Abstract
White adipose tissue (WAT) is essential for lipid storage and systemic energy homeostasis. Understanding adipocyte formation and stability is key to developing therapies for obesity and metabolic disorders. Through a high-throughput cDNA screen, we identified PATZ1, a POZ/BTB and AT-Hook Containing Zinc Finger 1 protein, as an important adipogenic transcription factor. PATZ1 is expressed in human and mouse adipocyte precursor cells (APCs) and adipocytes. In cellular models, PATZ1 promotes adipogenesis via protein-protein interactions and DNA binding. PATZ1 ablation in mouse adipocytes and APCs leads to a reduced APC pool, decreased fat mass, and hypertrophied adipocytes. ChIP-Seq and RNA-seq analyses show that PATZ1 supports adipogenesis by interacting with transcriptional machinery at the promoter regions of key early adipogenic factors. Mass-spec results show that PATZ1 associates with GTF2I, with GTF2I modulating PATZ1’s function during differentiation. These findings underscore PATZ1’s regulatory role in adipocyte differentiation and adiposity, offering insights into adipose tissue development.
Subject terms: Lipid signalling, Obesity
Adipogenesis is important for white adipose tissue function. Here the authors identify the transcription factor PATZ1 as a regulator of adipogenesis and adipocyte maintenance, and report that adipose-tissue specific loss of PATZ1 leads to reduced fat mass and larger adipocytes in mice.
Introduction
The formation of adipocytes from precursor cells involves a complex and highly orchestrated program of gene transcription1. Adipogenesis can be divided into two phases: commitment and terminal differentiation. The commitment phase involves the dynamic integration of cytoarchitecture, signaling pathways, and transcriptional regulators. Multiple signals from bone-morphogenetic factor (BMP), transforming growth factor (TGF), fibroblast growth factor (FGF), and WNT influence stem cell conversion to adipocytes2. Zinc-finger protein 423 (ZFP423), TCF7-like 1 (TCF7L1), and Krüppel-like factors (KLFs) are among the transcriptional regulators that have been shown to play essential roles in the adipocyte precursors’ commitment and differentiation3–5. Prior analysis on activator protein 1 (AP-1) factors6, activating transcription factors (ATFs)7, KLFs5,8, p300/CBP9,10, and E2F transcription factors11 have shed light on the early stages of adipogenesis. Genomic approaches have revealed that a network of these early factors modifies the chromatin environment of terminal factors like PPARγ to enable terminal adipocyte differentiation12,13. After the commitment phase, a cascade of transcriptional events is activated that induces the expression of metabolic genes and adipokines associated with terminal adipocyte differentiation1. Transcriptional events in the terminal phase have been well studied, and more than twenty different transcription factors have been identified as playing key roles2. Among them, peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer-binding proteins (C/EBPs) are the primary drivers of gene induction during terminal differentiation1,14.
PATZ1 has an N-terminal POZ/BTB domain for protein interaction, a central DNA binding AT-hook domain, and a second, C-terminal, zinc finger (ZF) DNA binding domain15. PATZ1 is an architectural transcriptional regulator that negatively or positively modulates the expression of genes by binding to promoter GC-rich regions. PATZ1 is known to regulate stem cell pluripotency and reprogramming, spermatogenesis, T-cell development, and cancer progression15–17. However, the role of PATZ1 in metabolism and adipose biology is still unexplored. Here, we present PATZ1 as a transcription factor involved in the regulation of APCs and adipocytes and promotes both early and late stages of adipocyte differentiation through interaction with the promoter regions of adipogenic factors.
Results
A high-throughput screen of cDNA modulators of adipogenesis identifies PATZ1 as an adipogenic factor
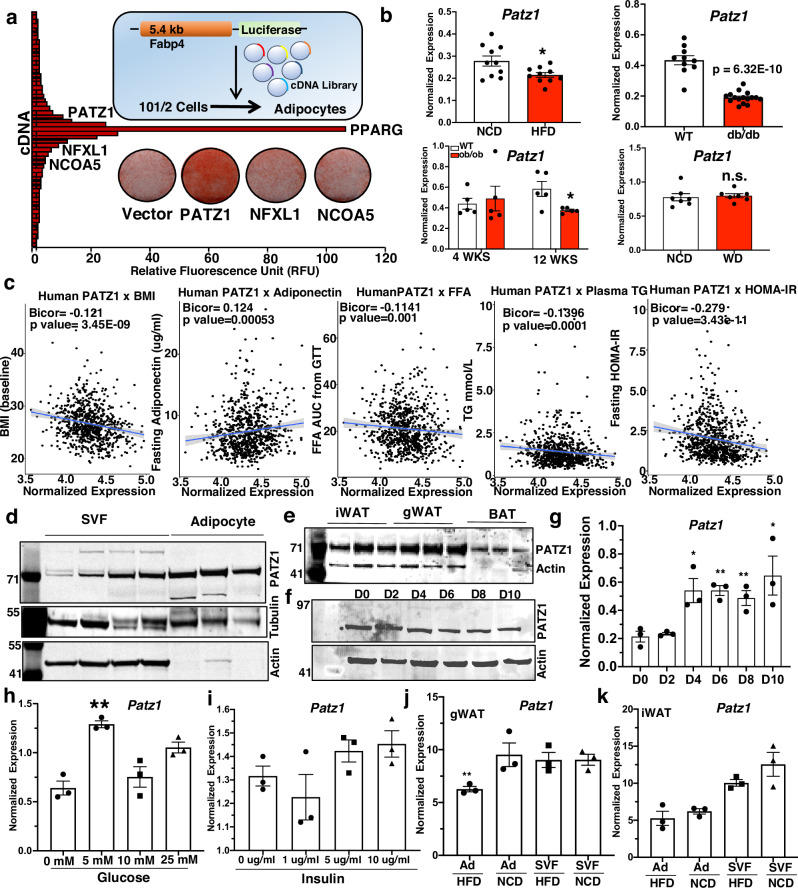
To identify novel factors involved in adipocyte differentiation, we previously performed a cDNA library screen in which 10T1/2 cells were retro-transfected simultaneously with a luciferase reporter driven by the −5.4 kb Fabp4 promoter and a collection of 18,292 individually spotted mammalian cDNA expression vectors (Fig. 1a)18. The day after transfection, cells were treated with insulin and a PPARγ agonist (rosiglitazone) to induce adipogenic differentiation, then luciferase activity was evaluated 4 days later. Several cDNAs were identified as activators of Fabp4-driven luciferase activity. PPARγ emerged as the most potent activator and several additional known adipogenic factors were also represented, including CCAAT/enhancer-binding proteins (C/EBPα and C/EBPδ), early B cell factor 1 (EBF1), and mitogen-activated protein kinase kinase 6 (MAPKK6). We recently reported adipogenic roles of paraspeckle protein 1 (PSPC1) and transducing-like enhancer protein 3 (TLE3), two other top candidates from this screen18–20. To identify other cDNA modulators of adipogenesis, we tested three potentially novel regulators: PATZ1, nuclear transcription factor X-Box binding like 1 (NFXL1), and nuclear receptor coactivator 5 (NCOA5). The cDNAs encoding these genes were cloned into a retrovirus vector and stably overexpressed in 10T1/2 cells, which were then differentiated into white adipocytes. Among others, cells stably expressing PATZ1 showed the highest adipogenic potential, as indicated by Oil-Red-O staining (Fig. 1a).
Fig. 1. Identification of PATZ1 as an adipogenic factor.
a Diagram of the cDNA library screen, the plot of relative fluorescence unit (RFU), and top hits of cDNA, including PATZ1, NFXL1, and NCOA5. Oil-red-O staining of D6 differentiated 10T1/2 cells expressing Vector, PATZ1, NFXL1, and NCOA5. b Real-time qPCR analysis of Patz1 in mice fed a normal chow diet (NCD) (n = 10) and HFD for 12 weeks (n = 10), WT (n = 10) and 10- week-old db/db (n = 16), 4 weeks (n = 5)- or 12-week-old (n = 5) ob/ob mice, or mice fed NCD (n = 7) mice fed western diet (WD) (n = 7) for 10 weeks. c Correlation of human WAT PATZ1 levels with indicated traits from the METSIM study. Correlations were assessed from the midweight bicorrelation coefficient and corrected p value using the R package WGCNA. d Western blot showing PATZ1 levels in SVF and adipocyte fraction from iWATs of 10-week-old chow-fed mice. Tubulin, Actin, and ponceau stain were used as loading controls. Blots from three biological replicates. e Western blot showing PATZ1 protein levels in mouse iWAT, gWAT, and BAT. Actin was used as a loading control. Blots from three biological replicates. f Western blot showing PATZ1 levels in differentiated 10T1/2 cells for indicated times. HMGB1 was used as a loading control. Representative blot from two independent experiments. g qPCR showing PATZ1 levels in differentiated 10T1/2 cells for indicated times without rosiglitazone. n = 3 per timepoint. h, i Real-time qPCR showing PATZ1 levels in 24 h glucose (H) or insulin (I) treated 10T1/2 cells. n = 3 per glucose or insulin concentration. j, k qPCR showing PATZ1 levels in gWAT ( J) or iWAT (K) SVF or adipocytes (Ad) from HFD or NCD fed 10-week-old mice. n = 3 per condition. g–k Results are from three independent experiments. b, g–k, Unpaired Student’s t-test. Data were mean ± s.e.m. *p < 0.05; **p < 0.01, n.s. not significant. b comparing HFD to NCD, db/db to WT, 12 weeks ob/ob to WT, WD to NCD. g comparing to D0. h Comparing to 0 mM. i Comparing to 0 ug/ml. j comparing Ad HFD to Ad NCD. d–f Loading controls were run on the same blot. Source data are provided as a Source Data file.
To understand how obesity impacts the expression of PATZ1, we analyzed Patz1 expression in normal chow diet (NCD), ob/ob, db/db, high-fat diet (HFD), and western diet (WD) fed mice. Consistent with the expression of adipogenic factors like PPARγ20, we noticed decreased Patz1 transcript levels in these models (Fig. 1b). PATZ1 levels in WATs were similar in fed and fasted conditions (Fig. S1a). We next determined the correlation between WAT PATZ1 levels and human metabolic traits within the Metabolic Syndrome in Men (METSIM)21,22 population data (N = ~10,000). We observed that PATZ1 negatively correlated with body mass index (BMI), plasma free fatty acid (FFA) levels, plasma triglyceride (TG) levels, and insulin resistance (HOMA-IR) (Fig. 1c). We found a positive correlation between PATZ1 and adiponectin levels (Fig. 1c). Single-cell RNA-seq (scRNA-seq)23 data from mouse inguinal WAT (iWAT) and gonadal WAT (gWAT) and human subcutaneous WAT (SAT) and visceral WAT (VAT) showed that Patz1 was expressed by both APCs and adipocytes in both species (Fig. S1b–e). Consistent with the scRNA-seq data, fractionation of WAT showed that PATZ1 was expressed in both the stromal vascular fraction (SVF) and adipocyte portions of WAT (Fig. 1d). PATZ1 protein was expressed in inguinal WAT (iWAT) and gonadal WAT (gWAT), and to a lesser extent in BAT (Fig. 1e). Analysis of Tabula Senis data24 showed that Patz1 was similarly expressed in the WATs and BATs of male and female mice (Fig. S1f). The levels of mouse PATZ1 proteins increased during the first few hours and remained relatively unchanged after 24 hours of adipocyte differentiation of 10T1/2 cells (Fig. 1f and S1g). We did not detect PPARγ DNA binding25 regions in the PATZ1 locus, and adipocyte differentiation in the absence of PPARγ ligand did not alter human and mouse PATZ1 protein levels despite increased mRNA expression (Fig. 1g and S1h, i).
PATZ1 was reported to be upregulated during hyperglycemia in endothelial cells26; treating glucose-starved 10T1/2 cells with increasing concentrations of glucose and insulin showed an increase in Patz1 levels (Fig. 1h, i and S1j, k). Gene expression analysis on SVF and adipocytes from HFD and NCD showed differential regulation of PATZ1 in iWAT and gWAT (Fig. 1j, k). A recent study using transcriptomics analysis to identify transcription factors in the upstream gene regulatory networks that drive adipocyte formation in human ASCs identified PATZ1, among other transcription factors27. Our analysis of single-cell assay for transposase accessible chromatin-seq (scATAC-seq) analysis in human adipocytes28 showed an open chromatin configuration, consistent with active transcription of PATZ1 in human adipocytes (Fig. S1l). Altogether, the expression pattern of PATZ1 is consistent with a potential function in adipocyte gene expression.
PATZ1 is a driver of the adipogenic program in vitro
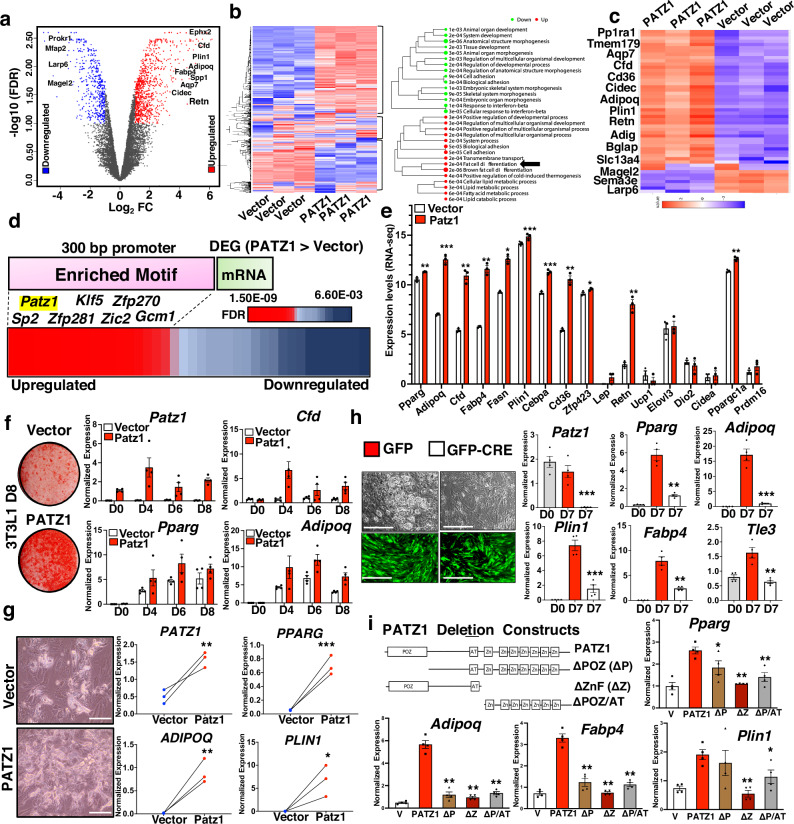
To further elucidate the role of PATZ1 in adipogenesis, we generated 10T1/2 cells stably overexpressing PATZ1 (PATZ1) or vector-expressing control cells (Vector) and differentiated them into white adipocytes for 8 days. We then performed global RNA-seq on three independent replicates of differentiated Vector and PATZ1 cells to unbiasedly evaluate PATZ1’s role in the late stages of adipogenesis. Principle component analysis (PCA) showed a similar clustering pattern of the replicates from both PATZ1 and Vector cells (Fig. S2a). The scatter plot of the total transcript analysis showed comparable expression patterns among Vector cells (R = 0.989) and PATZ1 cells (R = 0.959) (Fig. S2b). Both groups of samples (Vector and PATZ1) had comparable overall distribution and density of the transformed data, implying that PATZ1 overexpression did not cause overt genomic transcript shifts during adipocyte differentiation (Fig. S2c). To probe for transcriptomic differences between Vector and PATZ1 cells, we performed differential gene expression (DEG) analysis and found 1131 genes upregulated and 543 genes downregulated in PATZ1 cells compared to Vector cells (Fig. S2d). Volcano plot analysis of DEGs between PATZ1 and Vector cells showed an increase in late adipocyte genes such as Ephx2, Cfd, Plin1, Aqp7, Retn, Fabp4, and Adipoq and a marked decrease in anti-obesity and anti-adipogenic genes such as Mfap229, Magel229,30, and Prokr131 in PATZ1 cells (Fig. 2a). We isolated upregulated, downregulated, and similarly expressed DEGs and represented them as heatmaps and found a marked increase in “fat cell differentiation” pathway genes in PATZ1 cells (Fig. 2b). We also noticed an upregulation of “Brown fat cell differentiation” and “positive regulation of cold-induced thermogenesis” pathways in PATZ1 cells (Fig. 2b). Sub-setting global DEGs from the heatmap showed a high enrichment of adipogenic genes in PATZ1 cells (Fig. 2c). Gene ontology and pathway analysis from most RNA-seq are unbiased but could be driven by a few genes that may over-represent a specific pathway. Based on our subset of genes, we performed a rigorous test by binning DEGs in four clusters (A–D) and performing pathway analysis using individual genes to define clusters. The results were represented as a heatmap (Fig. S2e). Single gene cluster analyses were plotted as a tSNE plot, showing a highly specific clustering of pathways (A–D) based on binning highly expressed genes in PATZ1 cells (Fig. S2f). Cluster A pathways were upregulated in PATZ1 cells and showed a definite increase in the white fat cell differentiation program (Fig. S2f down).
Fig. 2. PATZ1 promotes adipocyte differentiation in vitro.
a Volcano plot of gene expression differences between PATZ1 and Vector cells. The log2FC of PATZ1/Vector was plotted as a function of −log10 (FDR) with selected genes indicated as upregulated in PATZ1 cells. b Heatmap showing upregulated, downregulated, and similarly expressed genes in Vector and PATZ1 cells (Left) and GO analysis of top genes upregulated and downregulated in PATZ1 cells (Right). Black arrows indicate “Fat cell differentiation”. c Heatmap showing top binned genes that are upregulated and downregulated in PATZ1 and Vector cells. d TF motif analysis 300 bp upstream of DEGs that are greater in PATZ1 cells. e Expression values of indicated genes represented as average FPKM values from PATZ1 or Vector cells. Vector: n = 3 and PATZ1: n = 3. f Oil-Red-O staining (Left) and qPCR analysis of indicated genes from control and PATZ1-overexpressing differentiated 3T3-L1 cells (Right). Vector: n = 4 and PATZ1: n = 4. Representative oil-Red-O staining from three independent experiments. g Bright-field microscopy (Left) and qPCR analysis (Right) of indicated genes from control and PATZ1 overexpressing human abdominal subcutaneous hADSCs differentiated for 12 days. Vector: n = 3 and PATZ1: n = 3. Representative microscopy images from three biological replicates. Scale bars, 100 μm. h (Right) Bright-field (top) and fluorescence at 509 nm (bottom) microscopy showing lipid droplets (top) and GFP fluorescence (bottom) in 7 days differentiated SVFs derived from PATZ1F/F mice infected with adenovirus (Adeno) expressing GFP or GFP-Cre and (Left) qPCR showing mRNA levels of indicated genes from PATZ1 F/F SVFs at D0 or D7 infected with Adeno GFP or Adeno GFP-Cre. GFP levels (bottom panel) show comparable viral infection. D0: n = 4, GFP: n = 4, and GFP-Cre: n = 4. Scale bars, 100 μm. i Domain structure of PATZ1 constructs with indicated deletion and qPCR showing indicated mRNA levels in D4 differentiated 3T3-L1 cells expressing indicated PATZ1 constructs. n = 4 per condition. e, g Results are from three independent experiments. e, g, h, i Unpaired Student’s t-test. Data were mean ± s.e.m. ***p < 0.001; **p < 0.01; *p < 0.05. e, g comparing PATZ1 to Vector; h comparing GFP-Cre to GFP; i all comparisons were made to PATZ1. Source data are provided as a Source Data file.
Next, we calculated enriched transcription factor binding motifs at the promoter regions of DEGs upregulated in PATZ1 cells. We set 300 bp upstream of DEGs as promoter regions and found a highly significant enrichment of PATZ1 motifs (FDR = 3.4E-09) among other transcription factors (Fig. 2d), suggesting a PATZ1-driven adipogenic transcriptional program in PATZ1 expressing cells. Gene network analysis showed an interaction between transcriptional programs of adipogenic genes when we binned both the top 10 and Top 20 DEGs (Fig. S2g). To further unbiasedly validate the adipogenic role of PATZ1, we performed a directed gene expression (fragments per kilobase of transcript per million mapped reads, FPKM) analysis of WAT genes and BAT genes in PATZ1 and Vector cells. We found an increase in white adipocyte genes (Fig. 2e). Brown adipocyte genes were comparable between PATZ1 and Vector cells except for Ppargc1a in PATZ1 cells, which could be one of the drivers of the “brown fat differentiation” GO pathway annotation in Fig. 2e, b.
To further test the role of PATZ1 in adipogenesis in other cellular models, we overexpressed PATZ1 in another adipogenic cell line (3T3-L1) via retroviral transduction. Control and PATZ1-overexpressing 3T3-L1 cells were differentiated for 8 days using an adipogenic cocktail (See Methods). Oil-Red-O staining on D6 showed that PATZ1 expression increased the capacity of 3T3-L1 cells to differentiate (Fig. 2f). Real-time quantitative PCR (qPCR) during the differentiation time course (D0, D4, D6, D8) confirmed an increase in adipocyte genes in cells overexpressing PATZ1 (Fig. 2f and S2h). We next tested if PATZ1 also promoted adipogenesis of primary preadipocytes isolated from mouse and human WAT. We isolated primary SVFs from mouse iWAT, treated confluent cells with either Control or PATZ1 retrovirus, and differentiated the cells from D0–D11. As shown in Fig. S2i, PATZ1 overexpression also markedly enhanced primary APC adipogenesis on D8 and D11, as demonstrated by a marked increase in Oil-Red-O staining. Since PATZ1 is also expressed in human WAT adipocytes, we reasoned that PATZ1 may also be involved in human adipocyte-derived stem cell (hADSC) differentiation. We used frozen primary hADSC isolated from abdominal WAT of human subjects and performed adipogenic differentiation studies. We found that PATZ1 increased lipid-laden adipocytes and the expression of adipogenic genes such as PPARG, ADIPOQ, and PLIN1 (Fig. 2g). To test if PATZ1 loss affects adipogenesis, we generated 10T1/2 cells stably expressing control or Patz1 shRNA. Results in Fig. S2j showed that PATZ1 loss reduced the adipogenic potential of 10T1/2 cells via Oil-Red-O staining. We also isolated SVFs from iWAT from Patz1f/f mice and treated cells with GFP- or GFP-Cre-expressing adenovirus. As shown in Fig. 2h, Cre-expressing cells had almost no visible lipid droplets, and qPCR analysis showed a complete knockout of PATZ1 and a decrease in mature adipocyte genes.
PATZ1 is a modular protein consisting of a protein interaction POZ domain and two DNA-binding AT-hook and Zinc finger domains. To test the importance of these domains in PATZ1 function, we generated PATZ1 mutants lacking POZ domain (ΔPOZ), zinc finger domains (ΔZnF), or POZ and AT-hook domains (ΔPOZ/AT) and generated 10T1/2 cells stably expressing these mutants (Fig. 2i). Our qPCR analysis on differentiated 10T1/2 cells showed that overexpression of full-length PATZ1 increased expression of Pparg, Adipoq, Fabp4, and Plin1. However, cells expressing all three mutants had markedly reduced expression of Pparg, Adipoq, Fabp4, and Plin1, suggesting that both protein interaction and DNA-binding functions of PATZ1 are essential for its adipogenic function (Fig. 2i). To test the impact of PATZ1 exogenous overexpression during adipocyte differentiation, we infected adenoviral receptor-expressing (hCAR) 10T1/2 cell with GFP (Ad-GFP) or PATZ1 (Ad-PATZ1) adenovirus on D2, D4, and D6 of differentiation. As shown in Fig. S2k, compared to stable overexpression models, PATZ1 transduction during differentiation caused an increase in late adipocyte genes on D4 compared to D2 and D6, indicating a possible role of PATZ1 in both priming cells during the early stages and late stages of adipocyte differentiation. There were no differences in beige maker genes between PATZ1 overexpressing cells, but PATZ1 stable overexpression caused a markedly high glucose uptake in differentiated adipocytes (Fig. S2l, m). Our data show that PATZ1 enhances adipocyte differentiation and glucose uptake, and both protein interaction and DNA binding functions are important.
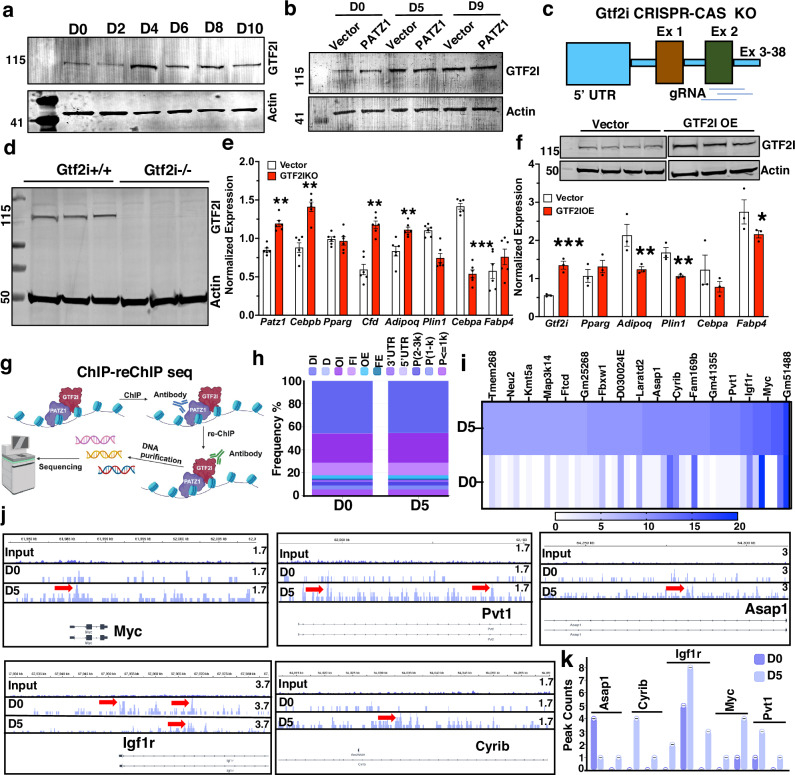
Our results show that both DNA-binding and protein-interaction domains are important for PATZ1’s adipogenic function. To further identify the PATZ1 interactome in differentiated 10T1/2 cells that could influence adipogenesis, we used an unbiased global PATZ1 immunoprecipitation mass spectrometry (IP-MS). Among all the interacting partners, GTF2I showed (i) the highest protein abundance index (EmPAI), (ii) the highest PATZ1/IgG abundance ratio, and (iii) the highest abundance p value (Fig. S3a). A recent meta-analysis on 1.4 million multi-ancestry participants showed that variants and risk alleles rs67755137-G (p = 2 × 10−8) and rs13238568-G (p = 2 × 10−9) for type 2 diabetes were mapped to Gtf2i locus32. Another study exploring 220 deep phenotype genome-wide association studies in non-European populations (~800,000) identified Gtf2i rs35473599-A (p = 3 × 10−9) as a risk allele for type 2 diabetes33. However, the role of GTF2I in the etiology of obesity and type 2 diabetes is still unexplored. GTF2I was expressed by WAT APCs and adipocytes but is markedly upregulated during adipogenesis, indicating a possible role for GTF2I in adipocyte differentiation (Fig. 3a and S3b). To test if PATZ1 regulated GTF2I, we measured GTF2I levels in control and PATZ1 overexpressing differentiated 10T1/2 cells and found that GTF2I was consistently upregulated during differentiation (Fig. 3b). However, GTF2I levels were comparable between PATZ1 and Vector cells (Fig. 3b). To test the role of GTF2I, we generated 10T1/2 cells with CRISPR-Cas9 knockdown of Gtf2i (Fig. 3c, d) and 10T1/2 cells stably overexpressing GTF2I (Fig. 3f). Knockdown of GTF2I markedly increased adipogenic genes, and overexpression caused downregulation of adipogenic genes, including PATZ1 (Fig. 3e, f), and knockdown of GTF2I did not change the expression of early adipogenic genes (Fig. S3c). To unbiasedly probe for DNA regions co-occupied by PATZ1 and GTF2I complex, we performed genome-wide chromatin-immunoprecipitation (ChIP)-re-ChIP sequencing using sequential PATZ1 and GTF2I antibodies on D0 and D5 differentiation of 10T1/2 cells (Fig. 3g). Binding distribution analysis showed similar frequency percentages of PATZ1 and GTF2I interaction in the indicated genomic loci between D0 and D5 (Fig. 3h), but the total peak proportion, peak counts, and fraction of all mapped reads (fRiP) scores were markedly higher on D5 (Fig. S3d). Annotated significant peak scores and peak locations showed an interaction between PATZ1 and GTF2I on the regulatory loci of known positive and negative adipocyte modulators such as Myc34–36, Pvt137,38, Igf1r39–42, Asap143, and Cyrib (Fig. 3i–k). Therefore, our data suggest a model in which GTF2I could form interactions with PATZ1 at the promoter regions of regulators to control adiposity and late differentiation.
Fig. 3. PATZ1 binding partner GTF2I regulates adipocyte differentiation.
a Immunoblot analysis of GTF2I protein during differentiation of 10T1/2. Actin was used as a loading control. Representative blot from three independent experiments. b Immunoblot analysis of GTF2I protein during differentiation of 10T1/2 expressing Vector, PATZ1, or PATZ1 transduced with GTF2I siRNA. Actin was used as a loading control. Representative blot from two independent experiments. c, d Model showing gRNA designed target Gtf2i exon 2 (c) and immunoblot analysis of GTF2I in WT (Gtf2i+/+) or GTF2I knockout (Gtf2i−/−) 10T1/2 cells (d). Representative blot from two independent experiments. e Realtime qPCR of indicated genes from D5 differentiated 10T1/2 vector or GTF2IKO cells. Vector: n = 3 and GTF2IKO: n = 3. f Realtime qPCR of indicated genes from D5 differentiated 10T1/2 vector or GTF2I overexpression (GTF2IOE) cells. The inset shows an immunoblot of GTF2I in Vector or GTF2IOE cells. Actin was used as a loading control. Representative blot from two independent experiments. Vector: n = 3 and GTF2IOE: n = 3. g Cartoon representation of the ChIP-reChIP sequencing experiment. h Functional annotation of peaks from D0 and D5 samples. DI distal intergenic, D downstream, OI other intron, FI first intron, OE other exon, FE first exon, UTR untranslated region, P promoter. i Heatmap showing annotated peaks regions on D0 and D5. j Bedgraph images of peaks on Input, D0, and D5 samples. Red arrows indicated significant “called” peaks. k Bar graphs showing consensus peak counts on the promoter and intronic regions of indicated gene locus on D0 and D5. n = 1 per timepoint. e, f Results are from three independent experiments. e, f Unpaired Student’s t-test. Data were mean ± s.e.m. ***p < 0.001; **p < 0.01; *p < 0.05. e, f Comparing to Vector. g Created in BioRender. Rajbhandari, P. (2020) BioRender.com/n87p485. Source data are provided as a Source Data file.
PATZ1 ablation in adipocytes controls white and beige adiposity
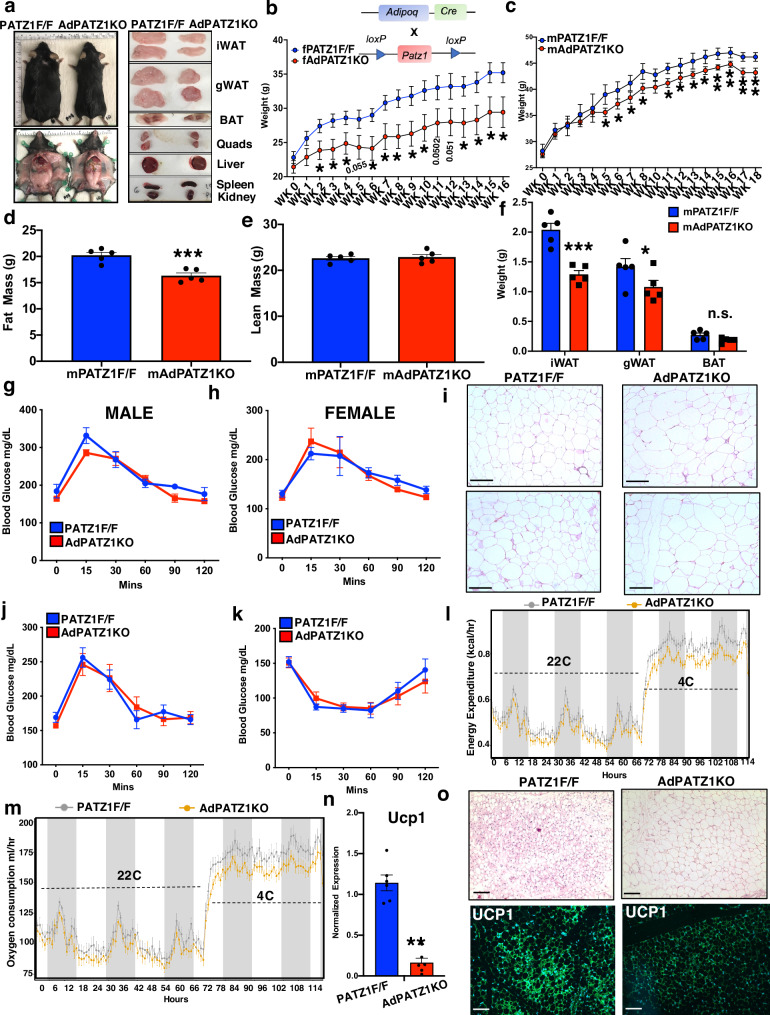
To test the role of PATZ1 in mouse adipose tissues, we generated an adipocyte-specific knockout of PATZ1 (AdPATZ1KO) by crossing Patz1f/f (PATZ1F/F) mice and Adipoq-Cre mice (Fig. S4a). On HFD, upon visual inspection after laparotomy, AdPATZ1KO mice appeared leaner and the size of iWATs and gWATs were reduced compared to controls, with no visible changes in quadriceps (quad), BAT, liver, spleen, or kidney (Fig. 4a). HFD-fed female AdPATZ1KO (fAdPATZ1KO) weighed less than controls and body composition analysis using magnetic resonance imaging (MRI) showed reduced fat mass in fAdPATZ1KO mice with no change in lean mass (Fig. 4b and S4b, c). Female mice also showed reduced iWAT and gWAT weights (Fig. S4d). To test whether PATZ1 adipogenic action was sex-dependent, we performed HFD studies and found that male AdPATZ1KO (mAdPATZ1KO) mice also displayed reduced fat mass and WAT weights, and small but statistically significant decreases in body weight compared to controls (Fig. 4c–f and S4e). On the 12th week of the HFD regiment, our GTTs showed similar glucose tolerance between AdPATZ1KO and control mice (Fig. 4g, h). However, histological analysis of WAT from AdPATZ1KO mice showed exceedingly larger adipocyte size compared to controls, indicating a potential increased lipid loading capacity of adipocytes (Fig. 4i). Gene expression analysis showed comparable expression of adipogenic genes in AdPATZ1KO and control mice except for Adipoq, Elf4, and Atf1 (Fig. S4f). These data show that PATZ1 regulates adiposity and protects mice from diet-induced obesity. Although PATZ1 did not affect beige adipogenesis in vitro, we wanted to test whether PATZ1 has any role in beige adipocyte biogenesis in vivo. We performed a cold exposure experiment on chow-fed AdPATZ1KO and control mice in temperature-controlled metabolic chambers. Mice were of similar weights on the chow-diet and had comparable GTT and ITT profiles (Fig. 4j, k). Metabolic chambers showed comparable EE and VO2 at 22 °C, but AdPATZ1KO mice showed markedly less EE and VO2 at 4 °C with no change in RER and food consumption (Figs. 4l, m and S4g, h). Gene expression showed highly reduced Ucp1 in the iWAT of AdPATZ1KO mice compared to controls (Fig. 4n). Furthermore, histological analysis showed markedly more multilocular adipocytes and UCP1 protein in the iWATs of control mice compared to AdPATZ1KO mice (Fig. 4o and S4i). Overall, our comprehensive in vitro and in vivo data suggest that PATZ1 is also involved in mature adipocyte biology, and PATZ1 absence could affect adiposity and beige adiposity in WAT.
Fig. 4. PATZ1 ablation in adipocytes protects mice from diet-induced obesity and reduced cold-induced beiging.
a (Left) External appearance of representative fPATZ1F/F and AdPATZ1KO mice after 16 weeks on HFD and (Right) gross appearance of tissues from PATZ1F/F and AdPATZ1KO mice after 16 weeks on HFD. b Body weight of fAdPATZ1KO and fPATZ1F/F female mice fed HFD for indicated weeks. fAdPATZ1KO: n = 7 and fPATZ1F/F: n = 5. c Body weight of mAdPATZ1KO and mPATZ1F/F male mice fed HFD for indicated weeks. mAdPATZ1KO: n = 5 and mPATZ1F/F: n = 5. d, e Body fat (D) and lean mass (E) in male PATZ1F/F and AdPATZ1KO mice after 18 weeks on HFD determined by EchoMRI. N = 5,5. f Average weight of individual white and brown adipose fat pads from male PATZ1F/F and AdPATZ1KO mice after 18 weeks on HFD. mAdPATZ1KO: n = 5 and mPATZ1F/F: n = 5. g, h GTT of male (g) and female (h) AdPATZ1KO and PATZ1F/F after 8 weeks of HFD. male AdPATZ1KO: n = 5, male PATZ1F/F: n = 5 and female AdPATZ1KO: n = 7, female PATZ1F/F: n = 5. i H&E staining of iWATs of 16 weeks HFD-fed AdPATZ1KO and PATZ1F/F mice. Scale bars, 100 μm. j, k GTT ( j) and ITT (k) of chow-fed 11-week-old AdPATZ1KO and PATZ1F/F mice. AdPATZ1KO: n = 9 and PATZ1F/F: n = 7. l, m Energy Expenditure (kcal/hr) (l) and Oxygen consumption (ml/hr) (m) of chow-fed 11-week-old AdPATZ1KO and PATZ1F/F mice were analyzed by Sable Promethion System metabolic chambers. Twelve-hour light/dark cycles, 114-h total duration; each light/gray bar represents 12-h duration. 0–114 h duration was at 22 °C (0–66 h) and at 4 °C (66–114 h). AdPATZ1KO: n = 9 and PATZ1F/F: n = 7. n Realtime qPCR of iWAT Ucp1 from cold-exposed mice mentioned in (l, m). AdPATZ1KO: n = 6 and PATZ1F/F: n = 6. o H&E and UCP1 immunofluorescence analysis of iWATs from cold-exposed mice mentioned in l and m. Scale bars, 100 μm. b–f, n Unpaired Student’s t-test. Data were mean ± s.e.m. ***p < 0.001; **p < 0.01; *p < 0.05; n.s. not significant. b–f, n, comparing to PATZ1F/F. Source data are provided as a Source Data file.
PATZ1 ablation in BAT precursors does not affect BAT activity
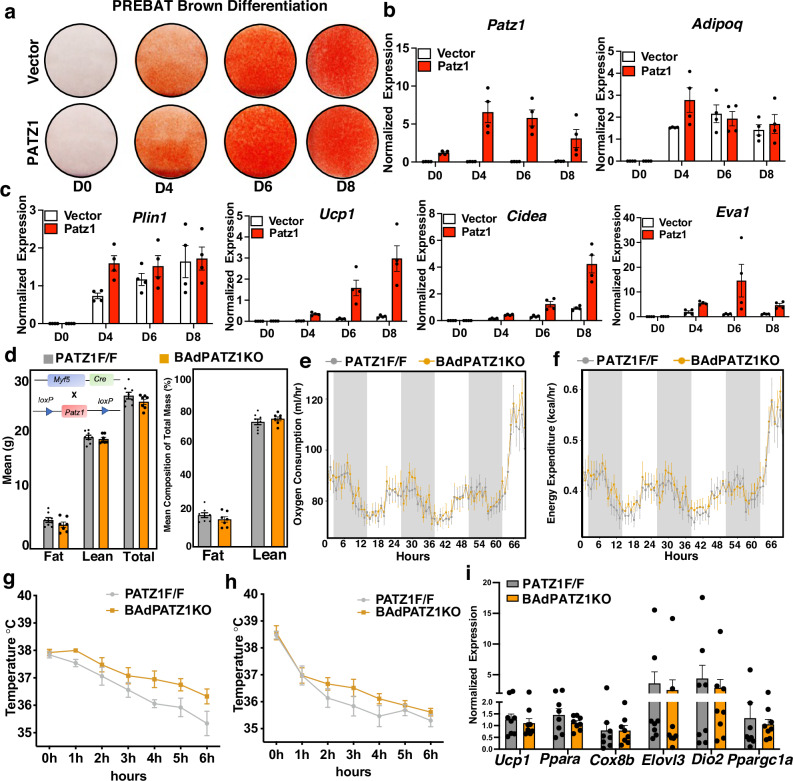
We next asked whether PATZ1 was an adipogenic factor in BAT. We first generated stable BAT preadipocytes (PREBAT) overexpressing PATZ1 or Vector and differentiated these cells into mature brown adipocytes using a brown adipogenic cocktail (See methods). As shown in Fig. 5a, b, PATZ1 did not alter the brown adipogenic potential of PREBAT cells, as shown by Oil-Red-O staining and qPCR analysis for Adipoq and Plin1. However, transcript analysis showed upregulation of thermogenic genes such as Ucp1 and Cidea (Fig. 5c). To test the in vivo role of PATZ1 in BAT development and thermogenesis, we crossed PATZ1f/f mice with myogenic factor 5-Cre (myf5-Cre) to generate PATZ1 BAT knockdown (BAdPATZ1KO) in BAT precursor cells. We asked whether BAT Patz1 deletion might result in an altered systemic energy balance. Indirect calorimetry using metabolic chambers revealed no differences in oxygen consumption (VO2) and energy expenditure (EE) between BAdPATZ1KO and control mice at 10 weeks of age (Fig. S5a, b). BAdPATZ1KO mice showed a slight reduction in lean mass and body weight compared to controls (Fig. S5a). Since MYF5 is also expressed in myogenic precursors, PATZ1 could potentially be involved in muscle development at an early age. However, body composition analysis showed no differences between BAdPATZ1KO and control mice at 24 weeks of age. (Fig. 5d). To test if BAdPATZ1KO mice showed age-dependent changes in systemic energy balance, we performed indirect calorimetry in 24-week-old BAdPATZ1KO and control mice. Metabolic chamber analysis at RT or cold showed no difference in VO2 and EE between the genotypes (Fig. 5e, f). BAdPATZ1KO and control mice raised at thermoneutral temperature (30 °C) also showed comparable body weights and indirect calorimetric measurements (Fig. S5c, d). BAT plays a vital role in non-shivering thermogenesis during cold exposure. To test if PATZ1 deletion in BAT affects cold tolerance, we exposed both male and female BAdPATZ1KO and control mice to cold stress (4 °C) for 6 h and measured their rectal temperature. As shown in Fig. 5g, h, both genotypes showed similar temperature changes, and we did not notice heightened or attenuated cold tolerance in BAdPATZ1KO mice compared to controls. In agreement with our cold exposure and indirect calorimetry data, BAT thermogenic gene expression was comparable between BAdPATZ1KO and controls (Fig. 5i). These data suggest PATZ1 may be dispensable for BAT development and function.
Fig. 5. PATZ1 deletion in BAT precursor does not affect BAT activity.
a Oil-Red-O staining of immortalized brown preadipocyte (PREBAT) cells stably expressing Vector or PATZ1, brown-differentiated for indicated days. b, c Real-time qPCR of indicated genes from Vector or PATZ1 overexpressing PREBAT cells brown differentiated for indicated days. Vector: n = 4 and Patz1: n = 4. d Body fat, lean mass, and total mass in female PATZ1F/F and BAdPATZ1KO mice at 24 weeks of age determined by EchoMRI. PATZ1F/F: n = 8 and BAdPATZ1F/F: n = 7. e, f Oxygen consumption (ml/hr) (H) and energy expenditure (kcal/hr) (I) of mice in (D) were analyzed by Sable Promethion System metabolic chambers. Twelve-hour light/dark cycles, 60-h total duration; each light/gray bar represents 12-h duration. 0–60 h duration was at 22 °C and the last 6 h was at 4 °C. PATZ1F/F: n = 8 and BAdPATZ1F/F: n = 7. g, h Rectal temperature of males (g) and females (h) PATZ1F/F and BAdPATZ1KO mice exposed to 4 °C for 6 h. Female PATZ1F/F: n = 5 and BAdPATZ1F/F: n = 4; male PATZ1F/F: n = 8 and BAdPATZ1F/F: n = 8. i Realtime qPCR of indicated genes from BAT derived from cold-exposed PATZ1F/F and BAdPATZ1KO mice. PATZ1F/F: n = 8 and BAdPATZ1F/F: n = 8. Data were mean ± s.e.m. Source data are provided as a Source Data file.
PATZ1 promotes an early adipogenic program
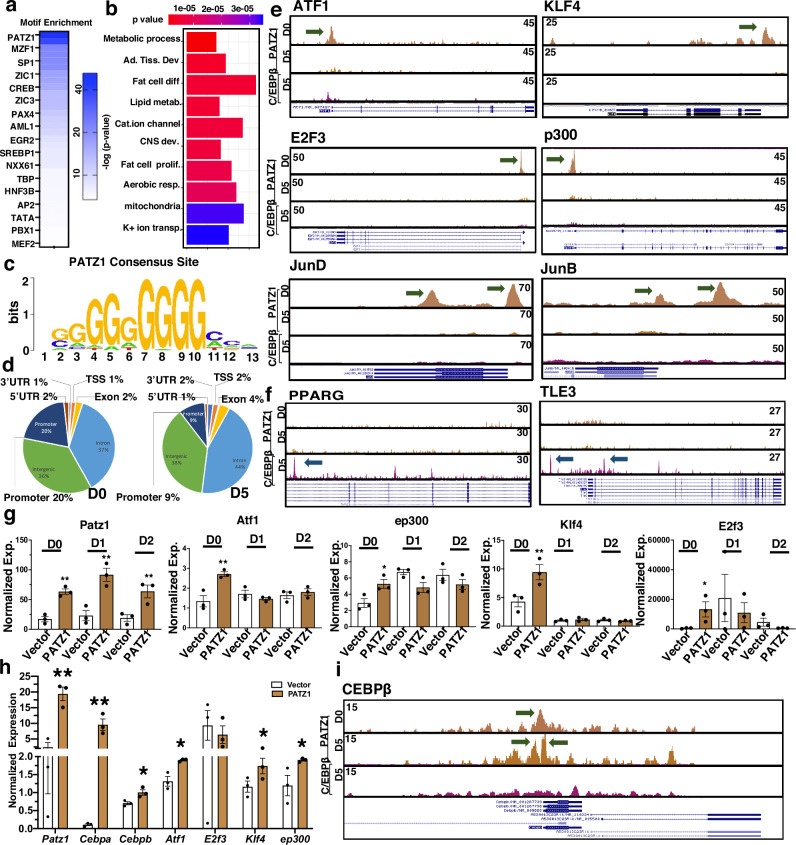
To characterize the DNA binding dynamics of PATZ1 during adipocyte differentiation, we performed genome-wide binding of PATZ1 using ChIP-seq (See Methods) during D0 and D5 of 10T1/2 adipogenic differentiation. Motif enrichment analysis of the ChIP-seq data showed strong enrichment for the PATZ1 motif, validating our immunoprecipitation method (Fig. 6a). The specific role of PATZ1 in binding genes related to adipogenesis was confirmed through over-representation tests of genes residing within 1 kB of PATZ1 ChIP-Seq peaks (p value <1e-3) which showed a significant representation in binding genes related to adipogenesis, respiration and lipid metabolic processes (Fig. 6b). Further analysis showed that PATZ1 bound to a consensus site of a stretch of guanines (G) and that genome-wide PATZ1 binding was enriched at intergenic and intronic regions (Fig. 6c). The percentage of promoter occupancy of PATZ1 markedly decreased from 20% at D0 to 9% at D5, indicating a substantial loss of PATZ1 promoter interactions during differentiation (Fig. 6d), despite no changes in PATZ1 protein levels (Fig. 1f and S1g). Further analysis of top PATZ1 peaks showed that PATZ1 binds preferentially to genes known to play key roles in the early stages of adipocyte differentiation, including ATF-17, AP-1 factors6 and KLF45, E2F311, and INSR (Fig. 6e and S6a, b). We identified a robust peak on the promoter regions of p300/CBP9, which are important for adipose formation (Fig. 6e). As a control, we included ChIP-seq for C/EBPß44 to compare PATZ1 binding to a known transcriptional regulator at D0 and D5 of differentiation (Fig. 6e). Although PATZ1 caused an increase in the expression of markers of late differentiation such as Pparg and Tle3, we detected recruitment of C/EBPß, but not PATZ1 to the TSS of those genes (Fig. 6f). These findings were independently reproduced for D2, D0, D5, and D8 differentiated 10T1/2 cells using a separate PATZ1 antibody (Fig. S6c). Directed qPCR showed increased expression of early adipogenic factors (Atf1, ep300, Klf4, and E2f3) in PATZ1-expressing cells, supporting our ChIP-seq data (Fig. 6g). By D2 of differentiation, PATZ1 expression also caused a marked increase in the transcription of late adipocyte genes such as Pparg, Adipoq, and Plin1 (Fig. S6d). Interestingly, qPCR analysis in undifferentiated Vector or PATZ1-expressing cells showed a robust increase in early genes, including Cebpb (Fig. 6h). Our ChIP-seq data further showed a PATZ1 binding peak at the Cebpb promoter regions at both D0 and D5 (Fig. 6i), implicating a role of PATZ1 and Cebpb in both preadipocytes and mature adipocytes. These results show that PATZ1 may play a role in the early phases of adipocyte differentiation when major chromatin remodeling events occur45.
Fig. 6. PATZ1 binds to the promoter regions during early adipocyte differentiation.
a Motif enrichment analysis from PATZ1 ChIP-seq. b Gene ontology derived from top PATZ1 peaks. c, d ChIP-seq data derived consensus binding site of PATZ1 (c) and global occupancies of PATZ1 from PATZ1 ChIP-seq in 10T1/2 cells differentiated for D0 and D5 (d). e, f Bedgraph image peaks from PATZ1 and C/EBPß ChIP-seq data showing PATZ1 and C/EBPß peaks at the promoter regions of indicated genes on D0 and D5 differentiated 10T1/2 cells. Green arrows show PATZ1 peaks. g Real-time qPCR of indicated genes from D0-D2 differentiated 10T1/2 cells expressing PATZ1 or vector. Vector: n = 3 and PATZ1: n = 3. h Real-time qPCR of indicated genes from undifferentiated 10T1/2 cells expressing PATZ1 or vector. Vector: n = 3 and PATZ1: n = 3. i Bedgraph image peaks from PATZ1 and C/EBPß ChIP-seq data showing PATZ1 and C/EBPß peaks at the promoter regions of Cebpb on D0 and D5 differentiated 10T1/2 cells. g, h Results from three independent replicates. g, h Unpaired Student’s t-test. Data were mean ± s.e.m. **p < 0.01; *p < 0.05. g, h comparing to Vector. Source data are provided as a Source Data file.
To test our findings in vivo, we generated a mouse model with Patz1 deletion in APCs by crossing Patz1f/f with platelet-derived growth factor receptor α-chain (Pdgfra)-Cre (pAdPATZ1KO) (Fig. 7a). Our results showed that knocking down Patz1 in mouse WAT APCs caused a decrease in body weight, fat mass, WAT size, and WAT weights compared to controls (Fig. 7b–f). Although Pdgfra-Cre effectively targets the adipocyte lineage in adipose tissue46, Pdgfra expression is not restricted to APCs or WAT. However, we did not notice any developmental defects in pAdPATZ1KO mice, and lean masses were comparable between pADPATZ1KO and control mice (Fig. 7e and S7a, b). Previous studies indicated a potential role of PATZ1 in the spleen and liver47,48, and our morphological analysis showed no differences in the spleen and liver between pADPATZ1KO and control mice (Fig. S7b). Histological analysis of WAT showed markedly larger adipocytes in pADPATZ1KO mice, and FACS analysis showed the percentage of PDGFRα + APCs was reduced to half compared to control mice, suggesting a potential role of PATZ1 in the survival of APCs (Fig. 7g, h and S7c). However, there were no changes in cell proliferation markers, including ki67, in cells overexpressing or lacking PATZ1 (Fig. S7d, e). Consistent with the ChIP-seq and in vitro data (Fig. 6), gene expression of undifferentiated APCs showed significantly reduced expression of early factors in pADPATZ1KO mice compared to control (Fig. 7i and S7f–h). Indirect calorimetry data showed a reduced energy expenditure (EE) and increased respiratory exchange ratio (RER) in pAdPATZ1KO mice compared to controls with no change in food consumption (Fig. 7j, k and S7j, k). To determine the impact of PATZ1 deletion in APCs during diet-induced obesity, we fed an HFD to pAdPATZ1KO and PATZ1F/F mice for 9 weeks. Although preAdPATZ1KO mice weighed less during the initial phases of the HFD regimen, after 9 weeks of the HFD regimen, the final weight, fat mass, and lean mass between AdPATZ1KO and control mice were similar (Fig. 7l, m). GTT showed that pADPATZ1KO mice were more glucose intolerant than controls, and although WAT sizes and gene expression were similar, there was presence of markedly hypertrophied adipocytes in preAdPATZ1KO mice compared to controls (Fig. 7n–p and S7i, l). Overall, our results suggest a role of PATZ1 in promoting the survival of APCs and early adipocyte differentiation both in vitro and vivo, and the absence of PATZ1 in APC causes an increase in hypertrophied adipocytes in WAT.
Fig. 7. PATZ1 ablation from APCs decreased fat mass and adiposity.
a Real time qPCR of Patz1 in iWATs of 10-week-old pAdPATZ1KO and PATZ1F/F mice. pAdPATZ1KO: n = 7 and PATZ1F/F: n−7. b–f Body weight (b), fat mass (c), gross appearance of iWAT and gWAT (d), lean mass (e), and iWAT and gWAT weights (f ) of 10-week-old pAdPATZ1KO and PATZ1F/F mice. pAdPATZ1KO: n = 5 and PATZ1F/F: n = 7. g H&E-stained histological slide from iWAT of 10-week-old pAdPATZ1KO and PATZ1F/F mice. Scale bars, 200 μm. h FACS analysis showing percentages of PDGFRα + cells from iWAT of 10-week-old pAdPATZ1KO and PATZ1F/F mice. i Real time qPCR of Patz1 in iWAT APCs of 10-week-old pAdPATZ1KO and PATZ1F/F mice. pAdPATZ1KO: n = 3 and PATZ1F/F: n = 3. j, k Energy expenditure (kcal/hr) ( j) and respiratory quotient ratio (k) of 20-week-old chow-fed pAdPATZ1KO and PATZ1F/F mice were analyzed by Sable Promethion System metabolic chambers. Twelve-hour light/dark cycles, 72-h total duration; each light/gray bar represents 12-h duration. 0–72 h duration was at 22 °C. pAdPATZ1KO: n = 10 and PATZ1F/F: n = 6. l, m Body weight (l) ad fat mass and lean mass (m) of 9 weeks HFD-fed pAdPATZ1KO and PATZ1F/F mice. pAdPATZ1KO: n = 10 and PATZ1F/F: n = 6. n GTT was performed in 9 weeks HFD-fed pAdPATZ1KO and PATZ1F/F mice pAdPATZ1KO: n = 10 and PATZ1F/F: n = 6. o Gross appearance of iWATs, gWATs, BATs, and livers of 9 weeks HFD-fed pAdPATZ1KO and PATZ1F/F mice. p H&E-stained histological slide from iWAT of 9 weeks HFD-fed pAdPATZ1KO and PATZ1F/F mice. Scale bars, 100 μm. i Results from three independent experiments. a–c, e, f, i–l Unpaired Student’s t-test. n Two-way ANOVA. j, k ANCOVA. Data were mean ± s.e.m. **p < 0.01; *p < 0.05. b, c, e, f, i–l, n comparing to PATZ1F/F. Source data are provided as a Source Data file.
Discussion
Adipogenesis is a highly orchestrated event involving the interplay between transcriptional networks. In the present study, we have delineated the role of PATZ1 in transcriptional events controlling adipose tissue and adipocyte homeostasis. PATZ1 is expressed in both SVF and adipocyte portions of WAT. PATZ1 is not a PPARγ target gene, and its level remains relatively constant during adipocyte differentiation. Our data show PATZ1 promotes adipogenesis through a mechanism dependent on DNA binding and protein interaction. Knockout of PATZ1 in adipose tissues protects mice from obesity. Mechanistically, PATZ1 binds to the promoter regulatory regions of early adipogenic factors. We also identified another transcription factor affecting adipogenesis, promoter-binding transcription factor GTF2I, that attenuates PATZ1 adipogenic function.
Previous work has linked zinc finger proteins (ZFPs) to adipogenesis. Early commitment factors such as ZFP423 are known to drive adipogenesis and maintain white adiposity3,49,50. ZFP467 suppresses osteogenesis and promotes adipogenesis of fibroblast-like progenitors by enhancing the expression of another master regulator of adipogenic transcription, CCAAT/enhancer-binding protein-α (C/EBPα)51. ZFP KLF5 acts as a coactivator of C/EBPβ, enhancing PPARγ and C/EBPα transcription8. Architecturally similar ZFPs containing BTB/POZ domains, such as BCL6 and PLZF, have also been implicated in adipogenesis52–54. Our discovery of PATZ1 as a new adipogenic transcription factor adds to the diverse repertoire of ZFP transcription factors in regulating various early and late stages of adipogenesis.
Genome-wide chromatin accessibility approaches, transcriptomics (RNA-seq and scRNA-seq), and chromatin-immunoprecipitation-seq (ChIP-seq) have provided mechanistic insights into the role of TFs driving the initial stage of adipogenesis55. DNase I hypersensitive (DHS) regions based on ultradeep sequencing and ChIP-seq have revealed genomic footprinting and hotspots at enhancers and super-enhancers of PPARγ occupied by early adipogenic factors such as AP-1, GR, ATFs, and KLFs12. This approach has revealed a network of the first wave of commitment factors that modify the chromatin environment of terminal factors like PPARγ to enable terminal adipocyte differentiation. Genome-wide glucocorticoid receptor (GR) Chip-seq in differentiated 10T1/2 cells56 and FAIRE-seq (formaldehyde-assisted isolation of regulatory elements-seq) in differentiated 3T3-L1 cells57 showed an abundance of the PATZ1 consensus region, hinting towards a plausible role of PATZ1 in adipose biology. We performed an unbiased ChIP-seq study to probe for PATZ1 DNA binding capacity during adipogenesis. We observed that the PATZ1 cistromic region includes GC-rich motifs. PATZ1 promoter occupancy revealed marked changes between D0 and D5. Loss of PATZ1 promoter binding at D5 of adipocyte differentiation despite stable PATZ1 expression levels suggests a differentiation-dependent DNA-binding capacity of PATZ1. PATZ1-binding peaks were primarily present at the promoter regions of genes known to play key roles in the early stages of adipocyte differentiation, including ATF-17, AP-1 factors6, KLF45, and E2F311. We also identified a robust peak of PATZ1 binding on the promoter region of p300, which is known to be critical for adipocyte formation9. Further, our directed qPCR studies showed an increase in the expression of these factors in PATZ1-expressing cells, directly supporting the role of PATZ1 binding in increasing their gene expression. Of particular interest was that PATZ1 increased the expression of late differentiation markers (Pparg, Tle3, and Plin1), but we did not detect the recruitment of PATZ1 to the TSS of those genes. These findings suggest that PATZ1 plays a key role in the early regulatory wave of adipogenesis, ultimately promoting terminal differentiation. Our results suggest that PATZ1 plays a vital role in the early and commitment phases of adipocyte differentiation when major chromatin remodeling events occur45.
WAT expansion in obesity relies on continuously recruiting adipocyte progenitor cells (APCs). APCs’ proliferation, self-renewal, and differentiation into preadipocytes and then adipocytes drive AT remodeling. APCs, found within adipose stromal cells (ASCs) similar to mesenchymal stromal cells (MSCs) in other organs, include distinct sub-populations marked by PDGFRα58. Our data from the APC knockout of PATZ1 showed that the absence of PATZ1 caused a marked decrease in body weight, fat mass, and adipose size but not lean mass. Histological analysis of chow-fed mice showed increased adipocyte size, and FACS analysis showed a markedly decreased percentage of the PDGFRα population. qPCR analysis of APCs shows significantly fewer early adipogenic genes. Indirect calorimetry showed reduced EE and increased RER, indicating a defect in fat metabolism. This data shows that PATZ1 is crucial for the transcription program in APCs and the APC population.
Interestingly, after prolonged HFD feeding, preAdPATZ1KO mice weighed comparably to control mice with similar fat and lean mass. However, the size of WAT depots of preAdPATZ1KO mice were visibly larger than controls, and histological analysis showed markedly increased hypertrophic adipocytes. This hypertrophy was consistent with highly glucose-intolerant AdPATZ1KO mice compared to controls. Our data from in vitro and in vivo studies show a crucial role of PATZ1 in APC homeostasis, an its disruption may perturbation of PATZ1 could lead to metabolic abnormalities and WAT dysfunction.
Our animal data also suggest a role of PATZ1 in adipocytes since the adult Adipoq-Cre mouse model will not deplete PATZ1 in uncommitted APCs and WAT MSCs. Adipocyte-specific deletion of the Patz1 gene resulted in decreased WAT mass and protection from diet-induced obesity in both sexes fed an HFD (60% kcal fat). Histological analysis of WAT showed a markedly higher proportion of larger adipocytes in AdPATZ1KO mice than in control HFD. Although GTT between AdPATZ1KO and controls were comparable, hypertrophic adipocytes could lead to a decrease in glucose tolerance and insulin resistance under prolonged HFD. However, gene expression analysis of adipogenic factors was similar between AdPATZ1KO and PATZ1F/F mice, suggesting a more complex mechanism of PATZ1 in controlling mature adiposity and needs further investigation. PATZ1 expression was downregulated under obesity, yet AdPATZ1KO was protected from diet-induced obesity compared to control mice. The expression pattern of PATZ1 in obese models resembled that of PPARγ and adiponectin20, and the phenotype of AdPATZ1KO mice could arise from depot differences and differential expression of PATZ1 in the SVF and adipocyte portion of iWAT and gWAT in obesity (Fig. 1j, k), where PATZ1 was only decreased in the gWATs of mice under HFD. Our cold exposure experiments in AdPATZ1KO showed reduced energy expenditure and WAT beiging, implicating a potential role of PATZ1 in cold-induced beige adipogenesis. Recent studies have demonstrated the importance of adipocytes in de novo beige adipogenesis59, and adipocytes lacking PATZ1 could be defective in eliciting beige adipogenesis and/or transdifferentiation. Overall, our data suggest that PATZ1 could have a role in maintaining adiposity by regulating gene programs in mature adipocytes.
To better understand how PATZ1 contributes to adipogenesis, we performed PATZ1 IP-MS on differentiated 10 T/2 cells. PATZ1 is known to interact with other transcriptional regulators known to affect adipose homeostasis, including BCL660, p5317, NCoR61, and Bach162–65. However, our IP-MS results did not show enrichment of these proteins. Among all the interacting partners, GTF2I showed the highest abundance in the PATZ1 complex. GTF2I is a transcription factor involved in both basal transcription and signal-induced transduction activation or repression66. Therefore, GTF2I is not considered a general transcription factor but rather a protein that regulates transcription of a selected number of genes67. Like PATZ1, GTF2I can inhibit and activate genes depending on stimuli and cellular context. The role of GTF2I in adipose biology and adipogenesis is still unexplored. Still, another member of the GTF2I family of TFs, GTF2IRD1, is known to bind PRDM16 and suppress the adipose tissue fibrosis gene program68. ChIP-seq studies have shown a high preference for GTF2I to bind to the regulatory regions of its target genes69, and its occupancy overlaps with activating H3K4me3 peaks that mark transcriptionally active regions. Strikingly, genome sequences under GTF2I bound peaks are enriched for motifs for transcriptional regulators, including PATZ169. We showed that GTF2I is upregulated during adipocyte differentiation. Surprisingly, we found that GTF2I interaction with PATZ1 could form a repressive complex to inhibit adipogenesis since ablation of GTF2I enhanced, while overexpression decreased the expression of adipogenic genes. PATZ1 overexpression did not cause upregulation of GTF2I, suggesting a potential interaction-based dynamic of PATZ1 and GTF2I in regulating adipogenesis. This was further supported by our ChIP-reChIP seq data, where we found potential PATZI-GTF2I complex on known regulators of adipocyte differentiation and adipocyte function. Our findings reveal PATZ1 and GTF2I as two critical regulators of adipogenesis and adiposity.
Methods
All experiments were conducted in accordance with the Mount Sinai Institutional Animal Care and Research Advisory Committee and Institutional Review Boards of Mount Sinai School of Medicine
Reagents, plasmids, and viruses
Dexamethasone (D2915), 3-isobutyl-1-methylxanthine (IBMX, I7018), PPARγ agonist GW1929 (G5668), and Oil-Red-O (O0625) were from Sigma-Aldrich. Insulin (12585-014) was from Thermo Scientific Fisher. cDNA encoding PATZ1, NFXL1, and NCOA5 was cloned from the cDNA library into pBabe-puro retroviral vector using Gateways Cloning System (Thermo). PATZ1 domain deletion mutants (ΔPOZ, ΔZnF, and ΔPOZ/AT) were cloned out of WT PATZ1 cDNA using site-specific and domain-spanning primers and cloned in the pBabe retroviral vectors. pBABE-plamids were transfected into retrovirus packaging Pheonix E cells for 48 h. Target cells (immortalized beige/brown preadipocytes) were plated at 50% confluency 24 h post-transfection. Forty-eight hours after transfection media from transfected Phoenix-E cells were harvested and spun down for 5 min at 5000 rpm to pellet cells and debris. Retrovirus-containing supernatant was carefully removed and plated onto target cells with 1:1000 polybrene for overnight. The next day, the media was replaced with regular growth media, and cells were incubated for an additional 24 h. About 4.5 μg/ml puromycin selection was performed to select for cells stably expressing PATZ1. Patz1 shRNA sequences were designed using the BLOCK-iT RNAi designer tool (Invitrogen). Sense and antisense oligos were annealed and cloned into pENTR/U6 plasmid (Invitrogen). Using LR recombination (Invitrogen), shRNA constructs were subcloned into a Gateway-adapted pBabe-Puro plasmid. Only sense strands are shown here. Adenovirus was amplified, purified, and tittered by Viraquest. PATZ1 was cloned into the pDONR221 entry vector using the Gateway system. The sequences in the entry clones were then transferred by LR recombination into the pAd/CMV/V5-DEST Gateway vector for viral particle production. All adenoviruses, including Ad-GFP and Ad-CRE, were amplified, purified, and titered by Viraquest.
Animals
C57BL/6 WT male and female mice (#000664), Adipoq-CRE (#028020), and Myf5-Cre (#007893), Pdgfra-Cre (#013148) were acquired from Jackson Laboratory. Patz1f/f (PATZ1 F/F) mice were generated as described previously70. All mice were maintained in a pathogen-free barrier-protected environment (12:12 h light/dark cycle, 22–24 °C) at the Mount Sinai animal facilities. Mice were fed with either a Chow diet (Purina, 5001) or 60% kcal HFD (Research Diets, D12492) and had unrestricted access to food and water. Animal experiments were conducted in accordance with the Mount Sinai Institutional Animal Care and Research Advisory Committee. We did not use randomization in our allocation process since the experimental groups were primarily separated based on genotype or sex. For animal experiments, mice were separated based on genotype or sex, so the investigators were not blinded.
Human participants
Human participants—Abdominal subcutaneous adipose tissues were sampled from needle aspiration from a volunteer (Sex: Female, age: 30–40 years) who was free of diabetes, cancers, and endocrine or inflammatory diseases by medical history. All participants provided written informed consent, and the study was approved by the Institutional Review Boards of Mount Sinai School of Medicine.
Cell culture
Murine preadipocyte cell lines 3T3-L1 and 10T1/2, and primary SVFs were cultured in DMEM supplemented with 10% fetal calf serum (FCS) for 3T3-L1 and 10% fetal bovine serum (FBS) for 10T1/2. For in vitro differentiation, preadipocytes were grown to confluence in DMEM with 10% FBS plus insulin (5 μg/ml). Confluent cells were induced to differentiate with dexamethasone (1 μM), IBMX (0.5 μM), insulin (5 μg/ml), and with or without GW1929 for 2 days, followed by insulin and GW1929 alone. Growth media was exchanged every 2 days during differentiation. All stable cells were generated using the pBabe retroviral system71. Retrovirus was obtained by transfection of Phoenix-E cells with the pBabe vector and harvest of growth media 72 h later. Preadipocytes were transduced with retrovirus overnight and selected with antibiotics. For adenoviral overexpression of GFP or PATZ1, differentiated 10T1/2 cells expressing hCAR were transduced with Ad-GFP or Ad-PATZ1 on indicated days for 24 h, and cells were harvested for transcript analysis by qPCR. For adenoviral CRE mediated knock down of PATZ1, SVFs from iWATs of PATZ1F/F mice were infected with Ad-GFP or Ad-CRE for 24 h and differentiated into white adipocytes and samples were harvested on D0 or D7. Human ADSCs were isolated with collagenase digestion of adipose tissue samples, cultured, and differentiated following published protocols72. Frozen primary human ADSCs were derived from abdominal scWAT as described previously73 and were transduced with Vector or PATZ1 retrovirus and differentiated using published protocol73. For human PATZ1 expression analysis, abdominal subcutaneous ASCs (P4, were cultured and differentiated as previously described (7 days of differentiation cocktail and 7 days of lipid filling (maintenance media). Cells were from a female volunteer (BMI :33.7 kg/m2, age: 30–40 years). Cells were harvested to prepare lysates 2 days after confluence and after full differentiation (day 14 of culture). During the maintenance phase, cells were refed every 2 days with freshly prepared maintenance media (DMEM:F12 supplemented 10 nM, insulin 10 nM, and dexamethasone). For the glucose study, 10T1/2 cells were maintained in DMEM without glucose and supplemented with 2% FBS. Cells were then treated with various concentrations of glucose (Corning) and harvested 6 h, 16 h (ON), and 24 h after glucose treatment. CRISPR-Cas9 technique was used to knockout GTF2I from 10T1/2 cells using gRNA targeting exon 3 and retroviral stable expression system was used to generated GTF2I overexpression cells using methods described above. Glucose uptake assay was conducted using a Glucose Uptake Assay Kit (Abcam #ab136955) according to the manufacturer’s instruction.
Isolation of and immortalization of primary white and brown preadipocytes
C57BL/6 WT male mice (8–10-week-old) were euthanized using isoflurane. The procedure was performed as previously described44. Briefly, euthanized mice were sprayed with 70% ethanol. Around 100–300 mg of iWAT or BAT was dissected, placed in ice-cold 1X PBS, blotted dry, and minced. Minced fat (600–800 mg) was digested in 3 ml of buffer (PBS, collagenase D or B, Dispase II, and CaCl2) at 37 °C for 45 min. After digestion, the mixture was resuspended with media, filtered, and centrifuged. The pellet was plated on collagen-coated plates and cultured until 70% confluence. Stromal vascular fractions were differentiated into beige or brown adipocytes or immortalized using a retrovirus expressing large T-antigen and selected with hygromycin.
Gene expression analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse transcribed with the High-Capacity cDNA synthesis kit (Thermo). cDNA was quantified by real-time PCR using SYBR Green Master Mix (Diagenode) on a Quant Studio 5 384 well plate. Gene expression levels were determined by using a standard curve. Each gene was normalized to the housekeeping gene 36B4 and was analyzed in duplicate. Primers used for real-time PCR are listed in Supplementary Table 1.
Protein analysis
Whole cell lysate was extracted using RIPA lysis buffer (Boston Bioproducts) supplemented with a complete protease inhibitor cocktail (Roche). Proteins were diluted in Nupage loading dye (Invitrogen), heated at 95 °C for 5 min, and run on 4–12% NuPAGE Bis-Tris Gel (Invitrogen). Proteins were transferred to hybond ECL membrane (GE Healthcare) and blotted with PATZ1 (sc-390577, Santa Cruz Biotechnology), (PATZ1 (A10053, Abclonal), GTF2I (PA5-17642, Thermo), Hmg1 (ab18256, Abcam), actin (A2066, Sigma), αTubulin (CP06, Calbiochem), GAPDH (D16H11, Cell Signaling) antibodies.
RNA-seq
Total RNA was prepared as described in ref. 74. Strand-specific libraries were generated from 500 ng total RNA using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). cDNA libraries were single-end sequenced (50 bp) on an Illumina HiSeq 2000 or 4000. Reads were aligned to the mouse genome (NCBI37/mm9) with TopHat v1.3.3 and allowed one alignment with up to two mismatches per read. mRNA RPKM values were calculated using Seqmonk’s mRNA quantitation pipeline. All FPKMs represent an average of three biological replicates for in vitro studies for library construction. A gene was included in the analysis if it met all of the following criteria: The maximum FPKM reached 10 at any timepoint, and the gene length was >200 bp as determined by the DESeq2 package in R Bioconductor. P values were adjusted using the Benjamini–Hochberg procedure of multiple hypothesis testing75.
Co-immunoprecipitation (Co-IP) and mass spectrometry
Cells expressing PATZ1 were lysed with ice-cold lysis buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.6% Triton X-100, plus a complete protease inhibitor (Roche). Cell lysates were pre-cleared with empty Protein A/G PLUS agarose beads (Santa Cruz) and then immunoprecipitated with beads prelinked with anti-Flag M2 antibody (F1804, Sigma) overnight at 4 °C. Beads were washed with 50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1% Triton X-100, then eluted with 3X Flag peptides (F4799, Sigma). In specific experiments, the RNase cocktail enzyme (AM2286, Ambion) was used to treat cell lysates before immunoprecipitation. IP Eluents from Flag-PATZ1 cells and control cells were processed by UCLA Pasarow Mass Spectrometry Laboratory. The results were analyzed using the Mascot software.
Mature Adipocyte and SVF fractionation and nuclei isolation
Cells were washed with 1X PBS and incubated in TEN buffer (10 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 7.5). After swelling on ice for 15 min in a solution containing 10 mM HEPES, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, and protease inhibitors, cells were lysed with a 0.4% NP-40 alternative. The cytoplasmic fraction was collected after centrifugation at 12,000×g for 5 min. The nuclear pellet was resuspended in 20 mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, and protease inhibitors, followed by centrifugation. RNA was extracted from both fractions using a Purelink RNA mini kit, with ribonuclease inhibitors added to all buffers for RNA protection.
Chromatin-immunoprecipitation (ChIP)-seq (ChIP-Seq) and ChIP-reChip seq—
ChIP experiments were performed according to standard protocols18,76. Lysed cells were sonicated using a Bioruptor (Diagenode) according to the manufacturer’s protocol, and chromatin was immunoprecipitated with antibodies against PATZ1 (sc-390577, Santa Cruz Biotechnology) for D0 and D5, sequential PATZ1 and GTF2I (PA5-17642, Thermo Fisher) for ChIP-reChip seq, PATZ1 (A10053, Abclonal) for D2, D0, D5, D8, and IgG (PP64, Millipore) overnight at 4 °C in the presence of Protein A beads (GE Healthcare). ChIP-Seq libraries were prepared using the Kapa LTP Library Preparation Kit (Kapa Biosystems). ChIP-Seq was performed as described in refs. 44,74. To confirm the robustness of peaks, the same BAM files were analyzed via HOMER, where 84% of the peaks (marked by genomic coordinates) overlapped. About 77,599 significant peaks (P < 1e-3) were detected in D0, and 131,553 significant peaks (P < 1e-3) were detected in D5. While little significant peaks overlapped between d0 and d5 (132) at the genomic coordinate level, 452 genes were observed to intersect between the two. We note that, possibly due to sequencing depth or other parameters, the most significant results were observed in D0, evident by comparing a highly stringent threshold (P < 1e-10) where 696 peaks were observed in D0. Reads were aligned to the mouse genome (NCBI37/mm9) with Bowtie. Uniquely mapped reads were used for peak calling and annotation using HOMER77. Peaks were called if they passed a false discovery rate of 0.01 and were enriched over input. Peaks were annotated to the nearest TSS. ChIP-reChip seq data were analyzed using the Pluto Biosciences (Denver, CO) software platform. Briefly, ChIP-seq pipeline paired_end FASTQ files were processed using the nf-core-chipseq pipeline (v2.0.0). Adapter sequences were removed with Trim Galore. Reads were aligned with Bowtie to GRCm38 (NCBI, p.6, release 108, updated annotations), and duplicates were marked and removed from all samples with picard. “peaks” refer to regions where at least one of the target samples significantly exceeds its matched control for a given genomic region. Here, peaks were called with MACS (peak mode: narrow) using a p value threshold of 0.01 and an effective genome size of 2,650,000,000. Consensus peaks were merged across all samples using BEDTools and annotated to the nearest gene transcriptional start site using HOMER. Peak counts were generated using featureCounts and added to the figure. PATZ1-GTF2I ChIP-reChip seq, D0;D5: unique mapped reads- 2,487,257; 3,644,176; Mapped: 338,601;448,900, MACS2 PEAK TSS COUNT-6540; 6163; paired-end.
Cold exposure studies
For the 4 °C cold exposure experiment, PATZ1 F/F or BAdPATZ1KO mice at 8–10 weeks of age were singly or doubly housed at 4 °C in a non-bedded cage with access to food and water for the time points indicated in the figure legend. At the end of the experiment, BATs were resected for gene expression analysis. The core body temperature of WT and Il10−/− mice was measured at room temperature using a rectal probe (BAT-10) purchased from Physitemp.
High-fat diet studies
For diet study, 10 weeks of age mice were fed a 60% high-fat diet (Research Diets) for the indicated times. Mice weights and body composition were measured every week and food was replaced weekly.
Indirect calorimetry and body composition measurements
Indirect calorimetry was performed using Promethion Metabolic Chambers (Sable Systems). Animals were placed individually in chambers for 3 consecutive days at ambient temperature (22.5 °C) or 6 h in cold temperature (4 °C) with 12 h light/dark cycles. Animals had free access to food and water. Respiratory measurements were made in 6-min intervals after an initial 7–9 h acclimation period. Energy expenditure (EE) was calculated from oxygen consumption (VO2) and respiratory exchange ratio (RER) using the Lusk equation, EE in Kcal/h = (3.815 + 1.232 X RER) X VO2 in ml/min. Food intake was measured in metabolic chambers. Body composition (fat and lean mass) was determined using the EchoMRI Body Composition Analyzer. Indirect calorimetry data were analyzed by CALR web-based software78.
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
GTT and ITT were performed as previously described in ref. 44. Briefly, for GTT, mice were fasted for 6 h and then administered an intraperitoneal injection of glucose (2 g/kg). For ITT, after the same fasting period, mice received an intraperitoneal injection of insulin (1 U/kg). Blood glucose levels were measured at the time points specified in the figure legends using the ACCUCHEK active glucometer (Roche).
Tissue hematoxylin and eosin (H&E) staining and immunohistochemistry
Tissues (4–5-microns thickness) were placed in cassettes and submerged in 10% formalin solution overnight. Tissue cassettes were washed with tap water for 15 min and stored in 70% EtOH at room temperature. Paraffin embedment and H&E staining was performed at the Pathology Core Laboratory at Mount Sinai.
Mouse and human population-based investigation of Patz1
Data analysis in mouse and human populations was conducted as previously described in ref. 44. Briefly, data79,80 from 106 inbred mouse strains fed a high-fat, high-sucrose diet for 8 weeks was analyzed for global adipose tissue expression using Affymetrix HT_MG430A arrays, and correlated with phenotypic data. Similarly, human adipose tissue expression from the METSIM study21,22 was analyzed using Affymetrix U219 arrays. Correlations were calculated using the midweight bicorrelation coefficient and adjusted p values, processed through the R package WGCNA81.
Statistics
All data were presented as mean ± SEM and analyzed using Microsoft Excel and Prism (GraphPad). Unpaired Student’s t-test was used for single variable comparison between two groups. One-way ANOVA followed by the Dunnett post hoc test was used for multiple comparisons versus the control group. Two-way ANOVA followed by Bonferroni posttests was used to examine interactions between multiple variables. Statistical significance for indirect calorimetry study was determined by using multiple regression analysis (ANCOVA). Measurements were taken from distinct samples. Data were presented as ±SEM p < 0.05 was statistically significant and is presented as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, or ∗∗∗∗p < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
P.R. is supported by R01DK136035, DP1DK140003, Irma T. Hirschl/Monique Weill-Caulier Foundation Scholar Award, and NIDDK-supported Einstein-Sinai Diabetes Research Center (DRC) Pilot & Feasibility Award. M.S. is supported by NIH-NIDDK grants DK130640 and DK097771. W.E. has been supported for work on MAZR/PATZ1 by the Austrian Science Fund projects P19930, P23641, and P34407. J.P.W. is supported by R01CA196263. P.T. is supported by DK120851. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Author contributions
S.P., K.G., CH.O., J.S., N.S., and S.S. performed animal and in vitro differentiation experiments under the supervision of P.R. R.Y. performed human ADSC experiment under the supervision of S.K.F. and P.R. PATZ1 F/F mice were generated by S.S. and W.E. S.P., J.W., and P.R. performed ChIP-Seq and IP-MS under the supervision of J.P.W., P.T., and P.R. M.S. supervised C.M.N. and processed the ChIP-seq and RNA-seq data. H.W. and C.J.V. designed and supervised the cDNA library screen under the supervision of P.T. P.T. and P.R. conceived the project, and P.T. and P.R. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Yoshihiro Matsumura and the other, anonymous, reviewers for their contribution to the peer review of this work.
Data availability
Source data for all figures are provided with the paper. NGS data generated for this paper has been uploaded to the Gene Expression Omnibus under accession numbers GSE273317, GSE273318, GSE273319, and GSE273320. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52917-y.
References
- 1.Rosen, E. D. & MacDougald, O. A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol.7, 885–896 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Cristancho, A. G. & Lazar, M. A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol.12, 722–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta, R. K. et al. Transcriptional control of preadipocyte determination by Zfp423. Nature464, 619–623 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristancho, A. G. et al. Repressor transcription factor 7-like 1 promotes adipogenic competency in precursor cells. Proc. Natl Acad. Sci. USA108, 16271–16276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birsoy, K., Chen, Z. & Friedman, J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab.7, 339–347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinent, M. et al. Adipose triglyceride lipase and hormone-sensitive lipase are involved in fat loss in JunB-deficient mice. Endocrinology152, 2678–2689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, K. E. et al. Depletion of cAMP-response element-binding protein/ATF1 inhibits adipogenic conversion of 3T3-L1 cells ectopically expressing CCAAT/enhancer-binding protein (C/EBP) alpha, C/EBP beta, or PPAR gamma 2. J. Biol. Chem.281, 40341–40353 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Oishi, Y. et al. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab.1, 27–39 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Namwanje, M. et al. The depot-specific and essential roles of CBP/p300 in regulating adipose plasticity. J. Endocrinol.240, 257–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mink, S., Haenig, B. & Klempnauer, K. H. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell Biol.17, 6609–6617 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fajas, L. et al. E2Fs regulate adipocyte differentiation. Dev. Cell3, 39–49 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Siersbæk, R. et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep.7, 1443–1455 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Siersbæk, R. et al. Molecular architecture of transcription factor hotspots in early adipogenesis. Cell Rep.7, 1434–1442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell156, 20–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedele, M. et al. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J. Biol. Chem.275, 7894–7901 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Ma, H. et al. The dosage of Patz1 modulates reprogramming process. Sci. Rep.4, 7519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentino, T. et al. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis.4, e963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva, ClaudioJ. et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab.13, 413–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson, S. et al. Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism. Genes Dev.33, 747–762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, J. et al. RNA-binding protein PSPC1 promotes the differentiation-dependent nuclear export of adipocyte RNAs. J. Clin. Invest.127, 987–1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laakso, M. et al. The metabolic syndrome in men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res.58, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stancakova, A. et al. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes58, 1212–1221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature603, 926–933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almanzar, N. et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature583, 590–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siersbaek, M. S. et al. Genome-wide profiling of peroxisome proliferator-activated receptor gamma in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol. Cell Biol.32, 3452–3463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, G. et al. PATZ1 down-regulates FADS1 by binding to rs174557 and is opposed by SP1/SREBP1c. Nucleic Acids Res.45, 2408–2422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambele, M. A. & Pepper, M. S. Identification of transcription factors potentially involved in human adipogenesis in vitro. Mol. Genet. Genom. Med.5, 210–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell184, 5985–6001.e5919 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craft, C. S. et al. The extracellular matrix protein MAGP1 supports thermogenesis and protects against obesity and diabetes through regulation of TGF-β. Diabetes63, 1920–1932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bischof, J. M., Stewart, C. L. & Wevrick, R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum. Mol. Genet.16, 2713–2719 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Szatkowski, C. et al. Prokineticin receptor 1 as a novel suppressor of preadipocyte proliferation and differentiation to control obesity. PLoS ONE8, e81175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vujkovic, M. et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet.52, 680–691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet.53, 1415–1424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freytag, S. O. & Geddes, T. J. Reciprocal regulation of adipogenesis by Myc and C/EBPα. Science256, 379–382 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Abdesselem, H. et al. SIRT1 limits adipocyte hyperplasia through c-Myc inhibition. J. Biol. Chem.291, 2119–2135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deisenroth, C. et al. MYC is an early response regulator of human adipogenesis in adipose stem cells. PLoS ONE9, e114133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, L., Zhang, D., Qin, Z. Y., Li, J. & Shen, Z. Y. The role and possible mechanism of long noncoding RNA PVT1 in modulating 3T3-L1 preadipocyte proliferation and differentiation. IUBMB Life72, 1460–1467 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Squillaro, T., Peluso, G., Galderisi, U. & Di Bernardo, G. Long non-coding RNAs in regulation of adipogenesis and adipose tissue function. eLife9, e59053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boucher, J. et al. Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nat. Commun.3, 902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klöting, N. et al. Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes57, 2074–2082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scavo, L. M., Karas, M., Murray, M. & Leroith, D. Insulin-like growth factor-I stimulates both cell growth and lipogenesis during differentiation of human mesenchymal stem cells into adipocytes. J. Clin. Endocrinol. Metab.89, 3543–3553 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Smith, P. J., Wise, L. S., Berkowitz, R., Wan, C. & Rubin, C. S. Insulin-like growth factor-I is an essential regulator of the differentiation of 3T3-L1 adipocytes. J. Biol. Chem.263, 9402–9408 (1988). [PubMed] [Google Scholar]
- 43.Schreiber, C. et al. Loss of ASAP1 in mice impairs adipogenic and osteogenic differentiation of mesenchymal progenitor cells through dysregulation of FAK/Src and AKT signaling. PLoS Genet. 15, e1008216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajbhandari, P. et al. IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell172, 218–233 e217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siersbæk, R. et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J.30, 1459–1472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berry, R. & Rodeheffer, M. S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol.15, 302–308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng, Z. L. et al. PATZ1 (MAZR) co-occupies genomic sites with p53 and inhibits liver cancer cell proliferation via regulating p27. Front. Cell Dev. Biol.9, 586150 (2021). [DOI] [PMC free article] [PubMed]
- 48.Andersen, L. et al. The transcription factor MAZR/PATZ1 regulates the development of FOXP3+ regulatory T cells. Cell Rep.29, 4447–4459.e4446 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Hepler, C. et al. Directing visceral white adipocyte precursors to a thermogenic adipocyte fate improves insulin sensitivity in obese mice. Elife6, e27669 (2017). [DOI] [PMC free article] [PubMed]
- 50.Shao, M. et al. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab.23, 1167–1184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quach, J. M. et al. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J. Biol. Chem.286, 4186–4198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei, S. et al. Emerging roles of zinc finger proteins in regulating adipogenesis. Cell Mol. Life Sci.70, 4569–4584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu, X. et al. Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol6, 160065 (2016). [DOI] [PMC free article] [PubMed]
- 54.Wei, S., Zhang, M., Zheng, Y. & Yan, P. ZBTB16 overexpression enhances white adipogenesis and induces brown-like adipocyte formation of bovine white intramuscular preadipocytes. Cell Physiol. Biochem.48, 2528–2538 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Siersbæk, R., Nielsen, R. & Mandrup, S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab.23, 56–64 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Yu, C.-Y. et al. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS ONE5, e15188–e15188 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waki, H. et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet.7, e1002311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee, Y.-H., Petkova, AneliaP., Mottillo, EmilioP. & Granneman, JamesG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab.15, 480–491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe, I. et al. Lipolysis-derived linoleic acid drives beige fat progenitor cell proliferation. Dev. Cell57, 2623–2637.e2628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Senagolage, M. D. et al. Loss of transcriptional repression by BCL6 confers insulin sensitivity in the setting of obesity. Cell Rep.25, 3283–3298.e3286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakaguchi, S. et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat. Immunol.11, 442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho, J. H., Kim, M. J., Kim, K. J. & Kim, J. R. POZ/BTB and AT-hook-containing zinc finger protein 1 (PATZ1) inhibits endothelial cell senescence through a p53 dependent pathway. Cell Death Differ.19, 703–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minamino, T. et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med15, 1082–1087 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Li, P. et al. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity. Cell147, 815–826 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsumoto, M. et al. Genomewide approaches for BACH1 target genes in mouse embryonic fibroblasts showed BACH1-Pparg pathway in adipogenesis. Genes Cells21, 553–567 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Roy, A. L. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene274, 1–13 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Fan, A. X. et al. Genomic and proteomic analysis of transcription factor TFII-I reveals insight into the response to cellular stress. Nucleic Acids Res.42, 7625–7641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hasegawa, Y. et al. Repression of adipose tissue fibrosis through a PRDM16-GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab.27, 180–194.e186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kopp, N. D. et al. Functions of Gtf2i and Gtf2ird1 in the developing brain: transcription, DNA binding and long-term behavioral consequences. Hum. Mol. Genet.29, 1498–1519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abramova, A. et al. The transcription factor MAZR preferentially acts as a transcriptional repressor in mast cells and plays a minor role in the regulation of effector functions in response to FcεRI stimulation. PLoS ONE8, e77677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hummasti, S. & Tontonoz, P. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol. Endocrinol.20, 1261–1275 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Lee, M. J. & Fried, S. K. Optimal protocol for the differentiation and metabolic analysis of human adipose stromal cells. Methods Enzymol.538, 49–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee, M. J., Jash, S., Jones, J. E. C., Puri, V. & Fried, S. K. Rosiglitazone remodels the lipid droplet and britens human visceral and subcutaneous adipocytes ex vivo. J. Lipid Res.60, 856–868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tong, A. J. et al. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell165, 165–179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol.57, 289–300 (1995). [Google Scholar]
- 76.Villanueva, C. J. et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab.17, 423–435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mina, A. I. et al. CalR: a Web-based analysis tool for indirect calorimetry experiments. Cell Metab.28, 656–666 e651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parks, B. W. et al. Genetic architecture of insulin resistance in the mouse. Cell Metab.21, 334–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parks, B. W. et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab.17, 141–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for all figures are provided with the paper. NGS data generated for this paper has been uploaded to the Gene Expression Omnibus under accession numbers GSE273317, GSE273318, GSE273319, and GSE273320. Source data are provided with this paper.