [(H), left] and RFS [(H), right)] of patients with HCC with low or high levels of NOD1+CD68+ cells. Statistical analysis was performed using the Mann-Whitney U test in (B), the Student’s t test in [(C) and (D)], the chi-square test in (G), and the log-rank test in (H). *P < 0.05, **P < 0.01, and ***P < 0.001. Data were presented as median with interquartile range (IQR) in (B) and mean with SD in (D). NK, natural killer.
To validate our findings, we performed a multiplex immunofluorescence (mIF) assay to substantiate the predominant colocalization of NOD1 with CD68+ TAMs, as opposed to CD11c+ dendritic cells and CD3+ T cells, in human HCC tissues (Fig. 1C). Furthermore, we collected tumor samples from 10 patients with HCC for flow cytometric analysis (gating strategies in fig. S2A). The expression of NOD1 on TAMs was significantly higher than on other critical immune cell types within HCC TME (Fig. 1D). Notably, HCC with high NOD1+ TAM infiltrations exhibited higher proportions of activated CD8+ T cells, as evidenced by the increased expressions of Ki67, perforin, and granzyme B (GZMB), as well as decreased expression of inhibitory receptor Cytotoxic T-Lymphocyte Antigen 4 (CTLA4), a phenomenon not observed in CD4+ T cells (Fig. 1E). Moreover, the infiltrations of B cells, natural killer cells, and regulatory T cells (Tregs) showed no discernible differences between the two groups (fig. S3).
Subsequently, we evaluated the level of NOD1+ TAMs and CD8+ T cells in a cohort of 125 patients with HCC from our center by mIF. Patients with high-NOD1+ TAMs exhibited significantly higher infiltrations of CD8+ T cells (Fig. 1, F and G). Correlation analysis revealed that high levels of NOD1+ TAM were associated with multiple tumors but not with other clinical characteristics (Table 1). Kaplan-Meier analysis indicated that high levels of NOD1+ TAM were associated with both favorable overall survival (OS) and recurrence-free survival (RFS) (Fig. 1H). Multivariate Cox regression analysis showed that the NOD1+ TAM level was an independent indicator for both OS and RFS in patients with HCC (Table 2).
Table 1. The association between NOD1+ CD68+ cell infiltration and clinicopathological characteristics in the HCC cohort.
HBsAg, hepatitis B surface antigen; AFP, α-fetoprotein; MVI, microvascular invasion; TNM, tumor-nodes-metastasis; BCLC, Barcelona Clinic Liver Cancer.
| Variable | NOD1+CD68+cell-low | NOD1+CD68+cell-high | P value |
|---|---|---|---|
| (n = 91) | (n = 34) | ||
| Age (years) | 0.810 | ||
| ≤65 | 74 (81.3%) | 27 (79.4%) | |
| >65 | 17 (18.7%) | 7 (20.6%) | |
| Gender | 0.809 | ||
| Male | 76 (83.5%) | 29 (85.3%) | |
| Female | 15 (16.5%) | 5 (14.7%) | |
| HbsAg | 0.138 | ||
| Negative | 18 (19.8%) | 11 (32.4%) | |
| Positive | 73 (80.2%) | 23 (67.6%) | |
| HCV | 0.618 | ||
| Negative | 89 (97.8%) | 33 (97.1%) | |
| Positive | 2 (2.2%) | 1 (2.9%) | |
| AFP (ng/ml) | 0.496 | ||
| ≤400 | 64 (70.3%) | 26 (76.5%) | |
| >400 | 27 (29.7%) | 8 (23.5%) | |
| Cirrhosis | 0.700 | ||
| Absent | 16 (17.6%) | 7 (20.6%) | |
| Present | 75 (82.4%) | 27 (79.4%) | |
| Ascites | 0.386 | ||
| Absent | 85 (93.4%) | 33 (97.1%) | |
| Present | 6 (6.6%) | 1 (2.9%) | |
| Tumor size (cm) | 0.545 | ||
| ≤5 | 59 (64.8%) | 24 (70.6%) | |
| >5 | 32 (35.2%) | 10 (29.4%) | |
| Tumor number | 0.021* | ||
| Single | 80 (87.9%) | 24 (70.6%) | |
| Multiple | 11 (12.1%) | 10 (29.4%) | |
| Tumor encapsulation | 0.286 | ||
| Absent | 81 (89.0%) | 33 (97.1%) | |
| Present | 10 (10.7%) | 1 (2.9%) | |
| Tumor differentiation | 0.177 | ||
| I and II | 58 (63.7%) | 26 (76.5%) | |
| III and IV | 33 (36.3%) | 8 (23.5%) | |
| MVI | 0.473 | ||
| Absent | 58 (63.7%) | 24 (70.6%) | |
| Present | 33 (36.3%) | 10 (29.4%) | |
| Vascular invasion | 0.256 | ||
| Absent | 80 (87.9%) | 32 (94.1%) | |
| Present | 11 (12.1%) | 2 (5.9%) | |
| TNM stage | 0.648 | ||
| I and II | 47 (51.6%) | 16 (47.1%) | |
| III and IV | 44 (48.4%) | 18 (52.9%) | |
| BCLC stage | 0.427 | ||
| 0 + A | 49 (53.8%) | 21 (61.8%) | |
| B + C | 42 (46.2%) | 13 (38.2%) |
Table 2. Univariate and multivariate analyses for OS and RFS in the HCC cohort.
*P < 0.05. HR, hazard ratio; CI, confidence interval; HBsAg, hepatitis B surface antigen; AFP, α-fetoprotein; MVI, microvascular invasion; TNM, tumor-nodes-metastasis; BCLC, Barcelona Clinic Liver Cancer.
| Variable | OS | RFS | ||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||
| P value | HR (95% CI) | P value | P value | HR (95% CI) | P value | |
| Age, years (>65 versus ≤65) | 0.1035 | 0.6089 | ||||
| Gender (male versus female) | 0.8522 | 0.6201 | ||||
| HbsAg (positive versus negative) | 0.0490* | 2.5 (1.2–5.3) | 0.020* | 0.2139 | ||
| AFP, ng/ml (>400 versus ≤400) | 0.0103* | 0.0076* | ||||
| Cirrhosis (present versus absent) | 0.0526* | 0.1682 | ||||
| Ascites (present versus absent) | 0.3934 | 0.0541* | ||||
| Tumor size, cm (>5 versus ≤5) | 0.0261* | 0.1057 | ||||
| Tumor number (multiple versus single) | 0.2267 | 0.0044* | 2.5 (1.5–4.2) | 0.001* | ||
| Tumor encapsulation (absent versus present) | 0.1917 | 0.0302* | 2.4 (1.1–5.0) | 0.025* | ||
| Tumor differentiation (III and IV versus I and II) | 0.6205 | 0.6489 | ||||
| Vascular invasion (present versus absent) | <0.0001* | <0.0001* | ||||
| MVI (present versus absent) | 0.0088* | 0.0472* | ||||
| TNM stage (III and IV versus I and II) | 0.0021* | 0.0005* | ||||
| BCLC stage (B + C versus 0 + A) | 0.0067* | 2.5 (1.7–3.6) | <0.0001* | 0.010* | 1.9 (1.4–2.7) | <0.0001* |
| NOD1+CD68+cell (high versus low) | 0.0188* | 0.5 (0.3–0.9) | 0.036* | 0.0224* | 0.5 (0.3–0.8) | 0.005* |
NOD1 activation induces TAMs to acquire an immunostimulatory phenotype capable of supporting CD8+ T cell response in vitro
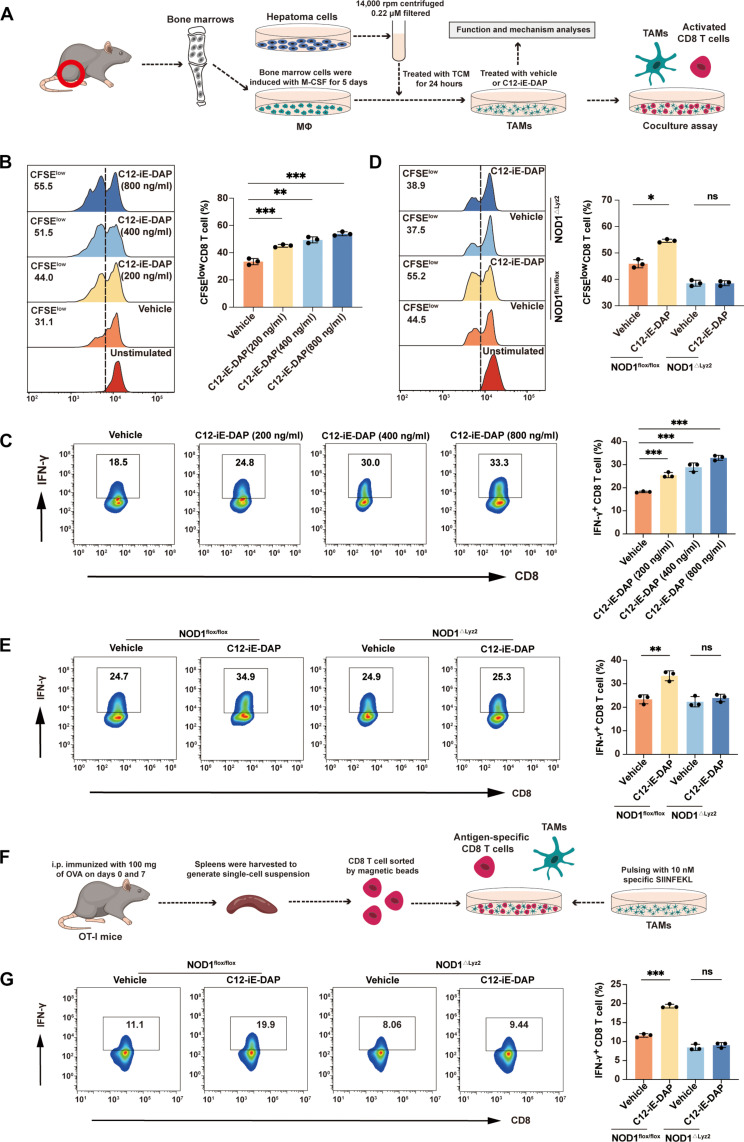
To investigate the impact of NOD1 activation on TAM phenotype in vitro, we used TAM-like macrophages by treating bone marrow–derived macrophages (BMDMs) with tumor conditioned medium (TCM) from hepatoma cells (Fig. 2A) (29). Treatment with C12-iE-DAP, an acylated derivative of the iE-DAP dipeptide for NOD1 stimulation, resulted in a reduction of immunosuppressive gene expression and an increase in immunostimulatory gene expression in TAMs (fig. S4, A to C). When cocultured TAMs with splenic CD8+ T cells (Fig. 2A), C12-iE-DAP–treated TAMs demonstrated a significantly enhanced ability to activate CD8+ T cells, as indicated by increased T cell proliferation (Fig. 2B and fig. S4D) and up-regulated cytotoxic effector molecule, interferon-γ (IFN-γ) (Fig. 2C and fig. S4E). This immunostimulatory effect of C12-iE-DAP was eliminated in TAMs generated from NOD1△Lyz2 (macrophage-specific NOD1-deficient) mice, indicating a dependency on NOD1 (Fig. 2, D and E). This finding was further substantiated by a coculture assay using mouse antigen–specific OT-I CD8+ T cells (Fig. 2, F and G, and fig. S4F).
Fig. 2. NOD1 activation induces TAMs to acquire an immunostimulatory phenotype capable of supporting CD8+ T cell response.
(A) Schematic representation of the in vitro CD8+ T cell coculture assay. (B and D) Flow cytometric analysis of carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled CD8+ T cell proliferation after cocultured with TAMs that were generated from wild-type (WT) mice and treated with either vehicle or C12-iE-DAP (200, 400, and 800 ng/ml) for 24 hours (B) or from NOD1flox/flox or NOD1△Lyz2 mice and treated with either vehicle or C12-iE-DAP (400 ng/ml) for 24 hours (D). (C and E) Flow cytometry analysis of IFN-γ+ CD8+ T cells after cocultured with TAMs that were generated from WT mice and treated with vehicle or C12-iE-DAP (200, 400, and 800 ng/ml) for 24 hours (C) or from NOD1flox/flox or NOD1△Lyz2 mice and treated with either vehicle or C12-iE-DAP (400 ng/ml) for 24 hours (E). (F) Schematic representation of the in vitro antigen-specific CD8+ T cell coculture assay. i.p., intraperitoneal. (G) Flow cytometric analysis of IFN-γ+ OT-I CD8+ T cells after cocultured with TAMs that were generated from NOD1flox/flox or NOD1△Lyz2 mice and treated with either vehicle or C12-iE-DAP and then pulsed with SIINFEKL (OVA257-264). Statistical analysis was performed using the Student’s t test. Data were presented as mean with SD. ns, no significance; *P < 0.05, **P < 0.01, and ***P < 0.001. M-CSF, macrophage colony-stimulating factor.
Macrophage NOD1 activation delays HCC progression in a CD8+ T cell–dependent manner in vivo
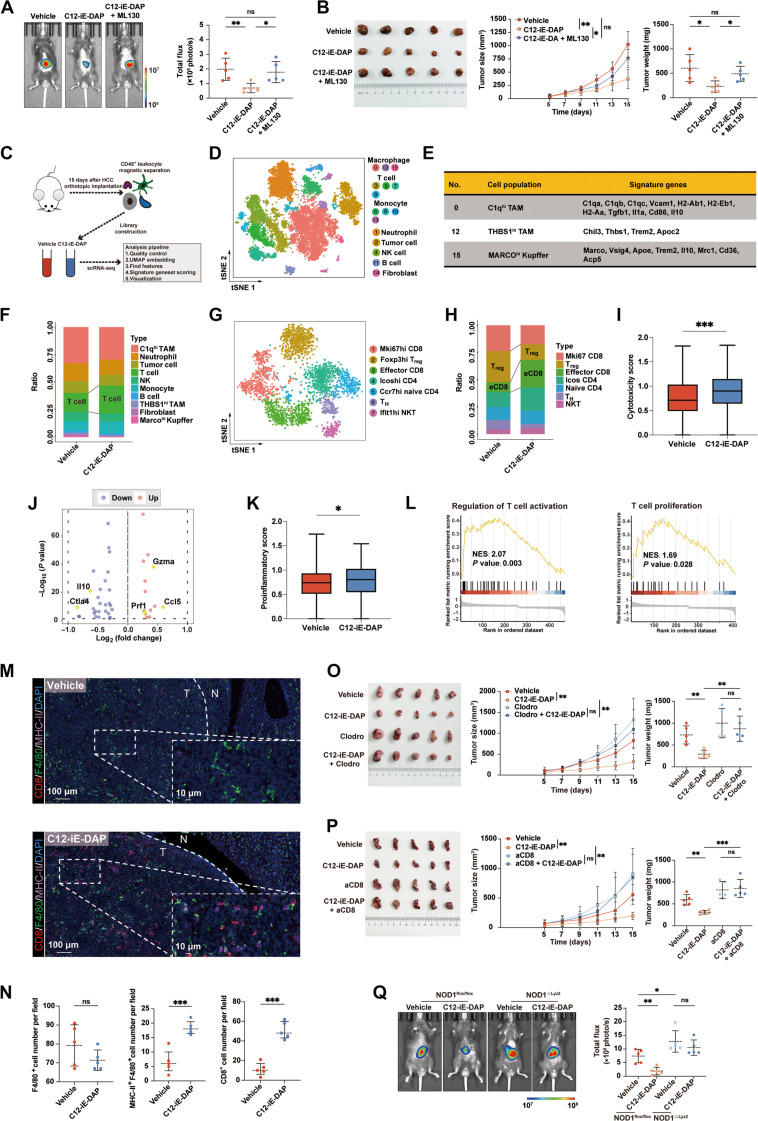
Next, we investigated whether NOD1 activation affects tumor growth in vivo. We established orthotopic and subcutaneous HCC models in C57BL/6 mice and observed that administration of C12-iE-DAP intraperitoneally significantly inhibited HCC growth, which was abolished by ML130, a NOD1 selective inhibitor (Fig. 3, A and B).
Fig. 3. In vivo activation of macrophage NOD1 delays HCC progression by enhancing CD8+ T cell immunity.
(A and B) Orthotopic and subcutaneous HCC tumors treated with either C12-iE-DAP, C12-iE-DAP + ML130, or vehicle (n = 5 each). (C) Schematic of the scRNA-seq experiment design. (D) t-Distributed stochastic neighbor embedding (tSNE) plot clustering with cell annotation for CD45+ cells. (E) Signature genes of identified clusters of macrophages. (F) Proportions of different immune cell types. (G) tSNE plot clustering with cell annotation for T cells. (H) Proportions of different T cell types. (I) The cytotoxicity score in CD8+ T cells between two groups. (J) Volcano plot showing differentially expressed genes between CD8+ T cells in the vehicle or C12-iE-DAP–treated HCC. (K) The proinflammatory score in NOD1+ C1qhi TAMs between two groups. (L) Gene set enrichment analysis (GSEA) analysis. (M and N) mIF analysis for CD8, F4/80, and MHC-II markers in the vehicle- or C12-iE-DAP–treated orthotopic HCC tumors. (O and P) Subcutaneous HCC tumors treated with either vehicle or C12-iE-DAP in macrophage-depleted (O) or CD8-depleted (P) mice (n = 5, each). (Q) Orthotopic HCC tumors growing in NOD1flox/flox versus NOD1△Lyz2 mice treated with either vehicle or C12-iE-DAP (n = 5 each). Statistical analysis was performed using the Student’s t test [or the Mann-Whitney U test in (I) and (K)]. Data were presented as mean with SD [or median with IQR in (I) and (K)]. *P < 0.05, **P < 0.01, and ***P < 0.001. UMAP, Uniform Manifold Approximation and Projection; TH, T helper; NES, normalized enrichment score.
We further isolated CD45+ immune cells from orthotopic HCC tumors grown in mice treated with either vehicle or C12-iE-DAP for single-cell RNA-seq (scRNA-seq) (Fig. 3C). A total of 16 cell clusters were identified, including 4 clusters (3, 5, 7, and 9) designated as T cells and 3 clusters (0, 12, and 15) designated as macrophages displaying various features (Fig. 3, D and E, and fig. S5A). C12-iE-DAP–treated HCC exhibited a higher T cell infiltration compared to the control HCC (Fig. 3F). Upon detailed T cell clustering (Fig. 3G and fig. S5, B and C), we observed an increase in the effector CD8+ T cells and a decrease in Tregs in C12-iE-DAP–treated HCC compared to control HCC (Fig. 3H). In addition, CD8+ T cells in C12-iE-DAP–treated HCC displayed significantly higher cytotoxicity scores and higher expressions of immunostimulatory genes such as Gzma, Ccl5, and Prf1, along with lower expressions of immunosuppressive genes such as Il10 and Ctla4, compared to the vehicle group (Fig. 3, I and J).
Among macrophages, clusters 12 (THBS1hi TAMs) and 15 (MARCOhi Kupffers) were characterized by weak antigen presentation and immune activation capabilities, indicated by low expressions of major histocompatibility complex (MHC)–related and immune-costimulatory genes. Cluster 0 (C1qhi TAMs) exhibited high MHC-related gene expression (H2-Ab1, H2-Eb1, and H2-Aa) as well as coexpression of both M1 marker genes (CD86, Vcam1, and Il1a) and M2 marker genes (Tgfb1 and Il10) (Fig. 3E), which were identified as “double-agent” immune regulatory TAMs (30) and chosen for further analyses. The NOD1+ subset of C1qhi TAMs from the C12-iE-DAP–treated HCC exhibited significantly higher proinflammatory scores than those from the vehicle-treated group (Fig. 3K). Moreover, gene set enrichment analysis (GSEA) revealed that NOD1+ C1qhi TAMs in C12-iE-DAP–treated HCC were notably enriched in functions related to T cell activation and T cell proliferation (Fig. 3L).
Following this, the mIF assay confirmed a significant increase in MHC-II+ M1-like macrophages and CD8+ T cells in orthotopic HCC tumors upon C12-iE-DAP treatment (Fig. 3, M and N). In addition, flow cytometric analysis (gating strategies depicted in fig. S2B) revealed that C12-iE-DAP treatment polarized TAMs toward an M1-like phenotype, as evidenced by an elevated MHC-II to CD206 ratio, along with increased infiltration of CD8+ T cells expressing high levels of IFN-γ, GZMB, and PD-1 (fig. S6). Notably, these effects were reversed upon administration of ML130, a selective NOD1 inhibitor.
As expected, the depletion of macrophages or CD8+ T cells from the TME significantly eliminated the antitumor effects of C12-iE-DAP (Fig. 3, O and P, and fig. S7). Notably, C12-iE-DAP treatment did not significantly inhibit HCC growth in severe combined immunodeficiency (SCID) mice characterized by a severe deficiency in both T and B lymphocytes (fig. S8A), as well as BALB/c nude mice characterized by a lack of thymus (fig. S8, B to E). Furthermore, we observed that HCC growth in NOD1△Lyz2 mice was significantly accelerated and could not be inhibited by C12-iE-DAP, highlighting the essential role of NOD1 activation in macrophages for antitumor immunity in HCC (Fig. 3Q). In summary, our findings demonstrated that in vivo activation of NOD1 delays HCC progression, and this antitumor effect is contingent on the presence of macrophages and CD8+ T cells.
NOD1 activation disrupts fatty acid oxidation in TAMs by regulating PLIN5
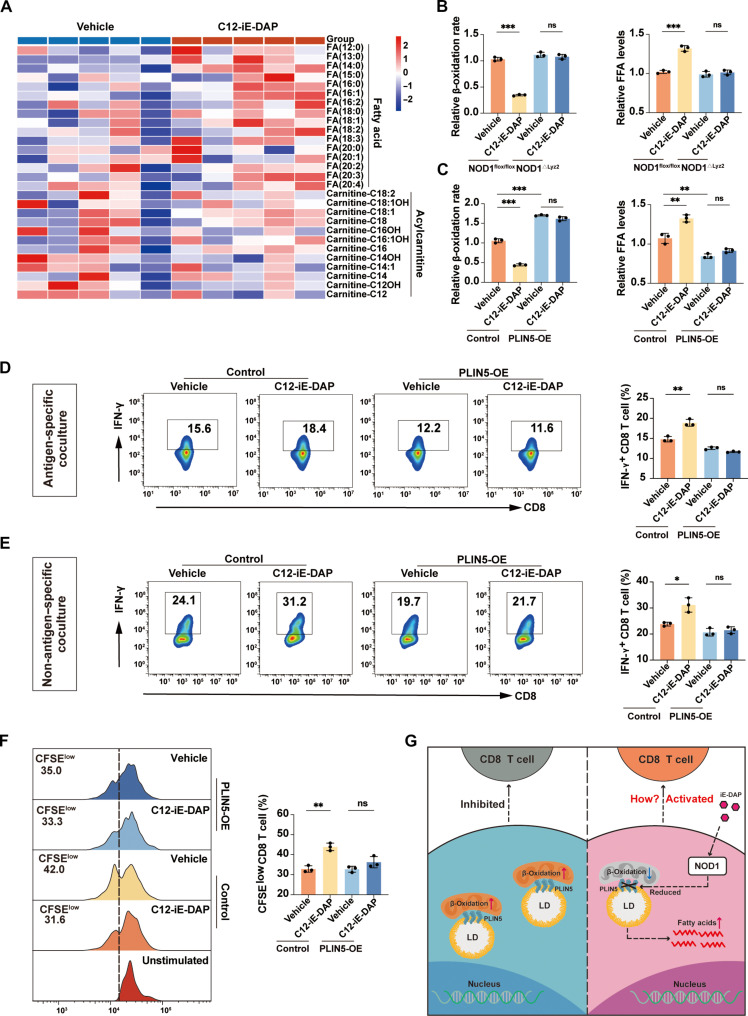
To investigate the mechanism by which NOD1 promoted the TAM phenotype switch, we performed GSEA enrichment analysis of scRNA-seq data from patients with HCC (GSE140228), which suggested that NOD1 exhibited a noteworthy negative correlation with pathways related to FAO (fig. S9A). Increasing evidence indicated that FAO is critical for governing the generation and function of TAMs (31). We conducted a metabolomic analysis of TAMs treated with either vehicle or C12-iE-DAP. C12-iE-DAP–treated TAMs exhibited elevated free fatty acid (FFA) levels compared to vehicle-treated TAMs. In contrast, acylcarnitine, the main form of long-chain FAs transported into the mitochondria, showed a relative decrease upon C12-iE-DAP treatment, suggesting potential impairment in shuttling FAs into the mitochondria for oxidation (Fig. 4A). In contrast, there were no significant differences in the levels of diacylglycerol and major structural lipids between the two groups, indicating that the elevated FFAs were not redirected into these crucial lipid components (fig. S9B). Colorimetric assays further confirmed the decreased β-oxidation rates and increased FFA levels in C12-iE-DAP–treated TAMs compared to control TAMs. Notably, these effects were absent in TAMs generated from NOD1△Lyz2 mice (Fig. 4B). These findings suggested that NOD1 activation significantly decreases FAO and increases FFA levels in TAMs.
Fig. 4. NOD1 activation decreases PLIN5 expression to regulate the FAO and immunostimulatory function in TAMs.
(A) Heatmap of representative FFAs and acylcarnitines in TAMs treated with either vehicle or C12-iE-DAP (400 ng/ml) for 24 hours. (B and C) β-Oxidation rate and FFA levels were measured in TAMs with different treatments. (D) Flow cytometric analysis of IFN-γ+ OT-I CD8+ T cells after cocultured with TAMs that were transfected with either control or PLIN5 overexpression plasmids, followed by treatment with either vehicle or C12-iE-DAP (400 ng/ml), and then pulsed with SIINFEKL (OVA257-264). (E) Flow cytometric analysis of IFN-γ+ CD8+ T cells after cocultured with TAMs transfected with either control or PLIN5 overexpression plasmids, followed by treatment with either vehicle or C12-iE-DAP (400 ng/ml) for 24 hours. (F) Flow cytometry analysis of CFSE-labeled CD8+ T cell proliferation after cocultured with TAMs transfected with either control or PLIN5 overexpression plasmids, followed by treatment with either vehicle or C12-iE-DAP (400 ng/ml) for 24 hours. (G) Schematic diagram depicting the regulatory role of NOD1 on TAMs. Statistical analysis was performed using the Student’s t test. Data were presented as mean with SD. *P < 0.05, **P < 0.01, ***P < 0.001. LD, lipid droplet; OE, overexpression.
Next, we explored the molecules downstream of NOD1 that disrupt FAO in TAMs. By taking the intersection of the gene sets involved in the top four lipid metabolism–related pathways determined by GSEA of scRNA-seq data from patients with HCC (GSE140228), we identified PLIN5 and Peroxisome Proliferator-Activated Receptor αlpha (PPARA) as potential downstream genes (fig. S10, A and B). Quantitative reverse transcription polymerase chain reaction (PCR) and Western blot analyses confirmed that C12-iE-DAP treatment significantly reduced the mRNA and protein levels of PLIN5 and PPARα in TAMs (fig. S10, C and D). Recent studies suggested that PLIN5 plays an indispensable role in facilitating the flux of FA from lipid droplets (LDs) to the mitochondria for oxidation (32, 33). Therefore, we overexpressed PLIN5 in TAMs and observed that PLIN5 overexpression reversed the C12-iE-DAP–induced FAO disruption and FFA accumulation (fig. S11A and Fig. 4C), while contrasting effects were observed in PLIN5-silenced TAMs (fig. S11, B and C). These results indicate that NOD1 may disrupt FAO within TAMs by PLIN5.
We then investigated whether PLIN5 mediates the NOD1-induced immunostimulatory phenotype of TAMs. Analyses of scRNA-seq data from patients with HCC (GSE140228) revealed that PLIN5− TAMs had significantly enhanced phagocytic, proinflammatory, and T cell activation regulatory capabilities compared to PLIN5+ TAMs (fig. S11D). Moreover, in vitro coculture experiments demonstrated that PLIN5 overexpression in TAMs reversed their ability to promote CD8+ T cell activation upon C12-iE-DAP treatment in both antigen-specific (Fig. 4D) and non-antigen–specific (Fig. 4, E and F) cocultures. Conversely, PLIN5-silenced TAMs enhanced their ability to promote CD8+ T cell proliferation (fig. S11, E and F) and increased their cytotoxicity (fig. S11, G and H). Collectively, these results indicate that NOD1 may induce immunostimulatory phenotype in TAMs by PLIN5 (Fig. 4G).
Next, we investigated the mechanism by which NOD1 regulates the expression of PLIN5. It has been reported that PPARα (34) could form a heterodimer with another nuclear receptor, retinoid X receptor (RXR), and bind to the peroxisome proliferator response element (PPRE) in the promoter region of target genes, initiating their transcription. Notably, previous studies have shown that the first intron of the PLIN5 gene contains a PPRE (35), and PLIN5 can be induced in the liver in a PPARα-dependent manner (36–38). In our GSEA analysis of TCGA-LIHC data, we revealed that the PPAR signaling pathway was significantly down-regulated in patients with HCC with high NOD1 expression (fig. S10F). In addition, PPARA expression was significantly positively correlated with PLIN5 in the TCGA-LIHC dataset (fig. S10G). Furthermore, treatment with a PPARα agonist promoted PLIN5 expression and reversed the reduction in PLIN5 expression induced by the NOD1 agonist in TAMs (fig. S10H). Therefore, we propose that NOD1 activation primarily reduces PLIN5 expression by down-regulating PPARα.
NOD1 stimulation leads to self-oligomerization and the recruitment of receptor-interacting serine/threonine-protein kinase 2 (RIPK2) through homotypic Caspase Recruitment Domain (CARD)-CARD interactions, resulting in the activation of the mitogen-activated protein kinase (MAPK) p38 signaling pathway (39). Phosphorylated p38 is known to negatively regulate PPARα (40). In the GSEA analysis of the RNA-seq data from TCGA-LIHC, we demonstrated that MAPK signaling pathway was significantly up-regulated in NOD1-high HCC compared to NOD1-low HCC (fig. S10I). In addition, we confirmed elevated levels of phosphorylated RIPK2 and phosphorylated p38 in TAMs following NOD1 activation (fig. S10J). Moreover, treatment with the specific p38 MAPK inhibitor SB203580 reversed the down-regulation of PPARα and PLIN5 induced by the NOD1 agonist in TAMs (fig. S10K). Together, our findings demonstrated that NOD1 may regulate the expression of PLIN5 via the p38/PPARα pathway.
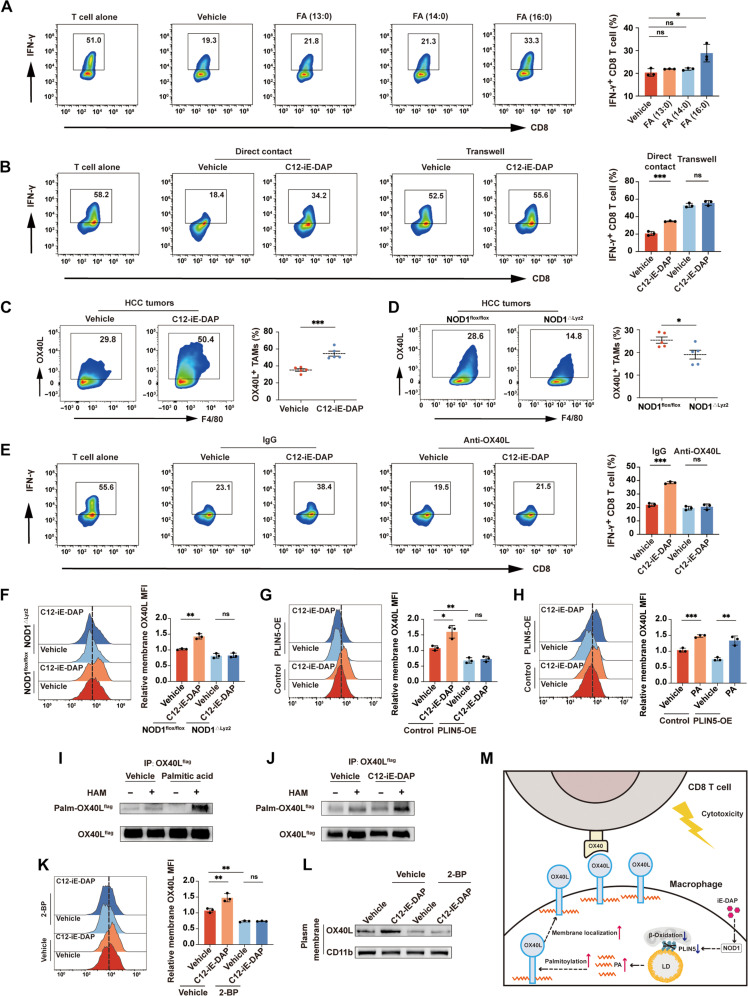
Increased palmitic acid promotes membrane localization of costimulatory ligand OX40L in TAMs to activate CD8+ T cells
Our metabolomic analysis of TAMs revealed that saturated FAs such as tridecanoic acid [FA (13:0)], myristic acid [FA (14:0)], and palmitic acid [FA (16:0)] were among the most significantly elevated FFAs in C12-iE-DAP–treated TAMs compared with vehicle-treated TAMs (fig. S12). Previous studies have highlighted that the overload of specific types of FFAs could induce a phenotypic switch in macrophages (41). We thereby treated TAMs with tridecanoic acid, myristic acid, and palmitic acid, respectively, followed by coculturing with splenic CD8+ T cells in vitro. Our results suggested that palmitic acid significantly enhanced the ability of TAMs to promote CD8+ T cell activation, whereas tridecanoic acid and myristic acid did not (Fig. 5A).
Fig. 5. NOD1-PLIN5–induced elevation of palmitic acid promotes macrophage OX40L membrane localization and activates CD8+ T cells.
(A) Flow cytometric analysis of IFN-γ+ CD8+ T cells cocultured with TAMs treated separately with 125 μM tridecanoic acid, myristic acid, or palmitic acid for 24 hours. (B) Flow cytometric analysis of IFN-γ+ CD8+ T cells cocultured with TAMs treated with either vehicle or C12-iE-DAP (400 ng/ml) in direct contact or separately in transwells. (C and D) Flow cytometric analysis of OX40L+ TAMs in orthotopic HCC tumors growing in WT mice treated intraperitoneally with either vehicle or C12-iE-DAP (C) or growing in NOD1flox/flox versus NOD1△Lyz2 mice (D). (E) Flow cytometric analysis of IFN-γ+ CD8+ T cells cocultured with TAMs pretreated with either immunoglobulin G (IgG) or anti-OX40L (5 μg/ml) and then treated with either vehicle or C12-iE-DAP (400 ng/ml). (F to H) Flow cytometric analysis of membrane OX40L in TAMs with different treatments. (I and J) OX40L palmitoylation was detected in TAMs using immunoprecipitation and acyl-biotin exchange (IP-ABE) assay. (K and L) Flow cytometric (K) and Western blot (L) analyses of membrane OX40L expression on TAMs pretreated with either vehicle or 2-bromopalmitate (2-BP; 50 μM), followed by treatment with either vehicle or C12-iE-DAP (400 ng/ml). (M) Schematic diagram depicting the regulatory role of NOD1 on TAMs. Statistical analysis was performed using the Student’s t test. Data were presented as mean with SD. *P < 0.05, **P < 0.01, and ***P < 0.001. MFI, mean fluorescence intensity; PA, palmitic acid; HAM, hydroxylamine.
The coculture experiments using vehicle- or C12-iE-DAP–treated TAMs and splenic CD8+ T cells, either in direct contact or separately in a transwell (29), demonstrated that promoting CD8+ T cell activation by the NOD1 on TAMs was largely dependent on direct cell-cell contact (Fig. 5B). Considering palmitic acid plays a pivotal role in regulating cellular localization of membrane proteins (42), coupled with the fact that macrophages can regulate T cell function through membrane costimulatory or inhibitory ligands (29, 43), we screened the membrane expression of nine key ligands and found that NOD1 activation significantly up-regulated the membrane expression of OX40L, a costimulatory molecule, in TAMs (fig. S13). In murine orthotopic HCC models, HCC treated with C12-iE-DAP exhibited higher proportions of OX40L+ TAM than controls, while significantly lower proportions of OX40L+ TAM were observed in HCC tumors growing in NOD1△Lyz2 mice compared to NOD1flox/flox mice (Fig. 5, C and D). Notably, in vitro coculture assay showed that the effects of C12-iE-DAP on CD8+ T cell activation were nullified by the OX40L-neutralizing antibody (Fig. 5E), indicating the critical role of OX40L in NOD1-induced immunostimulatory phenotype. Consistently, in vivo administration of OX40L blocking antibody significantly reversed the antitumor effect of the C12-iE-DAP (fig. S14).
Both flow cytometry and Western blotting analysis demonstrated that NOD1 activation increased OX40L expression on the TAM’s membrane, without influencing total protein and mRNA levels of OX40L. This effect was absent in TAMs generated from NOD1△Lyz2 mice or TAMs overexpressing PLIN5 (Fig. 5, F and G, and fig. S15). In addition, treatment with palmitic acid could reverse the reduction of membrane OX40L expression induced by PLIN5 overexpression (Fig. 5H). Palmitic acid–mediated protein palmitoylation enhances protein hydrophobicity and targets them to the plasma membrane (42). By using the acyl-biotin exchange (ABE), we demonstrated that treatment with C12-iE-DAP or palmitic acid significantly increased OX40L palmitoylation in TAMs (Fig. 5, I and J). Notably, treatment of TAMs with the palmitoylation inhibitor, 2-bromopalmitate (2-BP), effectively rever-sed the C12-iE-DAP–induced increase in OX40L membrane expression (Fig. 5, K and L). Together, our findings suggested that NOD1 elevates palmitic acid levels in TAMs, consequently increasing the membrane localization of OX40L through palmi-toylation, thereby activating CD8+ T cells (Fig. 5M).
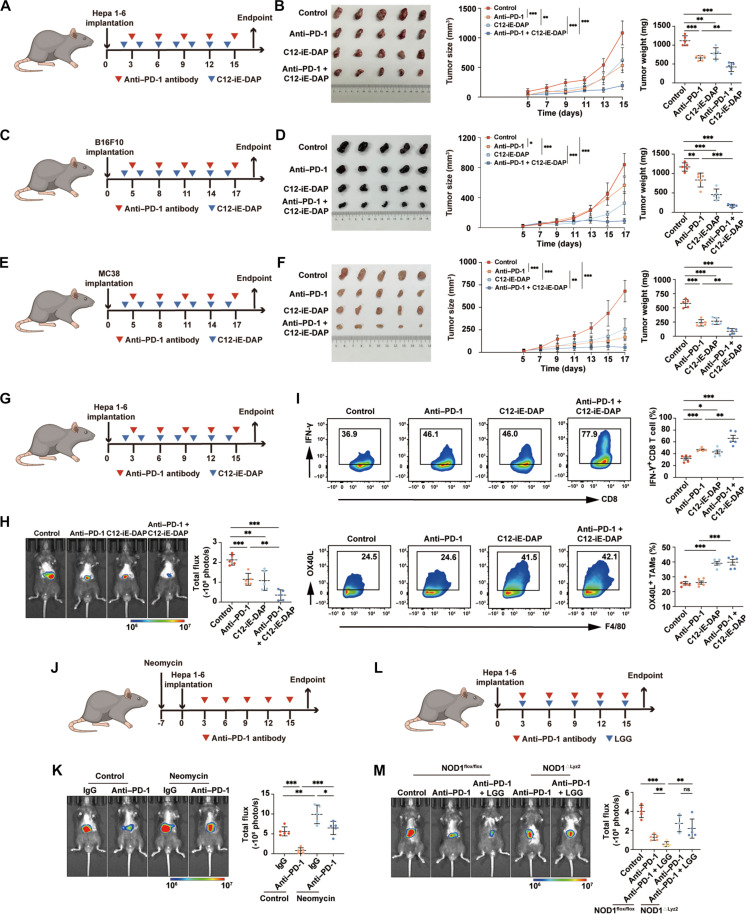
Activation of NOD1 enhances the therapeutic efficacy of anti–PD-1 treatment
To investigate the potential synergistic antitumor effects of NOD1 activation in conjunction with anti–PD-1 antibodies, we established subcutaneous HCC models in C57BL/6 mice. The combination of C12-iE-DAP with anti–PD-1 antibodies led to greater inhibition of HCC growth compared to the anti–PD-1 monotherapy group (Fig. 6, A and B). Subsequently, we used two distinct tumor models with different immunogenicities and sensitivities to anti–PD-1 therapies to validate our findings. The B16F10 melanoma is a poorly immunogenic tumor model that demonstrates poor responsiveness to anti–PD-1 therapy (44). Conversely, the MC38 colon carcinoma represents an immunogenic colon carcinoma model that exhibits partial responsiveness to anti–PD-1 therapy (45). Our results demonstrated that treatment with either C12-iE-DAP or anti–PD-1 alone could inhibit tumor growth. The combined treatment of both agents resulted in a more profound suppression of tumor growth (Fig. 6, C to F, and fig. S16).
Fig. 6. Activation of NOD1 enhances the therapeutic efficacy of anti–PD-1 treatment.
(A and B) Subcutaneous Hepa 1-6 tumors in C57BL/6 mice treated intraperitoneally with either anti–PD-1, C12-iE-DAP, anti–PD-1 + C12-iE-DAP, or isotype control (n = 5 each). Tumor growth curves and tumor weights are shown. (C and D) Subcutaneous B16F10 tumors in C57BL/6 mice treated intraperitoneally with either anti–PD-1, C12-iE-DAP, anti–PD-1 + C12-iE-DAP, or isotype control (n = 5 each). Tumor growth curves and tumor weights are shown. (E and F) Subcutaneous MC38 tumors in C57BL/6 mice treated intraperitoneally with either anti–PD-1, C12-iE-DAP, anti–PD-1 + C12-iE-DAP, or isotype control (n = 5 each). Tumor growth curves and tumor weights are shown. (G and H) Orthotopic Hepa 1-6 tumors in C57BL/6 mice treated intraperitoneally with either anti–PD-1, C12-iE-DAP, anti–PD-1 + C12-iE-DAP, or isotype control (n = 5 each). (I) Flow cytometry analysis of IFN-γ+ CD8+ T cells and OX40L+ TAMs in orthotopic Hepa 1-6 tumors (n = 5 each). (J and K) Orthotopic Hepa 1-6 tumors growing in neomycin-treated mice or untreated mice, followed by treatment of anti–PD-1 antibodies (n = 5 each). (L and M) Orthotopic HCC tumors growing in NOD1flox/flox or NOD1△Lyz2 mice and treated with either anti–PD-1, anti–PD-1 + LGG, or isotype control (n = 5 each). Statistical analysis was performed using the Student’s t test. Data were presented as mean with SD. *P < 0.05, **P < 0.01, and ***P < 0.001. LGG, Lactobacillus rhamnosus GG.
Next, we validated our finding by using orthotopic HCC models. Consistently, treatments with C12-iE-DAP significantly enhanced the therapeutic efficacy of anti–PD-1 treatment (Fig. 6, G and H). Furthermore, flow cytometric analysis indicated that orthotopic HCC receiving combination treatments exhibited significantly higher infiltrations of OX40L+ TAMs and IFN-γ+ CD8+ T cells, in comparison with the anti–PD-1 monotherapy group (Fig. 6I).
We analyzed gut microbiome metagenomic sequencing data of anti–PD-1–treated patients with HCC reported by Zheng et al. (46). The relative abundance of Lactobacillus, a primary iE-DAP–bearing probiotic known to activate NOD1 (47, 48), was significantly higher in fecal samples from anti–PD-1 responders compared to those from nonresponders (fig. S17). Subsequently, we treated mice with neomycin, which has been confirmed to inhibit iE-DAP–bearing Lactobacillus in previous studies (47, 48). We found that neomycin treatment markedly diminished the effectiveness of anti–PD-1 antibodies in orthotopic HCC models (Fig. 6, J and K). In contrast, orally administering Lactobacillus rhamnosus GG (LGG), one of the most widely used probiotic strains (49), significantly inhibited HCC growth in NOD1flox/flox mice, while this effect was not statistically significant in NOD1△Lyz2 mice (fig. S18). Furthermore, LGG oral administration significantly improved the responsiveness of HCC to anti–PD-1 therapy, an effect that was abolished in NOD1△Lyz2 mice (Fig. 6, L and M). Together, our findings demonstrated that activation of NOD1 could substantially enhance the therapeutic efficacy of anti–PD-1 treatment.
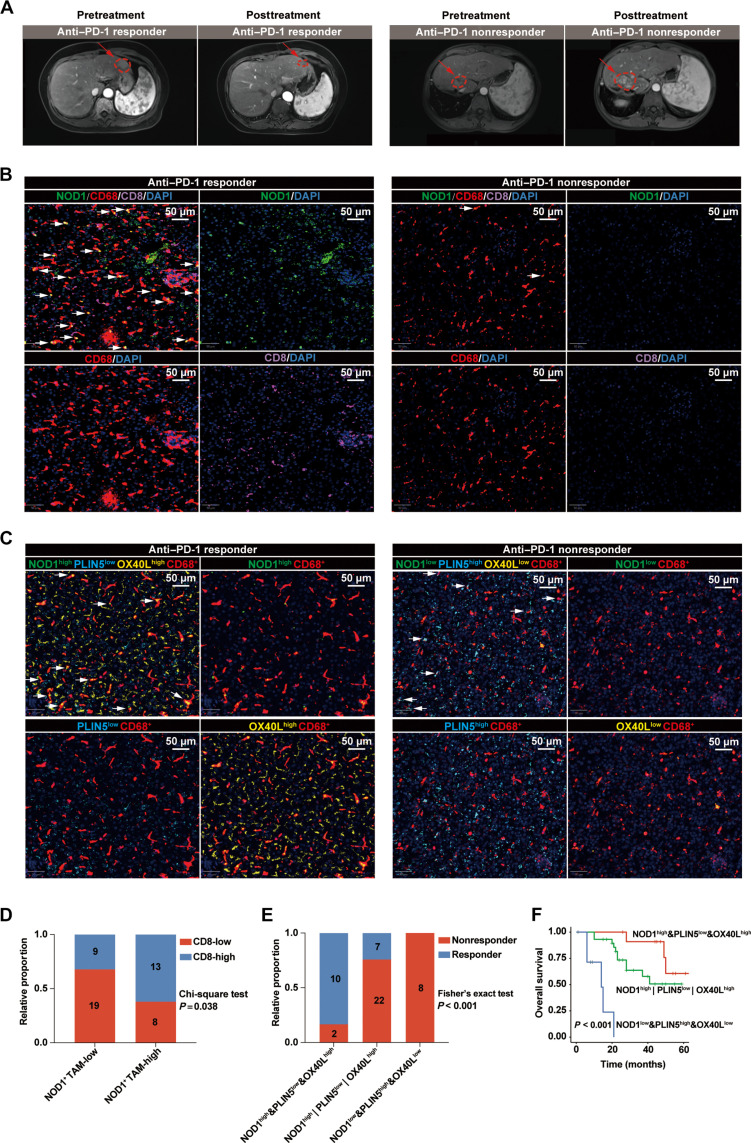
Patients with HCC with high levels of NOD1+ TAMs exhibited favorable responses and improved survival outcomes to anti–PD-1 treatments
To validate our findings clinically, we gathered tumor specimens from a cohort of 49 patients with HCC who underwent anti–PD-1 treatments (Fig. 7A). Our mIF analyses revealed that anti–PD-1–treated patients with HCC with high levels of NOD1+ TAMs exhibited a significant augmentation in the infiltration of CD8+ T cells (Fig. 7, B and D). Furthermore, patients with HCC with TAMs characterized by NOD1high&PLIN5low&OX40Lhigh demonstrated the most favorable responses and survival outcomes to anti–PD-1 treatments (Fig. 7, C, E, and F).
Fig. 7. Activated NOD1/PLIN5/OX40L axis within TAMs is positively correlated with improved clinical outcomes in patients with HCC receiving anti–PD-1 treatments.
(A) Representative magnetic resonance imaging depicting patients with HCC classified as either responders (n = 17) or nonresponders (n = 32) to anti–PD-1 treatment. (B) The representative mIF images of NOD1, CD68, and CD8 markers in tumor tissues from patients with HCC who either responded or did not respond to anti–PD-1 treatment. White arrows indicate NOD1+ TAMs. Scale bars, 50 μm. (C) The representative mIF images of NOD1, PLIN5, OX40L, and CD68 markers in tumor tissues from patients with HCC who either responded or did not respond to anti–PD-1 treatment. White arrows indicate NOD1highPLIN5lowOX40Lhigh TAMs (left) or NOD1lowPLIN5highOX40Llow TAMs (right). Scale bars, 50 μm. (D) The levels of CD8+ T cells among anti–PD-1–treated patients with HCC with low- (n = 28) or high- (n = 21) NOD1+ CD68+ cells. (E) The distributions of treatment responses among anti–PD-1–treated patients with HCC exhibited either NOD1high&PLIN5low&OX40Lhigh (n = 12), NOD1low&PLIN5high&OX40Llow (n = 8), or others (n = 29). (F) Kaplan-Meier analysis comparing the OS of anti–PD-1–treated patients with HCC exhibited either NOD1high&PLIN5low&OX40Lhigh, NOD1low&PLIN5high&OX40Llow, or others. Statistical analysis was performed using the chi-square test in (D), the Fisher’s exact test in (E), and the log-rank test in (F).
DISCUSSION
Anti–PD-1 therapy has benefited only a fraction of patients with HCC. Ongoing efforts are directed toward complementing ICB therapies with interventions designed to repolarize TAMs. In this study, we found that NOD1 activation induces TAMs to acquire an immunostimulatory phenotype capable of supporting CD8+ T cell immunity. Activation of NOD1 serves as a promising strategy to augment the therapeutic effectiveness of anti–PD-1 treatment. Specifically, we demonstrated that NOD1 activation attenuated PLIN5-mediated FAO and resulted in palmitic acid accumulation in TAMs, which promoted membrane localization of palmitoylated OX40L and thereby activating CD8+ T cells.
NOD1, an extensively studied member of the NLR family, exhibits significant functional pleiotropy within the immune cells of the gastrointestinal tract. Chen et al. (50) illustrated NOD1’s capacity to suppress colitis-associated tumorigenesis by augmenting T cell–mediated IFN-γ responses. Conversely, Maisonneuve et al.’s (51) study suggested that NOD1 might foster colorectal carcinogenesis by modulating the immunosuppressive functions of myeloid cells. Nevertheless, the involvement of NOD1 in regulating HCC immunity remains elusive. In this study, our investigation unveils the predominant expression of NOD1 in TAMs within the HCC TME. Although the upstream regulatory mechanisms of NOD1 expression in HCC are not yet clarified, IFN-γ, an important cytokine that promotes the macrophage immunostimulatory phenotype, has been shown to activate NOD1 mRNA transcription via the IFN regulatory factor-1 binding motif (52).
In HCC microenvironment, NOD1 may be stimulated through various ways. First, patients with HCC are more prone to intestinal dysbiosis and increased intestinal permeability, which leads to the translocation of microbes and microbial products, including cell wall components, into the HCC microenvironment (17). Second, increasing evidence suggests that intratumoral microbes form a significant component of the TME. These microbes are intricately involved in tumorigenesis, progression, and sensitivity to therapy in HCC (53, 54). Third, high levels of circulating peptidoglycan can be detected even in healthy individuals (55), which provides a physiological basis for the activation of NOD1 receptors in extraintestinal organs. In our animal experiments, we observed that HCC growth in NOD1△Lyz2 mice was significantly accelerated compared to NOD1flox/flox mice, indicating that NOD1 in the HCC microenvironment can be activated and exert antitumor effects. Furthermore, in vivo activation of NOD1 by C12-iE-DAP delays HCC progression in a macrophage-dependent manner, as demonstrated through experiments using clodronate liposomes and NOD1△Lyz2 mice. In addition, patients with HCC exhibiting high infiltration of NOD1+ TAMs demonstrated augmented responses and improved survival outcomes to anti–PD-1 treatments. Using HCC, B16F10 melanoma, and MC38 colon carcinoma models characterized by distinct immunogenicities and sensitivities to anti–PD-1 therapies, we substantiate that the administration of the NOD1 agonist amplifies sensitivity to ICB treatment. Recent studies underscore the growing interest in oral supplementation of commensal bacteria as a potential strategy to modulate tumor response to ICB therapy (56). LGG emerges as one of the most extensively studied and widely used probiotics, known for its potential therapeutic benefits in the management of colorectal cancer and melanoma through the cyclic GMP-AMP synthase (cGAS)/Stimulator of Interferon Genes (STING) pathway (49, 57). In this study, we illustrated a significant synergistic effect resulting from the combination therapy involving live LGG and anti–PD-1 antibodies in HCC. Notably, this effect was partly reliant on the presence of macrophage-specific NOD1, as evidenced by its absence in NOD1Lyz2 mice. These results suggest a previously unappreciated role for NOD1 in bolstering antitumor T cell immunity in HCC. However, our future research still necessitates the development of a selective in vivo approach to target TAM-specific NOD1, potentially through the utilization of nanomaterials (58, 59).
Mechanistically, we discovered that PLIN5, an LD surface protein, is the downstream effector for NOD1 to induce metabolic and phenotypic shifts in TAMs. PLIN5 is highly expressed in oxidative tissues such as the liver, heart, and skeletal muscle, facilitating the flux of FAs from LDs to the mitochondria for oxidation (32, 33). Recently, Bosch et al. (60) revealed that down-regulation of PLIN5 reduces the tethering of LDs to mitochondria while increasing interactions between LDs and bacteria, thereby enhancing host defenses against microbial pathogens. This indicates that PLIN5 also plays a role in innate immunity regulation. Although increased PLIN5 expression has been observed in multiple cancers, including HCC (61, 62), suggesting its potential role as an oncogene, its function in cancer remains unclear. Our findings revealed that NOD1 activation decreased PLIN5 levels in TAMs, disrupting FAO and contributing to the accumulation of FFA, particularly palmitic acid, thereby enhancing their ability to support CD8+ T cell activation. Consistent with our results, recent studies have shown that inhibiting FAO can promote the transformation of TAMs into an immune-activated phenotype. Our results reveal a previously unrecognized mechanism by which NOD1 endows TAMs with an immunostimulatory phenotype.
It is becoming increasingly clear that fatty acids, besides serving as energy reservoirs, play an essential role in the ability of TAMs to function as potent APCs (63, 64). Previous research has indicated that palmitic acid–mediated protein palmitoylation plays a crucial role in modulating antitumor immunity. Yao et al. elucidated that palmitoylation stabilized the inhibitory ligand PD-L1 on tumor cell surfaces, contributing to immune evasion in cancer (65). In this study, we have shown that the increased palmitic acid in TAMs induced by NOD1 activation promotes the membrane localization of the costimulatory molecule OX40L through palmitoylation, demonstrating that palmitoylation also plays a significant role in the function of membrane costimulatory molecules in TAMs. Although several palmitoyl acyltransferases, Aspartate-Histidine-Histidine-Cysteine (DHHC) proteins, have been shown to play important roles in therapy resistance (66) and tumor progression (67) in HCC, the specific DHHC enzyme responsible for the palmitoylation of OX40L remains to be further elucidated. It has been established that augmenting the interaction between OX40L, primarily expressed on APCs, and OX40, expressed on activated T cells, contributes to CD8+ T cell activation and improved ICB efficacy (68). On the one hand, OX40L signaling enhances the antigen-presenting and proinflammatory capacities of APCs (69–71). On the other hand, OX40 plays a critical role in maintaining cytokine production, survival, expansion, and homeostasis of effector T cells (72). These results, together with our findings, deepen our understanding of the intricate relationship between macrophage FA metabolism and CD8+ T cell–mediated antitumor immunity.
In summary, our study highlights the critical role of NOD1+ TAMs in boosting CD8+ T cell–mediated immunity in HCC. Activation of NOD1 effectively improved the efficacy of anti–PD-1 therapy.
MATERIALS AND METHODS
Clinical tissue samples
Tumor specimens were collected from 49 patients with HCC treated with anti–PD-1 antibodies between February 2018 and September 2022. The response to anti–PD-1 therapy was evaluated according to the Response Evaluation Criteria in Solid Tumors guidelines (73). The final follow-up occurred in December 2023. OS was defined as the duration between the initiation of treatment and the date of death or the last follow-up. RFS was defined as the period from treatment commencement to local recurrence, distant metastasis, or death.
For flow cytometric analysis, we acquired fresh tumor tissues from 10 consecutive patients diagnosed with HCC who underwent curative resection. For HCC tissue microarray construction, we included a total of 125 formalin-fixed, paraffin-embedded HCC tissues. These tissues were collected from consecutive HCC patients who underwent curative resection at our center. Baseline and clinical characteristics of patients were retrospectively retrieved from the electronic medical record. Written informed consent was obtained from all patients, and samples were obtained with the approval of the Institutional Review Board (B2022-164).
Public database analysis
HCC RNA-seq data were obtained from publicly available databases, including TCGA-LIHC and the GEO with accession numbers GSE62044 and GSE14520. The abundance of M1-type macrophages and CTLs in tumor samples from TCGA-LIHC and GSE62044 was quantified using the CIBERSORT algorithm (25, 26) and the MCP-counter (27), respectively. Furthermore, assessment of the immune cell infiltrations in patients with myeloma, pancreatic cancer, cervical cancer, and esophageal cancer were performed using the CIBERSORT algorithm (25, 26) and the TIDE (28).
The gut microbiome metagenomic sequencing data for anti–PD-1 responding and nonresponding patients with HCC were obtained from National Center for Biotechnology Information BioProject (accession number: PRJNA505228) and subjected to analysis (46). Species-level bacterial counts were exported, and relative abundance values were calculated by normalizing raw counts to the total number of reads in the samples. The relative abundance values of iE-DAP–bearing bacteria were compared between samples from anti–PD-1 responders versus nonresponders.
Cell lines
The mouse HCC cell line Hepa 1-6, the mouse melanoma cell line B16F10, and the mouse colon cancer cell line MC38 were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Invitrogen).
Animals
Animal experiments were conducted according to the Shanghai Medical Experimental Animal Care Commission Guidelines and approved by the Ethical Committee on Animal Experiments of the Animal Care Committee of Zhongshan Hospital, Fudan University (B2022-554R). C57BL/6 and BALB/c nude mice were obtained from Shanghai Laboratory Animal Center (SLAC) Laboratory Animal Co. Ltd., Shanghai, China. OT-I (T cell receptor transgenic on a B6 background) (#C001198), Nod1flox/flox (#S-CKO-00588), and Lyz2-Cre (#C001003) mice with a C57BL/6 genetic background were obtained from Cyagen Biosciences. SCID mice (#SM-015) were purchased from Shanghai Model Organisms Center. All mice were housed in a specific pathogen–free barrier facility at Fudan University. Male mice aged 6 to 8 weeks were used for all experiments.
Subcutaneous and orthotopic murine tumor models
For the subcutaneous model, 100 μl of a single-cell suspension of 5 × 106 Hepa 1-6 cells, 5 × 106 B16F10 cells, or 1 × 106 MC38 cells in 1× phosphate-buffered saline (PBS) were inoculated into the right flank of each mouse. Tumor volumes were measured every 2 days using the formula length × width2 × 0.5 (mm3). Tumor weight was assessed at the study endpoint, and tissue samples were collected for analysis.
For the orthotopic HCC model, mice were anesthetized before orthotopic tumor cell implantation. An 8-mm transverse skin incision was made in the upper abdomen, and 25 μl of a single-cell suspension of Hepa 1-6 cells (2 × 106 per mouse), prepared in Matrigel (Corning, #356234) and 1× PBS at a 1:1 ratio, was injected into the left hepatic lobe of the recipient C57BL/6 mouse using a microsyringe (74). The total radiant flux of the region of interest, which represented tumor size, was measured using an In Vivo Xtreme device (Bruker). Upon sacrifice, liver tissue was collected for flow cytometry and histological evaluation.
In vivo treatments
Mice were treated intraperitoneally with C12-iE-DAP (10 or 25 μg per injection, every other day, InvivoGen, #tlrl-c12dap) (16, 75), ML130 (50 μg per injection, every other day, MedChemExpress, #HY-18639) (76), and anti–PD-1 antibody (200 μg per injection, every third day, Bio X Cell, #BE0146) (77) after tumor cell implantation. To deplete macrophages, mice were injected with clodronate liposomes (100 μl per injection, Yeasen, #40337ES10) (77) beginning 3 days before tumor cell implantation and continuing every fourth day until the endpoint. For CD8+ T cell depletion, mice were treated intraperitoneally with anti-mouse CD8α antibodies (250 μg per injection, Bio X Cell, #BE0117) (77) starting the same day as tumor cell implantation and administered every third day until the endpoint. To blockade OX40L, mice were injected with anti-mouse OX40L antibodies (250 μg per injection, Bio X Cell, #BE0033-1) beginning 2 days before tumor cell implantation and continuing every third day until the endpoint. To inhibit iE-DAP–bearing Lactobacillus in the murine intestine, neomycin (Cayman Chemical Company, #1405-10-3) was provided in the drinking water (1 mg/ml) 1 week before tumor cell implantation, and the water bottle was replaced twice per week (47, 48). For LGG oral administration, selected mice were gavaged with 2 × 109 CFU LGG every third day (ATCC, 53103).
Immunohistochemistry and immunofluorescence
Immunohistochemistry (IHC) and immunofluorescence (IF) analyses were performed on paraffin-embedded sections of HCC tissues. The slides were deparaffinized in xylene and rehydrated through a series of graded alcohols. Antigen retrieval was performed by treating the slides with citrate buffer (pH 6.0). To block nonspecific binding, the slides were incubated with 5% bovine serum albumin (BSA) and stained with primary antibodies against F4/80 (Cell Signaling Technology, #70076 T), Ki67 (Abcam, #ab16667), and CD8 (Abcam, #ab217344) overnight at 4°C. For IHC, horseradish peroxidase (HRP)–conjugated secondary antibodies (Vector Laboratories) were used, and the slides were then incubated with diaminobenzidine substrate (Vector Laboratories) and counterstained with hematoxylin. For IF, sections were incubated with secondary antibodies conjugated with Alexa Fluor 488 and 594 (1:500 dilution, Invitrogen) for 1 hour at room temperature. The slides were mounted with a 4′,6-diamidino-2-phenylindole (DAPI)–containing mounting medium and imaged under a fluorescence microscope (Olympus). The cell apoptosis was detected using the In Situ Cell Death Detection Kit (Roche, #12156792910). The quantification of the positive area was performed using ImageJ software. Statistical analysis was conducted using GraphPad Prism software.
Preparation of single-cell suspensions
Before flow cytometric analysis and magnetic bead cell sorting, single-cell suspensions were prepared. Tumor tissues were dissected and finely minced on a programmable gentleMACS dissociator (Miltenyi Biotech) and digested with an enzyme solution containing collagenase IV (0.2 mg/ml) and deoxyribonuclease I (10 mg/ml; Sigma-Aldrich). The single-cell suspension was obtained by passing through a 70-mm cell mesh and resuspending in Hanks’ buffer. Mouse spleens were pressed through a 40-mm cell mesh using a syringe plunger to obtain a single-cell suspension. The red blood cells in cell preparations and whole blood were removed using Ammonium–chloride–potassium (ACK) Lysing Buffer (Gibco).
Flow cytometric analysis
To reduce nonspecific binding, cells were first incubated with the Fc receptor block for 10 min at room temperature. Subsequently, cells were stained with fluorescence-labeled monoclonal antibodies that specifically recognize desired surface markers for 30 min at 4°C. After surface staining, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS to enable intracellular staining. The cells were then incubated with fluorochrome-conjugated antibodies specific to the desired intracellular markers for 30 min at 4°C in the dark. The antibodies used for surface and intracellular staining are listed in table S1. Labeled cells were washed twice with fluorescence-activated cell sorting (FACS) buffer and analyzed using a BD FACSCantoII Analyzer (BD Biosciences) and FlowJo software (version 10.4).
mIF assay
The mIF assay was performed using the OPAL 780 REAGENT PACK (Akoya Biosciences, FP1501001KT) following the manufacturer’s protocol. Briefly, slides containing tissue sections were deparaffinized in xylene and rehydrated in graded ethanol solutions. Endogenous peroxidases were blocked with 0.3% hydrogen peroxide, and antigen retrieval was performed using citrate buffer. The slides were then blocked with 5% BSA to prevent nonspecific binding. Primary antibodies against F4/80 (Cell Signaling Technology, #70076T), CD68 (Abcam, #ab955), MHC-II (Abcam, #ab23990), CD11c (Abcam, #ab52632), CD3 (Abcam, #ab16669), CD8 (Abcam, #ab217344 for mouse and #ab237709 for human), NOD1 (Abcam, #ab217798), PLIN5 (Proteintech, #26951-1-AP), and OX40L (Invitrogen, #PA5-116057) were added to the slides and incubated for 60 min at 37°C in a humidified chamber. After washing with buffer, corresponding HRP-conjugated secondary antibodies were applied and incubated for 45 min at room temperature. The slides were rewashed to remove unbound antibodies before proceeding to the next step. Last, the slides were stained with DAPI solution at 37°C for 10 min in the dark to visualize nuclei. Images were acquired using the QuPath software.
Single-cell gene expression processing and analysis
Sequencing results were processed by Fastp (78) to trim primer sequence and low-quality bases, and then the processed sequence data were aligned to the Ensembl genome GRCm38 reference genome using SeekOne Tools to obtain a gene expression matrix. Raw count matrices were then merged and analyzed using the Seurat R package (v4.0.1) (79). Low-quality cells with <200 or >5000 genes detected and cells with mitochondrially encoded transcripts representing >15% were excluded from the analysis. Counts for the remaining cells were normalized against library size and regressed to correct for the unwanted cycling bias among proliferating cells, using S and G2M phase scores calculated by the CellCycleScoring function in the Seurat package. Scaled and centered read counts were used as gene expression for further analysis. To visualize inferred cell clusters, t-Distributed stochastic neighbor embedding was applied on the basis of the top 50 principal components. Automatic immune cell annotation was performed using the SingleR package. We conducted manual inspections to calibrate the automatic cell annotations by examining the most highly expressed marker genes between clusters and literature- and database-derived cell markers. The proinflammatory score was calculated in the C1qhi TAM population, and the cytotoxic score was calculated in the CD8+ T cell population at the single-cell level to assess immune cell phenotype shift. The genes for each score were defined in previous studies (80). The differentially expressed genes between CD8+ T cells from vehicle or C12-iE-DAP–treated HCC were determined by using the FindMarkers function of the Seurat pipeline.
To explore the immune microenvironment in human HCC, we reanalyzed the scRNA-seq data from Zhang et al. (81) (GSE140228). The cell-cell communication network was identified by the CellChat package (82). The scores for phagocytosis, proinflammatory, and regulation of T cell activation were calculated to determine the phenotype of TAMs (80). For GSEA functional analysis, only NOD1+ TAMs (with the expression of NOD1 larger than 0) were kept to deal with missing data points in scRNA-seq. NOD1+ TAMs were stratified into NOD1-low and NOD1-high TAMs based on the median value of NOD1 expression. All statistical analyses were conducted using R software (v4.2.2).
Generation of primary mouse macrophages
C57BL/6 mice were euthanized, and their femurs and tibias were flushed with DMEM medium (Gibco) to collect bone marrow cells using a syringe. After red blood cell lysis, bone marrow cells were cultured with DMEM (Gibco) containing 10% FBS (Gibco) and mouse macrophage colony-stimulating factor (20 ng/ml; Absin) for 5 days to obtain BMDMs. The BMDMs were then exposed to a TCM derived from hepatoma cells for 24 hours to promote their polarization toward a TAM phenotype.
Small interfering RNA and overexpression plasmid transfection
To knockdown PLIN5 expression, DNA oligos targeting PLIN5 were synthesized with the following sequences: siPLIN5-1 (GGGACTAGACAAATTGGAA) and siPLIN5-2 (GCTCTGTCTCGAAGTTTGA). For PLIN5 overexpression, the overexpression plasmid was constructed by cloning PLIN5 cDNA into the CMV-MCS-3XFlag-PGK-Puro vector. For OX40L overexpression, the overexpression plasmid was constructed by cloning OX40L cDNA into the PGMLV-CMV-3XFlag-PGK-Puro vector. Empty vectors were used as negative controls. BMDMs were transfected with either the small interfering RNAs or overexpression plasmid using Lipo8000TM Transfection Reagent following the manufacturer’s instructions (Beyotime, #C0533). Subsequent experiments were conducted on macrophages 48 hours after transfection.
RNA isolation and quantitative real-time PCR
To isolate total RNA, cells were subjected to the RNAeasy Animal RNA Isolation Kit with Spin Column (Beyotime, #R0027), and then the RNA was reverse-transcribed into cDNA using the 4 EZscript Reverse Transcription Mix II (EZBioscience, #EZB-RT2). Quantitative PCR (qPCR) analysis was performed on the resulting single-stranded cDNA using the 2 SYBR Green qPCR Master Mix (ROX2 plus) (EZBioscience, A0001-R2). β-Actin was used as the internal control, and each reaction was performed in triplicate to ensure accuracy. The primer sequences used in this study are listed in table S2.
Immunoblotting
To extract proteins, samples were treated with radioimmunoprecipitation assay lysis buffer and phenylmethanesulfonyl fluoride (Beyotime) and then centrifuged at 14,000 rpm for 20 min at 4°C. Protein quantification was performed using a Bicinchoninic Acid (BCA) protein assay (Beyotime). Subsequently, the proteins were mixed with 1× loading buffer and boiled for 8 to 10 min. Separation of proteins was carried out by 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and then electroblotted onto polyvinylidene difluoride (PVDF) membranes (0.2 μm, Millipore). The membranes were then blocked with Protein-Free Rapid Blocking Buffer (EpiZyme Biotechnology) for 15 min and incubated with primary antibodies overnight at 4°C, followed by incubation with secondary antibodies for 1 hour at room temperature. The primary antibodies used in this experiment were anti–β-actin antibody (Beyotime, #AF2811), anti-CD11b antibody (ABclonal Technology, #A1581), anti-PLIN5 antibody (Proteintech, #26951-1-AP), anti-OX40L antibody (Invitrogen, #PA5-116057), anti-PPARA antibody (Proteintech, #66826-1-Ig), anti-p38 MAPK antibody (Cell Signaling Technology, #8690), anti–phospho-p38 MAPK antibody (Cell Signaling Technology, #4511), anti–phospho-p38 MAPK antibody, anti-RIPK2 antibody (Affinity Biosciences, #DF6967), and anti–phospho-RIPK2 antibody (Affinity Biosciences, #AF0049). Protein bands were visualized using Ncm-ECL Ultra (New Cell and Molecular Biotech Co. Ltd.).
Coculture assays
CD8+ T cells were isolated from the spleens of wild-type C57BL/6 mice using the CD8 T Cell Isolation Kit (BioLegend, #480035) after red blood cell lysis. For the proliferation assay, purified CD8+ T cells were labeled with 2 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen) for 10 min at room temperature in the dark. The CD8+ T cells were then counted and plated into a flat-bottom 96-well culture plate that had been precoated with anti-CD3 (2 μg/ml; BioLegend, #100340) plus anti-CD28 (2 μg/ml; BioLegend, #102116) antibodies in complete RPMI 1640 medium supplemented with 10% FBS, interleukin-2 (100 U/ml; PeproTech, #212-12), 2 mM l-glutamine (STEMCELL Technologies, #07100), 50 μM β-mercaptoethanol (GENOM Biotech, #GNM21985-1), 1 mM sodium pyruvate (Beyotime, C0331), 100 μM MEM non-essential amino acid (STEMCELL Technologies, #07600), and 10 mM Hepes (STEMCELL Technologies, #07200). The CD8+ T cells were activated for 24 hours before coculture with macrophages.
Macrophages were seeded with activated CD8+ T cells at a ratio of 1:1. After coculture for another 36 hours, staining for IFN-γ and GZMB was performed to analyze T cell activation status, and CFSE dilution was analyzed to assess T cell proliferation.
Antigen presentation and T cell activation assay
On days 0 and 7, OT-I mice were intraperitoneally immunized with 100 μg of ovalbumin (OVA; Sigma-Aldrich, #A5503) that was adsorbed to 2 mg of an aqueous solution of alum (Thermo Fisher Scientific, #WK333962). On day 14, CD8+ T cells were isolated from the spleens of the immunized mice. Macrophages were plated in flat-bottom 96-well plates and then pulsed with 10 nM specific peptide SIINFEKL (OVA257-264) (Sigma-Aldrich, #S7951) for 4 hours. After being washed twice, the macrophages were cocultured with CD8+ T cells from the OVA-sensitized OT-I mice at a 1:1 ratio for 72 hours. The activation status of the CD8+ T cells was then assessed using flow cytometric analysis.
Metabolomic analysis
Metabolites were extracted from cell residues using a mixture of methanol, acetonitrile, and water. Quality control (QC) samples were prepared by pooling aliquots of all samples for data normalization. Metabolites were separated and detected using a Shimadzu Nexera X2 LC-30 AD system equipped with an Acquity UPLC HSS T3 column and a triple quadruple mass spectrometer. Positive and negative ionization modes were used to detect the metabolites. During data acquisition, QC samples were regularly injected for quality assurance. MultiQuant 3.0.2 software was used to extract the original multiple reaction monitoring data of metabolites and obtain the peak area of each metabolite. Discriminating metabolites were identified using the Mann-Whitney U test. Metabolites with a P value less than 0.05 were considered statistically significant. The identified differential metabolites were then used to perform cluster analyses using the R package.
Measurement of fatty acid β-oxidation rate using colorimetric assay
Mitochondria were isolated from 2 × 107 TCM-educated BMDMs using a Cell Mitochondria Isolation Kit (Beyotime, #C3601) following the manufacturer’s protocol and resuspended in ice-cold PBS. The fatty acid β-oxidation rate in mitochondrial suspensions was assessed using a commercially available colorimetric assay kit according to the manufacturer’s instructions (Shanghai Haling Biotechnology Co. Ltd., #1-119974-26). The absorbance values of the reaction mixture were measured at 0, 1, 2, 3, 4, and 5 min using a Multiskan Spectrum spectrophotometer (Thermo Fisher Scientific) at wavelengths of 420 and 470 nm. The fatty acid β-oxidation rate was calculated as micromoles of ferricyanide reduced per minute per milligram of protein.
Quantification of FFAs using colorimetric assay
The levels of FFAs in macrophages were quantified using a commercially available colorimetric assay kit following the manufacturer’s protocol (Mlbio, #ml092765). FAs were extracted from 1 × 107 TCM-educated BMDMs using an organic solvent extraction method in conjunction with ultrasonic cell disruption and agitation. The organic phase containing the extracted FFAs was then subjected to a reaction with cupric ions in the presence of triethanolamine to form fatty acid copper (copper soap) complexes. The copper soap complexes produced a purple-red colored solution, and the absorbance of the colored complex was measured at a wavelength of 550 nm using a Multiskan Spectrum spectrophotometer (Thermo Fisher Scientific). The FFA concentration in the sample was determined by comparing its absorbance to a standard curve generated from known concentrations of FFA standards.
Immunoprecipitation and ABE assay
To assess the protein palmitoylation of OX40L, we used the immunoprecipitation and acyl-biotin exchange (IP-ABE) assay (83). Total protein was extracted from cell lysates using a lysis buffer containing 1% Triton X-100, 150 mM NaCl, 50 mM tris-HCl (pH 7.4), 5 mM EDTA, and a protease inhibitor cocktail. After blocking free cysteines with N-ethylmaleimide (Aladdin Biochemistry Science and Technology, #E100553), OX40L-flag protein was precipitated using an anti-flag antibody (ABclonal Technology, #AE092) and protein A-Sepharose beads, and the samples were then resuspended with a stringent buffer. To cleave thioester bonds between palmitate and cysteine residues, lysis buffer (0.5 ml per sample) pH 7.2 was added to all −hydroxylamine (HAM) samples, and HAM buffer (0.5 ml per sample; Macklin Biochemical Co. Ltd., #H828371) was added to all +HAM samples. The samples were rotated at room temperature for 1 hour. Then, the samples were added with Biotin-BMCC buffer (Sangon Biotech, #C100222) and rotated for 1 hour at 4°C to label newly exposed cysteines with biotin. Labeled proteins were eluted from the beads by boiling them in SDS sample buffer, separated on SDS-PAGE gels, and electro-transferred to PVDF membranes. To detect palmitoylated OX40L, membranes were incubated with HRP-conjugated anti-streptavidin overnight at 4°C. To detect total OX40L, membranes were incubated with an anti-flag antibody and subsequent HRP-conjugated secondary antibody.
Statistical analysis
Statistical analyses were conducted using R software (v4.2.2) or GraphPad Prism (version 9.1.1; San Diego, CA, USA). Before the parametric or nonparametric tests, the D’Agostino-Pearson omnibus normality test and Brown-Forsythe test were performed. Group comparisons were evaluated using an unpaired Student’s t test when the normality and equal variance assumptions were met. Otherwise, a nonparametric test (Mann-Whitney U test) was applied. Paired Student’s t tests were used to compare paired data. Differences in constituent ratios between groups were assessed using either the chi-square test or Fisher’s exact test depending on the sample size and expected cell frequencies. Survival curves were plotted using the Kaplan-Meier method, and the log-rank test was used to assess the differences between groups. Independent prognostic factors were identified using the Cox proportional hazards regression model. Statistical significance was set at P value < 0.05. The results are reported as not significant (ns) or with asterisks indicating the level of significance: *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
Funding: This work was supported by The National Natural Science Foundation of China grant 82270135 (S.Z.), The National Natural Science Foundation of China grant 82273027 (L.D.), The National Natural Science Foundation of China grant 81972234 (L.D.), The National Natural Science Foundation of China grant 82103096 (Y.Z.), The Natural Science Foundation of Shanghai grant 21ZR1408900 (S.Z.), The Outstanding Youth Fund of Natural Science Foundation of Anhui Province grant 2308085J27 (S.Z.), The Science and Technology Innovation Action Plan of Shanghai Science and Technology Committee grant 22140901600 (R.X.), and The Shanghai Science and Technology Innovation Action Plan grant 23XD1422700 (R.X.).
Author contributions: Conceptualization: F.Z., S.Z., Q.J., L.D., R.X., J.C., and X.S.; methodology: F.Z., F.M., Z.L., X.L., and W.L.; resources: F.Z., S.Z., Q.J., L.D., W.T., and Y.Z.; software: F.Z.; investigation: F.Z., S.Z., Q.J., J.C., F.M., W.T., Z.L., and W.L.; project administration: F.Z., S.Z., and J.C.; visualization: F.Z., S.Z., Q.J., L.D., F.M., W.T., X.L., and Y.Z.; formal analysis: F.Z., Q.J., and J.C.; supervision: F.Z., S.Z., L.D., R.X., and X.S.; data curation: F.Z., S.Z., and Q.J.; validation: F.Z., S.Z., Q.J., and L.D.; writing—original draft: F.Z., S.Z., Q.J., L.D., and J.C.; writing—review and editing: F.Z., S.Z., and L.D.; funding acquisition: S.Z., L.D., R.X., and Y.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Tables S1 and S2
REFERENCES AND NOTES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Llovet J. M., Kelley R. K., Villanueva A., Singal A. G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R. S., Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 7, 6 (2021). [DOI] [PubMed] [Google Scholar]
- 3.El-Khoueiry A. B., Sangro B., Yau T., Crocenzi T. S., Kudo M., Hsu C., Kim T. Y., Choo S. P., Trojan J., Welling T. H. R., Meyer T., Kang Y. K., Yeo W., Chopra A., Anderson J., Dela Cruz C., Lang L., Neely J., Tang H., Dastani H. B., Melero I., Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yau T., Park J. W., Finn R. S., Cheng A. L., Mathurin P., Edeline J., Kudo M., Harding J. J., Merle P., Rosmorduc O., Wyrwicz L., Schott E., Choo S. P., Kelley R. K., Sieghart W., Assenat E., Zaucha R., Furuse J., Abou-Alfa G. K., El-Khoueiry A. B., Melero I., Begic D., Chen G., Neely J., Wisniewski T., Tschaika M., Sangro B., Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): A randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 23, 77–90 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Robinson M. W., Harmon C., O'Farrelly C., Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 13, 267–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H., Peng X., Yang S., Li X., Huang M., Wei S., Zhang S., He G., Liu J., Fan Q., Yang L., Li H., Targeting tumor-associated macrophages in hepatocellular carcinoma: Biology, strategy, and immunotherapy. Cell Death Discov. 9, 65 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lesokhin A. M., Hohl T. M., Kitano S., Cortez C., Hirschhorn-Cymerman D., Avogadri F., Rizzuto G. A., Lazarus J. J., Pamer E. G., Houghton A. N., Merghoub T., Wolchok J. D., Monocytic CCR2+ myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 72, 876–886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannarile M. A., Weisser M., Jacob W., Jegg A. M., Ries C. H., Rüttinger D., Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 5, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y. C., Wang X., Yu J., Ma F., Li Z., Zhou Y., Zeng S., Ma X., Li Y. R., Neal A., Huang J., A. To, Clarke N., Memarzadeh S., Pellegrini M., Yang L., Targeting monoamine oxidase A-regulated tumor-associated macrophage polarization for cancer immunotherapy. Nat. Commun. 12, 3530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molgora M., Esaulova E., Vermi W., Hou J., Chen Y., Lou J., Brioschi S., Bugatti M., Omodei A. S., Ricci B., Fronick C., Panda S. K., Takeuchi Y., Gubin M. M., Faccio R., Cella M., Gilfillan S., Unanue E. R., Artyomov M. N., Schreiber R. D., Colonna M., TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell 182, 886–900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barkal A. A., Brewer R. E., Markovic M., Kowarsky M., Barkal S. A., Zaro B. W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L. J., Weissman I. L., CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logtenberg M. E. W., Scheeren F. A., Schumacher T. N., The CD47-SIRPα immune checkpoint. Immunity 52, 742–752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G., Shaw M. H., Kim Y. G., Nunez G., NOD-like receptors: Role in innate immunity and inflammatory disease. Annu. Rev. Pathol. 4, 365–398 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Li D., Wu M., Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 6, 291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa M., Yang K., Hashimoto M., Park J. H., Kim Y. G., Fujimoto Y., Nunez G., Fukase K., Inohara N., Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J. Biol. Chem. 281, 29054–29063 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Griffin M. E., Espinosa J., Becker J. L., Luo J. D., Carroll T. S., Jha J. K., Fanger G. R., Hang H. C., Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373, 1040–1046 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi A., Debelius J., Brenner D. A., Karin M., Loomba R., Schnabl B., Knight R., The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu B., Ding G. Y., Fu P. Y., Zhu X. D., Ji Y., Shi G. M., Shen Y. H., Cai J. B., Yang Z., Zhou J., Fan J., Sun H. C., Kuang M., Huang C., NOD-like receptor X1 functions as a tumor suppressor by inhibiting epithelial-mesenchymal transition and inducing aging in hepatocellular carcinoma cells. J. Hematol. Oncol. 11, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udden S. N., Kwak Y. T., Godfrey V., Khan M. A. W., Khan S., Loof N., Peng L., Zhu H., Zaki H., NLRP12 suppresses hepatocellular carcinoma via downregulation of cJun N-terminal kinase activation in the hepatocyte. eLife 8, e40396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng Y. Y., He Y. H., Chen C., Xu T., Li L., Ni M. M., Meng X. M., Huang C., Li J., NLRC5 regulates cell proliferation, migration and invasion in hepatocellular carcinoma by targeting the Wnt/β-catenin signaling pathway. Cancer Lett. 376, 10–21 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Hu L., Tang W., Li D., Ma L., Liu H., Zhang S., Zhang X., Dong L., Shen X., Chen S., Xue R., Zhang S., Hepatic NOD2 promotes hepatocarcinogenesis via a RIP2-mediated proinflammatory response and a novel nuclear autophagy-mediated DNA damage mechanism. J. Hematol. Oncol. 14, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K., Smyth M. J., Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell. Mol. Immunol. 17, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J., Fan W., Liu T., Zhu B., Liu Y., Wang S., Wu J., Liu J., Zou F., Wei J., Liu L., Zhang X., Zhuang J., Wang Y., Lin H., Huang X., Chen S., Kuang M., Li J., TREM2+ macrophages suppress CD8+ T-cell infiltration after transarterial chemoembolisation in hepatocellular carcinoma. J. Hepatol. 79, 126–140 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Xun Z., Ma K., Liang S., Li X., Zhou S., Sun L., Liu Y., Du Y., Guo X., Cui T., Zhou H., Wang J., Yin D., Song R., Zhang S., Cai W., Meng F., Guo H., Zhang B., Yang D., Bao R., Hu Q., Wang J., Ye Y., Liu L., Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol. 78, 770–782 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Newman A. M., Liu C. L., Green M. R., Gentles A. J., Feng W., Xu Y., Hoang C. D., Diehn M., Alizadeh A. A., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X. S., TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becht E., Giraldo N. A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W. H., de Reyniès A., Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G. J., Brown M. A., Wucherpfennig K. W., Liu X. S., Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Xiao Y., Li Q., Yao J., Yuan X., Zhang Y., Yin X., Saito Y., Fan H., Li P., Kuo W. L., Halpin A., Gibbons D. L., Yagita H., Zhao Z., Pang D., Ren G., Yee C., Lee J. J., Yu D., The allergy mediator histamine confers resistance to immunotherapy in cancer patients via activation of the macrophage histamine receptor H1. Cancer Cell 40, 36–52.e39 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Che L. H., Liu J. W., Huo J. P., Luo R., Xu R. M., He C., Li Y. Q., Zhou A. J., Huang P., Chen Y. Y., Ni W., Zhou Y. X., Liu Y. Y., Li H. Y., Zhou R., Mo H., Li J. M., A single-cell atlas of liver metastases of colorectal cancer reveals reprogramming of the tumor microenvironment in response to preoperative chemotherapy. Cell Discov. 7, 80 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su P., Wang Q., Bi E., Ma X., Liu L., Yang M., Qian J., Yi Q., Enhanced lipid accumulation and metabolism are required for the differentiation and activation of tumor-associated macrophages. Cancer Res. 80, 1438–1450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason R. R., Watt M. J., Unraveling the roles of PLIN5: Linking cell biology to physiology. Trends Endocrinol. Metab. 26, 144–152 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Itabe H., Yamaguchi T., Nimura S., Sasabe N., Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 16, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bougarne N., Weyers B., Desmet S. J., Deckers J., Ray D. W., Staels B., De Bosscher K., Molecular actions of PPARα in lipid metabolism and inflammation. Endocr. Rev. 39, 760–802 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Mass Sanchez P. B., Krizanac M., Weiskirchen R., Asimakopoulos A., Understanding the role of perilipin 5 in non-alcoholic fatty liver disease and its role in Hepatocellular Carcinoma: A review of novel insights. Int. J. Mol. Sci. 22, 5284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I., LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta 1771, 210–227 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., Finck B. N., Bickel P. E., OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes 55, 3418–3428 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Kimmel A. R., Sztalryd C., Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr. Opin. Lipidol. 25, 110–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caruso R., Warner N., Inohara N., Nunez G., NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 41, 898–908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Huang Q., Long X., Zhang J., Huang X., Aa J., Yang H., Chen Z., Xing J., CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARα pathways. J. Hepatol. 63, 1378–1389 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Rosa Neto J. C., Calder P. C., Curi R., Newsholme P., Sethi J. K., Silveira L. S., The immunometabolic roles of various fatty acids in macrophages and lymphocytes. Int. J. Mol. Sci. 22, 8460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linder M. E., Deschenes R. J., Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Guerriero J. L., Macrophages: Their untold story in T cell activation and function. Int. Rev. Cell Mol. Biol. 342, 73–93 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Momin N., Mehta N. K., Bennett N. R., Ma L., Palmeri J. R., Chinn M. M., Lutz E. A., Kang B., Irvine D. J., Spranger S., Wittrup K. D., Anchoring of intratumorally administered cytokines to collagen safely potentiates systemic cancer immunotherapy. Sci. Transl. Med. 11, eaaw2614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicks K. C., Chariou P. L., Ozawa Y., Minnar C. M., Knudson K. M., Meyer T. J., Bian J., Cam M., Schlom J., Gameiro S. R., Tumour-targeted interleukin-12 and entinostat combination therapy improves cancer survival by reprogramming the tumour immune cell landscape. Nat. Commun. 12, 5151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Y., Wang T., Tu X., Huang Y., Zhang H., Tan D., Jiang W., Cai S., Zhao P., Song R., Li P., Qin N., Fang W., Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 7, 193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiao J., Liu J., Li Q., Zhang G., Pan C., Luo F., Zhang Q., Qi B., Zhao L., Yin P., Shang D., Gut microbiota-derived diaminopimelic acid promotes the NOD1/RIP2 signaling pathway and plays a key role in the rrogression of severe acute pancreatitis. Front. Cell. Infect. Microbiol. 12, 838340 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hergott C. B., Roche A. M., Tamashiro E., Clarke T. B., Bailey A. G., Laughlin A., Bushman F. D., Weiser J. N., Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si W., Liang H., Bugno J., Xu Q., Ding X., Yang K., Fu Y., Weichselbaum R. R., Zhao X., Wang L., Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut 71, 521–533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhan Y., Seregin S. S., Chen J., Chen G. Y., Nod1 limits colitis-associated tumorigenesis by regulating IFN-γ production. J. Immunol. 196, 5121–5129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maisonneuve C., Tsang D. K. L., Foerster E. G., Robert L. M., Mukherjee T., Prescott D., Tattoli I., Lemire P., Winer D. A., Winer S., Streutker C. J., Geddes K., Cadwell K., Ferrero R. L., Martin A., Girardin S. E., Philpott D. J., Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. Cell Rep. 34, 108677 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Hisamatsu T., Suzuki M., Podolsky D. K., Interferon-gamma augments CARD4/NOD1 gene and protein expression through interferon regulatory factor-1 in intestinal epithelial cells. J. Biol. Chem. 278, 32962–32968 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Li S., Xia H., Wang Z., Zhang X., Song T., Li J., Xu L., Zhang N., Fan S., Li Q., Zhang Q., Ye Y., Lv J., Yue X., Lv H., Yu J., Lu W., Intratumoral microbial heterogeneity affected tumor immune microenvironment and determined clinical outcome of HBV-related HCC. Hepatology 78, 1079–1091 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji J., Ji F., Bayarsaikhan E., Intratumoral microbiota in HCC: A new kid on the block? Hepatology 78, 1012–1014 (2023). [DOI] [PubMed] [Google Scholar]
- 55.Huang Z., Wang J., Xu X., Wang H., Qiao Y., Chu W. C., Xu S., Chai L., Cottier F., Pavelka N., Oosting M., Joosten L. A. B., Netea M., Ng C. Y. L., Leong K. P., Kundu P., Lam K. P., Pettersson S., Wang Y., Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 4, 766–773 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V., Spencer C. N., Nezi L., Reuben A., Andrews M. C., Karpinets T. V., Prieto P. A., Vicente D., Hoffman K., Wei S. C., Cogdill A. P., Zhao L., Hudgens C. W., Hutchinson D. S., Manzo T., Petaccia de Macedo M., Cotechini T., Kumar T., Chen W. S., Reddy S. M., Sloane R. S., Galloway-Pena J., Jiang H., Chen P. L., Shpall E. J., Rezvani K., Alousi A. M., Chemaly R. F., Shelburne S., Vence L. M., Okhuysen P. C., Jensen V. B., Swennes A. G., McAllister F., Marcelo Riquelme Sanchez E., Zhang Y., Le Chatelier E., Zitvogel L., Pons N., Austin-Breneman J. L., Haydu L. E., Burton E. M., Gardner J. M., Sirmans E., Hu J., Lazar A. J., Tsujikawa T., Diab A., Tawbi H., Glitza I. C., Hwu W. J., Patel S. P., Woodman S. E., Amaria R. N., Davies M. A., Gershenwald J. E., Hwu P., Lee J. E., Zhang J., Coussens L. M., Cooper Z. A., Futreal P. A., Daniel C. R., Ajami N. J., Petrosino J. F., Tetzlaff M. T., Sharma P., Allison J. P., Jenq R. R., Wargo J. A., Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Sung C. Y., Lee N., Ni Y., Pihlajamäki J., Panagiotou G., El-Nezami H., Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E1306–E1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ovais M., Guo M., Chen C., Tailoring nanomaterials for targeting tumor-associated macrophages. Adv. Mater. 31, e1808303 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Zhao C., Pang X., Yang Z., Wang S., Deng H., Chen X., Nanomaterials targeting tumor associated macrophages for cancer immunotherapy. J. Control. Release 341, 272–284 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Bosch M., Sanchez-Alvarez M., Fajardo A., Kapetanovic R., Steiner B., Dutra F., Moreira L., Lopez J. A., Campo R., Mari M., Morales-Paytuvi F., Tort O., Gubern A., Templin R. M., Curson J. E. B., Martel N., Catala C., Lozano F., Tebar F., Enrich C., Vazquez J., Del Pozo M. A., Sweet M. J., Bozza P. T., Gross S. P., Parton R. G., Pol A., Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Hashani M., Witzel H. R., Pawella L. M., Lehmann-Koch J., Schumacher J., Mechtersheimer G., Schnölzer M., Schirmacher P., Roth W., Straub B. K., Widespread expression of perilipin 5 in normal human tissues and in diseases is restricted to distinct lipid droplet subpopulations. Cell Tissue Res. 374, 121–136 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Asimakopoulou A., Vucur M., Luedde T., Schneiders S., Kalampoka S., Weiss T. S., Weiskirchen R., Perilipin 5 and lipocalin 2 expression in hepatocellular carcinoma. Cancers 11, 385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christofides A., Strauss L., Yeo A., Cao C., Charest A., Boussiotis V. A., The complex role of tumor-infiltrating macrophages. Nat. Immunol. 23, 1148–1156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan J., Horng T., Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 30, 979–989 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Yao H., Lan J., Li C., Shi H., Brosseau J. P., Wang H., Lu H., Fang C., Zhang Y., Liang L., Zhou X., Wang C., Xue Y., Cui Y., Xu J., Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 3, 306–317 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Sun Y., Zhang H., Meng J., Guo F., Ren D., Wu H., Jin X., S-palmitoylation of PCSK9 induces sorafenib resistance in liver cancer by activating the PI3K/AKT pathway. Cell Rep. 40, 111194 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Jeong D. W., Park J. W., Kim K. S., Kim J., Huh J., Seo J., Kim Y. L., Cho J. Y., Lee K. W., Fukuda J., Chun Y. S., Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma. Nat. Commun. 14, 6370 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silva C. A. C., Facchinetti F., Routy B., Derosa L., New pathways in immune stimulation: Targeting OX40. ESMO Open 5, e000573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karulf M., Kelly A., Weinberg A. D., Gold J. A., OX40 ligand regulates inflammation and mortality in the innate immune response to sepsis. J. Immunol. 185, 4856–4862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gwyer Findlay E., Danks L., Madden J., Cavanagh M. M., McNamee K., McCann F., Snelgrove R. J., Shaw S., Feldmann M., Taylor P. C., Horwood N. J., Hussell T., OX40L blockade is therapeutic in arthritis, despite promoting osteoclastogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 2289–2294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yanagita S., Hori T., Matsubara Y., Ishikawa T., Uchiyama T., Retroviral transduction of acute myeloid leukaemia-derived dendritic cells with OX40 ligand augments their antigen presenting activity. Br. J. Haematol. 124, 454–462 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Fu Y., Lin Q., Zhang Z., Zhang L., Therapeutic strategies for the costimulatory molecule OX40 in T-cell-mediated immunity. Acta Pharm. Sin. B 10, 414–433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lencioni R., Llovet J. M., Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin. Liver Dis. 30, 052–060 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Li Y. M., Liu Z. Y., Wang J. C., Yu J. M., Li Z. C., Yang H. J., Tang J., Chen Z. N., Receptor-interacting protein kinase 3 deficiency recruits myeloid-derived suppressor cells to hepatocellular carcinoma through the chemokine (C-X-C Motif) ligand 1-chemokine (C-X-C Motif) receptor 2 axis. Hepatology 70, 1564–1581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., Valvano M. A., Foster S. J., Mak T. W., Nuñez G., Inohara N., An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Correa R. G., Khan P. M., Askari N., Zhai D., Gerlic M., Brown B., Magnuson G., Spreafico R., Albani S., Sergienko E., Diaz P. W., Roth G. P., Reed J. C., Discovery and characterization of 2-aminobenzimidazole derivatives as selective NOD1 inhibitors. Chem. Biol. 18, 825–832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mirji G., Worth A., Bhat S. A., El Sayed M., Kannan T., Goldman A. R., Tang H. Y., Liu Q., Auslander N., Dang C. V., Abdel-Mohsen M., Kossenkov A., Stanger B. Z., Shinde R. S., The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci. Immunol. 7, eabn0704 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C., Li J., Cheng Y., Meng F., Song J. W., Fan X., Fan H., Li J., Fu Y. L., Zhou M. J., Hu W., Wang S. Y., Fu Y. J., Zhang J. Y., Xu R. N., Shi M., Hu X., Zhang Z., Ren X., Wang F. S., Single-cell RNA sequencing reveals intrahepatic and peripheral immune characteristics related to disease phases in HBV-infected patients. Gut 72, 153–167 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Q., He Y., Luo N., Patel S. J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., Peet G. W., Zhong G., Lu S., Zhu W., Mao Y., Xiao M., Bergmann M., Hu X., Kerkar S. P., Vogt A. B., Pflanz S., Liu K., Peng J., Ren X., Zhang Z., Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C. H., Myung P., Plikus M. V., Nie Q., Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brigidi G. S., Bamji S. X., Detection of protein palmitoylation in cultured hippocampal neurons by immunoprecipitation and acyl-biotin exchange (ABE). J. Vis. Exp. 72, e50031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Tables S1 and S2