Abstract
Osteosarcoma (OS) is the most frequent primary malignant bone tumour, whose heterogeneity represents a major challenge for common antitumour therapies. Inflammatory cytokines are known to be necessary for OS progression. Therefore, to optimise therapy, it is important to discover reliable biomarkers by identifying the mechanism generating OS and investigating the inflammatory pathways that support the undifferentiated state. In this work, we highlight the differences of epigenetic activities of IL-1β and TNFα, and the susceptibility of TET-1 enzymatic inhibition, in tumour progression of three different OS cell lines. Investigating DNA methylation of IL-6 promoter and determining its expression, we found that TET enzymatic inhibition influences proliferation induced by inflammatory cytokines in OS cell lines. Moreover, Bobcat 339 treatment blocks IL-1β epigenetic action on IL-6 promoter, while only partially those of TNFα as well as inhibits IL-1β-dependent epithelial–mesenchymal transition (EMT) process, but only partially those of TNFα. In conclusion, this work highlights that IL-1β and TNFα have different effects on DNA demethylation in OS cell lines, making DNA methylation a potential biomarker of disease. Specifically, in IL-1β treatment, TET-1 inhibition completely blocks tumour progression, while in TNFα actions, it is only partially effective. Given that these two inflammatory pathways can be therapeutic targets for treating these tumours, knowledge of their distinct epigenetic behaviours can be useful for developing precise and specific therapeutic strategies for this disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01745-4.
Keywords: Osteosarcoma, Inflammation, Epigenetics, Metastasis
Introduction
Osteosarcoma (OS) is the most common primary malignant tumour of bone, despite its low incidence, which has a characteristic bimodal distribution with a first peak in childhood, approximately 75%, and a second peak over the age of 70 [1–3]. The main challenge in the prognosis of OS is the tendency to produce systemic metastases, which reduces survival to less than 30% [4]. The heterogeneity of OS cells challenges the efficacy of chemotherapy, and it is, therefore, particularly important, for its optimisation, to characterise tumours specifically, including reliable biomarkers, to recognise the mechanism of relapse and to identify the type of cells that have generated OS [5].
In recent years, several studies have explored the relationship between the complex biological system of the bone microenvironment and tumour progression, with the aim of identifying potential targets for intervention and specific therapies [5]. Several studies have demonstrated the pivotal role of inflammatory cytokines in the progression of osteosarcoma by maintaining an undifferentiated state [6]. It is known in the literature that pro-inflammatory cytokines IL-1β and TNFα are able to activate IL-6 expression and release, recently correlated with metastatic process [7] in several tumoural cells, such as breast cancer [8–11], prostate cancer [12–14] and colorectal cancer [15–17], including OS cells [18, 19]. Furthermore, IL-1β and TNFα, being responsible of maintaining the undifferentiated state in OS cells, are necessary for tumour progression. In fact, pharmacological inhibition of TNFα has been shown to inhibit tumour growth and promote osteoblastic differentiation. Similarly, IL-1β treatment can abrogate osteogenesis and stimulate OS tumour development, while its inhibition can block this phenomenon [2]. Our previous study has shown that IL-1β plays a role in tumour progression and bone metastasis of breast cancer through epigenetic activities. Inhibition of ten-eleven translocation methyl cytosine dioxygenase-1 (TET-1) activities, implicated in the first step of methyl cytosine demethylation [20], has been found to block IL-1β-dependent proliferation, IL-6 expression, EMT process and bone homing in Mcf-7 cell line. This work has also highlighted how the other TET, TET-2, implicated in DNA demethylation is no related to IL-1β activities [8].
This work highlights the differences in epigenetic activities between IL-1β and TNFα, as well as the susceptibility of TET-1 enzymatic inhibition in the tumour progression of various OS cell lines.
Materials and methods
Reagents
Dulbecco’s modified Eagle’s high-glucose medium (DMEM), fetal bovine serum (FBS), L-glutamine and penicillin/streptomycin were bought from Lonza, while Bobcat339 hydrochloride (BC) was purchased from MedChemExpress (MCE, Monmouth Junction, NJ, USA).
Cell cultures
The OS cell lines KHOS (KHOS/NP, R-970-5, ATCC®CRL-1544™), MG63 (MG-63, ATCC®CRL-1427™) and Saos-2 (Saos-2, ATCC®HTB-85™), obtained from ATCC® (Manassas, VA, USA), were cultured at 37 °C, 5% CO2 in DMEM high glucose (Euroclone S.p.A., Pero, Milan, Italy) in the presence of 10% FBS (heat-inactivated) (Lonza, Verviers, Belgium), 1 mM sodium pyruvate (Euroclone), 2 mM glutamine, 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco, Invitrogen Corp., Carlsbad, CA, USA) as described elsewhere [21].
Viability assay (MTT assay)
Methyl thiazole tetrazolium (MTT) was used to evaluate cell viability of OS cell lines as well described elsewhere [22]. Briefly, each OS cell line was seeded with a density of 1 × 105 on 96-well plate and treated with 12.5 ng/mL and 25 ng/mL of IL-1β or TNFα for 24 and 48 h. Similarly, we evaluated the viability of the cells exposed to the final concentration of 33 or 75 μM of BC, and cells treated both with both BC and IL-1β or TNFα (25 ng/mL), at the same experimental time. The absorbance at 540 nm was measured by Bio-Rad microplate Reader (Bio-Rad Laboratories, Hercules, CA, USA). Viability is expressed in percentage respect to controls.
Genomic DNA (gDNA) isolation
Genomic DNAs of OS cell lines, under different treatments, were extracted using the PureLink Genomic DNA mini-kit (Invitrogen™, Waltham, MA, USA). gDNA was evaluated quantitatively with the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), qualitatively evaluated through electrophoretic analyses using 0.8% agarose gel stained with Gel Red staining (Biotium, Hayward, CA, USA) and photographed with a Chemi-Doc apparatus (Bio-Rad Laboratories, Hercules, CA, USA) as described elsewhere [23].
Methylation analysis of IL-6 promoter by MSRE-PCR
The methylation-sensitive restriction endonuclease–PCR (MSRE–PCR) analysis was carried out to evaluate the methylation status of six CpG-rich sites on the promoter of interleukin-6 (IL-6). Experiments were performed as described elsewhere [8, 24–26]. Amplicons analysed in 2% agarose gel electrophoresis were stained with Gel Red staining (Biotium, Hayward, CA, USA) and visualised using the Chemi-Doc instrument (Bio-Rad Laboratories, Hercules, CA, USA). Densitometric measurements of captured bands were carried out using the “Image Lab” application (version 5.2.1) of Bio-Rad Laboratories (Hercules, CA, USA).
ELISA assay
About 2 × 104 OS cells were seeded in the 96-well plate which were treated with IL-1β (25 ng/mL), TNFα (25 ng/mL) or BC (33 μM), or co-treatment with IL-1β (25 ng/mL) or TNFα (25 ng/mL) in the presence of BC (33 μM) for 48 h. Secreted IL-6, IL-1β and TNFα levels were measured in culture supernatants with the enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems Europe, Ltd., Abingdon Science Park, Abingdon, UK) according to the instructions of manufacturers.
Western blot analysis
The cells were lysed for 1 h in NP40 Cell lysis buffer containing 50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% Nonidet P40 (NP40) and 0.02% NaN3 (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA). To the cell lisates were added 1 mM PMSF (1 M, Sigma–Aldrich, St. Louis, MO, USA) and Protease Inhibitor Cocktail (100X, Sigma–Aldrich, St. Louis, MO, USA) to avoid protein degradation. For SDS-PAGE electrophoresis, the 4–12% Novex Bis–Tris SDS-acrylamide gels (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA) was used to separate the cell lysates (30 μg per lane). Protein electrophoresis was transferred on Nitrocellulose membranes (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA) through the iBlot 2 Dry Blotting System (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA) and immunoblotted with the primary antibodies. The following antibodies were used for Western blot: GAPDH (1:1000, sc-47724, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), Vimentin and Snail (1:500, Epithelial-Mesenchymal Transition (EMT) Antibody Sampler Kit, #9782, Cell Signaling Technology, Danvers, MA, USA), the secondary anti-mouse IgG HRP-linked antibody (1:2000, #7076, Cell Signaling Technology, Danvers, MA, USA) and anti-rabbit IgG HRP-linked antibody (1:2000, #7074, Cell Signaling Technology, Danvers, MA, USA).
Chemi-Doc apparatus (Bio-Rad Laboratories, Hercules, CA, USA) was used to capture chemo-luminescent bands, and the images were analysed using the “Image Lab” application (version 5.2.1) of Bio-Rad Laboratories (Hercules, CA, USA) [27].
Statistical analysis
The statistical analysis was conducted using R software v.4.3.3 [28] and specific packages. After checking for normal distribution of the data (Shapiro–Wilk test) and homogeneity of variance (Levene's test), one-way or two-way ANOVA tests were used to assess significant effects and/or factorial interactions, with “treatment” as a factor for one-way and two-way tests, and “experimental time” as a factor for two-way tests, followed by a pairwise comparison test (tested treatments versus untreated controls, and co-treatments with BC versus tested treatment). For data with non-normal distribution and heterogeneity of variance, we used the Kruskal–Wallis test followed by Dunn's test for comparisons. In both cases, we adjusted the p-values according to the Sidak–Holm method.
Results
TET-1 enzymatic inhibition influences proliferation induced by inflammatory cytokines in OS cell lines
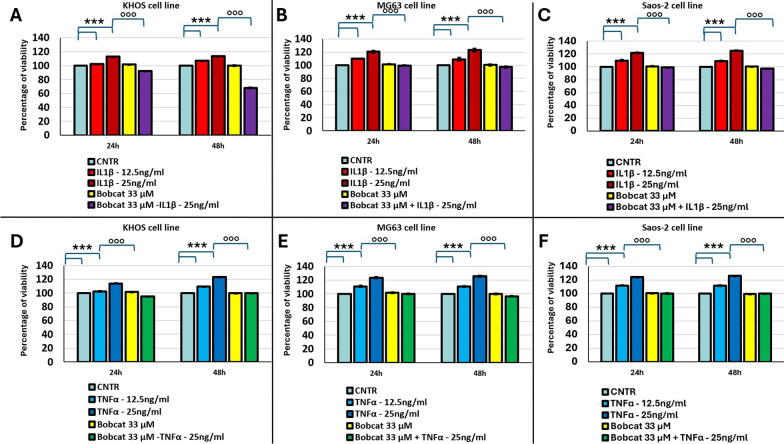
We have initially investigated how inflammation signals, mimicked by inflammatory cytokine IL-1β or TNFα treatments, can induce an increased proliferation of different OS cell lines, and if these signals are influenced by TET-1 enzymatic inhibition, obtained by BC treatment. Figure 1 shows that the proliferation of MG63, Saos-2 and KHOS cell lines is significantly enhanced when cells are treated with IL-1β or TNFα, in dose- and time-dependent manner (p < 0.0005). Moreover, while the treatment with BC alone has no effect in cell proliferation, the co-treatment of BC with pro-inflammatory cytokines, IL-1β and TNFα, inhibited cell proliferation induction in all OS cell lines (p < 0.0005, Fig. 1). Since the use of TET inhibitor to 75 μM, able to inhibit both TET-1 and TET-2, has been lethal for all OS cell line (see supplemental material), this dose was excluded from the study.
Fig. 1.
Percentages of viability of KHOS (A, D), MG63 (B, E) and Saos-2 (C, F) cell lines after 24 and 48 h submitted to the following treatments: (1) untreated (CNTR); treated with two concentrations of IL-1β (12.5 ng/ml and 25 ng/ml); BC 33 μM and co-treatment BC + IL-1β, 25 ng/ml (A–C); or (2) untreated (CNTR); treated with two concentrations of TNFα (12.5 ng/ml and 25 ng/ml); BC 33 μM and co-treatment BC + TNFα, 25 ng/ml (D–F) (Mean ± SD, n = 3). Pairwise comparisons among all tested treatments versus untreated controls (*), and co-treatments with BC versus the 25 ng/ml IL-1β or TNFα treatments (°) within each experimental time are reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005
BC treatment block epigenetic action of IL-1β on IL-6 promoter, while only partially those of TNFα
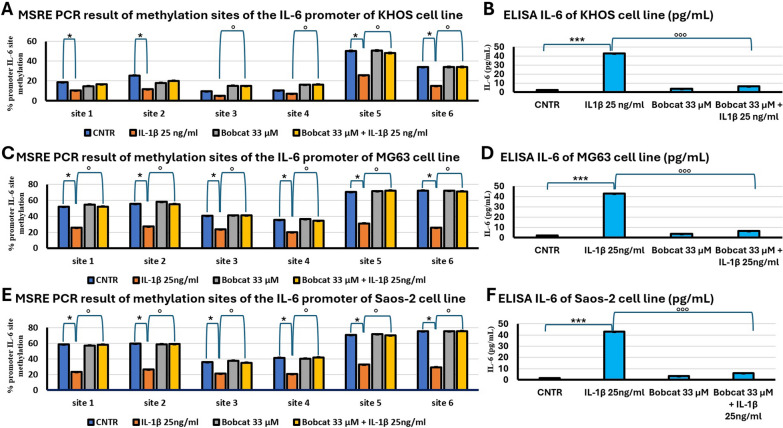
To evaluate if IL-1β treatment induce a switch in methylation level of IL-6 proximal promoter in the three OS cell lines, we have carried out MSRE-PCR analyses of the six sites present in IL-6 promoter, containing CpG islands. As shown in Fig. 2, IL-1β treatment reduces the percentage of methylation of all analysed sites (p < 0.05) and consequently enhances the expression and release of IL-6 in conditioned medium in all cell lines compared to untreated cells (p < 0.0005). Furthermore, it was also highlighted how BC in co-treatment with IL-1β is able to block IL-6 promoter demethylation (p < 0.05, Fig. 2A, C and E) with relative alterations of IL-6 release in all cell lines (p < 0.0005, Fig. 2B, D and F).
Fig. 2.
Methylation analysis of IL-6 promoter and relative releases. Percentage of methylation of six methyl-sensible restriction sites through MSRE-PCR analyses of IL-6 promoter of KHOS (A), MG63 (C) and Saos-2 (E) cell lines, after 48 h submitted to the following treatments: untreated (CNTR); IL-1β 25 ng/ml; BC 33 μM and BC 33 μM + IL-1β 25 ng/ml (Mean ± SD, n = 3, duplicates). ELISA of released IL-6 of KHOS (B), MG63 (D) and Saos-2 (F) cell lines after 48 h under the same treatments (Mean ± SD, n = 3, duplicates). Dunn’s test among tested treatments versus untreated controls (*) and co-treatments with BC versus IL-1β treatment (°) is reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005)
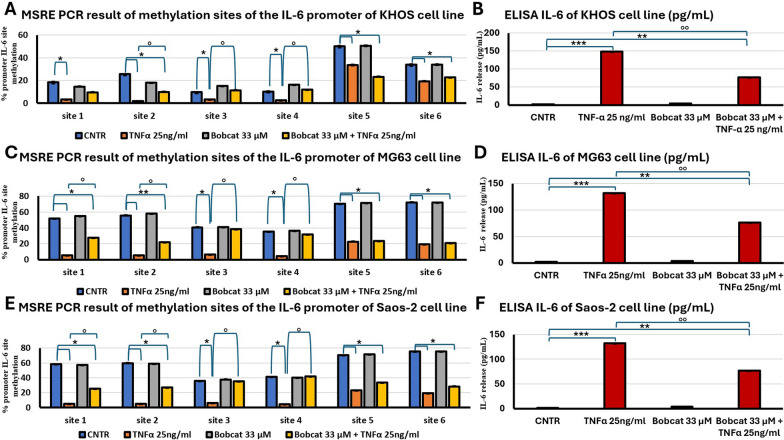
In parallel, the effects of TNFα and IL-1β in the methylation status of IL-6 promoter were investigated. The reduction of CpG islands was observed (p < 0.05, Fig. 3A, C and E), and an increase in the inflammatory cytokine IL-6 (p < 0.0005, Fig. 3B, D and F) relative release was revealed. Regarding the actions of BC, the co-treatment of TNFα with BC was able to block only partially the demethylation process induced by TNFα. In fact, BC treatment blocked only the sites 3 and 4 of IL-6 promoter in the demethylation; in the sites 1 and 2, the methylation levels were between TNFα treatment condition and untreated control, indicating a partially arrest of demethylation; while in sites 5 and 6, BC was not able to revert TNFα action (p < 0.05, Fig. 3A, C and E). The ELISA assay showed as TNFα treatment was able to induce the expression and secretion of IL-6 (p < 0.0005, Fig. 3B, D and F), while BC was able to block TNFα-dependent IL-6 secretion only partially (p < 0.005, Fig. 3B, D and F).
Fig. 3.
Methylation analysis of IL-6 promoter and relative releases. Percentage of methylation of six methyl-sensible restriction sites through MSRE-PCR analyses of IL-6 promoter of KHOS (A), MG63 (C) and Saos-2 (E) cell lines, after 48 h submitted to the following treatments: untreated (CNTR); TNFα 25 ng/ml; BC 33 μM and BC 33 μM + TNFα 25 ng/ml (Mean ± SD, n = 3, duplicates). ELISA of released IL-6 of KHOS (B), MG63 (D) and Saos-2 (F) cell lines after 48 h under the same treatments (Mean ± SD, n = 3, duplicates). Dunn’s test among tested treatments versus untreated controls (*) and co-treatments with BC versus the TNFα treatment (°) is reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005
BC treatment block IL-1β-dependent EMT process, but only partially those of TNFα
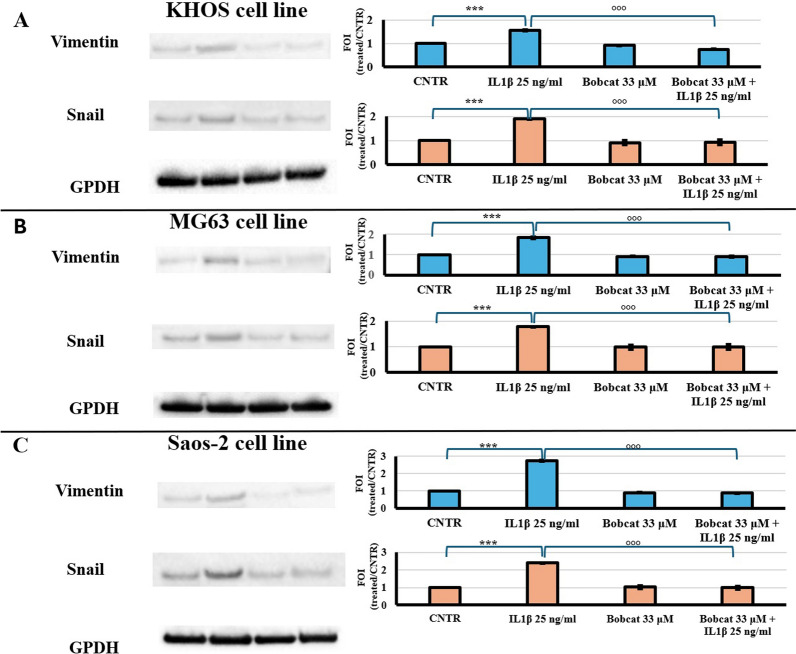
It has been described in the literature that IL-6 expression and release represents, in several tumoural systems including OS, the first step of metastasis inducing EMT process [7–14]. Western blot analyses confirm that co-treatment of IL-1β may block EMT process as indicated by the expression of Vimentin and Snail protein, in all OS cell lines (Fig. 4).
Fig. 4.
Western blot (WB) analysis of two markers (Vimentin and Snail) of EMT process of KHOS (A), MG63 (B) and Saos-2 (C) cell lines untreated (CNTR) and treated with IL-1β 25 ng/ml, BC 33 μM or BC 33 μM + IL-1β 25 ng/ml at 48 h. Representative images of WB of Vimentin and Snail and relative densitometric analysis (Mean ± SD, n = 3). Pairwise comparisons among tested treatments versus untreated controls (*), and co-treatments with BC versus the IL-1β treatment (°) are reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005
Subsequently, same analyses conducted in TNFα treatments showed that BC is able to reduce but not block the expression of markers of EMT process, as indicated in Fig. 5.
Fig. 5.
Western blot (WB) analysis of two markers (Vimentin and Snail) of EMT process of KHOS (A), MG63 (B) and Saos-2 (C) cell lines untreated (CNTR) and treated with TNFα 25 ng/ml, BC 33 μM or BC 33 μM + TNFα 25 ng/ml at 48 h. Representative images of WB of Vimentin and Snail and relative densitometric analysis (Mean ± SD, n = 3). Pairwise comparisons among tested treatments versus untreated controls (*) and co-treatments with BC versus the TNFα treatment (°) are reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005
Positive feedback blocked by BC in IL-1β treatment but only partially in those of TNFα
In the progression of inflammation, the first signal is often followed by the expression and release of other pro-inflammatory factors that amplify the initial signal. For this reason, it was carried out the analysis of TNFα release in tumour cells treated with IL-1β, and the production of IL-1β in tumour cells treated with TNFα, and their behaviour in the presence of the TET inhibitor, BC.
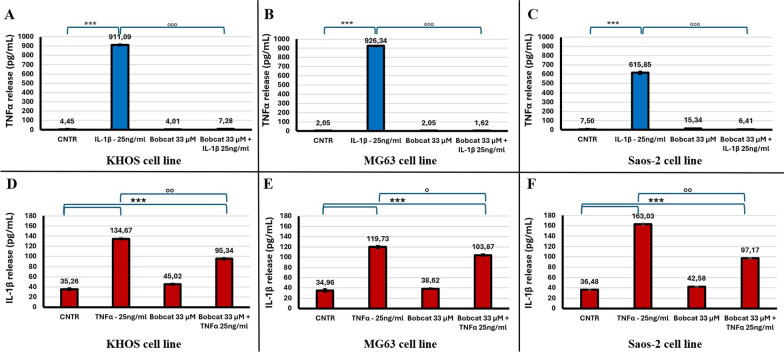
Regarding the expression and release of TNFα in IL-1β treated tumoural cells, after 48 h of IL-1β treatment, TNFα secreted levels were increased about 250, 500 and 82 times in KHOS, MG63 and Saos-2 cell lines, respectively, compared to the control condition. The co-treatment of IL-1β with BC, where it is blocked the demethylation activities of the TET, we observe a complete downregulation of TNFα release (p < 0.0005, Fig. 6A, B and C). The lack of release of TNFα determines the loss of signal enhancement carried out by this factor.
Fig. 6.
ELISA of released TNFα (A–C) and IL-1β (D–F) of KHOS (A, D), MG63 (B, E) and Saos-2 (C, F) cells after 48 h under the following treatments: untreated (CNTR); IL-1β 25 ng/ml or TNFα 25 ng/ml; BC 33 μM and BC 33 μM + IL-1β 25 ng/ml or BC 33 μM + TNFα 25 ng/ml (Mean ± SD, n = 3, duplicates). Dunn’s test among tested treatments versus untreated controls (*) and co-treatments with BC versus the IL-1β or TNFα treatments (°) is reported in the graphs: 1 symbol, p < 0.05; 2 symbols, p < 0.005 and 3 symbols, p < 0.0005
Regarding the release of IL-1β in these cell lines, we observe a basal release, both in untreated cells and BC treatment, indicating that BC has no effect in basal expression. In TNFα treatment, we observe an increase in IL-1β release of about 4, 3.5 and 4.5 times in KHOS, MG63 and Saos-2 cell lines, respectively. In the co-treatment BC/TNFα, we observe only a reduction of IL-1β release induced by TNFα treatment, leading to an increase in release relative to untreated cells of about 2.7, 3 and 2.7 times in KHOS, MG63 and Saos-2 cell lines, respectively (p < 0.005, Fig. 6D, E and F), leading to an enhancement of these signals.
Discussion
This study aimed at evaluating how primary inflammatory signals, represented by treatments with IL-1β and TNFα, are able to induce tumour progression in OS, and how these signals are linked to the regulation of DNA methylation. Current results demonstrated that these signals are influenced, albeit differently, by inhibitor of the enzymatic activity of TETs, BC [29]. These enzymes are responsible for multiple consecutive hydroxylations of methyl cytosine, which is reduced to uracil, with subsequent involvement of DNA repair system and replacement of uracil with a cytosine, resulting in a DNA demethylation in this position [20]. We had previously demonstrated that IL-1β inflammatory signal is able to induce tumour progression in a primary breast cancer cell line, MCF-7, modulating EMT process and inducing bone homing factor, leading to bone metastasis formation [8]. It was also pointed out sensitivity of IL-1β signal to the enzymatic inhibition of TET-1, implicated in metastasizing of MCF-7 cell line. BC treatment, despite not inducing to programmed cell death, is able to revert proliferation induction and block expression of EMT and bone homing factors, avoiding bone metastasis formation induced by IL-1β treatment [8].
Similarly, we evidenced that inflammation signals, represented by IL-1β or TNFα, induce the proliferation and activation of EMT process, determining tumour progression in three different OS cell lines. Furthermore, we investigated the influence of BC at 33 μM, determining the enzymatic inhibition only of TET-1, on these inflammatory signals. In particular, it was observed that BC treatment is able to block IL-1β-dependent proliferation, IL-6 expression and release, as well as EMT marker expression, showing a regulation of IL-1β tightly related to TET-1 activity and then to DNA methylation regulation.
Regarding TNFα signal, the same behaviour as IL-1β was not found. In fact, although BC treatment is able to revert proliferation induction, it cannot completely block the TNFα-dependent demethylation of IL-6 promoter, and consequently its release, as well as the induction of EMT process in almost all analysed experimental times.
In the progression of inflammation, the first signal, often represented by IL-1β or TNFα, is joined by other signals due to the expression of other factors (e.g. IL-6), which led to the amplification of the initial signal, through a positive feedback system. These factors are also known to be implicated in several processes leading to tumour progression, such as tumour proliferation, EMT process and inhibition of apoptosis [7–19, 30]. For this reason, we studied the expression and relative release of TNFα in IL-1β treated tumoural cells, as well as those of IL-1β in TNFα treated tumoural cells, and how these cells react to TET-1 enzymatic inhibition activities of BC. We observed that BC is able to block also the TNFα expression in IL-1β/BC co-treatment, indicating a tight relation with DNA methylation in IL-1β actions. Contrary, in TNFα/BC treatment, BC is unable to entirely prevent IL-1β expression and release, indicating that the block of TET-1 actions in DNA demethylation is partially ineffective, as indicated by the expression and release of IL-1β.
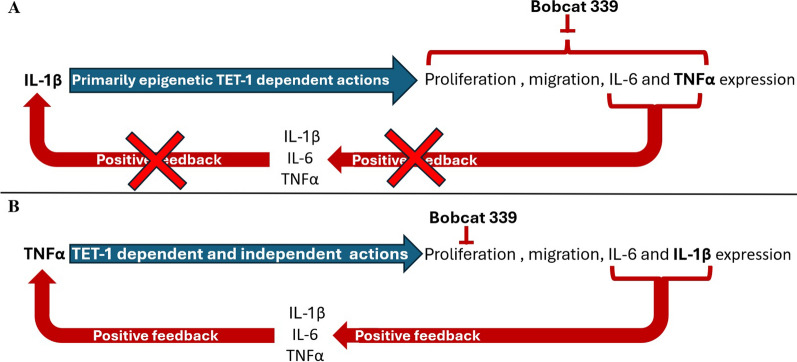
The regulation of DNA methylation is extremely important in the activation of several pathways enforcing IL-1β actions. It is described as several factors are activated by DNA demethylation, responsible of transduction signal of IL-1β. For example, interleukin-1 receptor associated kinase 1 (IRAK1) is regulated epigenetically, and its overexpression was found significantly associated with poor survival in several cancers [31–35]. Several molecules implicated in reduction of oxidative status such as plant flavonoids (e.g. resveratrol), or other metabolites as melatonin, could influence DNA methylation status, reducing inflammation status and then tumour progression and metastasis formation [26, 36–38]. Figure 7 reports a schematic draw in which is evidenced how IL-1β signal, acting through almost exclusively in epigenetic manner, is blocked by TET-1 inhibition by BC treatment (Fig. 7A), while TNFα acts through both epigenetic and non-epigenetic manners, where the block of TET-1 action by BC is able only to inhibit proliferation, but not the pro-inflammatory cytokines expression and releases, determining positive feedback in inflammation and in metastasis formation (Fig. 7B).
Fig. 7.
Schematic draw of feedback loop induced by IL-1β (A) and TNFα (B) and the influence of TET-1 inhibition in positive feedback of these signals
In conclusion, this work highlights the different behaviours of the two inflammatory signals, IL-1β and TNFα, which both could sustain OS tumour growth. BC co-treatment indicates that TET-1-dependent DNA demethylation is of great importance in IL-1β treatment, resulting in a complete block of tumour progression, while it is only partially effective in TNFα action, indicating an involvement of other molecular mechanism. Given that these two inflammatory pathways can be therapeutic targets for the treatment of these tumours, knowledge of their distinct epigenetic behaviours can be valuable for developing precise and specific curative strategies for these diseases.
Supplementary Information
Acknowledgements
This work was funded by the Italian Ministry of Health—Ricerca Corrente.
Author contributions
B.D.; N.F. and G.G. helped in conceptualisation. B.D.; N.F. and G.G. helped in methodology. B.D.; C.V.; R.L.; D.L.A. and G.G. helped in validation. B.D.; C.V.; R.L.; D.L.A. and G.G. helped in formal analysis. B.D. and C.V. worked in investigation. B.D.; C.V.; N.F. and G.G. helped in resources. B.D. and G.G. helped in data curation. B.D. and G.G. contributed to writing—original draft preparation. B.D.; C.S.; C.V.; U.O.; R.L.; D.L.A.; C.F.; P.S.; N.F. and G.G. contributed to writing—review & editing. B.D.; C.V.; U.O.; D.L.A. and R.L. helped in visualisation. B.D. worked in supervision. B.D. and G.G. worked in project administration. G.G. helped in funding acquisition.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors declared any conflict of interest.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Flores Naselli and Gianluca Giavaresi these have contributed equally to this work.
References
- 1.Beird HC, Bielack SS, Flanagan AM, Gill J, Heymann D, Janeway KA, et al. Osteosarcoma. Nat Rev Dis Primers. 2022;8(1):77. 10.1038/s41572-022-00409-y. [DOI] [PubMed] [Google Scholar]
- 2.Kim C, Davis LE, Albert CM, Samuels B, Roberts JL, Wagner MJ. Osteosarcoma in pediatric and adult populations: are adults just big kids? Cancers (Basel). 2023. 10.3390/cancers15205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortini M, Avnet S, Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–9. 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Tsujisaka R, Nakayama R, Sekita T, Asano N, Kikuta K, Oguro S, et al. Dedifferentiated osteosarcoma of the distal ulna: a case report. Case Rep Oncol. 2021;14(2):1228–36. 10.1159/000518266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020. 10.3390/ijms21196985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avnet S, Lemma S, Cortini M, Di Pompo G, Perut F, Lipreri MV, et al. The release of inflammatory mediators from acid-stimulated mesenchymal stromal cells favours tumour invasiveness and metastasis in osteosarcoma. Cancers (Basel). 2021. 10.3390/cancers13225855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amer MF, Mohamed A, Ismail A, Bayoumi LA, Shibel PEE, Elnaggar GN. Possible role of IL-6R/STAT3/MiRNA-34a feedback loop in osteosarcoma. Asian Pac J Cancer Prev. 2023;24(9):3269–74. 10.31557/APJCP.2023.24.9.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellavia D, Costa V, De Luca A, Cordaro A, Fini M, Giavaresi G, et al. The binomial “Inflammation-Epigenetics” in breast cancer progression and bone metastasis: IL-1β Actions are influenced by TET inhibitor in MCF-7 cell line. Int J Mol Sci. 2022. 10.3390/ijms232315422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haq ATA, Yang PP, Jin C, Shih JH, Chen LM, Tseng HY, et al. Immunotherapeutic IL-6R and targeting the MCT-1/IL-6/CXCL7/PD-L1 circuit prevent relapse and metastasis of triple-negative breast cancer. Theranostics. 2024;14(5):2167–89. 10.7150/thno.92922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil M, Desouky EM, Khaliefa AK, Hozyen WG, Mohamed SS, Hasona NA. Insights into the crosstalk between miR-200a/lncRNA H-19 and IL-6/SIRT-1 axis in breast cancer. J Interferon Cytokine Res. 2024. 10.1089/jir.2023.0216. [DOI] [PubMed] [Google Scholar]
- 11.Peyvandi S, Bulliard M, Yilmaz A, Kauzlaric A, Marcone R, Haerri L, et al. Tumor-educated Gr1+CD11b+ cells drive breast cancer metastasis via OSM/IL-6/JAK-induced cancer cell plasticity. J Clin Invest. 2024. 10.1172/JCI166847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Lu K, Hou Y, You Z, Shu C, Wei X, et al. YY1 complex in M2 macrophage promotes prostate cancer progression by upregulating IL-6. J Immunother Cancer. 2023. 10.1136/jitc-2022-006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadrkhanloo M, Paskeh MDA, Hashemi M, Raesi R, Motahhary M, Saghari S, et al. STAT3 signaling in prostate cancer progression and therapy resistance: an oncogenic pathway with diverse functions. Biomed Pharmacother. 2023;158: 114168. 10.1016/j.biopha.2022.114168. [DOI] [PubMed] [Google Scholar]
- 14.Seven D, Tecimel D, Bayrak Ö. NANOG dominates interleukin-6-induced sphere formation in prostate cancer. Urol Res Pract. 2023;49(6):376–80. 10.5152/tud.2023.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv H, Mu Y, Zhang C, Zhao M, Jiang P, Xiao S, et al. Comparative analysis of single-cell transcriptome reveals heterogeneity and commonality in the immune microenvironment of colorectal cancer and inflammatory bowel disease. Front Immunol. 2024;15:1356075. 10.3389/fimmu.2024.1356075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L, Yin Y, Yu Z, Xu N, Qiao W, Zhen X, et al. Toll-like receptor 6 inhibits colorectal cancer progression by suppressing NF-κB signaling. Heliyon. 2024;10(6): e26984. 10.1016/j.heliyon.2024.e26984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu A, Fang D, Liu Y, Shi X, Zhong Z, Zhou B, et al. Nuclear translocation of thioredoxin-1 promotes colorectal cancer development via modulation of the IL-6/STAT3 signaling axis through interaction with STAT3. Theranostics. 2023;13(14):4730–44. 10.7150/thno.85460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Cao Y, Xiao H, Feng J, Lin J. Bazedoxifene suppresses the growth of osteosarcoma cells by inhibiting IL-6 and IL-11/GP130 signaling pathway. J Pediatr Hematol Oncol. 2024;46(1):8–14. 10.1097/MPH.0000000000002782. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Chen C, Sun K, Shi Q, Wang B, Huang Y, et al. Tocilizumab (monoclonal anti-IL-6R antibody) reverses anlotinib resistance in osteosarcoma. Front Oncol. 2023;13:1192472. 10.3389/fonc.2023.1192472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin X, Hu L, Xu Y. Structure and function of TET enzymes. Adv Exp Med Biol. 2022;1389:239–67. 10.1007/978-3-031-11454-0_10. [DOI] [PubMed] [Google Scholar]
- 21.Volpes S, Cruciata I, Ceraulo F, Schimmenti C, Naselli F, Pinna C, et al. Nutritional epigenomic and DNA-damage modulation effect of natural stilbenoids. Sci Rep. 2023;13(1):658. 10.1038/s41598-022-27260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellavia D, Raimondo S, Calabrese G, Forte S, Cristaldi M, Patinella A, et al. Interleukin 3- receptor targeted exosomes inhibit. Theranostics. 2017;7(5):1333–45. 10.7150/thno.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragusa MA, Naselli F, Cruciata I, Volpes S, Schimmenti C, Serio G, et al. Indicaxanthin induces autophagy in intestinal epithelial cancer cells by epigenetic mechanisms involving DNA methylation. Nutrients. 2023. 10.3390/nu15153495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellavia D, Dimarco E, Caradonna F. Characterization of three different clusters of 18S–26S ribosomal DNA genes in the sea urchin P. lividus: genetic and epigenetic regulation synchronous to 5S rDNA. Gene. 2016;580(2):118–24. 10.1016/j.gene.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Caradonna F, Cruciata I, Schifano I, La Rosa C, Naselli F, Chiarelli R, et al. Methylation of cytokines gene promoters in IL-1β-treated human intestinal epithelial cells. Inflamm Res. 2018;67(4):327–37. 10.1007/s00011-017-1124-5. [DOI] [PubMed] [Google Scholar]
- 26.De Luca A, Bellavia D, Raimondi L, Carina V, Costa V, Fini M, et al. Multiple effects of resveratrol on osteosarcoma cell lines. Pharmaceuticals (Basel). 2022. 10.3390/ph15030342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caradonna F, Schiera G, Di Liegro CM, Vitale V, Cruciata I, Ferrara T, et al. Establishment and preliminary characterization of three astrocytic cells lines obtained from primary rat astrocytes by sub-cloning. Genes (Basel). 2020. 10.3390/genes11121502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. . Vienna ARFfSC, 2020. R: A Language and Environment for Statistical Computing. R Core Team: Vienna, Austria; 2021.
- 29.Chua GNL, Wassarman KL, Sun H, Alp JA, Jarczyk EI, Kuzio NJ, et al. Cytosine-based TET enzyme inhibitors. ACS Med Chem Lett. 2019;10(2):180–5. 10.1021/acsmedchemlett.8b00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimbo K, Park JS, Yokosuka K, Sato K, Nagata K. Positive feedback loop of interleukin-1beta upregulating production of inflammatory mediators in human intervertebral disc cells in vitro. J Neurosurg Spine. 2005;2(5):589–95. 10.3171/spi.2005.2.5.0589. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Duan X, Zhang Z. Interleukin-1 receptor-associated kinase 1 correlates with metastasis and invasion in endometrial carcinoma. J Cell Biochem. 2018;119(3):2545–55. 10.1002/jcb.26416. [DOI] [PubMed] [Google Scholar]
- 32.Ye ZH, Gao L, Wen DY, He Y, Pang YY, Chen G. Diagnostic and prognostic roles of IRAK1 in hepatocellular carcinoma tissues: an analysis of immunohistochemistry and RNA-sequencing data from the cancer genome atlas. Onco Targets Ther. 2017;10:1711–23. 10.2147/OTT.S132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Liu S, Deng P, Liang Y, Xiao R, Tang LQ, et al. Targeting the IRAK1-S100A9 axis overcomes resistance to paclitaxel in nasopharyngeal carcinoma. Cancer Res. 2021;81(5):1413–25. 10.1158/0008-5472.CAN-20-2125. [DOI] [PubMed] [Google Scholar]
- 34.Rahemi S, Nematollahi-Mahani SN, Rajaie A, Fallah H. Inhibitor of interleukin-1 receptor-associated kinases 1/4, can increase the sensitivity of breast cancer cells to methotrexate. Int J Mol Cell Med. 2019;8(3):200–9. 10.22088/IJMCM.BUMS.8.3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uo T, Sprenger CC, Plymate SR. Androgen receptor signaling and metabolic and cellular plasticity during progression to castration resistant prostate cancer. Front Oncol. 2020;10: 580617. 10.3389/fonc.2020.580617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannino G, Caradonna F, Cruciata I, Lauria A, Perrone A, Gentile C. Melatonin reduces inflammatory response in human intestinal epithelial cells stimulated by interleukin-1β. J Pineal Res. 2019;67(3): e12598. 10.1111/jpi.12598. [DOI] [PubMed] [Google Scholar]
- 37.Guo Y, Wu R, Gaspar JM, Sargsyan D, Su ZY, Zhang C, et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis. 2018;39(5):669–80. 10.1093/carcin/bgy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das D, Karthik N, Taneja R. Crosstalk between inflammatory signaling and methylation in cancer. Front Cell Dev Biol. 2021;9: 756458. 10.3389/fcell.2021.756458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.