Abstract
3-Hydroxy-2,2-dimethyl-N-[4({[5-(dimethylamino)-1-naphthyl]sulfonyl}amino)-phenyl]propanamide (BAY 38-4766) is a novel selective nonnucleoside inhibitor of cytomegalovirus (CMV) replication with an excellent safety profile. This compound and structural analogues inhibit neither viral DNA synthesis nor viral transcription and translation. Accumulation of dense bodies and noninfectious enveloped particles coincides with inhibition of both concatemer processing and functional cleavage at intergenomic transitions, pointing to interference with viral DNA maturation and packaging of monomeric genome lengths. Resistant virus populations, including a murine CMV (MCMV) isolate with 566-fold-decreased drug sensitivity, were selected in vitro. Sequencing of the six open reading frames (ORFs) known to be essentially involved in viral DNA cleavage and packaging identified mutations in ORFs UL56, UL89, and UL104. Construction of MCMV recombinants expressing different combinations of murine homologues of mutant UL56, UL89, and UL104 and analysis of drug susceptibilities clearly demonstrated that mutant ORFs UL89 exon II (M360I) and M56 (P202A I208N) individually confer resistance to BAY 38-4766. A combination of both mutant proteins exhibited a strong synergistic effect on resistance, reconstituting the high-resistance phenotype of the in vitro mutant. These findings are consistent with genetic mapping of resistance to TCRB (2,5,6-trichloro-1-β-d-ribofuranosyl benzimidazole) (P. M. Krosky et al., J. Virol. 72:4721–4728, 1998) and provide further indirect evidence that proteins encoded by UL89 and UL56 function as two subunits of the CMV terminase. While these studies also suggest that the molecular mechanism of BAY 38-4766 is distinct from that of benzimidazole ribonucleosides, they also offer an explanation for the excellent specificity and tolerability of BAY 38-4766, since mammalian DNA does not undergo comparable maturation steps.
Human cytomegalovirus (HCMV), a member of the Herpesviridae family, is of minimal consequence in immunocompetent persons but causes significant morbidity and mortality in immunocompromised patients (1). HCMV is frequently transmitted perinatally and is the leading cause of neurological disease and hearing loss in congenitally infected newborns (3). Prior to the advent of highly active retroviral therapy, retinitis and life-threatening disease occurred in HCMV patients with late-stage AIDS as a consequence of primary infection or reactivation of latent infection (29, 40). Approximately 15 to 70% of kidney, liver, bone marrow, and heart/lung transplant recipients are affected by HCMV hepatitis and pneumonia, resulting in decreased graft and patient survival (21). Additionally, HCMV infection might be involved in the development of atherosclerosis (26, 45) and restenosis following angioplasty (58).
Current systemic chemotherapy of HCMV infection is limited to ganciclovir (GCV) (17), cidofovir (CDV) (34), and foscarnet (15). Despite strong antiviral potential, these drugs are associated with poor oral bioavailability, multiple side effects such as dose-limiting bone marrow and kidney toxicity, and development of single- and double-drug resistance (22, 25, 50). All of these drugs inhibit viral replication through interaction with the virally encoded DNA polymerase (20, 28, 39), leading to potential cross-resistance. The antisense phosphorothioate deoxyoligonucleotide Fomivirsen, recently developed for the treatment of HCMV retinitis (4), can only be applied intravitreally, so that a high medical need for the discovery of novel HCMV inhibitors with unique mechanisms of action still exists.
3-Hydroxy-2,2-dimethyl-N-[4({[5-(dimethylamino)-1-naphthyl]sulfonyl}amino)-phenyl]propanamide (BAY 38-4766) is a novel selective nonnucleoside inhibitor of cytomegalovirus (CMV) replication that specifically targets cleavage of viral high-molecular-weight DNA concatemers and packaging of monomeric genome lengths into procapsids.
The maturation of Herpesviridae is a multistep process that has been only partially characterized (54). Current evidence suggests that viral DNA is packaged into a procapsid consisting of major capsid protein (UL86), minor capsid protein (UL85), minor capsid protein-binding protein (UL46), smallest capsid protein (UL47/48), assembly protein (UL80.5), and proteinase precursor protein (UL80a) (23). The translocation of concatenated viral DNA into procapsids and its cleavage at packaging sites is not understood. Recent studies with herpes simplex virus type 1 (HSV-1) mutants defective in UL6, UL15, UL25, UL28, UL32, or UL33 suggest that these genes are essentially involved in viral DNA cleavage and packaging, since cells infected with these mutants produce only B capsids (2, 5, 35, 36, 41, 46, 52, 56). The respective homologues of these genes in HCMV are UL104, UL89, UL77, UL56, UL52, and UL51 (14). By analogy to gp17, a known ATP-dependent endonuclease from bacteriophage T4 (7, 8), the HCMV UL89 gene may encode an endonucleolytic subunit of a putative HCMV terminase. Recent studies by Bogner et al. (12) suggest that the gene product of HCMV open reading frame (ORF) UL56 has specific nuclease activity that cleaves substrates bearing the a sequence, as well as specific binding affinity to packaging elements.
MATERIALS AND METHODS
Viruses, cells, and drugs.
The HCMV laboratory strain AD169 (ATCC catalog no. VR 538; American Type Culture Collection, Manassas, Va.) and the clinical isolate Hellebrand (He) (A. Eis, Department of Microbiology, University of Bonn, Bonn, Germany) were propagated on human embryonic lung fibroblasts (HELF), normal human dermal fibroblasts (NHDF; CellSystems, St. Katharinen, Germany), or human newborn embryonic foreskin fibroblasts (HS68 cells; ATCC no. CRL 1635), passages 5 to 30. HELF and NHDF were cultivated in Eagle's minimal essential medium (EMEM) supplemented with 1% (vol/vol) l-glutamine (200 mM; 29.2 mg/ml), 1% (vol/vol) penicillin-streptomycin (10.000 U of penicillin and 10,000 μg of streptomycin/ml in physiological saline), and 10% (vol/vol) fetal calf serum (FCS), referred to below as EMEM/10. HS68 cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) FCS (DMEM/10). Murine CMV (MCMV) strain Smith (ATCC no. VR 194) was propagated on NIH 3T3 cells (ATCC no. CRL 1658) maintained in EMEM/10. The following reference drugs were used: intravenous formulations of Cymevene (GCV) from Syntex, Foscavir (foscarnet) from Astra, and Vistide (CDV) from Gilead as 50 mM solutions in 0.9% saline. (1R-1α, 2β, 3α)-9-(2,3-bis-hydroxymethylcyclobutyl)guanine (lobucavir [LBV]) was a gift from Bristol-Myers-Squibb (BMS 180194); 5,6-dichloro-2-isopropylamino-1-β-l-ribofuranosyl-1H-benzimidazole (benzimidavir) was provided by U. Groth (Konstanz, Germany); and 2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (BDCRB), BAY 38-4766, and its close structural analogues were synthesized at Medicinal Chemistry (Bayer AG, Wuppertal, Germany). The last four compounds were used as 50 mM solutions in dimethyl sulfoxide.
RNA isolation and Northern blot analyses.
Confluent monolayers of HELF were infected with HCMV AD169 at a multiplicity of infection (MOI) of 1. After removal of the viral inoculum, cells were washed twice with EMEM/10 and once with medium containing Cytoglobin (1:100) to inactivate nonadsorbed viruses (Bayer AG). At 24 and 72 h postinfection (p.i.), total cellular RNAs were isolated using the RNeasy kit (Qiagen, Hilden, Germany) as instructed by the manufacturer. Total RNA (2.5 μg per lane) was size fractionated under denaturing conditions, transferred to a nylon membrane, and hybridized as described previously (24). An RNA probe specific for UL99 was generated as described under “Hybridization probes” below, and luminescence was detected as described previously (24).
Electron microscopy and ultrastructural investigation.
HS68 cells were seeded on 4-well Lab-Tec chamber slides and grown to confluency. Following infection with HCMV AD169 at an MOI of 1, untreated control cells and cells treated with 0.67 μM BAY 36-8888 were cultivated for 3 to 4 days. In order to harvest cells, culture medium was replaced by 2% glutaraldehyde fixative in phosphate-buffered saline. Samples were stored at 4°C until further preparation. Primary fixation was followed by secondary fixation in a 2% osmium tetroxide-buffer mixture, followed by dehydration and subsequent embedding in Epon. For orientation within the cells, 1-μm semithin sections were prepared and stained by Richardson's method (methylene blue). Additionally, blocks were trimmed for ultrathin sections and contrasted with uranyl acetate and lead citrate. Examination was performed with a Philips electron microscope.
Pulsed-field gel electrophoresis and Southern blot analyses.
HELF were infected with HCMV AD169 at an MOI of 1 and cultivated in the presence of either BAY 38-4766 at 0.5, 1, 5, or 25 μM or BDCRB at 25 μM. At 3 to 4 days p.i., cells were washed twice in phosphate-buffered saline buffer and embedded in agarose plugs. Blocks were incubated twice for 24 h at 50°C in 1 ml of ESP buffer (0.5 M EDTA [pH 9.5], 1% sodium lauryl sarcosine, 0.1% proteinase K), washed three times for 2 h in 15 ml of TE buffer (0.1 M Tris-Cl [pH 8], 1 mM EDTA), and stored at 4°C until use. Fractions of agarose plugs were loaded onto a 1% agarose gel. Lambda phage DNA concatemers (New England Biolabs, Frankfurt am Main, Germany) were used as molecular size standards. Electrophoretic separation was performed on a CHEF-DRII system (Bio-Rad, Munich, Germany) at a constant voltage of 200 V for 22 h at 14°C in 0.5× Tris-borate-EDTA buffer (pH 8.2) with pulse times increasing linearly from 5 to 70 s. Subsequently, DNA was immobilized on nylon membranes and probed with HindIII-digested genomic HCMV DNA entirely labeled with digoxigenin (DIG). Blot propagation and detection were performed according to the user's guide for the DIG system for filter hybridization (Roche, Mannheim, Germany).
Functional viral DNA cleavage assay.
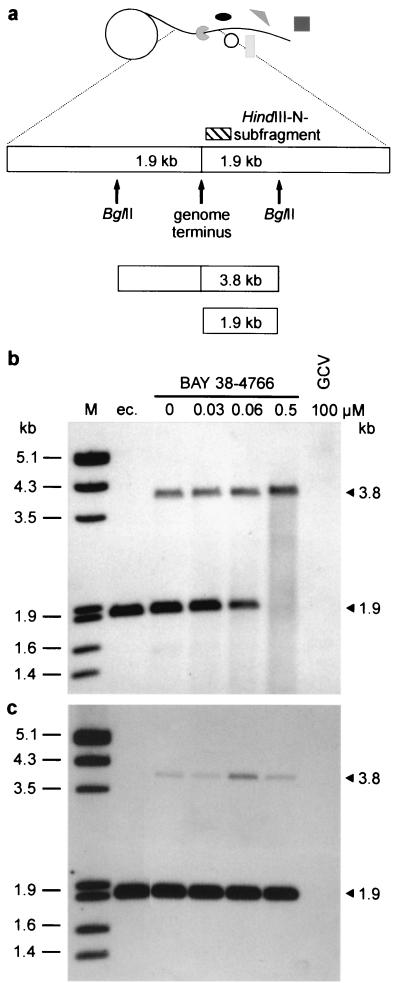
NIH 3T3 cells were infected with MCMV strain Smith at an MOI of 0.05 and maintained at 0, 0.03, 0.06, or 0.5 μM BAY 38-4766, or 100 μM GCV, for 3 to 4 days. After three cycles of freeze-thawing of infected cells, the supernatant was harvested and cell debris was removed by low-speed centrifugation (1,000 × g, 10 min, 4°C). High-molecular-weight protein-DNA-complexes were collected by ultracentrifugation for 1 h at 45,000 × g and 4°C. Pellets were resuspended using a Potter-Elvehjem homogenizer and further purified by a centrifugation step through a discontinuous gradient of 15 and 40% sucrose for 1 h at 150,000 × g and 4°C. To extract viral DNA, the Qiagen Blood and Cell Culture DNA kit was used according to the manufacturer's instructions. DNA was quantified by dot blot hybridization using a DIG-labeled DNA probe of the MCMV HindIII M subfragment. Viral DNA (0.5 μg per lane) was digested with 20 U of BglII overnight, size fractionated on a 0.6% agarose gel by electrophoresis, and analyzed by Southern blotting. Cleaved and uncleaved terminal DNA fragments were visualized by a DIG-labeled MCMV HindIII N subfragment.
Hybridization probes.
Full-length UL99 cDNA amplified by PCR was purified and cloned into the pGEM9Zf(−) vector (Promega, Mannheim, Germany). An RNA hybridization probe was generated in vitro as a runoff transcript from template UL99 using the DIG-RNA labeling kit (Roche). For synthesis of a random-labeled HCMV AD169 probe, DNA was purified from extracellular virus particles as described under “Preparation of viral DNA and sequencing analyses” below and disintegrated by HindIII treatment, followed by phenol-chloroform extraction (49). Template subfragments of the MCMV HindIII M fragment and the HindIII N fragment cloned into pGEM7Zf(+) (19) were obtained by digestion of constructs with HindIII/BamHI and HindIII/AflII, respectively, and gel extraction (QiaQuick gel extraction kit; Qiagen). Random-labeling of DNA probes was performed with the DIG High Prime kit (Roche). All probes were diluted in hybridization buffer to a final concentration of 10 ng/ml.
Selection of BAY 35-5014-resistant HCMV AD169, BAY 38-4766-resistant HCMV He, and BAY 38-4766-resistant MCMV Smith.
A modification of a previously described procedure was used (53). Drug-resistant HCMV strains were selected by infecting confluent HELF monolayers with HCMV strains AD169 and He at an MOI of ∼0.05. Selection was initiated in the presence of BAY 35-5014 and BAY 38-4766, respectively, starting at 50% effective concentrations (EC50). Two populations of resistant viruses (termed AD169-rt and He-rt) were isolated by serial passages of progeny virus from the culture overlay medium in the presence of increasing compound concentrations (twofold steps) to a final concentration of 15 μM. In addition, a BAY 38-4766-resistant MCMV strain Smith (termed MCMV-rt) was serially passaged on NIH 3T3 cells (MOI, ∼0.005) to a final inhibitor concentration of 25 μM. The resistant populations of HCMV and MCMV grew at concentrations ∼50- and ∼500-fold above the EC50, respectively. Derived progeny viruses were plaque purified three times by limiting dilution in the presence of the respective compounds. The stability of resistance was tested by serial passages of plaque-purified viruses without selective pressure (10 to 12 times).
Preparation of viral DNA and sequencing analyses.
AD169-rt, He-rt, and MCMV-rt were propagated on confluent monolayers of NHDF (MOI, ∼0.01) and NIH 3T3 cells (MOI, ∼0.001), respectively. After 100% cytopathic effect was reached, released virions were collected by differential centrifugation, and viral DNA was extracted from ultracentrifugation pellets according to the protocol for the functional viral DNA cleavage assay described above. ORFs UL51, UL52, UL56, UL77, UL89, and UL104, and the respective MCMV homologues, were amplified by PCR from DNA of resistant and wild-type strains using PfuTurbo DNA polymerase (Stratagene, Amsterdam, The Netherlands) according to standard protocols. PCR products were purified using the QiaQuick PCR purification kit (Qiagen). Double-stranded sequencing was performed by Qiagen, and data were evaluated with Align Manager (Scientific and Educational Software, Durham, N.C.). A complete list of primers, PCR constituents, and cycles is available on request. Nucleotide and amino acid sequences were obtained from GenBank, National Center for Biotechnology Information, Bethesda, Md.
Generation of lacZ-tagged MCMV recombinants expressing mutant M89 and/or mutant M104.
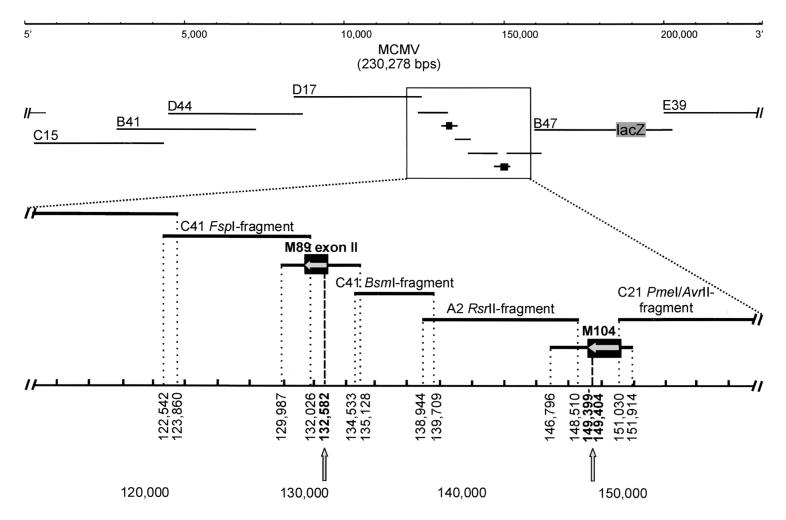
A cosmid library was constructed as described previously (30). MCMV Smith DNA was extracted from extracellular virions, partially digested with Sau3A and was ligated to prepared arms of cosmid vector SuperCos A1 (Stratagene), which had been modified by insertion of an oligonucleotide linker incorporating PmeI recognition sequences (italicized) flanking a unique BamHI site (boldfaced) (5′-GATCGTTTAAACGGATCCGTTTAAAC-3′) into the original BamHI site. For cloning of intergenomic transitions, circular virus DNA was separated from nuclei of infected NIH 3T3 cells by high-salt precipitation (27). Cosmid clones comprising genome transitions were identified in a colony screen using a DIG-labeled MCMV HindIII N subfragment. From a library of 37 characterized clones, a set of seven cosmids reconstituting the complete genome was chosen (C15, nucleotides [nt] 2950 to 44019; B41, nt 28710 to 72510; D44, nt 45430 to 87370; D17, nt 84330 to 123860; C41, nt 117260 to 160770; B47, nt 158980 to 202330; and E39, nt 199060 to 6300).
To construct the mutagenesis plasmid, the tetracycline gene was excised from vector pBR322 (New England Biolabs) by StyI/EcoRI digestion and cloned into the FspI site of the shuttle plasmid pST76K-SR, a derivative of pST76K (47), thereby disrupting the kanamycin resistance gene and resulting in pST76K-SR-T. A complete reporter gene recombination cassette comprising the lacZ gene of control vector pSV-β-galactosidase (Promega) flanked with MCMV DNA sequences homologous to the intended integration site on the target cosmid (nt 186748 to 189836 and nt 189836 to 191947) was transferred to the I-SceI site of pST76K-SR-T.
Mutagenesis of cosmid B47 was performed by a two-step replacement procedure in Escherichia coli as described previously (13, 42). E. coli bacteria that already contained cosmid clone B47 were transformed with the mutagenesis plasmid, and selection steps for formation of cointegrates were performed at 43°C on Luria-Bertani agar plates containing tetracycline and ampicillin at 10 μg/ml each. Mutant cosmids were selected at 30°C on fresh Luria-Bertani agar plates supplemented with ampicillin at 25 μg/ml, 5% sucrose, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg/ml).
Gaps around nt 132582 and nt 149399 to 149404 were created by subcloning of cosmid clones as follows. A C41 FspI fragment (nt 122542 to 132026), a C41 BsmI fragment (nt 134533 to 139709), an A2 RsrII fragment (nt 138944 to 148510), and a C21 PmeI/AvrII fragment (nt 151030 to 161414) were excised from parental cosmids and cloned into the pCR-blunt vector (Invitrogen, Groningen, The Netherlands). MCMV sequences with homology to the gene regions nt 129987 to 135128 and nt 146796 to 151914 were synthesized via PCR from wild-type and BAY 38-4766-resistant MCMV templates, respectively, in a volume of 50 μl including 25 ng of DNA, deoxynucleoside triphosphates at 500 μM each, primers at 30 pmol each, 10% dimethyl sulfoxide, and 5 U of NativePfu DNA polymerase (Stratagene) in a 30-cycle program (94°C for 10 s, 59°C for 20 s, and 72°C for 13 min). Sequences of PCR products were verified by sequencing analysis. Constructs were digested with restriction enzymes to release intact inserts from pCR-blunt vector backbones, and the digested constructs were combined with PmeI-digested cosmid clones C15, B41, D44, D17, B47-lacZ, and E39 in equimolar amounts. Following phenol-chloroform extraction and ethanol precipitation, 5 × 105 NIH 3T3 cells/6 wells seeded 1 day earlier were transfected with 4 μg of the DNA mixtures using a 2:1 ratio of TransFast reagent to DNA according to the manufacturer's instructions (Promega). Transfected cultures were refed every 3 to 4 days, and virus plaques were observed approximately 12 days posttransfection. Virus stocks were prepared from infected cells and stored at −140°C until characterization. Desired mutations in ORFs M89 exon II and M104 were verified by sequencing of PCR products amplified from cell-free virion supernatant (53). Virus stocks were quantified by measuring the number of reporter molecules expressed in infected NIH 3T3 cells. For this purpose, 96-well plates were prepared with 50 μl of serial twofold virus dilutions in quadruplicate, and 150 μl of EMEM/10 containing 3 × 104 cells was added to each well. After incubation of the plates for 5 days, β-galactosidase activity in cell extracts was determined by using the β-galactosidase enzyme assay system as instructed by the manufacturer (Promega). The virus inoculum representing the 50% tissue culture infective dose (TCID50; referring to an MOI of 0.002) was determined graphically by plotting relative absorption units against the logarithm of virus dilutions.
EC50 for each recombinant virus were determined at least three times with infectious progeny obtained from two independent virus reconstitutions by performing β-galactosidase-based antiviral assays as described below.
Generation of lacZ-tagged MCMV recombinants expressing mutant M56.
A modification of a previously described transient-transfection-plus-infection assay was used (16). A PCR fragment (nt 85995 to 92028) comprising the major part of the M56 coding region was synthesized from wild-type MCMV (MCMV-wt) and MCMV-rt templates and cloned into the pCR-blunt vector. The sequences of the PCR products were verified by sequencing analyses. The 6-kb inserts were released from the vector backbone by restriction enzyme digestion prior to transfection. NIH 3T3 cells (5 × 105/6-well plate) were transfected with 4 μg of phenol-chloroform-extracted and ethanol-precipitated DNA using the TransFast reagent as described above. Immediately posttransfection, cells were washed with 5 ml of EMEM and infected with either lacZ-tagged MCMV with wild-type M89 and M104 (MCMV-M89/M104-wt), MCMV with mutant M89 (MCMV-M89-mt), MCMV-M104-mt, or MCMV-M89/M104-mt at 0.1 PFU per cell for 1 h at 37°C. Cells were washed twice with EMEM and incubated with 3 ml of EMEM/10. Maximum cytopathic effect was usually observed 2 days p.i. Heterogeneous virus populations were harvested from supernatants of cells at day 3 p.i., clarified by centrifugation (1,000 × g, 10 min, 4°C), and stored at −140°C. For selection of recombinant virus progeny expressing mutant M56, NIH 3T3 cells were grown to confluency in 10-mm dishes and infected with 10,000 to 60,000 infectious virions (MOI, 0.001 to 0.006). Subsequently, cultures were overlaid with EMEM/10–0.5% methylcellulose containing BAY 38-4766 at an EC100 of 0.4 μM for MCMV-M89/M104-wt and MCMV-M104-mt and an EC100 of 4 μM for MCMV-M89-mt and MCMV-M89/M104-mt. In parallel, NIH 3T3 cells were infected with progeny harvested from cells transfected with the wild-type M56 DNA fragment. Selection was performed for 7 days, with one medium change after 4 days. Control dishes were observed for the absence of plaques. For each virus mutant, two individual plaques were isolated from the cell monolayer by trypsin treatment, amplified on fresh monolayers in the presence of drug, and further plaque purified twice in the absence of drug. The desired mutations in ORFs M56, M89 exon II, and M104 were verified by sequencing of PCR products amplified from virus stocks as described above. Drug susceptibilities of M56 mutants were determined by β-galactosidase-based antiviral assays (see below) with progenies from 2 × 2 plaques selected from independent transient-transfection-plus-infection assays.
Drug susceptibility assays.
We used both conventional plaque reduction assays and β-galactosidase enzyme assays to determine drug susceptibility profiles of viruses. Plaque reduction assays have been described elsewhere (16). HCMV-infected cell cultures were stained after 10 days, with an overlay change after 5 days, and MCMV assay cultures were stained after 5 days, using neutral red dye or Giemsa's solution, respectively. Drug concentrations producing a 50% reduction in plaque formation (EC50) were determined from three assays using Sensovir '98 software (Tobias Haller, Institute of Virology, University of Ulm, Ulm, Germany). The sensitivities of lacZ-tagged MCMV recombinants were determined by β-galactosidase enzyme assays. Fifty microliters of serial twofold drug dilutions or EMEM without inhibitor was added to wells of a 96-well plate. Quadruplicate wells were maintained for each drug concentration and for the untreated virus control. Following addition of a 50-μl viral inoculum, corresponding to the TCID50, to each well (MOI, 0.002), NIH 3T3 cells were trypsinized and diluted to a concentration of 3 × 105/ml. One hundred microliters of the cell suspension was dispensed to each well, and the plates were incubated for 5 days at 37°C. β-Galactosidase activity was assayed, and EC50 were determined from semilogarithmic plots as described above.
RESULTS
Inhibitory effect of BAY 38-4766 compared to those of reference drugs.
BAY 38-4766 (Fig. 1) exhibits potent anti-HCMV activity in vitro and in vivo, with an attractive selectivity index up to 300 μM for human cells (P. Eckenberg, J. Reefschlaeger, W. Bender, S. Goldmann, M. Haerter, S. Hallenberger, J. Trappe, and O. Weber, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 940, p. 321, 1999; J. Reefschlaeger, W. Bender, O. Weber, S. Hallenberger, I. Buerger, P. Eckenberg, S. Goldmann, M. Haerter, A. Paessens, and J. Trappe, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 942, p. 321, 1999). It also exhibits antiviral activity against various monkey CMV strains (data not shown). The most pronounced inhibitory effects have been found against rodent CMV strains, with a >10-fold-decreased EC50 for MCMV. Resistant CMV strains were selected in vitro by growth in the presence of increasing concentrations of BAY 38-4766. Resistance indices (RI) determined by plaque reduction assays revealed that HCMV He-rt and MCMV Smith-rt, both resistant to BAY 38-4766, exhibited drug susceptibilities comparable to those of the respective wild-type strains to all CMV inhibitors tested: the nucleoside drugs GCV, CDV, and LBV, the pyrophosphate analogue foscarnet, and the benzimidazoles benzimidavir and BDCRB (Table 1). Similar results were obtained with HCMV AD169 resistant to BAY 35-5014 (data not shown). Cross-resistance was demonstrated only for members of the nonnucleoside compound class, including BAY 35-5014 and BAY 36-8888 (data not shown). These data suggest that BAY 38-4766 exerts its inhibitory effect within the replicative cycle of CMV, in distinction from all reference drugs tested. In contrast to the highly resistant MCMV-rt (RI = 566) which was selected over multiple passages for only 2.5 months, the He-rt isolate was selected for more than 6 months and reached an RI of only 30.
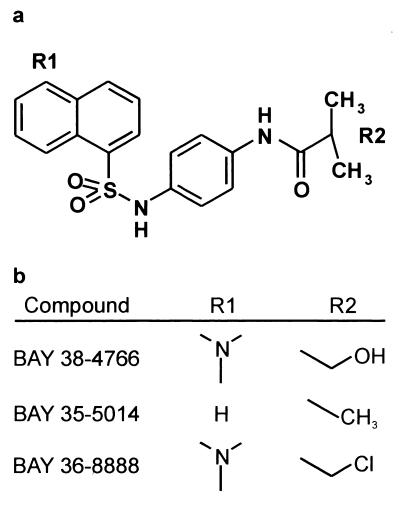
FIG. 1.
Structure of BAY 38-4766 and related compounds.
TABLE 1.
Antiviral activities of BAY 38-4766 and reference drugs against HCMV He and MCMV Smitha
| Drug | Activity against HCMV
|
Activity against MCMV
|
||||
|---|---|---|---|---|---|---|
| EC50 (μM)b ± SD
|
RIc | EC50 (μM)b ± SD
|
RIc | |||
| He-wt | He-rt | Smith-wt | Smith-rt | |||
| BAY 38-4766 | 1.21 ± 0.08 | 36.32 ± 0.39 | 30 | 0.05 ± 0.02 | 28.3 ± 1.93 | 566 |
| GCV | 6.91 ± 0.91 | 2.93 ± 0.31 | 0.4 | 27.8 ± 11.3 | 27.5 ± 2.5 | 1 |
| Foscarnet | 94.77 ± 9.38 | 94.01 ± 9.98 | 1 | 195 ± 5 | 230 ± 50 | 1.2 |
| CDV | 0.81 ± 0.39 | 0.6 ± 0.14 | 0.7 | 0.38 ± 0.1 | 0.31 ± 0.12 | 0.8 |
| LBV | 6.24 ± 0.7 | 3.36 ± 0.42 | 0.5 | 1.9 ± 0.37 | 2.45 ± 1.45 | 1.3 |
| Benzimidavir | 0.33 ± 0.07 | 0.22 ± 0.04 | 0.7 | 28.5d | 30d | 1.1 |
| BDCRB | 1.22 ± 0.27 | 2.03 ± 0.85 | 1.7 | 31d | 34d | 1.1 |
Determined by plaque reduction assay.
Average values determined from three assays in duplicate with each drug.
Calculated as the EC50 of the resistant strain divided by the EC50 of the wild-type strain.
Average values determined from one assay in duplicate because benzimidazole ribonucleosides are not active against MCMV.
Viral transcription and protein synthesis in the presence of BAY 38-4766.
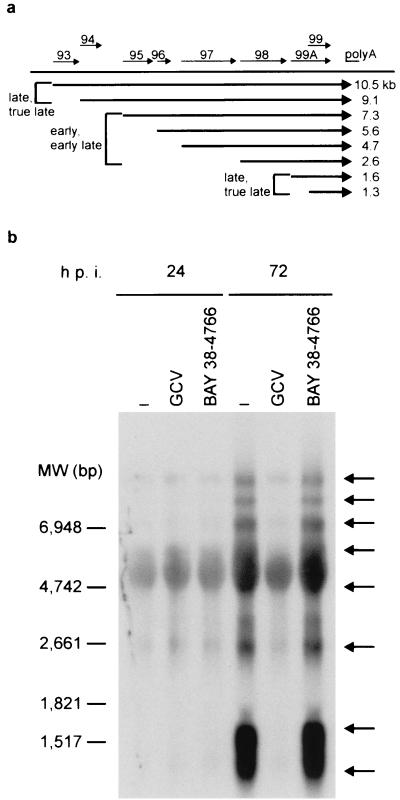
To investigate the time point of the inhibitory effect of BAY 38-4766 during the replicative cycle of HCMV, time-of-addition experiments under one-step conditions were performed. After addition of BAY 38-4766 at 0, 6, 12, or 24 h p.i., no infectious virus could be recovered from infected host cells. Addition of BAY 38-4766 at 36, 48, 60, or 72 h p.i. allowed the production of increasing amounts of progeny virus, as determined by retitration (data not shown). Thus, the time frame of maximal antiviral activity was found to be between 0 and <36 h p.i., indicating that viral DNA synthesis (12 to 16 h p.i.) is not affected. Northern blot analysis of late transcription confirmed these results (Fig. 2). A family of 3′-coterminal transcripts containing coding sequences for HCMV ORFs UL93 through UL99 is transcribed with early/early-late and late/true-late kinetics (55). Consistent with the previous observations, the transcriptional patterns of UL99 at 72 h p.i. in the presence and absence of our compound were identical., while late/true-late transcripts with sizes of 10.5, 9.1, 1.6, and 1.3 kb were absent from HCMV-infected cells grown in the presence of GCV. These findings strongly suggest that BAY 38-4766 does not inhibit viral DNA replication but that antiviral activity extends to late gene expression. De novo synthesis of viral mRNAs (TRL4, UL7, US9, US10, US11, and UL18) coding for different β and γ genes was not affected by BAY 38-4766, demonstrating that viral mRNA synthesis is not inhibited per se (data not shown). In addition, metabolic labeling of released virions demonstrated that, in contrast to the effects of GCV, HCMV-infected cells treated with and without our inhibitor produced abundant virus particles with nearly identical protein patterns (data not shown). Thus, neither overall transcription nor translation is affected by BAY 38-4766.
FIG. 2.
Northern blot analysis of UL99 transcription pattern. (a) Map of a family of 3′-coterminal transcripts containing ORFs UL93 to UL99 of HCMV. Synthesis of UL93 (10.5 kb), UL94 (9.1 kb), UL99A (1.6 kb), and UL99 (1.3 kb) transcripts proceeds during the late/true-late stage of the replicative cycle, whereas UL95, UL96, UL97, and UL98 are transcribed with early/early-late kinetics, resulting in 7.3-, 5.6-, 4.7-, and 2.6-kb mRNAs, respectively. (b) HELF were infected with HCMV AD169 and maintained either without inhibitor (−) or in the presence of the EC100 of BAY 36-8888 (0.67 μM) or of GCV (50 μM). At 24 and 72 h p.i., total RNA was harvested from infected cells and size fractionated under denaturing conditions using formaldehyde. Following transfer of RNA to nylon membranes, virus-specific transcripts (indicated by arrows) were visualized using a DIG-labeled, full-length antisense UL99 RNA probe and subsequent detection of luminescence. MW, molecular weight.
Ultrastructural investigation of infected cells.
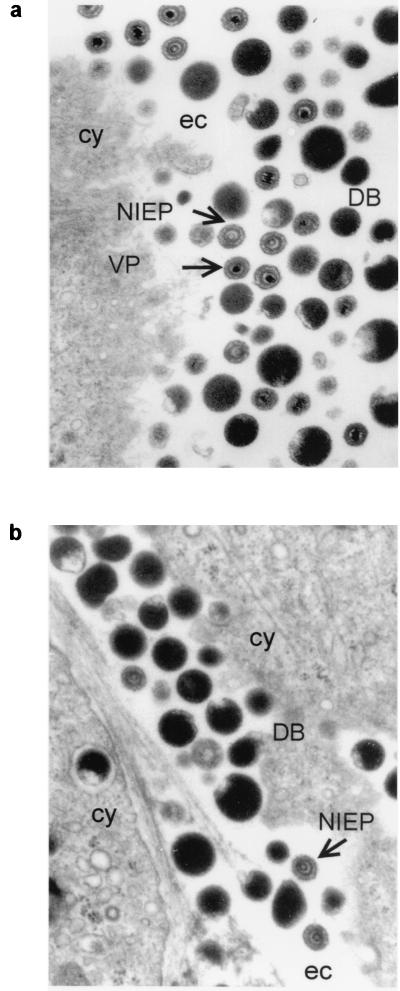
Ultrathin sections of HS68 cells infected with HCMV were examined by electron microscopy. Cells grown without inhibitor released three types of virus particles from the cell surface: mature infectious viral particles (VPs), noninfectious enveloped particles (NIEPs), and dense bodies (DBs) (Fig. 3a). VPs characteristically contain inner electron-dense DNA cores, whereas NIEPs contain electron-translucent cores due to the lack of DNA. DBs lack a nucleocapsid and are composed of an envelope containing glycoproteins and abundant tegument protein pp65 (1). In contrast to untreated cells, cells treated with BAY 36-8888 at an EC100 of 0.67 μM produced large numbers of DBs and NIEPs (Fig. 3b). VPs were not observed within these cells or at the cell surface. HCMV-infected cells treated with GCV as a control showed none of the stages in virion morphogenesis or DBs (data not shown).
FIG. 3.
Electron micrographs of AD169-infected HS68 cells incubated without inhibitor (a) or with the EC100 of BAY 36-8888 (0.67 μM) (b). After 3 to 4 days, cells were fixed in glutaraldehyde and prepared for electron microscopy. cy, cytoplasmic; ec, extracellular space.
Inhibition of viral DNA maturation.
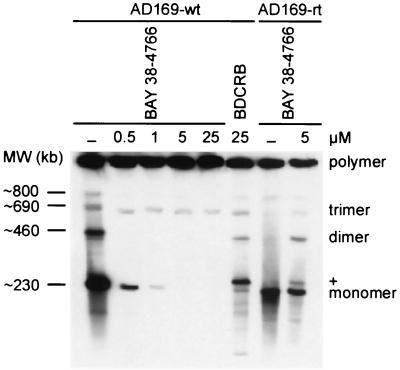
Because neither viral DNA synthesis nor transcription and translation were affected by BAY 38-4766, essential maturation steps following de novo DNA synthesis were analyzed within the replicative cycle of CMV. HELF infected with both AD169-wt and AD169-rt were maintained in the presence of selected drug concentrations. Intact nuclear high-molecular-weight DNA was separated by pulsed-field gel electrophoresis and analyzed by Southern blotting (Fig. 4). Processing of wild-type replicative concatemers to monomeric genome lengths decreased with increasing dosages of BAY 38-4766. Interestingly, the monomer+ form, also observed by Underwood et al. (53), was produced in the presence of 25 μM BDCRB but not in the presence of the EC100 of our compound. As expected, high-molecular-weight DNA of AD169-rt was almost properly processed in the presence of 5 μM BAY 38-4766, whereas negligible amounts of the monomer+ form were obtained with this virus isolate. To further explore the maturation of viral genome termini, a functional viral DNA cleavage assay was used (Fig. 5). Newly synthesized concatemeric MCMV DNA, which has been cleaved to unit length molecules, exhibits free termini that carry 1.9-kb BglII fragments on either side. Thus, the presence of one of these fragments shows that cleavage of concatemeric molecules to genome lengths has occurred. Functional processing of viral concatemers was visualized by Southern hybridization using a DIG-labeled terminal HindIII N subfragment probe which detects cleaved 1.9-kb BglII fragments, as well as 3.8-kb BglII fragments, representing genomic transitions of unprocessed viral DNA (Fig. 5a). In contrast to GCV, BAY 38-4766 did not inhibit de novo DNA synthesis of MCMV but decreased the amount of processed wild-type DNA in a dose-dependent manner (Fig. 5b). MCMV-rt, however, was not sensitive to inhibition by BAY 38-4766, as evidenced by cleavage of concatemeric DNA intermediates in the presence of the compound (Fig. 5c). In conclusion, BAY 38-4766 inhibits replication of CMV by interfering with the processing of concatemeric viral DNA.
FIG. 4.
Effect of BAY 38-4766 on concatemer processing of HCMV DNA. HELF were infected with AD169-wt and AD169-rt at an MOI of 1 and were cultivated in the absence (−) or presence of selected inhibitor concentrations. At 3 to 4 days p.i., cells were embedded in agarose plugs and digested with proteinase K. After electrophoretic separation, genomic DNA was subjected to Southern blot analysis using a DIG-labeled DNA probe synthesized with HindIII-digested AD169 DNA template fragments. MW, molecular weights derived from concatemeric phage lambda genomic DNA; polymer, wild-type replicative concatemers; monomer, monomeric genome lengths; +, monomer+ form of HCMV DNA (270 kb).
FIG. 5.
Functional viral DNA cleavage of MCMV-wt and MCMV-rt grown with increasing concentrations of BAY 38–4766. At 3 to 4 days p.i., MCMV-infected NIH 3T3 cells were lysed by three freeze-thaw cycles, and nuclear high-molecular-weight protein-DNA-complexes comprising viral DNA were sedimented by differential centrifugation followed by a sucrose step gradient. Subsequently, DNA was extracted and quantified by dot blot hybridization using a DIG-labeled MCMV HindIII M fragment. Equal amounts of viral DNA were digested with BglII, size fractionated on an agarose gel, and analyzed by Southern blotting. Extracellular viral DNA served as a control for mature linear virus genomes. (a) Model of rolling circle and location of BglII restriction sites at genomic transitions. Following restriction digestion, concatemeric viral DNA yields a 3.8-kb BglII fragment, whereas previous processing at the genome terminus results in a shortened 1.9-kb fragment. Both fragments were visualized by using a DIG-labeled DNA probe of the terminal HindIII N subfragment (hatched rectangle; genome position, nt 543 to 802). (b and c) LumiImager scans of wild-type (b) and resistant (c) MCMV. Selected drug concentrations and BglII fragments are indicated. ec., extracellular DNA extracted from released virions, representing the processed monomeric form of MCMV DNA; M, molecular size standard (phage lambda DNA cleaved with EcoRI and HindIII).
Genetic mapping of resistance to BAY 38-4766.
To further evaluate the concept that the novel nonnucleoside inhibitor class prevents viral DNA maturation, PCR products of genes essentially involved in viral DNA cleavage and packaging were amplified from resistant and parental CMV strains and directly sequenced. Confirming our previous observations, multiple mutations were detected in ORFs UL56, UL89, and UL104 of drug-resistant isolates (Table 2), whereas no sequence alterations were found in ORFs UL51, UL52, and UL77. In order to explore the significance of these mutations for the development of resistance, mutations identified in the murine homologues of UL56, UL89, and UL104 were transferred to the wild-type genome of MCMV. The BAY 38-4766-resistant MCMV isolate was the most resistant strain (RI = 566) and therefore is expected to contain mutations in all target proteins potentially inhibited by our compound. The genome of MCMV Smith was cloned into cosmid vectors, and the original fragment containing the M89 and M104 genes was replaced by cosmid subfragments and PCR fragments of the resistant and wild-type strains (16), allowing for the combinatorial introduction of mutations identified in ORFs M89 exon II and M104 (Fig. 6). Cotransfection of 12 fragments into NIH 3T3 cells resulted in the generation of homogeneous populations of infectious recombinants, with plaques usually detectable within 12 days p.i. Additionally, we inserted a lacZ reporter gene into the HindIII J fragment of cosmid B47 by a two-step replacement strategy in E. coli (13, 42). Viral growth kinetics demonstrated that the introduction of lacZ had no deleterious effect on viral growth in cultured cells (data not shown). Transient-transfection-plus-infection-assays were used to introduce the mutations identified in the M56 ORF into genomes of reconstituted lacZ-tagged virus progeny. In order to simplify drug susceptibility assays for the MCMV mutants constructed, we have established an easy virus assay, directly comparable to the conventional plaque reduction assay, based on the quantification of β-galactosidase activity in extracts prepared from infected cells (data not shown). In total, seven mutants expressing various mutations in the M56, M89, and M104 genes were tested for resistance to BAY 38-4766 in β-galactosidase enzyme assays (Table 3). Drug susceptibilities clearly demonstrated that each of the mutant ORFs M89 exon II (M360I) and M56 (P202A I208N) individually conferred significant resistance to wild-type MCMV, as indicated by the RIs of 15.3 and 56.6, respectively. A recombinant expressing a combination of mutant M89 and mutant M56 exhibited a strong synergistic effect, resulting in a final drug resistance profile comparable to that of the highly resistant MCMV isolate selected over a period of 2.5 months in vitro. In contrast, the deletion in M104 (T609 and A610 deleted), alone or in context with mutant M89 and/or mutant M56, had no effect on the development of resistance.
TABLE 2.
Mutations identified within genomes of drug-resistant virus isolates
| ORF | Mutation(s) in:
|
||
|---|---|---|---|
| HCMV
|
MCMV Smith-rtb | ||
| AD169-rta | He-rtb | ||
| UL56 | A662V | P202A I208N | |
| UL89 exon I | F142T S143E T144A S237R | ||
| UL89 exon II | D358N | N353S Y386H | M360I |
| UL104 | F602I | ΔT609 ΔA610 | |
AD169 resistant to BAY 35-5014 was obtained by in vitro selection in the presence of increasing inhibitor concentrations.
Virus strain resistant to BAY 38-4766.
FIG. 6.
Generation of MCMV recombinants expressing mutant M89 and/or mutant M104 by homologous recombination of 12 overlapping DNA fragments. A detailed view of the regions around M89 exon II and M104 is given. Subfragments of cosmids from a library generated from wild-type DNA are named according to terminal restriction sites. Rectangles represent coding regions of M89 and M104, with orientations indicated by horizontal arrows and positions of mutations indicated by vertical arrows. Nucleotide numbers of fragment termini correspond to nucleotides of the MCMV Smith genome. Gaps created by cosmid subcloning were spanned by respective PCR fragments of resistant and wild-type MCMV which were cotransfected with the remaining DNA fragments, providing different combinations of M89 exon II and M104 sequences.
TABLE 3.
Resistance of lacZ-tagged MCMV recombinants to BAY 38-4766
| MCMV strain | EC50 (μM)a | RIb | SD (μM) |
|---|---|---|---|
| M89/M104-wtc | 0.03 | ±0.01 | |
| M104-mtd | 0.03 | 1 | ±0.01 |
| M89-mt | 0.46 | 15.3 | ±0.03 |
| M89/M104-mt | 0.59 | 19.8 | ±0.12 |
| M56-mt | 1.7 | 56.6 | ±0.28 |
| M56/M104-mt | 1.69 | 56.5 | ±0.47 |
| M56/M89-mt | 17.18 | 572.5 | ±0.85 |
| M56/M89/M104-mt | 18.35 | 611.8 | ±0.68 |
| rte | 28.3 | 566f | ±1.93 |
Average values determined from β-galactosidase-based assays.
Calculated as the EC50 of the mutant strain divided by the EC50 of the wild-type strain.
MCMV recombinant expressing wild-type proteins.
MCMV recombinant expressing mutant protein.
Highly resistant MCMV isolate selected in vitro in the presence of increasing concentrations of BAY 38-4766.
RI calculated from plaque reduction assay data.
DISCUSSION
BAY 38-4766 is a member of a novel nonnucleoside inhibitor class with attractive antiviral activity and selectivity against CMV (Eckenberg et al., 39th ICAAC; Reefschlaeger et al., 39th ICAAC) and a molecular mechanism of action distinct from those of established drugs currently used for HCMV therapy. BAY 38-4766 and structural analogues inhibit neither viral DNA synthesis nor viral transcription and translation. DBs and NIEPs lacking a DNA core accumulate within infected cells in the presence of the compound. These findings coincide with detection of unprocessed high-molecular-weight DNA and inhibition of functional cleavage at viral intergenomic transitions in inhibitor-treated cells, pointing to interference with viral DNA cleavage and packaging.
Herpesviruses share common features with double-stranded DNA bacteriophages with regard to DNA maturation events. Initiation of packaging of phage DNA involves a specific interaction with the prohead in a process mediated by a phage-encoded terminase protein (48). Two general principles for packaging of concatemeric DNA into a virus head have been proposed: The first suggests a site-specific packaging in which the cos sequence of lambdoid phages plays an important role in initiation and termination of packaging (44). The second principle implies poorly characterized headful packaging initiating at specific pac sites (e.g., phage T4), but with volume measurement of the prohead followed by a termination step at a random DNA sequence (10). In a similar way, herpesvirus DNA replication results in the formation of large head-to-tail DNA concatemers (6), and maturation into unit length molecules involves site-specific cleavage at pac motifs localized to the a sequence (51). Current evidence suggests that cleavage and packaging of viral DNA are linked processes (33). Since the formation and processing of DNA concatemers are highly specific for the replication of herpesviruses lacking a mammalian counterpart, inhibition of these steps offers an alternative selective principle for antiviral therapy with potential broad-spectrum activity (J. Reefschlaeger, W. Bender, S. Hallenberger, O. Weber, P. Eckenberg, S. Goldmann, M. Haerter, I. Buerger, J. Trappe, J. A. Herrington, D. Haebich, and H. Ruebsamen-Waigmann, submitted for publication). In 1998, Underwood et al. reported on BDCRB, a benzimidazole ribonucleoside which inhibits maturation of polygenomic concatemeric HCMV DNA to unit genome length (53). Moreover, Krosky and collaborators have shown that emergence of the 2,5,6-trichloro-1-β-d-ribofuranosyl benzimidazole (TCRB)-resistant HCMV strain Towne was synergistically caused by amino acid substitutions residing in UL89 exon II and UL56, indicating that these gene products are potential antiviral-drug targets (32). During our mechanistic studies investigating the mode of action of BAY 38-4766 and derivatives, we made the intriguing observation that our drugs exert a virus-specific intervention similar to that of benzimidazole ribonucleosides, although they belong to different chemical-structure classes. Not only is the maturation of viral DNA inhibited by BAY 38-4766, but also drug resistance maps to murine homologues of both ORF UL89 exon II and ORF UL56.
From HSV-1 mutants it is known that six ORFs (UL6, UL15, UL25, UL28, UL32, and UL33) are essentially involved in viral DNA maturation (2, 5, 35, 36, 41, 46, 52, 56). The functional roles of the respective HCMV cleavage and packaging homologues have not been identified in detail, except for UL56, the HCMV homologue of HSV UL28 (11), which possibly binds the pac motif and has specific nuclease activity (12). UL89, the HCMV homologue of HSV UL15, is highly conserved among the Herpesviridae, and its potential in vivo function has been deduced from significant amino acid sequence similarities between corresponding herpesvirus proteins and the gp17 large subunit of the bacteriophage T4 terminase complex (18). The fact that mutations in murine homologues of ORFs UL89 exon II and UL56 synergistically confer resistance to BAY 38-4766 is consistent with the proposed role of each of these proteins in viral DNA maturation and suggests an interaction between them. This conclusion is supported by colocalization studies with the HSV homologues of UL56 (UL28) and UL89 (UL15) demonstrating that UL28 is localized to the cytoplasm of transfected cells and enters the nucleus when coexpressed with UL15 (31).
HSV-1 null mutants defective in one of the six cleavage and packaging proteins, including the HSV homologues of UL89 and UL56, exhibit the same phenotype as that induced by BAY 38-4766 (52, 56). Therefore, we hypothesize that our compound either inhibits these gene products independently or interferes with formation of a putative complex. The codon alteration M360I of the murine UL89 exon II homologue, manifesting decreased drug sensitivity of MCMV, is located close to substitutions identified in UL89 exon II of HCMV He-rt and HCMV AD169-rt. Hence, this region of the UL89 gene product seems to play a predominant role in the development of CMV drug resistance. In contrast to ORF UL89 exon II, no mutations analogous to P202A and I208N have been found in ORF UL56 of the resistant HCMV isolates. These findings indicate that codon alterations in this region of the murine homologue of the UL56 gene product have arisen as a consequence of the stringent conditions applied to MCMV during in vitro selection, resulting in a highly resistant strain. Therefore, we propose for the mode of action of BAY 38-4766 that the gene product of UL89 is directly targeted and that mutations in UL56 compensate for restricted activities of UL89.
With respect to interference with UL89 and UL56 viral DNA cleavage and packaging proteins, BAY 38-4766 displays a mechanistic phenotype which is identical to that induced by benzimidazole ribonucleosides (32, 53). However, despite the fact that mutations actually conferring resistance to BAY 38-4766 and TCRB are clustered in the UL89 and UL56 gene products, the molecular mode of action of our nonnucleoside inhibitor is distinct from that of benzimidazole ribonuncleosides: First, HCMV strains resistant to BAY 38-4766 and a related analogue demonstrated no cross-resistance to BDCRB. Second, MCMV is very susceptible to BAY 38-4766, whereas BDCRB is not active against MCMV, suggesting that no target binding pocket exists for BDCRB. In addition, differences have been detected in the processing of viral concatemeric DNA to monomeric genome lengths. BAY 38-4766 added at the EC100 to cells infected with wild-type AD169 did not result in the production of the monomer+ form observed at the EC100 of BDCRB. Studies by Krosky et al. have shown that this apparently higher-molecular-weight DNA species first appeared at drug concentrations around the EC50 and increased with increasing drug concentrations (32). Interestingly, we detected only negligible amounts of the monomer+ form in cells infected with AD169-rt and treated with BAY 38-4766 at the EC100. Again, these data demonstrate the similar mechanistic phenotypes of the two drugs but substantiate our hypothesis that the two inhibitors act with different molecular mechanisms. Due to the fact that the monomer+ form migrates slightly more slowly than the monomer form, it is possible that it represents a maturation intermediate which has been processed incorrectly. Although the structure and identity of the monomer+ form are unknown, it might represent dead-end products resulting from inexact maturation events taking place preferentially in the presence of BDCRB.
Transfer of the murine UL104 mutant protein to the wild-type genome has shown that the deletion of T609 and A610 is not related to the resistance of MCMV to BAY 38-4766. However, the fact that a substitution in the UL104 carboxy portion is also found in HCMV AD169 resistant to a close structural analogue of BAY 38-4766 attracts further attention to this region. Although they have no effect on the resistance phenotype, these additional mutations may compensate for the conformational changes in mutant UL89 and UL56 DNA cleavage and packaging proteins. The precise role of UL104 in the cleavage and encapsidation of replicated viral DNA is not understood. In cells infected with mutants lacking the HSV homologue of UL104 (UL6), specific capsid association of the HSV homologue of UL89 (UL15) is not observed, indicating that UL6 is required for docking of the putative terminase complex (57).
Additional considerations with regard to the potential functions of HCMV UL56, UL89, and UL104 arise from the DNA packaging pathway of T4 bacteriophages. Encapsidation of concatemeric DNA into bacteriophage proheads results from an ATP-driven process mediated by a multicomponent protein-machine consisting of terminase protein, portal vertex dodecamer, and accessory proteins (48). Portal-bound terminase, composed of the gp17 large subunit and the gp16 small subunit, appears to be essential for DNA translocation and processing of the concatemer (43). The gp17 subunit exhibits nonspecific endonucleolytic activity (7, 8), as well as in vitro packaging activity (37, 48) and ATPase activity (38). The small subunit gp16 enhances gp17 activities (38) and specifically forms a nucleoprotein structure at the packaging initiation site that helps to direct the terminase large subunit initiating its activities (9). By analogy with bacteriophages, genetic mapping of resistance to BAY 38-4766 provides further indirect evidence for the hypothesis that UL89 and UL56 function as a two-subunit cytomegalovirusl terminase. Furthermore, UL104 could serve the function of a gp20 portal vertex equivalent in HCMV capsids.
To summarize, our newly discovered compound class points the way toward a switch in strategy for developing HCMV inhibitors, with the aim of achieving a quality different from that of established DNA polymerase inhibitors. Intervention with viral DNA maturation arrests the replicative cycle at the DNA cleavage and packaging step, leading to an accumulation of empty procapsids and unprocessed concatemeric DNA. The fact that the compound class described in this report addresses a DNA maturation step which does not occur in eukaryotic cells may facilitate the development of alternative highly specific and well-tolerated therapeutic strategies. In addition, members of this class can be used to improve understanding of the still uncharacterized process of HCMV DNA maturation.
ACKNOWLEDGMENTS
We thank Christian Dilk, Kirsten Geiger, Bettina Schneider, and Anja Stamm for excellent technical assistance. The shuttle plasmid pST76K-SR was a kind gift of M. Messerle (Max von Pettenkofer-Institut für Hygiene und Medizinische Mikrobiologie, Ludwig-Maximilians-Universität München, D-81377 Munich, Germany).
REFERENCES
- 1.Alford C A, Britt W J. Cytomegalovirus. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 227–255. [Google Scholar]
- 2.Al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 3.Arvin A M, Alford C A. Chronic intrauterine and perinatal infections. In: Galasso G J, Whitley R J, Merigan T C, editors. Antiviral agents and viral diseases of man. New York, N.Y: Raven Press; 1990. pp. 497–580. [Google Scholar]
- 4.Azad R F, Driver V B, Tanaka K, Crooke R M, Anderson K P. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porat T, Rixon F J. Replication of herpesvirus DNA. IV. Analysis of concatemers. Virology. 1979;94:61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya S P, Rao V B. A novel terminase activity associated with the DNA packaging protein gp17 of bacteriophage T4. Virology. 1993;196:34–44. doi: 10.1006/viro.1993.1452. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya S P, Rao V B. Structural analysis of DNA cleaved in vivo by bacteriophage T4 terminase. Gene. 1994;146:67–72. doi: 10.1016/0378-1119(94)90834-6. [DOI] [PubMed] [Google Scholar]
- 9.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 10.Black L W. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays. 1995;17:1025–1030. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- 11.Bogner E, Reschke M, Reis B, Mockenhaupt T, Radsak K. Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology. 1993;196:290–293. doi: 10.1006/viro.1993.1477. [DOI] [PubMed] [Google Scholar]
- 12.Bogner E, Radsak M, Stinski M. The gene product of human cytomegalovirus ORF UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72:2259–2264. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borst E-M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chee M S, Bankeir A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnel T, Hutchinson C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S P, Tomlinson P, Weston K M, Barrel B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 15.Chrisp P, Clissold S P. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 16.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crumpacker C S. Ganciclovir. Drug Ther. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 18.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 19.Ebeling A, Keil G M, Knust E, Koszinowski U H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983;47:421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erriksson B, Öberg B, Wahren B. Pyrophosphate analogs as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim Biophys Acta. 1982;696:115–123. doi: 10.1016/0167-4781(82)90018-5. [DOI] [PubMed] [Google Scholar]
- 21.Falagas M E, Paya C, Ruthazer R, Badley A, Patel R, Wiesner R, Griffith J, Freeman R, Rohrer R, Werner B G, Snydman D R. Significance of cytomegalovirus for long-term survival after orthotopic liver transplantation: a prospective derivation and validation cohort analysis. Transplantation. 1998;66:1020–1028. doi: 10.1097/00007890-199810270-00010. [DOI] [PubMed] [Google Scholar]
- 22.Field A K, Biron K K. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev. 1994;7:1–13. doi: 10.1128/cmr.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 24.Hallenberger S, Moulard M, Sordel M, Klenk H D, Garten W. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J Virol. 1997;71:1036–1045. doi: 10.1128/jvi.71.2.1036-1045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada K, Eizuru Y, Isashiki Y, Ihara S, Minamishima Y. Genetic analysis of a clinical isolate of human cytomegalovirus exhibiting resistance against both ganciclovir and cidofovir. Arch Virol. 1997;142:215–225. doi: 10.1007/s007050050072. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix M G, Salimans M M, van Boven C P, Bruggeman C A. High prevalence of latently present cytomegalovirus in arterial walls of patients suffering from grade III atherosclerosis. Am J Pathol. 1990;136:23–28. [PMC free article] [PubMed] [Google Scholar]
- 27.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 28.Ho H-T, Woods K L, Bronson J J, DeBoeck H, Martin J C, Hitchcock M J M. Intracellular metabolism of the antiherpes agent (S)-1-[3-hydroxy-2-(phosphonylmethoxy)propyl]cytosine. Mol Pharmacol. 1992;41:197–202. [PubMed] [Google Scholar]
- 29.Jabs D A. Ocular manifestations of HIV infection. Trans Am Ophthalmol Soc. 1995;93:623–683. [PMC free article] [PubMed] [Google Scholar]
- 30.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alteration of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koslowski K M, Shaver P R, Casey II J T, Wilson T, Yamanaka G, Sheaffer A K, Tenney D J, Pederson N E. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J Virol. 1999;73:1704–1707. doi: 10.1128/jvi.73.2.1704-1707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krosky P M, Underwood M R, Turk S R, Feng K W-H, Jain R K, Ptak R G, Westerman A C, Biron K K, Townsend L B, Drach J C. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol. 1998;72:4721–4728. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladin B F, Blankenship M L, Ben-Porat T. Replication of herpesvirus DNA. V. The maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J Virol. 1980;33:1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lalezari J P, Stagg R J, Jaffe H S, Hitchcock M J, Drew W L. A preclinical and clinical overview of the nucleotide-based antiviral agent cidofovir (HPMPC) Adv Exp Med Biol. 1996;394:105–115. doi: 10.1007/978-1-4757-9209-6_12. [DOI] [PubMed] [Google Scholar]
- 35.Lamberti C, Weller S K. The herpes simplex type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 36.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leffers G, Rao V B. A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J Mol Biol. 1996;258:839–850. doi: 10.1006/jmbi.1996.0291. [DOI] [PubMed] [Google Scholar]
- 38.Leffers G, Rao V B. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 39.Mar E, Chiou J, Cheng Y-C, Huang E. Inhibition of cellular DNA polymerase α and human cytomegalovirus-induced DNA polymerase by the triphosphates of 9-(2-hydroxyethoxymethyl)guanine and 9-(1,3-dihydroxy-2-propoxymethyl)guanine. J Virol. 1985;53:776–780. doi: 10.1128/jvi.53.3.776-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenzie R, Travis M W D, Dolan S A, Pittaluga S, Feuerstein I M, Shelhamer J, Yarchoan R, Masur H. The cause of death in patients with human immunodeficiency virus infection: a clinical and pathological study with emphasis on the role of pulmonary disease. Medicine. 1991;70:326–343. doi: 10.1097/00005792-199109000-00004. [DOI] [PubMed] [Google Scholar]
- 41.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 44.Murialdo H. Bacteriophage λ DNA maturation and DNA packaging. Annu Rev Biochem. 1991;60:125–133. doi: 10.1146/annurev.bi.60.070191.001013. [DOI] [PubMed] [Google Scholar]
- 45.Nieto F J, Adam E, Sorlie P, Farzadegan H, Melnick J L, Comstock G W, Szklo M. Cohort study of cytomegalovirus infection as a risk factor for carotid intimal-medial thickening, a measure of subclinical atherosclerosis. Circulation. 1996;94:922–927. doi: 10.1161/01.cir.94.5.922. [DOI] [PubMed] [Google Scholar]
- 46.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 47.Posfai G, Koob M D, Kirkpatrick H A, Blattner F R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao V B, Black L W. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sarasini A, Baldanti F, Furione M, Percivalle E, Brerra R, Barbi M, Gerna G. Double resistance to ganciclovir and foscarnet of four human cytomegalovirus strains recovered from AIDS patients. J Med Virol. 1995;47:237–244. doi: 10.1002/jmv.1890470309. [DOI] [PubMed] [Google Scholar]
- 51.Spaete R R, Mocarski E S. The α sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985;54:817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Underwood M R, Harvey R J, Stanat S C, Hemphill M L, Miller T, Drach J C, Townsend L B, Biron K K. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J Virol. 1998;72:717–725. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin's scientific legacy. Washington, D.C.: ASM Press; 1995. pp. 189–213. [Google Scholar]
- 55.Wing B A, Huang E S. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y F, Leon M B, Waclawiw M A, Popma J J, Yu Z X, Finkel T, Epstein S E. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med. 1994;335:624–630. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]