Abstract
Murine coronavirus mouse hepatitis virus (MHV) produces a genome-length mRNA, mRNA 1, and six or seven species of subgenomic mRNAs in infected cells. Among these mRNAs, only mRNA 1 is efficiently packaged into MHV particles. MHV N protein binds to all MHV mRNAs, whereas envelope M protein interacts only with mRNA 1. This M protein-mRNA 1 interaction most probably determines the selective packaging of mRNA 1 into MHV particles. A short cis-acting MHV RNA packaging signal is necessary and sufficient for packaging RNA into MHV particles. The present study tested the possibility that the selective M protein-mRNA 1 interaction is due to the packaging signal in mRNA 1. Regardless of the presence or absence of the packaging signal, N protein bound to MHV defective interfering RNAs and intracellularly expressed non-MHV RNA transcripts to form ribonucleoprotein complexes; M protein, however, interacted selectively with RNAs containing the packaging signal. Moreover, only the RNA that interacted selectively with M protein was efficiently packaged into MHV particles. Thus, it was the packaging signal that mediated the selective interaction between M protein and viral RNA to drive the specific packaging of RNA into virus particles. This is the first example for any RNA virus in which a viral envelope protein and a known viral RNA packaging signal have been shown to determine the specificity and selectivity of RNA packaging into virions.
Within the “soup” of a virally infected cell, viral genome and viral proteins specifically and selectively coalesce into progeny viruses. The process of packaging the viral genome, or surrounding the nucleic acid with protein and possibly an envelope, is a critical step in production of new virus. Within their hosts, RNA viruses manufacture genomic RNA, antigenomic RNA, and, in some cases, subgenomic-length RNAs all in the presence of ubiquitous host cell mRNAs, tRNAs, and rRNAs. Occasional packaging of nongenomic viral RNAs and cellular RNAs results in noninfectious viruses, and yet this packaging seems to occur at constant rates which are characteristic for different species of viruses. Unchecked packaging of cellular nucleic acid into viral particles would be expected to overwhelm the ability of intracellular viral genomic RNA to associate with limited viral and host assembly factors. Each virus, therefore, probably has developed a defensive strategy for specific and selective packaging of intracellular genomic RNA into virus particles. RNA packaging signals required for viral RNA packaging are known for several RNA viruses (1, 3, 6, 9, 26, 28, 48, 65, 66, 81), and for some of these, the packaging signal is all that is needed (1, 66, 80, 82).
A critical step for the selective packaging of viral genomic RNA in those RNA viruses with icosahedral and spherical cores is the binding of core protein to intracellular genomic RNA; only the viral RNAs that associate with core protein are packaged into virus particles. The case for negative-strand RNA viruses with a helical nucleocapsid structure seems to be that both genomic and antigenomic RNAs form an intracellular helical nucleocapsid structure. For some reason, only the genomic-length RNA is selectively packaged.
Coronavirus is an enveloped virus containing a large positive-stranded RNA genome of about 28 to 31 kb (13, 22, 32, 43, 44, 47, 64). In infected cells, the virus produces an intracellular form of genomic RNA, mRNA 1, and six to eight species of subgenomic mRNAs (42, 45). These virus-specific mRNAs comprise a nested set with a common 3′ terminus (42, 45, 72, 73) and a common leader sequence of approximately 60 to 80 nucleotides (nt) at the 5′ end (41, 71). Only the genomic-length RNA, mRNA 1, is efficiently packaged into coronavirus particles. The subgenomic mRNAs generally are not incorporated into virus particles (43, 53, 55) or are incorporated at a low efficiency (15, 35, 70, 84); in the case of the prototypic coronavirus, mouse hepatitis virus (MHV), incorporation of MHV subgenomic mRNAs into MHV particles usually is undetectable (55).
MHV assembly occurs at the smooth membranes of the intermediate compartment, between the endoplasmic reticulum and the Golgi complex (40, 76). MHV contains three envelope proteins, M (formerly known as E1), E, and S. S protein is dispensable for packaging of viral nucleocapsid and viral assembly (36, 39, 69), but M protein and E protein both are essential for viral envelope formation and release; coronavirus-like particles are assembled and released from cells that express both E and M proteins (12, 79). M protein, the most abundant glycoprotein in the virus particle and in infected cells, is characterized as having three domains; these include a short N-terminal ectodomain, a triple-spanning transmembrane domain, and a C-terminal endodomain (2). E protein is a transmembrane protein with its N-terminal two-thirds spanning the lipid bilayer twice (50) and the C-terminal region exposed in the virion interior (19, 67). E protein is present only in minute amounts in infected cells and in the virus envelope (31, 46, 67, 77, 83), and yet E protein affects coronavirus morphogenesis (24) and has an ability to produce membrane vesicles containing E protein (19, 49). The viral genomic RNA and N protein form the helical nucleocapsid structure, which exists inside the viral envelope (23, 75).
In MHV-infected cells, MHV N protein not only binds to mRNA 1 to form a ribonucleoprotein (RNP) complex (mRNA 1-RNP complex) but also binds to all subgenomic mRNAs to form subgenomic mRNP complexes (4, 59). M protein selectively interacts only with the mRNA 1-RNP complex in infected cells (59). This interaction occurs in a pre-Golgi compartment and does not require the presence of S and E proteins (59). The selective and specific interaction between M protein and mRNA 1-RNP complex likely determines the specific and selective packaging of mRNA 1 into MHV particles. Previous studies, using MHV defective interfering (DI) RNAs, identified a short MHV cis-acting RNA element (packaging signal) that is necessary for specific packaging of MHV DI RNAs into MHV particles (11, 26, 78). The packaging signal is located 21 kb from the 5′ end of mRNA 1 and is not present in the subgenomic mRNAs (26, 78). When non-MHV RNA transcripts containing the packaging signal are expressed in MHV-infected cells, they are packaged into MHV particles, and non-MHV RNA transcripts lacking the packaging signal are not packaged (82); the MHV packaging signal is sufficient for packaging RNA into MHV particles (82). How the packaging signal determines the selective packaging of RNAs into MHV particles is not known.
We hypothesized that the packaging signal, present in mRNA 1, mediates the selective and specific interaction between M protein and the mRNA 1-RNP complex to drive the specific packaging of the mRNA 1-RNP complex into MHV particles. The present study showed that N protein associated with MHV DI RNAs and expressed non-MHV transcripts alike, in either the presence or the absence of the packaging signal, to form an RNP complex in infected cells. M protein, however, selectively interacted only with the RNP complex containing the packaging signal, and only these RNPs were efficiently packaged into virus particles. The packaging signal determined the selective interaction between M protein and the mRNA 1-RNP complex that led to the selective and specific packaging of mRNA 1 into virus particles.
MATERIALS AND METHODS
Viruses and cells.
The plaque-cloned A59 strain of MHV was the helper virus (42). MHV was propagated in mouse DBT cells (33). Recombinant vaccinia virus vTF7-3, which expresses T7 polymerase (27), was grown, and its titers were determined, in RK13 cells.
Plasmid construction.
The construction of MHV DI clones DF1-2, FA1, FA2, FA4, FA992A, and FB1 is described elsewhere (26). We constructed clone FA4+PS by inserting the 0.6-kb NsiI-XbaI fragment from FB1 into the 5-kb AccI-XbaI large fragment of DF1-2. A recombinant PCR procedure was used to generate DFΔPS and ΔFA2. DF1-2 was incubated with two oligonucleotides, 969 (5′-GCTTCTACCCACTGTTTG-3′), which binds to antigenomic-sense DF1-2 at nt 2144 to 2162 from the 5′ end, and 10161 (5′-GATAGTGCCACGTGCTAGCGGTTCAAGGCTCCCTG-3′), which binds to genomic-sense DF1-2 at nt 2949 to 3052 from the 5′ end, under the PCR conditions described previously (26). Another PCR product was obtained by incubating DF1-2 with oligonucleotide 10162 (5′-GCCTTGAACCGCTAGCACGTGGCACTATC-3′), which hybridizes to antigenomic-sense DF1-2 at nt 2955 to 3052 from the 5′ end, and oligonucleotide 130 (5′-TTCCAATTGGCCATGATCAA-3′), which hybridizes to genomic-sense DF1-2 at nt 3532 to 3551 from the 5′ end. The two PCR products of the expected sizes were mixed, and a second round of PCR was performed using oligonucleotides 969 and 130 as the primers. The recombinant PCR product was digested with NsiI-MscI, and the resulting 1.3-kb fragment was cloned into the NsiI-MscI large fragment of DF1-2 to generate DFΔPS. FA2 was incubated with oligonucleotide 10100 (5′-GTTGTCTGATATCTATGCTGT-3′), which binds to antigenomic-sense FA2 at nt 1285 to 1305 from the 5′ end, and oligonucleotide 10161, under the same PCR conditions as described previously (26). Another PCR product was obtained by incubating FA2 with the oligonucleotides 10162 and 130. The two PCR products of the expected sizes were mixed, and a second round of PCR was performed using oligonucleotides 10100 and 130 as the primers. The recombinant PCR product was digested with SpeI-MscI, and the consequent 1.2-kb fragment was inserted into the SpeI-MscI large fragment of FA2 to generate ΔFA2.
RNA transcription and transfection.
Plasmids linearized by XbaI were transcribed in vitro by T7 RNA polymerase (52), and 5 μg of the RNA transcript was transfected using lipofection, as described previously (52). The resultant viruses were harvested 11 h posttransfection.
DNA transfection.
We infected subconfluent monolayers of DBT cells with vTF7-3 at a multiplicity of infection of 5 for 1 h at 37°C. At 1 h postinfection (p.i.), we transfected the cells with 10 μg of plasmid DNA using a lipofection procedure (37) and at 4 h p.i. superinfected the cells with MHV at a multiplicity of infection of 5. Harvesting of viruses and preparation of cytoplasmic protein lysates were performed at 12 h post-MHV infection.
Purification of viruses.
Supernatant from virus-infected cells was collected at 12 h post-MHV infection and briefly centrifuged to remove cell debris. Released viruses were partially purified using ultracentrifugation on a discontinuous sucrose gradient consisting of 60, 50, 30, and 20% sucrose as described previously (39). After centrifugation at 26,000 rpm for 3 h at 4°C in a Beckman SW28 rotor, virus particles at the interface of 30 and 50% sucrose were collected and further purified on a discontinuous sucrose gradient of 60, 50, 30, and 20% sucrose at 26,000 rpm for 18 h at 4°C. Purified viruses were pelleted through a 20% sucrose cushion in a Beckman SW28 rotor rotating at 26,000 rpm for 2.5 h at 4°C.
Preparation of virion RNA and intracellular RNA.
Virion RNA was extracted from purified viruses using established methods (53). The intracellular virus-specific RNA was extracted from cytoplasmic lysates as described previously (54).
Immunoprecipitation of MHV-specific RNAs.
MHV-specific RNAs were coimmunoprecipitated using an anti-M protein monoclonal antibody, J1.3; an anti-N protein monoclonal antibody, J3.3 (25); or a non-MHV monoclonal antibody, H2KkDk (anti-H2K antibody), which reacts with major histocompatibility complex class I antigen, as described previously (59).
Agarose gel electrophoresis of RNA and Northern (RNA) blotting.
RNAs were denatured and separated on a 1% agarose electrophoretic gel containing formaldehyde as described previously (51). For Northern blot analysis, the nonradiolabeled RNAs were separated on a 1% denaturing agarose gel and then transferred onto nylon filters (51). Northern blot analysis was performed using two digoxigenin-labeled random-primed probes (Boehringer), one corresponding to 85 to 474 nt from the 5′ end of MHV genomic RNA and the other specific to the chloramphenicol acetyltransferase (CAT) gene (59, 82); the separated RNAs were visualized using the DIG luminescent detection kit (Boehringer) according to the manufacturer's protocol. RNA was quantitated using densitometric scanning. The packaging efficiency for a given RNA species was calculated as the ratio of the amount of that RNA from virions divided by the amount of that RNA from cells.
RESULTS
Direct comparison of packaging efficiencies of MHV DI RNAs.
We used a series of MHV DI RNAs to determine whether the MHV packaging signal present in MHV RNA mediates the specific interaction between M protein and viral RNP complex that leads to specific packaging of MHV RNA into MHV particles. Our previous studies of MHV DI RNAs identified the packaging signal as a 190-nt sequence (190-nt packaging signal) located about 21 kb from the 5′ end of the MHV genome (26). Subsequently, we showed that DI RNAs containing a 69-nt sequence (69-nt packaging signal), which is part of the 190-nt packaging signal, are also packaged into MHV particles. Site-directed mutagenesis of the packaging signal showed that the secondary structure formed by the 69-nt sequence is important for the packaging activity (26). In our previous report, the packaging efficiency of DI RNA containing the 69-nt packaging signal was not directly compared with that of the DI RNA containing the 190-nt packaging signal, because all the experiments were performed using passaged virus samples, which could have allowed the amplification of poorly packaged DI RNA (26).
In the present study, we directly compared the packaging efficiencies of transfected DI RNAs containing the 190-nt packaging signal (DF1-2, FA1, FA992A, and FA2) with the efficiencies of those containing the 69-nt packaging signal (FB1 and FA4+PS) and those lacking the packaging signal (FA4, DF1-2ΔPS, and ΔFA2) (Fig. 1). MHV-infected cells were transfected with the same amount of in vitro-synthesized, capped DI RNA transcripts. The culture fluid containing the released virus particles was harvested at 12 h p.i. Released viruses were purified using sucrose gradient centrifugation, and viral RNAs were extracted from purified virus particles. To examine the intracellular level of these DI RNAs, intracellular RNA was also extracted at 12 h p.i. Northern blot analysis using a probe that specifically hybridizes with DI RNAs and mRNA 1 showed that, for each set of experiments, the levels of MHV genomic RNA in the released virus were similar across the different samples (Fig. 2). Levels of the DI RNAs were similar in the DI RNA-transfected, MHV-infected cells, too (Fig. 2). A very low level of intracellular DI RNA was detected in DI RNA-transfected, mock-infected cells (data not shown), demonstrating that the majority of intracellular DI RNA signal represented replicating DI RNA. Some weak additional bands, designated by asterisks in Fig. 2, were probably other, spontaneously generated DI RNA species.
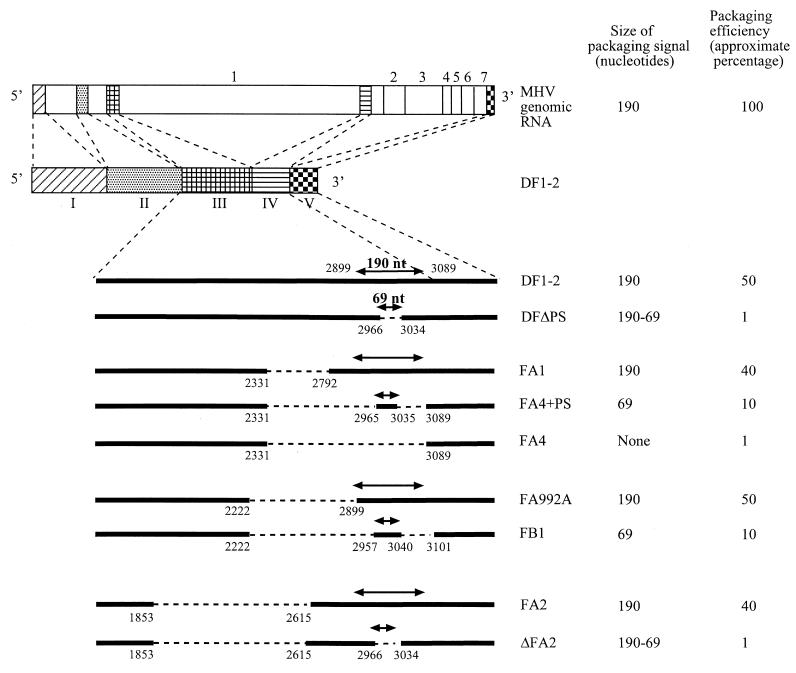
FIG. 1.
Schematic diagrams of the structures of MHV genomic RNA and DI RNAs. The five domains of DF1-2 (domains I through V) are indicated below the diagram of DF1-2; the locations of these domains on MHV genomic RNA are shown as shaded boxes. The numbers 1 through 7 represent the seven genes of MHV. The deleted regions in DI RNAs are shown as dashed lines. The exact locations of the deleted regions are shown as nucleotides numbered from the 5′ end of DF1-2. The locations of the 190-nt packaging signal and the 69-nt packaging signal in DI RNAs are also indicated. DFΔPS and ΔFA2 both had a deletion of the 69-nt packaging signal within the 190-nt packaging signal. The packaging efficiency for a given RNA species was calculated as the ratio of amount of that RNA from virions divided by the amount of that RNA from cells. The packaging efficiencies of different DI RNAs are reported as approximate percentages of the packaging efficiency of MHV genomic RNA.
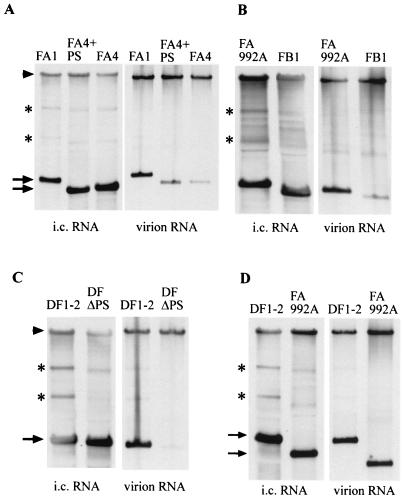
FIG. 2.
Comparison of packaging efficiencies of MHV DI RNAs containing the 190-nt packaging signal (FA1, FA992A, and DF1-2) with the efficiencies of those containing the 69-nt packaging signal (FA4+PS and FB1) and those lacking the packaging signal (FA4 and DFΔPS). The same amount of in vitro-synthesized RNA of each DI clone was independently transfected into MHV-infected cells. Released virus particles were harvested at 12 h p.i. and purified by sucrose gradient centrifugation. Viral RNAs were extracted from purified virus particles. Intracellular (i.c.) RNAs were also extracted at 12 h p.i. from cytoplasmic protein lysates. Intracellular RNAs and virion RNAs were analyzed using Northern blot analysis with a probe that binds to MHV genomic RNA (or mRNA 1) and DI RNAs. The arrowheads indicate MHV genomic RNA (mRNA 1). The arrows indicate DI RNAs of expected sizes. Panels A to D represent separate experiments, each of which was repeated in triplicate.
The relative amounts of RNA packaged into particles varied among the different DI RNAs. We analyzed viral and cellular RNAs in parallel for each experiment in independently transfected cells and calculated the relative packaging efficiencies of the different DI RNAs as the ratio of the amount of a DI's RNA from virions divided by the amount from cells. We also calculated the packaging efficiency of MHV genomic RNA using the same method. The packaging efficiency of different DI RNAs was presented as an approximate percentage of the packaging efficiency of MHV genomic RNA (Fig. 1). Figure 2A shows the data from an experiment comparing DIs containing the 190-nt packaging signal (FA1), the 69-nt packaging signal (FA4+PS), and no packaging signal (FA4); packaging efficiencies of FA1 and FA4+PS were about 40- and 10-fold higher, respectively, than that of FA4. The sequence of FA4 RNA in extracellular virions was the same as that of input FA4 RNA (data not shown). Similarly, direct comparison of the packaging efficiencies of FA992A, with the 190-nt packaging signal, and FB1, containing the 69-nt packaging signal, showed that FA992A was packaged about fivefold more efficiently than was FB1 (Fig. 2B). Figure 2C shows DF1-2, containing the 190-nt packaging signal, and DFΔPS RNA, lacking the 69-nt packaging signal; the packaging efficiency of DF1-2 was about 40-fold higher than that of DFΔPS RNA. Figure 3D shows a similar example: FA2, containing the 190-nt packaging signal, was packaged about 40 times more efficiently than was ΔFA2, which lacked the 69-nt packaging signal. Direct comparison of the packaging efficiencies of DF1-2 and of FA992A, which has a deletion of a 0.68-kb sequence upstream of the 190-nt packaging signal of DF1-2, showed that the two DI RNAs were packaged with similar efficiencies (Fig. 2D), demonstrating that inclusion of an additional MHV sequence 5′ to the 190-nt packaging signal did not improve the packaging efficiency. All results were reproduced consistently in triplicate experiments.
FIG. 3.
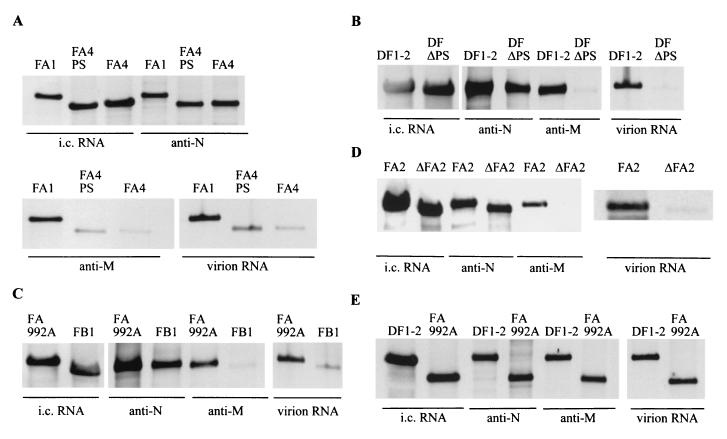
Specific binding of M protein to replicating DI RNAs containing the packaging signal in MHV-infected cells. Cell lysates were prepared from DI RNA-transfected, MHV-infected cells at 12 h p.i. Anti-N protein monoclonal antibody (anti-N) and anti-M protein monoclonal antibody (anti-M) were independently added to equal volumes of cell lysates, and immunoprecipitation was performed. RNA was extracted from the immunoprecipitated samples. Intracellular (i.c.) RNAs and virion RNAs were extracted as described in the legend to Fig. 2. Extracted RNAs were analyzed on Northern blots using a probe that binds to DI RNAs. Only the section of the autoradiogram with the DI RNAs is shown. Each panel shows representative data from triplicate experiments.
These studies clearly showed that DI RNAs containing the 190-nt packaging signal (DF1-2, FA1, FA992A, and FA2) were efficiently packaged into virus particles. DI RNAs containing the 69-nt packaging signal were also packaged into virus particles; however, their packaging efficiency was about four- to fivefold lower than the efficiency of those containing the 190-nt packaging signal (Fig. 2A and B). DI RNA lacking the 190-nt packaging signal (FA4) and those lacking the 69-nt packaging signal (DF1-2ΔPS and ΔFA2) were packaged very poorly into MHV particles. The presence of a low level of DI RNAs lacking the packaging signal in virus particles was not surprising, as MHV DI RNAs lacking the packaging signal replicate in cells infected with passaged virus samples, initially obtained from DI RNA-transfected, MHV-infected cells (26); DI RNAs lacking the packaging signal are packaged nonspecifically with a low efficiency.
Specific interaction of M protein with intracellular DI RNP complex containing the packaging signal.
We wanted to know whether the packaging signal mediates a specific interaction between M protein and DI RNA, complexed with N protein. With that purpose, we looked at how helper virus-derived N protein associates with replicating DI RNA and at whether helper virus-derived M protein specifically recognizes DI RNP containing the packaging signal. Cell extracts, prepared at 12 h p.i. from DI RNA-transfected, MHV-infected cells, were immunoprecipitated with an anti-N protein monoclonal antibody, J3.3, or an anti-M protein monoclonal antibody, J1.3; MHV-specific RNAs were extracted from the immunoprecipitated samples. Intracellular RNAs were also extracted from the cytoplasmic protein lysates at 12 h p.i. as described previously (59). The RNAs were analyzed on a Northern blot using the same probe, which recognizes DI RNA and mRNA 1, that was used for the data in Fig. 2. Coimmunoprecipitation analysis, using anti-N protein antibody, showed efficient coimmunoprecipitation of all the DI RNAs (Fig. 3), demonstrating that all those DI RNAs associated with N protein to form DI RNP complex, whether or not that RNA had the packaging signal. This result was expected because all MHV mRNAs, including subgenomic mRNAs, are also associated with N protein in infected cells (4, 59). Coimmunoprecipitation analysis, using anti-M protein antibody, showed efficient coimmunoprecipitation of only the DI RNAs containing the 190-nt packaging signal (DF1-2, FA1, FA992A, and FA2) (Fig. 3). Anti-M protein antibody also coimmunoprecipitated a lesser amount of DI RNAs containing the 69-nt-long packaging signal (FA4+PS and FB1), whereas it coimmunoprecipitated only a trace amount of DI RNA lacking the packaging signal (FA4, DF1-2ΔPS, and ΔFA2) (Fig. 3). The non-MHV monoclonal antibody, anti-H2K antibody, did not coimmunoprecipitate any DI RNAs (data not shown). Densitometric analysis of the autogradiograms showed an excellent correlation between the amount of DI RNAs detected in virus particles and the amount of intracellular DI RNAs coimmunoprecipitated by anti-M protein antibody (Fig. 3). Results from triplicate experiments were consistent.
These data demonstrated that N protein associated with all the DI RNAs to form DI RNP complexes, regardless of the presence or absence of the packaging signal, whereas M protein selectively interacted only with DI RNP complexes containing the packaging signal. The profile of M protein interaction with DI RNPs was strikingly similar to the packaging profile of these DI RNPs. This experimental evidence strongly suggested that the M envelope glycoprotein bound only those DI RNP complexes having a packaging signal in a process that brought about the efficient packaging of those same complexes into MHV particles.
Specific interaction of M protein with non-MHV RNA carrying the packaging signal.
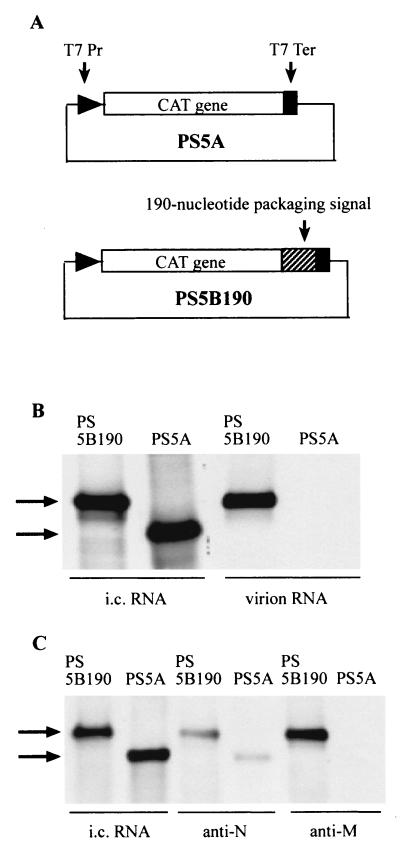
When non-MHV RNA transcripts containing the MHV packaging signal are expressed in MHV-infected cells, they are packaged into MHV particles, while expressed non-MHV RNA transcripts lacking the packaging signal are not packaged (82). Analogously, when RNA transcripts containing the bovine coronavirus (BCV) packaging signal are expressed in BCV-infected cells, the expressed RNA transcripts are packaged into BCV particles (17). We speculated that, in MHV-infected cells, M protein would specifically interact with expressed non-MHV RNA transcripts if they had been constructed with an MHV packaging signal and that this specific interaction would allow packaging of the packaging signal-positive, non-MHV transcripts into MHV particles. To test this hypothesis, we used two plasmids, PS5B190 and PS5A, for the expression of non-MHV RNA transcripts in MHV-infected cells (82). PS5A contains the CAT gene, without a poly(A) sequence, under the control of the T7 promoter and the T7 terminator and has no MHV-specific sequence (Fig. 4A). PS5B190 contains the 190-nt MHV packaging signal inserted downstream of the CAT gene (Fig. 4A). RNA transcripts from these plasmids were expressed using the recombinant vaccinia virus vTF7-3 (27). Briefly, vTF7-3-infected DBT cells were transfected with PS5A or PS5B190 and superinfected with MHV, and virus particles were harvested 12 h post-MHV superinfection. The released virus particles were purified on a sucrose gradient, and viral RNAs were extracted from purified virus particles as described previously (82). Intracellular RNAs were also extracted 12 h post-MHV infection. RNA was analyzed on Northern blots using a CAT-sequence-specific probe (82). As we had observed before (82), the PS5A and PS5B190 transcripts were expressed at similar levels in MHV-infected cells, and yet only PS5B190 RNA was packaged into MHV particles (Fig. 4B). In three independent experiments, the efficiency of packaging of PS5B190 was consistently about 100-fold higher than that of PS5A RNA.
FIG. 4.
Specific binding of M protein to non-MHV RNA transcripts containing the packaging signal. (A) Schematic diagrams of the structures of plasmids PS5A and PS5B190. T7 Pr, T7 promoter; T7 Ter; T7 terminator. (B) Northern blot analysis of expressed RNA transcripts in RNA-expressing, MHV-infected cells (intracellular [i.c.] RNA) and packaged RNA transcripts in MHV particles (virion RNAs). PS5A RNA transcripts or PS5B190 RNA transcripts were independently expressed in MHV-infected cells. Intracellular RNAs and virion RNAs were analyzed on Northern blots using a probe that binds to the CAT sequence. The arrows indicate expressed RNA transcripts. (C) Specific binding of M protein to the expressed PS5B190 RNA transcripts in MHV-infected cells. Cell lysates were prepared from MHV-infected cells expressing non-MHV RNA transcripts at 12 h post-MHV infection. Anti-N protein monoclonal antibody (anti-N) and anti-M protein monoclonal antibody (anti-M) were independently added to equal volumes of cell lysates, and immunoprecipitation was performed. RNA was extracted from the immunoprecipitated samples. Intracellular (i.c.) RNAs and coimmunoprecipitated RNAs were analyzed using Northern blot analysis with a probe that binds to the CAT sequence. The arrows indicate expressed RNA transcripts.
We went on to look at whether N protein can bind expressed non-MHV RNA transcripts to form an RNP complex in MHV-infected cells. For this analysis, cell extracts prepared from MHV-infected cells expressing the non-MHV RNA transcripts were immunoprecipitated with the anti-N protein monoclonal antibody. Then, the RNA was extracted from the immunoprecipitates and analyzed on a Northern blot using the same CAT-specific probe. Coimmunoprecipitation analysis showed that N protein associated with both PS5A and PS5B190 transcripts (Fig. 4C). The amount of coimmunoprecipitated PS5B190 RNA was only slightly (about 1.5-fold) higher than the amount of PS5A RNA. N protein bound to both non-MHV RNA transcripts to form an RNP complex; therefore, the presence or absence of the packaging signal did not determine the binding of N protein to the expressed non-MHV RNA transcripts. These results were consistent with a recent report that BCV N protein binds to expressed RNA transcripts containing the CAT sequence in BCV-infected cells (18). Anti-M protein monoclonal antibody coprecipitated only PS5B190 RNA (Fig. 4C); it did not coimmunoprecipitate the PS5A RNA (Fig. 4C). The amount of coimmunoprecipitated PS5B190 RNA was consistently about 100-fold greater than that of PS5A RNA in three independent experiments. In control experiments, using the same cell extracts, anti-H2K antibody coimmunoprecipitated neither PS5A RNA nor PS5B190 RNA (data not shown). These data demonstrated that M protein selectively interacted with a nonreplicating, non-MHV RNA transcript containing the 190-nt MHV packaging signal and that it did not interact with non-MHV RNA that lacked the MHV packaging signal.
We concluded that the packaging signal determined the specific and selective interaction between M protein and specific intracellular RNP complexes and that this interaction is responsible for the selective and efficient packaging of MHV RNAs containing the packaging signal into MHV particles.
DISCUSSION
The present study tested the possibility that the packaging signal determines the selective interaction between M protein, a membrane glycoprotein, and the viral RNP complex containing the packaging signal. In MHV-infected cells, the MHV nucleocapsid protein, N, bound to MHV DI RNAs and the expressed non-MHV RNA transcripts, PS5A and PS5B190, regardless of the presence or absence of the packaging signal. In marked contrast, M protein selectively interacted only with MHV DI RNPs and non-MHV RNA transcripts, both carrying the 190-nt packaging signal, in MHV-infected cells. The efficiency of interaction of M protein with RNA correlated with the packaging efficiency of RNA into MHV particles. Previously, we demonstrated that M protein selectively interacts only with MHV mRNA 1 and a self-replicating MHV DI RNA, DIssA, both of which contain the 190-nt packaging signal, and also showed that M protein does not interact with MHV subgenomic mRNAs (59); both mRNA 1 and DIssA were efficiently packaged into MHV particles. Collectively, these data convincingly showed that, in the infected cell, M protein specifically interacted with RNPs containing the packaging signal, which then were selectively packaged into virus particles.
Analysis of packaging of DI RNAs into MHV particles showed that the 190-nt packaging signal conferred a higher packaging efficiency on DI RNAs than did the 69-nt packaging signal. The effect of the size of the packaging signal on the efficiency of packaging of RNA into virus particles has not been previously demonstrated for coronaviruses. For retroviruses, the size of the packaging signal affects the relative packaging efficiency of nonretroviral RNAs carrying the retroviral packaging signal (1, 7). Computer prediction of the secondary structure of the 190-nt packaging signal (mfold version 2.3) showed a stable stem-loop structure, which was identical in MHV-A59 and MHV-JHM strains (data not shown). The reason for the efficient interaction of M protein with RNPs containing the 190-nt packaging signal could be the formation of a favorable secondary structure. The RNA secondary structure of the 69-nt packaging signal probably was not optimal for interaction with M protein. Indeed, computer prediction of the secondary structure of the 69-nt packaging signal, in the context of MHV genomic RNA, showed that the structure was different from that of the 190-nt packaging signal (data not shown). This suggested that the sequences flanking the 69-nt region, within the 190-nt packaging signal, are important for the formation of a specific secondary structure, which may allow the RNA to interact efficiently with M protein. Our previous mutagenic analysis of the 69-nt packaging signal revealed that the secondary structure of the 69-nt packaging signal is important for its biological function (26). However, that study was less quantitative than the present study, because we examined the packaging efficiencies of DI RNAs, each of which contained a mutated 69-nt packaging signal, using passaged virus samples. Direct comparison of the packaging efficiencies of a series of DI RNAs, each of which contains a mutated 190-nt packaging signal with a different RNA secondary structure, will reveal the importance of the RNA secondary structure of the packaging signal for RNA packaging activity.
MHV genomic RNA was packaged about 2 to 2.5 times more efficiently than were DI RNAs containing the 190-nt packaging signal (Fig. 1). We know that helper virus mRNA synthesis is strongly inhibited in DI RNA-replicating cells (55); hence, the production of MHV structural proteins is most probably reduced significantly in DI RNA-replicating, MHV-infected cells. Accordingly, the availability of helper virus-derived trans-acting factors required for RNA packaging may be limited in DI RNA-replicating cells, and this situation probably affected the production of DI particles; we speculate that the environment for RNA packaging was not optimized for DI RNA packaging in DI RNA-replicating, MHV-infected cells. Another possibility, for a higher level of MHV genomic RNA packaging, is that some unidentified sequences, which are missing in DI RNAs and are present only in genomic RNA, may enhance the activity of the packaging signal to promote efficient MHV genomic RNA packaging.
For many viruses, viral nucleocapsid recognition of an RNA packaging signal generally begins encapsidation of viral genomic RNA. This interaction is assumed to ensure specificity of packaging of genomic-length RNA into the virus particle. For example, in alphaviruses, the capsid protein specifically recognizes the packaging signal and is the basis for the specific encapsidation of viral genomic RNA (30, 63, 80). In retrovirus human immunodeficiency virus type 1, the NCp7 domain of Gag polyprotein has been shown elsewhere to specifically recognize the human immunodeficiency virus packaging signal and is principally responsible for the specific encapsidation of the unspliced genomic RNA into the virus particles (8, 10, 20, 29). In the case of hepatitis B virus, viral RNA packaging occurs through the specific binding of P protein to the encapsidation signal, followed by addition of multiple C proteins to viral RNA to form the nucleocapsid (5, 16, 34).
Like coronavirus, the viral genome in many negative-strand animal RNA viruses is packaged in the form of a helical nucleocapsid structure. In influenza virus, the packaging signal, which overlaps with cis-acting viral RNA replication signals, has been identified previously (48), and yet how the packaging signal drives the packaging of specific influenza virus RNA is not known. Both the genomic and antigenomic RNAs of influenza virus form the helical nucleocapsid structure in the nucleus, which is the site of viral RNA synthesis. The influenza virus mRNAs do not associate with N protein. The nucleocapsid-containing genomic RNA, but not the antigenomic RNA, is exported from the nucleus to the cytoplasm. M1 protein and NEP (NS2) protein may play a role in the nuclear export of viral nucleocapsids (14, 56, 60, 62). The mechanism of this selective transport of the nucleocapsid, containing the genomic RNA, from the nucleus to the cytoplasm is unclear. This selective transport of specific nucleocapsids appears to be important for influenza virus RNA packaging, because envelopment of nucleocapsid occurs at the cytoplasmic membrane. The nucleocapsid of rhabdovirus, a negative-strand RNA virus, also has helical nucleocapsid symmetry. Genomic and antigenomic RNAs form helical nucleocapsids in the cytoplasm of the infected cells, while viral subgenomic RNAs do not form this structure (21, 65). Of the two helical nucleocapsid species in rhabdoviruses, only the nucleocapsid containing the genomic RNA is efficiently packaged into virus particles. A short cis-acting RNA element, at the 5′ end of the genome of vesicular stomatitis virus, a prototypic rhabdovirus, is key to the packaging of that viral RNA (65, 81). Another cis-acting viral element(s) also may be involved in the packaging of nucleocapsid (81). Rhabdovirus matrix protein interacts with the viral helical nucleocapsid (38, 61), and this interaction probably is important for the packaging of helical nucleocapsid into virus particles, although the mechanism of the selective recognition of nucleocapsid containing the genomic RNA by the matrix protein is unknown. For the negative-strand RNA viruses carrying the genome in a helical nucleocapsid, association of nucleocapsid protein with RNA appears to be a prerequisite for RNA packaging, but a mechanism for selective packaging of specific intracellular helical nucleocapsids is not described.
What we have learned about MHV is that binding of MHV N protein to RNA does not determine the specificity and selectivity in packaging of MHV RNA, because N protein associates with all MHV RNAs (4, 59) and any expressed non-MHV RNAs in MHV-infected cells. The observation that MHV N protein bound to all MHV mRNAs and non-MHV RNA transcripts in infected cells was not unexpected, because N protein is reported to bind in vitro with sequences other than the leader and the packaging signal within the MHV genome (18, 74) and RNAs of nonviral origin (57, 68, 74). The formation of intracellular RNP complex is not the determinant of selectivity in MHV RNA packaging; rather, the selective interaction between M protein and RNA containing the packaging signal, complexed with N protein, was critical for the specificity and selectivity in RNA packaging. This finding is remarkable in that MHV M protein is a transmembrane viral envelope protein. To our knowledge, for any enveloped virus this is the first example of a viral envelope protein determining the selectivity and efficiency of incorporation of viral RNA into virus particles.
A major question that remains to be addressed is how M protein selectively recognizes packaging signal-containing RNAs, including MHV mRNA 1, DIssA, various DI RNAs, and expressed non-MHV RNA transcripts that contain the packaging signal. One possible explanation is rooted in the earlier step of helical nucleocapsid formation. N protein binding to the packaging signal might induce a specific conformational change in N protein that could serve as a nucleation event for the cooperative binding of N protein to the rest of the RNA, thereby generating the helical nucleocapsid structure. If an RNA lacks the packaging signal, then binding of N protein may not induce a putative nucleation event-generating conformational change. Indeed, an in vitro binding assay showed that MHV N protein binds to the 190-nt packaging signal but not to the 69-nt packaging signal (58). It is unknown whether binding of N protein to the 190-nt packaging signal induces any conformational change in N protein. Nevertheless, the finding that N protein binds to the 190-nt packaging signal but not to the 69-nt packaging signal (58) was consistent with our present data that the 190-nt packaging signal conferred a relatively higher packaging efficiency than did the 69-nt packaging signal. Among a pool of intracellular viral RNP complexes, M protein may efficiently interact only with one specific helical nucleocapsid structure that is ordained by the packaging signal; in this way, both N protein and M protein would contribute to the selective packaging of specific RNA species into the virus particle. Another possible explanation for the selective interaction of M protein with the packaging signal-loaded RNP complex is that M protein may specifically bind the packaging signal directly. An initial M protein-packaging signal interaction might be further stabilized by the subsequent association of M protein with N protein in the RNP complex. In fact, a direct RNA-independent interaction between M protein and N protein does occur in MHV-infected cells (59). This stable interaction could lead to the incorporation of the RNP complex into the virus particle. This possible mechanism of RNA packaging that would involve direct binding of an RNA packaging signal by a viral membrane protein has not been described for any other virus, and yet several data are consistent with this possibility. In the absence of N protein, M protein cosediments with genomic RNA in vitro (75). We observed that only a small amount of PS5B190 transcripts was coimmunoprecipitated by anti-N protein antibody (Fig. 4C), while the same transcripts were efficiently coimmunoprecipitated by anti-M protein antibody (Fig. 4C), implying that the expressed PS5B190 transcripts that were not associated with N protein probably bound to M protein in MHV-infected cells. Furthermore, we have recently observed that the expressed M protein bound to the expressed PS5B190 transcripts, but not PS5A transcripts, in the absence of N protein (K. Narayanan and S. Makino, unpublished data).
ACKNOWLEDGMENTS
We thank John Fleming for monoclonal antibodies against MHV proteins and Paul Gottlieb for anti-H2K monoclonal antibody. We also thank Yoshihiro Kawaoka, Chiaho Shih, Amiya Banerjee, and Andy Ball for helpful information and comments.
This work was supported by Public Health Service grant AI29984 from the National Institutes of Health.
REFERENCES
- 1.Adam M A, Miller A D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks J D, Kealoha B O, Linial M L. An Mψ-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J Virol. 1999;73:8926–8933. doi: 10.1128/jvi.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric R S, Nelson G W, Fleming J O, Deans R J, Keck J G, Casteel N, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J, Bartos H, Nassal M. Experimental confirmation of a hepatitis B virus (HBV) epsilon-like bulge-and-loop structure in avian HBV RNA encapsidation signals. Virology. 1997;227:500–504. doi: 10.1006/viro.1996.8329. [DOI] [PubMed] [Google Scholar]
- 7.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Hammarskjold M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 10.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos E C, Dobbe J C, Luytjes W, Spaan W J. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J Virol. 1997;71:5684–5687. doi: 10.1128/jvi.71.7.5684-5687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos E C, Luytjes W, van der Meulen H V, Koerten H K, Spaan W J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boursnell M E, Brown T D. Sequencing of coronavirus IBV genomic RNA: a 195-base open reading frame encoded by mRNA B. Gene. 1984;29:87–92. doi: 10.1016/0378-1119(84)90169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bui M, Wills E G, Helenius A, Whittaker G R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavanagh D, Shaw K, Zhao X. Analysis of messenger RNA within virions of IBV. Adv Exp Med Biol. 1993;342:123–128. doi: 10.1007/978-1-4615-2996-5_20. [DOI] [PubMed] [Google Scholar]
- 16.Cohen B J, Richmond J E. Electron microscopy of hepatitis B core antigen synthesized in E. coli. Nature. 1982;296:677–679. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 17.Cologna R, Hogue B G. Identification of a bovine coronavirus packaging signal. J Virol. 2000;74:580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cologna R, Spagnolo J F, Hogue B G. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology. 2000;277:235–249. doi: 10.1006/viro.2000.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corse E, Machamer C E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das T, Chakrabarti B K, Chattopadhyay D, Banerjee A K. Carboxy-terminal five amino acids of the nucleocapsid protein of vesicular stomatitis virus are required for encapsidation and replication of genome RNA. Virology. 1999;259:219–227. doi: 10.1006/viro.1999.9768. [DOI] [PubMed] [Google Scholar]
- 22.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escors D, Ortego J, Laude H, Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming J O, Shubin R A, Sussman M A, Casteel N, Stohlman S A. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology. 1989;168:162–167. doi: 10.1016/0042-6822(89)90415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimura T, Esteban R, Esteban L M, Wickner R B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 29.Geigenmuller U, Linial M L. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J Virol. 1996;70:667–671. doi: 10.1128/jvi.70.1.667-671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geigenmuller-Gnirke U, Nitschko H, Schlesinger S. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J Virol. 1993;67:1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godet M, L'Haridon R, Vautherot J F, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guy J S, Brian D A. Bovine coronavirus genome. J Virol. 1979;29:293–300. doi: 10.1128/jvi.29.1.293-300.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano N, Fujiwara K, Hino S, Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 34.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes K V, Doller E W, Sturman L S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joo M, Banerjee S, Makino S. Replication of murine coronavirus defective interfering RNA from negative-strand transcripts. J Virol. 1996;70:5769–5776. doi: 10.1128/jvi.70.9.5769-5776.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaptur P E, Rhodes R B, Lyles D S. Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J Virol. 1991;65:1057–1065. doi: 10.1128/jvi.65.3.1057-1065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K H, Narayanan K, Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J Virol. 1997;71:3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klumperman J, Locker J K, Meijer A, Horzinek M C, Geuze H J, Rottier P J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai M M, Baric R S, Brayton P R, Stohlman S A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai M M, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai M M, Stohlman S A. RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee H J, Shieh C K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomniczi B, Kennedy I. Genome of infectious bronchitis virus. J Virol. 1977;24:99–107. doi: 10.1128/jvi.24.1.99-107.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luytjes W, Krystal M, Enami M, Pavin J D, Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 49.Maeda J, Maeda A, Makino S. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology. 1999;263:265–272. doi: 10.1006/viro.1999.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda J, Repass J F, Maeda A, Makino S. Membrane topology of coronavirus E protein. Virology. 2001;281:163–169. doi: 10.1006/viro.2001.0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makino S, Lai M M. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989;63:5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makino S, Shieh C K, Keck J G, Lai M M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makino S, Taguchi F, Hirano N, Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984;139:138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makino S, Yokomori K, Lai M M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 57.Masters P S. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch Virol. 1992;125:141–160. doi: 10.1007/BF01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molenkamp R, Spaan W J. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neumann G, Hughes M T, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogden J R, Pal R, Wagner R R. Mapping regions of the matrix protein of vesicular stomatitis virus which bind to ribonucleocapsids, liposomes, and monoclonal antibodies. J Virol. 1986;58:860–868. doi: 10.1128/jvi.58.3.860-868.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owen K E, Kuhn R J. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qu F, Morris T J. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol. 1997;71:1428–1435. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raamsman M J, Locker J K, de Hooge A, de Vries A A, Griffiths G, Vennema H, Rottier P J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robbins S G, Frana M F, McGowan J J, Boyle J F, Holmes K V. RNA-binding proteins of coronavirus MHV: detection of monomeric and multimeric N protein with an RNA overlay-protein blot assay. Virology. 1986;150:402–410. doi: 10.1016/0042-6822(86)90305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rottier P J, Horzinek M C, van der Zeijst B A. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stern D F, Kennedy S I. Coronavirus multiplication strategy. I. Identification and characterization of virus-specified RNA. J Virol. 1980;34:665–674. doi: 10.1128/jvi.34.3.665-674.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stern D F, Kennedy S I. Coronavirus multiplication strategy. II. Mapping the avian infectious bronchitis virus intracellular RNA species to the genome. J Virol. 1980;36:440–449. doi: 10.1128/jvi.36.2.440-449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stohlman S A, Baric R S, Nelson G N, Soe L H, Welter L M, Deans R J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in Sac− cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 77.Tung F Y, Abraham S, Sethna M, Hung S L, Sethna P, Hogue B G, Brian D A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Most R G, Bredenbeek P J, Spaan W J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vennema H, Godeke G J, Rossen J W, Voorhout W F, Horzinek M C, Opstelten D J, Rottier P J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss B, Nitschko H, Ghattas I, Wright R, Schlesinger S. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J Virol. 1989;63:5310–5318. doi: 10.1128/jvi.63.12.5310-5318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whelan S P, Wertz G W. The 5′ terminal trailer region of vesicular stomatitis virus contains a position-dependent cis-acting signal for assembly of RNA into infectious particles. J Virol. 1999;73:307–315. doi: 10.1128/jvi.73.1.307-315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woo K, Joo M, Narayanan K, Kim K H, Makino S. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol. 1997;71:824–827. doi: 10.1128/jvi.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- 84.Zhao X, Shaw K, Cavanagh D. Presence of subgenomic mRNAs in virions of coronavirus IBV. Virology. 1993;196:172–178. doi: 10.1006/viro.1993.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]