ABSTRACT
Both abdominal radiotherapy and a nuclear event can result in gastrointestinal symptoms, including acute radiation syndrome (GI-ARS). GI-ARS is characterized by compromised intestinal barrier integrity increasing the risk for infectious complications. Physiologically relevant animal models are crucial for elucidating host responses and therapeutic targets. We aimed to determine the radiation dose requirements for creating GI-ARS in the Sinclair minipig. Male, sexually mature swine were randomly divided into sham (n = 6) and three lower hemibody radiation dosage groups of 8, 10, and 12 Gy (n = 5/group) delivered using linear accelerator-derived x-rays (1.9 Gy/min). Animals were monitored for GI-ARS symptoms for 14 days with rectal swab and blood collection at days 0–3, 7, 10, and 14 followed by necropsy for western blotting and histology. Dose-dependent increases in weight loss, diarrhea severity, and mortality (log-rank test, P = 0.041) were seen. Villi length was significantly reduced in all irradiated animals compared to controls (P < 0.001). Serum citrulline decreased and bacterial translocation increased after irradiation compared to controls. Increased NLRP3 levels in post-mortem jejunum were seen (P = 0.0043) as well as increased IL-1β levels in the 12 Gy group (P = 0.041). Radiation dose and survival were associated with significant gut microbial community shifts in beta diversity. Moreover, decedents had increased Porphyromonas, Campylobacter, Bacteroides, Parvimonas, and decreased Fusobacterium and decreased Aerococcus, Lactobacillus, Prevotella, and Streptococcus. Our novel Sinclair minipig model showed dose-dependent clinical symptoms of GI-ARS. These findings provide invaluable insights into the intricate interplay between GI-ARS, intestinal inflammation, and gut microbiota alterations offering potential targets for therapeutic and diagnostic interventions after radiation exposure.
KEYWORDS: Gut microbiota, gastrointestinal, acute radiation syndrome, NLRP3 inflammasome, swine
INTRODUCTION
Radiation exposure poses a significant threat to human health, and one of its most severe consequences is the development of acute radiation syndrome (ARS). While irradiation is a valuable tool in medical diagnostics and cancer treatment, an ever-increasing threat of radiological or nuclear attacks could also produce high-dose radiation exposures. In humans, ARS manifests in three subsyndromes based on the absorbed dose measured in gray (Gy): hematopoietic (H, >2–3 Gy), gastrointestinal (GI, 5–12 Gy), and cerebrovascular syndrome (10–20 Gy) (1). H-ARS is the most studied subsyndrome and is characterized by leukopenia, immunosuppression, increased susceptibility to infection, anemia, and hemorrhage (2). Like H-ARS, GI-ARS also results from detrimental effects of radiation on DNA within rapidly proliferating cells. GI-ARS encompasses a spectrum of symptoms ranging from nausea and vomiting to severe diarrhea and systemic complications (1). The pathophysiology of GI-ARS involves enterocyte loss and vascular injury contributing to significant mortality (2). While there are Food and Drug Administration (FDA)-approved countermeasures for H-ARS, none exist for GI-ARS underscoring the need to explore host responses to identify diagnostic markers and effective therapeutic targets.
Treating GI-ARS following a radiological/nuclear attack is currently a knowledge gap that may be informed by patients receiving abdominal radiotherapy who suffer from GI symptoms (3). Still, medical management of GI-ARS includes only supportive care such as antiemetics, fluids, and antibiotics (4). The use of H-ARS FDA-approved medical countermeasures (MCMs) to mitigate GI-ARS effects has been explored with limited efficacy (5,6). To this end, development and testing of novel MCMs requires well-characterized animal models. Historically, many animal models for studying GI-ARS have been rodent models which exhibit differences in radiosensitivity, immune responses, and gut flora compared to humans (7,8). Several large animal species have been used in GI-ARS models including nonhuman primates (NHPs) (9,10), and minipigs (11–13). Larger animals are generally more anatomically and physiologically similar to humans with minipigs offering ethical, financial, and handling advantages (14,15). While there has been experience in producing GI-ARS in Gottingen minipigs (11–13), the appropriate absorbed dose to produce this effect is yet to be identified in the Sinclair minipig, which has the advantage of leveraging natural skin pigmentation variation, and larger size (roughly 20–25 kg as opposed to 10–15 kg) allowing for more blood sampling (volume and time points) longitudinally.
To address this limitation, our aim was to determine the radiation dose requirements in the Sinclair minipig for creating GI-ARS. We focused on clinical and histological signs of GI-ARS, as well as l-citrulline as a GI function biomarker. We also examined bacteremia as a surrogate for sepsis, as reduced epithelial barrier integrity may result in bacterial translocation into the bloodstream (10). In addition, we evaluated inflammasome activation and longitudinal gut microbiota alterations. We hypothesized the clinical course and GI alterations in our Sinclair minipig model of GI-ARS would depend on the dose received. The presented Sinclair minipig GI-ARS model will aid in understanding the complexities of GI-ARS to progress the development and testing of MCMs, to include the involvement of cutaneous injuries (i.e., burns).
MATERIALS AND METHODS
Animals and irradiation
A supplemental methods section is given for further details of all experiments. Twenty-one sexually mature male Sinclair minipigs (22.1 ± 3.9 kg) were obtained from Sinclair Bio Resources (Auxvasse, MO) and acclimated to the facility for at least 72 h. Research was approved by the Uniformed Services University of the Health Sciences Institutional Animal Care and Use Committee and performed at an AAALAC fully accredited facility. All animal procedures were conducted in compliance with the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act (16,17). Anesthetized animals received an indwelling jugular catheter and were randomly assigned as sham controls (n = 6) or to one of three radiation exposure groups at either 8, 10, or 12 Gy (n = 5/dose group). Hemibody irradiation was performed distal to the diaphragm with a clinical linear accelerator (Elekta Infinity, Armed Forces Radiobiology Research Institute LINAC) delivering 4 MV photons for a single dose of 8, 10, or 12 Gy (dose rate approximately 1.9 Gy/min). Ionization chamber dosimetry with cylindrical water-filled polymethylmethacrylate phantoms of various diameters were used to calibrate LINAC output at the midplane (18). At the end of the irradiation (or catheter implantation for controls), animals were transported back to their home cages for recovery from anesthesia.
Monitoring and sample collection
Each day body temperature and weight, and scoring of appetite, mobility/posture, natural behavior, and diarrhea were evaluated using a pain and distress assessment score sheet (Fig. S1, http://links.lww.com/SHK/C2). Four of the following criteria qualified the animal for early euthanasia: hyperthermia (>42°C), anorexia (skipping 3 consecutive meals), anemia/pallor, severe vomiting or bloody diarrhea, lethargy, seizures, vestibular signs (falling, circling), or prolonged hemorrhage. Euthanasia was achieved through the jugular catheter with sodium pentobarbital (Euthasol, 100 mg/kg, Virbac, Carros, France). Harvested jejunum tissues were snap-frozen, and stored at −80°C, or preserved in 10% formalin for histology. In addition, mesenteric lymph node (MLN), liver, and spleen tissues were homogenized in phosphate-buffered saline for aerobic bacterial culture.
Blood analyses
Blood and rectal swabs were collected prior to sham injury and/or radiation (day 0) and on days 1, 2, 3, 7, 10, and 14. For analysis of blood chemistry, BD Vacutainer tubes with Lithium Heparin were used (Becton Dickinson, Franklin Lakes, NJ), centrifuged at 1,200g for 10 min and analyzed on the VITROS 350 (Ortho-Clinical Diagnostics, Raritan, NJ). At each blood draw, approximately 0.1 mL of blood was directly inoculated onto tryptic soy agar with 5% sheep blood (BA) and MacConkey agar (MAC) for examination of colony-forming units (Becton Dickinson). For detection of circulating cytokines (IL-1β, IL-6, and IL-18), porcine-specific ELISA kits were used according to the manufacturer’s instructions (Abcam, Cambridge, United Kingdom).
Histology
Formalin-fixed tissue was paraffin embedded, cut into 5-μm sections, and stained with hematoxylin and eosin. A board-certified veterinary pathologist blinded to groups scored stained slides for villi blunting/fusion, apoptosis, and crypt loss with the following scale: 0 = normal; 1 = slight; 2 = easily identified/limited severity; 3 = moderate; 4 = marked damage. Immunohistochemistry (IHC) was used to probe for Ki67 and zonula occludens-1 (ZO-1) (Ki67 and ZO-1, 1:200, MyBioSource, San Diego, CA). In addition, TUNEL staining was performed according to the manufacturer’s instructions (Abcam). ImageJ (National Institutes of Health, Bethesda, MD) was used to measure villi length and semiquantification using either the ‘Split Channels’ or ‘Color Deconvolution’ tools for fluorescent or brightfield microscopy images, respectively.
Assessment of l-citrulline
High-performance liquid chromatography was carried out on an Agilent 1200 system using an iHILIC Fusion column to measure levels of l-citrulline in isolated pig serum. The high-performance liquid chromatography solvents were A = 80 mM ammonium acetate (pH 5.0), B = 75% acetonitrile/25% 25 mM ammonium acetate. The Analyte to Internal Standard ratio (A/IS) equation is as follows: [Cit.] (ug/mL) = ((A/IS) o) X(16 ug/mL)/{(A/IS) SA – (A/IS) o} where o = sample with no added Cit and SA = sample with added Cit (Standard Addition sample).
Protein analysis
Western blotting was performed on jejunum lysates for the following targets: ZO-1, toll-like receptor 4 (TLR4), NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3), pro-caspase-1, and interleukins (IL-1β and IL-18). Conjugated secondary antibodies, IRDye 800CW, or IRDye 680RD (LI-COR, Lincoln, NE; 1:15,000) were used for detection and visualized on the Odyssey infrared imaging system (LI-COR).
16S rRNA sequencing and analysis
Rectal swabs were taken using sterile cotton tipped applicators and stored at −80°C. DNA was isolated using the QIAmp PowerFecal Pro kit (Qiagen, Germantown, MD) without modifications. Library preparation was performed using the “Illumina 16S Metagenomic Sequencing Library Preparation” protocol with 16S V4 region 515F–806R primers (18,19). DNA and libraries were quantified using Qubit 4.0 (ThermoFisher). Sequencing was performed on a NextSeq 500 using a Mid Output v2.5 2 × 150 bp kit (Illumina San Diego, CA). Bioinformatic analysis was performed through previously described pipelines (20), with DADA2 for feature table generation based on a median quality score of >25 (21–23), which excluded reverse reads. The resulting table were imported into Quantitative Insights Into Microbial Ecology (QIIME2, version 2023.7) (24), with taxonomy analyzed against the Greengenes database. Further details and tools on the analysis pipeline were informed from the literature, (51–55) and can be found in the supplemental information. Sequences were deposited in the NCBI Sequence Read Archive.
Statistics
Mortality was assessed using a Kaplan-Meier survival curve and Log-rank (Mantel-Cox) test. For longitudinal data, the control is normalized and represented by either the mean ± SD or median and interquartile ranges from baseline (day 0) data. Scored clinical outcomes (i.e., diarrhea, loss of appetite, behavior) analysis was conducted using area under the curve. Differences in terminal study endpoints (e.g., protein levels, IHC quantification, and histopathological analyses) were performed using a one-way analysis of variance (ANOVA) or Kruskal-Wallis test as appropriate. Statistical testing for longitudinal metrics such as clinical chemistries, cytokines, and alpha diversity were conducted with a two-way ANOVA with Tukey’s or Dunnett’s posttesting. Grubbs’ test was used for removing outliers in cytokine ELISA and microbiome data. A permutational analysis of variance (PERMANOVA) was to assess pairwise associations in beta diversity measures. Unless otherwise stated, values are represented as mean ± SD. Statistical analysis and figure generation was performed using Prism 10.0 (GraphPad Software, San Diego, CA) except for PCA plots from QIIME2 and differential abundance figures from R. Statistical significance was set as P < 0.05.
RESULTS
Clinical endpoints
Hemibody irradiation resulted in dose-dependent survival (Fig. 1A), wherein minipigs in the control and 8 Gy groups all survived to day 14, which was significantly different from the 12 Gy group (P = 0.041). Radiation also induced significant weight loss, decreased appetite, and higher diarrhea and behavior scores for 10 and 12 Gy groups (Fig. 1, B and C, S2A, http://links.lww.com/SHK/C3). The diarrhea was also accompanied by increased total protein and albumin levels (Fig. S2C–D, http://links.lww.com/SHK/C3), indicative of the overall dehydration that occurred in this model.
Fig. 1.
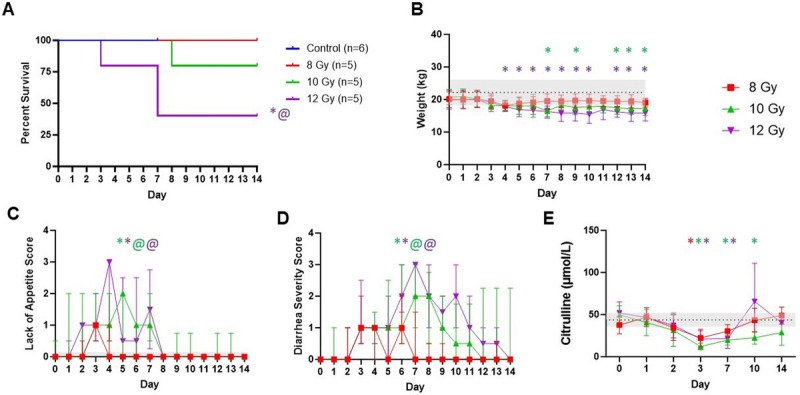
Impact of radiation dose on clinical outcomes. Longitudinal endpoints are shown as (A) survival, (B) weight, (C) loss of appetite, (D) diarrhea severity, and (E) LC-MS analysis for L-citrulline concentration in plasma over time. Control (n = 6), 8 Gy (n = 5), 10 Gy (n = 5), and 12 Gy (n = 5). Values for weight are represented as control mean ± SD. Loss of appetite and diarrhea severity are depicted as median and interquartile ranges. Statistical significance is indicated by text, color, and symbol. * Significant difference between control and radiation dose group. @ Significant difference between 8 Gy and another radiation dose group. The color of each symbol represents the dose group.
L-citrulline has been touted as a promising GI function biomarker for assessing radiation-induced intestinal epithelial damage (5). Irradiation induced decreases in l-citrulline concentration (Fig. 1E), regardless of the dose received reaching a low at day 3 which was significantly different compared to controls (8 Gy, P = 0.022; 10 Gy, P = 0.0004; 12 Gy, P = 0.015). Higher doses led to sustained lower l-citrulline on day 7 (10 Gy, P = 0.0098, 12 Gy P = 0.026), but overall l-citrulline proved insufficiently sensitive to distinguish doses. To evaluate radiation dose effects on organ function, biomarkers for kidney (blood urea nitrogen and creatinine), pancreatic (amylase and lipase), and liver (lactate dehydrogenase, alanine transaminase, alkaline phosphatase, and aspartate transaminase) function were also measured (Fig. S3, http://links.lww.com/SHK/C4). Elevation in markers of kidney and pancreatic function were observed after irradiation to varying degrees depending on the dose.
Radiation-induced GI epithelial damage
Histology showed that control jejunum exhibited normal crypts and villi and radiation induced structural damage. Specifically, villus length was significantly reduced in all radiation groups compared to control animals (8 Gy, P = 0.008; 10 Gy, P < 0.0001; 12 Gy, P < 0.0001) with no significant difference in length between doses (Fig. 2, A and B). The loss of villi and crypts was more apparent in animals that were euthanized early than any particular radiation dose. Blinded pathologist scoring showed villi blunting/fusion and mucosal apoptosis were significantly increased after 12 Gy irradiation compared to controls (Fig. 2, C–D, P = 0.03), while crypt loss was only observed in 10 and 12 Gy groups (Fig. 2E). We examined maintenance of epithelial barrier integrity with tight-junction protein zonula occludens-1 (ZO-1) evaluation (Fig. 2, F–G). A marked decrease of ZO-1 in 10–12 Gy groups was not significant due to high variation, especially in the 8 Gy group (P = 0.095). Moreover, Goblet cells are apparent in higher abundance in control intestinal epithelium when compared to irradiated intestines (Fig. 2G). Intestinal epithelial cell proliferation and apoptosis were evaluated by IHC targeting Ki67 and TUNEL staining with semiquantitation (Fig. 3, A and B). The control group exhibited a significantly higher Ki67-positive cell number than any irradiated group (P < 0.0001). Irradiated animals also had increased apoptosis than controls, significantly so between control and 12 Gy (P = 0.043).
Fig. 2.
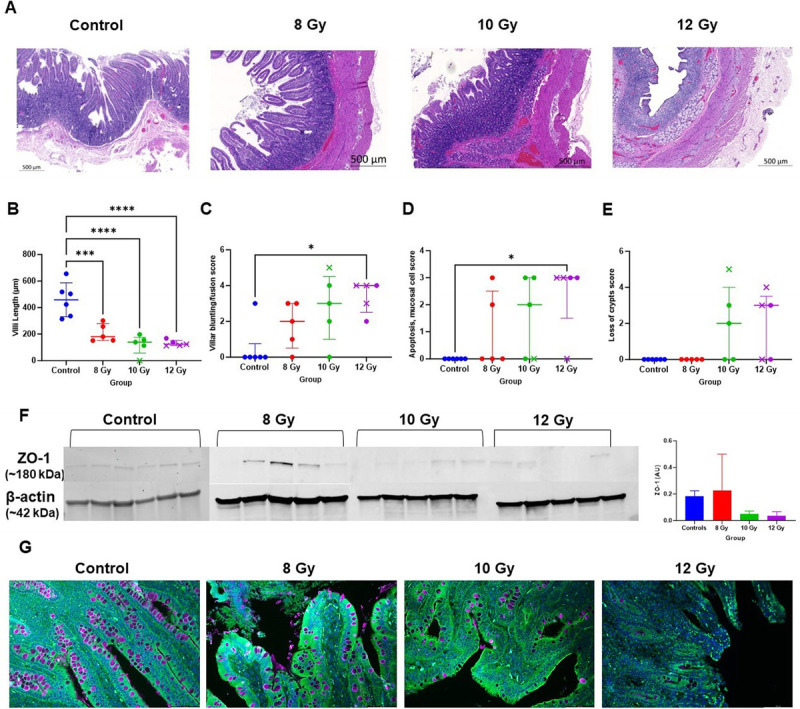
Radiation-induced alterations to the jejunum and epithelial barrier integrity. (A) Representative hematoxylin and eosin–stained images of jejunum from control and irradiated animals (20× objective, scale bar 500 μm). (B–E) Survivors are indicated with circles while decedents are represented by an x. (B) Villi length in μm. (C–E) Veterinary pathology scored metrics including villi blunting/fusion, mucosal apoptosis, and loss of crypts. Median ± interquartile range is indicated. (F) Western blots for protein expression of zonula occludens-1 (ZO-1) in jejunum tissue lysates in control and different irradiated dose group animals. Bars represent mean ± SD (G) Immunochemistry staining for ZO-1 expression (green), goblet cells using Wheat Germ Agglutinin (purple), and nuclei with DAPI (blue) (magnification, 20×). The color of each symbol represents the dose group. * P < 0.05, *** P < 0.0005, and **** P < 0.0001.
Fig. 3.
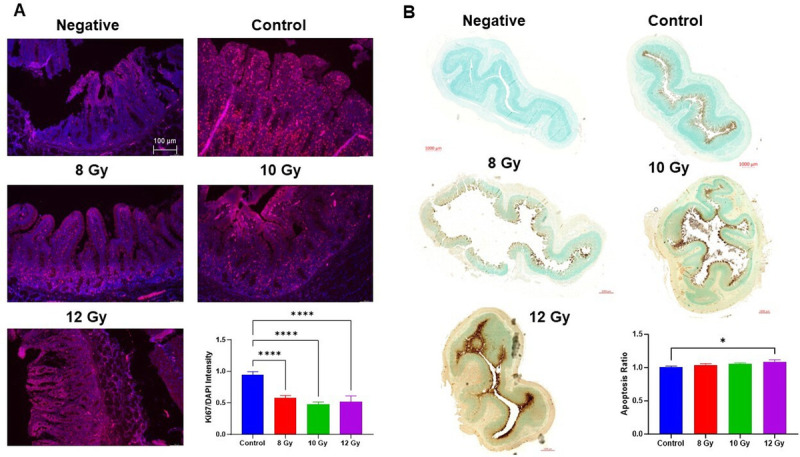
Immunohistochemistry of cell turnover. Stains depict negative controls, control, 8–12 Gy irradiated jejunum (A) Immunofluorescence with DAPI (blue) and Ki67 (red). Representative 10× images taken with Leica microscope, scale at 100 μm. (B) TUNEL stain using DAB to generate a brown color at the site of DNA fragmentation while methyl green was the counterstain for normal cells. Representative images from Zeiss AxioScan at 20x with scale bars of 1 mm. Barplots show quantified Ki67 and TUNEL positive ratio. Bars represent mean ± SD. *P < 0.05 and **** P < 0.0001 (one-way ANOVA, Tukey’s posttesting).
Bacteremia was found to varying extents in control and irradiated minipigs (Table 1). Colony-forming units/mL (CFU/mL) were highest at day 14, which was significant from baseline (8 Gy, P < 0.0001; 10 Gy, <0.0001; 12 Gy, P = 0.0002). A variety of organisms were isolated from blood to include Staphylococcus aureus identified by catalase and Staphylase test, and Escherichia coli by RapID One with Acinetobacter lwoffii, and Burkholderia cepacia confirmed with 16S amplicon sequencing. There was also a tendency for more gram-positive bacteria within the first 3 days and gram-negative organisms on days 7–14. Bacteria were commonly isolated from MLNs (89.5%) from all groups, while the liver and spleen only harbored bacteria after irradiation with higher doses resulting in higher frequency. Organisms cultured from these tissues included S. aureus (catalase and Staphylase test), E. coli (RapID One), Pseudomonas aeruginosa (oxidase and morphology), A. lwoffii, Staphylococcus xylosus, and Ralstonia solanacearum (16S sequencing).
Table 1.
Bacterial translocation into blood or tissues of irradiated minipigs
| Group | Blood (Avg. CFU/mL) | MLN | Liver | Spleen | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DO | D1 | D2 | D3 | D7 | D10 | D14 | ||||
| Control | 0 | 33 | 45 | 40 | 55 | ΝΑ | ΝΑ | 5/6 (83.3%) | 0/6 (0%) | 0/6 (0%) |
| 8 Gy | 0 | 156 | 120 | 114 | 18 | 54 | 304 | 4/5 (80%) | 3/5 (60%) | 2/5 (40%) |
| 10 Gy | 0 | 40 | 4 | 132 | 182 | 163 | 463 | 4/4* (100%) | 3/4* (75%) | 3/4* (75%) |
| 12 Gy | 0 | 90 | 70 | 34 | 50 | 30 | 270 | 4/4* (100%) | 4/5 (80%) | 4/5 (80%) |
Bacterial colony-forming units per milliliter of blood for each blood draw day and organisms isolated from tissues. Mesenteric lymph node, liver, and spleen.
*Indicates missing values for one animal. Countable range from tissues, 15–150 bacterial colonies.
Radiation-induced activation of the NLRP3 Inflammasome
Pathogen-associated molecular patterns can lead to TLR4-induced activation of the NLRP3 inflammasome, resulting in caspase-1 cleavage and subsequent inflammatory cytokine release (25). We examined radiation-induced changes in these protein levels in jejunum lysates (Fig. 4) and found TLR4 protein levels were significantly upregulated in the 10 Gy group compared to control and 8 Gy animals (control, P = 0.021; 8 Gy, P = 0.031). NLRP3 protein levels increased after irradiation (8 Gy, P = 0.02; 10 Gy, P < 0.0001; 12 Gy, P = 0.001) as did pro-caspase-1 levels (8–12 Gy, P < 0.0001) compared to controls. Consequently, there were also increased cleaved IL-1β tissue levels after irradiation; however, this was only significant in the 12 Gy group (P = 0.041).
Fig. 4.
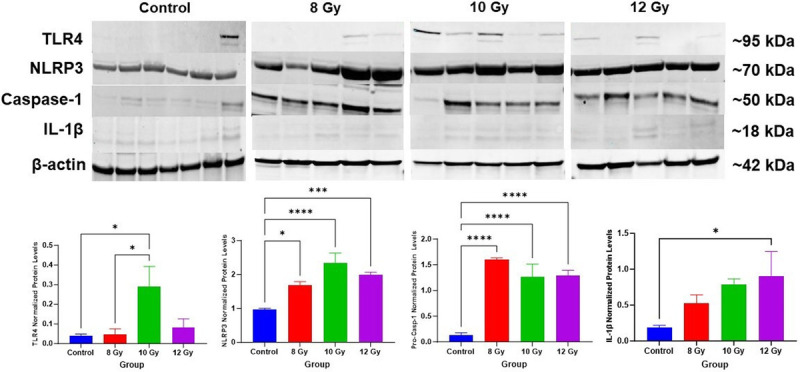
NLRP3 inflammasome activation in the gut. Representative blots and beta-actin (β-actin) of components in the NLRP3 inflammasome activation pathway including toll-like receptor 4 (TLR4), NLRP3, pro-caspase-1 (Casp-1), and interleukin-1 beta (IL-1β). Data are represented as mean ± SD. Quantification of western blots are shown below the images. *P < 0.05, *** P < 0.001, and **** P < 0.0001 (One-way ANOVA, Tukey’s posttesting).
Systemic inflammation was evaluated using IL-6, IL-1β, and IL-18 changes in plasma concentrations across time in irradiated and sham animals (Fig. 5). IL-6 increased over time in the 10 and 12 Gy groups (albeit with high variability) with no notable changes observed in 8 Gy, which was significant between 8 and 12 Gy on day 10 (P = 0.01). IL-1β was significantly elevated in the 8 Gy group by day 10 (P = 0.011) and in 10 Gy animals compared to control at days 10 (P = 0.045) and 14 (P = 0.015), with more modest increases in the 12 Gy group. There were significant differences in IL-18 at days 2–3 between control and 12 Gy (d2 P = 0.0083; d3 P = 0.034) and day 2 between the 10 and 12 Gy groups (P = 0.029).
Fig. 5.
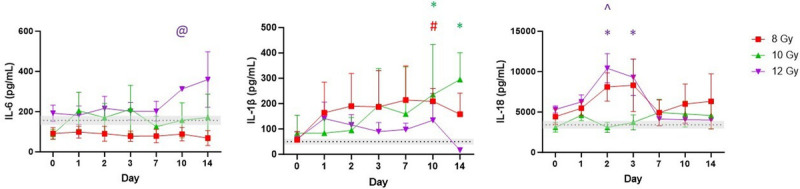
Select circulating cytokines (IL-6, IL-1β, and IL-18) across time. Average of baseline values of all treatments is set as a reference line and bars represent mean ± SEM. Asterisk denotes a significant difference between control and radiation dose group. @ Significant difference between 8 Gy and another radiation dose group. ^ represents a significant difference between 10 Gy and another radiation dose group. The color of each symbol represents the dose group.
Effects of high dose radiation exposure on the gut microbiota
A mean of 486,159 reads per sample were generated from 16S sequencing. After quality control there was a total frequency of 42,086,018 features retained with a mean frequency of 369,176 per sample (minimum 21,910 and max 1,707,549). Rarefaction was used to set the sampling depth at 21,500 for inclusion of all samples.
Shannon diversity and Faith’s PD were used to assess alpha diversity, which were not significantly altered after irradiation throughout the experiment (Fig. 6A). Beta diversity was evaluated with Bray-Curtis (BC, nonphylogenetic, Fig. S4A, http://links.lww.com/SHK/C5) and generalized UniFrac (GU, phylogenetic, Fig. 6B) metrics with both revealing that microbial communities are altered following irradiation. After 8 Gy irradiation, community composition was significantly different from baseline at day 1 and 3 (GU; d1, P = 0.008; d2, P = 0.095; d3, P = 0.029) with clustering closer to baseline from day 7 onward (GU; d7, P = 0.72; d10, P = 0.4; d14, P = 0.13). These shifts persisted longer with higher doses which were different from baseline at 10 Gy (d3, P = 0.01; d7, P = 0.022; d10, P = 0.013; d14, P = 0.022) and 12 Gy (d2, P = 0.01; d3, P = 0.067; d7, P = 0.048, d10, P = 0.52; d14, P = 0.49). Beta diversity differences were also apparent based on survival at day 7 (GU, P = 0.023).
Fig. 6.
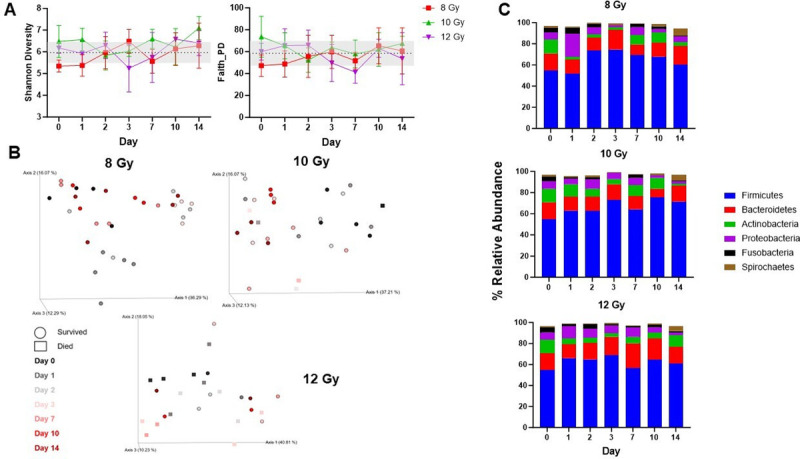
Effects of radiation on the gut microbiota. (A) Shannon and Faith’s Phylogenetic alpha diversity. Values are represented as mean ± SD. (B) Generalized UniFrac analysis evaluating microbial community composition at each irradiation dose over time. Each axis of the PCoA plot explains the variance in the data. Each dot (survivors) or square (decedents) is an individual sample and time points are color-coded. (C) Phyla level taxonomic classification showing mean relative abundance across time in each dose group. Phyla consisting of >1% of total bacterial composition are depicted.
Taxonomic classifications revealed that Firmicutes relative abundance increased from baseline in each dose group (Fig. 6C, S4B, http://links.lww.com/SHK/C5) and was significantly higher than baseline in 8 Gy animals at day 2 (P = 0.0034). Longitudinal decreases in Actinobacteria were observed for each dose with significant differences from baseline at days 3 (8 Gy P = 0.0097) and 14 (8 Gy P = 0.0036; 10 Gy P < 0.0001). Proteobacteria transiently increased on day 1 in the 8 and 12 Gy groups but consistently decreased thereafter.
Irradiation caused significant longitudinal alterations in beneficial gut commensal genera (Fig. S4C, http://links.lww.com/SHK/C5; Table S2, http://links.lww.com/SHK/C7). Prevotella and Lactobacillus were generally higher than baseline in each dose group, significantly at day 3 (Prevotella P = 0.016–0.026; Lactobacillus P = 0.021–0.049). Bifidobacterium and Faecalibacterium were significantly increased in each dose group across the first 7 days after irradiation, with Bifidobacterium higher at day 1 in 12 Gy irradiated animals than 8 Gy (P = 0.022). Megasphaera were significantly more abundant after irradiation with each dose. Opportunistic commensal organisms such as Fusobacterium and Bacteroides increased from baseline while Porphyromonas and parvimonas significantly decreased after 8 and 10 Gy irradiation at day 3 from baseline and 12 Gy (P < 0.0001).
We also examined genera based on survival and presence of diarrhea (Fig. 7, A and B). For mortality, there were no significant differences on days 1–2 after radiation; however, by day 3, there was significantly higher Fusobacterium (P = 0.003), Bacteroides (P < 0.0001), and Parvimonas (P = 0.0038) in decedents. In addition, Lactobacillus (P = 0.019) and Faecalibacterium were significantly reduced in decedents (P = 0.004) on day 7. In addition, we observed significant increases in Bifidobacterium (P < 0.0001), Megasphaera (P < 0.0001), Dialister (P < 0.0001), Lactobacillus (P < 0.0001), Prevotella (P = 0.0027), and Facecalibacterium (P = 0.007) with the presence of diarrhea. Alternatively, there were significant reductions in Treponema (P < 0.0001) and Peptoniphilus (P = 0.002) in those with diarrhea.
Fig. 7.
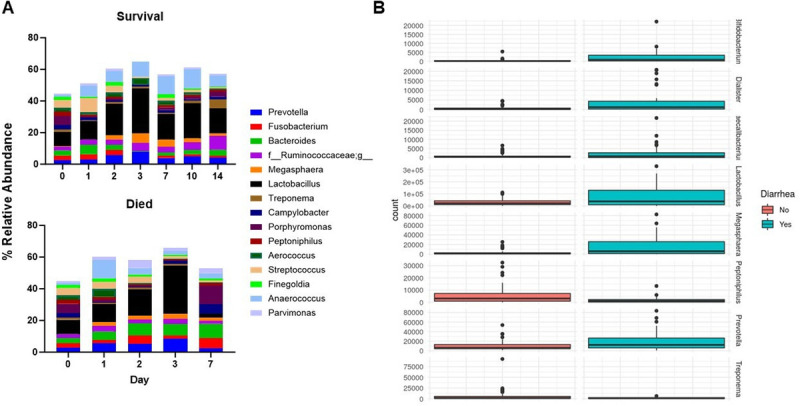
Association of survival and diarrhea prevalence with gut microbiota. (A) Genera mean relative abundance represented for survivors and decedents of irradiation. Top 15 genera at baseline are shown. Survivors at day 1 (n = 10), 2 (n = 11), 3 (n = 7), 7 (n = 10), 10 (n = 9), and 14 (n = 10). Decedents at day 1 (n = 4), 2 (n = 4), 3 (n = 4), and 7 (n = 3). (B) Select differentially abundant genera from DESeq2 analysis of diarrhea prevalence. Absence of diarrhea (n = 53) and diarrhea (n = 33).
DISCUSSION
GI-ARS would result after focused abdominal radiotherapy and nuclear/radiological mass casualty events that involve any bone marrow sparing due to, for example, shielding from concrete. To address the lack of MCMs, there is a need for additional GI-ARS animal models (10–13). Minipigs are advantageous due to their anatomical similarity to humans and ease of handling (15). Because the natural history of GI-ARS in Sinclair minipigs has not been previously reported, we sought to evaluate the effects of partial-body irradiation on GI symptoms, structure, cell turnover, and gut microbiota. We present a novel GI-ARS model with clinically relevant findings, impaired intestinal barrier integrity, and altered enterocyte turnover. Furthermore, we demonstrate that radiation exposure led to sustained activation of the NLRP3 inflammasome in the jejunum and altered gut microbiota. Higher doses led to irrecoverable shifts in beta diversity, and mortality was associated with increased Porphyromonas, Campylobacter, Bacteroides, Parvimonas, and Fusobacterium and decreased Aerococcus, Lactobacillus, and Streptococcus. To our knowledge, this is the first study to show inflammasome and microbiome changes over 14 days in a porcine model specific to GI-ARS.
Much of our results are consistent with previous total and partial-body irradiation models of GI-ARS in multiple animals including dose-dependent mortality (9–13,26–29). We were not able to pinpoint a specific overarching cause of mortality (e.g., sepsis) but decedent did display higher multiorgan dysfunction markers (i.e., creatinine). Reasons for unscheduled euthanasia were similar to previous reports and included weight loss, diarrhea, lethargy, and anorexia (11,13,28,29). The exception was respiratory distress, which we did not see but may have with longer time points given it is a delayed effect of irradiation (9). Most Gottingen studies have shown that doses >12 Gy are required for mortality, suggesting Sinclair minipigs are more sensitive to radiation, which would be the opposite of H-ARS after whole body irradiation. (15,30). Histologically, GI-ARS models across species consistently demonstrate dose-dependent GI damage with Measey et al. (2018) noting a trend toward intestinal epithelium recovery past day 14, which we did not investigate (10,11,26). Time may also explain some nuanced difference in enterocyte turnover between models as Kaur et al. (2020) demonstrated recovery of cell proliferation from 6–30 days after irradiation (13,27). However, we cannot rule out that low specificity on semiquantitative histological analysis may be affected by, for example, autofluorescence.
The gut microbiome serves immunological, barrier, and metabolic functions contributing to nutrient production and enterocyte proliferation and could provide a radiation signature for diagnostic information on potential dysbiosis or protective bacteria (31). The effects of high-dose ionizing radiation on the gut microbiota are relatively unexplored in large animal models with three studies published to date (32–35). For alpha diversity, NHP irradiation models found no differences post radiation, consistent with our findings, while a Gottingen minipig model observed alpha diversity decreases (33–35). In terms of beta diversity most studies evaluate a single time point following irradiation, while we extended this to show longitudinal microbial community differences over multiple time points out to 14 days (33–35). Moreover, we also saw beta diversity differences based on mortality on day 7, which speaks to the potential for beta diversity to inform dose received and outcomes, as has been seen in trauma patients (36).
Taxonomic changes may also identify changes in opportunistic pathogens versus beneficial bacteria, with genus level alterations providing more detail than higher taxonomic classifications. Treponema is an intracellular symbiont that is a more of a common gut bacterium in animals than humans, while Faecalibacterium is an important commensal in human gut microbiota not present in rodents (32,33). Our results revealed increased Treponema and Faecalibacterium in each radiation dose with higher Treponema abundances associated with survival. In a previous minipig model, Treponema increased and Faecalibacterium decreased after irradiation (33,34). We found radiation-induced increases in Prevotella, Bacteroides, and Bifidobacterium, to varying extents across time and doses, which is the opposite of a previous minipig model. However, that study showed increases in Lactobacillus postirradiation as did we, which differs from rodent studies (33,37,38). While the implications of these discordant findings are unclear, any differences in animal models should be considered alongside any available human data. For example, in radiotherapy patients, increased Bacteroides has been associated with GI mucositis (32,39).
Similarly, genus changes considered along with clinical symptoms (e.g., mortality, diarrhea) could provide actionable information for treatment of GI-ARS. Previously, Prevotella has correlated with increased survival in both minipigs and NHPs while Streptococcus decreases correlated with death in NHP (34). In our study, decedents had increased abundances of Porphyromonas, Campylobacter, Bacteroides, Parvimonas, and Fusobacterium with decreases in Aerococcus, Lactobacillus, Prevotella, and Streptococcus. There is limited literature on Fusobacterium postirradiation, but it has been found to promote the development of colorectal cancer and resistance to radiotherapy (40). Furthermore, Singh et al. (2022) highlights that Fusobacterium, Bacteroides, and Campylobacter correlated with increased reactive oxygen species (ROS) whereas Lactobacillus associated with reduced ROS load (41). Kalkeri et al. (2021) reported elevated Lactobacillus reuteri, Dialister, and Veillonella from radiation-induced diarrhea which aligns with our observations (35), while we also found alterations in Bifidobacterium, Megasphaera, Dialister, Lactobacillus, Prevotella, Facecalibacterium, Treponema, and Peptoniphilus. Interestingly, the organisms associated with mortality and diarrhea do not always align. For example, we observed increases in Lactobacillus, Bifidobacterium, and Dialister with diarrhea, but decreases in decedents, perhaps more consistent with their roles as short-chain fatty acid producers (34). The increased prevalence of obligate anaerobes in our study could be innate tolerance to the cellular conditions induced by radiation in the gut or a host defense response required for restoration of homeostasis via repairing damaged epithelium, reducing ROS, or producing short-chain fatty acid (35,38,42). Further study is warranted to unravel the mechanistic underpinning of these organisms and their role after irradiation.
Inflammasome activation can be caused by signals including ROS, DAMPs, K+ efflux, Ca2+, lysosomal rupture, and mitochondrial disruption (25). Furthermore, alterations in commensal gut flora, increased opportunistic pathogens, or microbial products (i.e., LPS) can activate NLRP3 within the gut, with subsequent caspase-1 activation and pro-inflammatory cytokine (IL-1β and IL-18) release (25). Hu et al. (2020) showed increased NLRP3 expression a day following 9 Gy of abdominal gamma irradiation in rats (43), while Stoeklein et al. (2015) exposed mice to 2 Gy total body radiation and found NLRP3 inflammasome activation was sustained for 7 days (44), but not 14 days as we found. Although we lack direct evidence of the cause of NLRP3 inflammasome activation, ROS and DAMPs are widely acknowledged as playing a role in radiation-induced gut damage (45). Radiation-induced ROS are produced in a continuous manner, which may explain our NLRP3 inflammasome activation 14 days after irradiation (46). Nevertheless, the link between radiation-induced activation (and potential pharmacologic inhibition) of the NLRP3 inflammasome and microbiota warrants further investigation. Moreover, while we focused on GI-ARS, a previous study implicated inflammasome activation in bone marrow and intestinal cells, suggesting a potential dual effect on H-ARS as well (47).
We acknowledge that there are limitations to this study. It is important to note that male swine were used for husbandry considerations and sex differences in radiosensitivity have been documented (48,49) although potentially not in minipigs (28,29). While using male swine offers certain advantages such as military relevance, wider availability, and larger size, future studies should use both male and female. Furthermore, while subject age is an important consideration in the response to radiation exposure (49), we chose to focus on sexually mature subjects. Other limitations include our sample size per group (n = 5–6), a single terminal endpoint for control animals (7 days) and irradiated animals (14 days). In addition, the broad-spectrum cephalosporin antibiotic given to our subjects is in line with supportive therapies, but we are not able to tease out the effects of irradiation or antibiotics on the gut flora. There are also technical limitations as such as the determination of certain circulating and tissue biomarkers due to the scarcity and specificity of porcine-specific antibodies. Blood and tissue collection were performed as sterilely as possible, but the opportunity for bacterial contamination still exists. Lastly, culture conditions were not supportive for the growth of anaerobes and fastidious organisms.
Despite these limitations, we showed a dose-dependent response in clinical outcomes consistent with GI-ARS in a Sinclair minipig model. Prediction of survival, or rather detection of physiological changes and intestinal epithelial injury that precede mortality, could be a critical component of medical management following radiation exposure. Moreover, these findings provide further evidence on the progression of GI-ARS and associate NLRP3 inflammasome activation and gut microbiota alterations after irradiation. The gut microbiome profiling has provided the elucidation of particular bacteria that are more resilient post-irradiation, and those that are associated with increased diarrhea and mortality. In the future, this Sinclair GI-ARS model can be useful for testing MCM efficacy and evaluating strategies such as NLRP3 inhibitors and modulation of gut microbiota as approaches to treat GI-ARS.
ACKNOWLEDGMENTS
The authors thank the staff in the Uniformed Services University of the Health Sciences Biomedical Instrumentation Center Histopathology and Microscopy cores, and the Department of Laboratory Animal Research.
Footnotes
first review completed 13 May 2024;
The authors report no conflicts of interest.
The opinions and assertions expressed herein are those of the author(s) and do not reflect the official policy or position of the Uniformed Services University of the Health Sciences or the Department of Defense.
Supplemental digital content is available for this article. Direct URL citation appears in the printed text and is provided in the HTML and PDF versions of this article on the journal’s Web site (www.shockjournal.com).
Contributor Information
Timothy S. Horseman, Email: timothy.horseman@usuhs.edu.
Babita Parajuli, Email: babita.parajuli.ctr@usuhs.edu.
Andrew M. Frank, Email: andrew.frank.ctr@usuhs.edu.
Alia Weaver, Email: alia.weaver@usuhs.edu.
David A. Schauer, Email: david.schauer@usuhs.edu.
Sean Moran, Email: sean.moran@usuhs.edu.
Joseph A. Anderson, Email: joseph.anderson1@usuhs.edu.
Gregory P. Holmes-Hampton, Email: gregory.holmes-hampton@usuhs.edu.
REFERENCES
- 1.López M, Martín M. Medical management of the acute radiation syndrome. Rep Pract Oncol Radiother. 2011;16(4):138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh VK, Seed TM. Pharmacological management of ionizing radiation injuries: current and prospective agents and targeted organ systems. Expert Opin Pharmacother. 2020;21(3):317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Reilly M Mellotte G Ryan B, et al. Gastrointestinal side effects of cancer treatments. Ther Adv Chronic Dis. 2020;11:204062232097035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenchegowda D Bolduc DL Kurada L, et al. Severity scoring systems for radiation-induced GI injury–prioritization for use of GI-ARS medical countermeasures. Int J Radiat Biol. 2023;99(7):1037–1045. [DOI] [PubMed] [Google Scholar]
- 5.MacVittie TJ Bennett AW Farese AM, et al. The effect of radiation dose and variation in Neupogen® initiation schedule on the mitigation of myelosuppression during the concomitant GI-ARS and H-ARS in a nonhuman primate model of high-dose exposure with marrow sparing. Health Phys. 2015;109(5):427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farese AM Bennett AW Gibbs AM, et al. Efficacy of Neulasta or Neupogen on H-ARS and GI-ARS mortality and hematopoietic recovery in nonhuman primates after 10 Gy irradiation with 2.5% bone marrow sparing. Health Phys. 2019;116(3):339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams JP Brown SL Georges GE, et al. Animal models for medical countermeasures to radiation exposure. Radiat Res. 2010;173(4):557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh VK Newman VL Berg AN, et al. Animal models for acute radiation syndrome drug discovery. Expert Opin Drug Discovery. 2015;10(5):497–517. [DOI] [PubMed] [Google Scholar]
- 9.MacVittie TJ Bennett A Booth C, et al. The prolonged gastrointestinal syndrome in rhesus macaques: the relationship between gastrointestinal, hematopoietic, and delayed multi-organ sequelae following acute, potentially lethal, partial-body irradiation. Health Phys. 2012;103:427–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacVittie TJ Farese AM Bennett A, et al. The acute gastrointestinal subsyndrome of the acute radiation syndrome: a rhesus macaque model. Health Phys. 2012;103:411–426. [DOI] [PubMed] [Google Scholar]
- 11.Elliott TB Deutz NE Gulani J, et al. Gastrointestinal acute radiation syndrome in Göttingen minipigs (Sus scrofa domestica). Comp Med. 2014;64:456–463. [PMC free article] [PubMed] [Google Scholar]
- 12.Moroni M Elliott TB Deutz NE, et al. Accelerated hematopoietic syndrome after radiation doses bridging hematopoietic (H-ARS) and gastrointestinal (GI-ARS) acute radiation syndrome: early hematological changes and systemic inflammatory response syndrome in minipig. Int J Radiat Biol. 2014;90:363–372. [DOI] [PubMed] [Google Scholar]
- 13.Kaur A Ten Have GAM Hritzo B, et al. Morphological and functional impairment in the gut in a partial body irradiation minipig model of GI-ARS. Int J Radiat Biol. 2020;96:112–128. [DOI] [PubMed] [Google Scholar]
- 14.Vodicka P Smetana K Jr. Dvoránková B, et al. The miniature pig as an animal model in biomedical research. Ann N Y Acad Sci. 2005;1049:161–171. [DOI] [PubMed] [Google Scholar]
- 15.Doyle-Eisele M Brower J Aiello K, et al. Developing and comparing models of hematopoietic-acute radiation syndrome in Göttingen and Sinclair minipigs. Int J Radiat Biol. 2021;97(sup1):S73–S87. [DOI] [PubMed] [Google Scholar]
- 16.Institute for Laboratory Animal Research : Guide for the Care and Use of Laboratory Animals. 8th ed. Washington, DC: National Academies Press, 2011. [Google Scholar]
- 17.American Veterinary Medical Association : AVMA Guidelines for the Euthanasia of Animals. 2013th ed. Schaumberg, IL: American Veterinary Medical Association, 2013. [Google Scholar]
- 18.Almond PR Biggs PJ Coursey BM, et al. AAPM's TG-51 protocol for clinical reference dosimetry of high-energy photon and electron beams. Med Phys. 1999;26(9):1847–1870. [DOI] [PubMed] [Google Scholar]
- 19.Caporaso JG Lauber CL Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1(Suppl 1)):4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson LR Sanders JG McDonald D, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straub D Blackwell N Langarica-Fuentes A, et al. Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (gene) amplicon sequencing pipeline. Front Microbiol. 2020;11:550420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed July 19, 2024.
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 24.Callahan BJ McMurdie PJ Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E Rideout JR Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Man SM. Inflammasomes in the gastrointestinal tract: infection, cancer and gut microbiota homeostasis. Nat Rev Gastroenterol Hepatol. 2018;15:721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booth C Tudor G Tudor J, et al. Acute gastrointestinal syndrome in high-dose irradiated mice. Health Phys. 2012;103(4):383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim S Jang W-S Lee S-J, et al. Development of a new minipig model to study radiation-induced gastrointestinal syndrome and its application in clinical research. Radiat Res. 2014;181:387–395. [DOI] [PubMed] [Google Scholar]
- 29.Measey TJ Pouliot M Wierzbicki W, et al. Pilot study of radiation-induced gastrointestinal injury in a hemi-body shielded göttingen minipig model. Health Phys. 2018;114(1):43–57. [DOI] [PubMed] [Google Scholar]
- 30.Measey TJ Pouliot M Wierzbicki W, et al. Expanded characterization of a hemi-body shielded göttingen minipig model of radiation-induced gastrointestinal injury incorporating oral dosing procedures. Health Phys. 2018;114(1):32–42. [DOI] [PubMed] [Google Scholar]
- 31.Kenchegowda D Legesse B Hritzo B, et al. Selective insulin-like growth factor resistance associated with heart hemorrhages and poor prognosis in a novel preclinical model of the hematopoietic acute radiation syndrome. Radiat Res. 2018;190(2):164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawford PA, Gordon JI. Microbial regulation of intestinal radiosensitivity. Proc Natl Acad Sci U S A. 2005;102:13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes A Oliveira A Soares R, et al. The effects of ionizing radiation on gut microbiota: what can animal models tell us?—a systematic review. Curr Issues Mol Biol. 2023;45(5):3877–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbonero F Mayta-Apaza AC Yu J-Z, et al. A comparative analysis of gut microbiota disturbances in the Gottingen minipig and rhesus macaque models of acute radiation syndrome following bioequivalent radiation exposures. Radiat Environ Biophys. 2018;57:419–426. [DOI] [PubMed] [Google Scholar]
- 35.Carbonero F Mayta A Bolea M, et al. Specific members of the gut microbiota are reliable biomarkers of irradiation intensity and lethality in large animal models of human health. Radiat Res. 2019;191(1):107–121. [DOI] [PubMed] [Google Scholar]
- 36.Kalkeri R Walters K Van Der Pol W, et al. Changes in the gut microbiome community of nonhuman primates following radiation injury. BMC Microbiol. 2021;21(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burmeister DM Johnson TR Lai Z, et al. The gut microbiome distinguishes mortality in trauma patients upon admission to the emergency department. J Trauma Acute Care Surg. 2020;88(5):579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M Dong Y Wu J, et al. Baicalein ameliorates ionizing radiation-induced injuries by rebalancing gut microbiota and inhibiting apoptosis. Life Sci. 2020;261:118463. [DOI] [PubMed] [Google Scholar]
- 39.Yamanouchi K Tsujiguchi T Sakamoto Y, et al. Short-term follow-up of intestinal flora in radiation-exposed mice. J Radiat Res. 2019;60(3):328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y Zhang Y Wei K, et al. Review: effect of gut microbiota and its metabolite SCFAs on radiation-induced intestinal injury. Front Cell Infect Microbiol. 2021;11:577236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y Huang Z Tang Z, et al. More than just a periodontal pathogen –the research progress on Fusobacterium nucleatum. Front Cell Infect Microbiol. 2022;12:815318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh V Ahlawat S Mohan H, et al. Balancing reactive oxygen species generation by rebooting gut microbiota. J Appl Microbiol. 2022;132(6):4112–4129. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y Fang Z Zhai Q, et al. Supernatants of Bifidobacterium longum and Lactobacillus plantarum strains exhibited antioxidative effects on A7R5 cells. Microorganisms. 2021;9(2):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu L Chen H Zhang X, et al. Rosiglitazone ameliorates radiation-induced intestinal inflammation in rats by inhibiting NLRP3 inflammasome and TNF-α production. J Radiat Res. 2020;61(6):842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoecklein VM Osuka A Ishikawa S, et al. Radiation exposure induces inflammasome pathway activation in immune cells. J Immunol. 2015;194(3):1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaga S Aziz M Murao A, et al. DAMPs and radiation injury. Front Immunol. 2024;15:1353990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azzam EI, Jay-Gerin JP, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327(1–2):48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B Jin C Li HB, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354(6313):765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alsbeih G Al-Meer RS Al-Harbi N, et al. Gender bias in individual radiosensitivity and the association with genetic polymorphic variations. Radiother Oncol. 2016;119(2):236–243. [DOI] [PubMed] [Google Scholar]
- 50.Patterson AM Vemula S Plett PA, et al. Age and sex divergence in hematopoietic radiosensitivity in aged mouse models of the hematopoietic acute radiation syndrome. Radiat Res. 2022;198(3):221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janssen S McDonald D Gonzalez A, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3(3):e00021–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bisanz JE. qiime2R: Importing QIIME2 artifacts and associated data into R sessions. Version 0.99. 2018. Available at: https://github.com/jbisanz/qiime2R. Accessed July 19, 2024.
- 53.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley statsref: statistics reference online. 2014;14:1–5. [Google Scholar]


