Abstract
SE translocation (SET) is a cancer‐promoting factor whose expression is upregulated in many cancers. High SET expression positively correlates with a poor cancer prognosis. SETBP1 (SET‐binding protein 1/SEB/MRD29), identified as SET‐binding protein, is the causative gene of Schinzel–Giedion syndrome, which is characterized by severe intellectual disability and a distorted facial appearance. Mutations in these genetic regions are also observed in some blood cancers, such as myelodysplastic syndromes, and are associated with a poor prognosis. However, the physiological role of SETBP1 and the molecular mechanisms by which the mutations lead to disease progression have not yet been fully elucidated. In this review, we will describe the current epidemiological data on SETBP1 mutations and shed light on the current knowledge about the SET‐dependent and ‐independent functions of SETBP1.
Keywords: cancer, neurological disease, PP2A, SET, SETBP1
SE translocation (SET) is a cancer‐promoting factor whose expression is upregulated in many cancers. SETBP1 (SET binding protein 1) is the causative gene for Schinzel‐Giedion syndrome, and mutations in the same region are also observed in various blood cancers. In this review, we will describe the current epidemiological data on SETBP1 mutations and shed light on the current knowledge about the SET‐dependent and ‐independent functions of SETBP1.
![]()
1. STRUCTURE OF SETBP1
SET is an oncoprotein—also known as I2PP2A or TAF1—and was identified as a SET‐CAN fusion protein in acute non‐lymphocytic leukemia, produced by the deletion of chromosome 9 (Adachi et al., 1994). SET is a multifunctional protein that acts as an inhibitor of protein phosphatase 2A (PP2A), a major serine/threonine protein phosphatase. Additionally, it also acts as an epigenetic modulator through its function as a histone chaperone and as an inhibitor of the histone acetyl‐transferase complex (INHAT) (Dacol et al., 2021). Furthermore, it is a tumor‐promoting factor, and increased SET expression has been reported in various types of cancers. This protein is composed of a coiled‐coil domain (CD) responsible for dimerization, an earmuff domain (ED), and an acidic region (AR) (Figure 1a) (Bayarkhangai et al., 2018).
FIGURE 1.
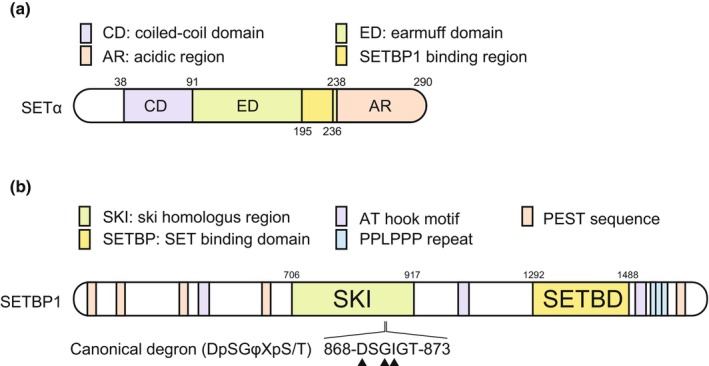
The structure of SET and SETBP1. (a) Schematic representation of SETα isoform. SETBP1 directly associates with 195–236 amino acid residues of SETα. (b) Schematic representation of SETBP1. The residues of canonical degron are located in the SKI region. Hotspot mutations observed in SGS patients are indicated by the arrowhead.
SETBP1 was identified as a SET‐binding protein using yeast two‐hybrid screening (Minakuchi et al., 2001). There are two transcriptional variants of SETBP1 that result in the synthesis of two isoforms, namely SETBP1a and SETBP1b. They are, approximately, 170 and 20 kDa, respectively. Exons 1–3 form a common region for both; SETBP1a is synthesized from six exons and SETBP1b from four exons. Previous studies have mainly focused on SETBP1a, and thus, the function of SETBP1b remains largely unknown (Coccaro et al., 2017). In this review, we will focus on SETBP1a and it will be referred to as SETBP1 hereafter.
SETBP1 is widely conserved ranging from insects to mammals; however, it is absent in yeast and nematodes. Human SETBP1 has a homologous region of the cancer‐promoting gene ski (SKI homologous region), six PEST sequences, three consecutive PPLPPP repeats, and three AT‐hook motifs (Figure 1b). Asp1292‐Lys1488 of SETBP1 is the SET‐binding domain (SETBD) that binds to Gln182‐Pro223 in the ED of SET (Minakuchi et al., 2001; Piazza et al., 2013).
SETBP1 proteins are ubiquitously expressed in the body and are most notably localized in the cell nuclei. The most well‐established function of SETBP1 is the suppression of PP2A activity via the stabilization of the SET proteins. SETBP1 binds directly to SET and accumulation of SETBP1 indirectly suppresses PP2A activity by protecting SET from cleavage by proteases and degradation via autophagy (Figure 2a) (Basurto‐Islas et al., 2013; Cristóbal et al., 2010; Fan et al., 2003; Kohyanagi et al., 2022; Piazza et al., 2013). SET also functions as an epigenetic modulator; although, this functional aspect has not received much attention with regard to SETBP1. This suggests that SETBP1 may also be involved in epigenetic regulation.
FIGURE 2.
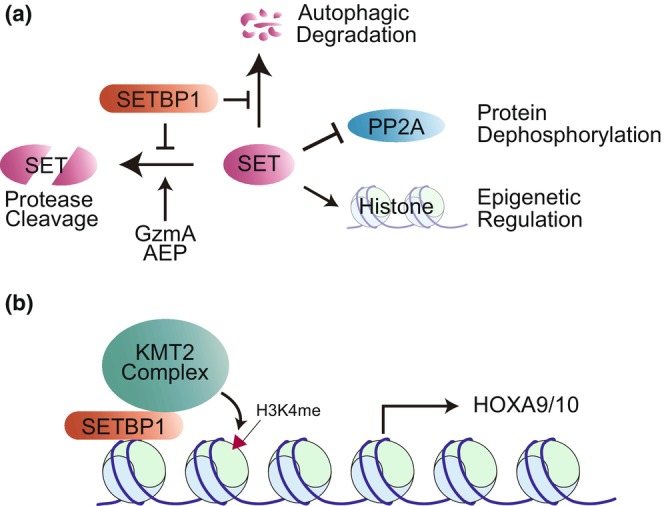
SET‐dependent and ‐independent functions of SETBP1. (a) SETBP1 stabilizes SET protein by blocking SET cleavage by proteases, such as granzyme A (GzmA) and asparaginyl endopeptidase (AEP), as well as autophagic degradation. As SET directly associates with PP2A and inhibits its phosphatase activity, SET accumulation by SETBP1 affects global phosphorylation signals. SET also functions as a histone chaperone. Therefore, SETBP1 may be involved in epigenetic regulation. (b) SETBP1 associates with AT‐rich promoter region and recruits the KMT2A complex. This causes methylation of Lys4 of histone H3 (H3K4me) and induces HOXA9/10 expression.
In contrast, the SET‐independent functions of SETBP1 have been thoroughly investigated in previous studies. In mouse myeloid progenitor cells, SETBP1 induces the expression of the homeobox proteins HOXA9 and HOXA10 and promotes their self‐renewal (Oakley et al., 2012; Sakaguchi et al., 2013). Additionally, SETBP1 binds to the AT‐rich promoter region of genomic DNA via its AT‐hook motif and recruits the KMT2A (MLL1) complex. This causes methylation of Lys4 in histone H3 and induces target gene expression (Figure 2b) (Nguyen et al., 2022; Piazza et al., 2018).
2. SETBP1 MUTATIONS IN NEUROLOGICAL DISEASES
Schinzel–Giedion syndrome (SGS), first described in 1978, is an autosomal dominant disorder characterized by severe intellectual disability, distorted facial features, skeletal abnormalities, and various congenital malformations of multiple organs (Schinzel & Giedion, 1978). It is a fatal disease that kills about half of the patients by 2 years and most by 10 years (Touge et al., 2001). De novo heterozygous mutations in the SETBP1 gene were identified in unrelated SGS patients with SGS. Furthermore, Sanger sequencing identified similar mutations in eight out of the nine SGS patients that were evaluated (Hoischen et al., 2010; Suphapeetiporn et al., 2011). These mutations were located within the SKI homologous region and were not found in parents of the patients or healthy individuals. Moreover, p.G870S was the most frequently found mutation with an incidence of approximately 27% (7/26 patients) (Leone et al., 2020). These findings suggest that SETBP1 is the major causative gene of this disease. Recently, SETBP1 mutations have also been observed in psychiatric disorders such as developmental epileptic encephalopathy, eating disorders, and autism (Table 1) (Alsubaie et al., 2020; Hu et al., 2022; Martínez‐Magaña et al., 2022).
TABLE 1.
SETBP1 mutation and diseases.
| Variants | Biochemical functions | Diseases | References |
|---|---|---|---|
| p.G15R | LoF | SGS | (Coe et al., 2014) |
| p.R143V | LoF | SGS | (Coe et al., 2014) |
| p.E244Dfs8 | ND | CML | (Romzova et al., 2021) |
| p.N272D | ND | GCT | (H. Wang et al., 2022) |
| p.L411G | LoF | SGS | (Coe et al., 2014) |
| p.W532 | LoF | SGS | (Coe et al., 2014) |
| p.R589* | LoF | ID | (Leonardi et al., 2020) |
| p.K592 | LoF | SGS | (Coe et al., 2014) |
| p.R625 | LoF | SGS | (Coe et al., 2014) |
| p.R626 | LoF | SGS | (Coe et al., 2014) |
| p.R627C | ND | AML | (Li et al., 2019) |
| p.E734Afs19 | LoF | ID | (Leonardi et al., 2020) |
| p.Q766R | ND | MDS | (Polprasert et al., 2022) |
| p.I822Y | LoF | SGS | (Coe et al., 2014) |
| p.E858K | ND | DEE, ccRCC, aCML | (Leonardi et al., 2020; Piazza et al., 2013) |
| p.E862K | ND | SGS, EP | (Acuna‐Hidalgo et al., 2017; Balciuniene et al., 2019) |
| p.S867G | ND | MPN | (Eder‐Azanza et al., 2019) |
| p.S867R | ND | SGS | (Acuna‐Hidalgo et al., 2017; Carvalho et al., 2015) |
| p.D868A | ND | SGS | (Hoischen et al., 2010) |
| p.D868N | PS | SGS, aCML, sAML, RAEB, CMML1, CMML2, CNL, JMML, PT, MDS | (Carvalho et al., 2015; Cui et al., 2014; Elliott et al., 2015; Gao et al., 2020; Hirao et al., 2022; Hoischen et al., 2010; Kwon et al., 2022; Li et al., 2019; Makishima et al., 2013; Montalban‐Bravo et al., 2021; Mori et al., 2022; Piazza et al., 2013; Polprasert et al., 2022; Volk et al., 2015; Wakamatsu et al., 2021; Yin et al., 2019) |
| p.D868Y | ND | CMML2 | (Makishima et al., 2013) |
| p.S869C | ND | SGS | (Landim et al., 2015) |
| p.S869G | ND | aCML | (Piazza et al., 2013) |
| p.S869N | ND | SGS | (Acuna‐Hidalgo et al., 2017) |
| p.S869R | ND | SGS | (Acuna‐Hidalgo et al., 2017) |
| p.G870C | ND | SGS | (López‐González et al., 2015) |
| p.G870D | ND | SGS, CNL, CL | (Acuna‐Hidalgo et al., 2017; Elliott et al., 2015; Hoischen et al., 2010; Mori et al., 2022) |
| p.G870N | ND | CNL | (Cui et al., 2014) |
| p.G870S | PS |
SGS, aCML, CMML, CNL, sAML, JMML PMF, PNH, MDS |
(Fontana et al., 2020; Herenger et al., 2015; Hirao et al., 2022; Hoischen et al., 2010; Kim et al., 2021; Ko et al., 2013; Kwon et al., 2022; Leone et al., 2020; Makishima et al., 2013; Montalban‐Bravo et al., 2021; Park et al., 2021; Piazza et al., 2013; Polprasert et al., 2022; Qian et al., 2021; Suphapeetiporn et al., 2011; Yun et al., 2020) |
| p.I871S | ND | SGS | (Sullivan et al., 2020; Takeuchi et al., 2015) |
| p.I871T | PS | SGS, aCML, CNL, sAML, FA | (Acuna‐Hidalgo et al., 2017; Hirao et al., 2022; Hoischen et al., 2010; Lestner et al., 2012; Makishima et al., 2013; Miyake et al., 2015; Pergande et al., 2020; Piazza et al., 2013; Yin et al., 2019) |
| p.T873I | ND | SGS | (Acuna‐Hidalgo et al., 2017) |
| p.D874N | ND | CNL | (Cui et al., 2014) |
| p.D880E | ND | sAML | (Makishima et al., 2013) |
| p.D880N | ND | CMML1 | (Makishima et al., 2013) |
| p.S893Y | ND | JMML | (Kim et al., 2021) |
| p.R972W | ND | BIA‐ALCL | (Fiore et al., 2020) |
| p.S1011 | LoF | SGS | (Coe et al., 2014) |
| p.S1076L | ND | ID, IAC | (Leonardi et al., 2020) |
| p.T1078M | ND | JMML | (Kim et al., 2021) |
| p.H1116R | ND | NSID | (Taşkıran et al., 2021) |
| p.A1193T | ND | AML | (Li et al., 2019) |
| p.V1377L | ND | BC | (Glentis et al., 2019) |
| p.E1466D | ND | AML | (Li et al., 2019) |
| p.P1563L | ND | AML | (Li et al., 2019) |
| Fusion with NOTCH3 | ND | MGM | (Khan et al., 2020) |
| Fusion with NPMI | ND | AML | (Martelli et al., 2021) |
Abbreviations: aCML, atypical chronic myelogenous leukemia; BC, breast cancer; BIA‐ALCL, breast implant‐associated anaplastic large cell lymphoma; ccRCC, clear cell renal carcinoma; CL, chronic leukocytosis; CMML1/2, chronic myelomonocytic leukemia 1/2; CNL, chronic neutrophilic leukemia; DEE, Developmental and epileptic encephalopathies; EP, epilepsy; FA, fetal akinesia; GCT, germ cell tumor; IAC, intestinal adenocarcinoma; ID, intellectual disability; ITAC, intestinal‐type adenocarcinoma; JMML, juvenile myelomonocytic leukemia; LoF, loss of function; MDS, myelodysplastic syndromes; MGM, meningioma; MPN, myeloproliferative neoplasms; ND, not determined; NSID, non‐specific intellectual disability; PMF, primary myelofibrosis; PNH, paroxysmal nocturnal hematuria; PS, protein stabilization; PT, persistent thrombocytopenia; RAEB, refractory anemia with ringed sideroblasts; sAML, secondary acute myeloid leukemia; SGS, Schinzel–Giedion syndrome.
SETBP1 mutations are involved in early‐stage neuronal degeneration and lead to SGS pathogenesis. It is known that the SKI homology region is a binding site for the ubiquitin ligase β‐TrCP1; these mutations reduce SETBP1 binding to β‐TrCP1. Furthermore, they inhibit SETBP1 degradation via the ubiquitin‐proteasome system (Piazza et al., 2013). Recently, iPS cells were established from SGS patients and used to demonstrate that SETBP1 mutations in neural progenitor cells accumulate DNA damage by upregulating SET expression and inhibiting p53 transcriptional activity (Banfi et al., 2021). SET‐mediated p53 inhibition is mediated by SET‐inhibitory effect on histone acetylation. SET is a component of the INHAT complex that masks lysine residues on histones and other proteins; this protects them from acetylation by p300/CBP and PCAF. It has also been reported to bind to non‐acetylated lysine residues at the C‐terminus of p53 and inhibit histone H3 acetylation. This results in the repression of p53 target gene transcription (Kim et al., 2012; Wang et al., 2016).
Furthermore, the loss‐of‐function mutations in SETBP1 have also been reported to be associated with intellectual disability, expressive language disorders, childhood aphasia, autism spectrum disorders, neurodevelopmental disorders, and developmental epileptic encephalopathy (Antonyan & Ernst, 2022; Coe et al., 2014; Filges et al., 2011; Jansen et al., 2021; Leonardi et al., 2020; Morgan et al., 2021; Rakhlin et al., 2020; Wong et al., 2022). These symptoms are also observed in SGS but are more severe. Therefore, appropriate SET regulation by SETBP1 may be important for normal neural development.
3. SETBP1 MUTATIONS IN CANCER
Elevated SETBP1 expression has been identified in many hematologic cancers. These include acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), myelodysplastic syndrome (MDS), and solid tumors, such as bladder cancer. Its expression is also associated with poor prognosis (Cristóbal et al., 2010; Eisfeld et al., 2020; Elena et al., 2016; Hwang et al., 2019; Jiang et al., 2020; Li et al., 2021; Lucas et al., 2018; Martín et al., 2020; Robinson et al., 2020; Tang et al., 2022). The major cause of elevated SETBP1 expression is mutations in and around the SKI region; however, there are also known cases that occur in the absence of SETBP1 mutations (discussed later).
SETBP1 mutations in cancer were first reported as NUP98‐SETBP1 fusion genes in patients with T‐cell acute lymphoblastic leukemia (Panagopoulos et al., 2007). Later, it was discovered that mutations in the SKI homologous region found in SGS patients were also present in cancer. Exome sequencing performed on atypical chronic myeloid leukemia (aCML) identified SETBP1 mutations in 19 of 78 cases, 92% of which were located in the SKI homologous region (Makishima et al., 2013). aCML patients with SETBP1 mutations showed signs of acute transformation in blood tests and had a shorter overall survival rate than the healthy controls (Meggendorfer et al., 2013). Specific mutations in Asp868, Ser869, Gly870, Ile871, and Asp880 within the SKI homology region have also been reported in approximately 15% of CMML, a type of MDS, and 17% of secondary AML that progressed from MDS (Han et al., 2022; Makishima et al., 2013; Nie et al., 2022). These were also associated with a shorter overall survival rate. These SETBP1 mutations have rarely been identified in non‐secondary acute myeloid leukemia (Badar et al., 2020; Makishima et al., 2013). A cohort study of 100 MDS patients showed that the SETBP1 mutation rate is higher in males; their prognosis is worse than that of females (Karantanos et al., 2021). Bone marrow fibrosis (BMF) is a known poor prognostic factor in MDS. SETBP1 mutations were identified at a higher rate in the BMF grade 3 group than in the grade 0–2 group (Melody et al., 2020). These findings suggest that mutations in the SKI region of SETBP1 are a potential cause for the transformation of MDS into leukemia (Makishima et al., 2013). SETBP1 mutations have also been identified in solid tumors including pancreatic neuroendocrine tumors, breast cancer, non‐small cell lung cancer, and gastric cancer (Ban et al., 2022; Coudray et al., 2018; Glentis et al., 2019; Zhang et al., 2019).
The co‐existence of certain genetic mutations along with SETBP1 mutations leads to a worse prognosis. Of the 368 MDS patients studied, 64 (17.4%) possessed mutations in the ASXL1 gene, encoding ASXL transcriptional regulator 1 (Inoue et al., 2015). Furthermore, 9.4% of the 64 patients with ASXL1 mutations also had SETBP1 mutations. In contrast, the SETBP1 mutation rate in ASXL1 mutation‐negative patients was only 0.7%. MDS with ASXL1 and SETBP1 co‐mutations had a higher rate of conversion to AML and a shorter overall survival rate compared to that of ASXL mutations alone (Inoue et al., 2015). In CMML, 36% (9/25) of patients with ASXL1 mutations carried SETBP1 mutations, while 81.8% (9/11) of patients with SETBP1 mutations were positive for ASXL1 mutations (Mason et al., 2016). These data suggest that ASXL1 mutation is followed by SETBP1 mutations, thus leading to disease progression. Mutation analysis of various myeloproliferative neoplasms classes also showed the prevalence of ASXL1 and SETBP1 co‐mutations in aCML (Palomo et al., 2020). Accumulating evidence has shown that ASXL1 mutations inhibit cancer cell differentiation and that SETBP1 mutations promote cell proliferation, thus leading to AML progression (Inoue et al., 2015; Makishima et al., 2013; Saika et al., 2018). It has also been reported that the presence of ASXL1 and SETBP1 mutations reduces histone H3 and H4 acetylation levels near the TGFβ target gene promoter region and inactivates the TGFβ pathway, thus inhibiting AML progression (Inoue et al., 2015; Makishima et al., 2013; Saika et al., 2018).
Association of SETBP1 with mutations other than ASXL1 has also been reported. SETBP1 mutations have been shown to be a poor prognostic factor in chronic neutrophilic leukemia (CNL) which is a rare myeloproliferative leukemia (Gao et al., 2022). Patients with colony‐stimulating factor 3 receptor (CSFR3) mutations, which account for more than half of the CNL patients, have a high frequency of SETBP1 mutations. The concomitant occurrence of these mutations exacerbates CNL by upregulating the expression of c‐Myc and its target genes (Anil et al., 2021; Carratt et al., 2022; Qian et al., 2021). Approximately, 85% of juvenile myelomonocytic leukemia (JMML) patients have JAK3 mutations. The SETBP1 p.G870N mutation co‐exists frequently with JAK3 mutations and is associated with a poor prognosis (Wakamatsu et al., 2021). In JMML, SETBP1 enhances NRAS gene expression, causing the activation of mitogen‐activated protein kinase signaling and repression of differentiation (Carratt et al., 2021). MDS with mutations in U2FA1 gene, which encodes U2 small nuclear RNA auxiliary factor 1, is associated with a poor prognosis and is prone to mutations in SETBP1 (H. Wang et al., 2020). The prognosis of MDS and other hematologic cancers patients with isochromosome 17q (i(17q)), a monosomy of the short arm and trisomy of the long arm of chromosome 17, is poor. SETBP1 mutation rates are higher in these cancers, and they frequently co‐exist with mutations in the serine and arginine‐rich splicing factor 2 (SRSF2) gene that regulates RNA splicing (Kanagal‐Shamanna et al., 2022).
4. INCREASED SETBP1 EXPRESSION IN CANCER
Mutations in the SKI region contribute to cancer malignancy by stabilizing the SETBP1 protein; on the other hand, increased SETBP1 protein not caused by mutations has also been reported. In AML, the t(12; 18)(p13; q12) translocation causes increased expression of SETBP1; the translocation is located near its cleavage point (Cristóbal et al., 2010). It is associated with a shorter overall survival rate. Owing to this translocation, elevated SETBP1 expression associated with decreased miR‐4319 expression has also been observed in patients who progressed from primary myelofibrosis to AML (Albano et al., 2012, p. 1). Additionally, SETBP1 expression was higher in younger patients with an adverse karyotype of AML (Lucas et al., 2018). According to The Cancer Genome Atlas Program (TCGA) database, patients with high SETBP1 expression in urothelial bladder carcinoma and stomach adenocarcinoma have a poor prognosis (Figure 3a,b).
FIGURE 3.
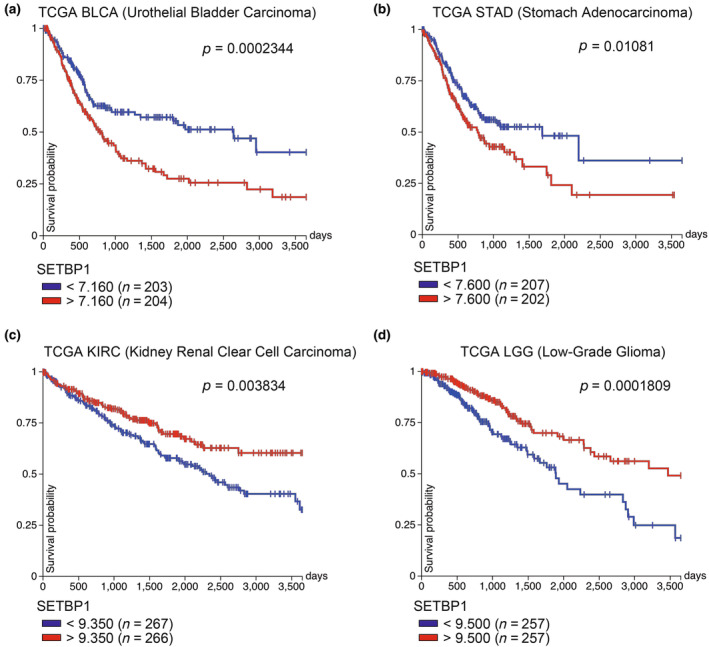
SETBP1 expression and prognosis of cancer patients. Kaplan–Meier curves for SETBP1 expression and 10‐year survival. TCGA dataset for (a) urothelial bladder carcinoma (BLCA), (b) stomach adenocarcinoma (STAD), (c) kidney renal clear cell carcinoma (KIRC), and (d) low‐grade glioma (LGG).
Internal tandem duplication caused by mutations in the FLT3 gene (FLT3‐ITD) leads to the constitutive activation of FMS‐like tyrosine kinase 3 (FLT3). FLT3‐ITD promotes AML development, in concert with oncogenic mutations and chimeric fusion genes, which are referred to as the “class‐defining” mutations. However, approximately 20% of FLT3‐ITD‐positive AML cases do not possess these class‐defining mutations. In some of these cancers, elevated SETBP1 expression appears to cooperate with FLT3‐ITD to trigger AML development (Pacharne et al., 2021).
On the other hand, lower SETBP1 expression also correlates with a poor prognosis. According to TCGA database, patients with low SETBP1 expression in kidney renal clear cell carcinoma and low‐grade glioma had a poor prognosis (Figure 3c, d). In non‐small cell lung cancers, decreased SETBP1 expression has been reported to cause epithelial‐mesenchymal transition and cell proliferation via ERK signaling (Li et al., 2020). It has also been reported that SETBP1 expression decreases in the stroma as breast cancer progresses from grades I to III (Uddin & Wang, 2022).
The molecular mechanisms that regulate SETBP1 transcription are not fully understood; however, some have recently been elucidated. Vascular endothelial zinc finger 1 (VEZF1/ZNF161), a transcription factor with a C2H2‐type zinc finger motif, binds to the SETBP1 promoter region and upregulates its expression in ovarian cancer (Figure 4) (Qiao et al., 2022). This is caused by the transcription of VEZF1 that is induced by a tripartite motif containing 29 (TRIM29). TRIM29 is upregulated in pancreatic, gastric, lung, bladder, and ovarian cancers; its expression levels are known to correlate with tumor size and grade.
FIGURE 4.
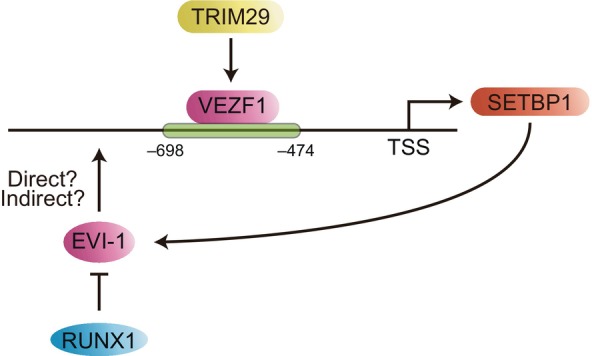
Regulation of SETBP1 transcription. Tripartite motif‐containing 29 (TRIM29) promotes the expression of the transcriptional factor VEZF1. VEZF directly binds to the SETBP1 promoter region and induces SETBP1 transcription. Transcriptional factor EVI‐1 directly or indirectly induces SETBP1 expression. The expression of EVI‐1 is positively and negatively regulated by SETBP1 and RUNX1, respectively.
Ecotropic virus integration site 1 protein homolog (EVI‐1) is an essential transcriptional factor for the proliferation of hematopoietic stem and leukemic cells. It is not clear whether EVI‐1 directly binds to the promoter region of SETBP1; however, SETBP1 expression is decreased by EVI‐1 deficiency (Goyama et al., 2008). RUNX1 is frequently mutated in human leukemia patients, and the mutated protein is involved in leukemogenesis through a dominant‐negative effect on the normal RUNX1. Analysis of the TCGA database revealed that SETBP1 expression was elevated in AML cells with RUNX1 mutations (Pacharne et al., 2021). EVI‐1 expression was also elevated in AML cells with RUNX1 mutations, suggesting that RUNX1 mutations may upregulate SETBP1 expression via EVI‐1 (Watanabe‐Okochi et al., 2008, p. 1). Furthermore, SETBP1 binds to the promoter region of EVI1 and promotes EVI‐1 expression (Piazza et al., 2018). Both EVI1 and SETBP1 upregulation promote bone marrow hematopoiesis (Ott et al., 2006). Cases of CNL with simultaneous EVI‐1 upregulation and SETBP1 mutations have also been reported (Altangerel et al., 2015). These observations suggest a dynamic interaction between EVI‐1 and SETBP1 in hematopoietic stem cells and leukemia cells.
Although the detailed molecular mechanisms are unclear, increased cell density promotes SETBP1 transcription in adherent cells (Kohyanagi et al., 2022). Elevated SETBP1 expression at high cell densities is a common phenomenon observed in many normal and cancer cell lines. Increased cell density causes acidification of the microenvironment, hypoxia, low nutritional status, and increased physical pressure. AMP‐activated protein kinase (AMPK) is a metabolic master switch that maintains cellular energy homeostasis. According to the Human Gene Database GeneCards, several AMPK‐downstream transcription factors, such as MEF2 and FOXO4, bind to the promoter region of SETBP1. Therefore, changes in nutritional status associated with increased cell density and decreased blood flow in cancer tissues may contribute to increased SETBP1 expression.
5. CONCLUSION AND PERSPECTIVE
Here, we described the impact of SETBP1 mutations in neurological diseases and cancer, in addition to their molecular mechanisms. Although numerous SETBP1 mutations have been reported in both diseases, the knowledge of why these mutations cause this pathology is limited. The function of SETBP1 is largely explained by the stabilization of SET and a concomitant decrease in PP2A activity. However, SET also functions as a histone modulator. SETBP1 also appears to have SET‐independent functions. Therefore, future research should not be limited only to the SETBP1/SET/PP2A axis.
While SETBP1 mutations have been identified in the SKI homology region in both SGS and cancer, differences exist between the two diseases. Mutations in Ile871 are more frequent in SGS than in cancer, and they have a relatively weak effect on SETBP1 protein cellular accumulation (Acuna‐Hidalgo et al., 2017). The Asp868 mutation, however, is more frequent in cancer and causes more pronounced SETBP1 accumulation. Additionally, SGS patients with Asp868 mutations are more susceptible to cancer than those with Ile871 mutations. These observations indicate that the threshold for causing cancer is higher than SGS. The phenotypic differences arising from the mutations within each residue require further analyses.
AUTHOR CONTRIBUTIONS
Takashi Ohama and Naoki Kohyanagi wrote this article.
FUNDING INFORMATION
This work was partially supported by JSPS KAKENHI (grant numbers 20H03151 and 22J23055).
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest regarding the publication of this paper.
Kohyanagi, N. , & Ohama, T. (2023). The impact of SETBP1 mutations in neurological diseases and cancer. Genes to Cells, 28(9), 629–641. 10.1111/gtc.13057
[The copyright line for this article was changed on 8 August 2023 after original online publication]
Communicated by: Eisuke Nishida
REFERENCES
- Acuna‐Hidalgo, R. , Deriziotis, P. , Steehouwer, M. , Gilissen, C. , Graham, S. A. , van Dam, S. , Hoover‐Fong, J. , Telegrafi, A. B. , Destree, A. , Smigiel, R. , Lambie, L. A. , Kayserili, H. , Altunoglu, U. , Lapi, E. , Uzielli, M. L. , Aracena, M. , Nur, B. G. , Mihci, E. , Moreira, L. M. A. , … van Bon, B. W. (2017). Overlapping SETBP1 gain‐of‐function mutations in Schinzel‐Giedion syndrome and hematologic malignancies. PLoS Genetics, 13(3), e1006683. 10.1371/journal.pgen.1006683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi, Y. , Pavlakis, G. N. , & Copeland, T. D. (1994). Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. The Journal of Biological Chemistry, 269(3), 2258–2262. [PubMed] [Google Scholar]
- Albano, F. , Anelli, L. , Zagaria, A. , Coccaro, N. , Casieri, P. , Minervini, A. , & Specchia, G. (2012). SETBP1 and miR_4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. Journal of Hematology & Oncology, 5, 48. 10.1186/1756-8722-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsubaie, L. M. , Alsuwat, H. S. , Almandil, N. B. , AlSulaiman, A. , AbdulAzeez, S. , & Borgio, J. F. (2020). Risk Y‐haplotypes and pathogenic variants of Arab‐ancestry boys with autism by an exome‐wide association study. Molecular Biology Reports, 47(10), 7623–7632. 10.1007/s11033-020-05832-6 [DOI] [PubMed] [Google Scholar]
- Altangerel, O. , Cao, S. , Meng, J. , Liu, P. , Haiyan, G. , Xu, Y. , & Zhao, M. (2015). Chronic neutrophilic leukemia with overexpression of EVI‐1, and concurrent CSF3R and SETBP1 mutations: A case report. Oncology Letters, 10(3), 1694–1700. 10.3892/ol.2015.3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil, V. , Gosal, H. , Kaur, H. , Chakwop Ngassa, H. , Elmenawi, K. A. , & Mohammed, L. (2021). Chronic neutrophilic leukemia: A literature review of the rare myeloproliferative pathology. Cureus, 13(6), e15433. 10.7759/cureus.15433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonyan, L. , & Ernst, C. (2022). Putative roles of SETBP1 dosage on the SET oncogene to affect brain development. Frontiers in Neuroscience, 16, 813430. 10.3389/fnins.2022.813430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badar, T. , Szabo, A. , Sallman, D. , Komrojki, R. , Lancet, J. , Padron, E. , Song, J. , & Hussaini, M. O. (2020). Interrogation of molecular profiles can help in differentiating between MDS and AML with MDS‐related changes. Leukemia & Lymphoma, 61(6), 1418–1427. 10.1080/10428194.2020.1719089 [DOI] [PubMed] [Google Scholar]
- Balciuniene, J. , DeChene, E. T. , Akgumus, G. , Romasko, E. J. , Cao, K. , Dubbs, H. A. , Mulchandani, S. , Spinner, N. B. , Conlin, L. K. , Marsh, E. D. , Goldberg, E. , Helbig, I. , Sarmady, M. , & Abou Tayoun, A. (2019). Use of a dynamic genetic testing approach for childhood‐onset epilepsy. JAMA Network Open, 2(4), e192129. 10.1001/jamanetworkopen.2019.2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, X. , Mo, S. , Lu, Z. , Jia, C. , Shao, H. , Yan, J. , Chang, X. , Mao, X. , Wu, Y. , Zhang, Y. , Fan, X. , Yu, S. , & Chen, J. (2022). Alternative lengthening of telomeres phenotype predicts progression risk in noninsulinomas in a Chinese cohort. Neuroendocrinology, 112(5), 510–522. 10.1159/000518413 [DOI] [PubMed] [Google Scholar]
- Banfi, F. , Rubio, A. , Zaghi, M. , Massimino, L. , Fagnocchi, G. , Bellini, E. , Luoni, M. , Cancellieri, C. , Bagliani, A. , Di Resta, C. , Maffezzini, C. , Ianielli, A. , Ferrari, M. , Piazza, R. , Mologni, L. , Broccoli, V. , & Sessa, A. (2021). SETBP1 accumulation induces P53 inhibition and genotoxic stress in neural progenitors underlying neurodegeneration in Schinzel‐Giedion syndrome. Nature Communications, 12(1), 4050. 10.1038/s41467-021-24391-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basurto‐Islas, G. , Grundke‐Iqbal, I. , Tung, Y. C. , Liu, F. , & Iqbal, K. (2013). Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease. The Journal of Biological Chemistry, 288(24), 17495–17507. 10.1074/jbc.M112.446070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarkhangai, B. , Noureldin, S. , Yu, L. , Zhao, N. , Gu, Y. , Xu, H. , & Guo, C. (2018). A comprehensive and perspective view of oncoprotein SET in cancer. Cancer Medicine, 7, 3084–3094. 10.1002/cam4.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carratt, S. A. , Braun, T. P. , Coblentz, C. , Schonrock, Z. , Callahan, R. , Smith, B. M. , Maloney, L. , Foley, A. C. , & Maxson, J. E. (2021). Mutant SETBP1 enhances NRAS‐driven MAPK pathway activation to promote aggressive leukemia. Leukemia, 35(12), 3594–3599. 10.1038/s41375-021-01278-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carratt, S. A. , Kong, G. L. , Curtiss, B. M. , Schonrock, Z. , Maloney, L. , Maniaci, B. N. , Blaylock, H. Z. , Baris, A. , Druker, B. J. , Braun, T. P. , & Maxson, J. E. (2022). Mutant‐SETBP1 activates transcription of Myc programs to accelerate CSF3R‐driven myeloproliferative neoplasms. Blood, 140(6), 644–658. 10.1182/blood.2021014777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, E. , Honjo, R. , Magalhães, M. , Yamamoto, G. , Rocha, K. , Naslavsky, M. , Zatz, M. , Passos‐Bueno, M. R. , Kim, C. , & Bertola, D. (2015). Schinzel‐Giedion syndrome in two Brazilian patients: Report of a novel mutation in SETBP1 and literature review of the clinical features. American Journal of Medical Genetics. Part A, 167A(5), 1039–1046. 10.1002/ajmg.a.36789 [DOI] [PubMed] [Google Scholar]
- Coccaro, N. , Tota, G. , Zagaria, A. , Anelli, L. , Specchia, G. , & Albano, F. (2017). SETBP1 dysregulation in congenital disorders and myeloid neoplasms. Oncotarget, 8(31), 51920–51935. 10.18632/oncotarget.17231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, B. P. , Witherspoon, K. , Rosenfeld, J. A. , van Bon, B. W. M. , Vulto‐van Silfhout, A. T. , Bosco, P. , Friend, K. L. , Baker, C. , Buono, S. , Vissers, L. E. L. M. , Schuurs‐Hoeijmakers, J. H. , Hoischen, A. , Pfundt, R. , Krumm, N. , Carvill, G. L. , Li, D. , Amaral, D. , Brown, N. , Lockhart, P. J. , … Eichler, E. E. (2014). Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature Genetics, 46(10), 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray, N. , Ocampo, P. S. , Sakellaropoulos, T. , Narula, N. , Snuderl, M. , Fenyö, D. , Moreira, A. L. , Razavian, N. , & Tsirigos, A. (2018). Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nature Medicine, 24(10), 1559–1567. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristóbal, I. , Blanco, F. J. , Garcia‐Orti, L. , Marcotegui, N. , Vicente, C. , Rifon, J. , Novo, F. J. , Bandres, E. , Calasanz, M. J. , Bernabeu, C. , & Odero, M. D. (2010). SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood, 115(3), 615–625. 10.1182/blood-2009-06-227363 [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Li, B. , Jiang, Q. , Xu, Z. , Qin, T. , Zhang, P. , Zhang, Y. , & Xiao, Z. (2014). CSF3R, ASXL1,SETBP1, JAK2 V617F and CALR mutations in chronic neutrophilic leukemia. Zhonghua Xue Ye Xue Za Zhi, 35(12), 1069–1073. 10.3760/cma.j.issn.0253-2727.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Dacol, E. C. , Wang, S. , Chen, Y. , & Lepique, A. P. (2021). The interaction of SET and protein phosphatase 2A as target for cancer therapy. Biochimica Et Biophysica Acta. Reviews on Cancer, 1876(1), 188578. 10.1016/j.bbcan.2021.188578 [DOI] [PubMed] [Google Scholar]
- Eder‐Azanza, L. , Hurtado, C. , Navarro‐Herrera, D. , Calavia, D. , Novo, F. J. , & Vizmanos, J. L. (2019). Analysis of genes encoding epigenetic regulators in myeloproliferative neoplasms: Coexistence of a novel SETBP1 mutation in a patient with a p.V617F JAK2 positive myelofibrosis. Molecular and Clinical Oncology, 10(6), 639–643. 10.3892/mco.2019.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld, A.‐K. , Kohlschmidt, J. , Mims, A. , Nicolet, D. , Walker, C. J. , Blachly, J. S. , Carroll, A. J. , Papaioannou, D. , Kolitz, J. E. , Powell, B. E. , Stone, R. M. , de la Chapelle, A. , Byrd, J. C. , Mrózek, K. , & Bloomfield, C. D. (2020). Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia, 34(12), 3215–3227. 10.1038/s41375-020-0872-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, C. , Gallì, A. , Such, E. , Meggendorfer, M. , Germing, U. , Rizzo, E. , Cervera, J. , Molteni, E. , Fasan, A. , Schuler, E. , Ambaglio, I. , Lopez‐Pavia, M. , Zibellini, S. , Kuendgen, A. , Travaglino, E. , Sancho‐Tello, R. , Catricalà, S. , Vicente, A. I. , Haferlach, T. , … Cazzola, M. (2016). Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood, 128(10), 1408–1417. 10.1182/blood-2016-05-714030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, M. A. , Pardanani, A. , Hanson, C. A. , Lasho, T. L. , Finke, C. M. , Belachew, A. A. , & Tefferi, A. (2015). ASXL1 mutations are frequent and prognostically detrimental in CSF3R‐mutated chronic neutrophilic leukemia. American Journal of Hematology, 90(7), 653–656. 10.1002/ajh.24031 [DOI] [PubMed] [Google Scholar]
- Fan, Z. , Beresford, P. J. , Oh, D. Y. , Zhang, D. , & Lieberman, J. (2003). Tumor suppressor NM23‐H1 is a granzyme A‐activated DNase during CTL‐mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell, 112(5), 659–672. 10.1016/S0092-8674(03)00150-8 [DOI] [PubMed] [Google Scholar]
- Filges, I. , Shimojima, K. , Okamoto, N. , Röthlisberger, B. , Weber, P. , Huber, A. R. , Nishizawa, T. , Datta, A. N. , Miny, P. , & Yamamoto, T. (2011). Reduced expression by SETBP1 haploinsufficiency causes developmental and expressive language delay indicating a phenotype distinct from Schinzel‐Giedion syndrome. Journal of Medical Genetics, 48(2), 117–122. 10.1136/jmg.2010.084582 [DOI] [PubMed] [Google Scholar]
- Fiore, D. , Cappelli, L. V. , Zumbo, P. , Phillips, J. M. , Liu, Z. , Cheng, S. , Yoffe, L. , Ghione, P. , Di Maggio, F. , Dogan, A. , Khodos, I. , de Stanchina, E. , Casano, J. , Kayembe, C. , Tam, W. , Betel, D. , Foa’, R. , Cerchietti, L. , Rabadan, R. , … Inghirami, G. (2020). A novel JAK1 mutant breast implant‐associated anaplastic large cell lymphoma patient‐derived xenograft fostering pre‐clinical discoveries. Cancers, 12(6), E1603. 10.3390/cancers12061603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, D. , Ramazzotti, D. , Aroldi, A. , Redaelli, S. , Magistroni, V. , Pirola, A. , Niro, A. , Massimino, L. , Mastini, C. , Brambilla, V. , Bombelli, S. , Bungaro, S. , Morotti, A. , Rea, D. , Stagno, F. , Martino, B. , Campiotti, L. , Caocci, G. , Usala, E. , … Gambacorti‐Passerini, C. (2020). Integrated genomic, functional, and prognostic characterization of atypical chronic myeloid leukemia. HemaSphere, 4(6), e497. 10.1097/HS9.0000000000000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J.‐P. , Zhai, L.‐J. , Gao, X.‐H. , & Min, F.‐L. (2022). Chronic neutrophilic leukemia complicated with monoclonal gammopathy of undetermined significance: A case report and literature review. Journal of Clinical Laboratory Analysis, 36(4), e24287. 10.1002/jcla.24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T. , Yu, C. , Xia, S. , Liang, T. , Gu, X. , & Liu, Z. (2020). A rare atypical chronic myeloid leukemia BCR‐ABL1 negative with concomitant JAK2 V617F and SETBP1 mutations: A case report and literature review. Therapeutic Advances in Hematology, 11, 2040620720927105. 10.1177/2040620720927105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glentis, S. , Dimopoulos, A. C. , Rouskas, K. , Ntritsos, G. , Evangelou, E. , Narod, S. A. , Mes‐Masson, A.‐M. , Foulkes, W. D. , Rivera, B. , Tonin, P. N. , Ragoussis, J. , & Dimas, A. S. (2019). Exome sequencing in BRCA1‐ and BRCA2‐negative Greek families identifies MDM1 and NBEAL1 as candidate risk genes for hereditary breast cancer. Frontiers in Genetics, 10, 1005. 10.3389/fgene.2019.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama, S. , Yamamoto, G. , Shimabe, M. , Sato, T. , Ichikawa, M. , Ogawa, S. , Chiba, S. , & Kurokawa, M. (2008). Evi‐1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell, 3(2), 207–220. 10.1016/j.stem.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Han, W. , Zhou, F. , Wang, Z. , Hua, H. , Qin, W. , Jia, Z. , Cai, X. , Chen, M. , Liu, J. , Chao, H. , & Lu, X. (2022). Mutational landscape of chronic myelomonocytic leukemia and its potential clinical significance. International Journal of Hematology, 115(1), 21–32. 10.1007/s12185-021-03210-x [DOI] [PubMed] [Google Scholar]
- Herenger, Y. , Stoetzel, C. , Schaefer, E. , Scheidecker, S. , Manière, M.‐C. , Pelletier, V. , Alembik, Y. , Christmann, D. , Clavert, J.‐M. , Terzic, J. , Fischbach, M. , De Saint Martin, A. , & Dollfus, H. (2015). Long term follow up of two independent patients with Schinzel‐Giedion carrying SETBP1 mutations. European Journal of Medical Genetics, 58(9), 479–487. 10.1016/j.ejmg.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Hirao, M. , Watanabe, K. , Tsukada, Y. , Kunieda, H. , Osada, M. , Yamazaki, K. , Denda, R. , Okamoto, S. , & Kikuchi, T. (2022). Rare evolution of CSF3R‐mutated chronic neutrophilic leukemia to t(4;12)(q12;p13) acute myeloid leukemia with SETBP1 mutation. Leukemia Research Reports, 17, 100311. 10.1016/j.lrr.2022.100311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen, A. , van Bon, B. W. M. , Gilissen, C. , Arts, P. , van Lier, B. , Steehouwer, M. , de Vries, P. , de Reuver, R. , Wieskamp, N. , Mortier, G. , Devriendt, K. , Amorim, M. Z. , Revencu, N. , Kidd, A. , Barbosa, M. , Turner, A. , Smith, J. , Oley, C. , Henderson, A. , … Veltman, J. A. (2010). De novo mutations of SETBP1 cause Schinzel‐Giedion syndrome. Nature Genetics, 42(6), 483–485. 10.1038/ng.581 [DOI] [PubMed] [Google Scholar]
- Hu, C. , Liu, D. , Luo, T. , Wang, Y. , & Liu, Z. (2022). Phenotypic spectrum and long‐term outcome of children with genetic early‐infantile‐onset developmental and epileptic encephalopathy. Epileptic Disorders: International Epilepsy Journal with Videotape, 24(2), 343–352. 10.1684/epd.2021.1394 [DOI] [PubMed] [Google Scholar]
- Hwang, S. M. , Kim, S.‐M. , Nam, Y. , Kim, J. , Kim, S. , Ahn, Y.‐O. , Park, Y. , Yoon, S.‐S. , Shin, S. , Kwon, S. , & Lee, D. S. (2019). Targeted sequencing aids in identifying clonality in chronic myelomonocytic leukemia. Leukemia Research, 84, 106190. 10.1016/j.leukres.2019.106190 [DOI] [PubMed] [Google Scholar]
- Inoue, D. , Kitaura, J. , Matsui, H. , Hou, H.‐A. , Chou, W.‐C. , Nagamachi, A. , Kawabata, K. C. , Togami, K. , Nagase, R. , Horikawa, S. , Saika, M. , Micol, J.‐B. , Hayashi, Y. , Harada, Y. , Harada, H. , Inaba, T. , Tien, H.‐F. , Abdel‐Wahab, O. , & Kitamura, T. (2015). SETBP1 mutations drive leukemic transformation in ASXL1‐mutated MDS. Leukemia, 29(4), 847–857. 10.1038/leu.2014.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, N. A. , Braden, R. O. , Srivastava, S. , Otness, E. F. , Lesca, G. , Rossi, M. , Nizon, M. , Bernier, R. A. , Quelin, C. , van Haeringen, A. , Kleefstra, T. , Wong, M. M. K. , Whalen, S. , Fisher, S. E. , Morgan, A. T. , & van Bon, B. W. (2021). Clinical delineation of SETBP1 haploinsufficiency disorder. European Journal of Human Genetics, 29(8), 1198–1205. 10.1038/s41431-021-00888-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Z. , Sun, X. , Wu, Z. , Alhatem, A. , Zheng, R. , Liu, D. , Wang, Y. , Kumar, D. , Xia, C. , You, B. , Wang, H. , Liu, C. , & Jiang, J.‐G. (2020). Cytogenetic and molecular landscape and its potential clinical significance in Hispanic CMML patients from Puerto Rico. Oncotarget, 11(47), 4411–4420. 10.18632/oncotarget.27824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagal‐Shamanna, R. , Orazi, A. , Hasserjian, R. P. , Arber, D. A. , Reichard, K. , Hsi, E. D. , Bagg, A. , Rogers, H. J. , Geyer, J. , Darbaniyan, F. , Do, K.‐A. , Devins, K. M. , Pozdnyakova, O. , George, T. I. , Cin, P. D. , Greipp, P. T. , Routbort, M. J. , Patel, K. , Garcia‐Manero, G. , … Bueso‐Ramos, C. (2022). Myelodysplastic/myeloproliferative neoplasms‐unclassifiable with isolated isochromosome 17q represents a distinct clinico‐biologic subset: A multi‐institutional collaborative study from the bone marrow pathology group. Modern Pathology, 35(4), 470–479. 10.1038/s41379-021-00961-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantanos, T. , Gondek, L. P. , Varadhan, R. , Moliterno, A. R. , DeZern, A. E. , Jones, R. J. , & Jain, T. (2021). Gender‐related differences in the outcomes and genomic landscape of patients with myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes. British Journal of Haematology, 193(6), 1142–1150. 10.1111/bjh.17534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. B. , Gadot, R. , Shetty, A. , Bayley, J. C. , Hadley, C. C. , Cardenas, M. F. , Jalali, A. , Harmanci, A. S. , Harmanci, A. O. , Wheeler, D. A. , Klisch, T. J. , & Patel, A. J. (2020). Identification of novel fusion transcripts in meningioma. Journal of Neuro‐Oncology, 149(2), 219–230. 10.1007/s11060-020-03599-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. S. , Lee, J. W. , Kang, D. , Yu, H. , Kim, Y. , Kang, H. , Lee, J.‐M. , Ahn, A. , Cho, B. , Kim, S. , Chung, N.‐G. , Kim, Y. , & Kim, M. (2021). Characteristics of RAS pathway mutations in juvenile myelomonocytic leukaemia: A single‐institution study from Korea. British Journal of Haematology, 195(5), 748–756. 10.1111/bjh.17861 [DOI] [PubMed] [Google Scholar]
- Kim, J.‐Y. , Lee, K.‐S. , Seol, J.‐E. , Yu, K. , Chakravarti, D. , & Seo, S.‐B. (2012). Inhibition of p53 acetylation by INHAT subunit SET/TAF‐Iβ represses p53 activity. Nucleic Acids Research, 40(1), 75–87. 10.1093/nar/gkr614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J. M. , Lim, B. C. , Kim, K. J. , Hwang, Y. S. , Ryu, H. W. , Lee, J. H. , Kim, J. S. , & Chae, J.‐H. (2013). Distinct neurological features in a patient with Schinzel‐Giedion syndrome caused by a recurrent SETBP1 mutation. Child's Nervous System, 29(4), 525–529. 10.1007/s00381-013-2047-2 [DOI] [PubMed] [Google Scholar]
- Kohyanagi, N. , Kitamura, N. , Tanaka, K. , Mizuno, T. , Fujiwara, N. , Ohama, T. , & Sato, K. (2022). The protein level of the tumor‐promoting factor SET is regulated by cell density. Journal of Biochemistry, mvab125, 295–303. 10.1093/jb/mvab125 [DOI] [PubMed] [Google Scholar]
- Kwon, A. , Ibrahim, I. , Le, T. , Jaso, J. M. , Weinberg, O. , Fuda, F. , & Chen, W. (2022). CSF3R T618I mutated chronic myelomonocytic leukemia: A proliferative subtype with a distinct mutational profile. Leukemia Research Reports, 17, 100323. 10.1016/j.lrr.2022.100323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landim, P. O. L. , Silva Junior, R. C. , Leitzke, L. , Lima, S. L. , Siqueira, H. H. , Galera, M. F. , Prates, M. T. , Dalbem, J. S. , Dos Reis, E. B. S. , & Campos, D. C. V. (2015). Refractory epilepsy and other neurological manifestations of Schinzel‐Gieidion syndrome. Journal of the Neurological Sciences, 357, e154. 10.1016/j.jns.2015.08.520 [DOI] [Google Scholar]
- Leonardi, E. , Bettella, E. , Pelizza, M. F. , Aspromonte, M. C. , Polli, R. , Boniver, C. , Sartori, S. , Milani, D. , & Murgia, A. (2020). Identification of SETBP1 mutations by gene panel sequencing in individuals with intellectual disability or with ‘developmental and epileptic encephalopathy’. Frontiers in Neurology, 11, 593446. 10.3389/fneur.2020.593446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone, M. P. , Palumbo, P. , Palumbo, O. , Di Muro, E. , Chetta, M. , Laforgia, N. , Resta, N. , Stella, A. , Castellana, S. , Mazza, T. , Castori, M. , Carella, M. , & Bukvic, N. (2020). The recurrent SETBP1 c.2608G > A, p.(Gly870Ser) variant in a patient with Schinzel‐Giedion syndrome: An illustrative case of the utility of whole exome sequencing in a critically ill neonate. Italian Journal of Pediatrics, 46(1), 74. 10.1186/s13052-020-00839-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestner, J. M. , Chong, W. K. , Offiiah, A. , Kefas, J. , & Vandersteen, A. M. (2012). Unusual neuroradiological features in Schinzel‐Giedion syndrome: A novel case. Clinical Dysmorphology, 21(3), 152–154. 10.1097/MCD.0b013e3283518f1e [DOI] [PubMed] [Google Scholar]
- Li, H.‐R. , Gao, J. , Jin, C. , Jiang, J.‐H. , & Ding, J.‐Y. (2020). Downregulation of SETBP1 promoted non‐small cell lung cancer progression by inducing cellular EMT and disordered immune status. American Journal of Translational Research, 12(2), 447–462. [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Cao, J. , Li, P. , Yao, Z. , Deng, R. , Ying, L. , & Tian, J. (2021). Construction of a novel mRNA‐signature prediction model for prognosis of bladder cancer based on a statistical analysis. BMC Cancer, 21(1), 858. 10.1186/s12885-021-08611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Lv, X. , Ge, X. , Yuan, D. , Ding, M. , Zhen, C. , Zhao, W. , Liu, X. , Wang, X. , Xu, H. , Li, Y. , & Wang, X. (2019). Mutational spectrum and associations with clinical features in patients with acute myeloid leukaemia based on next‐generation sequencing. Molecular Medicine Reports, 19(5), 4147–4158. 10.3892/mmr.2019.10081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐González, V. , Domingo‐Jiménez, M. R. , Burglen, L. , Ballesta‐Martínez, M. J. , Whalen, S. , Piñero‐Fernández, J. A. , & Guillén‐Navarro, E. (2015). Schinzel‐Giedion syndrome: A new mutation in SETBP1. Anales De Pediatria (Barcelona, Spain: 2003), 82(1), e12–e16. 10.1016/j.anpedi.2014.06.017 [DOI] [PubMed] [Google Scholar]
- Lucas, C. M. , Scott, L. J. , Carmell, N. , Holcroft, A. K. , Hills, R. K. , Burnett, A. K. , & Clark, R. E. (2018). CIP2A‐ and SETBP1‐mediated PP2A inhibition reveals AKT S473 phosphorylation to be a new biomarker in AML. Blood Advances, 2(9), 964–968. 10.1182/bloodadvances.2017013615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima, H. , Yoshida, K. , Nguyen, N. , Przychodzen, B. , Sanada, M. , Okuno, Y. , Ng, K. P. , Gudmundsson, K. O. , Vishwakarma, B. A. , Jerez, A. , Gomez‐Segui, I. , Takahashi, M. , Shiraishi, Y. , Nagata, Y. , Guinta, K. , Mori, H. , Sekeres, M. A. , Chiba, K. , Tanaka, H. , … Maciejewski, J. P. (2013). Somatic SETBP1 mutations in myeloid malignancies. Nature Genetics, 45(8), 942–946. 10.1038/ng.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli, M. P. , Rossi, R. , Venanzi, A. , Meggendorfer, M. , Perriello, V. M. , Martino, G. , Spinelli, O. , Ciurnelli, R. , Varasano, E. , Brunetti, L. , Ascani, S. , Quadalti, C. , Cardinali, V. , Mezzasoma, F. , Gionfriddo, I. , Milano, F. , Pacini, R. , Tabarrini, A. , Bigerna, B. , … Falini, B. (2021). Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML. Blood, 138(25), 2696–2701. 10.1182/blood.2021012732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín, I. , Navarro, B. , Serrano, A. , Villamón, E. , Calabuig, M. , Solano, C. , Chaves, F. J. , Yagüe, N. , Orts, M. , Amat, P. , Fuentes, A. , Seda, E. , García, F. , Hernández‐Boluda, J. C. , & Tormo, M. (2020). Impact of clinical features, cytogenetics, genetic mutations, and methylation dynamics of CDKN2B and DLC‐1 promoters on treatment response to azacitidine. Annals of Hematology, 99(3), 527–537. 10.1007/s00277-020-03932-8 [DOI] [PubMed] [Google Scholar]
- Martínez‐Magaña, J. J. , Hernandez, S. , Garcia, A. R. , Cardoso‐Barajas, V. , Sarmiento, E. , Camarena, B. , Caballero, A. , Gonzalez, L. , Villatoro‐Velazquez, J. A. , Medina‐Mora, M. E. , Bustos‐Gamiño, M. , Fleiz‐Bautista, C. , Tovilla‐Zarate, C. A. , Juárez‐Rojop, I. E. , Nicolini, H. , & Genis‐Mendoza, A. D. (2022). Genome‐wide analysis of disordered eating behavior in the Mexican population. Nutrients, 14(2), 394. 10.3390/nu14020394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, C. C. , Khorashad, J. S. , Tantravahi, S. K. , Kelley, T. W. , Zabriskie, M. S. , Yan, D. , Pomicter, A. D. , Reynolds, K. R. , Eiring, A. M. , Kronenberg, Z. , Sherman, R. L. , Tyner, J. W. , Dalley, B. K. , Dao, K.‐H. , Yandell, M. , Druker, B. J. , Gotlib, J. , O'Hare, T. , & Deininger, M. W. (2016). Age‐related mutations and chronic myelomonocytic leukemia. Leukemia, 30(4), 906–913. 10.1038/leu.2015.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggendorfer, M. , Bacher, U. , Alpermann, T. , Haferlach, C. , Kern, W. , Gambacorti‐Passerini, C. , Haferlach, T. , & Schnittger, S. (2013). SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia, 27(9), 1852–1860. 10.1038/leu.2013.133 [DOI] [PubMed] [Google Scholar]
- Melody, M. , Al Ali, N. , Zhang, L. , Ramadan, H. , Padron, E. , Sallman, D. , Sweet, K. , Lancet, J. , List, A. , Bennett, J. M. , & Komrokji, R. (2020). Decoding bone marrow fibrosis in myelodysplastic syndromes. Clinical Lymphoma, Myeloma & Leukemia, 20(5), 324–328. 10.1016/j.clml.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Minakuchi, M. , Kakazu, N. , Gorrin‐Rivas, M. J. , Abe, T. , Copeland, T. D. , Ueda, K. , & Adachi, Y. (2001). Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia‐associated protein SET. European Journal of Biochemistry, 268(5), 1340–1351. 10.1046/j.1432-1327.2001.02000.x [DOI] [PubMed] [Google Scholar]
- Miyake, F. , Kuroda, Y. , Naruto, T. , Ohashi, I. , Takano, K. , & Kurosawa, K. (2015). West syndrome in a patient with Schinzel‐Giedion syndrome. Journal of Child Neurology, 30(7), 932–936. 10.1177/0883073814541468 [DOI] [PubMed] [Google Scholar]
- Montalban‐Bravo, G. , Kanagal‐Shamanna, R. , Sasaki, K. , Masarova, L. , Naqvi, K. , Jabbour, E. , DiNardo, C. D. , Takahashi, K. , Konopleva, M. , Pemmaraju, N. , Kadia, T. M. , Ravandi, F. , Daver, N. , Borthakur, G. , Estrov, Z. , Khoury, J. D. , Loghavi, S. , Soltysiak, K. A. , Pierce, S. , … Garcia‐Manero, G. (2021). Clinicopathologic correlates and natural history of atypical chronic myeloid leukemia. Cancer, 127(17), 3113–3124. 10.1002/cncr.33622 [DOI] [PubMed] [Google Scholar]
- Morgan, A. , Braden, R. , Wong, M. M. K. , Colin, E. , Amor, D. , Liégeois, F. , Srivastava, S. , Vogel, A. , Bizaoui, V. , Ranguin, K. , Fisher, S. E. , & van Bon, B. W. (2021). Speech and language deficits are central to SETBP1 haploinsufficiency disorder. European Journal of Human Genetics: EJHG, 29(8), 1216–1225. 10.1038/s41431-021-00894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, N. , Ohwashi‐Miyazaki, M. , Yoshinaga, K. , Ogasawara, T. , Marshall, S. , Shiseki, M. , Sakura, H. , & Tanaka, J. (2022). Genetic alterations in patients with chronic leucocytosis and persistent thrombocytosis. Journal of Genetics, 101, 11. [PubMed] [Google Scholar]
- Nguyen, N. , Gudmundsson, K. O. , Soltis, A. R. , Oakley, K. , Roy, K. R. , Han, Y. , Gurnari, C. , Maciejewski, J. P. , Crouch, G. , Ernst, P. , Dalgard, C. L. , & Du, Y. (2022). Recruitment of MLL1 complex is essential for SETBP1 to induce myeloid transformation. IScience, 25(1), 103679. 10.1016/j.isci.2021.103679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Y. , Shao, L. , Zhang, H. , He, C. K. , Li, H. , Zou, J. , Chen, L. , Ji, H. , Tan, H. , Lin, Y. , & Ru, K. (2022). Mutational landscape of chronic myelomonocytic leukemia in Chinese patients. Experimental Hematology & Oncology, 11(1), 32. 10.1186/s40164-022-00284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, K. , Han, Y. , Vishwakarma, B. A. , Chu, S. , Bhatia, R. , Gudmundsson, K. O. , Keller, J. , Chen, X. , Vasko, V. , Jenkins, N. A. , Copeland, N. G. , & Du, Y. (2012). Setbp1 promotes the self‐renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood, 119(25), 6099–6108. 10.1182/blood-2011-10-388710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, M. G. , Schmidt, M. , Schwarzwaelder, K. , Stein, S. , Siler, U. , Koehl, U. , Glimm, H. , Kühlcke, K. , Schilz, A. , Kunkel, H. , Naundorf, S. , Brinkmann, A. , Deichmann, A. , Fischer, M. , Ball, C. , Pilz, I. , Dunbar, C. , Du, Y. , Jenkins, N. A. , … Grez, M. (2006). Correction of X‐linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1‐EVI1, PRDM16 or SETBP1. Nature Medicine, 12(4), 401–409. 10.1038/nm1393 [DOI] [PubMed] [Google Scholar]
- Pacharne, S. , Dovey, O. M. , Cooper, J. L. , Gu, M. , Friedrich, M. J. , Rajan, S. S. , Barenboim, M. , Collord, G. , Vijayabaskar, M. S. , Ponstingl, H. , De Braekeleer, E. , Bautista, R. , Mazan, M. , Rad, R. , Tzelepis, K. , Wright, P. , Gozdecka, M. , & Vassiliou, G. S. (2021). SETBP1 overexpression acts in the place of class‐defining mutations to drive FLT3‐ITD‐mutant AML. Blood Advances, 5(9), 2412–2425. 10.1182/bloodadvances.2020003443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo, L. , Meggendorfer, M. , Hutter, S. , Twardziok, S. , Ademà, V. , Fuhrmann, I. , Fuster‐Tormo, F. , Xicoy, B. , Zamora, L. , Acha, P. , Kerr, C. M. , Kern, W. , Maciejewski, J. P. , Solé, F. , Haferlach, C. , & Haferlach, T. (2020). Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood, 136(16), 1851–1862. 10.1182/blood.2019004229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos, I. , Kerndrup, G. , Carlsen, N. , Strömbeck, B. , Isaksson, M. , & Johansson, B. (2007). Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12). British Journal of Haematology, 136(2), 294–296. 10.1111/j.1365-2141.2006.06410.x [DOI] [PubMed] [Google Scholar]
- Park, S. , So, M.‐K. , Cho, M.‐S. , Kim, D.‐Y. , & Huh, J. (2021). Coexistence of primary Myelofibrosis and paroxysmal nocturnal hemoglobinuria clone with JAK2 V617F, U2AF1 and SETBP1 mutations: A case report and brief review of literature. Diagnostics (Basel, Switzerland), 11(9), 1644. 10.3390/diagnostics11091644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergande, M. , Motameny, S. , Özdemir, Ö. , Kreutzer, M. , Wang, H. , Daimagüler, H.‐S. , Becker, K. , Karakaya, M. , Ehrhardt, H. , Elcioglu, N. , Ostojic, S. , Chao, C.‐M. , Kawalia, A. , Duman, Ö. , Koy, A. , Hahn, A. , Reimann, J. , Schoner, K. , Schänzer, A. , … Cirak, S. (2020). The genomic and clinical landscape of fetal akinesia. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 22(3), 511–523. 10.1038/s41436-019-0680-1 [DOI] [PubMed] [Google Scholar]
- Piazza, R. , Magistroni, V. , Redaelli, S. , Mauri, M. , Massimino, L. , Sessa, A. , Peronaci, M. , Lalowski, M. , Soliymani, R. , Mezzatesta, C. , Pirola, A. , Banfi, F. , Rubio, A. , Rea, D. , Stagno, F. , Usala, E. , Martino, B. , Campiotti, L. , Merli, M. , … Gambacorti‐Passerini, C. (2018). SETBP1 induces transcription of a network of development genes by acting as an epigenetic hub. Nature Communications, 9(1), 2192. 10.1038/s41467-018-04462-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza, R. , Valletta, S. , Winkelmann, N. , Redaelli, S. , Spinelli, R. , Pirola, A. , Antolini, L. , Mologni, L. , Donadoni, C. , Papaemmanuil, E. , Schnittger, S. , Kim, D.‐W. , Boultwood, J. , Rossi, F. , Gaipa, G. , De Martini, G. P. , di Celle, P. F. , Jang, H. G. , Fantin, V. , … Gambacorti‐Passerini, C. (2013). Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nature Genetics, 45(1), 18–24. 10.1038/ng.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polprasert, C. , Kongkiatkamon, S. , Niparuck, P. , Rattanathammethee, T. , Wudhikarn, K. , Chuncharunee, S. , Kobbuaklee, S. , Suksusut, A. , Lanamtieng, T. , Lawasut, P. , Asawapanumas, T. , Bunworasate, U. , & Rojnuckarin, P. (2022). Genetic mutations associated with blood count abnormalities in myeloid neoplasms. Hematology (Amsterdam, Netherlands), 27(1), 765–771. 10.1080/16078454.2022.2094134 [DOI] [PubMed] [Google Scholar]
- Qian, Y. , Chen, Y. , & Li, X. (2021). CSF3R T618I, SETBP1 G870S, SRSF2 P95H, and ASXL1 Q780* tetramutation co‐contribute to myeloblast transformation in a chronic neutrophilic leukemia. Annals of Hematology, 100(6), 1459–1461. 10.1007/s00277-021-04491-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, H.‐Y. , Zhang, Q. , Wang, J.‐M. , Jiang, J.‐Y. , Huyan, L.‐Y. , Yan, J. , Li, C. , & Wang, H.‐Q. (2022). TRIM29 regulates the SETBP1/SET/PP2A axis via transcription factor VEZF1 to promote progression of ovarian cancer. Cancer Letters, 529, 85–99. 10.1016/j.canlet.2021.12.029 [DOI] [PubMed] [Google Scholar]
- Rakhlin, N. , Landi, N. , Lee, M. , Magnuson, J. S. , Naumova, O. Y. , Ovchinnikova, I. V. , & Grigorenko, E. L. (2020). Cohesion of cortical language networks during word processing is predicted by a common polymorphism in the SETBP1 gene. New Directions for Child and Adolescent Development, 2020(169), 131–155. 10.1002/cad.20331 [DOI] [PubMed] [Google Scholar]
- Robinson, J. E. , Greiner, T. C. , Bouska, A. C. , Iqbal, J. , & Cutucache, C. E. (2020). Identification of a splenic marginal zone lymphoma signature: Preliminary findings with diagnostic potential. Frontiers in Oncology, 10, 640. 10.3389/fonc.2020.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romzova, M. , Smitalova, D. , Hynst, J. , Tom, N. , Loja, T. , Herudkova, Z. , Jurcek, T. , Stejskal, L. , Zackova, D. , Mayer, J. , Racil, Z. , & Culen, M. (2021). Hierarchical distribution of somatic variants in newly diagnosed chronic myeloid leukaemia at diagnosis and early follow‐up. British Journal of Haematology, 194(3), 604–612. 10.1111/bjh.17659 [DOI] [PubMed] [Google Scholar]
- Saika, M. , Inoue, D. , Nagase, R. , Sato, N. , Tsuchiya, A. , Yabushita, T. , Kitamura, T. , & Goyama, S. (2018). ASXL1 and SETBP1 mutations promote leukaemogenesis by repressing TGFβ pathway genes through histone deacetylation. Scientific Reports, 8(1), 15873. 10.1038/s41598-018-33881-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, H. , Okuno, Y. , Muramatsu, H. , Yoshida, K. , Shiraishi, Y. , Takahashi, M. , Kon, A. , Sanada, M. , Chiba, K. , Tanaka, H. , Makishima, H. , Wang, X. , Xu, Y. , Doisaki, S. , Hama, A. , Nakanishi, K. , Takahashi, Y. , Yoshida, N. , Maciejewski, J. P. , … Kojima, S. (2013). Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nature Genetics, 45(8), 937–941. 10.1038/ng.2698 [DOI] [PubMed] [Google Scholar]
- Schinzel, A. , & Giedion, A. (1978). A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. American Journal of Medical Genetics, 1(4), 361–375. 10.1002/ajmg.1320010402 [DOI] [PubMed] [Google Scholar]
- Sullivan, J. A. , Stong, N. , Baugh, E. H. , McDonald, M. T. , Takeuchi, A. , & Shashi, V. (2020). A pathogenic variant in the SETBP1 hotspot results in a forme‐fruste Schinzel‐Giedion syndrome. American Journal of Medical Genetics. Part A, 182(8), 1947–1951. 10.1002/ajmg.a.61630 [DOI] [PubMed] [Google Scholar]
- Suphapeetiporn, K. , Srichomthong, C. , & Shotelersuk, V. (2011). SETBP1 mutations in two Thai patients with Schinzel‐Giedion syndrome. Clinical Genetics, 79(4), 391–393. 10.1111/j.1399-0004.2010.01552.x [DOI] [PubMed] [Google Scholar]
- Takeuchi, A. , Okamoto, N. , Fujinaga, S. , Morita, H. , Shimizu, J. , Akiyama, T. , Ninomiya, S. , Takanashi, J. , & Kubo, T. (2015). Progressive brain atrophy in Schinzel‐Giedion syndrome with a SETBP1 mutation. European Journal of Medical Genetics, 58(8), 369–371. 10.1016/j.ejmg.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Tang, F. , Li, Z. , Lai, Y. , Lu, Z. , Lei, H. , He, C. , & He, Z. (2022). A 7‐gene signature predicts the prognosis of patients with bladder cancer. BMC Urology, 22(1), 8. 10.1186/s12894-022-00955-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşkıran, E. Z. , Karaosmanoğlu, B. , Koşukcu, C. , Ürel‐Demir, G. , Akgün‐Doğan, Ö. , Şimşek‐Kiper, P. Ö. , Alikaşifoğlu, M. , Boduroğlu, K. , & Utine, G. E. (2021). Diagnostic yield of whole‐exome sequencing in non‐syndromic intellectual disability. Journal of Intellectual Disability Research, 65(6), 577–588. 10.1111/jir.12835 [DOI] [PubMed] [Google Scholar]
- Touge, H. , Fujinaga, T. , Okuda, M. , & Aoshi, H. (2001). Schinzel–Giedion syndrome. International Journal of Urology, 8(5), 237–241. 10.1046/j.1442-2042.2001.00291.x [DOI] [PubMed] [Google Scholar]
- Uddin, M. N. , & Wang, X. (2022). Identification of key tumor stroma‐associated transcriptional signatures correlated with survival prognosis and tumor progression in breast cancer. Breast Cancer (Tokyo, Japan), 29(3), 541–561. 10.1007/s12282-022-01332-6 [DOI] [PubMed] [Google Scholar]
- Volk, A. , Conboy, E. , Wical, B. , Patterson, M. , & Kirmani, S. (2015). Whole‐exome sequencing in the clinic: Lessons from six consecutive cases from the Clinician's perspective. Molecular Syndromology, 6(1), 23–31. 10.1159/000371598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu, M. , Okuno, Y. , Murakami, N. , Miwata, S. , Kitazawa, H. , Narita, K. , Kataoka, S. , Ichikawa, D. , Hamada, M. , Taniguchi, R. , Suzuki, K. , Kawashima, N. , Nishikawa, E. , Narita, A. , Nishio, N. , Kojima, S. , Muramatsu, H. , & Takahashi, Y. (2021). Detection of subclonal SETBP1 and JAK3 mutations in juvenile myelomonocytic leukemia using droplet digital PCR. Leukemia, 35(1), 259–263. 10.1038/s41375-020-0817-x [DOI] [PubMed] [Google Scholar]
- Wang, D. , Kon, N. , Lasso, G. , Jiang, L. , Leng, W. , Zhu, W.‐G. , Qin, J. , Honig, B. , & Gu, W. (2016). Acetylation‐regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature, 538(7623), 118–122. 10.1038/nature19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Chen, Y. , Lin, H. , Ni, W. , Zhang, Q. , Lan, J. , & Jin, L. (2022). Acute mast cell leukemia preceded by malignant mediastinal germ cell tumor: A case report and literature review. Cancer Management and Research, 14, 1783–1794. 10.2147/CMAR.S363508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Guo, Y. , Dong, Z. , Li, T. , Xie, X. , Wan, D. , Jiang, Z. , Yu, J. , & Guo, R. (2020). Differential U2AF1 mutation sites, burden and co‐mutation genes can predict prognosis in patients with myelodysplastic syndrome. Scientific Reports, 10(1), 18622. 10.1038/s41598-020-74744-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe‐Okochi, N. , Kitaura, J. , Ono, R. , Harada, H. , Harada, Y. , Komeno, Y. , Nakajima, H. , Nosaka, T. , Inaba, T. , & Kitamura, T. (2008). AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood, 111(8), 4297–4308. 10.1182/blood-2007-01-068346 [DOI] [PubMed] [Google Scholar]
- Wong, M. M. , Kampen, R. A. , Braden, R. O. , Alagöz, G. , Hildebrand, M. S. , Barnett, C. , Barnett, M. , Brusco, A. , Carli, D. , de Vries, B. B. , Dingemans, A. J. , Elmslie, F. , Ferrero, G. B. , Jansen, N. A. , van de Laar, I. M. , Moroni, A. , Mowat, D. , Murray, L. , Novara, F. , … Fisher, S. E. (2022). SETBP1 variants outside the degron disrupt DNA‐binding and transcription independent of protein abundance to cause a heterogeneous neurodevelopmental disorder. medRxiv. 10.1101/2022.03.04.22271462 [DOI]
- Yin, B. , Chen, X. , Gao, F. , Li, J. , & Wang, H. W. (2019). Analysis of gene mutation characteristics in patients with chronic neutrophilic leukaemia. Hematology (Amsterdam, Netherlands), 24(1), 538–543. 10.1080/16078454.2019.1642554 [DOI] [PubMed] [Google Scholar]
- Yun, J. W. , Yoon, J. , Jung, C. W. , Lee, K.‐O. , Kim, J. W. , Kim, S.‐H. , & Kim, H.‐J. (2020). Next‐generation sequencing reveals unique combination of mutations in cis of CSF3R in atypical chronic myeloid leukemia. Journal of Clinical Laboratory Analysis, 34(2), e23064. 10.1002/jcla.23064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Zhang, M. , Ge, S. , Huang, W. , Lin, X. , Gao, J. , Gong, J. , & Shen, L. (2019). Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K‐Akt signaling in gastric cancer. Cancer Medicine, 8(10), 4766–4781. 10.1002/cam4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]


