Abstract
Differences in virion RNA dimer stability between mature and protease-defective (immature) forms of human immunodeficiency virus type 1 (HIV-1) suggest that maturation of the viral RNA dimer is regulated by the proteolytic processing of the HIV-1 Gag and Gag-Pol precursor proteins. However, the proteolytic processing of these proteins occurs in several steps denoted primary, secondary, and tertiary cleavage events and, to date, the processing step associated with formation of stable HIV-1 RNA dimers has not been identified. We show here that a mutation in the primary cleavage site (p2/nucleocapsid [NC]) hinders formation of stable virion RNA dimers, while dimer stability is unaffected by mutations in the secondary (matrix/capsid [CA], p1/p6) or a tertiary cleavage site (CA/p2). By introducing mutations in a shared cleavage site of either Gag or Gag-Pol, we also show that the cleavage of the p2/NC site in Gag is more important for dimer formation and stability than p2/NC cleavage in Gag-Pol. Electron microscopy analysis of viral particles shows that mutations in the primary cleavage site in Gag but not in Gag-Pol inhibit viral particle maturation. We conclude that virion RNA dimer maturation is dependent on proteolytic processing of the primary cleavage site and is associated with virion core formation.
The human immunodeficiency virus type 1 (HIV-1) positive-stranded RNA genome, which forms a dimer within the virion, is part of a ribonucleoprotein (RNP) complex encased in a capsid shell and enveloped in the viral membrane studded with glycoproteins. HIV-1 virions are initially produced as immature particles that undergo maturation during or shortly after budding from the plasma membrane (12). The immature virion does not contain the internal core structure found in mature virions but instead has a thick electron-dense spherical structure directly underneath the virion envelope (31). Conversion of immature to mature virions is termed maturation, and it requires proteolytic cleavage of the HIV-1 structural polyproteins Gag and Gag-Pol by viral protease (PR). This proteolysis is initiated at the membrane of the infected cell during virion budding and release and improves the release of progeny viruses (30).
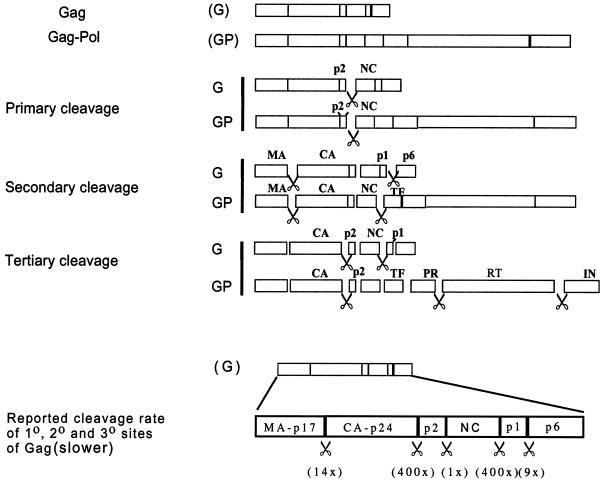
The main structural proteins of retroviruses are synthesized as a single polyprotein, which in the case of HIV-1 is called Gag or Pr55gag (48). The viral replication enzymes PR, reverse transcriptase (RT), and integrase (IN) are synthesized as a second polyprotein, Gag-Pol, which is N terminally colinear with Gag and is derived from ribosomal frameshifting at a rate of 5% (28). These polyproteins associate with other viral proteins and genomic RNA to form the immature virion. Within the virion, Gag is cleaved into matrix (MA), capsid (CA), nucleocapsid (NC), p6, and two spacer peptides, p2 and p1. Gag-Pol also contains MA and CA but its C-terminal cleavage products are p2, NC, transframe protein (TF), PR, RT, and IN (Fig. 1). The substrate specificity and the kinetics of cleavage by HIV-1 protease have previously been described (26, 41, 44). Cleavage sites differ in their amino acid composition, but in all cases the minimum substrate length required to permit cleavage by PR is roughly 7 amino acids (aa) (7). Many of these sites are distributed close to the CA region encoded by Pr55gag. Cleavage of CA from its neighbors within Gag polyprotein is necessary for core condensation and conical capsid shell formation during virion maturation (50, 51).
FIG. 1.
Schematic representation of sequential proteolytic processing of HIV-1 Gag (G) and Gag-Pol (GP) precursor proteins by viral PR and the initial proteolysis rate of the primary (1°), secondary (2°), and tertiary (3°) cleavage sites of Gag (slower) adapted from Pettit et al. (42, 43). The scissors symbol represents corresponding locations cut by PR in HIV-1. In this study, constructs containing mutations that do not allow the processing of the primary (p2/NC), secondary (MA/CA and p1/p6), and tertiary (CA/p2) cleavage sites, from two different sources, were analyzed for their protein profiles and genomic RNA dimer maturation.
Cleavage of Gag and Gag-Pol is essential for maturation and HIV-1 infectivity; mutation or inhibition of PR abolishes production of infectious viruses (42, 51). The order and contribution of individual cleavages have been studied in a cell-free system, as well as in tissue culture systems. These studies have revealed that in HIV-1, the proteolytic processing of Gag occurs in an orderly fashion with primary, secondary, and tertiary sites being sequentially cleaved by PR (Fig. 1) (43, 51). Gag proteolysis is thought to be regulated, in part, by the cleavage of the p2 spacer peptide (43). This peptide has also been shown in a cell-free system to act as the morphologic switch region during maturation of the virus particle (19). Moreover, using the protease inhibitor Ro 31-8959, Lindhofer et al. (35) have observed production of intermediate products via autolytic processing of Gag-Pol precursor protein, suggesting that the cleavage of Gag-Pol also occurs in a defined sequence (Fig. 1).
During viral particle maturation, the packaging and rearrangement of the genomic RNA rely on RNA-protein interactions (3, 36). A chaperone activity for Gag protein has been described (16). In addition, the NC sequence within Gag protein binds to genomic RNA and facilitates RNA packaging (5, 13, 23). In HIV-1, at least one zinc finger of the NC is required for efficient RNA packaging to occur (4, 5, 11). Mutations in the first zinc finger markedly reduce genomic RNA packaging into virions, while mutations in the second zinc finger only decrease RNA packaging by 30% (22).
Genomic RNA packaged in HIV-1 virions is dimeric. The inactivation of viral protease decreases the stability of the RNA dimer, suggesting that proteolytic processing and RNA dimer maturation are interrelated (18, 19). In addition, deletion of the dimer initiation sequence, a region essential for RNA dimerization, results in delayed processing of p2 from CA and is associated with a reduction in virus infectivity (33, 34). Other authors have suggested that noncoding viral RNA leader sequences may also accelerate proteolytic processing of the precursor proteins (47; for a review, see reference 50). In other retroviruses, e.g., Rous sarcoma virus, mutation of NC leads to the production of noninfectious Rous sarcoma virus, which contains unstable virion RNA dimers (39; for a review, see reference 31). Moreover, whether in its mature (p7) or precursor (p15) form, HIV-1 NC induces maturation of retroviral dimeric RNA in a cell-free system (16, 17, 40). From these studies it appears likely that the NC sequence within Gag or the mature NC itself stabilizes HIV-1 RNA dimers. However, the involvement of HIV-1 NC in RNA dimerization during viral assembly has not been directly examined.
We report here that a mutation in the primary cleavage site (p2/NC) of the HIV-1 genome which prevents the cleavage of CA-p2 from NC markedly decreases genomic RNA dimer stability. Mutations in the secondary (MA/CA and p1/p6) and tertiary (CA/p2) cleavage sites do not alter RNA dimer stability, suggesting that stable RNA dimer formation occurs either simultaneously or immediately after proteolysis at the primary cleavage site of Gag. In addition, using a cotransfection system, we show that for HIV-1 the cleavage of the p2/NC site in Gag is more important than cleavage of the same site in Gag-Pol for RNA dimerization and the condensation of the virion core. This study suggests that the free N terminus of NC is likely to be important for virion RNA dimer maturation and virion core capsid formation.
MATERIALS AND METHODS
DNA plasmids.
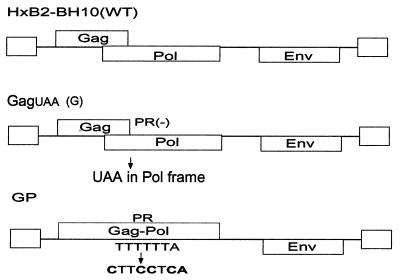
The cleavage site HIV-1 mutants MA/CA, p1/p6, and CA/p2, containing a P1 Ile substitution that inhibits cleavage, were previously described by Pettit et al. (42). A full-length HIV-1 proviral clone, NL4.3, was used as a control. The single cleavage site mutant CA2 (p2/NC), double cleavage site mutant CA5 (CA/p2 and a CA cryptic site mutation), and triple cleavage site mutant CA6 (p2/NC-CA/p2-cryptic site mutation) have been previously described by Wiegers et al. (51; see Table 1 for details). For the cotransfection study, the full-length wild-type (WT) HIV-1 plasmid used was HXB2-BH10 (49). The Gag-Pol expression plasmid (GP) was constructed using PCR stitch mutagenesis as previously described (37). The frameshift mutation in GP allows continuous expression of Gag-Pol and bypasses the Gag termination codon. The GagUAA (G) plasmid was also constructed by using PCR stitch mutagenesis in order to introduce a stop codon within the frame of the Pol protein. This stop codon insertion terminates Gag-Pol synthesis in aa 13 of PR. As a result, this mutation does not allow the synthesis of full-length Gag-Pol and the expression of a functional PR. HXB2-BH10 was used for the construction of both the G and the GP clones.
TABLE 1.
Cleavage site mutants used in this studya
| Mutant | Cleavage site(s) mutated | Source of mutants and/or references |
|---|---|---|
| pNL 4.3 | NIH reagent program | |
| MA/CA | Secondary | Pettit and Swanstrom, unpublished |
| CA/p2 | Tertiary | Pettit and Swanstrom, unpublished |
| p1/p6 | Secondary | Pettit and Swanstrom, unpublished |
| CA2 | Primary (p2/NC) | Wiegers et al. (36) |
| CA5 | Tertiary (CA/p2 and cryptic site) | Wiegers et al. (36) |
| CA6 | Primary, tertiary, and cryptic site |
For detailed information about the construction of the mutants see the references cited in column 3, and for sequential classification of sites see Fig. 1.
The DNA regions containing mutations within the primary cleavage site (p2/NC) or in primary and late cleavage sites (p2/NC and CA/p2) were removed from CA2 or CA6 mutants and cloned into the G and GP plasmids via SpeI and ApaI restriction enzyme sites.
Virus production.
The production of WT and mutant HIV-1 viral particles was achieved by transfection of 10 μg of proviral DNA of each plasmid into 293T cells using a calcium phosphate method as previously described (29).
The production of HIV-1 particles containing mutations in the cleavage sites of Gag or Gag-Pol was achieved by cotransfection of either intact G or GP DNA plasmids with the mutated counterparts maintaining the 20:1 GagUAA/GP ratio (10 μg of G to 0.5 μg of GP) found in natural infections. Cotransection of intact G and GP vectors resulted in the 20:1 Gag/Gag-Pol protein expression ratio in the virus-producing cells. An enhanced green fluorescent protein (EGFP; Clontech) reporter plasmid (2 μg) was added to the DNA mixture to determine the transfection efficiency.
Supernatants were collected 36 h posttransfection and centrifuged for 30 min at 4°C and 3,000 rpm (Beckman) to remove cellular debris. The clarified supernatants were either frozen at −70°C or used immediately for further analysis. Cells were washed twice with either phosphate-buffered saline (PBS) or 1× Tris-buffered saline (TBS) buffer (50 mM Tris, pH 7.4; 150 mM NaCl), followed by protein extraction using lysis buffer containing 1× TBS, 10 μl of Nonidet P-40/ml, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin. Cell lysates were collected and stored at −20°C for later use.
Intracellular viral protein analysis.
Cell lysates were rapidly frozen and thawed three times to weaken the cellular membrane. Cell debris was subsequently removed by centrifugation for 30 min at 4°C at 3,000 rpm (Beckman). The transfection efficiency of the samples was determined by measuring the level of EGFP from the reporter plasmid using a Bio Imaging Analyzer (Fuji Photo Film Co.). Intracellular viral protein from each sample normalized for equivalent levels of EGFP was mixed with 3 μl of sample buffer (100 mM Tris, pH 6.8; 3% sodium dodecyl sulfate [SDS]; 33% glycerol; 0.03% bromophenol blue), denatured for 10 min at 95°C, and resolved by SDS–10% polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred to a nitrocellulose membrane (Amersham) for Western blot analysis. The membrane was blocked for 2 h in 3% casein dissolved in 2× TBS containing 0.3% Tween 20 (TBST) and probed overnight with pooled HIV-1-seropositive patient sera or anti-p24/CA monoclonal antibody (MAb) (NEN). After three washes with 1× TBST buffer the membrane was incubated with anti-human or anti-mouse horseradish peroxidase-conjugated secondary antibody (Dako) for 2 h at room temperature. An enhanced chemiluminescence (ECL) technique was used for detection of HIV-1 proteins present in the intracellular lysates (Amersham). Results were visualized by autoradiography.
Virion purification and protein analysis.
Clarified supernatants from transfected cells were purified and concentrated by ultracentrifugation through a 20% sucrose cushion in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0) by using a Beckman L-90 ultracentrifuge (SW41 rotor) at 35,000 rpm for 1 h at 4°C. Pellets were resuspended in 50 μl of TBS lysis buffer.
Analysis of virion protein profile.
Equal amounts of virion protein normalized by dot blotting (46) from each sample were mixed with 3 μl of sample buffer containing 5 mM β-mercaptoethanol and heated for 10 min at 95°C. Virion proteins for analysis of the HIV-1 protein pattern and detection of p24-CA were then resolved by SDS–10% PAGE as described above. The resolved virion protein samples were transferred onto nitrocellulose membranes for Western blot analysis as described above. For the detection of p7-NC protein, normalized and denatured virion proteins were resolved in a 16.5% Tricine gel (Bio-Rad) under electrophoresis conditions with a Tris-Tricine buffer. Resolved proteins were transferred to a polyvinylidene difluoride membrane. An ECL technique was used for the detection of mature and intermediate NC proteins present in the virions (Amersham). Results were visualized by autoradiography.
Analysis of virion RNA dimerization.
Virion pellets were resuspended in 500 μl of dimeric RNA lysis buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1% SDS, 50 mM NaCl, and 10 U of proteinase K), lysed for 30 min at room temperature, phenol-chloroform extracted, and isolated for melting-curve analysis as previously described (18, 19).
Similar amounts of genomic RNA were used to analyze the stability of the virion RNA dimer in each preparation by heating the samples at the indicated temperatures for a period of 10 min, followed by a quick chill in ice. Heat-denatured dimeric and monomeric RNAs were separated by electrophoresis in a 1% native agarose gel in 0.5× Tris-borate-EDTA buffer and transferred overnight onto a nitrocellulose (Hybond N) membrane (Amersham). The membrane containing the RNA samples was air dried for 2 h at room temperature and exposed to UV light for 90 s to allow cross-linking to occur. The membrane was blocked for 1 h at 42°C with 10 ml of hybridization buffer (30). Dimeric and monomeric RNAs were incubated overnight with a 32P-labeled riboprobe, which is complementary to the 5′ end of the HIV-1 genomic RNA sequences, as previously described (46). The riboprobe was synthesized by linearizing the pGEM7z HIV-1 plasmid with BamHI, followed by T7 RNA polymerase-TP (NEN). After the probing, the membrane was washed once for 30 min with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS buffer and twice for 30 min with 0.2× SSC–0.1% SDS buffer. The results were visualized by autoradiography. Migration of WT RNA as well as RNA derived from the cotransfection of the intact G and GP plasmids served as controls to determine the effect of cleavage site mutations on RNA dimerization.
TSEM.
Transfected 293T cells were harvested at appropriate times posttransfection and processed for thin-section electron microscopy (TSEM) as described previously (32). Briefly, cells were washed in cacodylate (CAC) buffer (pH 7.2) and pelleted by centrifugation at 500 × g for 5 min. The cell pellet was then fixed in 2% glutaraldehyde-CAC buffer for 1 h at 4°C. After several washes in CAC buffer, the pellet underwent secondary fixation in 1% osmium tetroxide-CAC buffer for 1 h at 4°C. Pellets were rinsed in distilled water, dehydrated in graded ethanol, cleared in propylene oxide, and embedded in Spurr resin (Ladd Research, Inc., Williston, Vt.). Ultrathin sections with silver and gold interference color were mounted on uncoated 200-mesh copper grids and stained with uranyl acetate and lead citrate. The sections were then examined by using a CM12 electron microscope (Philips, Eindhoven, The Netherlands).
RESULTS
Mutations in the Gag and Gag-Pol cleavage sites of HIV-1 inhibit proteolysis by PR.
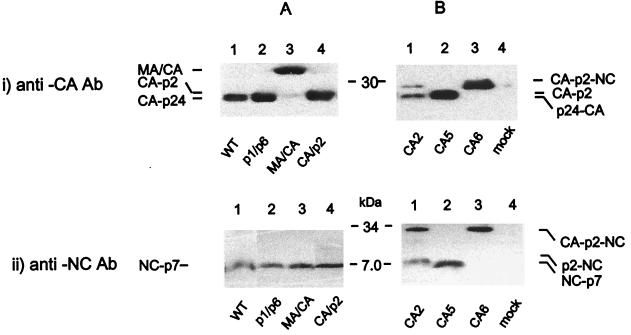
Cells transfected with either the WT, NL4.3, or the cleavage site mutants of this full-length molecular clone had protein profiles similar to those previously described (51; S. Pettit and R. Swanstrom, unpublished data). Virion protein profiles of cleavage site mutants probed with anti-p24 MAb were in agreement with previous reports (51; Pettit and Swanstrom, unpublished) (Fig. 2). Briefly, the WT (NL4.3) and the p1/p6 mutant expressed similar levels of CA-p24 (Fig. 2, panel i/A, lanes 1 and 2). Mutant MA/CA yielded an MA-CA intermediate product with a relative molecular mass of 39 kDa (Fig. 2, panel i/A, lane 3) as a result of a point mutation that hinders the release of the N terminus of the capsid protein from matrix. As previously observed (51; Pettit and Swanstrom, unpublished), mutants CA/p2 and CA5 yielded similar levels of a CA-p2 intermediate (CA extended by 14 aa) and no mature CA-p24 (Fig. 2, panel i/A, lane 4, and panel i/B, lane 2). The CA2 mutant expressed less CA-p24 and a CA-NC protein complex (Fig. 2, panel i/B, lane 1) due to inefficient processing of the N terminus of NC. This resulted in the formation of a p33 CA-p2-NC processing intermediate. The presence of CA-p24 suggests that the p2/NC mutation hindered but did not block the release of an N-terminally extended NC protein from the CA-p2-NC (p25) intermediate. As expected, no CA-p24 protein was detected for CA6 (Fig. 2, panel i/B, lane 3) due to point mutations in the primary site (p2/NC), the tertiary site (CA/p2), and a further cryptic cleavage site. These mutations completely blocked the release of the NC from the CA-p2 protein complex, resulting in virions containing predominantly a CA-p2-NC processing intermediate.
FIG. 2.
Impact of mutations within the primary, secondary, and late-proteolytic processing cleavage sites of HIV-1 on the protein composition of purified virions. Viral proteins were semiquantified by Western blot analysis. Purified virions were resolved by using 10% Tris-glycine SDS-PAGE (i) and by 16.5% Tris-Tricine SDS-PAGE (ii). Resolved proteins were probed with anti-p24 MAb or anti-p7 polyclonal antibody with specific activity to NC protein, as described in Materials and Methods.
WT, p1/p6, MA/CA, CA/p2, and CA5 mutants expressed similar levels of p7-NC in the virions (Fig. 2iiA, lanes 1 to 4, and iiB, lane 2). The mutation in the p2/NC cleavage site resulted in the release of a p9 (NC-p2) intermediate and no mature p7-NC, whereas combined mutations in the p2/NC and CA/p2 cleavage sites blocked processing of p2 from NC and CA from p2, giving rise to a p33 protein complex (Fig. 2iiB, lanes 1 and 3, respectively). The p33 protein complex was also observed in the p2/NC mutant virions, supporting the results obtained from the detection of CA-p24 described above.
A primary cleavage site mutation in Gag/Gag-Pol leads to reduced stability of the HIV-1 RNA dimers.
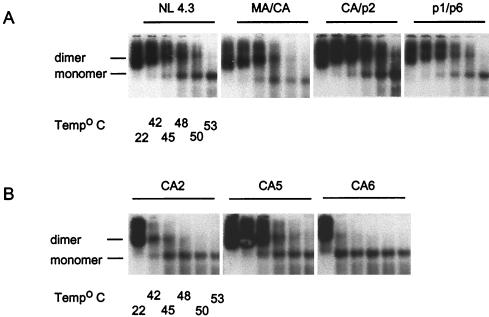
Dimeric RNA was observed in all mutant viruses, regardless of the nature of the mutation introduced in the full-length HIV-1 genome (Fig. 3). However, the stability of the dimeric RNA was markedly reduced when mutations were introduced in the primary cleavage site alone (CA2, Fig. 3B) or in both primary and tertiary cleavage sites (CA6, Fig. 3B). In these preparations, virion RNA dimers began to dissociate into monomers at temperatures as low as 42°C; in fact, almost all of the virion RNAs were detected as monomers when the samples were heated to 48°C. In contrast, HIV-1 mutants containing mutations in secondary (MA/CA and p1/p6, Fig. 3A) or tertiary proteolytic cleavage sites (CA/p2, Fig. 3A, and CA5, Fig. 3B) exhibited the same RNA dimer stability as that of the WT counterpart (NL 4.3, Fig. 3A); in these preparations, virion RNA dimers could still be detected at higher dissociation temperatures (48 to 50°C).
FIG. 3.
Effects of mutations within the primary, secondary, and late cleavage sites on virion RNA dimerization. The impact of mutations within proteolytic processing cleavage sites on genomic RNA dimerization was determined by using melting-curve and electrophoretic analysis of WT and mutant dimers. Virion RNA was resuspended in RNA dimerization buffer and heat denatured for 10 min at the indicated temperatures. Dimers and monomers were electrophoresed in a 1% native agarose gel and probed with an HIV-1 riboprobe, as described in Materials and Methods. (A) RNA dimerization analysis of genomic RNA isolated from WT HIV-1 (NL4.3), CA/p2, MA/CA, and p1/p6 mutant virions. (B) RNA dimerization analysis of genomic RNA isolated from CA2, CA5, and CA6 mutant virions.
Cleavage site mutations in Gag affect the HIV-1 virion protein profiles much more than the same mutations in Gag-Pol.
We have shown in Fig. 3 that a single (p2/NC) or a triple mutation (p2/NC, CA/p2, and in a cryptic site) block full processing of CA and NC proteins and destabilize virion RNA dimers. The relative contribution to this effect of the respective cleavage sites in Gag versus the corresponding sites in Gag-Pol is unknown. To answer this question, we have introduced either the p2/NC cleavage site mutation (CA2) or the combined p2/NC, CA/p2, and a cryptic site mutation (CA6) into either G or the GP expression vector (Fig. 4). The resultant DNA constructs were designated GCA2, GCA6, GPCA2, and GPCA6, respectively. The Gag (G) expression construct was then transfected with GPCA2 alone or in combination with GPCA6 mutant, while the Gag-Pol (GP) expression construct was transfected with GCA2 alone or in combination with GCA6.
FIG. 4.
Schematic representation of WT control and G and GP plasmid DNA used in cotransfections for the production of viral particles containing mutations in cleavage sites in either Gag or Gag-Pol. WT HXB2-BH10 was used for the construction of both the GagUAA (G) and the GP clones. The G plasmid was constructed by using PCR stitch mutagenesis in order to introduce a stop codon within the frame of the Pol protein. The Gag-Pol expression plasmid (GP) has been previously described (37). The frameshift mutation introduced here allows continuous expression of Gag-Pol and bypasses the Gag termination codon. The DNA regions containing mutations within the primary cleavage site (p2/NC) or in primary and tertiary cleavage sites (p2/NC and CA/p2) were removed from CA2 or CA6 mutants and cloned into the G and GP plasmids via SpeI and ApaI restriction enzyme sites, respectively. The resultant mutants were termed GCA2, GCA6, GPCA2, and GPCA6 (see also Table 2). The G plasmid was then cotransfected with either GPCA2 alone or in combination with GPCA6 while the GP plasmid was cotransfected with either GCA2 alone or in combination with GCA6 in 293T cells.
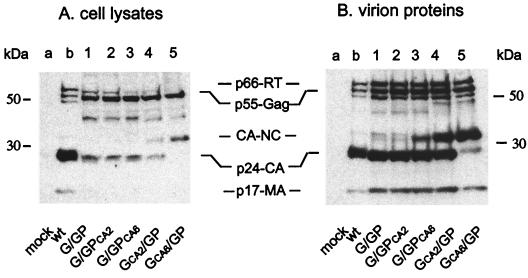
Since cotransfection of the WT Gag and WT Gag-Pol expression vectors (G/GP) produced viral particles with the same intracellular viral protein and virion protein profiles as WT virus (Fig. 5, lanes b), G/GP viruses were used as controls in all of the subsequent cotransfection studies (Fig. 5, lanes 1). Intracellular viral proteins and virion proteins derived from cells cotransfected with GCA2/GP constructs yielded levels of CA-p24 and CA-NC that were similar to those observed for the CA2 mutation in a proviral clone (Fig. 5, lanes 4), while G/GPCA2 intracellular and virion proteins yielded undetectable levels of CA-NC (Fig. 5, lanes 2). Accordingly, almost exclusively, CA-NC was observed upon cotransfection of GCA6 /GP (Fig. 5, lanes 5), while significantly lower levels of intracellular viral protein and virion CA-NC protein complex were yielded upon cotransfection with G/GPCA2 or G/GPCA6 (Fig. 5, lanes 2 and lanes 3, respectively).
FIG. 5.
Effect of a primary or combined primary and late cleavage site mutations within Gag or Gag-Pol on intracellular viral proteins and virion protein profiles of HIV-1. Intracellular viral proteins were standardized by EGFP, and total virion protein levels were standardized by dot blot analysis (data not shown). Cotransfected cell lysates (A) and purified virions (see Materials and Methods) (B) were resolved by SDS–10% PAGE. Resolved proteins were probed using sera from HIV-1-infected individuals, as described in Materials and Methods. Lanes 1, show G/GP cellular viral proteins and virion protein profiles, respectively. Lanes 2 and lanes 3 show cellular viral proteins and virion protein profiles of mutant viruses containing a primary CA2 (p2/NC) or a combined primary and late cleavage site mutation CA6 (p2/NC, CA/p2, and a cryptic site mutation) in Gag-Pol, respectively. Lanes 4 and lanes 5 show cellular viral proteins and virion protein profiles of mutant viruses containing a primary CA2 (p2/NC) or a combined primary and late cleavage site mutation CA6 (p2/NC and CA/p2) in Gag, respectively. Lanes a and lanes b show mock- and WT-transfected controls. The amounts of WT HIV-1 intracellular and virion proteins were independent of the other proteins tested and were used in these analysis only to compare HIV-1 protein patterns probed by HIV-1 sera.
The primary cleavage site in Gag is more important for RNA dimerization than the corresponding site in Gag-Pol.
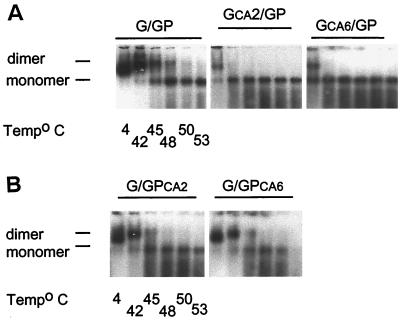
Genomic RNA isolated from viral particles derived from cotransfection of plasmids G and GP was dimeric and exhibited the same stability as virion RNA dimers isolated from WT NL4.3 virus (compare Fig. 6 and Fig. 3A, respectively). Genomic RNA isolated from viral particles containing the primary cleavage site mutation in Gag (GCA2/GP) started to dissociate into monomers at room temperature, while dimeric RNA from viral particles containing the corresponding mutation in Gag-Pol (G/GPCA2) was as heat stable as WT G/GP RNA dimers, with a dissociation temperature of 45 to 48°C (Fig. 6A and B, respectively). The introduction of the combined mutations in Gag (GCA6/GP) further decreased dimer stability with more than 50% of the RNA dimers dissociating into monomers at room temperature (Fig. 6A), while the corresponding mutations in Gag-Pol (G/GPCA6) caused a negligible decrease in the dimer stability of the RNA (Fig. 6B). These results show that the primary cleavage site in Gag-Pol plays a less important role in dimer RNA stability than the corresponding site in Gag.
FIG. 6.
Effect of cleavage site mutations in either Gag or Gag-Pol on virion RNA dimerization. The impact of mutations in the proteolytic processing cleavage sites within Gag or Gag-Pol on genomic RNA dimerization was examined by melting curve and electrophoretic analysis of WT and mutant dimers. Dimers and monomers were electrophoresed in 1% native agarose gel and probed with an HIV-1 riboprobe, as described in Materials and Methods. (A) RNA dimerization analysis of genomic RNA isolated from WT (G/GP) and mutant G/GPCA2 or G/GPCA6 virions. (B) RNA dimerization analysis of genomic RNA isolated from GCA2/GP and GCA6/GP mutant virions.
Mutations in the primary cleavage site of Gag (but not in Gag-Pol) inhibit core formation.
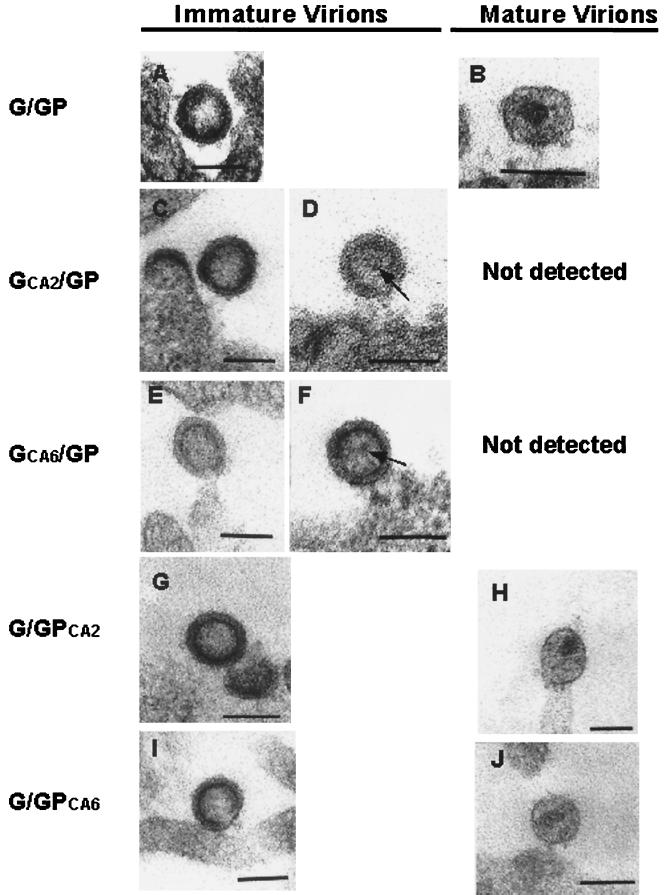
Immature (Fig. 7A) and mature (Fig. 7B) forms of virus detected on or near the surface of the cells transfected with G/GP DNA constructs were similar in morphology to their WT HIV-1 counterpart. The G/GP immature virions were observed as round or ovoid enveloped particles containing an inner ring and having a hollow appearance (Fig. 7A). In contrast, the mature G/GP virions appeared as round or ovoid spherical enveloped particles containing a mature electron-dense core, which appeared cone shaped or round depending on the angle of sectioning (Fig. 7B). In cells cotransfected with GCA2/GP (Fig. 7C and D) and GCA6/GP (Fig. 7E and F) DNA constructs, only immature forms of the virus were detected; at times, an internal structure similar to that reported by Hockley et al. (27) was observed within these immature virions (Fig. 7D and F). In preparations that were derived from cells cotransfected with G/GPCA2 and G/GPCA6 constructs, both immature (Fig. 7G and I) and mature (Fig. 7H and J) virions were detected; the ratios of immature to mature virions in these preparations were similar to those derived from the G/GP preparation (data not shown). Measurements of virus particles (Table 2) showed no significant differences between mutant and WT virions either in the mature or in the immature form.
FIG. 7.
Electron microscopic analysis of the mutant viral particles. 293T cells were cotransfected with G/GP (A and B), GCA2/GP (C and D), GCA6/GP (E and F), GCA2/GP (C and D), and GCA6/GP (E and F). DNA constructs were harvested at 36 h posttransfection and processed for TSEM as described in Materials and Methods. Both immature (A, G, and I) and mature (B, H, and J) forms of the virus were detected on or near the surface of the cells transfected with G/GP (control), G/GPCA2, or G/GPCA6. Only immature virions were observed within the GCA2/GP (C and D) and GCA6/GP (E and F) preparations. (D and F) At times, an internal structure (arrow) within the inner ring of the immature virion was seen. Two diameters were measured for each viral particle: the longest and shortest diameters, roughly at right angles; the average of the two values was then taken as the diameter of the viral particle. Bars, 100 nm.
TABLE 2.
Dimensions of mature and immature virions as determined by electron microscopya
| WT or mutant prepn | Avg virus diam (nm) ± SD
|
Avg distance (nm) ± SD between two outer layers in immature particles | Avg core diam (nm) ± SD in mature viral particles | |
|---|---|---|---|---|
| Immature | Mature | |||
| G/GP | 90.2 ± 18.2 | 85.9 ± 5.4 | 11.8 ± 2.4 | 31.9 ± 5.8 |
| GCA2/GP | 107.9 ± 18.8 | 13.9 ± 2.0 | ||
| GCA6/GP | 90.3 ± 16.4 | 11.9 ± 1.4 | ||
| G/GPCA2 | 100.3 ± 9.4 | 109.6 ± 18.1 | 12.4 ± 1.8 | 36.2 ± 4.4 |
| G/GPCA6 | 97.2 ± 6.4 | 99.8 ± 11.6 | 14 ± 0.5 | 33.3 ± 4.8 |
Each measurement is an average ± the standard deviation for four to seven particles.
DISCUSSION
This study provides direct evidence that the sequential processing of Gag and Gag-Pol proteins by viral protease regulates genomic RNA maturation (i.e., RNA dimerization and stability). Virion RNA dimer maturation was dependent on the proteolysis of the primary cleavage site of Gag but independent of the processing of the secondary and tertiary cleavage sites. Furthermore, the cleavage of the p2/NC site in Gag but not Gag-Pol was critical for the formation of stable RNA dimers and condensation of the virion core. Our study supports and extends previous reports (1, 2, 14, 16, 17, 40) showing that genomic RNA dimer formation is under the control of Gag protein and that formation of N terminally cleaved NC is required for the generation of stable RNA dimers.
The cleavage of the p2/NC site in HIV-1 is critical for virion RNA dimer stability.
Previous studies have suggested that Gag/Gag-Pol PR processing may influence RNA dimerization. Fu and coworkers (18, 19) have shown that PR activity is required for RNA dimer stability. Our study shows that mutations of the primary (p2/NC) Gag/Gag-Pol cleavage site dramatically decrease the stability of virion RNA dimers. In contrast, RNA dimer stability remains unaffected by mutations in the secondary and tertiary cleavage sites. Our findings suggest that the p2-NC intermediate found in virions as a result of altered processing in the p2/NC site is unable to substitute for fully processed NC in the formation of stable virion RNA dimers. Therefore, the release of the NC protein from p2, thus freeing the N terminus of the NC protein, is essential for stable genomic RNA dimer formation. Mature NCp7 has been reported to convert unstable RNA dimers into stable dimers in a cell free system by interacting with a “kissing loop complex,” suggesting that NCp7 is an important contributor to RNA dimer formation (16, 17, 40). A similar observation has also been made for Moloney murine leukemia virus (MoMLV)-derived genomic RNA in vitro (8).
We speculate that the interaction between genomic RNA and an NC intermediate (NC-p1-p6 or p15) is sufficient for the formation of the dimeric RNA complex. However, the data suggest that the free N terminus of NC in the context of p15 is critical for the stability of these dimers in vivo and is in agreement with earlier reports from studies in a cell-free system in HIV-1 (2), as well as in MoMLV and Harvey murine sarcoma virus (17, 38). In another study, Wiegers et al. (51) have shown that a mutation in the p2/NC site results in the production of viral particles with immature cores. In addition, our data suggest that the release of the NC from p2 leads to RNA maturation. Formation of the RNP complex and condensation of the core may follow this step. Taken together, these results point to a specific role of NC for stable RNA dimer formation and condensation.
The proteolytic processing of the p2/NC cleavage site in Gag (but not Gag-Pol) is essential for RNA dimer stability and virion core formation.
We introduced the p2/NC mutation alone or in combination with the CA/p2 mutation into Gag or Gag-Pol in order to determine the contribution of these cleavage sites in Gag versus Gag-Pol in RNA dimerization and virion core formation. Our data showed a significant decrease in CA-NC processing, a dramatic effect in RNA dimer stability and immature core formation when these mutations were introduced only in Gag. Gag-Pol protein is synthesized as a result of a −1 frameshifting event resulting in a 20:1 production ratio of Gag to Gag-Pol in the virus producing cell (28). Therefore, the ratio of CA and NC derived from Gag protein to CA and NC derived from Gag-Pol protein at the assembly site of new virions should correspond with that of the WT Gag/Gag-Pol ratio (i.e., 20:1). In agreement with this ratio, our data show that the cleavage of the p2/NC site in Gag is essential for RNA dimer stability and virion core formation and further suggest that Gag-Pol proteins contribute only modestly to RNA dimer stability.
A free N terminus of NC is important for HIV-1 core formation.
NC plays diverse roles in HIV-1 replication (13). The NC domain in the Gag polyprotein facilitates RNA packaging and is involved in reverse transcription of the viral genome and possibly at later stages of viral entry (45). A later report has confirmed that mutations in the basic residues of the N terminal of NC lead to production of viral particles containing defective cores (6). Assembly studies in a cell free system have demonstrated that the MA-CA-NC complex gives rise to spherical particles and that the formation of tubular cores (in Escherichia coli) and conical cores requires only the CA-NC portion of this protein in the presence of RNA (9, 10, 21, 24, 25). Our results clearly show that in HIV-1, hindering the processing of the N terminus of NC from CA protein prevented virus maturation. The data demonstrate a strong association between stable RNA dimer formation and core maturation, as well as the importance of the N terminus of NC in both events. The presence of the RNA dimers within these immature viral particles, however, suggests that virion genomic RNA initially dimerizes as a thermally less stable structure. RNA-NC structural studies have shown that the free amino terminus of NC resides in the groove of a stem-loop RNA structure (15) which implies that this aspect of NC-RNA interaction is likely to be an important determinant for virion RNA packaging. Fuller et al. (20) have suggested that RNA may function as a scaffold in the formation of the immature spherical shell. In this case, cleavage of NC leading to the release of the RNP complex may be required for the morphologic rearrangement as it permits removal of the NC-RNA scaffold from the condensing core structure. Concomitantly, freeing the N terminus of NC appears to be necessary and may be sufficient for the formation of stable RNA dimers, which can serve as a nucleating event in core maturation.
In summary, our data suggest that correct N-terminal processing of NC during viral assembly is a requirement for HIV-1 core formation and high stability of RNA dimers. Once NC is separated from p2, precise levels of mature CA and NC proteins are generated to form the conical capsid shell and facilitate the condensation of the electron-dense RNP core. The study implies that processing of Gag polyprotein is critical for RNA dimerization.
ACKNOWLEDGMENTS
We thank Jean-Luc Darlix for useful discussions and John Mills for comments and review of the manuscript.
Miranda Shehu-Xhilaga is a recipient of an NHMRC Dora Lush Ph.D. scholarship. Suzanne M. Crowe is supported by a grant from the Australian National Council of HIV/AIDS and Related Diseases, the Australian National Centre in HIV Virology Research, and the MBC Research Fund. Johnson Mak is a recipient of an NHMRC Peter Doherty postdoctoral fellowship. This work was supported in part by NIH grant RO1-AI25321 to Ronald Swanstrom.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodefieciency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barat C, Lullien V, Schatz O, Keith G, Darlix J L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acids Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthoux L, Pechoux C, Ottoman M, Morel G, Darlix J L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6225–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billich S, Knoop M T, Hansen J, Strop P, Sedlacek J, Mertz R, Moelling K. Synthetic peptides as substrates and inhibitors of human immunodeficiency virus-1 protease. J Biol Chem. 1988;263:17905–17908. [PubMed] [Google Scholar]
- 8.Bonnet-Mathoniere B, Girard P, Muriaux D, Paoletti J. Nucleocapsid protein 10 activates dimerization of the RNA of Moloney murine leukemia virus in vitro. Biochemistry. 1996;238:129–135. doi: 10.1111/j.1432-1033.1996.0129q.x. [DOI] [PubMed] [Google Scholar]
- 9.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding site for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 13.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Darlix J L, Gabus C, Nugeyre M T, Clavel F, Barre-Sinoussi F. cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 15.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y-X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 21.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conocal models for HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 22.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelick R J, Nigida J S M, Bess J J W, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson L E, Bowers M A, Sowder R, Serabyn S A, Johnson D G, Bess J J, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockley D J, Wood R D, Jacobs J P, Garrett A J. Electron microscopy of human immunodeficiency virus. J Gen Virol. 1988;69:2455–2469. doi: 10.1099/0022-1317-69-10-2455. [DOI] [PubMed] [Google Scholar]
- 28.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 29.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kräusslich H G, editor. Morphogenesis and maturation of retroviruses. Vol. 214. Heidelberg, Germany: Springer; 1996. [Google Scholar]
- 32.Lee J Y, Marshall J A, Bowden D S. Replication complexes associated with the morphogenesis of rubella virus. Arch Virol. 1992;122:95–106. doi: 10.1007/BF01321120. [DOI] [PubMed] [Google Scholar]
- 33.Liang C, Rong L, Cherry E, Kleiman L, Laughrea M, Wainberg M A. Deletion mutagenesis within the dimerization initation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J Virol. 1999;73:6147–6151. doi: 10.1128/jvi.73.7.6147-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindhofer H, von der Helm K, Nitschko H. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T lymphocytes. Virology. 1995;214:624–627. doi: 10.1006/viro.1995.0074. [DOI] [PubMed] [Google Scholar]
- 36.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Prasad V R, Parniak M A, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 38.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Méric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muriaux D, De Rocquigny H, Roques B, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 41.Oroszlan S, Luftig T B. Retroviral proteinase. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 42.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The P2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettit S C, Sheng N, Tritch R, Erickson-Vitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 44.Pettit S C, Simsic J, Loeb D, Everitt L, Hutchison III C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the p1 amino acids. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 45.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shehu-Xhilaga M, Crowe S M, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheng N, Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom R, Wills J W. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 49.Terwilliger E, Sodroski J G, Rosen C A, Haseltine W A. Effects of mutations within the 3′ open reading frame region of human T-cell lymphotropic virus type III (HTLV-III/LAV) on replication and cytopathogenicity. J Virol. 1986;60:754–760. doi: 10.1128/jvi.60.2.754-760.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–132. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 51.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]