Abstract
STUDY QUESTION
What is the molecular landscape underlying the functional decline of human testicular ageing?
SUMMARY ANSWER
The present study provides a comprehensive single-cell transcriptomic atlas of testes from young and old humans and offers insights into the molecular mechanisms and potential targets for human testicular ageing.
WHAT IS KNOWN ALREADY
Testicular ageing is known to cause male age-related fertility decline and hypogonadism. Dysfunction of testicular cells has been considered as a key factor for testicular ageing.
STUDY DESIGN, SIZE, DURATION
Human testicular biopsies were collected from three young individuals and three old individuals to perform single-cell RNA sequencing (scRNA-seq). The key results were validated in a larger cohort containing human testicular samples from 10 young donors and 10 old donors.
PARTICIPANTS/MATERIALS, SETTING, METHODS
scRNA-seq was used to identify gene expression signatures for human testicular cells during ageing. Ageing-associated changes of gene expression in spermatogonial stem cells (SSCs) and Leydig cells (LCs) were analysed by gene set enrichment analysis and validated by immunofluorescent and functional assays. Cell–cell communication analysis was performed using CellChat.
MAIN RESULTS AND THE ROLE OF CHANCE
The single-cell transcriptomic landscape of testes from young and old men was surveyed, revealing age-related changes in germline and somatic niche cells. In-depth evaluation of the gene expression dynamics in germ cells revealed that the disruption of the base-excision repair pathway is a prominent characteristic of old SSCs, suggesting that defective DNA repair in SSCs may serve as a potential driver for increased de novo germline mutations with age. Further analysis of ageing-associated transcriptional changes demonstrated that stress-related changes and cytokine pathways accumulate in old somatic cells. Age-related impairment of redox homeostasis in old LCs was identified and pharmacological treatment with antioxidants alleviated this cellular dysfunction of LCs and promoted testosterone production. Lastly, our results revealed that decreased pleiotrophin signalling was a contributing factor for impaired spermatogenesis in testicular ageing.
LARGE SCALE DATA
The scRNA-seq sequencing and processed data reported in this paper were deposited at the Genome Sequence Archive (https://ngdc.cncb.ac.cn/), under the accession number HRA002349.
LIMITATIONS, REASONS FOR CAUTION
Owing to the difficulty in collecting human testis tissue, the sample size was limited. Further in-depth functional and mechanistic studies are warranted in future.
WIDER IMPLICATIONS OF THE FINDINGS
These findings provide a comprehensive understanding of the cell type-specific mechanisms underlying human testicular ageing at a single-cell resolution, and suggest potential therapeutic targets that may be leveraged to address age-related male fertility decline and hypogonadism.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by the National Key Research and Development Program of China (2022YFA1104100), the National Natural Science Foundation of China (32130046, 82171564, 82101669, 82371611, 82371609, 82301796), the Natural Science Foundation of Guangdong Province, China (2022A1515010371), the Major Project of Medical Science and Technology Development Research Center of National Health Planning Commission, China (HDSL202001000), the Open Project of NHC Key Laboratory of Male Reproduction and Genetics (KF202001), the Guangdong Province Regional Joint Fund-Youth Fund Project (2021A1515110921, 2022A1515111201), and the China Postdoctoral Science Foundation (2021M703736). The authors declare no conflict of interest.
Keywords: single-cell, human testicular ageing, spermatogonial stem cells, Leydig cells, base-excision repair, redox homeostasis, pleiotrophin
Introduction
The testis is an essential male reproductive organ, acting as the source of sperm and a principal provider of testosterone (Makela et al., 2019). Therefore, the testis is critical for maintaining male fertility and ensuring endocrine homeostasis. However, testicular function experiences a gradual decline as men age. Previous studies have shown that ageing negatively affects sperm parameters, sperm DNA integrity, genomic DNA mutations, chromosomal structures, and epigenetic factors (Johnson et al., 2015; Matzkin et al., 2021). Despite this, it is increasingly common for men in developed countries to delay parenthood to an older age, which raises concerns about a decline in fertilizing capacity and the consequences for the offspring’s health (Laurentino et al., 2020). Additionally, ageing detrimentally impacts testosterone synthesis, leading to male hypogonadism. This condition manifests through a variety of symptoms including diminished libido, erectile dysfunction, infertility, muscle weakness, obesity, osteoporosis, cognitive decline, and other complications (Kaufman et al., 2019; Mularoni et al., 2020). Therefore, testicular ageing not only compromises male reproductive capabilities but also profoundly influences their general health and quality of life (Matzkin et al., 2021). Consequently, it is important to elucidate the mechanisms underlying testicular ageing and to identify interventions that might slow or postpone this process.
The testis is a multifaceted organ composed of diverse cell types, including germline cells at multiple developmental stages and a variety of somatic cell types (Guo et al., 2018). The propagation of the male germline is sustained by a specialized cell population known as spermatogonial stem cells (SSCs) (Sharma et al., 2019). SSCs have unlimited self-renewal capacity and, upon induction to differentiate, give rise to a series of germ cell stages that ultimately generate sperm in a highly orchestrated manner within the seminiferous tubules (Sharma et al., 2019). The testis niche plays an essential role in regulating the survival and differentiation of the male germline (Oatley and Brinster, 2012). In the adult testis, somatic niche cells, including Sertoli cells (SCs) (Chen and Liu, 2015), Leydig cells (LCs) (Oatley et al., 2009), peritubular myoid cells (PTMs) (Chen et al., 2014), etc, provide physical and biochemical support for successful spermatogenesis from SSCs. In particular, LCs are responsible for the biosynthesis of testosterone, which acts on target cells in the testes and elsewhere to promote spermatogenesis and male-associated characteristics (Zirkin and Papadopoulos, 2018). Previous studies have demonstrated that ageing testes experience substantial morphological changes in both germline and somatic cells, resulting in diminished functionality (Jiang et al., 2014; Santiago et al., 2019). However, the cellular and molecular alterations that underlie these changes have yet to be systematically explored and remain largely unknown.
For a heterogeneous tissue such as the testis, it is difficult to accurately reveal cell type-specific changes in gene expression using conventional bulk RNA-sequencing approaches. The advent of single-cell RNA sequencing (scRNA-seq) has revolutionized our ability to analyse transcriptional variations within tissues characterized by high heterogeneity at the single-cell resolution (Guo et al., 2018; Di Persio et al., 2021). Recently, several scRNA-seq studies have begun to lift the veil on the full compendium of gene expression phenotypes and changes in spermatogenic and somatic cells, demonstrating the potency of scRNA-seq profiling as a means to study human testes at the single-cell resolution (Guo et al., 2018, 2020; Mahyari et al., 2021). These datasets have revealed the previously obscured molecular heterogeneity among and between varied testicular cell types, reinvigorating the investigation of testicular biology and pathology (Zhao et al., 2020; Alfano et al., 2021; Di Persio et al., 2021; Mahyari et al., 2021; Matzkin et al., 2021; Nie et al., 2022). The extent of the impact of ageing on different testicular cell types in humans has yet to be fully explored and the essential molecular drivers underlying the functional decline in testicular cells during ageing remain unclear.
In this study, we employed human samples to conduct a comprehensive analysis of the transcriptome profiles at the single-cell level within aged human testicular tissue. We used scRNA-seq to identify gene expression signatures for five germline cell types and six somatic cell types. We examined the age-associated transcriptional changes within each major testicular cell type in young and old tissues, and reported a set of molecular mechanisms underlying human testicular ageing. Our analysis of ageing-associated gene expression changes revealed disturbances in DNA repair in old SSCs, suggesting a potential mechanism underlying the age-related increase in de novo germline mutations. Analysis of human LCs revealed ageing-associated upregulation of oxidative stress response genes, with subsequent experiments demonstrating that antioxidant treatments alleviated the cellular dysfunction of LCs and promoted testosterone production. The analysis of cell interactions uncovered that the interrupted pleiotrophin (PTN) signalling pathway is a contributing factor for impaired spermatogenesis in testicular ageing. These data may be used at both bench and bedside in future efforts to address age-related male fertility decline and hypogonadism.
Materials and methods
Animals
Male NOD/Prkdc−/−IL-2Rg−/− (NCG) mice (8 weeks old) were purchased from GemPharmatech (Nanjing, Jiangsu, China). All mice were maintained under controlled temperature (24 ± 1°C) and relative humidity (50–60%) with a standard 12-h light-and-dark cycle for the duration of the study. All procedures were approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University (Assurance No. 2019-013).
Ethics statement for the collection of human testicular tissues
In this study, we included a total of 10 testicular tissues from young donors and 10 testicular tissues from old donors. Among the young donors, Y1 was diagnosed with a testicular teratoma and Y2 was diagnosed with a testicular LC tumour. Normal tissues were collected from areas distant from the lesions in these cases. The remaining young testicular tissues were collected from eight young male donors who underwent microdissection testicular sperm extraction surgery due to obstructive azoospermia caused by vas deferens obstruction. Histological analysis confirmed normal spermatogenesis in the testicular samples from these young donors. Additionally, the samples obtained from the young donors with obstructive azoospermia were residual tissues after retrieving sperm for intracytoplasmic sperm injection, which did not cause any additional trauma to the donors. In the old group, O1 was diagnosed with a testicular cyst, while the remaining nine old donors underwent orchiectomy for prostate cancer without undergoing androgen deprivation therapy. All of the old donors were confirmed to have offspring, indicating that they had normal reproductive function when they were young. The detailed clinical characteristics of these donors are listed in Supplementary Table S1. We subjected six testicular tissues (Young = 3 samples, Old = 3 samples) to scRNA-seq. Informed consent was obtained from all the above-listed donors. The protocols were approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University (Assurance No. 2019-148).
Tissue processing
After being collected from the operating room, the samples were transported to the laboratory on ice in storage solution (Miltenyi Biotec, Shanghai, China) within 1 h. The tunica was removed and testicular tissues were minced and washed three times with phosphate-buffered saline (PBS) to eliminate the storage solution and blood. To ensure accuracy and stability, ∼200 mg of tissue was immediately applied for scRNA-seq. For histological or immunostaining analyses, tissue samples (∼500 mg) were fixed with 4% paraformaldehyde (PFA; Thermo Fisher Scientific, Wilmington, DE, USA). For reactive oxygen species (ROS) detection in testicular tissue, samples (∼500 mg) were quickly frozen in liquid nitrogen. The remaining testis tissues were cryopreserved for functional assessments, as previously described (Guo et al., 2018).
Sample preparation for scRNA-seq
For single-cell sequencing, testicular samples were minced and subjected to a standard two-step digestion procedure (Guo et al., 2018). First, the tissues were digested with dissociation buffer including 1 mg/ml type IV collagenase (Gibco, Grand Island, NY, USA) and 200 μg/ml DNase I (Roche, Indianapolis, IN, USA) dissolved in DMEM/F-12 (Gibco) at 37°C in a water bath for 15 min. The tissues were gently pipetted against the bottom of the tube with a Pasteur pipet, passed through a 40-μm filter, washed twice with PBS, and stored temporarily at 4°C. Given that this strategy might not have isolated all cells present in the spermatogenic tubules, we redigested the samples left on the filter with 0.25% trypsin-EDTA at 37°C in a water bath for 10 min. The digestion was terminated with buffer containing 10% foetal bovine serum (FBS; Thermo Fisher Scientific, Wilmington, DE, USA), and the samples were filtered through a 40-μm filter and centrifuged. Finally, the obtained cells were combined, passed through a 40-μm filter, and resuspended in PBS. Cell numbers were counted with a Cellometer Auto T4 automated cell counter (Nexcelom Bioscience, Lawrence, MA, USA) and resuspended at 1000 cells/μl in PBS containing 0.1% BSA for single-cell sequencing.
H&E staining
Fixed tissues were embedded in paraffin and sectioned at 4 μm. The sections were deparaffinized with xylene, rehydrated with an ethanol series (100%, 95%, 85%, 75%), and stained with haematoxylin and eosin (Sigma–Aldrich, St Louis, MO, USA). Images were collected with a Leica DMi8 microscope (Leica, Wetzlar, Germany).
Masson staining
Fixed tissues were embedded in paraffin and sectioned at 4 μm. After being deparaffinized with xylene and rehydrated with an ethanol series (100%, 95%, 85%, 75%), the sections were stained overnight with potassium bichromate solution and rinsed with running tap water for 5 min. The sections were then incubated sequentially in Weigert’s iron haematoxylin working solution for 10 min, Biebrich scarlet-acid fuchsin solution for 10 min, and phosphomolybdic-phosphotungstic acid solution for 15 min. Between each step, the slides were washed in distilled water. The sections were then transferred directly (without being rinsed) to aniline blue solution and stained for 10 min. After being briefly washed with distilled water, the sections were differentiated in 1% acetic acid solution for 5 min and then rinsed with distilled water. Finally, the sections were dehydrated and mounted using resinous mounting medium. Images were collected with a Leica DMi8 microscope (Leica).
Single-cell RNA-seq library construction, sequencing, and alignment
Single-cell suspensions were loaded to a 10× Chromium Controller instrument (10× Genomics, Pleasanton, CA, USA), and ∼5000 single cells were captured using a Chromium Single Cell 3’ Library & Gel Bead Kit (V3; 10× Genomics) according to the manufacturer’s instructions. The cDNA amplification and library construction procedures were performed according to standard protocols. The resulting libraries were sequenced on the Illumina sequencing platform by LC-BIO Co., Ltd (Hangzhou, China).
Cell Ranger (Version 6.1.2) was used to process the single-cell data, align the reads, and generate feature-barcode matrices. Homo_sapiens_GRCh38_96 was used as the reference genome. Then the matrices from different samples were aggregated using the ‘aggr’ function.
Quality control, dimension reduction, clustering, and cell-type identification
Basic data processing and visualization were performed with the Seurat package (Version 4.0.2) (Hao et al., 2021). Briefly, data were loaded using the ‘Read10x’ function, and a Seurat object was built. The data were log normalized and scaled. Variable genes were identified by the ‘FindVariableGenes’ function. Next, principal component analysis (PCA) was performed, and the top 20 principal components (PCs) were used for uniform manifold approximation and projection (UMAP) dimension reduction and clustering (resolutions = 0.5). We then performed quality control with the following criteria: (i) clusters displayed over 30% mitochondrial gene expression in more than half of the cells were identified; those with high SOX9 expression were retained, while the remaining clusters were entirely removed; and (ii) cells with a total number of expressed genes >800 and <4000, total unique molecular identifier (UMI) count <120 000 and mitochondria genes <60% were retained. We pre-processed the data again and removed doublet cells with DoubletFinder (Version 2.0.3). The 22 520 cells that remained were taken as having passed quality control and were used for subsequent analyses.
After quality control, data matrices from different donors were integrated by canonical correlation analysis (CCA) (Stuart et al., 2019). Briefly, the data were integrated using the top 4000 variable genes as integration anchors. Then, the first 30 PCs were used for UMAP dimension reduction, to construct a k-nearest neighbour (kNN) graph and to refine the edge weights between any two cells. The cells were then clustered using the Louvain algorithm for modularity optimization with the resolution parameter set to 0.6. Cell types were identified using the indicated marker genes (see Results section). Marker genes for each cluster were determined with ROC analysis using the ‘FindAllMarkers’ function. Only genes with |avg_logFC|>0.4, min.pct = 0.5, and P-value <0.05 were considered as marker genes. Gene enrichment analysis of cell type-specific markers, based on the Gene Ontology (GO) database, was carried out by employing clusterProfiler (Version 4.0.5).
Analysis of coefficient of variations
To assess the impacts of ageing on germline and somatic cells, we calculated age-related coefficient of variations (CVs) as previously described (Wang et al., 2020). Using the ‘FindVariableFeatures’ function, 3438 highly variable genes (HVGs) were identified among the top 10% of 34 378 total genes and were subsequently used for downstream ageing-associated transcriptional variation analysis.
For a given cell type, c, we defined the cell-paired-distance d for HVG x between the cells in young (denoted as i) and old groups (denoted as j) as:
where y and o are the cell numbers in the young and old groups, respectively, of cell type c.
Next, we calculated the arithmetic mean ( and SD () of dc, x. Consequently, the CV of cell-paired-distance, or transcriptional noise, can be represented as:
Profiling of differentially expressed genes
The function ‘FindMarkers’ in the R package, Seurat, which is based on the Wilcoxon rank-sum test, was used to identify differentially expressed genes (DEGs). Genes with an average log2-transformed difference >0.25 and P-value <0.05 were considered to be ageing-associated DEGs.
Gene set enrichment analysis
Enrichment analysis between the young and old groups was performed based on gene set enrichment analysis (GSEA) using the gseGO function in clusterProfiler (Version 4.0.5). Genes were ranked according to fold change, and the gene fold-change data frame was used as the input. The ‘ont’ parameter was set to ‘ALL’, indicating the simultaneous utilization of ‘BP’, ‘MF’, and ‘CC’ subontologies for analysis. The ‘OrgDb’ parameter employed ‘org.Hs.eg.db’, and the ‘keyType’ was set to ‘ENTREZID’. The remaining parameters were set to their default values. We filtered the obtained GO items with adjusted P-value <0.05 and |normalized enrichment score (NES)|>1. The results were visualized using ggplot2 (Version 3.3.5).
Analysis of gene regulation networks
Gene regulation network analysis was performed based on the single-cell regulatory network inference (SCENIC) workflow using default parameters. First, the quality-controlled, log-transformed UMI count matrix and transcription factors (TFs) were loaded as input. For the UMI count matrix, the DEGs between age groups were depicted as row names, and the cell barcodes of each cell type were represented as column names. Second, the correlation matrix of genes was constructed for network inference using the random forest-based algorithm applied by GENIE3 (Version 1.12.0) (Huynh-Thu et al., 2010). Reference TFs were downloaded from RcisTarget (https://resources.aertslab.org/cistarget). Third, based on the RcisTarget database, coexpression modules enriched for target genes of each candidate TF were detected by SCENIC (Version 1.2.4). The activity of each TF module in each cell was computed by AUCell (Version 1.12.0), and the regulation networks of TF modules with high NESs were visualized by Cytoscape (Version 3.8.2).
AUCell
To score individual cells for pathway activities, we used the R package AUCell (Version 1.12.0). First, a log-normalized expression matrix was used as input to compute gene-expression rankings in each cell by applying the ‘AUCell_buildRankings’ function with default parameters. The genes in each gene set are listed in Supplementary Table S2. The activity of canonical pathway gene sets for each cell was then computed by scoring the area under the curve (AUC) values based on gene expression rankings using the ‘AUCell_calcAUC’ function. The score of each cell was visualized with the R package application ggplot2 (Version 3.3.5).
Cell trajectory analysis
Pseudotime trajectory analysis was conducted using monocle3 (Version 1.3.1). The UMI count matrix and metadata were extracted from the quality-controlled Seurat object and loaded into monocle3 using the ‘new_cell_data_set’ function. The resulting monocle3 object was used for ‘UMAP’ dimensionality reduction. We then used the ‘learn_graph’ function to infer the developmental trajectory of the cells and the ‘order_cells’ function to define the root cell.
Cell–cell communication analysis
The analysis of cell–cell communication by ligand–receptor pairs was performed using CellChat (Version 1.1.3). First, the normalized data matrix and metadata were extracted from the quality-controlled Seurat object and loaded. The ‘secreting signal’ of CellChatDB was used as the ligand–receptor interaction database. Once the overexpressed genes were identified, the between-cell interaction pairs and interaction probabilities for each ligand–receptor pair were calculated. Then, the communication probability on a signalling pathway level was calculated by summarizing all related ligands/receptors using the function ‘computeCommunProbPathway’. Finally, the young and old CellChat objects were merged for comparison.
Immunofluorescence staining
Human testicular tissues were embedded in paraffin and sectioned at 4 µm. After being deparaffinized by xylene and rehydrated with an ethanol series (100%, 95%, 85%, 75%) at room temperature, the sections were incubated in citrate antigen-retrieval solution (Beyotime, Shanghai, China) in a hot water bath (96°C) for 20 min. For intracellular protein detection, the sections were permeabilized with 0.5% Triton X-100 (Sigma–Aldrich, St Louis, MO, USA) for 20 min. The tissue sections were then incubated with 3% BSA for 60 min at room temperature and incubated with primary antibodies overnight at 4°C. Following incubation, the tissue sections were rinsed five times with PBS and incubated with secondary antibodies for 60 min at room temperature. After being rinsed five times with PBS, the tissue sections were stained with 4,6-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA, USA) for 5 min, after which the DAPI was removed and replaced with mounting medium (DAKO; Glostrup, Denmark). The specific fluorescence was visualized and photographed using an LSM800 confocal microscope (Zeiss, Jena, Germany) or a Leica DMi8 microscope (Leica). The utilized primary and secondary antibodies are listed in Supplementary Table S3.
The cultured human testicular tissues were fixed in 4% PFA (Phygene, Fujian, China), dehydrated with 30% sucrose (Sangon Biotech, Shanghai, China), and sectioned at a thickness of 10 μm. Then, the samples were permeabilized with 0.3% Triton X-100 (Sigma–Aldrich) for 20 min. Following permeabilization, the samples were washed three times with PBS. Next, the samples were blocked with 3% BSA (Sigma–Aldrich) for 60 min at room temperature and incubated with primary antibodies at 4°C overnight. After incubation, the samples were washed three times with PBS and incubated with fluorescent secondary antibodies in the dark for 60 min at room temperature. Finally, the samples were stained with DAPI (Invitrogen) for 5 min at room temperature. Specific fluorescence was visualized under a Leica DMi8 microscope (Leica). The primary and secondary antibodies used are listed in Supplementary Table S3.
Isolation and culture of LCs
Primary human LCs were isolated from human testicular tissues as previously described (Luo et al., 2021). In brief, cryopreserved testicular tissues were thawed quickly and washed three times with PBS. Then, the testes were mechanically cut and enzymatically dissociated using 1 mg/ml type IV collagenase and 200 μg/ml DNase I dissolved in DMEM/F-12 for 20 min with slow shaking (100 cycles/min) at 37°C. The samples were filtered through a 40-μm filter and centrifuged at 300g for 4 min. The cell pellets were rinsed twice with PBS and resuspended in PBS containing 0.1% BSA for fluorescence-activated cell sorting (FACS) (MoFlo Astrios EQs, Beckman Coulter, CA, USA). Before LCs isolation, we used a two-step gating strategy to eliminate debris and cell doublets based on the side scatter and forward scatter parameters. Then, the cell samples were analysed in the combined fluorescence channels (405–448 and 640–671 nm) of flow cytometry, and the LCs population, distinguished from the main group, was isolated.
The obtained primary LCs were quantified by cell counting plate, and ∼20 000 cells were seeded per well in a 12-well plate. Then, LCs were cultured in medium containing DMEM/F12, 10% FBS, and 1% insulin–transferrin–sodium selenite (ITS; Thermo Fisher Scientific) at 35°C with 5% CO2. The ability of the cells to produce testosterone was assessed after 3 h of incubation with DMEM/F12 containing 0.1% BSA, 1 IU/ml hCG (R&D, Systems, Minneapolis, USA), 10 μM 22-HC (Sigma–Aldrich), and 1× ITS. The cell supernatants were collected and stored at −80°C until analysis. Testosterone production of LCs was standardized by cell number per well.
Culture of testicular tissues for testosterone measurement
For short-term tissue culture, we modified a previously described method (Li et al., 2016). Briefly, cryopreserved testicular tissues were thawed quickly and washed three times with PBS. Then, the testes were mechanically cut into small pieces (2–4 mm2 in size). Three pieces of tissues were plated per well of a 12-well plate and cultured in medium containing DMEM/F12, 0.1% BSA, and 1× ITS (Gibco) at 35°C with 5% CO2.
The ability of the tissues to produce testosterone was assessed after 3 h of incubation with DMEM/F12 (Gibco) containing 0.1% BSA, 1 IU/ml hCG (R&D), 10 μM 22-HC (Sigma–Aldrich), and 1× ITS (Gibco). The supernatants to be used for the testosterone assay were collected and stored at −80°C until analysis. Tissues were collected and lysed in cold RIPA buffer, followed by protein quantification for statistical analysis.
Antioxidant treatments in vitro
To elucidate the protective effect of antioxidants on cellular oxidative stress and testosterone production, primary LCs or testicular tissues from the old group were cultured with vehicle, 10 mM N-acetyl-L-cysteine (NAC; MedChemExpress, Shanghai, China), and 50 μM vitamin E (VE, MedChemExpress). After a 24-h culture period, LCs or tissues were used for subsequent analysis.
Renal subcapsular transplantation and antioxidant treatments in vivo
Male NCG mice (9 weeks old) were castrated one week before renal subcapsular transplantation. In brief, dorso-lateral incisions were made under anaesthesia to expose both kidneys of the castrated mice. A small incision was created in the kidney capsule, and the capsule was delicately elevated. An equal mass of testicular tissue (∼50 mg) mixed with Matrigel (Corning Life Sciences, USA) was transplanted under the capsule of each kidney. Then the wound was closed by absorbable surgical sutures, and moxifloxacin was administered to prevent infection. Subsequently, these mice were randomly assigned to one of three groups: the control group, the NAC group, and the VE group. Thereafter, mice in each group received daily intraperitoneal injection of either the vehicle, NAC (300 mg/kg, MedChemExpress), or VE (100 mg/kg, MedChemExpress) for 7 days. Then, all animals were sacrificed, and the subcapsular grafts were collected for H&E staining and quantitative RT-PCR analysis, and the serum was collected for testosterone detection.
Testosterone measurements
Testosterone concentrations were assayed as previously reported by our group (Xia et al., 2022). The cell or tissue supernatants were collected at the indicated timepoints and stored at −80°C until analysis. Testosterone levels were measured using a chemiluminescent immunoassay (CLIA) system (Architect system; Abbott GmbH & Co. KG, Germany). The coefficient of variation of this CLIA system is 1.9–5.1% for intra-assay precision and 2.5–5.2% for inter-assay precision. The lowest detectable dose of testosterone was 0.01 ng/ml.
ROS detection
ROS production in LCs of human testicular samples was detected using a reactive oxygen species assay kit (Thermo Fisher Scientific). Concurrently, LCs were marked by Boron-dipyrromethene-fluorecein (BODIPY-FL, Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, human testicular samples were rapidly frozen in liquid nitrogen and cryosectioned at 10 μm thickness using a frozen slicer (Leica CM1950). Then, the slices were washed three times with PBS, and incubated with PBS containing 1 μM dihydroethidium-phycoerythrin (DHE-PE) and 1 μM BODIPY-FL for 30 min at 37°C in the dark. Nuclei were counterstained with DAPI (Invitrogen). Specific fluorescence was visualized under a Leica DMi8 microscope (Leica).
ROS production in primary LCs was detected using the reactive oxygen species assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, primary LCs were isolated by FACS and incubated with DMEM/F12 medium containing 1 μM DHE for 30 min at 37°C in the dark, and fluorescence intensity (Ex = 495 nm, Em = 520 nm) was measured by flow cytometry (CytoFLEX; Beckman Coulter, CA, USA) or photographed under a Leica DMi8 microscope (Leica).
RNA extraction, cDNA synthesis, and quantitative RT-PCR
Total RNA of transplanted human testicular samples was extracted by a RNeasy Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. The purity of RNA was determined by a NanoDrop 1000 (Thermo Fisher Scientific) and reverse transcription was carried out by a NovoScript®1st Strand cDNA Synthesis Kit (Novoprotein, Shanghai, China). Furthermore, quantitative RT-PCR was performed by LightCycler®480 SYBR Green I Master (Roche, Indianapolis, IN, USA) and signals were detected in a Light Cycler 480 Detection System (Roche). A melting curve for primer validation was generated to confirm a single peak and rule out the non-specific product or primer dimer formation. All expression levels of target genes were calculated by the △Ct method and expressed relative to GAPDH. The primer sequences were listed in Supplementary Table S4.
Culture of human testicular tissues for spermatogenesis evaluation
For spermatogenesis evaluation, human testicular tissues were cultured as previously reported (Song et al., 2022). Briefly, human testicular tissues were bluntly dissected using sterile microforceps. Then, equal mass of seminiferous tubules was placed on 12-well plates and cultured in the medium consisting of StemPro-34 serum free medium with StemPro supplement (Gibco), 1% Knockout serum replacement (Gibco), 0.1 mM non-essential amino acid (Gibco), 2 mM Glutamax (Gibco), 5 mg/ml BSA (Sigma–Aldrich), and 1% N2 supplement (Thermo Fisher Scientific) at 35°C with 5% CO2. The medium was changed every other day. To investigate the role of PTN, human testicular tissues from young donors were incubated with either 2.5 μg/ml PTN antibody (R&D) or 2.5 μg/ml IgG (R&D). Additionally, testicular tissues from old donors were incubated with either 50 ng/ml recombinant PTN protein (R&D) or PBS. All samples were incubated in 35°C for 4 days, and then these tissues were collected for histological analysis.
Statistical analysis
Bioinformatics data were subjected to statistical analysis using Student’s t-tests in R (Version 4.0.3), with P or adjusted P values presented in each figure. The statistical analysis for age-dependent CV differences between different cell types was processed with SPSS 26.0 Software (IBM, Armonk, NY, USA) using the Kruskal–Wallis test. The validation data were statistically analysed using GraphPad Prism v8 (GraphPad Software, La Jolla, CA, USA). Statistical differences between samples were assessed with Student’s t-tests or one-way ANOVA. In all figures, differences were considered significant when P < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Construction of a single-cell transcriptomic atlas of the human testis
To investigate human testicular ageing, we obtained human testicular biopsies from 10 young individuals (24–31 years old) and 10 old individuals (61–87 years old) (Supplementary Table S1). Histologically, the area occupied by seminiferous tubules was significantly decreased in old men compared with that in young men (Supplementary Fig. S1A). In addition, age-related thickening of boundary tissue was typically found in old testicular tissues (Supplementary Fig. S1B). These data are in agreement with previous observations (Johnson et al., 1988; Mularoni et al., 2020) and confirmed that this cohort of tissues could be used to examine age-related changes in testicular function.
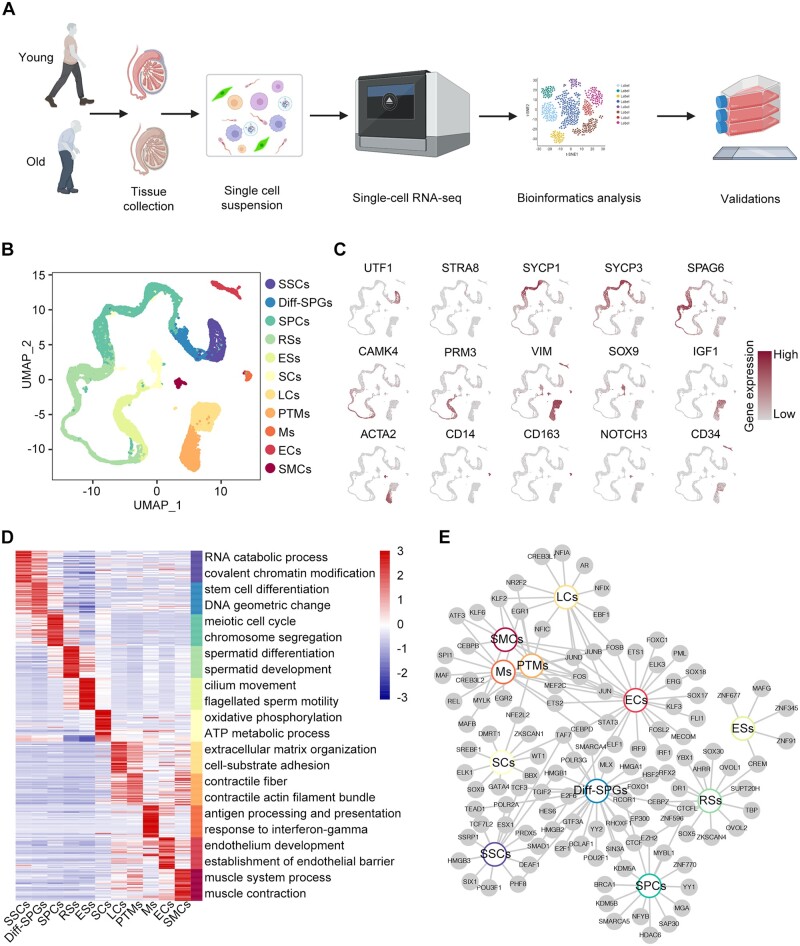
To profile gene expression changes specific to each cell type during testicular ageing at single-cell resolution, we performed scRNA-seq on young (Y1, Y2, and Y3) and old (O1, O2, and O3) testicular biopsies using the 10× Genomics platform (Fig. 1A). Data from 11 444 and 11 076 cells passed quality control and were included in the subsequent analysis for young and old samples, respectively (Supplementary Fig. S2A–E; Supplementary Table S5). UMAP was applied to visualize the comprehensive landscape of human testicular cells, identifying 11 distinct cell types based on the expression of specific marker genes (Guo et al., 2018, 2021; Alfano et al., 2021): SSCs (UTF1+, ID4+), differentiating spermatogonia (Diff-SPGs; STRA8+, DMRT1+), spermatocytes (SPCs; SYCP1+, SYCP3+, and DAZL+), round spermatids (RSs; SPAG6+, CAMK4+), elongating spermatids (ESs; PRM3+, HOOK1+, and TNP1+), SCs (SOX9+, AMH+, and WT1+), LCs (IGF1+, CFD+, and LUM+), PTMs (ACTA2+, MYH11+, NOTCH3−), macrophages (Ms; CD14+, CD163+, and C1QA+), endothelial cells (ECs; CD34+, NOSTRIN+), and smooth muscle cells (SMCs; ACTA2+, MYH11+, NOTCH3+) (Fig. 1B and C; Supplementary Fig. S2F). Investigation of the top 30 marker genes uncovered distinctive transcriptional signatures and enriched pathways across different cell types, aligning with their unique biological functionality (Fig. 1D; Supplementary Table S5). For example, SPCs showed enrichment in GO terms related to ‘meiosis’, while PTMs exhibited enrichment in terms associated with ‘myofiber’ functions (Fig. 1D).
Figure 1.
Construction of a single-cell transcriptomic atlas of human testis. (A) Flow chart of scRNA-seq and bioinformatics analysis of the human testis. (B) UMAP plot showing the distribution of 11 different cell types in the human testis, with annotation. (C) UMAP plot showing the expression profiles of the indicated cell type-specific marker genes for the assessed cell types in the human testis. The colour key, ranging from grey to brown, indicates low to high gene expression levels, respectively. (D) Heatmap showing the expression profiles of the top 30 cell type-specific marker genes of different cell types, with their enriched functional annotations on the right. The value for each gene represents scaled data. (E) Network plot showing transcriptional regulators of cell type-specific marker genes (P-value < 0.05, |logFC| > 0.4, min.pct = 0.5) for different cell types in the human testis. UMAP, uniform manifold approximation and projection; SSCs, spermatogonial stem cells; Diff-SPGs, differentiating spermatogonia; SPCs, spermatocytes; RSs, round spermatids; ESs, elongating spermatids; SCs, Sertoli cells; LCs, Leydig cells; PTMs, peritubular myoid cells; Ms, macrophages; ECs, endothelial cells; SMCs, smooth muscle cells.
We further established a regulatory network for TFs, highlighting core TFs exclusive to individual cell types alongside hub TFs that are commonly shared across at least two cell types (Fig. 1E; Supplementary Table S5). Prominent genes within the cell type-specific TFs networks included TCF7L2 for SSCs, SOX9 for SCs, and CREB3L1 for LCs. TCF3 was identified in the networks of both SSCs and Diff-SPGs, suggesting that TCF3 plays essential roles in spermatogonia maintenance and specification. FOS was identified in LCs, PTMs, Ms, ECs, and SMCs, indicating that it functions as a broad-acting transcriptional regulator (Fig. 1E).
We next endeavoured to explore the cell type-specific alterations in gene expression patterns associated with ageing. Intriguingly, our findings revealed comparable transcriptional profiles of marker genes across each cell type between the young and old groups (Supplementary Fig. S3A), suggesting that the ageing process exerts minimal effect on cell identity. In addition to these classic markers, we identified a set of novel markers of testicular cells: SIX1 and LIN7B for SSCs, BEND2 for SPCs, ABHD4 and TCEAL5 for SCs, KRT17 for PTMs, ADAP2 for Ms, PCAT19 and LCN6 for ECs, and RRAD and RERGL for SMCs (Supplementary Fig. S3B).
Ageing-related cellular alterations along the trajectories of spermatogenesis
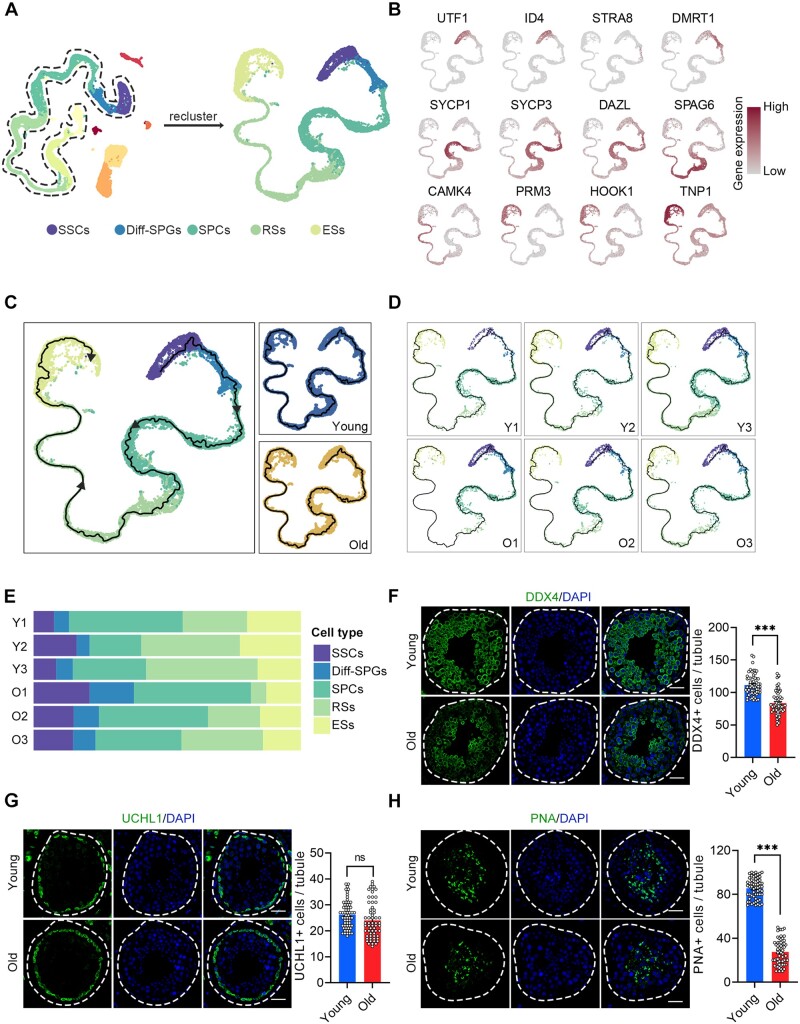
We next performed a focused analysis of age-associated changes in germ cell clusters. To investigate the cellular composition of germ cells, we identified the following five broad developmental stages based on their marker genes: SSCs (UTF1+, ID4+), Diff-SPGs (STRA8+, DMRT1+), SPCs (SYCP1+, SYCP3+, and DAZL+), RSs (SPAG6+, CAMK4+), and ESs (PRM3+, HOOK1+, and TNP1+) (Fig. 2A and B). Subsequently, we inferred spermatogenesis trajectories using orthogonal pseudotime analysis with monocle3 (Cao et al., 2019). Germ cells were parsed by developmental stage to examine their relative compositions at different ages. These analyses revealed complete spermatogenesis in all samples (Fig. 2C–E). However, the proportions of germ cells at the RSs stage and beyond tended to decline with age (Fig. 2E), indicating that germ cell differentiation was heterogeneously impacted by ageing. For validation, we stained DEAD-box helicase 4 (DDX4), a germ cell marker, and found reduced signals in the testes of the old group (Fig. 2F). Further analysis revealed that the numbers of UCHL1+ SSCs were equivalent between the young and old groups (Fig. 2G), indicating that alterations in SSCs quantity may not significantly contribute to the ageing-associated deficiency of spermatogenesis. The number of peanut agglutinin (PNA)+ RSs and ESs was significantly lower in the testes of old versus young individuals (Fig. 2H). This indicates that germ cell differentiation was markedly impacted by ageing, corroborating findings from earlier histological studies (Paniagua et al., 1991; Kimura et al., 2003).
Figure 2.
Ageing-related cellular alterations along the trajectories of spermatogenesis. (A) UMAP projection of germ cells reveals the developmental progression of spermatogenesis in the young and old human testis. (B) Expression patterns of known spermatogenic markers projected onto the UMAP plot. The colour key, ranging from grey to brown, indicates low to high gene expression levels, respectively. (C) Pseudotime ordering of germ cells by monocle3 in UMAP space. Cells are coloured according to the cell type and group. Arrows indicate the developmental stages from SSCs to ESs. (D) Pseudotime ordering of germ cells split by the donors of origin. (E) Relative proportions of cells at different spermatogenic stages in the samples analysed. (F) Immunostaining and quantification of DDX4 in young and old human testis sections. Left, representative image of DDX4 in human testis sections. Right, quantification of the number of DDX4-positive cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. (G) Immunostaining and quantification of UCHL1 in young and old human testis sections. Left, representative image of UCHL1 in human testis sections. Right, quantification of UCHL1 + cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ns = not significant. (H) Immunostaining and quantification of PNA in young and old human testis sections. Left, representative image of PNA in human testis sections. Right, quantification of PNA + cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. Y, young; O, old.
Ageing-related molecular alterations along the trajectories of spermatogenesis
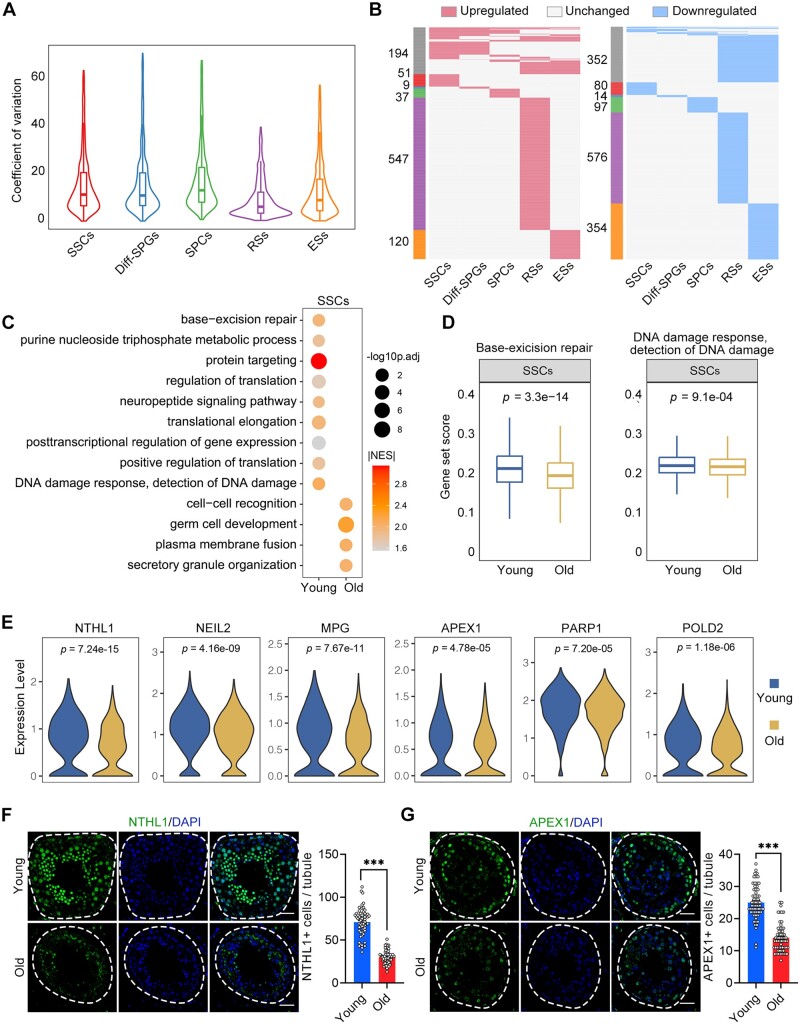
We next focused on transcriptional alterations linked to ageing within the identified germline cell types. Ageing has been associated with elevated transcriptional noise (Nikopoulou et al., 2019). Here, calculation of the age-related CV revealed that SPCs, Diff-SPGs, and SSCs exhibited higher transcriptional noise than later-stage germline cells (Fig. 3A; Supplementary Table S6), revealing that ageing triggered greater variability in early-stage germline cells compared to their late-stage counterparts. We further identified 174, 102, 90, 644, and 214 upregulated genes and 112, 46, 127, 891, and 665 downregulated genes (|avg_logFC|>0.25 and P-value <0.05) in the SSCs, Diff-SPGs, SPCs, RSs, and ESs germline subtypes, respectively, in the old versus young groups (Fig. 3B; Supplementary Table S7). Notably, only ∼22% of the DEGs overlapped between two or more cell populations, with the majority of DEGs being specific to particular cell types.
Figure 3.
Ageing-related molecular alterations along the trajectories of the spermatogenesis. (A) CV analysis showing the ageing-associated transcriptional noise in germ cells. (B) Heatmaps showing the distribution of upregulated (red) and downregulated (blue) DEGs (log2FC > 0.25, min.diff.pct = 0.1, P-value < 0.05) between old and young germ cells. Genes that are not differentially expressed are indicated in grey, and the numbers of DEGs are indicated. The grey bars on the left of the heatmaps denote DEGs shared by at least two cell types, and the others are cell-specific DEGs. (C) Representative GO terms of DEGs in old and young human SSCs. Dot size indicates the range of −log10(adjusted P-value). Colour keys, ranging from grey to orange to red, indicate the absolute value of the NES. (D) Gene set score analysis for the ‘BER’ and ‘DNA damage response, detection of DNA damage’ signalling pathways of SSCs from the young and old groups. Wilcoxon rank sum test was used; P-value is indicated. (E) Violin plot showing the expression levels of BER-associated genes for SSCs in young and old samples. (F) Immunostaining and quantification of NTHL1 in young and old human testis sections. Left, representative image of NTHL1 in human testis sections. Right, quantification of NTHL1-positive cells in seminiferous tubules of young and old human testis sections. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. (G) Immunostaining and quantification of APEX1 in young and old human testis sections. Left, representative image of APEX1 in human testis sections. Right, quantification of APEX1+ cells in seminiferous tubules of young and old human testis sections. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. CV, coefficient of variation; NES, normalized enrichment score; GO, gene ontology; DEGs, differentially expressed genes; BER, base-excision repair.
We therefore performed GSEA analysis based on the GO database to explore the ageing-associated alterations in the cellular functions of each germ cell type (Fig. 3C; Supplementary Fig. S4A–D; Supplementary Table S8). Given SSCs represent the sole germline stem cells and play essential roles in long-term spermatogenesis (Di Persio et al., 2021), we delved into the ageing-associated changes in gene expression pattern of SSCs. We observed that the genes downregulated in SSCs of the old group were linked to ‘base-excision repair (BER)’ and ‘DNA damage response, detection of DNA damage’ (Fig. 3C), which are crucial DNA repair mechanisms for addressing DNA damage (Ray Chaudhuri and Nussenzweig, 2017). Consistently, the gene set scores for ‘BER’ and ‘DNA damage response, detection of DNA damage’ signalling pathways were lower in the old SSCs than in the young SSCs (Fig. 3D; Supplementary Table S2). Furthermore, several BER-promoting genes, such as NTHL1, NEIL2, MPG, APEX1, PARP1, and POLD2, exhibited transcriptional downregulation in SSCs obtained from old testes (Fig. 3E). Corresponding to these observed changes in mRNA levels, immunofluorescence staining confirmed that the protein levels of NTHL1 and APEX1 were decreased in testicular tissues of the old group compared to the young group (Fig. 3F and G), further supporting the idea that the DNA repair function of SSCs is compromised during human testicular ageing.
Variations in the transcriptional landscapes of somatic cells during human testicular ageing
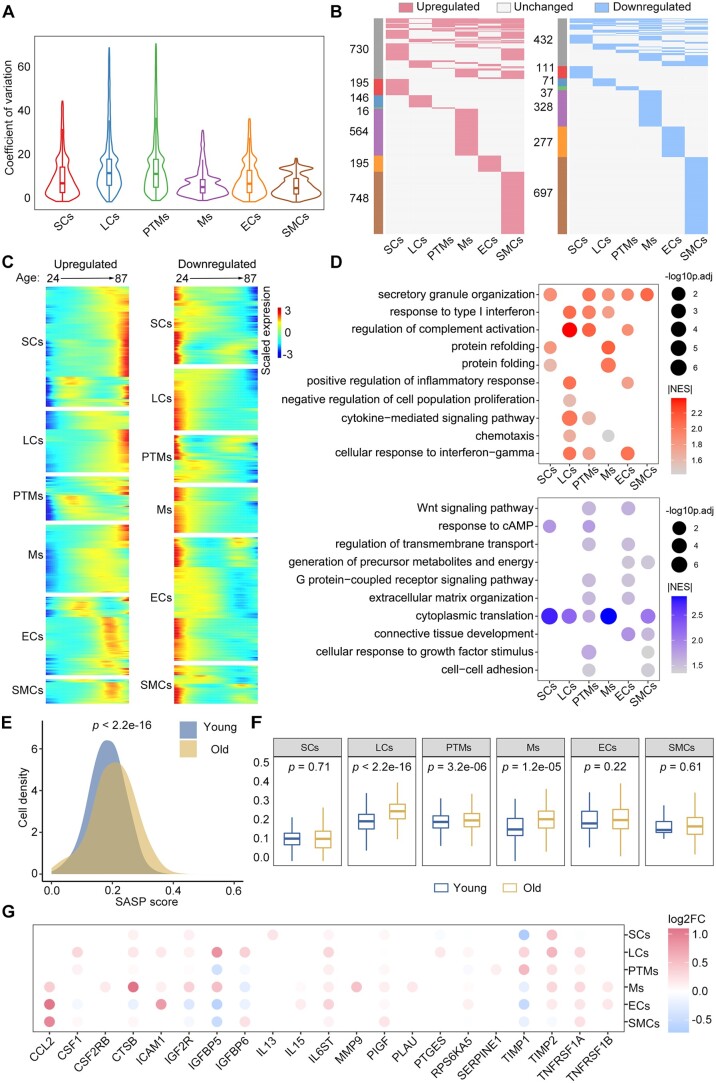
We next quantified the main somatic cell types, including LCs, SCs, and PTMs, in young and old testes by immunofluorescence analysis (Supplementary Fig. S5A–C). Interestingly, we observed no significant differences in the quantities of these cell types between the testes of young and old individuals. To further explore the mechanisms of testicular somatic ageing at the cellular level, we compared the gene expression patterns in somatic cells between the groups. By calculating the age-relevant CV (Wang et al., 2020), we found that the ageing-accumulated transcriptional noise was significantly higher in LCs and PTMs than in the other somatic cell types (Fig. 4A). This finding suggested that LCs and PTMs may be more vulnerable to age-related stress. We next explored ageing-associated DEGs in somatic cells between the groups. Thousands of genes were identified as DEGs (|avg_logFC|>0.25 and P-value <0.05) across at least one somatic cell type in the human testis during ageing process (Fig. 4B). Further analysis enabled us to identify 621, 316, 228, 944, 447, and 1274 upregulated genes and 280, 225, 171, 567, 447, and 913 downregulated genes in SCs, LCs, PTMs, Ms, ECs, and SMCs, respectively, in the old versus young testes (Fig. 4B; Supplementary Table S7). Notably, only ∼26% of the DEGs overlapped between two or more cell populations, indicating that the effects of ageing are largely somatic cell type-specific in this setting.
Figure 4.
Changes in the transcriptional profiles of somatic cells during human testicular ageing. (A) CV analysis showing ageing-associated transcriptional noise in somatic cells. (B) Heatmaps showing the distribution of upregulated (red) and downregulated (blue) DEGs (|log2FC| > 0.25, min.diff.pct = 0.1, P-value < 0.05) between old and young human somatic cells. Genes not differentially expressed are in grey, and the numbers of DEGs are indicated. (C) Heatmaps showing the age-related upregulated (left) and downregulated (right) DEGs of somatic cell types. The colour key, ranging from blue to red, indicates low to high gene expression levels. (D) Representative shared GO terms of age-related upregulated (top) and downregulated (bottom) DEGs in different somatic cell types. Dot size indicates the range of −log10(adjusted P-value). The colour keys, ranging from grey to red (top) or from grey to blue (bottom), indicate the absolute values of the NES. (E) Density plot showing the distribution of cells with different SASP scores in the young and old groups. The Wilcoxon rank sum test was used; P-value is indicated. (F) Box plot showing SASP scores in different types of somatic cells. Wilcoxon rank sum test was used; P-values are indicated. (G) Dot plot showing the log2-transformed fold change of SASP-related genes in somatic cells from young versus old groups. Only genes showing a statistically significant difference between the young and old groups are shown. Dot colour indicates the log2-transformed fold change. CV, coefficient of variation; GO, gene ontology; DEGs, differentially expressed genes; NES, normalized enrichment score; FC, fold change; SASP, senescence-associated secretory phenotype.
To uncover DEGs that show consistent increases or decreases in somatic cells with ageing, we arranged datasets of individual samples according to chronological age and analysed the expression profiles of 2594 DEGs that were upregulated with age and 1953 DEGs that were downregulated with age in six somatic cell types (Fig. 4C). GSEA analysis revealed that the genes upregulated with age were mainly associated with cytokine and stress responses, unfolded proteins, and inflammatory response (Fig. 4D; Supplementary Table S8). In comparison, the genes downregulated with age were mainly related to translation, response to growth factors, and tissue development (Fig. 4D; Supplementary Table S8). As a ubiquitous characteristic of senescent cells, the senescence-associated secretory phenotype (SASP) typically gives rise to a low-grade inflammatory state (Baker and Petersen, 2018). We questioned whether the somatic niche in the old testis might present an elevated SASP environment. Indeed, our analysis revealed that somatic cells exhibited much higher SASP gene set scores (Fig. 4E; Supplementary Table S2). Notably, LCs, PTMs, and Ms showed elevated SASP scores with age (Fig. 4F and G), indicating that these cell types had strong contributions to the age-related inflammatory state in the human testes.
Next, we performed an integrative comparative analysis of previously identified ageing-associated DEGs (Fig. 4B; Supplementary Table S7) alongside genes related to ageing or longevity from the GenAge database (Tacutu et al., 2018), and further found that many ageing/longevity-associated genes were differentially expressed in one or more somatic cell types (Supplementary Fig. S5D). For instance, CDKN1A, a known marker of senescent cells, was found to be upregulated in LCs, PTMs, and ECs, whereas several cell survival-related genes (PLCG2, IGFBP2, and GSTP1) were downregulated in three or more somatic cell populations during testicular ageing (Supplementary Fig. S5D).
Dysfunction of LCs during human testicular ageing
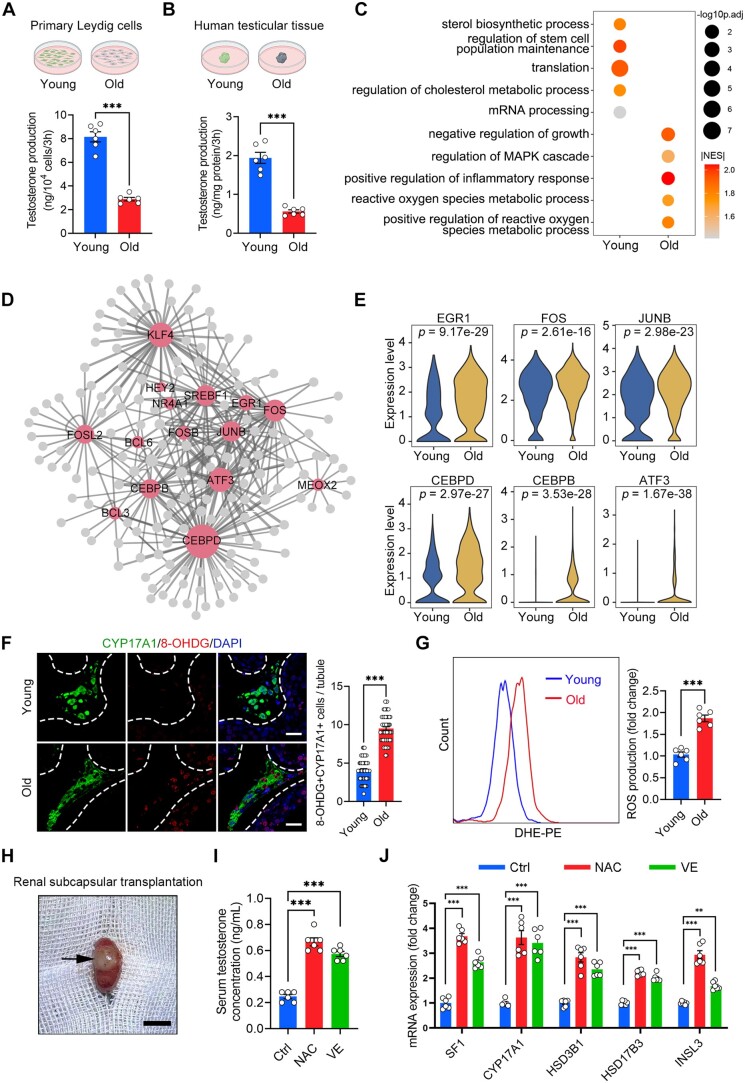
LCs produce testosterone and are thus critical for reproductive function and general health (Zirkin and Papadopoulos, 2018). LCs dysfunction causes testosterone deficiency, arrested spermatogenesis, and infertility (Xia et al., 2022). For functional validation, we isolated primary LCs from human testicular tissues of both groups (Supplementary Fig. S6A and B). Assessment of testosterone levels in the medium revealed that LCs from the young group produced significantly more testosterone than those from the old group (Fig. 5A). Consistently, when culturing small fragments of testicular tissue in vitro, we observed significantly reduced testosterone production in the old group compared to the young group (Fig. 5B). These results demonstrate that the fundamental function of LCs appears to become compromised during ageing.
Figure 5.
Dysfunction of Leydig cells during human testicular ageing. (A, B) Quantification of testosterone production in primary LCs (A) and human testicular tissue (B). Young, n = 6 samples; Old, n = 6 samples. Data are expressed as mean ± SEM. Significance was determined by Student’s t-test. ***P < 0.001. (C) Representative GO terms of DEGs in old and young human LCs. Dot size indicates the range of −log10(adjusted P-value). Colour keys, ranging from grey to orange to red, indicate the absolute values of the NES. (D) Network visualization of potential key transcriptional regulators of upregulated DEGs in LCs. Node size is positively correlated with the number of directed edges. Edge width is positively correlated with the NES. Transcription factors are highlighted in red; others are highlighted in grey. (E) Violin plot showing the expression levels of age-associated upregulated transcriptional factors. Wilcoxon rank sum test was used; P-value is indicated. (F) Immunostaining and quantification of 8-OHDG and CYP17A1 in young and old human testis sections. Left, representative image of 8-OHDG and CYP17A1 in human testis sections. Right, quantification of the proportion of 8-OHDG+ LCs (CYP17A1+). Scale bars, 25 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. (G) Quantification of ROS production in young and old primary LCs, as measured by flow cytometry. Young, n = 6 samples; Old, n = 6 samples. Data are expressed as mean±SEM. Significance was determined by Student’s t-test. ***P < 0.001. (H) Representative image of renal subcapsular transplantation with human testicular tissue. Arrows in the figure indicate the grafts. Scale bars, 1 cm. (I) Quantification of serum testosterone levels in Ctrl and antioxidant-treated (NAC and VE) groups. Ctrl, n = 6 samples; NAC (300 mg/kg), n = 6 samples; VE (100 mg/kg), n = 6 samples. Data are expressed as mean±SEM. Significance was determined by one-way ANOVA. ***P < 0.001. (J) Quantification of expression of steroidogenesis-related genes in the Ctrl and antioxidant-treated (NAC and VE) groups. Ctrl, n = 6 samples; NAC (300 mg/kg), n = 6 samples; VE (100 mg/kg), n = 6 samples. Data are expressed as mean±SEM. Significance was determined by one-way ANOVA. ***P < 0.001. GO, gene ontology; DEGs, differentially expressed genes; NES, normalized enrichment score; LCs, Leydig cells; ROS, reactive oxygen species; DHE-PE, Dihydroethidium-phycoerythrin; Ctrl, control group; NAC, N-acetylcysteine group; VE, vitamin E group.
To further characterize the ageing-associated changes of gene expression in LCs during human testicular ageing, we performed GSEA analysis based on GO database. Our results revealed that the genes upregulated with age were mainly associated with response to ROS and cell inflammation (Fig. 5C). Meanwhile, the downregulated DEGs showed enrichment in GO terms such as ‘sterol biosynthetic process’ and ‘regulation of cholesterol metabolic process’; these findings are consistent with the results of old LCs exhibiting decreases in testosterone production (Fig. 5A–C). To determine critical regulators involved in LCs ageing, we devised gene-regulatory networks grounded in ageing-associated DEGs. We noticed that several TFs, such as CEBPD, FOSL2, JUNB, KLF4, ATF3, and EGR1, were among the top upregulated genes in old human LCs (Fig. 5D; Supplementary Table S9). These findings are in agreement with the prominent upregulation of the response to oxidative stress found in our GSEA analysis, as EGR1 is a well-known central regulator of the oxidative stress response (Dai et al., 2021). Additional oxidative stress regulators, such as CEBPD (Wang et al., 2018), ATF3 (Feng et al., 2021), and JUNB (Chen et al., 2019), were also upregulated (Fig. 5E), further supporting the notion that redox homeostasis is impaired in the old human LCs.
Consistent with the transcriptomic changes, we observed an increase of the recognized oxidative stress marker 8-OHDG (Wang et al., 2020; Zou et al., 2021) in old CYP17A1+ LCs compared with those from the young group (Fig. 5F). Additionally, we performed staining to detect the ROS marker DHE and the LCs marker BODIPY on testicular tissue (Supplementary Fig. S6C). The results showed that DHE signals in BODIPY+ LCs had a higher intensity in the old tissue compared to the young tissue, further confirming LCs in old testis produced more ROS. Consistently, primary human LCs from the old group generated more ROS than those from the young group (Fig. 5G).
Antioxidants restore testosterone production by LCs from ageing human testes
Next, we investigated whether the suppression of oxidative stress could counteract the cellular dysfunction of LCs. As small-molecule agents, such as NAC and VE, have been reported to reduce oxidants (Chen et al., 2005; Zhao et al., 2017), we tested the effect of these antioxidants on the function of primary LCs and testicular samples from the old group. As expected, primary LCs treated with NAC or VE exhibited reductions in ROS levels (Supplementary Fig. S6D and E). Moreover, the antioxidant treatments recovered testosterone production in isolated primary LCs and old human testicular samples (Supplementary Fig. S6F and G), suggesting that antioxidative strategies restore the function of old LCs.
To verify these findings, we transplanted old human testicular tissue under the renal capsule of the immunodeficient NCG mice (Fig. 5H), and treated the mice with vehicle, NAC or VE daily. Seven days later, we found the antioxidant treatments restored testosterone production and increased the mRNA expression of functional LCs-related genes (SF1, CYP17A1, HSD3B1, HSD17B3, and INSL3) (Fig. 5I and J; Supplementary Fig. S6H). Collectively, these in vitro and in vivo experiments provided more convincing data to support the idea that antioxidants can effectively rejuvenate LCs.
Cell–cell communication changes during human testicular ageing
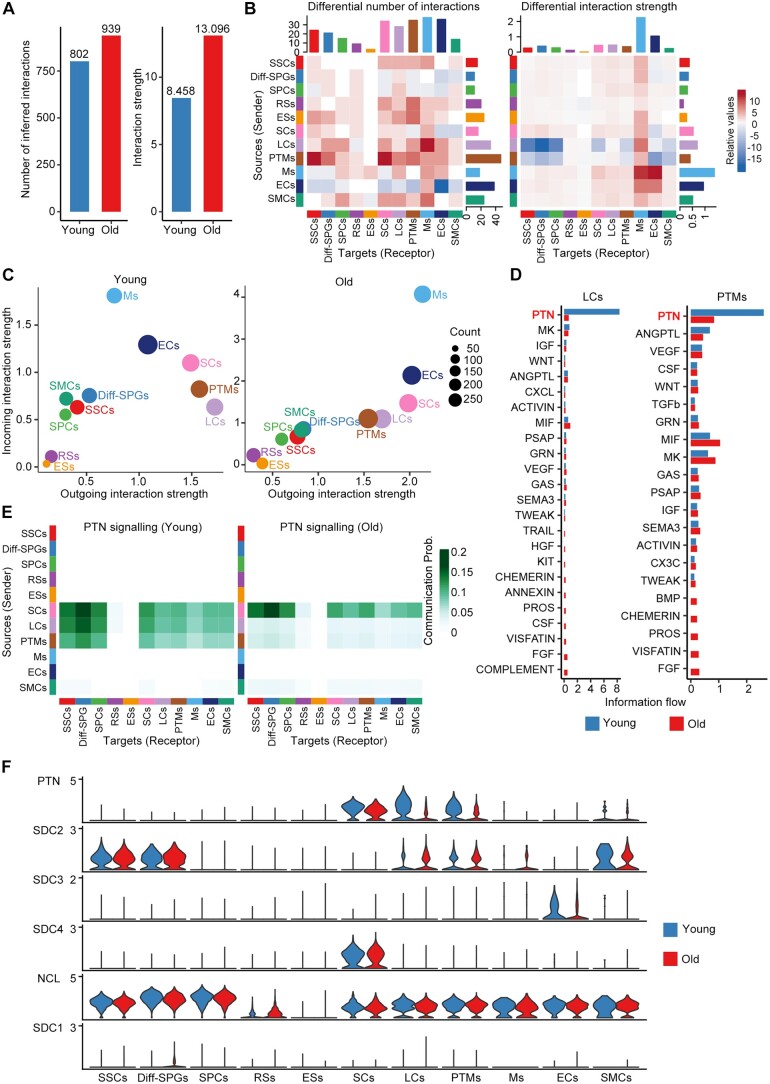
The maintenance of multicellular organism morphology and function largely relies on cell–cell communication. Therefore, using the ligand–receptor interaction tool CellChat (Jin et al., 2021), we next systematically inferred changes in communication networks between testicular cells at the single-cell level during ageing. To our surprise, the interaction number and interaction strength were enhanced in the old group compared to the young group (Fig. 6A; Supplementary Table S10). We then assessed differential interactions of testicular cells as either signal sources or signal targets during ageing. LCs and PTMs showed similar and relative decreases in their communicating roles during ageing; meanwhile, there was a global trend for increasing interactions in old testes compared to young testes (Fig. 6B). Consistently, investigation of the outgoing and incoming signalling patterns revealed relative declines in the outgoing interaction strengths for old LCs and PTMs compared to their young counterparts (Fig. 6C). These results encouraged us to further study the potential role of LCs and PTMs in testicular ageing.
Figure 6.
Cell–cell communication changes during human testicular ageing. (A) Bar plot showing the number (left) or strength (right) of interactions in the cell–cell communication network, as analysed by CellChat. (B) Heatmap showing the different number (left) or strength (right) of interactions in the cell–cell communication network between young and old groups. Red or blue colours represent increased or decreased signalling, respectively, in old groups compared to young. (C) The outgoing (sending signals) and incoming (receiving signals) interaction strength of the cell populations in young (left) and old (right) groups. ‘Count’ indicates the number of inferred links (both outgoing and incoming) associated with each cell group. (D) Information flow of significant signalling pathways sent by LCs (left) or PTMs (right) and received by all cell populations in the young (blue columns) and old (red columns) groups. The top signalling pathway, PTN, is labelled in red. (E) Heatmap showing the inferred intercellular communication networks of the PTN signalling pathways in young (left) and old (right) cell populations. Colour key, ranging from white to green, indicates the between-cell communication probability for the PTN signalling pathway. (F) Violin plot showing the expression levels of ligand–receptor pairs for the PTN signalling network in young and old cell populations. LCs, Leydig cells; PTM, peritubular myoid cells; PTN, pleiotrophin.
When we compared the information flow for each signalling pathway sent by LCs or PTMs between the young and old groups, we found that the PTN signalling pathway ranked the highest in flow for young testes but declined prominently during ageing (Fig. 6D). Based on this finding, we investigated the PTN signalling pathway network among testicular cells. In the young group, PTN signalling was mainly sent by SCs, LCs, and PTMs and received by SSCs, Diff-SPGs, SPCs, and somatic cells. In the old group, in comparison, PTN signalling was dramatically lower between LCs, PTMs, and other target cells (Fig. 6E). Considering growth factors and morphogen signals play important roles in spermatogenesis, we next inferred the age-related changes in the communication networks of target germ cells. Similar to the above-described findings, PTN was prominent among the incoming signalling pathways of SSCs, Diff-SPGs, and SPCs (Supplementary Fig. S7A), indicating the protentional roles of PTN signalling in spermatogenesis. Our analysis identified four significant ligand–receptor pairs for PTN signalling: PTN-NCL, PTN-SDC2, PTN-SDC3, and PTN-SDC4. Among them, PTN-NCL contributed the most strongly to PTN signalling in both the young and old groups (Supplementary Fig. S7B). The expression level of PTN was downregulated in LCs and PTMs of the old group compared to that in the young group, whereas NCL, SDC2, SDC3, and SDC4 showed substantial expression with age in most of the tested cell populations (Fig. 6F).
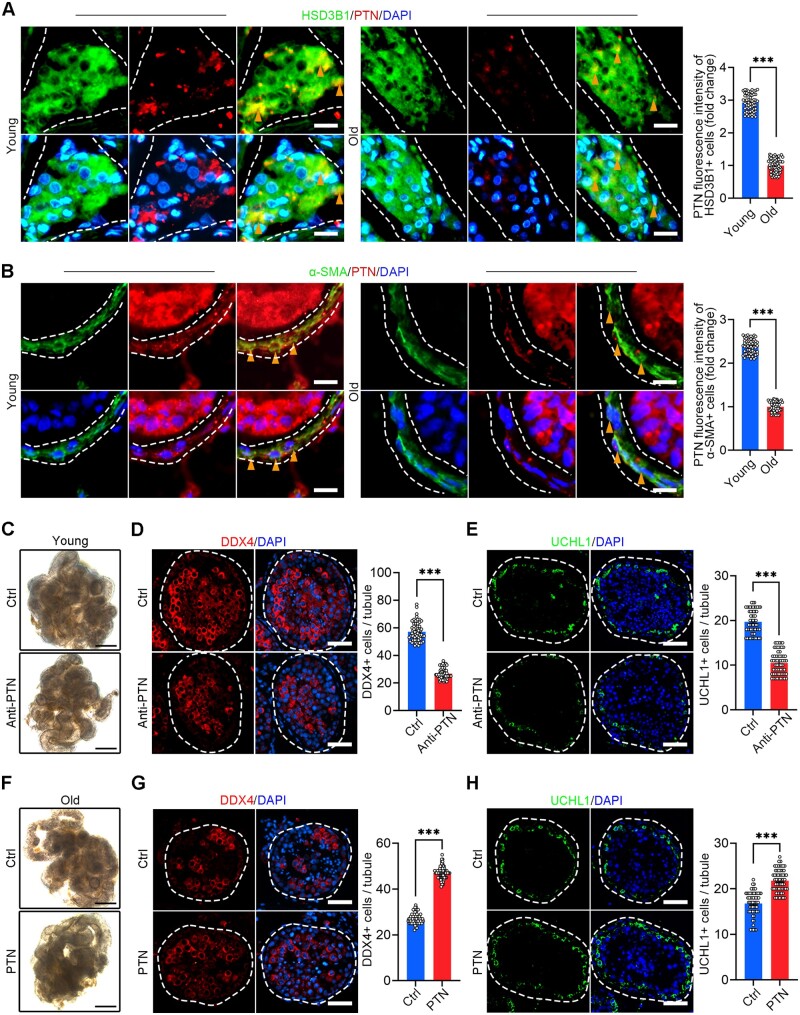
For validation, we performed co-immunostaining of PTN with the LCs marker HSD3B1 or the PTMs marker α-SMA, and we found the PTN signals of the old testes were much weaker than those of the young testes (Fig. 7A and B). To assess the direct effect of the decreased PTN signalling on spermatogenic impairment in human testicular ageing, we blocked the PTN signalling pathway by the anti-PTN antibody in cultured primary testicular tissue from young donors. We found the germ cells marker DDX4 significantly declined, and the number of SSCs (UCHL1+ cells) and differentiated SSCs (STRA8+ cells) also decreased in the anti-PTN group (Fig. 7C–E; Supplementary Fig. S7C). Moreover, we treated the cultured human testicular tissues from old donors with PTN recombinant protein in vitro for 4 days, and then we performed immunostaining assay to assess germ cells. Similarly, we found that germ cells marker DDX4 significantly recovered, and the number of SSCs (UCHL1+ cells) and differentiated SSCs (STRA8+ cells) also increased in the PTN group (Fig. 7F–H; Supplementary Fig. S7D). These results suggest that the downregulation of PTN and PTN signalling pathway during ageing may drive the age-related male fertility decline.
Figure 7.
Changes and roles of PTN signalling pathway in human testicular ageing. (A) Immunostaining and quantification of HSD3B1 and PTN in young and old human testis sections. Left, representative image of HSD3B1 and PTN in human testis sections. Right, quantification of the expression level of PTN in HSD3B1+ cells. Scale bars, 15 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. (B) Immunostaining and quantification of α-SMA and PTN in young and old human testis sections. Left, representative image of α-SMA and PTN in human testis sections. Right, quantification of the expression level of PTN in α-SMA+ cells. Scale bars, 15 μm. Bars represent the mean with SEM of 60 independent tubules per group (Young, n = 10 samples; Old, n = 10 samples). Significance was determined by Student’s t-test. ***P < 0.001. (C) Representative images of young men’s seminiferous tubules cultured in vitro of young men. Scale bars, 500 μm. (D) Immunostaining and quantification of DDX4 in young men’s seminiferous tubules cultured in vitro. Left, representative image of DDX4 in seminiferous tubules of Ctrl and Anti-PTN groups. Right, quantification of the number of DDX4+ cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Ctrl, n = 6 samples; Anti-PTN, n = 6 samples). Significance was determined by Student’s t-test. ***P < 0.001. (E) Immunostaining and quantification of UCHL1 in young men’s seminiferous tubules cultured in vitro. Left, representative image of UCHL1 in seminiferous tubules of Ctrl and Anti-PTN groups. Right, quantification of the number of UCHL1+ cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Ctrl, n = 6 samples; Anti-PTN, n = 6 samples). Significance was determined by Student’s t-test. ***P < 0.001. (F) Representative images of old men’s seminiferous tubules cultured in vitro. Scale bars, 500 μm. (G) Immunostaining and quantification of DDX4 in old men’s seminiferous tubules cultured in vitro. Left, representative image of DDX4 in seminiferous tubules of Ctrl and PTN groups. Right, quantification of the number of DDX4+ cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Ctrl, n = 6 samples; PTN, n = 6 samples). Significance was determined by Student’s t-test. ***P < 0.001. (H) Immunostaining and quantification of UCHL1 in old men’s seminiferous tubules cultured in vitro. Left, representative image of UCHL1 in seminiferous tubules of Ctrl and PTN groups. Right, quantification of the number of UCHL1-positive cells per seminiferous tubule. Scale bars, 35 μm. Bars represent the mean with SEM of 60 independent tubules per group (Ctrl, n = 6 samples; PTN, n = 6 samples). Significance was determined by Student’s t-test. ***P < 0.001. Ctrl, control group; Anti-PTN, anti-PTN antibody group; PTN, recombinant PTN protein group; PTN, pleiotrophin.
Discussion
In this study, we conducted a single-cell analysis of human testicular ageing, offering new perspectives on the mechanisms underlying this process. Our research provides significant advancements in four critical aspects. First, we detailed the gene expression profiles of 11 different human testicular cell types, encompassing both germline and somatic cells, and discovered several novel cell type-specific markers. Second, the analysis of age-associated gene expression changes revealed that DNA repair genes were downregulated in old human SSCs, potentially contributing to the age-related increase of de novo germline mutations in males. Third, human LCs exhibited ageing-associated upregulation of oxidative stress genes, and antioxidant treatments recovered the functions of old human LCs. Fourth, our results suggested that decreases in PTN signalling contribute to spermatogenic impairment in testicular ageing. Together, these observations provide novel insights into human testicular ageing and identify new potential targets for treating disorders associated with testicular ageing.
Although the fertility decline and testosterone deficiency caused by testicular ageing undermine reproductive and general health in ageing males, the impact of age on the human testis is still poorly understood (Kaufman et al., 2019; Santiago et al., 2019). Previous studies have mainly detailed the morphological changes occurring in the human testis during ageing, while the molecular mechanisms underlying testicular ageing have remained largely unknown (Perheentupa and Huhtaniemi, 2009; Jiang et al., 2014; Mularoni et al., 2020). Moreover, the few previous mechanistic reports have used analytical methods with inherent biases, limiting their ability to provide information for all cell populations (Han et al., 2021; Stockl et al., 2021). To address these gaps, we herein used scRNA-seq analysis, which enables the study of heterogeneous cells and facilitates the identification of cell states and cell type-specific gene changes during ageing or disease emergence (Wang et al., 2020; Di Persio et al., 2021; Guo et al., 2021; Yang et al., 2022). Recent scRNA-seq-based studies have reported transcriptomic changes in immune cells during ageing (Zheng et al., 2020) and in tissues such as the brain (Ximerakis et al., 2019), mammary gland (Li et al., 2020), and ovary (Wang et al., 2020). In earlier reports, scRNA-seq has also been employed to analyse human testes, further enhancing our understanding of testicular biology and related pathology (Guo et al., 2018, 2020, 2021; Alfano et al., 2021; Di Persio et al., 2021). For instance, scRNA-seq analyses of testes revealed profiles for human testis development (Guo et al., 2020, 2021), Klinefelter syndrome (Mahyari et al., 2021), idiopathic germ cell aplasia (Alfano et al., 2021), and azoospermia (Zhao et al., 2020). A recent study by Nie et al. (2022) applied scRNA-seq to investigate human testicular ageing, providing candidate molecular mechanisms for understanding the changes occurring during this process. However, the critical molecular drivers underlying the functional decline of germ cells and LCs during ageing remained unclear. Here, we used scRNA-seq to comprehensively delineate age-associated gene expression changes specific to each cell type in human testes. To our knowledge, this is the first scRNA-seq study to reveal the cell type particularly vulnerable to ageing and uncover the potential molecular drivers of testicular ageing by in-depth sequencing and reproducible bioinformatics tools.
Since human male germ cells have a unique germline-specific ageing process (Pohl et al., 2019; Laurentino et al., 2020), we first analysed germline cells and identified five cell categories based on their specific scRNA-seq signatures. Our findings showed that human germline ageing was linked to decreased RSs and ESs in old testes, consistent with previous reports that ageing males showed a decline in sperm parameters (Pohl et al., 2019, 2021). The results also suggest that there are cellular differences in spermatogenesis-related cells of the young and old groups. SSCs are known to be the foundational unit of fertility in all male mammals (Sharma et al., 2019; Tan and Wilkinson, 2020), but research has been slow to advance our knowledge of human SSCs biology, especially in the ageing process. Here, we characterized the molecular changes in human SSCs during testicular ageing. Surprisingly, we found that BER, a major DNA repair pathway (Ray Chaudhuri and Nussenzweig, 2017), was downregulated in old human SSCs. Consistently, immunostaining revealed age-related decreases in NTHL1 and APEX1, which are important BER-promoting genes (Galick et al., 2013; Li et al., 2018), within human germ cell populations. Cells face a range of endogenous and exogenous insults that induce DNA damage, which if left unrepaired, can impair genomic integrity and lead to the development of various diseases (Ray Chaudhuri and Nussenzweig, 2017). Among germline cells, differentiating germ cells exist only for the duration of one spermatogenic cycle, but SSCs are long-living stem cells that exist throughout the male’s life. This unique longevity exposes SSCs to age-accumulated DNA damage, making them susceptible to genetic alterations (Laurentino et al., 2020). Indeed, whole-genome sequencing studies have shown that de novo point mutations in children arise predominantly from male SSCs, and the mutational frequency increases with paternal age (Maher et al., 2016). These mutations have been linked to increased risks of breast cancer, developmental disorders, behavioural disorders, and neurological disease in the offspring of older men (Maher et al., 2016; Yatsenko and Turek, 2018). As BER is a major pathway for repairing oxidative base damage, alkylation damage, and abasic sites on DNA (Ray Chaudhuri and Nussenzweig, 2017), its dysregulation in SSCs is likely to contribute to the age-related increase of de novo germline mutations in males and may have further negative consequences for the offspring health. These possibilities warrant further thorough investigation in future studies.
Testicular somatic cells provide physical and biochemical support to ensure endocrine function and spermatogenesis (Guo et al., 2021). Based on the derived dataset, we uncovered thousands of DEGs that elucidate the intricate molecular shifts underlying the ageing of human testicular somatic cells. These massive ageing-related molecular changes occurred in old testicular somatic cells were likely contribute to a hostile milieu that impairs testicular function (Aitken, 2023). Among these somatic cells, LCs are primarily responsible for the production of testosterone (Zirkin and Papadopoulos, 2018). In the present study, we found that cultured testicular tissue or primary LCs from the old group produced lower testosterone than that from the young group. Further analysis revealed that the ageing-associated DEGs in old LCs were enriched for the ROS response, meanwhile the markers of DNA oxidation and ROS production were increased in old LCs compared to young LCs. We also observed an elevated inflammatory response in old LCs, which may reflect an accumulation of ROS-induced macromolecule damage (Forman and Zhang, 2021). Accordingly, LCs produce a large amount of ROS when testosterone is synthesized, making them vulnerable to ROS-induced damage (Cao et al., 2004; Zirkin and Papadopoulos, 2018). Moreover, recent evidence suggested that oxidative stress may be linked to LCs dysfunction and hypogonadism in rodents (Cao et al., 2004; Chen et al., 2005). Based on these findings, we speculated that oxidative stress could be targeted as a therapeutic avenue to prevent LCs dysfunction in ageing males. Several reports have examined the beneficial effects of antioxidants on the function of murine LCs, but mostly under pathological conditions (e.g. testicular torsion or diabetes) or following exposure to various toxic agents (Cao et al., 2004; Chen et al., 2005). To our knowledge, there is a lack of studies reporting successful antioxidant interventions that improve human LCs function. Our study bridges this gap by demonstrating that antioxidants restored testosterone production, extending from primary human LCs to include testicular samples from the old group. Most importantly, we further verified these findings in human testicular tissues-derived xenograft mice model. These in vitro and in vivo experiments provided more convincing data to support the idea that antioxidants can effectively restore the function of LCs from the old human testes. Based on the rejuvenating effects of antioxidants on old LCs, it is rational to hypothesize that scRNA-seq could represent a new platform for uncovering intervention targets and compounds for alleviating late-onset hypogonadism and testicular ageing in humans.
Testicular physiology broadly relies on cell–cell signalling, and imbalances in this signalling most likely contribute to various forms of spermatogenic impairment (Di Persio et al., 2021; Mahyari et al., 2021). Here, we found that LCs and PTMs showed decreases in their communication roles during ageing, which contrasted with the global tendency towards increased interaction in the old testes relative to the young testes. This finding suggests potential roles for LCs and PTMs in human testicular ageing. Moreover, we noted that the PTN signalling pathway was top-ranked in young LCs and PTMs and declined dramatically in the old group. PTN, an 18-kDa heparin-binding secretory growth/differentiation factor, has been previously reported to play important roles in neural development, angiogenesis, and self-renewal of haematopoietic stem cell (Deuel et al., 2002; Himburg et al., 2012). Here, we noticed that PTN was prominent among the incoming signalling pathways of SSCs, Diff-SPGs, and SPCs. It is possible that the continuous processes of self-renewal and differentiation in developing spermatogonia render them uniquely susceptible to the loss of PTN signalling. Consistently, a previous study showed that a dominant-negative PTN mutant caused testicular atrophy and apoptosis among spermatocytes at all stages of development; this suggested that PTN plays a vital role in normal spermatogenesis, and that interruption of PTN signalling may lead to sterility in males (Zhang et al., 1999). Using an in vitro tissue culture model, we showed that anti-PTN antibody decreased the expression of germline-related proteins in young testicular tissue. Most importantly, PTN recombinant protein significantly restored the expression of germline-related proteins in cultured testicular tissue from old donors. These results indicate that the downregulation of PTN and PTN signalling pathway during ageing may drive age-related male fertility decline. However, the effects and mechanisms of PTN on human testicular ageing are not yet fully understood and need to be further elucidated in future studies.
We acknowledge certain limitations in our current study. Due to the inherent challenges of obtaining human testicular tissue, sample size in the present study is constrained. Moreover, considering the observed variations in spermatogenesis among old individuals (Perheentupa and Huhtaniemi, 2009; Jiang et al., 2014; Mularoni et al., 2020), acquiring completely healthy and fertile testicular samples from this demographic remains exceptionally challenging. These factors may potentially impact our conclusions; thus, it is crucial to note that our study does not solely represent the alterations in the testes that occur during healthy ageing. Additionally, fixation of testes in 4% PFA is known to cause artefacts, including tissue shrinkage, particularly between seminiferous epithelium cells (Tu et al., 2011). These artefacts can potentially distort the morphological and structural assessment of the tissues, thus influencing our findings. Despite these limitations, we have taken steps to enhance the reliability of our results. The conclusions derived from sequencing analyses have been substantiated through experimental validation. However, future investigations with larger patient cohorts are still warranted to further validate and extend our results.
In summary, the present study provides a detailed single-cell transcriptomic landscape of the human testes across young and old individuals, significantly enhancing our understanding of the unique gene expression patterns specific to various testicular cell types. Notably, this study offers novel perspectives into the intricate molecular mechanisms that underpin testicular ageing in humans, which could help the field work towards developing targeted interventions to protect against testicular ageing and/or suggest new tools to rejuvenate old germline and LCs.
Supplementary Material
Acknowledgements
We thank the donors and their families for their dedication. We thank Dr Fangjian Zhou and Dr Yonghong Li for coordinating the human testicular tissue collection. We are grateful to Yan Guo, Hong Chen, Xiangqian Guo, and Xikun Han for their constructive suggestions. We acknowledge the use of Biorender to create schematic images.
Contributor Information
Kai Xia, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China; Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China.
Peng Luo, Reproductive Medicine Centre, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China; Guangdong Key Laboratory of Reproductive Medicine, Guangzhou, Guangdong, China.
Jiajie Yu, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Siyuan He, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China.
Lin Dong, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China.
Feng Gao, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China; Reproductive Medicine Centre, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Xuren Chen, Reproductive Medicine Centre, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Yunlin Ye, Department of Urology, Sun Yat-Sen University Cancer Centre, Guangzhou, China.
Yong Gao, Reproductive Medicine Centre, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China; Guangdong Key Laboratory of Reproductive Medicine, Guangzhou, Guangdong, China.
Yuanchen Ma, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China.
Cuifeng Yang, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Yadong Zhang, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Qiyun Yang, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Dayu Han, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Xin Feng, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Zi Wan, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Hongcai Cai, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Qiong Ke, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China; Guangdong Key Laboratory of Reproductive Medicine, Guangzhou, Guangdong, China.
Tao Wang, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China; National-Local Joint Engineering Research Center for Stem Cells and Regenerative Medicine, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, China.
Weiqiang Li, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China; Guangdong Key Laboratory of Reproductive Medicine, Guangzhou, Guangdong, China; National-Local Joint Engineering Research Center for Stem Cells and Regenerative Medicine, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, China.
Xiang’an Tu, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Xiangzhou Sun, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Chunhua Deng, Department of Urology and Andrology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China.
Andy Peng Xiang, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, China; National-Local Joint Engineering Research Center for Stem Cells and Regenerative Medicine, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, China.
Data availability
Requests for further information and reagents should be directed to and will be fulfilled by the corresponding author. The scRNA-seq sequencing and processed data reported in this paper were deposited at the Genome Sequence Archive (https://ngdc.cncb.ac.cn/), under the accession number HRA002349. All other relevant data supporting the key findings of this study are presented in the paper or the supplementary files.
Authors’ roles
K.X., P.L., J.Y., and S.H. contributed equally as first authors. A.P.X. and C.D. conceived this study and designed the experiments. K.X., P.L., and F.G. collected and prepared clinical samples. J.Y. and S.H. performed the data analysis. P.L. and L.D. performed most of the biological experiments. Y.Y., X.F., X.S., and Q.Y. helped with analysis and interpretation of data. Y.M. provided technical help in single-cell sequencing analysis. X.C., C.Y., D.H., and Z.W. helped with immunofluorescence staining. F.G., Y.Z. Y.G., and H.C. helped with in vitro culture and phenotypic characterization of human Leydig cells. Q.K., T.W., W.L., X.T., and X.S. assisted with the experimental design and revised the manuscript. K.X., P.L., S.H., and L.D. orchestrated the data integration, prepared the paper, and wrote the manuscript. A.P.X. and C.D. revised the manuscript and supervised the study. All authors critically read and commented on the final paper.
Funding
National Key Research and Development Program of China (2022YFA1104100); National Natural Science Foundation of China (32130046, 82171564, 82101669, 82371611, 82371609, 82301796); Natural Science Foundation of Guangdong Province, China (2022A1515010371); Major Project of Medical Science and Technology Development Research Center of National Health Planning Commission, China (HDSL202001000); Open Project of NHC Key Laboratory of Male Reproduction and Genetics (KF202001); Guangdong Province Regional Joint Fund-Youth Fund Project (2021A1515110921, 2022A1515111201); China Postdoctoral Science Foundation (2021M703736).
Conflict of interest
None declared.
References
- Aitken RJ. Male reproductive ageing: a radical road to ruin. Hum Reprod 2023;38:1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano M, Tascini AS, Pederzoli F, Locatelli I, Nebuloni M, Giannese F, Garcia-Manteiga JM, Tonon G, Amodio G, Gregori S et al. Aging, inflammation and DNA damage in the somatic testicular niche with idiopathic germ cell aplasia. Nat Commun 2021;12:5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Petersen RC. Cellular senescence in brain aging and neurodegenerative diseases: evidence and perspectives. J Clin Invest 2018;128:1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Leers-Sucheta S, Azhar S. Aging alters the functional expression of enzymatic and non-enzymatic anti-oxidant defense systems in testicular rat Leydig cells. J Steroid Biochem Mol Biol 2004;88:61–67. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu J, Luo L, Baig MU, Kim JM, Zirkin BR. Vitamin E, aging and Leydig cell steroidogenesis. Exp Gerontol 2005;40:728–736. [DOI] [PubMed] [Google Scholar]
- Chen LY, Brown PR, Willis WB, Eddy EM. Peritubular myoid cells participate in male mouse spermatogonial stem cell maintenance. Endocrinology 2014;155:4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Li X, Shi Q, Zhang Z, Xu S. Hydrogen sulfide exposure triggers chicken trachea inflammatory injury through oxidative stress-mediated FOS/IL8 signaling. J Hazard Mater 2019;368:243–254. [DOI] [PubMed] [Google Scholar]
- Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015;149:R159–R167. [DOI] [PubMed] [Google Scholar]
- Dai X, Liao R, Liu C, Liu S, Huang H, Liu J, Jin T, Guo H, Zheng Z, Xia M et al. Epigenetic regulation of TXNIP-mediated oxidative stress and NLRP3 inflammasome activation contributes to SAHH inhibition-aggravated diabetic nephropathy. Redox Biol 2021;45:102033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel TF, Zhang N, Yeh HJ, Silos-Santiago I, Wang ZY. Pleiotrophin: a cytokine with diverse functions and a novel signaling pathway. Arch Biochem Biophys 2002;397:162–171. [DOI] [PubMed] [Google Scholar]
- Di Persio S, Tekath T, Siebert-Kuss LM, Cremers JF, Wistuba J, Li X, Meyer Zu Horste G, Drexler HCA, Wyrwoll MJ, Tuttelmann F et al. Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis. Cell Rep Med 2021;2:100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Li Y, Jin X, Gong R, Xia Z. ATF3 regulates oxidative stress and extracellular matrix degradation via p38/Nrf2 signaling pathway in pelvic organ prolapse. Tissue Cell 2021;73:101660. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat Rev Drug Discov 2021;20:689–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, Sweasy JB. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc Natl Acad Sci USA 2013;110:14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, Guo Y, Takei Y, Yun J, Cai L et al. The adult human testis transcriptional cell atlas. Cell Res 2018;28:1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Nie X, Giebler M, Mlcochova H, Wang Y, Grow EJ, Kim R, Tharmalingam M, Matilionyte G, Lindskog C et al. ; DonorConnect. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell 2020;26:262–276.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Sosa E, Chitiashvili T, Nie X, Rojas EJ, Oliver E, Plath K, Hotaling JM, Stukenborg J-B, Clark AT et al. ; DonorConnect. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 2021;28:764–778.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Hong SH, Lee SJ, Hong SP, Cho C. Transcriptome analysis of testicular aging in mice. Cells 2021;10:2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg HA, Harris JR, Ito T, Daher P, Russell JL, Quarmyne M, Doan PL, Helms K, Nakamura M, Fixsen E et al. Pleiotrophin regulates the retention and self-renewal of hematopoietic stem cells in the bone marrow vascular niche (vol 2, pg 964, 2012). Cell Reports 2012;2:1774–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS One 2010;5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhu WJ, Li J, Chen QJ, Liang WB, Gu YQ. Quantitative histological analysis and ultrastructure of the aging human testis. Int Urol Nephrol 2014;46:879–885. [DOI] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Abdo JG, Petty CS, Neaves WB. Effect of age on the composition of seminiferous tubular boundary tissue and on the volume of each component in humans. Fertil Steril 1988;49:1045–1051. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Dunleavy J, Gemmell NJ, Nakagawa S. Consistent age-dependent declines in human semen quality: a systematic review and meta-analysis. Ageing Res Rev 2015;19:22–33. [DOI] [PubMed] [Google Scholar]
- Kaufman JM, Lapauw B, Mahmoud A, T’Sjoen G, Huhtaniemi IT. Aging and the male reproductive system. Endocr Rev 2019;40:906–972. [DOI] [PubMed] [Google Scholar]
- Kimura M, Itoh N, Takagi S, Sasao T, Takahashi A, Masumori N, Tsukamoto T. Balance of apoptosis and proliferation of germ cells related to spermatogenesis in aged men. J Androl 2003;24:185–191. [DOI] [PubMed] [Google Scholar]
- Laurentino S, Cremers JF, Horsthemke B, Tuttelmann F, Czeloth K, Zitzmann M, Pohl E, Rahmann S, Schroder C, Berres S et al. A germ cell-specific ageing pattern in otherwise healthy men. Aging Cell 2020;19:e13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Shapiro H, Tsiobikas C, Selfors LM, Chen H, Rosenbluth J, Moore K, Gupta KP, Gray GK, Oren Y et al. Aging-associated alterations in mammary epithelia and stroma revealed by single-cell RNA sequencing. Cell Rep 2020;33:108566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang X, Lu X, Dai N, Zhang S, Cheng Y, Zhang L, Yang Y, Liu Y, Yang Z et al. APE1 deficiency promotes cellular senescence and premature aging features. Nucleic Acids Res 2018;46:5664–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang Z, Jiang Z, Guo J, Zhang Y, Li C, Chung J, Folmer J, Liu J, Lian Q et al. Regulation of seminiferous tubule-associated stem Leydig cells in adult rat testes. Proc Natl Acad Sci USA 2016;113:2666–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Feng X, Deng R, Wang F, Zhang Y, Li X, Zhang M, Wan Z, Xiang AP, Xia K et al. An autofluorescence-based isolation of Leydig cells for testosterone deficiency treatment. Mol Cell Endocrinol 2021;535:111389. [DOI] [PubMed] [Google Scholar]
- Maher GJ, McGowan SJ, Giannoulatou E, Verrill C, Goriely A, Wilkie AO. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc Natl Acad Sci USA 2016;113:2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahyari E, Guo J, Lima AC, Lewinsohn DP, Stendahl AM, Vigh-Conrad KA, Nie X, Nagirnaja L, Rockweiler NB, Carrell DT et al. Comparative single-cell analysis of biopsies clarifies pathogenic mechanisms in Klinefelter syndrome. Am J Hum Genet 2021;108:1924–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela JA, Koskenniemi JJ, Virtanen HE, Toppari J. Testis development. Endocr Rev 2019;40:857–905. [DOI] [PubMed] [Google Scholar]
- Matzkin ME, Calandra RS, Rossi SP, Bartke A, Frungieri MB. Hallmarks of testicular aging: the challenge of anti-inflammatory and antioxidant therapies using natural and/or pharmacological compounds to improve the physiopathological status of the aged male gonad. Cells 2021;10:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni V, Esposito V, Di Persio S, Vicini E, Spadetta G, Berloco P, Fanelli F, Mezzullo M, Pagotto U, Pelusi C et al. Age-related changes in human Leydig cell status. Hum Reprod 2020;35:2663–2676. [DOI] [PubMed] [Google Scholar]
- Nie X, Munyoki SK, Sukhwani M, Schmid N, Missel A, Emery BR, Stukenborg J-B, Mayerhofer A, Orwig KE, Aston KI et al. Single-cell analysis of human testis aging and correlation with elevated body mass index. Dev Cell 2022;57:1160–1176.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikopoulou C, Parekh S, Tessarz P. Ageing and sources of transcriptional heterogeneity. Biol Chem 2019;400:867–878. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev 2012;92:577–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 2009;136:1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, Nistal M, Saez FJ, Fraile B. Ultrastructure of the aging human testis. J Electron Microsc Tech 1991;19:241–260. [DOI] [PubMed] [Google Scholar]
- Perheentupa A, Huhtaniemi I. Aging of the human ovary and testis. Mol Cell Endocrinol 2009;299:2–13. [DOI] [PubMed] [Google Scholar]
- Pohl E, Gromoll J, Wistuba J, Laurentino S. Healthy ageing and spermatogenesis. Reproduction 2021;161:R89–R101. [DOI] [PubMed] [Google Scholar]
- Pohl E, Hoffken V, Schlatt S, Kliesch S, Gromoll J, Wistuba J. Ageing in men with normal spermatogenesis alters spermatogonial dynamics and nuclear morphology in Sertoli cells. Andrology 2019;7:827–839. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017;18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Silva JV, Alves MG, Oliveira PF, Fardilha M. Testicular aging: an overview of ultrastructural, cellular, and molecular alterations. J Gerontol A Biol Sci Med Sci 2019;74:860–871. [DOI] [PubMed] [Google Scholar]
- Sharma S, Wistuba J, Pock T, Schlatt S, Neuhaus N. Spermatogonial stem cells: updates from specification to clinical relevance. Hum Reprod Update 2019;25:275–297. [DOI] [PubMed] [Google Scholar]
- Song K, Yang XY, An G, Xia XY, Zhao JX, Xu XH, Wan C, Liu TY, Zheng Y, Ren SF et al. Targeting APLN/APJ restores blood-testis barrier and improves spermatogenesis in murine and human diabetic models. Nat Commun 2022;13:7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockl JB, Schmid N, Flenkenthaler F, Drummer C, Behr R, Mayerhofer A, Arnold GJ, Frohlich T. Age-related alterations in the testicular proteome of a non-human primate. Cells 2021;10:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, Craig T, Diana E, Lehmann G, Toren D, Wang J et al. Human Ageing Genomic Resources: new and updated databases. Nucleic Acids Res 2018;46:D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Wilkinson MF. A single-cell view of spermatogonial stem cells. Curr Opin Cell Biol 2020;67:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Yu L, Zhang H. Morphology of rat testis preserved in three different fixatives. J Huazhong Univ Sci Technolog Med Sci 2011;31:178–180. [DOI] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, Liu Z, Min Z, Hu H, Jing Y et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell 2020;180:585–600.e19. [DOI] [PubMed] [Google Scholar]
- Wang SM, Lim SW, Wang YH, Lin HY, Lai MD, Ko CY, Wang JM. Astrocytic CCAAT/Enhancer-binding protein delta contributes to reactive oxygen species formation in neuroinflammation. Redox Biol 2018;16:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Wang F, Lai X, Dong L, Luo P, Zhang S, Yang C, Chen H, Ma Y, Huang W et al. AAV-mediated gene therapy produces fertile offspring in the Lhcgr-deficient mouse model of Leydig cell failure. Cell Rep Med 2022;3:100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci 2019;22:1696–1708. [DOI] [PubMed] [Google Scholar]
- Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, Losada PM, Chen MB, Schaum N, Khoury N et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 2022;603:885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko AN, Turek PJ. Reproductive genetics and the aging male. J Assist Reprod Genet 2018;35:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yeh HJ, Zhong R, Li YS, Deuel TF. A dominant-negative pleiotrophin mutant introduced by homologous recombination leads to germ-cell apoptosis in male mice. Proc Natl Acad Sci USA 1999;96:6734–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Yao C, Xing X, Jing T, Li P, Zhu Z, Yang C, Zhai J, Tian R, Chen H et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat Commun 2020;11:5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Feng H, Sun W, Liu K, Lu JJ, Chen X. Tert-butyl hydroperoxide (t-BHP) induced apoptosis and necroptosis in endothelial cells: roles of NOX4 and mitochondrion. Redox Biol 2017;11:524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu X, Le W, Xie L, Li H, Wen W, Wang S, Ma S, Huang Z, Ye J et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell 2020;11:740–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkin BR, Papadopoulos V. Leydig cells: formation, function, and regulation. Biol Reprod 2018;99:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Long X, Zhao Q, Zheng Y, Song M, Ma S, Jing Y, Wang S, He Y, Esteban CR et al. A single-cell transcriptomic atlas of human skin aging. Dev Cell 2021;56:383–397.e8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for further information and reagents should be directed to and will be fulfilled by the corresponding author. The scRNA-seq sequencing and processed data reported in this paper were deposited at the Genome Sequence Archive (https://ngdc.cncb.ac.cn/), under the accession number HRA002349. All other relevant data supporting the key findings of this study are presented in the paper or the supplementary files.