Abstract
Background
Microglia (MG) are myeloid cells of the central nervous system that support homeostasis and instigate neuroinflammation in pathologies. Single-cell RNA sequencing (scRNA-seq) revealed the functional heterogeneity of MG in mouse brains. Microglia are self-renewing cells and inhibition of colony-stimulating factor 1 receptor (CSF1R) signaling depletes microglia which rapidly repopulate. The functions of repopulated microglia are poorly known.
Methods
We combined scRNA-seq, bulk RNA-seq, immunofluorescence, and confocal imaging to study the functionalities and morphology of repopulated microglia.
Results
A CSRF1R inhibitor (BLZ-945) depleted microglia within 21 days and a number of microglia was fully restored within 7 days, as confirmed by TMEM119 staining and flow cytometry. ScRNA-seq and computational analyses demonstrate that repopulated microglia originated from preexisting progenitors and reconstituted functional clusters but upregulated inflammatory genes. Percentages of proliferating, immature microglia displaying inflammatory gene expression increased in aging mice. Morphometric analysis of MG cell body and branching revealed a distinct morphology of repopulated MG, particularly in brains of old mice. We demonstrate that with aging some repopulated MG fail to reach the homeostatic phenotype. These differences may contribute to the deterioration of MG protective functions with age.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-024-03242-0.
Keywords: Microglial heterogeneity, CSF1R inhibitors, Microglia repopulation, ScRNA-seq, Transcriptomics, Aging
Background
Microglia (MG) are myeloid cells in the central nervous system (CNS) which together with perivascular, choroid plexus, and meningeal macrophages play a crucial role in maintaining CNS homeostasis [1, 2]. Microglia are instrumental in maintaining brain homeostasis during development, aging, and disease. Fate mapping, multiple marker imaging, and single-cell transcriptomic studies demonstrated that during disease, aging or injury, microglia undergo context-dependent transcriptional, morphological, and functional reprogramming acquiring beneficial or deleterious functions [3–6].
Microglia are long-living cells, and their survival and proliferation are controlled by colony-stimulating factor-1 (CSF-1) [7]. Microglia depletion protocols have been widely exploited to better understand their roles in CNS homeostasis. CSF1R inhibitors (e.g., PLX3397 which crosses the blood–brain barrier, BBB) delivered for several weeks in the diet deplete MG which rapidly repopulate the CNS reaching similar cell densities, morphologies, and gene expression profiles as those of intact MG [8]. The brain volume, blood-brain barrier (BBB) and neuroinflammation markers were not affected by short-term (7 days) or long-term (2 months) depletion of MG with PLX3397. The absence of the MG did not affect mouse behavior or reduce performance in standard anxiety, locomotion, learning or memory tasks [9]. Fate-mapping approaches and parabiosis experiments excluded the blood origin of repopulated MG and demonstrated that all repopulated MG originate from the few surviving MG by proliferation [10]. Transient nestin expression was detected in the new MG, but none of the repopulated MG originated from nestin-positive non-microglial cells [10]. Repopulated MG had larger cell bodies and less complex extension branching, which resolved over time. LPS (lipopolysaccharide)-induced profiles of inflammatory gene expression in repopulated MG were similar to control ones. The mice subjected to three rounds of depletion/repopulation (7/7 days) reached a complete repopulation, but failed to replenish the MG after one more round of depletion [11].
Advances in single-cell RNA sequencing (scRNA-seq) allow the integration of comprehensive information about different molecular states, the inference of functional phenotypes, and the deduction of putative relationships between individual cells. ScRNA-seq studies revealed considerable heterogeneity of brain myeloid cells (mostly MG) in naïve mice, including several functional states of MG [12].
We sought to determine whether the observed MG heterogeneity is reconstituted after repopulation. For this purpose, we employed scRNA-seq to track transcriptomic changes in individual cells and confocal imaging to determine the morphology of repopulated MG in young and old mice. We demonstrated that after depletion the remaining MG proliferate, replenish the brain after cessation of CSF1Ri treatment, and restore the functional heterogeneity. However, the reestablishment of the homeostatic MG phenotype is less effective as the brain ages and morphological alterations suggest MG abnormalities in older mice.
Materials and methods
Animals and treatment
All experimental procedures involving animals were approved by the First Local Ethics Committee for Animal Experimentation in Warsaw (#836/2019; #1364P1/2022). Young (3 months old), aging (12 months old) or aged (18–22 months old) female and male GFP-M::Cx3cr1-CreERT2fl/fl::Rosa26-tdTomatotg/tg mice were housed at the Nencki Institute, Poland. The animals were kept in individually ventilated cages, with free access to food and water, at a temperature of 21–23 °C, and 50–60% humidity, under a 12 h/12 h day and night cycle. The animals were habituated for 3–4 days by being placed individually in a cage for 3 h, where a small amount (~ 100 µL) of the peanut butter (PB) was provided. The mice were fed with BLZ-945 (MedChemExpress, HY-12768) at a dose of 200 mg/kg b.w. daily in 100 µL PB (Peanut butter, GoOn, Santé); the controls received PB. BLZ-945 was delivered for 14 or 21 days to establish the kinetics of depletion; repopulation was studied 7 days after cessation of the treatment.
Tissue dissociation and isolation of CD11b+ by flow cytometry
The mice were anesthetized with 3% isoflurane (Iso-Vet), an overdose of ketamine (Biowet Pulawy), and xylazine (Sedazin, Biowet) (160 mg/kg and 20 mg/kg of body weight, respectively) via i.p. injection, followed by perfusion with cold PBS. The brains were collected and placed in cold HBSS (without Ca2+ and Mg2+). In the first experiment, a whole hemisphere was isolated for flow cytometry experiments, and the remaining hemisphere was stored in cold PFA for 48 h. Subsequently, the brains were transferred to 30% sucrose (w/v) in PBS for 48 h and frozen in Tissue Freezing Medium (Leica) at − 80 °C.
For flow cytometry, tissue was minced, transferred onto c-tubes containing 1950 µL DMEM supplemented with 50 µL of Deoxyribonuclease I (Sigma Aldrich). Dissociation was performed with the MACS dissociator with heaters (Militenyi Biotec) for 22 min and later the samples were passed through 70 nm and 40 nm strainers into 50 mL cold HBSS with ions to stop the enzymatic reaction (EasyStrainer™, BioOne). The cell suspension was centrifuged (10 min, 4 ºC, 300×g), the supernatant was removed and the pellet was resuspended in 25 mL of Myelin Gradient Buffer with 5 mL of cold PBS overlayed, followed by centrifugation (950×g, 4 ºC, 20’ with acc. 0 and deacc. 0). The pellet was resuspended in 1 mL of BD buffer (Dulbecco’s Phosphate-Buffered, Stain Buffer, FBS, BD Pharmigen™). The cells were counted using the Nucleocounter (Chemometec). The cells were suspended in the Stain Buffer (BD Pharmingen) with anti-mouse CD16/CD32 Fc Block™ 1:200 (BD Pharmigen) for 10 min. Next, anti-mouse CD11b antibody 1:250 (M1/70, BD Pharmigen) was added and cells were incubated for 20 min at 4 °C, then washed with Stain Buffer. The cells were sorted using purity precision mode on FACS Aria flow cytometer (BD FACSAria Cell sorter BD-Biosciences) into 200 μL of sterile PBS. The sorted sample was mixed by inverting the Eppendorf tubes and placed on ice, for no longer than 30 min.
Total RNA isolation from CD11b+ cells
For RNA isolation, sorted CD11b+ cells were stored in a − 80 °C freezer. The samples were thawed on ice and total RNA was isolated with the RNAeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA concentration was immediately determined using a spectrophotometer NanoDrop2000 (ThermoScientific).
Single-cell RNA-seq (scRNA-seq) experiments
Directly after sorting, the quantity and viability of CD11b+ cells were determined, and a cell suspension containing 5,000 target cells was used for further processing. Preparation of gel beads in emulsion and libraries were performed with Chromium Controller and Single-Cell Gene Expression v2 Chemistry (10 × Genomics) according to the Chromium Single-Cell 3’ Reagent Kits v2 User Guide provided by the manufacturer. Libraries’ quality and quantity were verified with a High-Sensitivity DNA Kit (Agilent Technologies, USA) on a 2100 Bioanalyzer (Agilent Technologies, USA). Next, sequencing was performed in the rapid run flow cell and paired-end sequenced (read 1–26 bp, read 2–100 bp) on a HiSeq 1500 (Illumina, San Diego, CA 92122, USA). To minimize batch effects, four scRNA-seq libraries (Rep1 to Rep4) were constructed, each combining 4 animals across 4 experimental conditions (control young, control old, repopulated young, repopulated old). For each scRNA-seq library, samples from individual experimental conditions were tagged using TotalSeq™ anti-mouse hashtag antibodies. The hashtag assignment was as follows: control young—#Ab 1, control old—#Ab 2, repopulated young—#Ab 3, repopulated old—#Ab 4:
| Antibody | Barcode | Source | Type |
|---|---|---|---|
| TotalSeq™-B0301 anti-mouse Hashtag 1 Antibody (Ab #1) | ACCCACCAGTAAGAC | Biolegend | HTO |
| TotalSeq™-B0302 anti-mouse Hashtag 2 Antibody (Ab #2) | GGTCGAGAGCATTCA | Biolegend | HTO |
| TotalSeq™-A0303 anti-mouse Hashtag 3 Antibody (Ab #3) | CTTGCCGCATGTCAT | Biolegend | HTO |
| TotalSeq™-B0304 anti-mouse Hashtag 4 Antibody (Ab #4) | AAAGCATTCTTCACG | Biolegend | HTO |
RNA sequencing
For single-cell and bulk RNAseq experiments, the quality and integrity of total RNA were assessed with an Agilent 2100 Bioanalyzer using an RNA 6000 Nano Kit (Agilent Technologies, Ltd.). For bulk RNA-seq strand-specific polyA-enriched RNA libraries were prepared using the KAPA Stranded mRNA Sample Preparation Kit according to the manufacturer’s protocol (Kapa Biosystems, MA, USA). Briefly, mRNA molecules were enriched from 500 ng of total RNA using poly-T oligo-attached magnetic beads (Kapa Biosystems, MA, USA). Obtained mRNA was fragmented and the first-strand cDNA was synthesized using a reverse transcriptase. Second cDNA synthesis was performed to generate double-stranded cDNA (dsDNA). Adenosines were added to the 3′ ends of dsDNA and adapters were ligated (adapters from NEB, Ipswich, MA, USA). Following the adapter ligation, the uracil in a loop structure of the adapter was digested by the USER enzyme from NEB (Ipswich, MA, USA). Adapters containing DNA fragments were amplified by PCR using NEB starters (Ipswich MA, USA). Library evaluation was done with Agilent 2100 Bioanalyzer using the Agilent DNA High Sensitivity chip (Agilent Technologies, Ltd.). The mean library size was 300 bp. Libraries were quantified using a Quantus fluorometer and QuantiFluor double-stranded DNA System (Promega, Madison, Wisconsin, USA). Libraries were run in the rapid run flow cell and were paired-end sequenced 2 × 76 bp on HiSeq 1500 (Illumina, San Diego, USA) in a case of scRNAseq and 2 × 151 bp on NovaSeq 6000 (Illumina, San Diego, USA) in a case of RNAseq.
Single-cell RNA-seq data preprocessing and normalization
Raw scRNA-seq data (1671 million raw reads, BCL files) were demultiplexed into libraries (Rep1 to Rep4) and converted to FASTQ files using CellRanger v3.0.1 (10 × Genomics) (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation) and bcl2fastq v2.20.0.422 (Illumina). Sequencing results were mapped to the mouse genome GRCm38 (mm10) downloaded from the 10 × Genomics website and quantified using CellRanger. The total number of cells identified by the CellRanger was 30,904. The median number of genes detected per cell was 2310, and the median unique molecular identifiers per cell was 68,228.
Further data analysis was performed in R using Seurat v4.2.0. [13]. The data were first demultiplexed by their hashtag oligonucleotides, which specified the experimental conditions. In accordance with the hashtag oligonucleotides, duplicates (two cells in one droplet) and negatives (empty droplets) were filtered out; the thresholds for finding duplicates and negatives were set for each hashtag separately using MULTIseqDemux() function. The libraries were first processed independently, following a standard Seurat workflow. Unless otherwise specified, all other quantitative parameters were set to default values.
To filter out possible empty droplets and low-quality cells, all cells with < 200 transcripts were excluded from the analysis. In addition, dead cells or cells of poor quality, recognized as those with > 5% of their transcripts coming from mitochondrial genes, were excluded from the downstream analysis. After applying these filters, 19,997 cells were present in the dataset. Gene expression measurements for each cell were normalized to the total number of transcripts in the cell using the “LogNormalize” method of the NormalizeData function multiplied by a default scale factor of 10,000, and the normalized values were log-transformed. For each library, the 2000 most highly variable genes were identified, using variance stabilizing transformation (“vst”). Finally, the libraries were merged into a single Seurat object using the merge() function.
The normalized counts were scaled using Seurat function ScaleData(), with all genes given as “features” argument. Dimensionality reduction was performed in the Seurat package with the PCA algorithm in the first 30 dimensions and data visualization in 2D space was performed with the UMAP algorithm. Clustering was performed via a method, that constructs a shared nearest neighbor graph by calculating the neighborhood overlap between every cell and its k.param nearest neighbors by using FindNeighbours(). FindClusters() was used with 0.5 resolution, which produced 16 separate cell clusters, by identifying clusters of cells by a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm (Louvain method).
Single-cell RNA-seq downstream analysis
For follow-up downstream analysis, we identified the cell populations present in the data. The comparison was made using ProjecTILs-3.0 R library [14] on the dataset from [15]. To further compare the experimental conditions, we set differentially expressed genes using FindMarkers() with negative binomial regression (test.use = ”negbinom”) in each condition, and compared the corresponding cell clusters. These genes were used to look for the enriched Gene Ontology terms, and pathways from KEGG and Reactome databases to identify processes, that were activated or suppressed in the compared populations. Enrichment analysis and visualizations were made using pathfindR (v.2.3.1) [16] library, which uses a protein–protein interaction network to find significant paths, that are activated during ongoing processes. To obtain an interaction network from the dataset, genes with their p-value are mapped into a subnetwork space and sorted according to the score they gain and the number of significant genes. The sorted list of significant genes was used for an enrichment analysis. Enriched terms with adjusted p values larger than the given threshold were discarded and those with the lowest adjusted p value for each term were retained. The process was computed multiple times in parallel to obtain stable results. The enriched terms were used to compute the distances between themselves, and then hierarchical clustering was implemented.
Trajectory interference
To investigate the dynamics in the MG repopulation processes and identify trajectories of cell differentiation, we used the scVelo (v0.3.2) library [17]. We compared the amount of mRNA and kinetics by using a ratio between spliced and unspliced fragments. First, velocyto (v.1.0) was used to calculate spliced and unspliced count matrices. Additional metadata was exported from the Seurat analyzed object. The velocity of RNA was estimated using dynamical modeling. The obtained results were mapped using the UMAP plot.
Identification of significant ligand–receptor pairs
We used the CellChat R library (v.1.5.0) [18] to analyze intercellular communication from MG repopulation process data via combined social network analysis and pattern recognition, on reduced dimensions of the analyzed dataset. This approach allowed us to predict patterns of incoming and outgoing signals from identified cell clusters and precisely determine their functionality.
Bulk RNA-seq analysis
Quality control assessment was performed using FastQC and the report was generated using MultiQC. Trimming adapters were performed. The sequences were aligned to the mm10/GRCm38 reference genome using STAR built-in functions from the pipeline RNA-seq-STAR-deseq2 v2.0.0. (https://github.com/snakemake-workflows/rna-seq-star-deseq2). Preprocessing, such as filtering out genes with null variation, normalization, scaling pseudocounts to counts per million, and dimension reduction to linear visualization was performed in the R environment using edgeR library. To expand information about genes, as different database annotations, gene descriptions were done using biomaRt library. During preprocessing, one sample from the controls was excluded because of significantly greater variance, as an outlier from other samples from the same condition. The linear regression model was fitted to the data using limma library. Comparisons were made between repopulated and control groups. After testing the FDR correction of the p-value was performed. The differentially expressed genes were chosen, if their log2FC absolute value was higher than one, and the adjusted p-value was lower > 0.05. We obtained 736 differentially expressed genes, 453 upregulated and 283 downregulated genes, which were used as input to the pathfindR function to search for activated or deactivated pathways in the analyzed dataset. We used the KEGG pathway database, Reactome, as well as Gene Ontologies, where we focused on Biological Processes. We selected genes of interest, whose expression was shown on boxplots.
Immunofluorescence
For immunostainings, the mice were anesthetized and transcardially perfused using PBS and 4% paraformaldehyde (PFA). The brains were removed, fixed in 4% PFA overnight, and then placed in 30% sucrose for 2–3 days before being embedded in Tissue-Tek O.C.T Compound. 10 µm thick sections were cut on a cryostat (Microm HM525, Thermo Scientific) and 3 sections were placed on a glass slide (Polysine™ slides, ThermoScientific). The slides were stored at – 80 ºC for further processing. The cryosections were first blocked in PBS containing 10% donkey serum in 0.1% Triton X-100 solution for 2 h and then incubated in 3% donkey serum buffer overnight at 4 °C with guinea pig anti-TMEM119 antibody (1:1000, Synaptic System). After that, the sections were washed in PBS and incubated with the secondary antibody Alexa Fluor 488 (1:1000, Life Technologies) for 2 h at room temperature. Finally, the sections were mounted in a mounting medium with DAPI (Vectashield Vibrance, Vector laboratories). Images were obtained using a Zeiss LSM800 Airyscan confocal microscope.
To quantify MG morphology in the immunofluorescent images, confocal images (21-μm z-stack at 3-μm intervals, Zeiss LSM 800 Airyscan, 40 × /1.3 oil objective) were acquired. The number of cell somas per frame was used to normalize all process endpoints and process lengths. In total, we analyzed 10 cells from young animals and 22 cells from the brains of old mice. Sholl analysis was automatically performed using the Imaris Software (Oxford Instruments, UK) for each cell by counting the number of intersections between MG branches and each increasing circle. We determined the number of intersections per cell and the maximum branch length. From these parameters, a ramification index was calculated to quantify the cell branching density.
Statistical analysis
For the animal and transcriptomic experiments the number of animals per group was n = 4. All quantitative data were analyzed using a one-way ANOVA or two-way ANOVA with a significance level of p < 0.05 and were performed using GraphPad Prism (version 9.3).
Results
Repopulated microglia reconstitute transcriptional heterogeneity but show upregulation of inflammatory genes
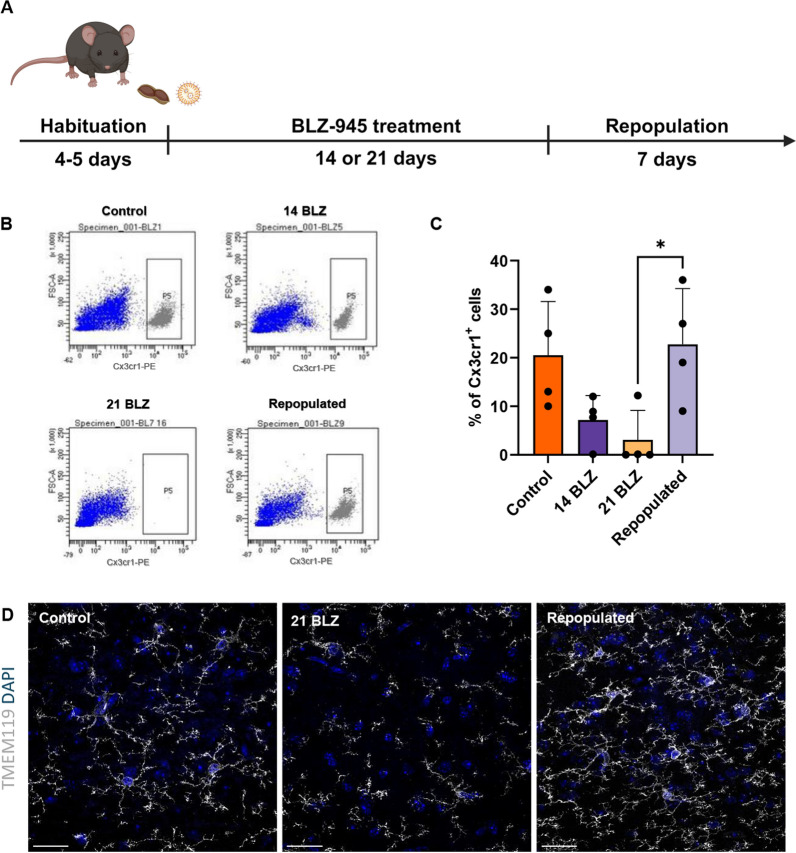
The mice were fed daily with 200 mg/kg BLZ495 (delivered in peanut butter to ensure that they consumed the whole amount) (Fig. 1A). The brains of 3-month-old mice were dissected at day 14 or 21 after BLZ-495 and 7 days after the treatment cessation. Myeloid cells were immunosorted using flow cytometry (fluorescent Cx3cr1 + cells) and we evaluated the kinetics of MG depletion. Representative flow cytometry graphs show changes in the abundance of MG under different conditions. On day 21 of treatment, we noted complete depletion of MG, and after 7 days without CSF1Ri the percentages of MG were restored to those in control animals (Fig. 1B, C). In parallel, the brains were collected for immunostaining, and MG were visualized with TMEM119 staining (a MG marker that shows processes and cell bodies). A few TMEM119+ cells were detected in the brains of mice treated with BLZ-945, mostly appearing as fragments of microglial processes. Repopulated MG were visible on the day 7th after BLZ-495 administration was stopped (Fig. 1D).
Fig. 1.
Kinetics of microglia depletion and repopulation. A Scheme of the experimental workflow. Mice were given orally peanut butter (PB) or BLZ-945 (200 mg/kg) for 14 or 21 days or were fed with BLZ-945 for 21 days and left for 7 days. B Representative graphs show the quantification of MG (Cx3cr1 + cells) from the brains of control or BLZ-945-fed mice as determined by flow cytometry. Mice were perfused with PBS, their brains removed, and subjected to enzymatic digestion. C Fluorescent Cx3cr1 + cells were evaluated by flow cytometry (n = 4), and statistical significance was calculated with one-way ANOVA with multiple comparisons (Tukey’s test df = 12, p = 0.0384). D TMEM119 staining of MG in the brains of controls, mice fed with BLZ-945 for 21 days, and mice fed with BLZ-945 for 21 days and then left for 7 days. Mice after indicated treatments were perfused with PFA, brains were removed, fixed and subjected to immunofluorescence (IF) staining followed by cell visualization with confocal microscopy. Representative IF images show Tmem119+ MG in the controls, loss of MG after 21 days of the treatment, with the remnants of filaments visible, and repopulated, branched MG 7 days after stopping BLZ-945 administration. Scale bar 50 µm
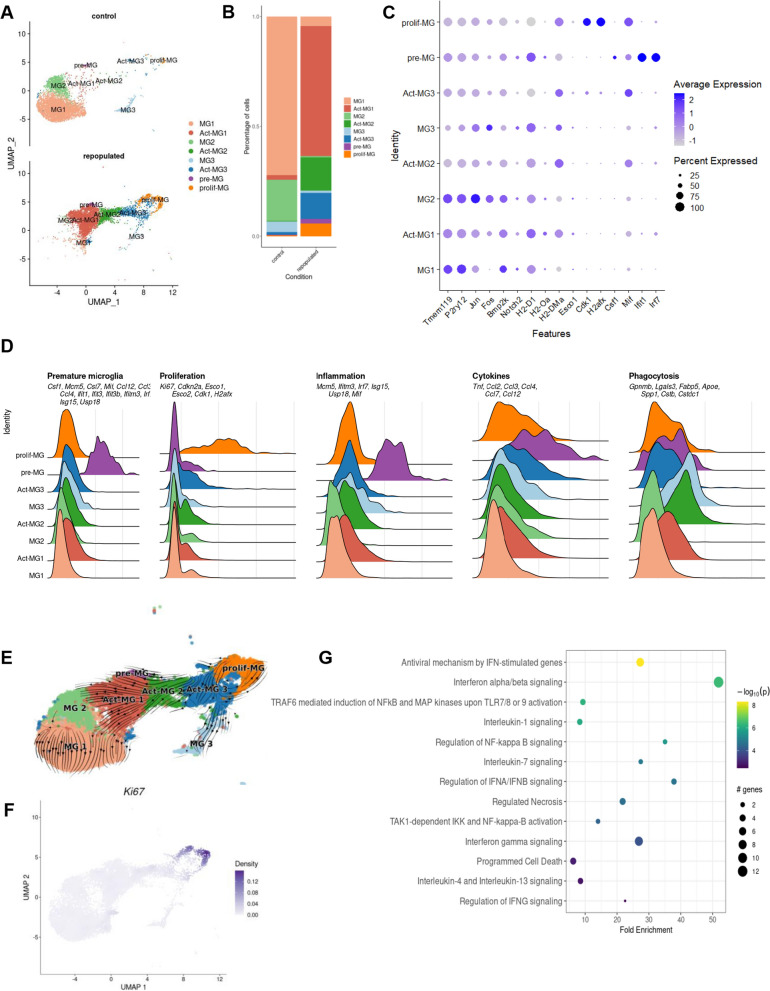
Single-cell RNA sequencing (scRNA-seq) provides an excellent means to characterize the identities and functionalities of cells under physiological and pathological conditions. We evaluated whether the repopulated MG restore the functional heterogeneity detected in intact brains and whether these processes undergo similarly in young and aging mice. We performed scRNA-seq on CD11b+cells immunosorted from the brains of young and aging mice either control or repopulated; CD11b + cells were sorted as previously described [12]. The cells were filtered and clusters of cells were visualized using Uniform Manifold Approximation and Projection (UMAP) (Fig. 2A). Overall, we sequenced 30,904 single cells from 16 samples and identified 16 cell clusters/types/states across all conditions. We clustered the cells into 16 clusters using Seurat and further merged these clusters (Supplemental Fig. S1A-B) into 8 cell subtypes that were annotated with the top differentially expressed markers and canonical markers from previous studies [12, 15]. Individual replicates are shown in Supplemental Fig. S1C. A majority of immune cells in the control and repopulated brains were homeostatic MG (Fig. 1A). MG clusters were investigated in greater depth and rare clusters associated with monocytes, CNS-associated macrophages, T cells, NK cells, and Cd24a + cells were excluded from further analysis (Supplemental Fig. S1A).
Fig. 2.
Characterization of the functional heterogeneity of repopulated microglia with single-cell transcriptomics. UMAP plot of scRNA-seq data showing cell clustering of control and repopulated MG. For each condition, four scRNA-seq replicates were combined. Cell clusters are colored and based on the annotations from [12, 15]. We identified MG1 homeostatic microglia, Act-MG1 activated microglia, MG2 transcriptionally active cells, Act-MG2 activated transcriptionally active cells, MG3 signaling inhibitors and transcriptional repressors, Act-MG3 activated signaling inhibitors and transcriptional repressors, pre-MG premature microglia, prolif-MG proliferating microglia. B The proportion of cells assigned into each cell cluster in two experimental groups (young controls and repopulated). C Dot plot showing the average expression levels of signature genes from previous studies [12] for the cell types or processes as defined above. D Ridge plots showing signature gene score profiles in each cell cluster associated with the specific cell types or processes: premature MG, proliferation, inflammation, cytokines, and phagocytosis. E UMAP plot overlaid with RNA velocity vectors (as quantified by scVelo), demonstrating expression dynamics across cell clusters of MG. Notably, all resulting clusters are derived from proliferating MG. F UMAP plot showing weighted kernel density [21] of scaled Ki67 gene expression. G Selected enriched Reactome terms in a set of the differentially expressed genes in the premature MG (pre-MG) cluster compared with other cell clusters from A
The identified clusters MG1, MG2, MG3, Act-MG1, Act-MG2 and Act-MG3 corresponded to Hom-MG1, Hom-MG2, Hom-MG3, Act-MG1, Act-MG2 and Act-MG3 clusters detected with scRNAseq and CITEseq (Cellular Indexing of Transcriptomes and Epitopes by Sequencing) in glioma associated microglia and have been described previously [14, 15]. The main difference in subtype composition between repopulated and control brains is the appearance of repopulated MG cells in an “active” state (Act-MG1, Act-MG2, and Act-MG3 clusters) and the emergence of premature and proliferating MG subtypes (Fig. 2B, Supplemental Table 1). The presence of Act-MG1-3 clusters among repopulated MG is surprising as they resemble functionalities of MG isolated from tumor-bearing brains, and reflect phagocytic, migratory and extracellular matrix-degrading activities of Act-MG. The proliferating MG cluster (prolif-MG) is characterized by high expression of the cell cycle genes (e.g. Ki-67, Esco1/2, Cdk1, H2afx). Interestingly, the premature MG (pre-MG) cluster showed high expression of inflammation-related genes (e.g. Mcm5, Ifitm3, Irf7, Isg15, Usp18, and Mif) and cytokine encoding genes (Tnf, Ccl2, 3, 4, 7, and 12) suggesting its active role in directing repopulation-related cell migration (Fig. 2A–D).
Trajectory inference has advanced scRNA-seq research by allowing to study dynamic changes in gene expression. This enables the discovery of expression changes associated with specific cells along the trajectory or those that are differentially expressed between distinct differentiation steps [19]. In the reduced dimensional space, a cell pseudo-time for a given lineage/state is the distance between a cell state and the origin of the lineage. These advancements allow for studies of the dynamics of biological processes, such as differentiation patterns from progenitors to more differentiated cellular states [20]. Pseudo-time ordering revealed a trajectory that originates in prolif-MG and progresses through active MG1-3 states, leading to homeostatic microglia MG1-3 clusters (Fig. 2E). The prolif-MG cluster exhibited high expression of Ki-67 (a marker of proliferation) (Fig. 2F). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of biological processes in the pre-MG cluster showed high expression of inflammation-related genes in the interferon-, TLR, and NFkB signaling pathways and cytokine encoding genes suggesting their intermediate inflammatory state before they become homeostatic MG (Fig. 2G).
Defective reconstitution of the homeostatic phenotype by repopulated microglia from older mice
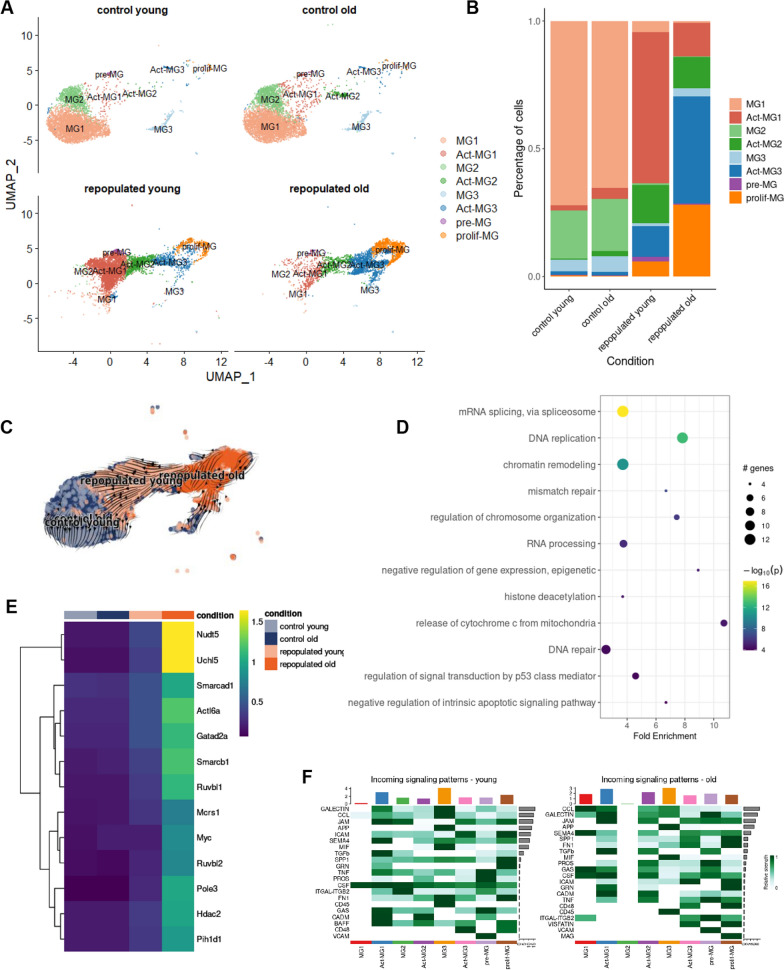
We compared the composition and heterogeneity of repopulated MG in young (3 months old) and aging (12 months old) mice. The previously identified MG clusters were detected in scRNA-seq samples from young and older mice, and their contribution was similar in naïve mice independently of age (Fig. 3A, Supplementary Fig. S2). However, the contribution of repopulated cells from a specific cluster differed between young and older mice (Fig. 3B). We found increased percentages of active cell states (Act-MG1-3) among repopulated MG in older mice. The repopulated MG from older mice were enriched in the active MG3 and prolif-MG clusters, while the repopulated MG from young mice had more act-MG1 cells. Trajectory analysis revealed that repopulated MG from older mice were enriched in prolif-MG clusters and did not reconstitute intermediate act-MG and homeostatic MG clusters at the same speed as those in young mice (Fig. 3A–C).
Fig. 3.
Dysfunctional microglial repopulation in aging mice. A UMAP plot of scRNA-seq data showing the clustering of control and repopulated MG from brains of young and aging mice. For each condition, four scRNA-seq replicates were combined. The cluster annotations as in the Fig. 2A. B Fraction of cells assigned into each cell cluster, separately for each of four conditions. C UMAP plot overlaid with RNA velocity vectors (as quantified by scVelo), demonstrating expression dynamics across experimental conditions. D Selected enriched KEGG pathways in the set of differentially expressed genes in the activated MG3 (Act-MG3) cluster compared to other cell clusters. E Heatmap of average expression of upregulated genes from the chromatin remodeling KEGG pathway and enriched in activated-MG3 cell cluster, divided by experimental conditions. F Communication probability via selected ligand-receptor pairs (y-axis) for each directed pair of cell clusters (x-axis)
To compare the expression of selected marker genes across four conditions, we plotted the normalized expression levels and a fraction of cells expressing them, separately for each of the 8 cell clusters. We verified that the selected genes presented similar expression profiles across the experimental conditions, confirming their suitability as marker genes (Supplemental Fig. S3). The KEGG analysis of genes upregulated in prolif-MG clusters of repopulated MG revealed that the enriched genes were associated with proliferation (mRNA splicing, DNA replication, and chromatin remodeling) (Fig. 3D, Supplemental Fig. S4). The profiles of selected differentially expressed genes are shown on the heatmap (Fig. 3E, Supplemental Fig. S4).
The identification of ligand–receptor pairs in scRNA-seq data can be used to infer intercellular communication from the coordinated expression of their cognate genes [22]. We explored the dataset for expression of the genes associated with the interacting proteins in the gene expression matrix and used those values as inputs to compute a communication score for each ligand-receptor (L-R) pair using the scoring function of the CellPhoneDB v.2.0. Putative L-R interactions between various subpopulations of repopulated MG show abundant incoming and outgoing interactions from pre-MG and act-MG1 involving genes encoding galectins, CCLs, ICAM, semaphorin 4, MIF (Fig. 3F, supplemental Fig. S5). C–C motif chemokine ligands (CCLs) are known to drive the chemotaxis of myeloid and lymphoid cells [23]. Galectins [24] and intercellular cell adhesion molecule (ICAM)-1 [25] control the migration of immune cells in brain parenchyma during inflammation. Macrophage migration inhibitory factor (MIF) is a pleiotropic protein, which participates in inflammatory and immune responses [26]. Similar subpopulations have increased incoming signaling patterns. Pre-MG is most active in sending signals in the brains of both young and aging mice (Fig. 3F, Supplemental Fig. S5).
Distinct gene expression in repopulated microglia from aged mice
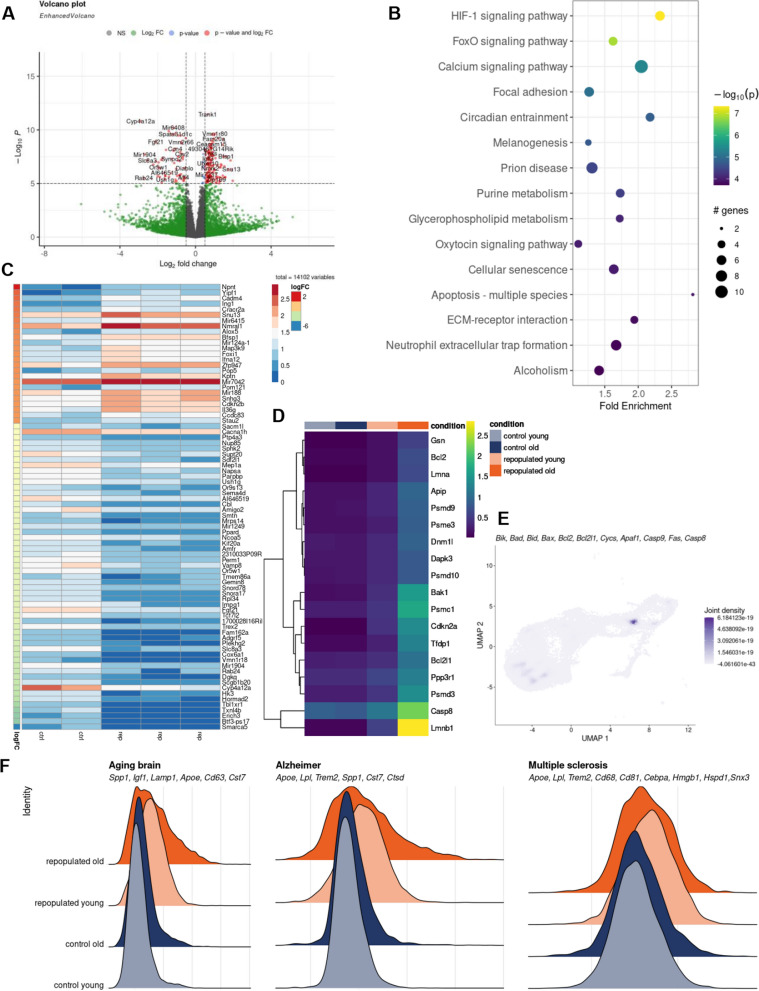
The observed differences in the dynamics of repopulated MG in aging mice prompted us to examine the transcriptional patterns of control and repopulated MG from the brains of aged mice (18–22 months old). Bulk RNA sequencing can generate large amounts of data at a lower cost than scRNA-Seq and detects more differentially expressed genes, which allows better identification associated functions or pathways. We analyzed bulk RNA-seq data from CD11b+ immunosorted cells from brains of the repopulated and control groups using linear regression models. The sample variance was estimated using Principal Component Analysis (PCA) dimension reduction and one sample, which was a significant outlier compared to the other samples, was excluded (Supplemental Fig. S6A). The volcano plot shows a number of differentially expressed genes (DEGs) in MG from aged mice (Fig. 4A). A KEGG analysis of genes upregulated in repopulated MG in old mice revealed several pathways, which are enriched within upregulated DEGs. Interestingly, several of those pathways are related to cell death, apoptosis, senescence, along with HIF1, Ca2+, and FoxO signaling (Fig. 4B). Bax expression is upregulated in MG from old mouse brains and Bax levels are upregulated in clusters of repopulated old MG in the scRNAseq dataset (Supplemental Fig. S6B, C). The expression of several apoptotic genes was greater in clusters of repopulated MG in aging mice (Supplemental Fig. S6B, C). The heatmap illustrates the expression of the top 50 DEGs which shows that numerous genes expressed in homeostatic MG in controls have lower expression in repopulated MG (Fig. 4C). It suggests that MG do not reach the mature state. We evaluated the expression of selected genes from those pathways in the scRNAseq dataset, and found that expression of proapoptotic genes (Lmnb1, Casp8, Psmd3, Bak1) was increased in repopulated MG from older mice (Fig. 4D). The expression of genes related to the apoptosome and cell death markers was restricted to the acti-MG3 cluster in older mice (Fig. 4E). We explored gene signatures that are upregulated in MG during aging, neurodegenerative or neuroinflammatory processes, and we found many of these genes are upregulated in repopulated MG in old mice (Fig. 4F). The observed differences in transcriptomic profiles of repopulated MG suggest the emergence of the transient, inflammatory phenotype, particularly in repopulated MG from old mice. With age, repopulated MG display elevated expression of apoptosis-related genes which is indicative that some cells may undergo cell death.
Fig. 4.
Transcriptional characteristics of repopulated microglia from aged mice. Volcano plot showing the expression fold change for all genes in the control and repopulated MG from aged mice (18–22 months old), along with p-value for differential expression. The mice were given orally PB or BLZ-945 (200 mg/kg) for 21 days and left for 7 days. Mice were perfused with PBS and their brains were removed, subjected to enzymatic digestion, and CD11b+ cells were immunosorted by FACS, total RNAs were isolated and subjected to RNAseq (n = 4/group). B Top 15 enriched KEGG pathways in the set of differentially expressed genes (p-value and log2 FC criteria) from A. C Heatmap of the top 50 differentially upregulated genes with the highest logFC value and adjusted p-value < 0.05 in the bulk RNAseq dataset. D Heatmap of average expression of the upregulated genes from “Apoptosis—multiple species” KEGG pathway in scRNA-seq data. E UMAP plot showing weighted kernel density of scaled gene expression of selected apoptosis marker genes in sc-RNAseq data from Fig. 3A. F Ridge plots showing expression scores of selected MGl neurodegeneration genes characteristics for the aging brain, Alzheimer's disease, and multiple sclerosis [31] in the scRNA-seq dataset
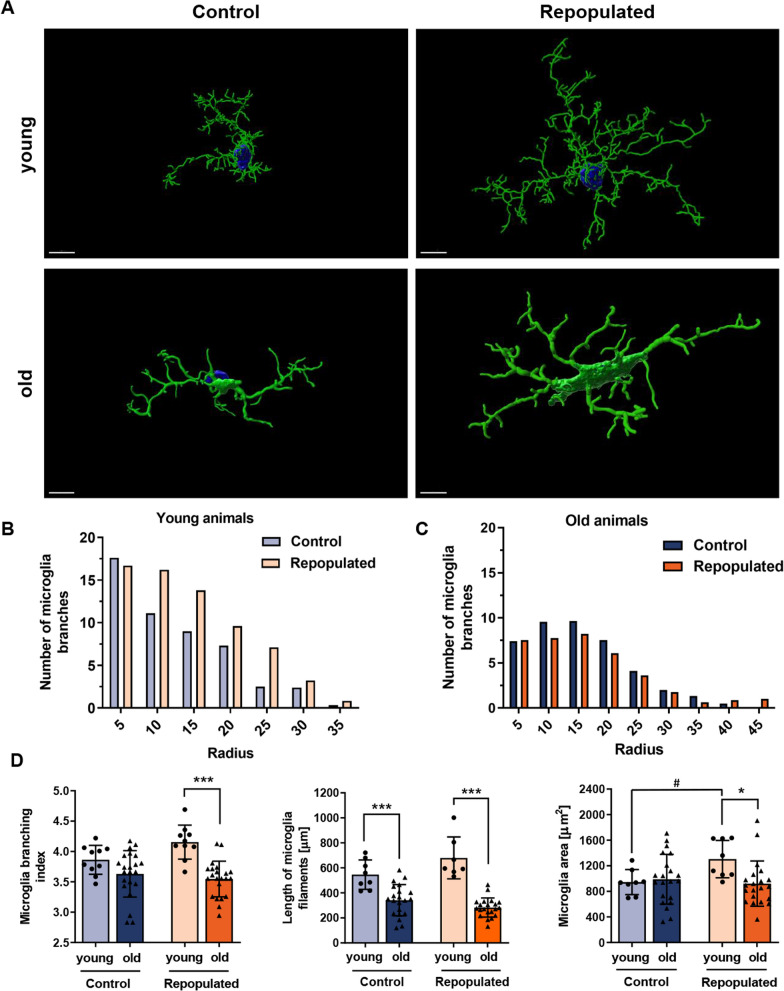
Repopulated microglia in older mice display different morphology than control microglia
MG in the CNS have mostly ramified morphology typical for mature cells with homeostatic functions [27]. Even unchallenged MG show numerous extensions and motility of microglial processes. Morphological alterations reflect MG plasticity and adaptation to changing or pathological conditions [28, 29]. Confocal microscopy and 3D imaging with specialized programs such as Imaris software allow the tracing of cell morphology and quantification of various parameters such as a cell volume, length of microglial processes, a number of terminal points, branch points and cell segments [30]. The changes in gene expression reported above suggested different functions of repopulated MG in the aging brain.
We performed immunofluorescence staining for TMEM119 and evaluated the morphology of repopulated, TMEM119+ MG in the brains of young (3 months, n = 6) and old (16–22 months, n = 7) mice using confocal microscopy and 3D reconstruction of cell shapes. When compared with the control MG, the repopulated MG present a distinct and more complex morphology, with numerous branches and more elongated microglial processes with more branch points (Fig. 5A). Using Sholl analysis, the radius of MG branches was quantified, and we found more cells with greater radii among repopulated MG in the brains of young mice. These differences were not detected among the repopulated MG in the brains of the old mice which presented less complex branching (Fig. 5B, C). We determined more morphological parameters, and found that repopulated MG in young mice had a larger area, but the branching and filament lengths are similar to MG in control mice (n = 4/group). In contrast, repopulated MG in old mice had significantly shorter filaments and less branching (Fig. 5D). These results show that the morphological complexity of MG is restored after repopulation in the brains of young but not old mice.
Fig. 5.
Different morphology of repopulated microglia from aged mice. A Representative confocal microscopy images of TMEM119+ MG. 3D models were created with the Imaris Software (Oxford Instruments, UK). Scale bar 7 µm. Notably, in young mice (3 months, n = 6) repopulated MG are more branched, with numerous elongated filaments with secondary processes located distal to the cell body. MG after repopulation have larger cell bodies. In old mice (18–22 months, n = 7) the repopulated MG are branched similarly to those in control mice and had fewer branches/processes than in the brains of young mice. B, C Quantification of control or repopulated MG branching with a Sholl analysis (radius 5 µm) in young (B) and old (C) mice, respectively. D Comparison of the MG branching (branching index and length of filaments) in brains of young and old mice (for branching, Sidak’s Test df = 60, p = 0.0001, for the length of MG filaments, Sidak’s Test control group df = 53, p = 0,0001, repopulated group df = 53, p = 0,0001); and the area covered by TMEM119+ MG (Sidak’s Test df = 54, p = 0.0193)
Discussion
MG functions are defined by intrinsic (ontogeny, sex or genetic background) and extrinsic (age, spatial location, and environment) factors [31]. Core markers define the MG as a subpopulation of myeloid cells distinct from other myeloid cells, but they are not informative about the functional “state” of the MG, which depends on the context. MG are highly responsive to alterations in their local environment, and their morphology, ultrastructure and molecular profile are dynamic, which results in many different cell states that cannot be resolved with classical technologies. We present the scRNAseq-based characterization of MG states after depletion/repopulation. We demonstrate that the repopulated MG recapitulates all the states/phenotypes occurring in the control MG. However, we detected transiently activated MG phenotypes (Act-MG1-3) that resemble proliferating, phagocytic, migratory MG detected in brain tumor niches [15]. Expression of selected genes expressed in main clusters is presented; of note is expression of Cstb, Ctsd, Spp1, ApoE in Act-MG1-3 (Supplemental Fig.S7C). While surprising, it can be explained by the fact that newly generated MG must migrate through the parenchyma to various regions of the brain, degrade the extracellular matrix along the way, and this migration is driven by cytokines and chemokines [32]. Based on gene expression analysis of scRNA-seq data, we found increased expression of genes encoding IL-1β, TNF-α and IL-6 cytokines in repopulated microglia, as well as CCL2, CCL3, CCL4 and CXCL10, which are involved in the inflammatory and migratory processes after depletion (Supplemental Fig. S3B). Newly generate MG, arising from precursor MG (pre-MG), display elevated expression of inflammation-related genes, mostly from interferon-related pathways. This may reflect mild inflammation preceding the acquisition of homeostatic functions by mature MG. Ligand-receptor interaction analysis shows the putative signals outgoing from pre-MG to other clusters, highlighting their decisive role in driving repopulation. We confirmed that all new MG clusters originate from the remaining pre-MG.
Under control conditions, the same MG clusters are detected in the brains of young and aging mice showing similar functions of those cells, irrespectively of age. However, among the repopulated cells from the brains of aging mice, we detected more proliferating MG and reduction of homeostatic MG clusters suggesting that those cells do not reach mature states after repopulation. A comparison of bulk RNA-seq results on repopulated CD11b+ cells from the brains of young and aged mice (16–18 months old) revealed pronounced downregulation of homeostatic MG genes and upregulation of genes related to cell death and apoptosis in MG from aged mice. scRNAseq results show that repopulated MG from young mice have increased expression of inflammation-related genes but there are clusters of homeostatic MG present. In repopulated MG from older mice gene signatures upregulated in MG during aging, neurodegenerative or neuroinflammatory processes are increased and homeostatic MG1-2 clusters are reduced suggesting that those cells do not mature into homeostatic MG. These results imply that MG from older and aged mice are deficient in attaining homeostatic phenotypes, which may affect their ability to support neuronal homeostasis. MG proliferation is balanced by the opposing force of MG apoptosis. Age-dependent changes in MG have been characterized by an enrichment of defined regulatory factors and gene expression profiles [33, 34]. MG can adopt an aberrant dystrophic or senescent phenotype, and aged MG have been frequently identified in neurodegenerative diseases [35, 36]. Aged MG upregulate MHC, CD86, CD68 and CR3 proteins, and pattern recognition receptors such as TLRs and Clec7a [37].
Microglial morphology is a good indicator of cell states as morphological transformation is associated with the activation of MG. We demonstrate that under physiological conditions the morphology of MG in the brains of young and aged mice is similar, except for shorter processes in the latter. However, Sholl analysis of shapes of 3-D reconstructed MG demonstrates that after repopulation MG from aged mice have shorter processes and less branching, features which indicates alterations of their functions. Pronounced morphological changes in MG such as deramification, cytoplasmic fragmentation, and shortening of cellular processes in the aged brain (even in the absence of forthright pathology) have been detected in human postmortem brain samples [38, 39]. While the presented data on MG repopulation do not mimic the physiological turnover of MG, they provide a glimpse into the mechanism underlying those processes. Compared with other myeloid cells, MG turnover is slow. In the human brain, approximately 28% of MG are replaced every year (an average of 2% are in the S phase on a given day) and in rodents, an average of 0.69% of MG are in the S phase at a given time [40]. Immunostaining studies revealed that MG proliferation and apoptosis are spatially and temporally coupled as proliferating cells were found to be the closest neighbors to dying cells. The apoptotic cascade controlling microglial death is dependent on the pro-apoptotic protein BIM [40], which is in agreement with our data showing increased Bim expression in aged MG. As MG are both immunological defenders and neuroprotective cells in the CNS, the consequences of MG dysfunction might involve increased susceptibility to brain infections and a failure to cope with neurodegenerative processes.
Supplementary information
Acknowledgements
We would like to thank Bartłomiej Gielniewski from the Sequencing Core Facility.
Author contributions
ZŁS: Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; PR: Data curation; Formal analysis; Visualization; Methodology; Writing—original draft. MBL, AK, NO, AML, MF: Investigation; Methodology; AJ: Data curation; Formal analysis; Visualization; Supervision; Funding acquisition; Methodology; Writing—original draft. BK: Conceptualization; Supervision; Visualization; Funding acquisition; Methodology; Writing—original draft; Writing—review and editing. All authors reviewed the manuscript.
Funding
This work was supported by the National Centre for Research and Development, Poland ERA-NET-NEURON/18/2018 (B.K.), JPco-fuND2 Programme no. 2022/04/Y/NZ5/00122, the National Science Centre, Poland (B.K.), the Polish National Agency for Academic Exchange Polish Returns 2019 (A.J.) and European Research Council ERC-CoG 865618 (M.F.).
Data availability
The single-cell and bulk RNAseq RNA-seq datasets produced in this study are available in the European Nucleotide Archive database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271559, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271560). The computer code produced in this study is available at https://github.com/rosapatrycja/microglia_repopulation_2024.
Declarations
Ethics approval and consent to participate
Animal studies were approved by the First Local Ethics Committee for Animal Experimentation in Warsaw (#836/2019; #1364P1/2022).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zuzanna M. Luczak-Sobotkowska and Patrycja Rosa contributed equally.
Contributor Information
Aleksander Jankowski, Email: aleksander.jankowski@uw.edu.pl.
Bozena Kaminska, Email: b.kaminska@nencki.edu.pl.
References
- 1.Kettenmann H, Kirchhoff F, Verkhratsky A. Microglia: new roles for the synaptic stripper. Neuron. 2013;77:10–8. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18:225–42. [DOI] [PubMed] [Google Scholar]
- 3.Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–81. [DOI] [PubMed] [Google Scholar]
- 4.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochocka N, Kaminska B. Microglia diversity in healthy and diseased brain: Insights from single-cell omics. Int J Mol Sci. 2021;22:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sousa C, Golebiewska A, Poovathingal SK, Kaoma T, Pires-Afonso Y, Martina S, Coowar D, Azuaje F, Skupin A, Balling R, et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018;19: e46171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green KN, Crapser JD, Hohsfield LA. To kill a microglia: a case for CSF1R inhibitors. Trends Immunol. 2020;41:771–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elmore MRP, Lee RJ, West BL, Green KN. Characterizing newly repopulated microglia in the adult mouse: impacts on animal behavior, cell morphology, and neuroinflammation. PLoS ONE. 2015;10(4): e0122912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, Wang J, Zhao L, Liang YX, Wu T, et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci. 2018;21:530–40. [DOI] [PubMed] [Google Scholar]
- 11.Najafi AR, Crapser J, Jiang S, Ng W, Mortazavi A, West BL, Green KN. A limited capacity for microglial repopulation in the adult brain. Glia. 2018;66:2385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochocka N, Segit P, Walentynowicz KA, Wojnicki K, Cyranowski S, Swatler J, Mieczkowski J, Kaminska B. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat Commun. 2021;12:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573-87.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun. 2021;12:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochocka N, Segit P, Wojnicki K, Cyranowski S, Swatler J, Jacek K, Grajkowska W, Kaminska B. Specialized functions and sexual dimorphism explain the functional diversity of the myeloid populations during glioma progression. Cell Rep. 2023;42: 111971. [DOI] [PubMed] [Google Scholar]
- 16.Ulgen E, Ozisik O, Sezerman OU. PathfindR: an R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet. 2019;10:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–14. [DOI] [PubMed] [Google Scholar]
- 18.Jin S, Plikus MV, Nie Q. Cell Chat for systematic analysis of cell-cell communication from single-cell and spatially resolved transcriptomics. bioRxiv. 2023. 10.1101/2023.11.05.565674.38234835 [Google Scholar]
- 19.Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrnes LE, Wong DM, Subramaniam M, Meyer NP, Gilchrist CL, Knox SM, Tward AD, Ye CJ, Sneddon JB. Lineage dynamics of murine pancreatic development at single-cell resolution. Nat Commun. 2018;9:3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alquicira-Hernandez J, Powell J. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics. 2021;37:2485–7. [DOI] [PubMed] [Google Scholar]
- 22.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. Cell PhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 23.Gschwandtner M, Derler R, Midwood KS. More than just attractive: how CCL2 influences myeloid cell behavior beyond chemotaxis. Front Immunol. 2019;10:2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soares LC, Al-Dalahmah O, Hillis J, Young CC, Asbed I, Sakaguchi M, O’neill E, Szele FG. Novel galectin-3 roles in neurogenesis, inflammation and neurological diseases. Cells. 2021;10:3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyck R, Enzmann G. The physiological roles of ICAM-1 and ICAM-2 in neutrophil migration into tissues. Curr Opin Hematol. 2015;22:53–9. [DOI] [PubMed] [Google Scholar]
- 26.Nishihira J. Macrophage migration inhibitory factor (MIF): its essential role in the immune system and cell growth. J Interferon Cytokine Res. 2000;20(9):751–62. [DOI] [PubMed] [Google Scholar]
- 27.Tay TL, Savage JC, Hui CW, Bisht K, Tremblay MÈ. Microglia across the lifespan: from origin to function in brain development, plasticity and cognition. J Physiol. 2017;595:1929–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–8. [DOI] [PubMed] [Google Scholar]
- 29.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8. [DOI] [PubMed] [Google Scholar]
- 30.Bosch LFP, Kierdorf K. The shape of μ—how morphological analyses shape the study of microglia. Front Cell Neurosci. 2022;16: 942462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paolicelli RC, Sierra A, Stevens B, Tremblay ME, Aguzzi A, Ajami B, Amit I, Audinat E, Bechmann I, Bennett M, et al. Microglia states and nomenclature: a field at its crossroads. Neuron. 2022;110:3458–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10:1387–94. [DOI] [PubMed] [Google Scholar]
- 33.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207-23.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antignano I, Liu Y, Offermann N, Capasso M. Aging microglia. Cell Mol Life Sci. 2023;80:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelova DM, Brown DR. Microglia and the aging brain: are senescent microglia the key to neurodegeneration? J Neurochem. 2019;151:676–768. [DOI] [PubMed] [Google Scholar]
- 37.Niraula A, Sheridan JF, Godbout JP. Microglia priming with aging and stress. Neuropsychopharmacology. 2017;42:318–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Streit WJ, Xue Q-S. The brain’s aging immune system. Aging Dis. 2010;1(3):254–61. [PMC free article] [PubMed] [Google Scholar]
- 39.Streit WJ, Sammons NW, Kuhns AJ, Sparks DL. Dystrophic microglia in the aging human brain. Glia. 2004;45:208–12. [DOI] [PubMed] [Google Scholar]
- 40.Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 2017;18:391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell and bulk RNAseq RNA-seq datasets produced in this study are available in the European Nucleotide Archive database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271559, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271560). The computer code produced in this study is available at https://github.com/rosapatrycja/microglia_repopulation_2024.