Abstract
Coinfection of pathogenic bacteria and viruses is associated with multiple diseases. During the COVID-19 pandemic, the co-infection of other pathogens with SARS-CoV-2 was one of the important determinants of the severity. Although primarily a respiratory virus gastric manifestation of the SARS-CoV-2 infection was widely reported. This study highlights the possible consequences of SARS-CoV-2 -Helicobacter pylori coinfection in the gastrointestinal cells. We utilized the transfection and infection model for SARS-CoV-2 spike Delta (δ) and H. pylori respectively in colon carcinoma cell line HT-29 to develop the coinfection model to study inflammation, mitochondrial function, and cell death. The results demonstrate increased transcript levels of inflammatory markers like TLR2 (p < 0.01), IL10 (p < 0.05), TNFα (p < 0.05) and CXCL1 (p < 0.05) in pre-H. pylori infected cells as compared to the control. The protein levels of the β-Catenin (p < 0.01) and c-Myc (p < 0.01) were also significantly elevated in pre-H. pylori infected group in case of co-infection. Further investigation of apoptotic and necrotic markers (Caspase-3, Caspase-8, and RIP-1) reveals a necroptotic cell death in the coinfected cells. The infection and coinfection also damage the mitochondria in HT-29 cells, further implicating mitochondrial dysfunction in the necrotic cell death process. Our study also highlights the detrimental effect of pre-H. pylori exposure in the coinfection model compared to post-exposure and lone infection of H. pylori and SARS-CoV-2. This knowledge could aid in developing targeted interventions and therapeutic strategies to mitigate the severity of COVID-19 and improve patient outcomes.
Keywords: SARS-CoV-2, Helicobacter pylori, Co-infection, Inflammation, Gastrointestine, Necroptosis
1. Introduction
The enduring consequences of SARS-CoV-2 infection remain a subject of profound concern for both the scientific community and individuals worldwide [1]. The global outbreak of SARS-CoV-2 has brought increased awareness to the issue of bacterial co-infection and their intricate involvement in the pathology of coronavirus disease 2019 (COVID-19) [2]. SARS-CoV-2 is a positive-sense ssRNA virus of genome size ∼30 kb [3]. This virus releases its viral genome into human host cells by attaching its spike protein to the host cell's membrane protein ACE2 (Angiotensin-converting enzyme 2) [4]. Further fusion of viral and host membrane facilitates the entry of its viral RNA into the host cell. This process is crucial for the virus to infect cells and initiate the synthesis of proteins essential for the production of new viral particles [5]. Despite being a respiratory virus, multiple evidences substantiate the virus's ability to infect other organs [6]. The ACE2 receptors are ubiquitous within the human body, particularly overexpressed on intestinal epithelial cells of the gut [7,8]. The prevalence of ACE 2 receptors in lung epithelial and gastroenteric cells indicates that the respiratory and gastrointestinal systems serve as primary entry points for the virus [7]. The clinical spectrum of COVID-19 ranges from asymptomatic or mild respiratory infection to fulminant pneumonia with acute respiratory distress syndrome or multi-organ failure resulting in a fatal outcome [9]. During COVID-19, hospitalized patients also reported GI-related symptoms like abdominal pain, diarrhoea, nausea, vomiting, anorexia, etc [10]. Previous studies have demonstrated that multiple coronaviruses could transit and thrive within the gastrointestinal system [11]. Several clinical studies found that SARS-CoV-2 RNA is detectable in the stool samples of COVID-19 patients. Strong evidence emerged when samples from the patient's gastrointestinal tissues, collected through endoscopy, tested positive for SARS-CoV-2 RNA [12]. Exploring viral fusion and entry mechanisms in gastrointestinal cells is essential for elucidating the pathogenesis underlying primary manifestations of the disease in the digestive system [13]. Studies suggest that such GI-related symptoms might be related to gut dysbiosis caused by SARS-CoV-2 infection in the GI tract [14].
In nature, viruses undergo persistent genetic transformations, marked by the ongoing assimilation of changes in their genetic code through processes like genetic mutations or viral recombination during the replication of their genomes. Similarly, SARS-CoV-2 is prone to persistent mutational changes, resulting in variations that differ from those observed in the wild-type strain of nCoV-2 [15]. During the pandemic, many variants of n-CoV2 were identified worldwide. WHO labeled a variant as VOC (variant of concern) based on specific attributes, including increased transmissibility, severe disease, increased hospitalizations, and reduced effectiveness of vaccines or prevailing treatments. The 5 VOCs that were first entitled with this status were – Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) [15]. SARS-CoV-2 spike is one of the virus's essential structural proteins, and mutation in this alters the dynamics of infection and pathogenesis [16]. The primary determinant for the emergence of VOCs are the mutations associated with spike protein. Studies have used the spike protein of different VOCs to understand the inflammatory response associated with each variant of concern [17]. In India, the delta wave (2nd wave) had devastating consequences with high hospitalization and fatality compared to the 1st wave (WT, alpha, beta, and gamma) and 3rd wave (omicron). Delta is also known to be associated with higher hospitalization due to digestive complications [18].
Bacterial co-infection is a common complication of viral infections with increasing morbidity and mortality in conjunction with more burden on healthcare resources [[19], [20], [21]]. Serious bacterial infections may be missed when all attention is focused on COVID-19. Therefore, recognition of co-infection in patients with COVID-19 is of utmost importance. It enables stewardship for the proper use and effective delivery of anti-microbial agents [19]. Helicobacter pylori, a gut bacterial pathogen, is notoriously known for its widespread infection, affecting over 50 % of the global population [22]. This bacterium is primarily associated with gastritis, gastric adenocarcinoma, MALT (mucosa-associated lymphoid tissue), and other gut-associated diseases [23]. Post infection it prompts cellular changes that hinder cell motility, impedes cellular proliferation, and influences apoptosis [24]. Naturally, it does not harm the gut, but external factors like viral infection, bacterial infection, or other stimulants manifest a disbalance in the colony of gut-residing H. pylori [25]. Often, this microbial pathogen is known to interact with different viruses such as Epstein-Barr virus (EBV), Human Immunodeficiency virus (HIV), and SARS-CoV-2 [26,27]. H. pylori-EBV coinfections are known to enhance the severity of inflammatory reactions causing chronic gastritis and gastric cancer while H. pylori-HIV coinfection causes an enhanced systemic immune activation by increasing CD4+ T cell count elucidating a high co-endemic setting [26,27]. This leads to an imbalance in the gut microbiota, potentially exacerbating the detrimental impact of external pathogenic agents, such as SARS-CoV-2, within the human gastrointestinal system. Studies have also suggested the possible entry of SARS-CoV-2 into gastric epithelial cells, leading to a disruption in the equilibrium of the gut microenvironment [28]. In this aspect, understanding the pathogenesis associated with SARS-CoV-2-H. pylori coinfection in the gastrointestinal system is important.
In the current study, we assessed the impact of concurrent H. pylori infection on the pathogenesis of COVID-19. The work aims to decipher the molecular mechanism involved in the severity posed by the co-infection of the gut pathogen and SARS-CoV-2. We explored the in-vitro effects of co-infection with H. pylori and the delta variant of SARS-CoV-2 on the HT-29 colon carcinoma cell line. As both pathogens are known to infect intestinal epithelial cells, we chose colon epithelial cells for our study. The findings suggest that coinfection with H. pylori exacerbates inflammation in colon cells. Further, the investigation of cell death type, reveals the necroptotic cell death in the coinfected HT-29 cells. Additionally, we found mitochondrial damage in the lone as well as coinfected model, further implicating mitochondrial dysfunction in the necrotic cell death process.
2. Methodology
2.1. Mammalian cell culture, plasmid construct, and transfection
Colon epithelial (HT-29) cells were obtained from the National Centre for Cell Science (NCCS), Pune, India, within 1 year of the study. As per standard procedure, the NCCS provides STR analysis and mycoplasma contamination -free report with the cell. The cells were cultured in Dulbecco's Modified Eagle's medium (DMEM; Thermo Scientific, USA) supplemented with 10 % Fetal Bovine Serum (FBS; Thermo Scientific, USA) along with 50 U/ml penicillin and 100 μg/ml streptomycin (Himedia, Mumbai, India), in a humidified atmosphere with 5 % CO2 at 37 °C (Forma, Steri-cycle i160, Thermo Scientific, Waltham, USA). The cells were tested frequently for mycoplasma contamination throughout the study period. The study used two plasmid types: pcDNA3.1 Myc-tag (PA3M, vector control) and pcDNA3.3-SARS-2-B.1.617.2 (Spike Delta strain) (Addgene, Watertown, Massachusetts). Transfections were given in a range of increasing concentrations of pcDNA3.3-SARS-2-B.1.617.2 plasmid DNA (0–3 μg) and the dose for further experiments was identified at 3 μg concentration (Suppl. Fig. 1) in a 60 mm culture dish using Invitrogen Lipofectamine™ 3000 Transfection Reagent at a confluency of 70 % [29].
2.2. Helicobacter pylori culture and infection
Previously described H. pylori was used for this study, we used two clinical strains of H. pylori: HB1, obtained from the biopsy sample, and HJ9, obtained from the patient's juice sample [30]. H. pylori was incubated in a microaerophilic chamber (Whitley DG 250) containing specific growth conditions (i.e., 85 % N2, 10 % CO2, and 5 % O2) at 37 °C. The growth media of H. pylori is complete brain heart infusion media (BHI, Cat. No. 237500- BD Brain Heart Infusion broth), containing 10 % Fetal Bovine Serum (FBS Hi-media, Cat. No. RM-10432) with 3× H. pylori selective antibiotics (5 mg/L cefsulodin, 10 mg/L vancomycin, 5 mg/L amphotericin B, 5 mg/L trimethoprim) as described previously [31]. For infection, MOI-100 was used.
2.3. Coinfection methodology
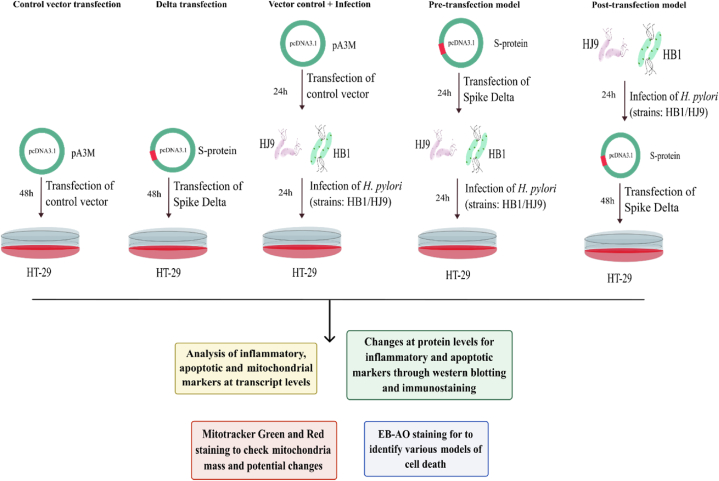
In the current study, we devised a model incorporating a single SARS-CoV-2 and H. pylori infection and the coinfection of both pathogens. As discovered by dose optimization experiments, we kept the time-point for incubating Delta-transfected cells to 48 h post-transfection (hpt). This time point was kept constant for coinfected samples as well. For H. pylori infection, 24 hpi was the time-point for incubation of alone infection samples, i.e., HB1 and HJ9, as the bacteria require a minimum of 12 h to establish their infection in the cells. As represented in Fig. 1, for infection in samples III, IV, and V, HB1 and HJ9 were used to infect the HT-29 cells at MOI = 100. After 24 h of incubation, sample III, i.e., HB1-infected HT-29 cells and HJ9-infected HT-29 cells, were collected for further experiments. For sample IV, i.e., transfection with Delta plasmid followed by infection with HB1 and transfection with Delta plasmid followed by infection with HJ9, samples were collected after 48 hpi. For sample V, i.e., infection with HB1 followed by transfection with Delta plasmid and infection with HJ9 followed by transfection with Delta plasmid, the samples were collected after 54 hpi.
Fig. 1.
Model to study the coinfection of SARS-CoV-2 and Helicobacter pylori. Colon carcinoma cell line HT-29 was used in the study. The cells were transfected with pcDNA3.1 Myc-tag and pcDNA3.3 SARS-CoV-2-spike delta for 48 h in the control/pA3M and Delta/S-protein group respectively. For the pre-transfection model pcDNA3.1 SARS-CoV-2-spike delta was transfected to HT-29 cells and 24 h post-transfection H. pylori strain (HB1/HJ9) was added with MOI 100 for 24 h. For the post-transfection model H. pylori (HB1/HJ9) was infected for 24 h followed by transfection of pcDNA3.1 SARS-CoV-2-spike delta for 48 h. After completion of incubation period samples were subjected to analysis of inflammatory and apoptotic markers by qRT-PCR and Western blot. EB/AO and mito-tracker staining were also performed post- incubation period.
2.4. Transcript level expression analysis through real-time polymerase chain reaction (qRT-PCR)
The real-time PCR (qRT-PCR) was performed to analyze transcript profiles for inflammatory, apoptotic, and mitochondrial markers in colon cells. The treated cells were collected and washed with 1× phosphate buffer saline (PBS) for RNA extraction through the TRIzol (Sigma-Aldrich, St. Louis, USA) method. The cDNA synthesis was performed using Takara PrimeScript (Takara, Shiga, Japan) according to the manufacturer's protocol. Further, qRT-PCR was performed using SYBR green real-time master mix (Applied Biosystems, Waltham, USA) using Agilent AiraMX qRT-PCR system (Agilent Technologies, Santa Clara, USA). The cycle was programmed at 10 min, 95 °C followed by 40 cycles of 15 s at 95 °C, 20 s at 58 °C, 20 s at 72 °C steps. The relative expression of SARS-CoV-2 Spike Delta, inflammatory (IL6, IL10, IL1β, IFN β2, IFN β3, CXCL1, CXCL2, IFN-γ, TNFα), apoptotic (BAK, FADD, BIDD, Bcl2, Bax, Caspase9, Apaf-1), and mitochondrial (CytC, ABAD, PDH, AKGDH, SDH, MDH, ANT, ATP synthase, MFN1, MFN2, OPA1, DRP1, MFF, MiD49, MiD51, Fis1, MAVS) markers were analyzed using specific primers alongside human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the reference gene. The analysis was performed using the del Ct method to compare the fold change values.
2.5. Western blot
The Western blot was performed as described previously [32]. In brief, the harvested cells were washed with PBS and lysed using radioimmunoprecipitation assay (RIPA) buffer (VWR, Radnor, USA) along with protease and phosphatase inhibitors. Protein quantification was done using Bradford reagent (Himedia, Mumbai, India) followed by separation of proteins with SDS-PAGE and transferred using 0.45 μm nitrocellulose membrane (Bio-Rad, Hercules, USA). The blocking was performed using 4.5 % BSA followed by incubation with primary antibody with washing. Further, HRP-conjugated secondary antibodies were incubated for 1 h, and the blot was visualized using chemiluminescent detection based on ECL western blotting substrate (Bio-Rad, ChemiDoc™ XRS + System with Image Lab™ Software, Bio-Rad, Hercules, USA). The analysis was performed using ImageJ software (National Institutes of Health, USA). Primary antibodies against NFkB p65 (#8242, 1:1000), β-catenin (#14903, 1:1000), cleaved caspase 3 (#9664, 1:1000), and RIPK1 (#3493, 1:1000) were purchased from Cell Signaling Technology, Danvers, USA. Antibody for Caspase-8 (Product code:10-1019, 4 μg/ml) was purchased from Abgenex, Bhubaneswar, India. c-Myc (#MA1-980, 1:1000) and Anti-GAPDH (#MA5-15738, 1:2000) were purchased from Invitrogen, Waltham, USA.
2.6. Immunofluorescence assay
The cells were seeded onto coverslips, and after infection and transfection, they were fixed with 4 % paraformaldehyde for 30 min. Permeabilization was done using 0.2 % Triton-X100 for 30 min. Further, blocking was performed using 1 % BSA (Himedia, Mumbai, India) followed by incubation with primary antibodies (RIPK1, c-Myc, b-catenin) for 2 h at room temperature. The cells were further washed and incubated with secondary antibodies (1:1000 dilution) with different fluorophores along with counterstaining of cell nuclei by 4′, 6′-diamidino-2-phenylindole (DAPI). The coverslips were transferred onto the slides with an antifade mounting medium and observed under a confocal microscope (FluoView 1000, Olympus, Tokyo, Japan). The protocol has been discussed in detail previously [29]. The image analysis was performed using ImageJ software (National Institutes of Health, USA), and fluorescent intensities were calculated and plotted compared to vector control.
2.7. EB/AO to analyze cell death
To study apoptotic, necrotic, and living cells in the samples, EB/AO dual staining was performed using ethidium bromide and acridine orange (100 μg/mL each) for 5 min at 37 °C followed by washing with PBS. The images were taken using a fluorescence microscope (Olympus IX83, Olympus, Tokyo, Japan) at 20× objective magnification. Manual analysis was performed to count the cell stages using color characteristics. A total of 1000 cells were counted to determine the percentage of live, necrotic, and apoptotic cells [29].
2.8. Mitotracker red and green assay to analyze mitochondrial functioning
After the infection or transfection period was completed, according to the coinfection model, cells were treated with MitoTracker Red (200 nM) in 500 μL of plain DMEM incubated for 40 min at 37 °C. After incubation, cells were washed with PBS, followed by MitoTracker Green (100 nM) treatment in 500 μL of DMEM for 40 min. After completion of the incubation period, cells were rewashed with PBS, and images were taken under Olympus IX83 fluorescent microscope aided with CellSens imaging software at 20 × objective magnification [33].
2.9. Statistical analysis and graphical representation
All the in vitro experiments were performed in triplicates. Data were presented as means ± standard deviation (SD) of three data points. The statistical analyses were performed, and the graphs were plotted using GraphPad Prism 8 software. The statistical significance was assessed by performing unpaired t-tests. The level of significance (α) was considered to be 5 % at the 95 % confidence interval. p-values of <0.05, <0.01 and < 0.0001 were considered statistically significant and represented as ∗, ∗∗, and ∗∗∗ for higher expression and #, ## and ### for down-regulation respectively.
3. Results
3.1. Augmented inflammatory response against coinfection scenario in colon cells
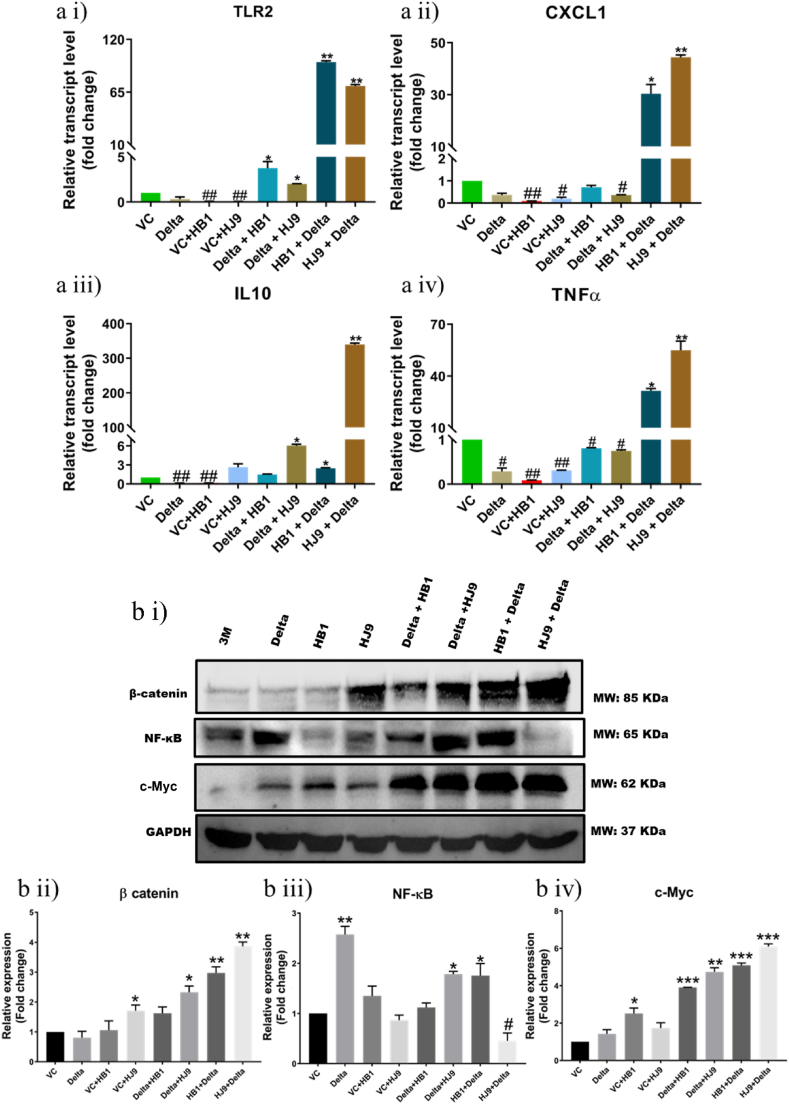
H. pylori has several virulence factors that assist the colonization of the pathogen and evade the host's immune response [34]. These virulence factors activate the host's immune system, causing elevated levels of several inflammatory molecules, and resulting in acute and chronic inflammation. Likewise, SARS-CoV-2 S protein is also a potent viral PAMP that upon sensing by TLR2, activates the NF-κB pathway, leading to the expression of inflammatory mediators in innate immune and epithelial cells. To confirm the infection of H. pylori and successful transfection of SARS-CoV-2 spike delta qRT-PCR of 16s rRNA and Spike was performed using gene specific primers (Suppl. Fig. 1 a i and ii). Additionally, immunostaining of H. pylori flagellin and Spike was performed post completion of the incubation period (Suppl Figure 1 bi). The results depict successful infection and transfection in the respective samples, in the coinfection model, co-expression of both the proteins was observed (Suppl Figure 1 b i, ii and iii). To understand the profile of inflammatory genes' transcripts in colon (HT-29) cells after exposure to H. pylori, SARS-CoV-2 delta alone, and coinfection scenario, we performed qRT-PCR for some crucial inflammatory genes (IL6, IL10, IL1β, IFN β2, IFN β3, CXCL1, CXCL2, IFN-γ, TNFα). Interestingly, the relative transcript expression of pro-inflammatory molecules such as TNFα (p < 0.05), CXCL1 (p < 0.05), IL10 (p < 0.05), and Toll-like receptor TLR2 (p < 0.01) was found to be significantly enhanced in H. pylori and SARS-CoV-2 delta co-infected colon cells compared to control and alone infected cells (Fig. 2 a i-iv). However, the most robust inflammatory responses were observed in cells exposed to HJ9+Delta (p < 0.01), where spike expression was also significantly higher (p < 0.0001) (Fig. 2 a i-iv and Suppl. Fig. 1 a i). This suggests that pre-existing H. pylori bacterial infection in colon cells could potentially create a favorable environment for the subsequent infection of the SARS-CoV-2 virus.
Fig. 2.
Coinfection of H. pylori and SARS-CoV-2 Delta modulates inflammation markers at transcript and protein levels. To determine the level of inflammatory markers and its regulators in the pcDNA 3.3 SARS-CoV-2 spike Delta transfected, H. pylori infected and their coinfection model qRT PCR (a) and Western blot (bi) of selected markers was performed. Relative transcript expression of TLR2 (a i), CXCL1 (a ii), IL10 (a iii) and TNFα (a iv) in HT29 cells. Relative protein expression of β-catenin (b ii), NF-κB (b iii) and c-Myc (b iv). The experiment was performed in triplicates, and the results are shown as the mean ± SD of three data points. Unpaired T-tests were applied to determine the statistical significance. p values of <0.05, <0.01 and < 0.001 were considered statistically significant and represented with #/∗, ##/∗∗ and ###/∗∗∗, denoting downregulation/upregulation respectively. VC- Vector control (pcDNA3.1 Myc-tag).
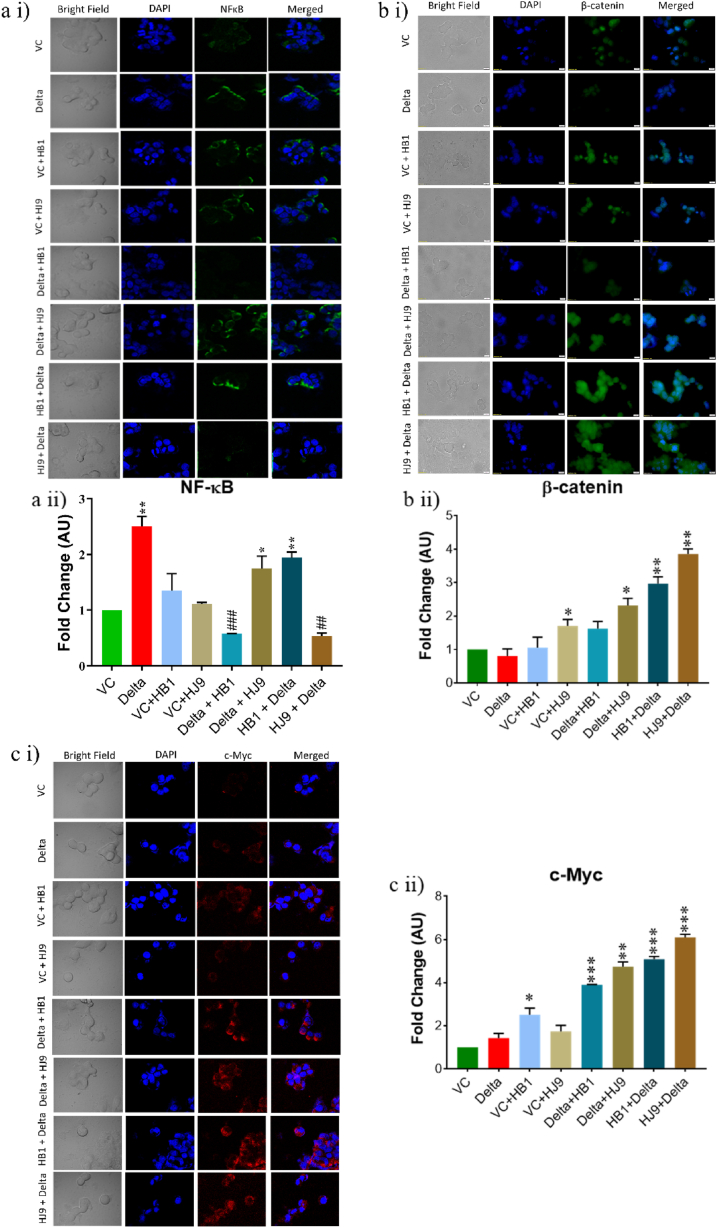
Further, we investigated selected inflammatory markers such as NF-κB and its downstream effector molecules such as β-catenin and c-Myc through Western blot and immunofluorescence analysis (Fig. 2, Fig. 3). Findings demonstrated a significantly enhanced expression of NF-κB (p < 0.05) in SARS-CoV-2 spike transfected and delta + HJ9 and HB1+delta samples, compared to vector control (Fig. 2 b i and iii). However, we found a downregulation of NF-κB (p < 0.05) in HJ9+delta panel. In the immunofluorescence study NF-κB expression was significantly elevated in the spike, delta + HJ9 and HB1+ delta samples (p < 0.05) (Fig. 3 a i and ii). Further, similar to the Western blot result a significant upregulation of β-catenin and c-Myc was observed in post transfection model (p < 0.01) (Fig. 3b and c). This interesting finding substantiates our hypothesis that the co-infection of SARS-CoV-2 and H. pylori induces the activation of NF-κB-mediated inflammation in colon cells which is further carried out by β-catenin and c-Myc.
Fig. 3.
Inflammation in the SARS-CoV-2- H. pylori coinfection model is mediated by β-catenin and c-Myc. Immunofluorescence of NF-κB, β-catenin and c-Myc was performed with pcDNA3.3 SARS-CoV-2 spike Delta transfected, H. pylori infected and their coinfection model in HT-29 cells. Representative immunofluorescence image and graphical representation of NF-κB (a i and ii), β-catenin (b i and ii), c-Myc (c i and ii) level. Quantification was performed using Image J software. The experiment was performed in triplicates, and the results are shown as the mean ± SD of three data points. Unpaired T-tests were applied to determine the statistical significance. p values of <0.05, <0.01 and < 0.001 were considered statistically significant and represented with #/∗, ##/∗∗ and ###/∗∗∗, denoting downregulation/upregulation respectively. Scale bar 10 μM. VC- Vector control (pcDNA3.1 Myc-tag), Delta-pcDNA3.3 SARS-CoV-2-spike delta.
3.2. Attenuation in the mitochondrial potential and mass post-exposure of H. pylori and SARS-CoV-2 delta
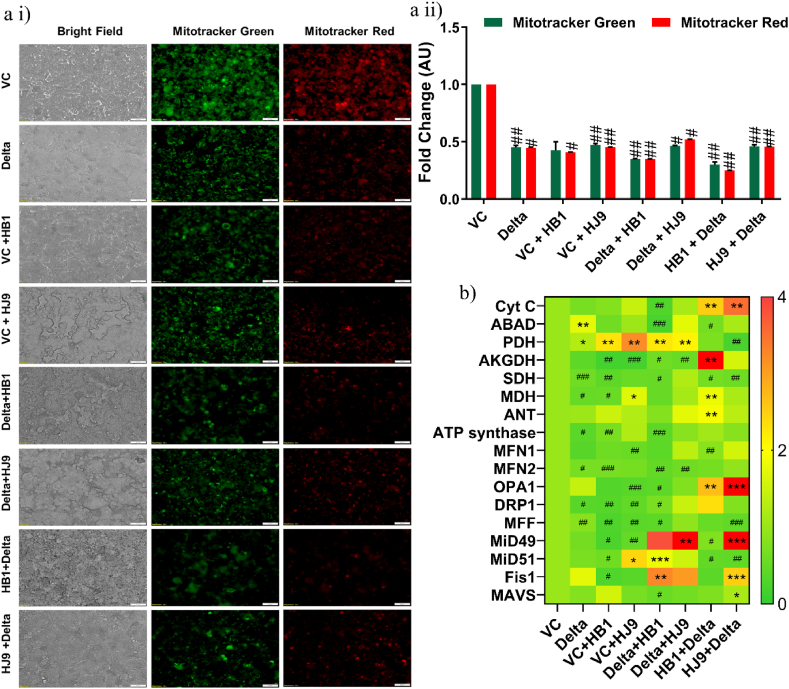
Along with the inflammatory responses, mitochondrial damage is a characteristic feature of viral infection [35]. Mitochondria are closely involved in SARS-CoV-2 replication and mitochondrial homeostasis is disrupted by SARS-CoV-2 in the virus-cell confrontation. We assessed the mitochondrial functionality to investigate the possibility of similar pathology in alone and coinfected colon cells. As a result, we found a significant reduction in mitochondrial potential and mass (p < 0.05) in infected and co-infected cells compared to the control cells (Fig. 4 a i and ii). Additionally, we checked the transcript expression of several mitochondrial markers, including fission and fusion genes. As a result, we found a significant upregulation of mitochondrial fission markers such as MiD49 and Fis1 (p < 0.001) in cells exposed to HJ9+Delta compared to other panels. On the contrary, we observed reduced expression of the fusion marker, MFN1 (p < 0.05) in HB1+Delta infected cells (Fig. 4 b).
Fig. 4.
Altered mitochondrial mass and potential observed in colon cells upon coinfection of H. pylori and Spike Delta. Mitotracker Red and Green Assay representing the mitochondrial membrane potential and mass respectively in coinfection model (a i). Quantitative representation of Mitotracker stained HT-29 cells was performed using Image J software (a ii). Relative transcript expression of mitochondrial markers with pcDNA 3.3 SARS-CoV-2 spike Delta transfected, H. pylori infected and their coinfection model in HT-29 cells (b). Quantification was performed using Image J software. The experiment was performed in triplicates, and the results are shown as the mean ± SD of three data points. Unpaired t-tests were applied to determine the statistical significance. p values of <0.05, <0.01 and < 0.001 were considered statistically significant and represented with #/∗, ##/∗∗ and ###/∗∗∗, denoting downregulation/upregulation respectively. Scale bar 50 μM. VC- Vector control (pcDNA3.1 Myc-tag), Delta-pcDNA3.3 SARS-CoV-2-spike delta. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
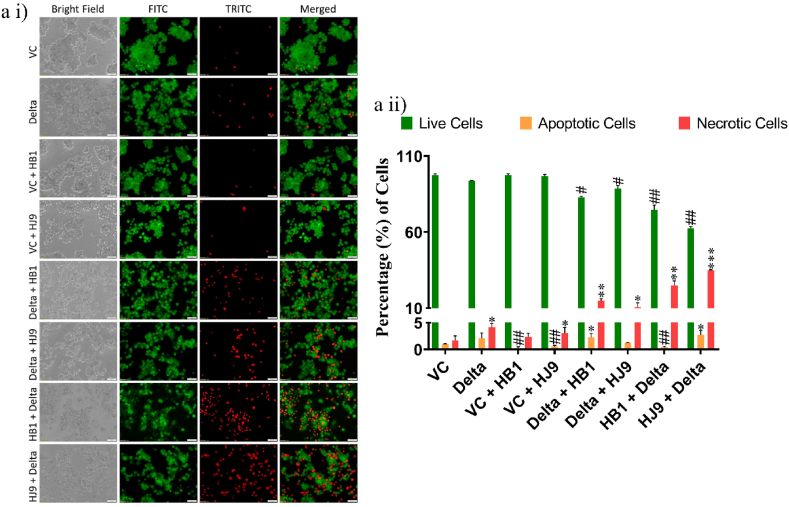
3.3. H. pylori and spike delta coinfection induce cell death in colon cells
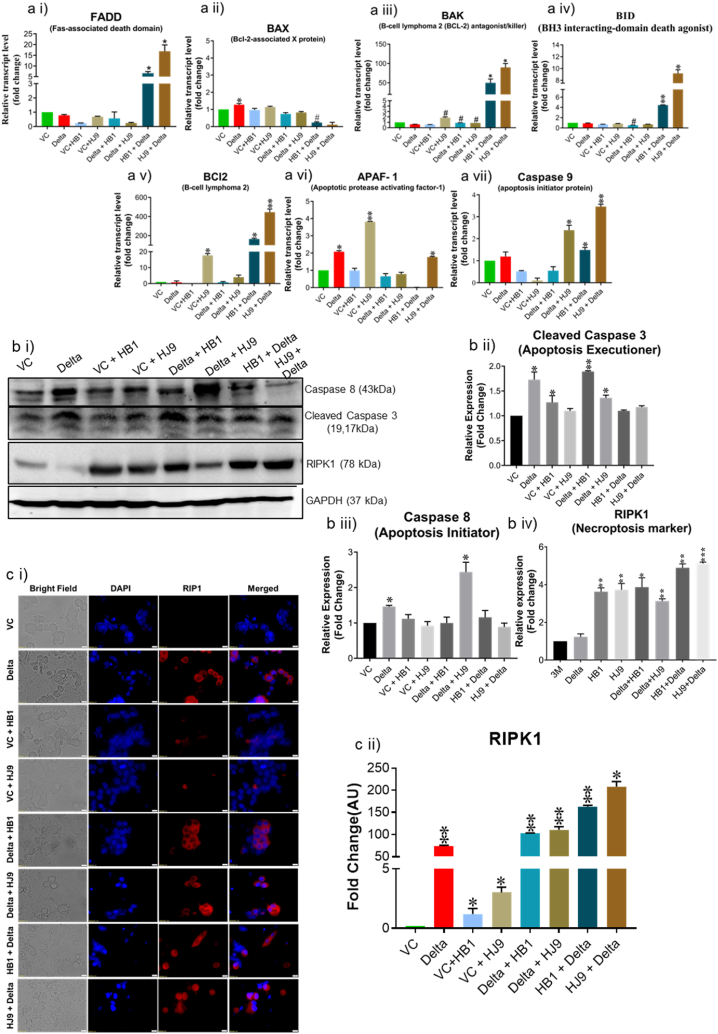
As SARS-CoV-2 immensely displays characteristic cytokine storms, it also induces cell death in specific cells, causing tissue degradation and spreading the virus [36]. To understand the association of cell death in our coinfection model, we performed EB/AO dual staining. When compared to alone infection samples, coinfected cells showed reduced viability. In the pre transfection group (Delta + HB1 and Delta + HJ9) samples, the necrotic cells increase to 14 % and 10 %, respectively. Meanwhile, post-transfection models (HB1+Delta and HJ9+Delta), display an even higher necrotic cell percentage, reaching up to 24 % and 34 %, respectively (Fig. 5). Previously, studies have shown the correlation between apoptosis and COVID-19 infection. However, the mechanism of cell death in the SARS-CoV-2 and H. pylori coinfection is yet to be elucidated. In addition to EB/AO staining, we performed transcript and protein analysis to decipher this mechanism. At transcript levels, we observed an enhanced level of pro-apoptotic genes (bid, bak, and caspase 9) and anti-apoptotic genes (bcl-2) (p < 0.01) (Fig. 6 a i-vii). The necroptotic marker, RIPK1, was significantly enhanced in Delta + HB1 and Delta + HJ9 (p < 0.05 and p < 0.01) and HB1+Delta and HJ9+Delta (p < 0.01 and p < 0.001) at protein levels (Fig. 6 b i and iv). Similar modulation is observed at Caspase 8 and 9 downregulation (Fig. 6 b i-iii). Similarly, we observed significant changes in RIPK1 expression in immunofluorescence images wherein prior infection of H. pylori (HB1 and HJ9) and then Delta transfection resulted in higher RIPK1 expression (p < 0.01 and p < 0.05) when compared to Delta transfection and then H. pylori infection to colon epithelial cells (p < 0.01) (Fig. 6 c i and ii). As we observed enhanced mitochondrial fission machinery in post transfected panels evidenced by elevated expression of mitochondrial fission genes (mid 49 and fis1) specially in HJ9+Delta experimental panel. On the contrary, reduced expression of mitochondrial fusion marker (MFN1) were observed. Interestingly, we found the significant upregulated expression of pro-apoptotic markers associated with intrinsic pathway of apoptosis mediated by mitochondria (bid, bak, and caspase 9) in the same set of post-transfected sample. These finding supports the previous literature that mitochondrial fission is an early and critical event in apoptosis, occurring prior to caspase activation and membrane blebbing. This process is closely associated with, and may even precede, the release of cytochrome c, as fragmented mitochondria are frequently observed retaining cytochrome c. Several key proteins involved in the mitochondrial fission machinery, including Drp1, Fis1, and Endophilin B1, have been implicated in the regulation of apoptosis, underscoring their role in programmed cell death progression (10.1101/gad.1658508; 10.3390/ijms22084260) Additionally, we observed the elevated expression of FADD, a marker associated with extrinsic pathway of apoptosis in the same set of sample. These findings provide substantial evidence for the theory that mitochondrial dysfunction particularly the excessive fission of mitochondria, plays a pivotal role in mediating cellular apoptosis.
Fig. 5.
SARS-CoV-2 Spike Delta and H. pylori coinfection induces cell death in colon cells. To understand the effect of SARS-CoV-2-H. pylori coinfection on cell death, dual acridine orange and ethidium bromide staining of HT-29 cells was performed. (a i) Representative image of EB/AO-stained HT-29 cells. (a ii) Quantitative representation of EB/AO stained HT29 cells. The experiment was performed in triplicate and total 1000 cells were counted in each set for determination of live, apoptotic and necrotic cells, the results are shown as the mean ± SD of three data sets. Unpaired T-tests were applied to determine the statistical significance. p values of <0.05, <0.01 and < 0.001 were considered statistically significant and represented with #/∗, ##/∗∗ and ###/∗∗∗, denoting downregulation/upregulation respectively. The scale bar 50 μM. VC- Vector control (pcDNA3.1 Myc-tag), Delta-pcDNA3.3 SARS-CoV-2-spike delta. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Coinfection of H. pylori and Spike Delta in colon cells induces necrotic cell death. To decipher the molecular mechanism underlying cell death induced by SARS-CoV-2- H. pylori coinfection model we determined the transcript level of apoptosis related markers, protein level of apoptotic and necroptotic markers in HT-29 cells. Relative transcript level of apoptosis marker FADD (ai), BAX (a ii), BAK (a iii), BID (a iv), BCL2 (a v), APAF-1 (a vi) and caspase-9 (a vii). Representative Western blot image of Caspase8, cleaved Caspase 3, RIPK1 and GAPDH in HT-29 cells (b i). Graphical representation of protein level of Caspase8 (b ii), cleaved Caspase 3 (b iii) and RIPK1 (b iv). Representative immunofluorescence image (c i) and graphical representation (c ii) of RIPK1. The experiment was performed in triplicates, and the results are shown as the mean ± SD of three data points. Unpaired T-tests were applied to determine the statistical significance. p < 0.05 was considered significant in all the cases. p-values of <0.05, <0.01 and < 0.0001 were represented with ∗, ∗∗ and ∗∗∗ respectively for significant upregulation and #, ##, and ### for significant downregulation. The scale bar 10 μM. VC- Vector control (pcDNA3.1 Myc-tag), Delta-pcDNA3.3 SARS-CoV-2-spike delta.
4. Discussion
Besides the lung, virus can infect multiple organs, including the gastrointestinal tract [37]. As the GI tract is home to a colossal microbiome, gut dysbiosis can alter SARS-CoV-2 pathogenesis [38]. One of the potent pathogens that causes gut dysbiosis is H. pylori, which can interfere with viral infection [39]. H. pylori enters the gastric cells through type IV secretion system further releasing its secretory proteins such as CagA, VacA, etc. As the infection is enhanced, gut microbial diversity gets altered due to numerous factors including changes in pH, metabolites, and host pathways ultimately causing leaky gut. This gut barrier disruption gives opportunity to several other pathogens to infect the intestine including viruses such as SARS-CoV-2. Furthermore, it potentially induces favorable circumstances wherein the coinfection of both pathogens can harm the host immensely [40]. However, the molecular mechanism of this coinfection pathogenesis is yet to be elucidated. Our study uses colon cells (HT-29) to explain the pathogenesis of H. pylori and SARS-CoV-2 Spike Delta-mediated inflammation and cell death.
One of the most pronounced effects of COVID-19 infection is cytokine storm and associated multi-organ failure [41]. The NF-κB pathway serves as a common inflammatory conduit in the pathogenesis of both H. pylori and SARS-CoV-2 infections [42]. In the case of SARS-CoV-2, the interaction between the Spike protein and ACE2 receptors triggers a signaling cascade involving TLR2, ultimately leading to an NF-κB-mediated inflammatory response [43]. Likewise, H. pylori induces inflammation by binding to TLR2 receptors and activating the NF-κB pathway [44].
In our findings, when colon cells are co-infected with H. pylori and SARS-CoV-2, there is a synergistic effect, resulting in an escalated inflammatory response. This heightened immune activation could contribute to the observed gastrointestinal symptoms in individuals with COVID-19. The concurrent activation of the NF-κB pathway by both pathogens in co-infected cells highlights a potential molecular interplay that may exacerbate inflammatory processes and shed light on the underlying mechanisms of gastrointestinal manifestations in COVID-19 patients with co-infections. In the co-infection model involving HB1+Delta and HJ9+Delta, characterized by a significant elevation in spike expression, there was a concurrent upregulation of transcripts for IL-10, TNFα, CXCL1, and TLR2 in comparison to other samples. This observation suggests that the preceding H. pylori infection in colon cells may create a permissive environment for SARS-CoV-2 infection. Moreover, this model mimics the natural scenario wherein bacteria pre-colonize the colon cells, and further, a foreign pathogen like SARS-CoV-2 invades and establishes its infectious cycle. In our study, we observed an upregulation of NF-κB in the sample transfected with the SARS-CoV-2 spike compared to the vector control. On the contrary, co-infected samples exhibited a reduced expression of NF-κB except for the HB1 coinfected group. This finding is consistent with previously reported results indicating a reduction in NF-κB expression after 12 h of H. pylori infection except for HB1 [45]. Besides NF-κB we have determined elevated expression of β-catenin and c-Myc in the pre-H. pylori exposed co-infected samples. Study by Melano et al. showed β-catenin expression is stronger in Spike Vpp-infected cells at 24 h post infection (hpi) compared to mock-infected cells [46]. In H. pylori infected cells, β-catenin expression and nuclear localization increase significantly compared to control [45]. The higher expression of β-catenin in pre-H. pylori exposed co-infected samples, suggests that it might facilitate the viral entry in the H. pylori exposed cells. Previous studies have also suggested the role of β-catenin in virus entry and establishment of infection [46,47]. c-Myc mediated effect of SARS-CoV-2 is not well described. Yet Deshpande et al. have shown the Orf7b induces lung injury via c-Myc mediated cell death [48]. The increased level of c-Myc in coinfection scenario and its further association with the severity needs to be studied in detail.
In addition, viruses also cause changes in mitochondrial function to promote viral translation and assembly [49]. One theory is that virus–mitochondria interactions hamper mitochondria-associated antiviral signalling mechanisms [49]. SARS-CoV-2 needs host cells to generate molecules for viral replication and propagation [50]. SARS-CoV-2 can block the expression of both nuclear-encoded and mitochondrial-encoded mitochondrial genes, resulting in impaired host mitochondrial function [51]. Viral infections affect the function of mitochondria in cells to impact the cell's metabolism [52]. SARS-CoV-2 infection has been reported to cause mitochondrial damage through fragmentation and leaky membrane [53]. Our findings corroborate existing literature by revealing observed mitochondrial stress in colon cells when exposed to infection, particularly in cases of both H. pylori and SARS-CoV-2 co-infection. Consistent with antecedent literature, our study elucidates a noteworthy attenuation in mitochondrial membrane potential and mass within cells subjected to coinfection. This reduction serves as a signal for bioenergetic stress within the cell, potentially initiating the release of apoptotic factors and, consequently, leading to cell death. This observation suggests the possible activation of apoptotic pathways in the context of the co-infection model.
Bacterial and viral co-infections can perturb the dynamic balance of mitochondria, inducing mitochondrial autophagy, which in turn promotes pathogen replication and persistence (10.1128/mbio.02096-21). These pathogens can disrupt the equilibrium between mitochondrial fusion and fission, resulting in mitophagy and mitochondrial-dependent apoptotic cell death. The excessive fragmentation of mitochondria is essential for the efficient execution of the intrinsic apoptotic pathway, particularly for the proper timing of cytochrome c release and subsequent caspase activation.
Mitochondria play a crucial role in regulating cell death via the intrinsic apoptotic pathway. Upon apoptotic stimulation, the activation of mitochondrial membrane proteins using the Bcl-2 family protein channels triggers mitochondrial outer membrane permeability and releases apoptosis proteins (such as Cyt c, Smac, etc.) into the cytoplasm. Cyt c and apoptotic protease activating factor 1 (APAF1) interact, forming apoptosomes and activating procaspase-9, which cracks caspase-3 and caspase-7, thus inducing cell apoptosis [54]. Cell death in the form of apoptosis is considered as a preventive mechanism up to certain levels, as the host elicits an intrinsic immune response that possibly reduces the viral replication and further spread [55]. However, at higher levels, cell death induces an immune response known as cytokine storm, which causes tissue degradation, viral spread, and ultimately death. Different components of SARS-CoV-2 are known to initiate cell death [41]. Cell death induced by SARS-CoV has been observed in various tissues of infected individuals, and distinct components of the virus have been identified as instigators of this cellular demise process [56]. As a result of apoptosis, cytochrome c released from damaged mitochondria leads to activation of caspases. This marks the cell for apoptosis or programmed cell death [57]. In our co-infection model, a notable rise in the percentage of necrotic cells was observed as we transitioned from a single infection to co-infection. Based on our findings, the combined evidence from EB/AO staining and the transcriptional expression of apoptotic genes strongly suggests a predilection towards necroptotic cell death. Notably, we assessed the expression of RIPK1, an essential adaptor kinase intricately involved in both necrosis and apoptosis pathways. Consistent with previous experimental results, we observed an elevated expression of RIPK1 in samples undergoing co-infection with H. pylori and SARS-CoV-2. Previous studies have reported that RIPK1 kinase undergoes autophosphorylation during TNFα-induced necroptosis. This phosphorylation event activates RIPK1, enabling it to recruit RIP3 and form a necrosome complex. In our study, we identified a substantial upregulation of TNFα at the transcript level in H. pylori + SARS-CoV-2 co-infected samples. This finding strongly implies that TNFα-induced necrosis could be a prominent pathway during the co-infection. The EB/AO staining patterns strongly suggest that, during co-infection, a substantial proportion of cells undergo necrosis. This occurrence is likely attributed to an increased pathogen burden in co-infected cells, as compared to individuals infected or transfected. Consequently, our interpretation aligns with the previous indications derived from the expression profiles of apoptotic and anti-apoptotic markers, supporting the notion that co-infection involving SARS-CoV-2 and H. pylori in colon cells leans towards necrosis rather than apoptosis.
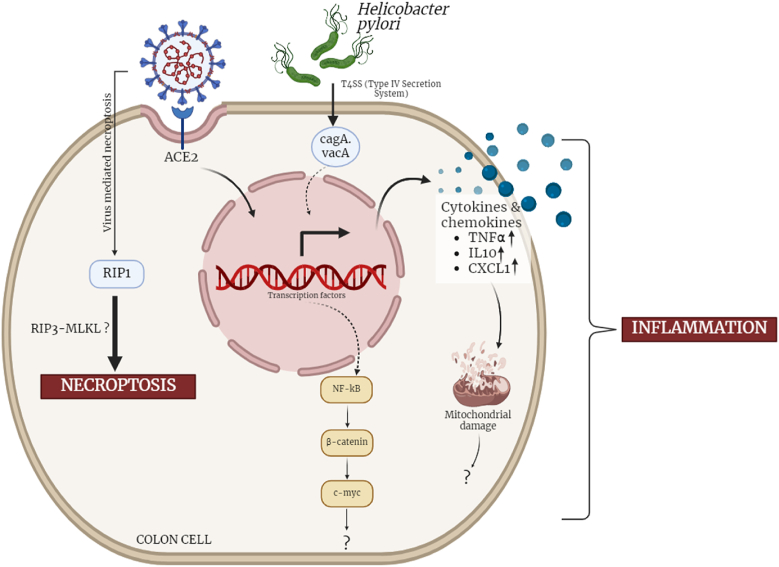
Conclusively, our study provided a mechanistic understanding of the H. pylori and SARS-CoV-2 coinfection induced necroptosis and inflammation. It highlights the involvement of inflammatory mediators NF-κB and its associated molecules like β-catenin and c-Myc in inducing inflammation. Furthermore, alterations in mitochondrial functions were also induced. Our study for cell death analysis revealed the prominence of necroptosis over apoptosis in the co-infection model marked by elevated expression of RIPK1 (Fig. 7). The present study also highlights the detrimental effect of pre-exposure of H. pylori in the coinfection model, as compared to post exposure of H. pylori and lone infections.
Fig. 7.
SARS-CoV-2-Helicobacter pylori co-infection induces, inflammation, mitochondrial damage and necrotic cell death in colon epithelial cells. In our effort to understand the plausible effect of SARS-CoV-2-H. pylori co-infection on gastrointestinal cells. We used a transfection + infection model for SARS-CoV-2 spike delta and H. pylori in colon epithelial cells (HT-29). Our findings highlight the NF-κB, β-catenin and c-Myc mediated inflammation in the colon cells. The co-infection model also elevates the level of other inflammatory chemokines and cytokines. Further we asked the status of mitochondria in coinfection model and the results depicts the mitochondrial dysfunction. The coinfection scenario also induces necrotic cell death in the colon cells possibly mediated by necroptotic marker RIPK1.
More than half of the global population is infected with H. pylori [53], emphasizing the need to explore the effects of viral interventions, such as SARS-CoV-2, in relation to the gastrointestinal symptoms observed during the COVID-19 pandemic. Our research uncovers a potential mechanistic pathway that may drive inflammation during co-infection. Further studies utilizing in vivo models could provide a deeper understanding of the inflammatory processes triggered by these infections. Such investigations hold promise in identifying therapeutic targets to alleviate GI symptoms in patients infected with COVID-19.
CRediT authorship contribution statement
Akrati Tandon: Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Budhadev Baral: Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Vaishali Saini: Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Meenakshi Kandpal: Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Amit Kumar Dixit: Writing – review & editing, Visualization, Validation. Hamendra Singh Parmar: Writing – review & editing, Visualization, Validation. Ajay Kumar Meena: Writing – review & editing, Visualization, Validation, Funding acquisition. Hem Chandra Jha: Writing – review & editing, Visualization, Validation, Supervision, Software, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
Authors have no conflict of interest to declare.
Acknowledgements
We are thankful to Indian Institute of Technology Indore for providing facilities and support. Dr. Ravinder Kumar, technical person at confocal facility, IIT Indore for his help and support in acquiring the confocal images for NFκB. We appreciate lab colleagues for their insightful discussions and advice. We acknowledge the insightful suggestion from Dr. Ajay Kumar Meena, Regional Ayurveda Research Institute, Gwalior. We acknowledge the Indian Council of Medical Research (Grant no. BMI/12(82)/2021 and ECD/CSTPU/Adhoc/COVID-19/28/2021-22) and Central Council for Research in Ayurvedic Sciences (Grant no. 1263/2022-23) Govt. of India for funding. Additionally, we acknowledge Center for Rural Development and Technology, IIT Indore (IITI/CRDT/2022-23/05) for funding. We are also thankful to University Grants Commission, Dept. of Biotechnology Govt. of India for fellowship to Budhadev Baral, Meenakshi Kandpal and Vaishali Saini in the form of research stipend. We also acknowledge DST-FIST support project No. SR/FST/LS-I/2020/621 for providing us with different instruments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37585.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losier A., Gupta G., Caldararo M., Dela Cruz C.S. The impact of coronavirus disease 2019 on viral, bacterial, and fungal respiratory infections. Clin. Chest Med. 2023;44:407–423. doi: 10.1016/j.ccm.2022.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr A.R., Perlman S. In: Coronaviruses [Internet] Maier H.J., Bickerton E., Britton P., editors. Springer New York; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23.http://link.springer.com/10.1007/978-1-4939-2438-7_1 [cited 2024 Feb 9] [Google Scholar]
- 4.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva R.A., Rouillé Y., Dubuisson J. International Review of Cytology. Elsevier; 2005. Interactions between virus proteins and host cell membranes during the viral life cycle; pp. 171–244.https://linkinghub.elsevier.com/retrieve/pii/S0074769605450068 [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavriatopoulou M., Korompoki E., Fotiou D., Ntanasis-Stathopoulos I., Psaltopoulou T., Kastritis E., et al. Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med. 2020;20:493–506. doi: 10.1007/s10238-020-00648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirbhate E., Pandey J., Patel V.K., Kamal M., Jawaid T., Gorain B., et al. Understanding the role of ACE-2 receptor in pathogenesis of COVID-19 disease: a potential approach for therapeutic intervention. Pharmacol. Rep. 2021;73:1539–1550. doi: 10.1007/s43440-021-00303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J., Navis G.J., Gordijn S.J., et al. Angiotensin‐converting enzyme 2 (ACE2), SARS‐CoV ‐2 and the pathophysiology of coronavirus disease 2019 (COVID ‐19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and multiorgan response. Curr. Probl. Cardiol. 2020;45 doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukharani N., Kumar J., Makheja K., Bai P., Sharma S., Subash K., et al. Gastrointestinal manifestation of COVID-19 in hospitalized patients. Cureus. 2021 doi: 10.7759/cureus.18024. https://www.cureus.com/articles/69591-gastrointestinal-manifestation-of-covid-19-in-hospitalized-patients [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., et al. The presence of SARS‐CoV‐2 RNA in the feces of COVID‐19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 12.Cherne M.D., Gentry A.B., Nemudraia A., Nemudryi A., Hedges J.F., Walk H., et al. Severe acute respiratory syndrome coronavirus 2 is detected in the gastrointestinal tract of asymptomatic endoscopy patients but is unlikely to pose a significant risk to healthcare personnel. Gastro Hep Advances. 2022;1:844–852. doi: 10.1016/j.gastha.2022.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durairajan S.S.K., Singh A.K., Saravanan U.B., Namachivayam M., Radhakrishnan M., Huang J.-D., et al. Gastrointestinal manifestations of SARS-CoV-2: transmission, pathogenesis, immunomodulation, microflora dysbiosis, and clinical implications. Viruses. 2023;15:1231. doi: 10.3390/v15061231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troisi J., Venutolo G., Pujolassos Tanyà M., Delli Carri M., Landolfi A., Fasano A. COVID-19 and the gastrointestinal tract: source of infection or merely a target of the inflammatory process following SARS-CoV-2 infection? WJG. 2021;27:1406–1418. doi: 10.3748/wjg.v27.i14.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abulsoud A.I., El-Husseiny H.M., El-Husseiny A.A., El-Mahdy H.A., Ismail A., Elkhawaga S.Y., et al. Mutations in SARS-CoV-2: insights on structure, variants, vaccines, and biomedical interventions. Biomed. Pharmacother. 2023;157 doi: 10.1016/j.biopha.2022.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almehdi A.M., Khoder G., Alchakee A.S., Alsayyid A.T., Sarg N.H., Soliman S.S.M. SARS-CoV-2 spike protein: pathogenesis, vaccines, and potential therapies. Infection. 2021;49:855–876. doi: 10.1007/s15010-021-01677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyrkalska S.D., Martínez-López A., Arroyo A.B., Martínez-Morcillo F.J., Candel S., García-Moreno D., et al. Differential proinflammatory activities of Spike proteins of SARS-CoV-2 variants of concern. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slabakova Y., Gerasoudis S., Miteva D., Peshevska-Sekulovska M., Batselova H., Snegarova V., et al. SARS-CoV-2 variant-specific gastrointestinal symptoms of COVID-19: 2023 update. Gastroenterol. Insights. 2023;14:431–445. [Google Scholar]
- 19.Saeed N.K., Al-Khawaja S., Alsalman J., Almusawi S., Albalooshi N.A., Al-Biltagi M. Bacterial co-infection in patients with SARS-CoV-2 in the Kingdom of Bahrain. WJV. 2021;10:168–181. doi: 10.5501/wjv.v10.i4.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashouri M., Nehzat Norozi Tehrani S., Karuppasamy Govindasamy, Zouhal H. Effect of Covid-19 on the lifestyles of vaccinated and unvaccinated elite athletes. A Cross-Country Analysis. HN. 2023;1:1–6. [Google Scholar]
- 21.Rassolnia A., Nobari H. The impact of socio-economic status and physical activity on psychological well-being and sleep quality among college students during the COVID-19 pandemic. Int J Sport Stud Health. 2024;7:1–12. [Google Scholar]
- 22.Bashir S.K., Khan M.B. In: Wu J., editor. vol. 2023. 2023. Overview of Helicobacter pylori infection, prevalence, risk factors, and its prevention; pp. 1–9. (Advanced Gut & Microbiome Research). [Google Scholar]
- 23.Keikha M., Sahebkar A., Yamaoka Y., Karbalaei M. Helicobacter pylori cagA status and gastric mucosa-associated lymphoid tissue lymphoma: a systematic review and meta-analysis. J. Health Popul. Nutr. 2022;41:2. doi: 10.1186/s41043-021-00280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palrasu M., Zaika E., Paulrasu K., Caspa Gokulan R., Suarez G., Que J., et al. Helicobacter pylori pathogen inhibits cellular responses to oncogenic stress and apoptosis. Blanke SR. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bravo D., Hoare A., Soto C., Valenzuela M.A., Quest A.F. Helicobacter pylori in human health and disease: mechanisms for local gastric and systemic effects. WJG. 2018;24:3071–3089. doi: 10.3748/wjg.v24.i28.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashyap D., Baral B., Jakhmola S., Singh A.K., Jha H.C. In: Damania B., editor. vol. 6. 2021. Helicobacter pylori and Epstein-Barr virus coinfection stimulates aggressiveness in gastric cancer through the regulation of gankyrin. (mSphere). 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abadi T., Teklu T., Wondmagegn T., Alem M., Desalegn G. CD4+ T cell count and HIV-1 viral load dynamics positively impacted by H. pylori infection in HIV-positive patients regardless of ART status in a high-burden setting. Eur. J. Med. Res. 2024;29:178. doi: 10.1186/s40001-024-01750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viana S.D., Nunes S., Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities – role of gut microbiota dysbiosis. Ageing Res. Rev. 2020;62 doi: 10.1016/j.arr.2020.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baral B., Saini V., Tandon A., Singh S., Rele S., Dixit A.K., et al. SARS-CoV-2 envelope protein induces necroptosis and mediates inflammatory response in lung and colon cells through receptor interacting protein kinase 1. Apoptosis [Internet] 2023 doi: 10.1007/s10495-023-01883-9. https://link.springer.com/10.1007/s10495-023-01883-9 [cited 2023 Sep 24]; Available from: [DOI] [PubMed] [Google Scholar]
- 30.Kashyap D., Baral B., Verma T.P., Sonkar C., Chatterji D., Jain A.K., et al. Oral rinses in growth inhibition and treatment of Helicobacter pylori infection. BMC Microbiol. 2020;20:45. doi: 10.1186/s12866-020-01728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonkar C., Verma T., Chatterji D., Jain A.K., Jha H.C. Status of kinases in Epstein-Barr virus and Helicobacter pylori Coinfection in gastric Cancer cells. BMC Cancer. 2020;20:925. doi: 10.1186/s12885-020-07377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshney N., Murmu S., Baral B., Kashyap D., Singh S., Kandpal M., et al. Unraveling the Aurora kinase A and Epstein-Barr nuclear antigen 1 axis in Epstein Barr virus associated gastric cancer. Virology. 2023;588 doi: 10.1016/j.virol.2023.109901. [DOI] [PubMed] [Google Scholar]
- 33.Kashyap D., Varshney N., Baral B., Kandpal M., Indari O., Jain A.K., et al. Helicobacter pylori infected gastric epithelial cells bypass cell death pathway through the oncoprotein Gankyrin. Advances in Cancer Biology - Metastasis. 2023;7 [Google Scholar]
- 34.Chang W.-L., Yeh Y.-C., Sheu B.-S. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J. Biomed. Sci. 2018;25:68. doi: 10.1186/s12929-018-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorouri M., Chang T., Hancks D.C. Mitochondria and viral infection: advances and emerging battlefronts. Forero A., Yount J., editors. mBio. 2022;13 doi: 10.1128/mbio.02096-21. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucia M., Ratajczak J., Bujko K., Adamiak M., Ciechanowicz A., Chumak V., et al. An evidence that SARS-Cov-2/COVID-19 spike protein (SP) damages hematopoietic stem/progenitor cells in the mechanism of pyroptosis in Nlrp3 inflammasome-dependent manner. Leukemia. 2021;35:3026–3029. doi: 10.1038/s41375-021-01332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Din A.U., Mazhar M., Waseem M., Ahmad W., Bibi A., Hassan A., et al. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed. Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.110947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L., Zhao M., Fu X. Gastric microbiota dysbiosis and Helicobacter pylori infection. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1153269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devi P., Khan A., Chattopadhyay P., Mehta P., Sahni S., Sharma S., et al. Co-Infections as modulators of disease outcome: minor players or major players? Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.664386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montazersaheb S., Hosseiniyan Khatibi S.M., Hejazi M.S., Tarhriz V., Farjami A., Ghasemian Sorbeni F., et al. COVID-19 infection: an overview on cytokine storm and related interventions. Virol. J. 2022;19:92. doi: 10.1186/s12985-022-01814-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kircheis R., Haasbach E., Lueftenegger D., Heyken W.T., Ocker M., Planz O. NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan S., Shafiei M.S., Longoria C., Schoggins J.W., Savani R.C., Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife. 2021;10 doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith S.M. Role of Toll-like receptors in Helicobacter pylori infection and immunity. WJGP. 2014;5:133. doi: 10.4291/wjgp.v5.i3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baral B., Kashyap D., Varshney N., Verma T.P., Jain A.K., Chatterji D., et al. Helicobacter pylori isolated from gastric juice have higher pathogenic potential than biopsy isolates. Genes & Diseases. 2023 doi: 10.1016/j.gendis.2023.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melano I., Chen H.-J., Ngwira L., Hsu P.-H., Kuo L.-L., Noriega L., et al. Wnt3a facilitates SARS-CoV-2 pseudovirus entry into cells. IJMS. 2023;25:217. doi: 10.3390/ijms25010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee S., Keshry S.S., Ghosh S., Ray A., Chattopadhyay S. Versatile β-catenin is crucial for SARS-CoV-2 infection. Das S., editor. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01670-22. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deshpande R., Li W., Li T., Fanning K.V., Clemens Z., Nyunoya T., et al. SARS-CoV-2 accessory protein Orf7b induces lung injury via c-Myc mediated apoptosis and ferroptosis. IJMS. 2024;25:1157. doi: 10.3390/ijms25021157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elesela S., Lukacs N.W. Role of mitochondria in viral infections. Life. 2021;11:232. doi: 10.3390/life11030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoraka S., Samarasinghe A.E., Ghaemi A., Mohebbi S.R. Host mitochondria: more than an organelle in SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1228275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X., Wu K., Zeng S., Zhao F., Fan J., Li Z., et al. Viral infection modulates mitochondrial function. IJMS. 2021;22:4260. doi: 10.3390/ijms22084260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao X., Nguyen V., Tsai J., Gao C., Tian Y., Zhang Y., et al. The SARS-CoV-2 spike protein induces long-term transcriptional perturbations of mitochondrial metabolic genes, causes cardiac fibrosis, and reduces myocardial contractile in obese mice. Mol. Metabol. 2023;74 doi: 10.1016/j.molmet.2023.101756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suen D.F., Norris K.L., Youle R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan C., Ma Z., Xie J., Li W., Su L., Zhang G., et al. The role of cell death in SARS-CoV-2 infection. Signal Transduct. Targeted Ther. 2023;8:357. doi: 10.1038/s41392-023-01580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan Y.-J., Lim S.G., Hong W. Regulation of cell death during infection by the severe acute respiratory syndrome coronavirus and other coronaviruses. Cell Microbiol. 2007;9:2552–2561. doi: 10.1111/j.1462-5822.2007.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.