Abstract
Expression of the structural proteins L1 and L2 of the human papillomaviruses (HPV) is tightly regulated. As a consequence, attempts to express these prime-candidate genes for prophylactic vaccination against papillomavirus-associated diseases in mammalian cells by means of simple DNA transfections result in insufficient production of the viral antigens. Similarly, in vivo DNA vaccination using HPV L1 or L2 expression constructs produces only weak immune responses. In this study we demonstrate that transient expression of the HPV type 16 L1 and L2 proteins can be highly improved by changing the RNA coding sequence, resulting in the accumulation of significant amounts of virus-like particles in the nuclei of transfected cells. Data presented indicate that, in the case of L1, adaptation for codon usage accounts for the vast majority of the improvement in protein expression, whereas translation-independent posttranscriptional events contribute only to a minor degree. Finally, the adapted L1 genes demonstrate strongly increased immunogenicity in vivo compared to that of unmodified L1 genes.
The human papillomaviruses (HPV) comprise a heterogeneous group of epitheliotropic DNA viruses. It is assumed that each of the more than 80 described HPV genotypes represents also a separate serotype (23, 24). The papillomavirus life cycle requires the infection of differentiating epithelia. In this environment, expression of the viral genes is controlled by the cell differentiation program (5, 17). Infections by human papillomaviruses are the major cause of uterine cancer in humans (22, 43, 44). It is estimated that worldwide half a million new cases of cervical cancer are caused by these viruses every year. The most important HPV type in this respect is HPV type 16 (HPV-16), accounting for approximately 50% of all cases of cervical cancer. Since the recognition of HPV infection as a major health burden, efforts have been undertaken to interrupt the cycle of papillomavirus infections in order to prevent virus-induced disease. Most promising for the prevention of papillomavirus-associated cancer seems to be the development of subviral vaccines that evoke protective immunity by the induction of neutralizing, capsid-directed antibodies. In fact, virus-like particles (VLP) based on the viral capsid protein L1 or L1 plus L2 are currently being developed for prophylactic and therapeutic vaccination against papillomavirus infections (19, 26, 27). Because they require costly production and purification protocols, it is predictable that it will require a long time for VLP-based vaccines to become affordable in the less-developed countries, which suffer most from papillomavirus-caused cancer. For the same reasons, production and purification of VLP-based vaccines likely have to be restricted to a very limited number of HPV serotypes. As an alternative approach, capsid-specific neutralizing antibodies could be induced by simple DNA vaccination strategies. Since production of DNA vaccines are standardized, it is feasible to produce vaccines against a larger number of different HPV serotypes.
A major hurdle in the use of in vivo expression techniques is the tight control of papillomavirus late gene expression (29). It has been demonstrated that expression of the structural genes is controlled by several means: the late viral promoters depend on the differentiation status of the cells, polyadenylation signals terminate transcripts before reaching the late region (3, 4, 11), and mRNAs encoding the structural proteins contain inhibitory elements that prevent nuclear export or destabilize the message (13, 14, 31, 36, 37). Finally, for bovine papillomavirus type 1 (BPV-1) it has been suggested that tRNA levels influence in a differentiation-dependent manner the translation of the L1 protein (40). In vivo, these elements prevent premature expression of the capsid genes in the undifferentiated epithelium. While the papillomavirus early proteins are expressed in all layers of the stratified epithelium, capsid gene expression is achieved only in differentiated cells of the outer epithelial layers (33). This feature of the papillomavirus life cycle evidently contributes to immune evasion and might be considered one of the prerequisites for viral persistence. As a further consequence, expression of the late viral proteins, be it in the context of the viral genome or under the control of strong heterologous promoters, cannot be achieved to significant levels after DNA transfections in vitro or after DNA uptake upon DNA vaccination in vivo (9, 28).
In order to allow genetic vaccination against the HPV-16 structural proteins using either simple viral expression systems or naked plasmid DNA, we improved the coding region of the HPV-16 L1 and L2 proteins for efficient translation in nondifferentiated human cells as has been previously demonstrated for BPV-1 in a similar approach by others (40). We demonstrate that the introduced modifications greatly influence the efficiency of protein translation but also, yet to a lesser extent, improve the availability of the L1 mRNA for expression.
MATERIALS AND METHODS
Cell lines and cell culture.
293T and 911 (10) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin per ml, and 100 U of streptomycin per ml at 37°C in 5% CO2. Seventy percent confluent 293T or 911 cells were transfected with 12 μg (per 10-cm-diameter dish) of the respective plasmid DNA. Transfections were carried out using the modified calcium phosphate precipitation protocol according to the method of Chen and Okayama (6).
Western blot analysis.
For detection of L1 protein expression after transfection of the various plasmids, cells were harvested 72 h posttransfection, washed with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM KH2PO4, 1.1 mM Na2HPO4 [pH 7.5]) and then lysed in 2 mM EDTA–100 mM Tris-HCl (pH 8.0)–4% sodium dodecyl sulfate–20% glycerol–10% 2-mercaptoethanol–0.02% bromophenol blue by heating to 96°C for 10 min. Aliquots were subjected to gel electrophoresis using 15% sodium dodecyl sulfate-polyacrylamide gels (38) and Western blotting onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). The filters were blocked overnight at 4°C in PBS containing 6% skim milk powder (blocking solution) and were then incubated for 1 h at room temperature with the monoclonal antibody CamVir-1 (18) or a polyclonal rabbit anti-L2 antiserum (M. Müller, unpublished results) diluted 1:50 in 1% bovine serum albumin in PBS with 0.01% thimerosal. The membranes were washed six times in PBS–0.1% Tween 20 for 5 min and then incubated with a peroxidase-coupled goat anti-mouse antibody (Dianova, Hamburg, Germany) diluted 1:5,000 in blocking solution for 1 h at room temperature. After being washed, detected L1 protein was visualized using an enhanced chemiluminescence detection kit (Amersham, Braunschweig, Germany).
Indirect immunofluorescence.
To detect L1 protein by indirect immunofluorescence, transfected cells grown on cover slides were fixed via incubation in −20°C methanol (10 min) and acetone (5 min) at 4°C and then air dried and rehydrated in PBS for 5 min. The cells were incubated with the monoclonal antibody CamVir-1 diluted 1:20 in PBS containing 1% skim milk powder for 1 h at room temperature. The cells were then washed five times in PBS for 5 min and incubated with Cy3-coupled anti-mouse antibody (Dianova) diluted 1:300 in PBS containing 1% skim milk powder for 1 h at room temperature. After being washed five times in PBS, the slides were air dried and embedded in Permafluor (Immunotech, Marseille, France).
DNA immunization.
Endotoxin-free plasmid DNA was prepared with the Endofree-Maxi-Kit (Qiagen, Hilden, Germany) and dissolved in PBS to a final concentration of 1 mg/ml. Immunizations were carried out twice within 4 weeks by intramuscularly (i.m.) injecting 0.05 mg of plasmid into each of the anterior tibialis muscles. During immunization, mice were anesthetized with Metofane (Janssen-Cilag, Neuss, Germany). After another 4 weeks mice were euthanatized by cervical dislocation, blood was collected by cardiac puncture, and the antibody titer was determined in an enzyme-linked immunosorbent assay (ELISA).
ELISA.
For the detection of HPV-16 L1-specific antibodies, microtiter plates were coated overnight with 50 μl of PBS containing 35 μg of VLP per ml. After blocking of the plates (5% skim milk in PBS for 1 h at 37°C), mouse sera were added in dilutions of 1:10 to 1:12,800 and incubated for 1 h at 37°C. To determine nonspecific binding, the same dilutions of the antisera were tested on plates coated with PBS only. After being washed, peroxidase-conjugated goat anti-mouse antibodies (Sigma) were added at a 1:4,000 dilution. After 1 h at 37°C, plates were washed and stained with ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate solution (1 mg/ml, containing 0.015% H2O2). Extinction at 405 nm was measured after 20 min in a Titertek automated plate reader.
Electron microscopy.
To visualize VLP after transient transfection, 911 cells were grown on 18-mm cover slides, washed with PBS, and incubated for 30 min with 2.5% glutaraldehyde in PBS containing 1 mM MgCl2. After being stained with osmium tetroxide, the cells were dehydrated with increasing concentrations of ethanol and embedded in epoxide resin. Ultrathin sections of the cells were stained with 1% uranyl acetate and lead citrate. The sections were examined with a Zeiss EM 10A microscope.
Codon improvement.
The genes encoding L1h, L1p, and L2h were synthesized as described earlier (15) in a template-free PCR using overlapping oligonucleotides (82-mers to 85-mers) spanning the entire L1 (L2) gene. Codons were adapted according to the codon usage tabulated from GenBank (21) (http://www.kazusa.or.jp/codon) for Homo sapiens or Solanum tuberosum. Deviations from the codon usage tabulated from GenBank were made to introduce recognition sites for endonucleases. The sequences of the synthetic genes are accessible from the EMBL nucleotide sequence database (16L1h gene, AJ313179; 16L1p gene, AJ313181; 16L2h gene, AJ313180). All genes were cloned in the XbaI and HindIII sites of pBK-CMV (Stratagene). For expression in mammalian cells, the genes were excised with NotI and SalI and cloned into the NotI and SalI sites of the pUF3 vector. To create bicistronic expression constructs, the L1 genes were excised from the pBK-CMV vector by XbaI and XhoI and inserted into the NheI and SalI sites of the vector pGEM-IRES-GFP. Expression of green fluorescent protein (GFP) dependent on the upstream-inserted L1 gene was compared to a biscistronic construct, containing the mouse ecotropic retrovirus receptor gene rec-1 (1). An expression construct containing the simian retrovirus cis-acting transactivation element (CTE), L1oriCTEa, was kindly provided by S. Schwartz (37). A similar clone, L1oriCTEb, was constructed by inserting the CTE element (kindly provided by B. Felber [35]) into the XbaI and ApaI sites of the pCDNA3.1 expression vector, downstream of the HPV-16 L1 gene.
Flow cytometry.
Transfected 293T cells were harvested with trypsin-EDTA and washed once with PBS. GFP expression of 10,000 viable cells was determined by flow cytometry using a FACSSort cytometer (Becton Dickinson) and Cellquest version 3.3. Autofluorescence of mock-transfected cells and the relative fluorescence of cells transfected with the bicistronic GFP expression constructs were measured.
RESULTS
Construction of codon-optimized L1 genes.
It was our objective to use the HPV-16 L1 gene in DNA vaccination for the induction of capsid-specific antibodies. Because several of our attempts to express amounts of HPV-16 L1 detectable by Western blotting upon transient transfection into 293T or HeLa cells failed, we intended to optimize expression of the L1 gene under the control of the human cytomegalovirus immediate-early promoter (pCMV).
To overcome the inefficient expression of HPV-16 L1 for vaccination purposes in cells which do not resemble differentiating keratinocytes, we synthesized in vitro two HPV-16 L1 genes (based on the HPV-16 isolate 114/K [16]) in which the codon usage was optimized for either plant cells (i.e., Solanum tuberosum L1p, EMBL accession no. AJ313181) or mammalian cells (H. sapiens L1h, accession no. AJ313179) (Table 1) (21). In both constructs, the majority of the codons were modified (51.5% modified codons for L1p and 78.8% for L1h), while the encoded protein sequence remained unchanged. Some deviations from strict usage of optimized codons were made to allow for the insertion of recognition sites for restriction endonucleases. In addition to optimized codon composition, these extensive changes are likely to inactivate all known and unknown negative regulatory elements present in the authentic L1 open reading frame (ORF), L1ori. In addition, all upstream and downstream noncoding sequences were removed in all the constructs analyzed in this study. While the human optimized L1 (L1h) shows a high GC content (64.1% GC), the plant optimized L1 (L1p) is comprised of a very AT-rich sequence (34.8% GC). In this respect, L1p resembles L1ori (38.1% GC). This closer relationship of L1p and L1ori is in part based on the fact that in L1p a smaller number of codons were modified than in L1h but also reflects the preferences for AT-rich codons in the L1ori and L1p genes.
TABLE 1.
Codon usage in HPV 16 -L1ori, L1h, L1p, L2ori, and L2h
| Amino acid | Codon | Frequency (%) of indicated codon in gene:
|
||||
|---|---|---|---|---|---|---|
| 16L2h | 16L2ori | 16L1h | 16L1p | 16L1ori | ||
| Ala | GCA | 0.0 | 3.0 | 0.0 | 0.2 | 2.8 |
| GCG | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| GCC | 6.1 | 0.4 | 5.9 | 0.0 | 1.2 | |
| GCT | 0.0 | 2.7 | 0.0 | 5.7 | 2.0 | |
| Arg | AGA | 0.0 | 0.4 | 0.0 | 0.0 | 0.8 |
| AGG | 4.4 | 0.8 | 3.8 | 3.8 | 0.8 | |
| CGA | 0.0 | 0.6 | 0.0 | 0.0 | 0.8 | |
| CGG | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | |
| CGC | 0.0 | 1.1 | 0.0 | 0.0 | 0.8 | |
| CGT | 0.0 | 1.5 | 0.0 | 0.0 | 0.4 | |
| Asn | AAC | 3.8 | 0.4 | 5.5 | 0.0 | 1.4 |
| AAT | 0.0 | 3.4 | 0.0 | 5.5 | 4.2 | |
| Asp | GAC | 6.3 | 1.5 | 5.3 | 0.0 | 1.8 |
| GAT | 0.0 | 4.9 | 0.0 | 5.3 | 3.6 | |
| Cys | TGC | 0.4 | 0.2 | 2.4 | 0.0 | 0.6 |
| TGT | 0.0 | 0.2 | 0.0 | 2.4 | 1.8 | |
| Gln | CAA | 0.0 | 1.7 | 0.0 | 3.8 | 2.2 |
| CAG | 2.3 | 0.6 | 3.8 | 0.0 | 1.6 | |
| Glu | GAA | 0.0 | 2.1 | 0.0 | 3.8 | 2.8 |
| GAG | 2.1 | 0.0 | 4.0 | 0.2 | 1.2 | |
| Gly | GGA | 0.0 | 2.1 | 0.0 | 6.9 | 1.6 |
| GGG | 0.0 | 1.3 | 0.0 | 0.0 | 0.6 | |
| GGC | 6.8 | 0.8 | 6.9 | 0.0 | 1.8 | |
| GGT | 0.2 | 2.7 | 0.0 | 0.0 | 3.0 | |
| His | CAC | 1.7 | 0.2 | 2.0 | 0.0 | 0.4 |
| CAT | 0.0 | 1.5 | 0.0 | 2.0 | 1.6 | |
| Ile | ATA | 0.0 | 3.0 | 0.0 | 0.0 | 2.0 |
| ATC | 8.5 | 0.0 | 4.4 | 0.2 | 0.0 | |
| ATT | 0.0 | 5.5 | 0.0 | 4.2 | 2.4 | |
| Leu | CTA | 0.0 | 0.4 | 0.0 | 0.0 | 1.4 |
| CTG | 5.9 | 0.0 | 8.5 | 0.0 | 1.0 | |
| CTC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| CTT | 0.0 | 0.8 | 0.0 | 8.5 | 0.6 | |
| TTA | 0.0 | 3.8 | 0.0 | 0.0 | 4.6 | |
| TTG | 0.0 | 0.8 | 0.0 | 0.0 | 1.0 | |
| Lys | AAA | 0.0 | 2.5 | 0.0 | 6.7 | 5.3 |
| AAG | 3.0 | 0.4 | 6.7 | 0.0 | 1.4 | |
| Met | ATG | 0.8 | 0.8 | 2.0 | 2.0 | 2.0 |
| Phe | TTC | 3.2 | 0.2 | 4.8 | 0.0 | 0.2 |
| TTT | 0.0 | 3.0 | 0.0 | 4.8 | 4.6 | |
| Pro | CCA | 0.0 | 3.0 | 0.0 | 7.3 | 3.0 |
| CCG | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | |
| CCC | 10.6 | 1.7 | 7.3 | 0.0 | 1.0 | |
| CCT | 0.0 | 5.7 | 0.0 | 0.0 | 3.4 | |
| Ser | AGC | 9.7 | 0.4 | 6.5 | 0.0 | 0.4 |
| AGT | 0.0 | 2.5 | 0.0 | 0.0 | 1.4 | |
| TCA | 0.0 | 2.1 | 0.0 | 6.5 | 1.4 | |
| TCG | 0.0 | 0.4 | 0.0 | 0.0 | 0.0 | |
| TCC | 0.0 | 0.4 | 0.0 | 0.0 | 0.8 | |
| TCT | 0.0 | 3.8 | 0.0 | 0.0 | 2.6 | |
| Thr | ACA | 0.0 | 7.0 | 0.0 | 0.0 | 3.6 |
| ACG | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | |
| ACC | 13.7 | 1.3 | 8.1 | 0.0 | 1.6 | |
| ACT | 0.0 | 5.5 | 0.0 | 8.1 | 2.8 | |
| Trp | TGG | 0.0 | 0.0 | 1.4 | 1.4 | 1.4 |
| Tyr | TAC | 4.7 | 0.6 | 4.4 | 4.4 | 1.4 |
| TAT | 0.0 | 4.0 | 0.0 | 0.0 | 3.0 | |
| Val | GTA | 0.0 | 2.7 | 0.0 | 0.0 | 2.0 |
| GTG | 5.7 | 0.8 | 6.3 | 0.0 | 0.6 | |
| GTC | 0.0 | 0.2 | 0.0 | 0.0 | 0.4 | |
| GTT | 0.0 | 1.9 | 0.0 | 6.3 | 3.4 | |
Expression of L1 after transient transfection.
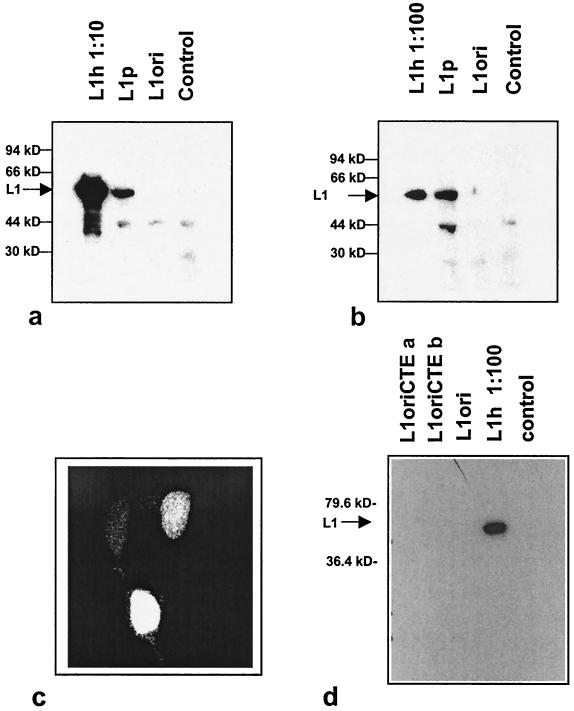
To evaluate the efficiency of the three constructs for L1 expression, the L1 ORFs were placed under the control of pCMV in the vector pUF3 (42) (Fig. 1a). The vector constructs were transfected into various mammalian cell lines, and L1 expression was analyzed by Western blotting (Fig. 2). In most experiments L1ori expression proved to be undetectable; only occasionally could a faint signal be observed. In contrast, L1 expressed from the construct L1p, carrying the plant optimized codons, was consistently readily detectable with an at least 100-fold-increased protein level as judged from the signal in Western blotting in gels where L1ori expression could be detected (for an example, see Fig. 4, which shows that L1ori could be visualized in longer exposures). A further increase of L1 expression was observed when the humanized L1 gene (L1h gene) was analyzed, resulting in additional ≈100-fold higher L1 protein levels in transfected cells (Fig. 2b). This accounts for a total increase in protein expression of 104- to 105-fold compared to expression from an L1ori-containing plasmid. This tremendous improvement of L1 protein expression by the L1p and L1h plasmids is unlikely to reflect differences in transfection efficiencies, because experiments with different plasmid preparations and different cell lines (911, 293T, and HeLa) gave the same results and the numbers of cells transfected by the L1p and L1h plasmids were the same, as detected by the nuclear staining of L1 in transfected cells (Fig. 2c).
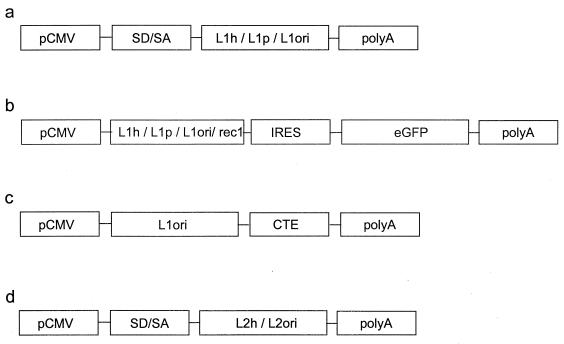
FIG. 1.
Expression constructs containing the L1 and L2 ORFs of HPV-16 with codons optimized for expression in human cells (L1h, L2h), plant cells (L1p), or with their original codons (L1ori, L2ori). In all constructs the expression of the capsid gene is driven by pCMV. To analyze the transient expressions of L1 and L2, the eukaryotic expression vector pUF3 was used (42) (a and d). This vector contains a small intron with a splice donor and splice acceptor site (SD/SA) located upstream of the respective capsid gene. (b) To analyze the influences of the various L1 genes on the expression of GFP (eGFP), bicistronic constructs in which the GFP gene was placed under the control of an IRES located downstream of the respective L1 gene were used. As a control, the L1 gene was replaced by the ecotropic retrovirus receptor gene rec1. (c) Expression construct containing L1ori in combination with the simian retrovirus CTE element cloned into the pcDNA 3.1 expression vector (Invitrogen).
FIG. 2.
Adaptation for codon usage improves the expression of HPV-16 L1. (a and b) Western blot analysis of L1 expression in 293T cells upon transfection with the L1h, L1p, and L1ori expression constructs. Extracts of transfected and untransfected cells (control) were analyzed by Western blotting using the L1-specific monoclonal antibody CamVir-I. To quantitatively compare levels of L1 expression from the various constructs, different amounts of extracts were loaded: 1/10 (a) and 1:100 (b) of the L1h extract compared to the L1p and L1ori extracts. (c) L1h expression in the nuclei of transiently transfected 911 cells by indirect immunofluorescence. (d) Western blot experiment to compare the expression of L1h to the expression of L1ori constructs containing the simian retrovirus CTE element for nuclear export of L1 mRNA.
FIG. 4.
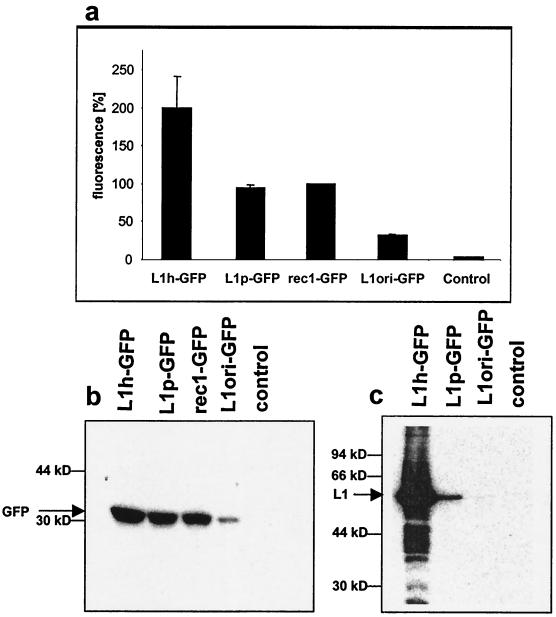
Influence of various HPV-16 L1 ORFs on the expression of a downstream-located GFP gene in a bicistronic expression construct. Cells were transfected with expression constructs containing genes encoding L1h, L1p, and L1ori, or a non-L1 gene (the ecotropic retrovirus receptor gene rec1) upstream of an IRES-GFP cassette. Expression of GFP was analyzed by FACS analysis (a) and Western blotting (b). Note that the total fluorescence of the rec1-GFP-transfected cells was set to 100%. (c) Western blot analysis of L1 expression levels in the same extracts as for panel b.
Previously it has been demonstrated that negative regulatory elements contained within the L1 gene interfere with nuclear export of the L1 message. This block in nuclear export can be overcome by adding the simian retrovirus CTE element to the L1 mRNA (36, 37). In order to relate the improved L1 expression by modification of the L1 codons to the improvement obtained by introduction of the CTE element, we compared L1 expression upon transient transfection of the L1h and the L1ori-CTE constructs (Fig. 1c and 2d). While the L1 protein could be readily detected in cells transfected with L1h, we were not able to detect L1 in cells transfected with one of two different L1ori-CTE constructs.
L1h-encoded protein assembles into VLP.
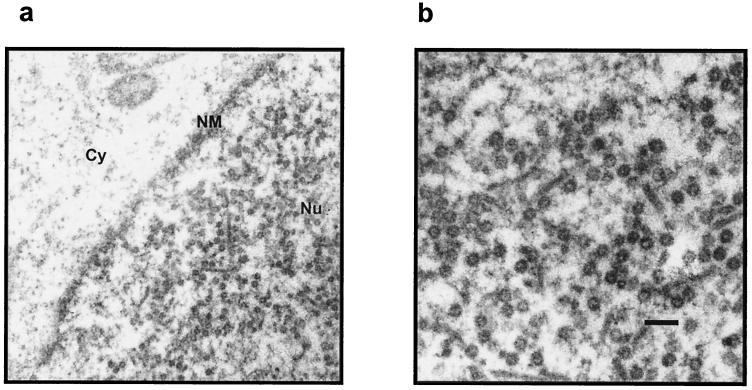
Based on the observed efficient expression of HPV-16 L1 from plasmids with humanized codons, we were interested in investigating whether the increase in protein expression leads to detectable amounts of assembled particles within transfected cells. For this purpose, 911 cells were transfected with the L1h expression construct and subsequently fixed with glutaraldehyde and ultrathin sections of the cells were analyzed by electron microscopy. As shown in Fig. 3, large quantities of assembled VLP could be detected but only within the nuclei. We were not able to detect VLP in the cytoplasms of L1h-transfected cells or within cells transfected with either L1ori or L1p. This suggests that capsid assembly is restricted to the nucleus and may depend on the intranuclear concentration of the L1 protein.
FIG. 3.
Transient expression of L1 from the L1h expression construct leads to the formation of VLP in the nuclei of transfected cells. Electron micrographs of ultrathin sections of 911 cells transfected with the L1h expression construct are shown. Bar, 0:2 μm. Cy, cytosol; NM, nuclear membrane; Nu, nucleus.
Translation-unrelated effects of codon adaptation.
We wished to determine whether the improved expression levels obtained by L1p and L1h were due to enhanced translation only or also to improvement of other posttranscriptional events. Therefore, we used several bicistronic constructs in which the gene for the GFP was placed downstream of either the respective L1 ORF or, for control purposes, the non-HPV gene rec1 (ecotropic retrovirus receptor gene). Translation of GFP initiates from an internal ribosomal entry site, located upstream of the GFP initiation codon (Fig. 1b). In this experimental system, the presence or absence of elements within the respective L1 gene and with a negative influence on mRNA stability or mRNA export from the nucleus should also influence GFP expression. In contrast, codon improvement for efficient translation of the L1 mRNA should not influence translation of GFP. The influences of the different L1 constructs on GFP expression were determined by fluorescence-activated cell sorter (FACS) analysis and Western blotting after transient transfection of the constructs with either L1ori, L1p, and L1h genes or rec1 upstream of the GFP expression cassette. FACS analysis of GFP-expressing cells (Fig. 4a) showed that L1p had a neutral cis effect on GFP expression compared to that of the rec1 gene but that L1ori indeed inhibited GFP expression by 60 to 70%. On the other hand, L1h stimulated GFP expression by a factor of roughly 2, a result which was confirmed by Western blot analysis of GFP expression (Fig. 4b). This result confirms the earlier proposed effects of L1ori sequences on mRNA stability and/or mRNA export (14, 36). However, when we measured L1 expression in the extracts of the same transfection experiments, we observed again a >1,000-fold increase of L1 expression from the humanized plasmid compared to that from the L1ori plasmid, strongly underlining that the major contribution to improved L1 expression by modification of codon usage has to be attributed to enhanced translation.
Codon usage improvement of HPV-16 L2.
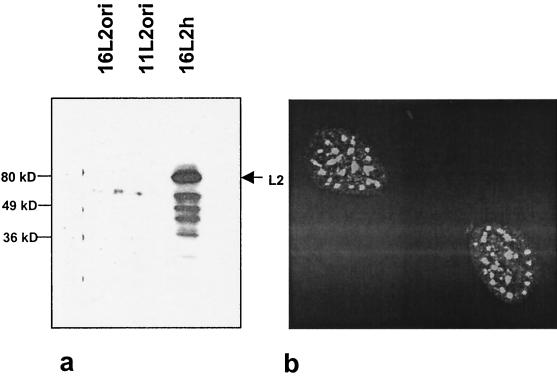
In previous studies it was demonstrated that HPV-16 L2 expression underlies a tight expression control similar to that of HPV-16 L1 (31). For vaccination studies, it was therefore desirable to determine whether HPV-16 L2 expression could also be improved by means of adapted codon usage. For this, we generated a humanized L2 ORF (L2h, EMBL accession no. AJ313180) using the same criteria that were applied for the generation of L1h (88.7% of the codons changed [Table 1]). The resulting expression construct (Fig. 1d) was transfected into 293T cells, and expression was analyzed by Western blotting using an L2-specific polyclonal antiserum (Fig. 5). While no L2 protein could be detected in cells transfected with unmodified L2ori, cells transfected with the L2h construct produced high levels of L2. The L2 protein was localized in a speckled pattern within the nuclei of transfected cells (Fig. 5b), as has been described earlier (7). Thus, similarly to that of HPV-16 L1, expression of HPV-16 L2 is negatively influenced by the primary structure of the L2 mRNA.
FIG. 5.
Expression of codon-optimized HPV-16 L2. 293T cells were transfected with expression constructs containing HPV-16 L2ori, HPV-11 L2ori or HPV-16 L2h. The L2 protein was subsequently detected by Western blotting in extracts of transfected cells by use of a polyclonal rabbit antiserum specific for HPV-16 and HPV-11 L2 (a) or by indirect immunofluorescence of transfected cells (b).
Codon usage improvement increases the efficacy of L1 DNA vaccines.
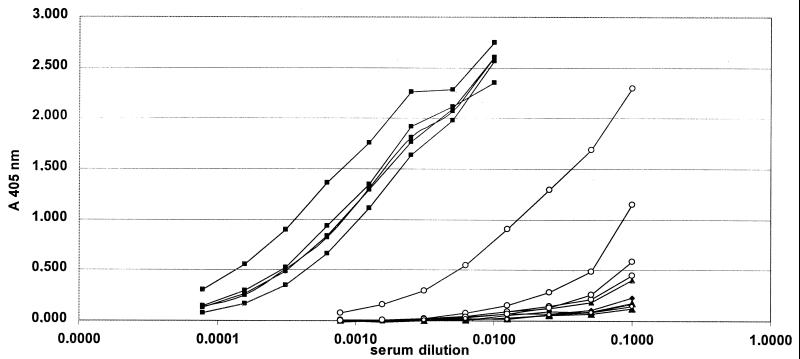
As it was our aim to prepare the HPV-16 L1 gene for use in DNA vaccination experiments, we wished to determine the ability of the humanized L1 gene to induce a humoral immune response after i.m. injection. For this purpose, mice were immunized i.m. twice in a 4-week interval with 100 μg of plasmid DNA per immunization. A total of 14 mice falling into three groups were analyzed: 4 mice immunized with a control construct harboring a non-L1 gene (VP22-E7) (M. Müller, unpublished), 5 mice immunized with L1h, and 5 mice immunized with L1ori. Four weeks after the second immunization, sera were collected and L1-specific antibody titers were determined using an HPV-16 VLP-specific ELISA (Fig. 6 and Table 2). While none of the mice immunized with the negative control plasmid developed L1-specific antibodies, high titers of anti-L1 antibodies could be observed for all five mice immunized with L1h. A positive ELISA signal was detectable even after 1:6,400 dilution of the sera of this group. In contrast, in the group of mice immunized with the L1ori constructs, only two of the five mice developed measurable anti-L1 antibodies, and the antibody titers for these two mice were 20- to 160-fold lower than those for the L1h group. The five mice of the L1h experimental group reached a mean titer above 1:6,400 compared to a mean titer of 1:80 in the L1ori group, indicating that codon optimization boosts the L1 expression not only in vitro but also in vivo. All sera were further analyzed for L1-specific antibodies by Western blotting. Seven of the 10 mice immunized with L1h were positive in this assay (four of these sera reacted weakly and three sera reacted strongly).
FIG. 6.
Induction of L1-specific antibodies by DNA immunization. Fifteen mice falling into four groups were immunized by injection of plasmid DNA. Anti-L1-specific antibodies were measured by an HPV-16 VLP-specific ELISA. ▪, five mice immunized with the expression plasmid containing the L1h gene; ○, five mice immunized with the plasmid containing the L1ori gene. The control group consisted of four mice immunized with an expression vector containing a non-L1 gene (VP22-E7) (▴) and one nonimmunized mouse (⧫).
TABLE 2.
Induction of capsid-specific antibodiesa
| Construct | No. of positiveb mice/ total no. in group | Mean titer of positive serac |
|---|---|---|
| None | 0/2 | |
| pUF3-VP22E7 | 0/4 | |
| pUF3-16L1ori | 1/5 | 1:80 |
| pUF3-16L1h | 10/10 | 1:7,100 |
DNA immunization using L1 expression constructs; results were compiled from two different experiments.
Positive mice scored positive in the VLP-specific ELISA.
Mean titers refer to seropositive mice only.
DISCUSSION
It was our aim to prepare the HPV-16 capsid proteins for use in vaccination protocols that are based either on simple vector systems, such as adeno-associated viruses, or on injection using naked plasmid DNA. In earlier studies, the use of HPV 6b L1 (28) or HPV-16 L1 (9, 30) DNA has proven difficult, presumably because of the low efficiency by which the HPV 6b L1 gene, as well as the L1 (and L2) genes of other papillomaviruses, can be expressed in cells for which HPV L1 expression is not adapted. In recent years there have been a number of reports concerning the limitations of papillomavirus capsid gene expression. Several genetic elements, among them differentiation-specific promoters and polyadenylation sites present on the HPV and BPV genomes lead to a block of mRNA transcription in nondifferentiated cells. However, even when placed under strong heterologous promoters, expression of the papillomavirus capsid genes proved to be difficult to achieve. Only by the use of virus vector systems, such as recombinant baculoviruses for insect cells or vaccinia viruses and Semliki Forest viruses for mammalian cells, is it possible to efficiently express papillomavirus capsid genes (12, 41). While vaccinia virus and Semliki Forest virus generate their mRNAs in the cytoplasm, thus circumventing the nuclear export process, this is not the case with recombinant baculoviruses or in the yeast systems, which also allow efficient production of papillomavirus late proteins (25). The mechanisms by which these expression systems overcome limitations of L1 production observed in undifferentiated tissue culture cells are unknown. However, it is conceivable that the tightly restricted L1 production is a means to escape the detection by the immune system in the in vivo situation. In order to vaccinate in vivo by expression of the L1 gene in a number of tissues it was a prerequisite to improve L1 gene expression in undifferentiated cells in vitro with the assumption that this would correlate with elevated expression of L1 in target tissues upon DNA vaccination in vivo.
Here we report the enhancement of HPV-16 L1 (and L2) capsid gene expression in cells in culture after transient transfection and in vivo after DNA injection into mouse muscle cells. We constructed L1 and L2 genes in which the codons were modified to codons frequently used in mammalian or plant genes (L1). A similar strategy was recently reported for the expression of BPV-1 L1 and L2 (40), although in that report the authors focused on modification of those codons that are extremely rare in human genes. Our data indicate that the resulting genes proved to express at drastically higher levels than those of their unmodified counterparts. Also, an HPV-16 L1 gene with codon usage adapted for plant cells expressed at much higher levels than those of the unmodified HPV-16 L1. Interestingly, the L1p gene exhibits a codon usage with preference of A or T in the third codon base, as does HPV-16 L1ori, indicating that optimal mammalian codons are not necessarily required for efficient gene expression. In fact, some of the codons (e.g., CTT for Leu, ACT for Thr, and CAA for Gln) described as possibly rate limiting for BPV-1 capsid gene expression are even overrepresented in the L1p gene. These different observations might reflect differences in the expression of BPV-1 versus HPV-16 L1 expression or might simply indicate that only a minority of the L1 codons actually negatively influences protein expression levels by providing rate-limiting steps for the translational machinery. If this is the case, knowledge of such critical and rate-limiting codons would facilitate the generation of expression-optimized L1 genes of other HPV types.
In addition to the enhancement of the codon usage for tRNA pools present in mammalian cells, the high degree of modifications of the L1 and L2 genes likely inactivates additional regulatory elements that control capsid gene expression. A number of such elements present on the L1 and L2 mRNA that interfere with mRNA posttranscriptional mechanisms have been described. To account for these translation-independent effects of the improved L1 expression, we further analyzed bicistronic constructs containing the GFP downstream of the respective L1 genes. For this, the GFP gene was placed under the control of an internal ribosome entry site (IRES). In these constructs, expression of the GFP gene should not depend on translation of the upstream L1 genes. In these experiments up to sixfold to sevenfold differences in GFP expression were observed, depending on the respective L1 gene placed upstream of the IRES element. Most notably, there is a significant inhibition of downstream GFP expression in the presence of the original L1 gene in cis. Since, however, expression levels of the various L1 genes differed by several orders of magnitude, we conclude that the modifications in codon usage dominantly influence the efficiency of L1 expression and that other events such as mRNA processing, stability, and nuclear export play only minor roles. This is in agreement with our observation that expression of the unmodified L1 protein was not significantly improved by including the simian retrovirus CTE element in the expression construct, although it has been previously reported that this element efficiently mediates nuclear export of the L1 mRNA (36).
Further, and most importantly, the codon usage-adapted L1 gene exhibits much improved immunogenicity in vivo upon DNA vaccination, indicating that L1 expression is also strongly improved in muscle cells. Our results indicate that high titers of VLP-specific antibodies can be induced by expression of the L1h gene, although at least one of the mice immunized with L1ori also produced measurable titers of L1-specific antibodies. This indicates that in vivo but not in vitro significant expression of the unmodified L1 protein can occur. Interestingly, in two independent studies it has been reported that anti-L1 antibodies can be induced by a polynucleotide vaccine. In both studies, the L1 was derived from cottontail rabbit papillomavirus. While Sundaram et al. (34) administered the DNA by the use of gold-particles, Donelly et al. (8) injected the expression constructs i.m. into various sites. Although it is possible that translation of the cottontail rabbit papillomavirus L1 gene is less tightly controlled than that of the HPV-16 L1 gene, it cannot be ruled out that different immunization protocols account for the different efficacies of unmodified L1 genes in mounting an immune response.
In conclusion, we believe that capsid-specific DNA vaccination could be an intriguing alternative to VLP vaccination in order to induce a prophylactic immune response against papillomavirus infections. Improvement of DNA vaccination efficacy by codon adaptation has been investigated earlier for the human immunodeficiency virus type 1 gp120 and for the tetanus toxoid (2, 20, 32, 39). Together with this report it has now been also demonstrated for two distantly related papillomaviruses that codon exchange significantly improves L1 (and L2) protein production. It is likely that this approach can easily be extended to other HPV types as well and will allow the cost-effective production of stable HPV vaccines.
ACKNOWLEDGMENTS
We are grateful to Georg Pougialis for excellent technical assistance. We thank Katja Parsche for help with the FACS analysis and H. zur Hausen for continued support and helpful discussions.
C.L. is supported by a grant of the HGF-Strategiefond I Infektionsabwehr und Krebsprävention.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker C C, Howley P M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987;6:1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day P M, Roden R B, Lowy D R, Schiller J T. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly J J, Martinez D, Jansen K U, Ellis R W, Montgomery D L, Liu M A. Protection against papillomavirus with a polynucleotide vaccine. J Infect Dis. 1996;173:314–320. doi: 10.1093/infdis/173.2.314. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy C, Buzoni-Gatel D, Touze A, Bout D, Coursaget P. Nasal immunization of mice with human papillomavirus type 16 (HPV-16) virus-like particles or with the HPV-16 L1 gene elicits specific cytotoxic T lymphocytes in vaginal draining lymph nodes. J Virol. 1999;73:9063–9071. doi: 10.1128/jvi.73.11.9063-9071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fallaux F J, Kranenburg O, Cramer S J, Houweling A, Van Ormondt H, Hoeben R C, Van Der Eb A J. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 11.Grassmann K, Rapp B, Maschek H, Petry K U, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heino P, Dillner J, Schwartz S. Human papillomavirus type 16 capsid proteins produced from recombinant Semliki Forest virus assemble into virus-like particles. Virology. 1995;214:349–359. doi: 10.1006/viro.1995.0044. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy I M, Haddow J K, Clements J B. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J Virol. 1990;64:1825–1829. doi: 10.1128/jvi.64.4.1825-1829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy I M, Haddow J K, Clements J B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C H, Oh Y, Lee T H. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene. 1997;199:293–301. doi: 10.1016/s0378-1119(97)00384-3. [DOI] [PubMed] [Google Scholar]
- 16.Kirnbauer R, Taub J, Greenstone H, Roden R, Dürst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 18.McLean C S, Churcher M J, Meinke J, Smith G L, Higgins G, Stanley M, Minson A C. Production and characterisation of a monoclonal antibody to human papillomavirus type 16 using recombinant vaccinia virus. J Clin Pathol. 1990;43:488–492. doi: 10.1136/jcp.43.6.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller M, Zhou J, Reed T D, Rittmüller C, Burger A, Gabelsberger J, Braspenning J, Gissmann L. Chimeric papillomavirus-like particles. Virology. 1997;234:93–111. doi: 10.1006/viro.1997.8591. [DOI] [PubMed] [Google Scholar]
- 20.Nagata T, Uchijima M, Yoshida A, Kawashima M, Koide Y. Codon optimization effect on translational efficiency of DNA vaccine in mammalian cells: analysis of plasmid DNA encoding a CTL epitope derived from microorganisms. Biochem Biophys Res Commun. 1999;261:445–451. doi: 10.1006/bbrc.1999.1050. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkin D M, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Roden R B, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden R B, Hubbert N L, Kirnbauer R, Christensen N D, Lowy D R, Schiller J T. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J Virol. 1996;70:3298–3301. doi: 10.1128/jvi.70.5.3298-3301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasagawa T, Pushko P, Steers G, Gschmeissner S E, Hajibagheri M A, Finch J, Crawford L, Tommasino M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 1995;206:126–135. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 26.Schiller J T. Papillomavirus-like particle vaccines for cervical cancer. Mol Med Today. 1999;5:209–215. doi: 10.1016/S1357-4310(99)01463-X. [DOI] [PubMed] [Google Scholar]
- 27.Schiller J T, Lowy D R. Papillomavirus-like particle vaccines. J Natl Cancer Inst Monogr. 2000;2000:50–54. doi: 10.1093/oxfordjournals.jncimonographs.a024258. [DOI] [PubMed] [Google Scholar]
- 28.Schreckenberger C, Sethupathi P, Kanjanahaluethai A, Müller M, Zhou J, Gissmann L, Qiao L. Induction of an HPV 6bL1-specific mucosal IgA response by DNA immunization. Vaccine. 2000;19:227–233. doi: 10.1016/s0264-410x(00)00173-0. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz S. Cis-acting negative RNA elements on papillomavirus late mRNAs. Semin Virol. 1998;8:291–300. [Google Scholar]
- 30.Smahel M, Sima P, Ludvikova V, Vonka V. Modified HPV16 E7 genes as DNA vaccine against E7-containing oncogenic cells. Virology. 2001;281:231–238. doi: 10.1006/viro.2000.0794. [DOI] [PubMed] [Google Scholar]
- 31.Sokolowski M, Tan W, Jellne M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J Virol. 1998;72:1504–1515. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stratford R, Douce G, Zhang-Barber L, Fairweather N, Eskola J, Dougan G. Influence of codon usage on the immunogenicity of a DNA vaccine against tetanus. Vaccine. 2000;19:810–815. doi: 10.1016/s0264-410x(00)00246-2. [DOI] [PubMed] [Google Scholar]
- 33.Stubenrauch F, Laimins L A. Human papillomavirus life cycle: active and latent phases. Semin Cancer Biol. 1999;9:379–386. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- 34.Sundaram P, Xiao W, Brandsma J L. Particle-mediated delivery of recombinant expression vectors to rabbit skin induces high-titered polyclonal antisera (and circumvents purification of a protein immunogen) Nucleic Acids Res. 1996;24:1375–1377. doi: 10.1093/nar/24.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan W, Felber B K, Zolotukhin A S, Pavlakis G N, Schwartz S. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J Virol. 1995;69:5607–5620. doi: 10.1128/jvi.69.9.5607-5620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan W, Schwartz S. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J Virol. 1995;69:2932–2945. doi: 10.1128/jvi.69.5.2932-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas J O, Kornberg R D. The study of histone–histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- 39.Uchijima M, Yoshida A, Nagata T, Koide Y. Optimization of codon usage of plasmid DNA vaccine is required for the effective MHC class I-restricted T cell responses against an intracellular bacterium. J Immunol. 1998;161:5594–5599. [PubMed] [Google Scholar]
- 40.Zhou J, Liu W J, Peng S W, Sun X Y, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J, Stenzel D J, Sun X Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]
- 42.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 44.zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]