Abstract
The most severe human immunodeficiency virus type 1 (HIV-1) epidemic is occurring in southern Africa. It is caused by HIV-1 subtype C (HIV-1C). In this study we present the identification and analysis of cumulative cytotoxic T-lymphocyte (CTL) responses in the southern African country of Botswana. CTLs were shown to be an important component of the immune response to control HIV-1 infection. The definition of optimal and dominant epitopes across the HIV-1C genome that are targeted by CTL is critical for vaccine design. The characteristics of the predominant virus that causes the HIV-1 epidemic in a certain geographic area and also the genetic background of the population, through the distribution of common HLA class I alleles, might impact dominant CTL responses in the vaccinee and in the general population. The enzyme-linked immunospot (Elispot) gamma interferon assay has recently been shown to be a reliable tool to map optimal CTL epitopes, correlating well with other methods, such as intracellular staining, tetramer staining, and the classical chromium release assay. Using Elispot with overlapping synthetic peptides across Gag, Tat, Rev, and Nef, we analyzed HIV-1C-specific CTL responses of HIV-1-infected blood donors. Profiles of cumulative Elispot-based CTL responses combined with diversity and sequence consensus data provide an additional characterization of immunodominant regions across the HIV-1C genome. Results of the study suggest that the construction of a poly-epitope subtype-specific HIV-1 vaccine that includes multiple copies of immunodominant CTL epitopes across the viral genome, derived from predominant HIV-1 viruses, might be a logical approach to the design of a vaccine against AIDS.
The extreme genetic diversity of human immunodeficiency virus type 1 (HIV-1) in the global AIDS epidemic coincides with an uneven distribution of HIV-1 strains that have been classified into groups and subtypes on the basis of their phylogenetic relationships (reviewed in reference 97). Causing most of HIV-1 infections worldwide, the most representative group of HIV-1—the M (main) group—is comprised of subtypes A through K. The most severe AIDS epidemic has occurred in sub-Saharan Africa, where an estimated 25.3 million adults and children lived with HIV-AIDS at the end of 2000 (110). The vast majority of HIV-1 infections in sub-Saharan Africa are caused by subtypes A and C (30). The HIV-1 subtype A epidemic, including CRF02_AG-IbNg-like strains (20, 21, 35, 55, 70, 75, 77, 79, 102, 108), dominates in West, Central, and East Africa and reached a plateau at the prevalence rate of 1.5 to 14% by the 1990s (111, 112). In contrast, countries in southern Africa and the horn of Africa have experienced the greatest burden of the AIDS epidemic caused predominantly by HIV-1 subtype C (HIV-1C). The extremely high prevalence rate of HIV-AIDS in Botswana (35.8%), Swaziland (25.3%), Lesotho (23.6%), Zimbabwe (25.1%), Zambia (20%), South Africa (19.9%), Namibia (19.5%), Malawi (16%), and Mozambique (13.2%) (111) urges the design of a vaccine that would be efficacious in these southern African countries.
The design of a vaccine for southern Africa would be impossible without a comprehensive characterization of HIV-1C at the full-length genome level. After the first nearly full-length genome sequencing of HIV-1C performed by Salminen et al. (101), work by several groups (39, 67, 73, 75, 82, 98) has led to the generation of an HIV-1C full-length genome sequence database that is comprised, to date, of 27 nonrecombinant HIV-1C isolates. Phylogenetic analyses of full-length sequences provide valuable information regarding diversity, variability, and consensus sequence across the entire viral genome and, together with biological and phenotypic characterization of HIV-1C variants, significantly facilitates vaccine design.
HIV-1-specific cytotoxic T-lymphocyte (CTL) and T-helper responses are thought to be important components of the immune response in the course and control of HIV-1 infection (reviewed in references 5, 15, 42, 58, 66, and 91). After the detection of high levels of CTLs in HIV-1 patients (113), evidence for the role of CTL in the control of viremia in HIV-1 infection was provided by a series of studies (13, 63) and was recently observed in the simian immunodeficiency virus model (56, 104). Viral escape from CTL recognition was shown to be an intimate part of HIV-1 pathogenesis, thus suggesting that the generation of viral escape mutants might be driven, at least in part, by CTL-mediated selection pressure (14, 41, 46, 84, 93). The role of HIV-1-specific T-helper responses in the control of viral infection was highlighted by showing a strong negative association between viral load and Gag-specific CD4+ T-helper-cell responses (99). A functional link between CTL and T-helper responses was demonstrated by providing evidence that levels of Gag-specific T-helper cells positively correlated with levels of Gag-specific CTLs and negatively correlated with levels of viral load in plasma (57). The finding of circulating HIV-1-specific CD8+ T cells without direct effector activity in the absence of CD4+ T-helper cells (106) underlines the significance of CTL–T-helper joint action. Thus, there is growing evidence that an induction of HIV-1-specific CTL and T-helper responses is an important goal in the development of an AIDS vaccine (47, 52).
Despite the fact that a large number of CTL epitopes across HIV-1 have been identified (62), the systematic mapping of CTL epitopes that are most relevant to epicenters of the global AIDS epidemic is still in its infancy. Most CTL epitope studies performed have been based on HIV-1B and HLA types that are most often found in the Caucasian population. A limited number of CTL studies have targeted non-B subtypes and identified CTL epitopes restricted by HLA types common in non-Caucasian ethnic groups (19, 27, 43, 45, 100), which illustrates that the body of literature relevant to CTL epitopes identified in populations that are worst affected by the AIDS epidemic is still quite small.
Traditionally, the magnitude and frequency of CTL responses have been studied on a per-patient basis. We attempted to analyze the cumulative magnitude of enzyme-linked immunospot (Elispot)-based CTL responses within a cohort of HIV-1-infected blood donors, reasoning that profiles of HIV-1-specific Elispot-based CTL responses could be extrapolated to the population of potential vaccinees. Cumulative analysis of Elispot-based CTL responses performed in this study was expressed as a sum of individual responses or as an average response per study subject. In this study, four HIV-1C proteins—Gag, Tat, Rev, and Nef—were targeted for the screening of potential CTL epitope-rich domains by gamma interferon (IFN-γ)-Elispot, an assay that was successfully applied to identify CTL responses (2, 6–8, 43, 45, 65, 100). The IFN-γ-Elispot assay was shown previously as a reliable method to map optimal CTL epitopes, which allowed testing of a wide spectrum of synthetic peptides and screening of a large number of HIV-infected individuals rapidly and effectively (42, 43, 45). Correlating with more-sensitive methods such as tetramer or intracellular staining (48, 65), the Elispot assay is often a method of choice in the preliminary screening of dominant CTL responses in population studies. The identified and characterized immunodominant regions within HIV-1C Gag, Tat, and Nef might then represent components for further consideration in a poly-epitope vaccine design.
MATERIALS AND METHODS
Study subjects.
Blood donors in Botswana are nonremunerated volunteers who pass a medical examination and sign a consent form before blood is collected. According to the existing protocol, blood donor specimens are tested routinely for HIV-1/2 status by the National Blood Transfusion Center in Gaborone, Botswana, and in the case of an HIV-seropositive test, the blood is discarded.
In this study, discarded units from HIV-seropositive blood donors were used. Sample collection was performed following and according to the guidelines of the Institutional Review Boards of the Ministry of Health of Botswana and the Harvard School of Public Health. Specimens were collected randomly by including in the study the first one to two donors who were tested as HIV seropositive from the daily set of blood donors from February 2000 to October 2000. HIV status was subsequently confirmed in two independent enzyme-linked immunosorbent assays (Ortho Ab-Capture, Ortho-Clinical Diagnostics, Raritan, N.J., and Murex HIV-1.2.0, Abbot-Murex, Murex Biotech Limited, Dartford, United Kingdom).
Table 1 describes relevant data for 74 study subjects. There were 53 male and 21 female subjects ranging in age from 16 to 52 years old (mean, 30.9 years, median, 30 years). Median plasma viral load was 35,082 copies/ml, ranging from <400 to 516,175 copies/ml. CD4 and CD8 data were available for 69 of 74 study subjects (93.2%). Medians for CD4 and CD8 count were 414 and 986 copies/ml, respectively. Viruses were successfully isolated in 56 of 74 cases (75.7%), and all of the isolated viruses were classified as R5. Near-full-length genome sequencing was performed for 22 of 56 viral isolates (39.3%). Samples were assigned for the Elispot screening of Gag-, Tat-, Rev-, or Nef-specific CTL responses based on the actual peripheral blood mononuclear cell (PBMC) viability and availability prior to any relevant information, including viral load, CD4 count, CD8 count, viral sequence, or HLA typing, becoming available.
TABLE 1.
Study subjectsa
| Statistic | Age (yr)b | Viral load (copies/ml)b | CD4 countc | CD8 countc |
|---|---|---|---|---|
| Mean | 30.9 | 93,873 | 430 | 1,040 |
| Median | 30 | 35,082 | 414 | 986 |
| Range | 16–52 | <400d–516,175 | 12–1,116 | 61–>2,000e |
Viruses were isolated in 56 of 74 subjects (75.7%). All 56 were of phenotype R5 (0 were of phenotype X4). Of 56 viral isolates, 22 (39.3%) were near full-length genome sequenced. PBMC from 46 study subjects (62.2%) were screened for HIV-1C Gag-specific CTL responses. PBMC from 48 study subjects (64.9%) were screened for HIV-1C Tat-specific CTL responses. PBMC from 47 study subjects (63.5%) were screened for HIV-1C Rev-specific CTL responses. PBMC from 45 study subjects (60.8%) were screened for HIV-1C Nef-specific CTL responses. Samples were randomly selected and screened in sets of 5 to 10 per HIV-1C protein based on PBMC availability and necessity to repeat Elispot assays.
Data were available for all 74 study subjects.
Data were available for 69 of 74 study subjects.
Viral load of <400 copies/ml (four cases) was treated as 400 for calculation purposes.
CD8 count of >2,000 (two cases) was treated as 2,000 for calculation purposes.
PBMC were separated from whole blood by Lymphocyte Separation Medium (ICN, Aurora, Ohio) within 3 to 4 h of blood collection and were washed in phosphate-buffered saline (PBS), resuspended in cell-freezing medium, aliquoted, and transported frozen to Boston, Mass.
Viral isolation, full-length genome amplification, cloning, and sequencing.
PBMC were thawed, washed twice in R10 medium (RPMI 1640, 10% fetal calf serum, 20 mM HEPES buffer [Sigma-Aldrich], antibiotics [50 U of penicillin-streptomycin/ml]), and short-term cocultured with donor PBMC. DNA was isolated using a QIAamp DNA blood kit (Qiagen, Chatsworth, Calif.). Long-range amplification was performed as described previously (81) using the 626/9690 primer set (26) with some modifications. The amplicon was gel purified with a QIAquick gel extraction kit (Qiagen) and cloned using a TOPO XL PCR cloning kit (Invitrogen, Carlsbad, Calif.). DNA plasmid purification and both-strand sequencing were performed as described previously (81, 82). In addition to the classical d-Rhodamine dye terminator sequencing using a DNA sequencer (model 373A; Applied Biosystems, Foster City, Calif.), Big Dye terminator sequencing using the ABI PRISM 3700 DNA analyzer (Applied Biosystems) was performed.
Viral phenotype.
The phenotype of viral isolates that were short-term cocultured was characterized as described previously (114) with modification (81), using the cell lines U87MG-CD4-CCR5, U87MG-CD4-CXCR4, and U87MG-CD4.
Phylogenetic analysis.
Nucleotide sequences were edited using Sequencher (Gene Codes Corp., Ann Arbor, Mich.). Multiple alignment was performed using ClustalW (107) and BioEdit (51). The pairwise evolutionary distances of nucleotide alignment were computed by the DNADIST program with the Kimura two-parameter model (33, 34). Pairwise distances between translated amino acid alignments were performed by the PROTDIST program with the PAM model (33, 34). To generate trees, alignments were globally gap stripped. A phylogenetic tree was drawn using the Njplot (90) and TreeView (87) programs. Nucleotide variability across viral genes was analyzed as an entropy function by the SWAN program (95). As shown previously (82), the effect of gaps did not alter the profiles of variability plots but hid the extreme regions with the highest level of variability. Thus, having performed analysis on data sets with and without gapped regions, the results shown are based on variability analyses that include gaps. Analysis of synonymous (dS) and nonsynonymous (dN) substitution rates across viral genes or regions was performed by the SNAP program (B. Korber, http://hiv-web.lanl.gov/ [online.]) based on the method of Nei and Gojobori (76), which incorporates a method described in reference 86.
HLA class I typing.
For the molecular HLA class I typing, DNA was isolated from PBMC as described previously (80). “Low-resolution” and “intermediate-resolution” HLA class I typing and subtyping were performed by sequence-specific primer PCR as described previously (80). The “high-resolution” HLA class I typing was carried out by using HLA-A and HLA-B sequencing-based typing kits (PE Biosystems, Foster City, Calif.) according to the manufacturer's instructions.
Viral load testing.
Plasma viral loads were analyzed by the Amplicor Monitor assay (version 1.5; Roche Diagnostics Corporation, Indianapolis, Ind.) according to the manufacturer's instructions.
Synthetic peptides.
PBMC were screened for CTL responses in the Elispot assay within HIV-1C Gag, Tat, Rev, and Nef using overlapping peptides of 15 to 20 amino acids that overlapped by 10 amino acids. Forty-nine HIV-1C Gag synthetic peptides that corresponded to the sequence of isolate 96ZM651.8 (accession number AF286224) and twenty HIV-1B Nef peptides that matched the sequence of isolate BRU (LAV-1, accession number K02013) were provided by the National Institute of Health AIDS Research and Reference Reagent Program. HIV-1C consensus amino acid sequences were generated based on the 30 full-length genome sequences from Botswana (75, 82; Novitsky et al., unpublished data). Consensus sequences were used to design screening peptides for HIV-1C Tat, Rev, and Nef as well as to design 9- to 12-mer peptides that overlapped by 8 to 11 amino acids for fine mapping of Gag- and Tat-specific CTL epitopes. Peptides spanning variable regions were represented by two to three variants. Peptides designed on the basis of HIV-1C consensus sequences were synthesized using 9-fluorenylmethoxy carbonyl chemistry commercially or at the Massachusetts General Hospital Peptide Synthesis Core Facility. Purity of peptides was established by high-performance liquid chromatography and in most cases was >85%.
Elispot assay.
MultiScreen 96-well membrane (Immobilon P, a hydrophobic polyvinylidene difluoride membrane)-bottomed plates (MAIP S45; Millipore) were coated with 100 μl (0.5 μg/ml in PBS) of anti-IFN-γ monoclonal antibody 1-D1K (Mabtech AB, Nacka, Sweden) and incubated at 4°C overnight. Before further use, plates were washed extensively with PBS that contained 0.5% fetal calf serum. Synthetic peptides were added directly to wells at a final concentration of 10 μM. Frozen PBMC were thawed, washed in R10 twice, and plated into the wells at a concentration of 25,000 to 100,000 cells/well. The plates were incubated at 37°C in 5% CO2 from 20 to 40 h and washed extensively with PBS. Biotinylated anti-IFN-γ monoclonal antibody 7-B6-1 (Mabtech AB) was added at a final concentration of 0.5 μg/ml and incubated at room temperature for 1.5 h. Following extensive washing with PBS, streptavidin-alkaline phosphatase conjugate (Mabtech AB) was added, and plates were incubated at room temperature for 45 min. Color development was performed by using an alkaline phosphatase conjugate substrate kit (premixed 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium solutions and color development buffer; Bio-Rad, Hercules, Calif.) at room temperature for 5 to 20 min. Color development was stopped by washing in tap water. IFN-γ-producing cells were counted by direct visualization and were expressed as spot-forming cells (SFC) per million PBMC. The number of specific IFN-γ-secreting T cells was calculated by subtracting the negative control value. The negative controls were <30 SFC/106 PBMC. Cases with higher background were repeated and, if higher background was consistent, such cases were excluded from the analysis. Only responses with a magnitude of >100 SFC/106 PBMC were considered to be positive responses in all screening tests. Wells that contained >50 spots/well were counted as 50. Phytohemagglutinin was used as a positive control. CD8+ specificity of IFN-γ release was confirmed by CD8 and CD4 depletion and enrichment experiments using magnetic CD8 MicroBeads and CD4 MicroBeads (MACS; Miltenyi Biotec, Auburn, Calif.), according to the manufacturer's protocol. Cumulative CTL responses were measured by summarizing the responses of per-patient analyses for each HIV-1C protein analyzed and were expressed as a sum of individual responses or as an average response per study subject. Comparison of cumulative magnitude of CTL responses among HIV-1C Gag, Tat, Rev, and Nef was performed by normalizing data for the amino acid diversity and number of study subjects screened (cumulative CTL responses observed per particular peptide were divided by the amino acid diversity of the region corresponding to the same peptide and divided by the number of study subjects screened).
Statistical analysis.
Statistical analysis and basic graphical delineation were done using SigmaPlot 5.0 (SPSS Inc.) and Microsoft Excel 2000 software (Microsoft Corp.). Additional graphical presentation was prepared using Adobe Illustrator (version 8.0) software. Two sample t tests were used to compare the mean values of diversity among samples and to the consensus sequence. Wilcoxon rank sum tests were used to compare the distribution of cumulative CTL responses to subtype C peptides versus subtype B peptides. A rank-based test was used for this comparison because it appropriately accounts for the censoring of within-well Elispot responses of >50 spots/well. The chi-square goodness-of-fit test was used to assess whether or not the probability of a subject mounting a CTL response to a synthetic peptide depended on the degree to which the synthetic peptide's sequence matched the subject's viral peptide sequence. To assess the relationship between HLA type and the probability of mounting a CTL response to certain synthetic peptides, relative response rates for particular HLA alleles versus all other HLA alleles were calculated, with 95% confidence intervals (95% CI). All tests were two tailed, and a cutoff of 0.05 was used to judge statistical significance.
RESULTS
Nonrecombinant HIV-1C is the predominant virus in Botswana.
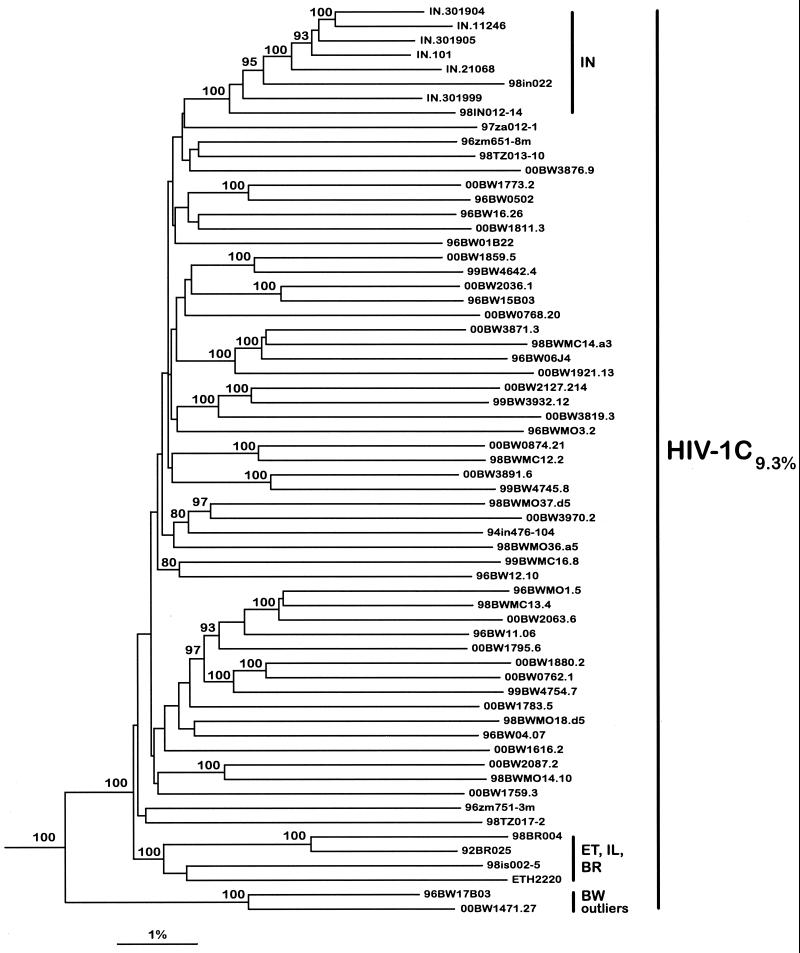
Elaborated phylogenetic analyses of HIV-1C full-length genome sequences have been described recently (82, 98). Figure 1 demonstrates phylogenetic relationships among 63 near-full-length genome sequences of HIV-1C isolates (Botswana [n = 45 {75, 82; Novitsky et al., unpublished data}], India [n = 9], Brazil [n = 2], Tanzania [n = 2], Zambia [n = 2], Ethiopia [n = 1], Israel [n = 1], and South Africa [n = 1]). Forty-five of the near-full-length genome sequences from Botswana clustered within HIV-1C. Two recombinant sequences were found among viral isolates from Botswana (81; Novitsky et al., unpublished data) (data not shown). It is worth noting the relatively high intersample nucleotide diversity, 9.3%, within 63 near-full-length genome HIV-1C sequences. The same degree of diversity was observed within 45 nonrecombinant HIV-1C sequences from Botswana. Taken together, phylogenetic analysis based on near-full-length genome analysis clearly demonstrates that the predominant virus in Botswana, HIV-1C, accounted for 45 of 47 (95.7%) sequences analyzed. As shown in Fig. 1, a neighbor-joining tree reflects the complexity of the phylogenetic relationship within the HIV-1C. From a vaccine standpoint the existence of multiple subclusters and/or lineages within HIV-1C prompts a comprehensive analysis of amino acid consensus sequences corresponding to different subclusters and assessing the extent of their variability and conservation.
FIG. 1.
Phylogenetic relationship of 45 near-full-length HIV-1C sequences from Botswana with other available nonrecombinant HIV-1C sequences from around the world. One isolate from Ethiopia (ETH2220), two from Brazil (92BR025 and 98BR004), nine from India (IN11246, IN301999, IN21068, IN301905, IN301904, IN101, 94IN476-104, 98IN012-14, and 98IN022), two from Zambia (96ZM651-8m and 96ZM751-3m), one from Israel (98IS002-5), two from Tanzania (98TZ013-10 and 98TZ017-2), and one from South Africa (97ZA012-1) were included in the analysis. HIV-1O isolate CM.ANT70 (accession number L20587) was used as an outlier. The neighbor-joining method and the Kimura two-parameter model were used. The bootstrap values of 80% or higher are shown at the nodes supporting branching order. Nucleotide diversity among the whole set of HIV-1C sequences was calculated using the DNADIST program.
All 56 viral isolates in this study demonstrated the R5 phenotype, confirming the HIV-1C preferential use of the CCR5 coreceptor described previously (1, 12, 109). A relatively high plasma viral load (median, 35,082 copies/ml) among healthy blood donors was observed in this study.
HLA class I typing.
Previously we reported the identification of major histocompatibility complex (MHC) class I antigen specificities that were observed at high frequencies in the Botswana population (80). In this study we extended the database of HLA class I types in Botswana and introduced high-resolution HLA typing for the HLA-A and HLA-B loci. Low-resolution or intermediate-resolution sequence-specific primer methods of HLA typing were used for the HLA-C locus. Table 2 enumerates HLA class I alleles found in this study and shows the alleles' relationship to the generic HLA types identified previously in Botswana (80).
TABLE 2.
Distribution and frequency of HLA-A and HLA-B alleles within the common generic HLA types among HIV-1-infected individuals in Botswana (n = 74)
| Locus and antigen specificities | HLA allele | Allele frequency (%) |
|---|---|---|
| HLA-A | ||
| A30 | A∗3001 | 10.8 |
| A∗3002 | 12.8 | |
| A∗3004 | 0.7 | |
| A02 | A∗02011 | 8.1 |
| A∗0202 | 2.7 | |
| A∗0205 | 6.1 | |
| A∗0214 | 0.7 | |
| A23 | A∗2301 | 9.5 |
| A68 | A∗68011 | 0.7 |
| A∗6802 | 9.5 | |
| A29 | A∗2901 | 0.7 |
| A∗2902 | 9.5 | |
| Others | 17 alleles | 28.2 |
| Total HLA-A | 29 alleles | 100.0 |
| HLA-B | ||
| B58 | B∗5801 | 6.8 |
| B∗5802 | 8.1 | |
| B72 | B∗1503 | 10.8 |
| B42 | B∗4201 | 10.8 |
| B44 | B∗44031 | 8.1 |
| B∗44032 | 4.1 | |
| B45 | B∗4501 | 8.8 |
| B∗4503 | 0.7 | |
| B71 | B∗1510 | 8.8 |
| B53 | B∗5301 | 7.4 |
| B∗5305 | 0.7 | |
| Others | 15 alleles | 24.9 |
| Total HLA-B | 26 alleles | 100.0 |
Within the HLA-A locus the most common antigen specificity, A30, was represented by alleles A*3001 (10.8%) and A*3002 (12.8%), with a few cases of HLA-A*3004 (0.7%). The HLA-A02 contained four alleles with a wide range of frequencies (A*02011, 8.1%; A*0202, 2.7%; A*0205, 6.1%; A*0214, 0.7%). HLA-A23 was uniformly represented by A*2301, while alleles within HLA-A68 and -A29 were distributed unevenly (A*68011, 0.7%, and A*6802, 9.5%; A*2901, 0.7%, and A*2902, 9.5%). Within the HLA-B locus, HLA-B58 included comparable proportions of B*5801 (6.8%) and B*5802 (8.1%) alleles. A similar distribution of alleles was observed for B44 (B*44031, 8.1%; B*44032, 4.1%) but not for B45 (B*4501, 8.8%; B*4503, 0.7%) or B53 (B*5301, 7.4% ; B5305, 0.7%). HLA-B42 was represented by B*4201 only (10.8%). Frequencies of HLA typing for the C locus in this study were similar to the frequencies of HLA-C antigen specificities described previously (80).
Immunodominant Elispot-based CTL epitope regions within HIV-1C Gag.
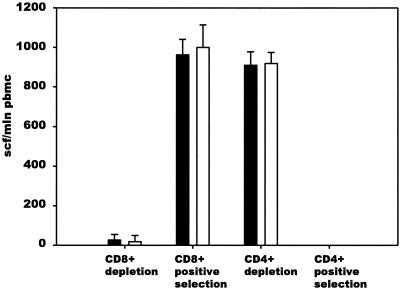
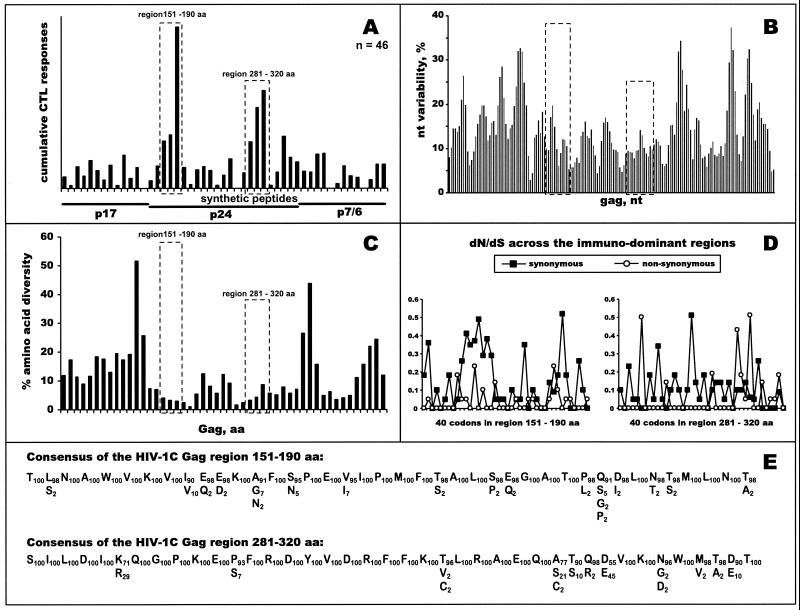
Forty-six HIV-1-infected blood donors from Botswana were screened for HIV-1C Gag-specific CD8+ T-cell responses by using overlapping 20-mer synthetic peptides in the Elispot assay. CD8+ T-cell specificity of IFN-γ responses to synthetic peptides was confirmed in the CD8 and CD4 depletion and enrichment experiments (Fig. 2). A profile of cumulative HIV-1C Gag-specific CTL responses is shown in Fig. 3A. Dominant HIV-1C Gag-specific CTL responses clustered within two regions of p24 that correspond to the amino acids 151 to 190 and amino acids 281 to 320 of the Gag protein according to the HXB2 numbering system (61). Forty amino acids of the second immunodominant region included the entire major homology region. Nineteen of 46 tested blood donors (41.3%) demonstrated CTL responses, with a magnitude of more than 100 SFC/106 PBMC, to one or more peptides within the first immunodominant region (peptides TLNAWVKVIEEKAFSPEVIP, EKAFSPEVIPMFTALSEGAT, and MFTALSEGATPQDLNTMLNT). The magnitude of response in the first region ranged from 103 to 1,447 SFC/106 PBMC (median, 495 SFC/106 PBMC; mean, 558 SFC/106 PBMC). The responses identified for the second immunodominant region (peptides SILDIKQGPKEPFRDYVDRF, EPFRDYVDRFFKTLRAEQAT, and FKTLRAEQATQEVKNWMTDT) were seen in 16 of 46 individuals (34.8%) and ranged from 100 to 1,250 SFC/106 PBMC (median, 500 SFC/106 PBMC; mean, 496 SFC/106 PBMC).
FIG. 2.
CD8+ T-cell specificity of IFN-γ responses to synthetic peptides. PBMC from donor 00BW1774 were CD8+ CD4+ depleted or enriched by using MACS CD8 MicroBeads and CD4 MicroBeads followed by Elispot assay with 20-mer Gag p24 peptide MFTALSEGATPQDLNTMLNT and 9-mer Gag p24 peptide TPQDLNTML (G180-TL9). Experiments were performed in triplicate. The extent of standard deviation is shown (error bars).
FIG. 3.
Profiles of HIV-1C Gag-specific CTL responses. (A) Forty-six subjects were screened in the IFN-γ-Elispot assay using 49 HIV-1C-based 20-mer overlapping synthetic peptides (x axis). Cumulative Elispot-based CTL responses were expressed as a sum of the per-patient responses to a particular peptide (y axis). Immunodominant regions are boxed with dashed lines. HIV-1C Gag p17, p24, and p7/p6 are delineated. (B) Nucleotide variability. Nucleotide variability was analyzed across the HIV-1C gag alignment of nucleotide sequences from Botswana isolates by using a sliding window of 30 nucleotides and increments of 10 nucleotides. Dashed boxes correspond to identified immunodominant regions within HIV-1C Gag p24. (C) Amino acid diversity across HIV-1C Gag. Fragments of amino acid alignment analyzed by the program PROTDIST from the PHYLIP package corresponded to 49 overlapping peptides used in the Elispot assay. Dashed boxes correspond to identified immunodominant regions within HIV-1C Gag p24. (D) dN/dS substitution rates: dN and dS were analyzed using the SNAP program and are shown per codon within identified immunodominant regions. (E) Consensus of immunodominant regions. The main row represents a consensus amino acid sequence of immunodominant regions within HIV-1C Gag. Residues beneath the main row represent amino acid variation at a particular position. Subscript numbers correspond to the percent occurrence of amino acids within HIV-1C Gag.
Figure 3B demonstrates nucleotide variability across the HIV-1C gag gene. Both immunodominant CTL regions (boxed) were relatively conserved (11.3 and 9.9% for the first and second regions, respectively, versus 14% for the entire gag). Mean values of diversity among samples within the immunodominant regions were 6.7 and 5.4% versus 8% for the entire HIV-1C gag. Mean values of diversity in comparison to the consensus sequence were 3.8 and 3.2% for the first and second immunodominant regions, respectively, compared to 5.4% for the gag gene (P < 0.001 for both comparisons). Translated amino acid diversity across HIV-1C Gag is shown in Fig. 3C. Diversity among samples within the first and second immunodominant regions was 3.5 and 5.9%, respectively, while across the entire HIV-1C Gag it was 10.3%. Mean values of diversity in comparison to the consensus amino acid sequence were 1.2 and 2.8% for the first and second immunodominant regions, respectively, compared to 5.4% for the Gag protein (P < 0.001 for both comparisons). Analysis of nonsynonymous and synonymous substitutions resulted in low dN/dS ratios overall in the first and second regions (0.05 and 0.13, respectively), which might suggest that the majority of nucleotide mutations in the immunodominant regions within the HIV-1C gag were under the purifying selection. Profiles of synonymous and nonsynonymous substitutions per codon across the two immunodominant regions are presented in Fig. 3D and confirmed the dominance of synonymous substitutions in those regions, although some codons in the second immunodominant region appeared to be under diversifying positive selection by showing a relatively high dN/dS ratio.
A consensus of amino acid sequences for the dominant CTL regions within HIV-1C Gag was generated (Fig. 3E). Twenty-four amino acid positions in the first immunodominant region (60%) and 28 positions in the second region (70%) were invariable. Among variable amino acid residues within the first immunodominant region, variation ranged from 2 to 10% per amino acid position, thus suggesting a high level of conservation. Polymorphism within the second region ranged from 2 to 45% per amino acid position, although a relatively high level of variation (>10%) was observed for only three amino acids (positions 286, K71/R29; 309, A77/S21/C2; and 312, D55/E45 [subscripted numbers represent the mean percentages of occurrence of the particular amino acids]). These results suggest the necessity to include multiple variants of the second immunodominant region in vaccine constructs.
Performing full-length genome sequencing of viral isolates and screening for CTL responses on the subset of the same study subjects allowed us to analyze the extent of match and mismatch between the autologous viral sequence and the sequence of the synthetic peptides for the observed CTL responses (more than 100 SFC/106 PBMC) across the entire Gag. A subset of 14 samples (out of 46 study subjects screened for Gag-specific CTL responses) was sequenced, and translated Gag amino acid sequences were aligned together with clone 96ZM651.8—a source sequence for the peptide synthesis (data not shown). Within the 49 detected CTL responses among 14 sequences from study subjects across the alignment, a complete match between viral and synthetic peptide sequence was found in 15 cases (30.6%), a single amino acid mismatch was also found in 15 cases (30.6%), a two-amino-acid mismatch was seen in 8 cases (16.3%), and a mismatch of three or more amino acids occurred in 11 cases (22.4%). There was not a trend of superior responsiveness to peptides with fewer mismatches (P = 0.42 [chi-square test]). The finding of CTL recognition in the case of mismatch between autologous viral sequence and pulsed synthetic peptide might suggest that mismatched amino acids were not in the position of functionally important anchor residues and/or that epitope presentation was not altered because of flexibility of the MHC class I molecules for the successful presentation of peptide to CD8+ cells (96), although a precise defining of CTL response might be required. Further analysis of match and mismatch data could be useful for fine CTL epitope mapping and the definition of HLA class I restriction.
The distribution of HIV-1C Gag-specific Elispot-based CTL responses was analyzed for subsets of common HLA alleles that were seen at increased frequency. The inclusion criteria was set to ≥30%, meaning that at least 30% of study subjects in the subset of carriers of a particular HLA class I allele should demonstrate peptide-specific CTL responses to be included in the analysis. The most responsive were carriers of allele HLA-A*4201; 7 of 11 subjects (64%) demonstrated CTL responses to the peptide MFTALSEGATPQDLNTMLNT, which contained the CTL epitope TPQDLNTML described previously (45). Four of eight individuals (50%) who were positive for HLA-A*02011 responded to the peptide SNFKGNKRMVKCFNCGKEGH, while three of six (50%) carriers of HLA-B*44031 showed CTL responses to the peptide EPFRDYVDRFFKTLRAEQAT—thus suggesting the existence of potential CTL epitopes within these peptides, which are restricted by A*02011 and B*44031, respectively. Other observed HLA class I alleles were associated with a low frequency of CTL responses across the HIV-1C Gag genome. The CTL epitope in Gag p17 RLSYNTVATLY (residues 76 to 86) was described previously as the strongest CTL response in two HLA-A*3002-positive individuals (43). In our study, only three of thirteen (23.1%) A*3002-positive subjects demonstrated moderate CTL responses to the peptide GTEELRSLYNTVATLYCVHE (residues 71 to 90), which contained the epitope RLSYNTVATLY. Interestingly, the breadth of HIV-1C Gag-specific CTL responses was uneven among subjects expressing different HLA class I alleles. Thus, while HLA-A*3002 and HLA-B*1510 alleles were likely to restrict multiple CTL epitopes across HIV-1C Gag, HLA-A*2902, -B*5301, and -B*5802 could not be associated with any HIV-1C Gag-specific CTL responses. If confirmed in a larger sample set, this finding suggests that genetic background may play an important role in the ability to generate broad CTL responses and to control (or not control) HIV-1 infection.
HIV-1C-specific Elispot-based CTL responses against Tat peptides.
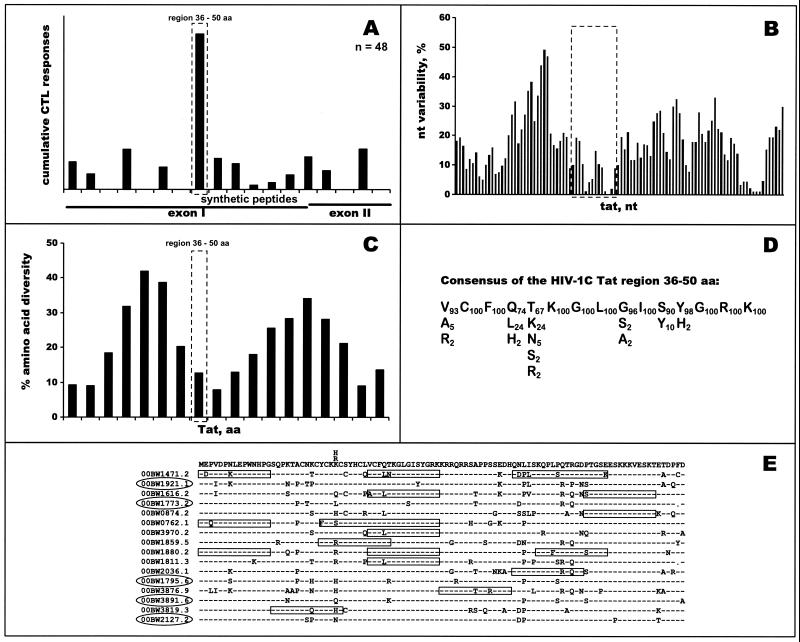
Figure 4A demonstrates profiles of cumulative HIV-1C Tat-specific Elispot-based CTL responses among 48 HIV-1-infected blood donors. Seventeen tested subjects (35.4%) showed CTL responses to the 15-mer peptide VCFQTKGLGISYGRK (amino acid positions 36 to 50, HXB2 numbering system [61]). The magnitude of responses ranged from 150 to 667 SFC/106 PBMC (median, 291 SFC/106 PBMC; mean, 316 SFC/106 PBMC). The identified immunodominant region in the HIV-1C tat has a low level of nucleotide variability (Fig. 4B). The mean value of nucleotide diversity among samples within the region was 5.3%, compared to 10.5% through the entire HIV-1C tat gene. The mean value of diversity in comparison to the consensus sequence was 3.0% in the immunodominant region compared to 5.9% in the tat gene (P < 0.001). Analysis of synonymous and nonsynonymous substitution rates revealed that the dN/dS ratio for the entire immunodominant region was 0.66, while the dN/dS ratio for the fourth and fifth codon was equal to 11 and 19, respectively, suggesting a high diversifying positive selection for these amino acids. As shown in Fig. 4C, diversity of translated amino acid sequences among samples was also significantly lower in the immunodominant region compared with the entire HIV-1C Tat, with mean values of 12.8% (95% CI, 12.2%, 13.4%) to 19.2% (95% CI, 18.8%, 19.6%). The mean value of amino acid diversity in comparison to the consensus sequence within the immunodominant region was 7.5%, compared to 11.0% for the entire Tat (P < 0.01). Figure 4D depicts the consensus across the immunodominant region of HIV-1C Tat and its amino acid variation. Being relatively conserved, the identified immunodominant region demonstrates leucine instead of glutamine at position 4 in 24% of samples and lysine instead of threonine at position 5 in 24% of samples, which suggests that these amino acid variations should be taken into account at the point of vaccine design.
FIG. 4.
Profiles of HIV-1C Tat-specific CTL response. (A) Forty-eight subjects were screened in the IFN-γ-Elispot assay using 18 HIV-1C-based 15-mer synthetic peptides overlapping by 10 amino acids (x axis). Cumulative Elispot-based CTL responses were expressed as a sum of the per-patient responses to a particular peptide (y axis). The immunodominant region is delineated with a dashed box. (B) Nucleotide variability. Nucleotide variability was analyzed across the HIV-1C tat alignment of nucleotide sequences from Botswana isolates by using a sliding window of 10 nucleotides and increments of 3 nucleotides. A dashed box corresponds to the identified immunodominant region within HIV-1C Tat. (C) Amino acid diversity across HIV-1C Tat. Fragments of amino acid alignment analyzed by the program PROTDIST from the PHYLIP package corresponded to the overlapping peptides that were used in the Elispot assay. The dashed box corresponds to the identified immunodominant region within HIV-1C Tat. (D) Consensus of immunodominant region. Amino acid consensus of the immunodominant region within HIV-1C Tat is shown as a sequence in the main row. Residues beneath the main row represent amino acid variation at a particular position. Subscript numbers correspond to the percentage of amino acid occurrence within HIV-1C Tat. (E) Tat-specific CTL responses among a subset of 16 sequenced study subjects. The top line of alignment represents contiguous sequence of 15-mer synthetic peptides used in the Elispot assay. Letters above the top line of alignment correspond to the synthetic peptide variants. Dashes throughout the alignment show the identical amino acids within viral isolates. Boxes across the alignment designate identified CTL responses. Boxes longer than 15 amino acids represent CTL responses to the overlapping peptides. Study subjects that did not demonstrate HIV-1C Tat-specific CTL responses in the Elispot assay are circled.
Alignment of translated amino acid sequences for 16 of 48 subjects who were tested for HIV-1C Tat-specific CTL responses is shown in Fig. 4E. The top line of the alignment corresponds to the sequence of synthetic peptides used. Nineteen Elispot-based CTL responses that were identified are boxed across the alignment. In five cases no Tat-specific responses were observed. Surprisingly, most of the CTL responses occurred when there was a mismatch between the autologous viral sequence and the synthetic peptide; complete matches were seen only in 4 of 19 cases (21%), while all remaining CTL responses (79%) were mismatched (one amino acid mismatch in 4 cases, two amino acid mismatches in 10 cases, and three amino acid mismatches in 1 case). Analyses performed with multiple peptide variants, whose sequences reflected naturally occurring viral sequences, followed by a comparison of responses to autologous viral sequences revealed the existence of mismatched Elispot-based CTL responses in some cases. The mismatched CTL responses could be characterized by (i) detectable CTL response to the peptide that mismatches to autologous viral sequence and (ii) an absence of CTL response to the peptide that completely matches the autologous viral sequence. For example, the translated amino acid sequence of the isolate 00BW3819.3 was SQPKTACNQCYCKHC. Despite the mismatch at position 24Q (HXB2 numbering system [61]), there was CTL recognition of the mismatched peptide SQPKTACNKCYCKRC (position 29 [italicized] had R instead of H) but not SQPKTACNKCYCKHC or SQPKTACNKCYCKKC. This finding might suggest CTL recognition of the progenitor viral variant that had R at position 29 and, perhaps, viral escape from CTL recognition by mutation R29H. In the case of isolate 00BW1859.5, there was CTL recognition of the peptide that was identical to the viral sequence YCKRCSYHCLVCFQT and also of the peptide with a single mismatch (R29K), but 00BW1859.5 had no recognition of another peptide with mismatch R29H, suggesting that a change from one basic amino acid to a similar one might not alter CTL recognition, while a mutation like R to H might be more dramatic and abrogate CTL recognition.
Screening for Elispot-based CTL responses across HIV-1C Rev.
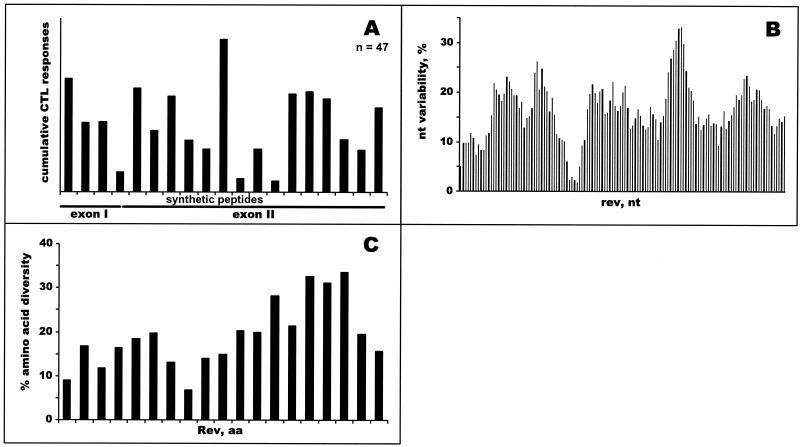
Analyses of HIV-1C Rev-specific CTL responses demonstrated scattering of cumulative responses across Rev without highlighting immunodominant regions (Fig. 5A). Compared with Gag and Tat, low variability and/or low diversity regions of Rev did not overlap with higher levels of cumulative CTL responses (Fig. 5B and C). Overall, 27 of 47 subjects (57%) demonstrated HIV-1C Rev-specific Elispot-based CTL responses of more than 100 SFC/106 PBMC. The magnitude among responders ranged from 120 to 1,074 SFC/106 PBMC (mean, 452 SFC/106 PBMC, median, 320 SFC/106 PBMC), demonstrating a wide extent of CTL response variation across the HIV-1C Rev and among the study subjects. Analysis of dN/dS substitution rates across the entire HIV-1C rev revealed dominance of synonymous substitutions over nonsynonymous. Averages of all pairwise comparisons were 0.08 and 0.16 for dN and dS, respectively, while the dN/dS ratio was equal to 0.5, which suggests relatively low selection pressure across HIV-1C Rev.
FIG. 5.
Profiles of HIV-1C Rev-specific CTL responses. (A) Forty-seven subjects were screened in the IFN-γ-Elispot assay using 19 HIV-1C-based 15-mer synthetic peptides overlapping by 10 amino acids (x axis). Cumulative Elispot-based CTL responses were expressed as a sum of the per-patient responses to a particular peptide (y axis). (B) Nucleotide variability. Nucleotide variability was analyzed across HIV-1C rev alignment of nucleotide sequences from Botswana isolates by using a sliding window of 20 nucleotides and increments of 3 nucleotides. (C) Amino acid diversity across HIV-1C Rev. Fragments of amino acid alignment analyzed by the program PROTDIST from the PHYLIP package corresponded to nineteen synthetic peptides that were used in the Elispot assay.
Dominant Elispot-based CTL epitope regions within HIV-1C Nef.
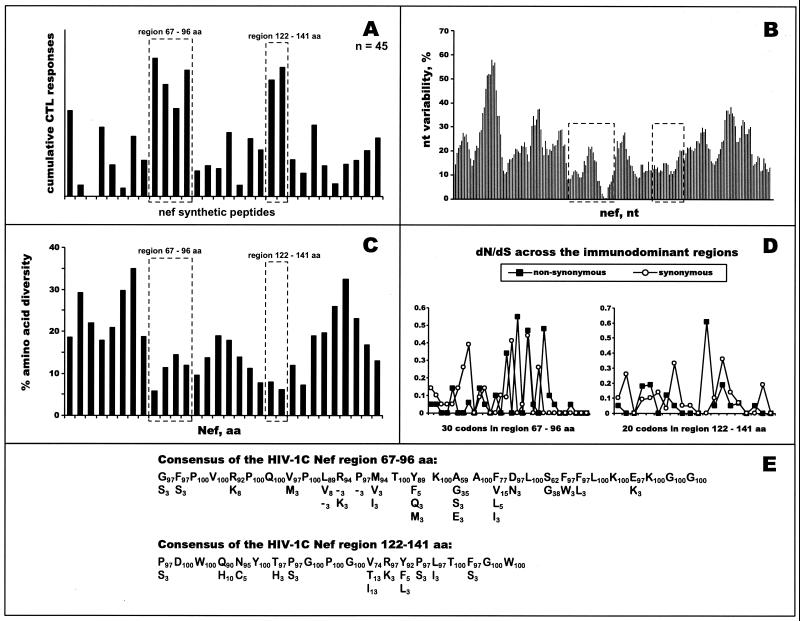
Figure 6A shows the profile of cumulative HIV-1C Nef-specific CTL responses determined among 45 HIV-1-infected blood donors. Thirty-seven of 45 subjects (82%) demonstrated CTL responses that exceeded 100 SFC/106 PBMC. Collectively, the magnitude of HIV-1C Nef-specific Elispot-based CTL responses across the entire HIV-1C Nef ranged from 120 to 2,430 SFC/106 PBMC (mean, 928 SFC/106 PBMC; median, 860 SFC/106 PBMC) and was comprised of from one to six (median, three) responses among a subset of responders. As shown in Fig. 6A, two immunodominant regions were identified. The first one spanned amino acid positions 67 to 96 (HXB2 numbering system [61]), while the second corresponded to amino acid positions 122 to 141 (61). Twenty-two subjects (49%) responded to one or more peptides from the first immunodominant region, demonstrating a magnitude of CTL responses from 124 to 728 SFC/106 PBMC (mean, 309 SFC/106 PBMC; median, 287 SFC/106 PBMC). Within the second immunodominant region, CTL responses were observed among 12 of 45 screened subjects (27%). The magnitude of CTL responses varied from 155 to 1.057 SFC/106 PBMC (mean, 352 SFC/106 PBMC; median, 290 SFC/106 PBMC).
FIG. 6.
Profiles of HIV-1C Nef-specific CTL responses. (A) Forty-five subjects were screened in the IFN-γ-Elispot assay using 30 HIV-1C-based 15- to 20-mer overlapping synthetic peptides (x axis). Cumulative Elispot-based CTL responses were expressed as a sum of the per-patient responses to a particular peptide (y axis). Immunodominant regions are boxed with dashed lines. (B) nucleotide variability. Nucleotide variability was analyzed across the HIV-1C nef alignment of nucleotide sequences from Botswana isolates by using a sliding window of 20 nucleotides and increments of 3 nucleotides. Dashed boxes correspond to identified immunodominant regions within HIV-1C Nef. (C) Amino acid diversity across HIV-1C Nef. Fragments of amino acid alignment analyzed by the program PROTDIST from the PHYLIP package corresponded to synthetic peptides that were used in the Elispot assay. Dashed boxes correspond to identified immunodominant regions within HIV-1C Nef. (D) dN/dS substitution rates. dN and dS were analyzed using the SNAP program and are shown per codon within identified immunodominant regions. (E) Consensus of immunodominant regions. Amino acid consensus of immunodominant regions within HIV-1C Nef are shown as sequences in the main rows. Residues beneath the main row represent amino acid variation at a particular position. Subscript numbers correspond to the percentage of amino acid occurrence within HIV-1C Nef.
Similarly to Gag and Tat, identified CTL immunodominant regions within the HIV-1C Nef overlapped with conserved regions on both the nucleotide (Fig. 6B) and amino acid (Fig. 6C) levels. The mean values of nucleotide diversity among samples within the first (6.4%) and second (6.5%) immunodominant regions were significantly lower than diversity across the entire HIV-1C nef gene (11.5%). The mean values of diversity in comparison to the consensus sequence were 3.9 and 3.5% for the first and second immunodominant regions, respectively, compared with 6.3% for the nef gene (P < 0.001 for both comparisons). The mean values of amino acid diversity among samples were 10.5 and 8.3% for the first and second regions, respectively, while diversity for the HIV-1C Nef was equal to 18.3%. The mean values of amino acid diversity in comparison to the consensus sequence were 6.3 and 4.5% for the first and second immunodominant regions, respectively, compared with 9.8% for the Nef (P < 0.001 for both comparisons). Rates of dN/dS substitutions within both the first (0.041/0.163) and the second (0.037/0.170) immunodominant regions suggested a higher level of synonymous mutations. Figure 6D demonstrates rates of synonymous and nonsynonymous substitutions across both immunodominant regions per codon and highlights spikes of nonsynonymous mutations for relatively variable amino acids.
The consensus of CTL immunodominant regions within HIV-1C Nef is shown in Fig. 6E. Amino acid diversity across the first immunodominant region was relatively low, demonstrating from 3 to 8% diversity per amino acid residue in most cases, while 12 of 30 amino acids (40%) were invariable. However, in four cases amino acids were more diverse—positions 81 (Y89/F5/Q3/M3), 83 (A59/G35/E3/S3), 85 (F77/V15/L5/I3), and 88 (S62/G38). Within most amino acid residues of the second immunodominant region, diversity did not exceed 10%. Nine amino acids in the second region were invariable (45%), while increased diversity was seen at position 133 (V74/T13/I13).
Magnitude of subtype-specific Elispot-based CTL responses in Nef is higher than subtype-nonspecific CTL responses.
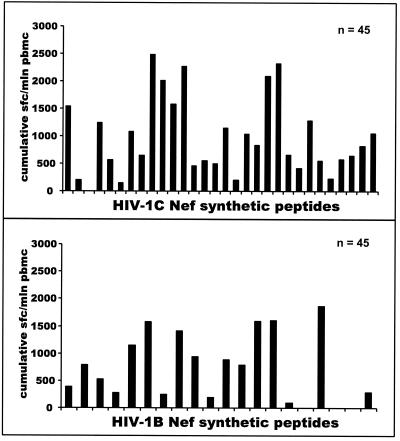
To compare subtype-specific CTL responses with subtype-independent responses, we screened PBMC from HIV-1C-infected blood donors using a set of HIV-1B Nef synthetic peptides. Figure 7 depicts differences between the profile of HIV-1C Nef-specific CTL responses and HIV-1B Nef-specific responses. The overall magnitude of Nef-specific Elispot-based CTL responses among HIV-1C-infected individuals was significantly higher relative to subtype C peptides than to subtype B peptides (mean and median of 763 and 780 SFC/106 PBMC, respectively, among responses to HIV-1C versus mean and median of 318 and 257 SFC/106 PBMC, respectively, among responses to HIV-1B [P < 0.0001; Wilcoxon test for the entire group of 45 samples], or a mean and median of 928 and 860 SFC/106 PBMC, respectively, among responses to HIV-1C versus mean and median of 398 and 279 SFC/106 PBMC, respectively, among responses to HIV-1B [P < 0.0001; Wilcoxon test for the subset of responders only]). The number of nonresponders was similar in both groups (eight and nine for HIV-1C and HIV-1B, respectively). HIV-1C-infected individuals targeted more CTL epitopes within HIV-1C Nef, compared with the HIV-1B Nef (median of three epitopes compared with a median of one epitope; mean of 3.1 epitopes compared with a mean of 1.42 epitopes [P < 0.0001; Wilcoxon test]).
FIG. 7.
Comparison of HIV-1C-specific CTL responses with HIV-1B-specific responses in Nef. Forty-five HIV-1C-infected subjects were screened in the IFN-γ-Elispot assay using 30 HIV-1C-based 15- to 20-mer overlapping synthetic peptides and 20 HIV-1B-based 20-mer peptides (x axis). Cumulative Elispot-based CTL responses were measured as a sum of the per-patient responses to a particular peptide and expressed as the number of cumulative SFC/106 PBMC (y axis).
Comparison of HIV-1C Gag-, Tat-, Rev-, and Nef-specific Elispot-based CTL responses.
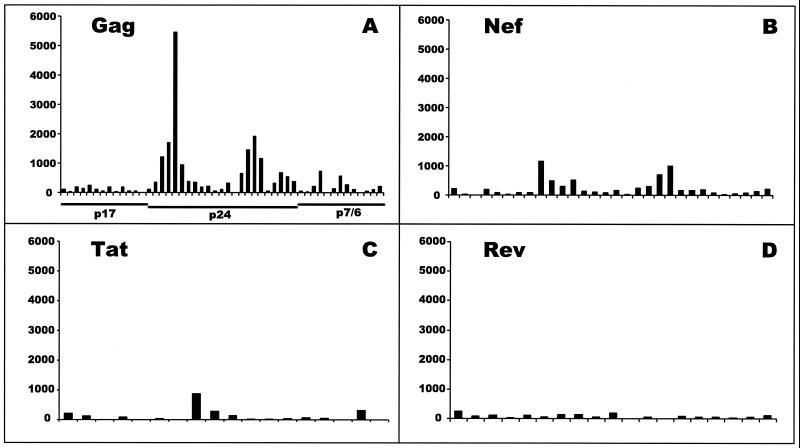
The cumulative magnitude, as well as the magnitude adjusted to the protein length for HIV-1C-specific Elispot-based CTL responses to the epitopes embedded within Gag, Tat, Rev, and Nef, are shown in Table 3. Although cumulative Elispot-based CTL responses to Gag-specific epitopes were the highest among the four HIV-1 proteins, when adjusted to the number of amino acids, scores were compatible among the four (Gag, 2.3; Tat, 3.1; Rev, 2.4; Nef, 3.7). This observation suggests a relatively equal density of CTL epitope distribution across HIV-1C Gag, Tat, Rev, and Nef when adjusted to the protein's length.
TABLE 3.
Cumulative and amino acid-adjusted magnitude of HIV-1C-specific CTL responses to the epitopes within Gag, Tat, Rev, and Nef
| HIV-1C protein | Cumulative CTL response
|
Amino acid-adjusted score
|
|||
|---|---|---|---|---|---|
| Mean | Median | No. of amino acids | Mean | Median | |
| Gag | 1,157 | 887 | 494 | 2.3 | 1.8 |
| Tat | 313 | 275 | 101 | 3.1 | 2.7 |
| Rev | 259 | 176 | 107 | 2.4 | 1.6 |
| Nef | 763 | 780 | 207 | 3.7 | 3.8 |
We hypothesized that the analysis of cumulative CTL responses normalized by amino acid diversity could be an alternative way to characterize CTL responses and to compare data obtained for different viral proteins. The magnitude of cumulative HIV-1C-specific CTL responses within Gag, Tat, Rev, and Nef was brought to the same scale (Fig. 8). Applied analysis highlighted CTL responses to the epitopes in the conserved regions. Peaks corresponded to the immunodominant regions that were described earlier in the study. The highest magnitude of Elispot-based CTL responses within Gag might suggest a dominant role for Gag-specific CTL epitopes in the natural course of HIV-1C infection compared to Tat-, Rev-, and Nef-specific epitopes. In regards to a prospective vaccine, this finding might have at least two implications. First, eliciting a high magnitude of Gag-specific CTL responses by including multiple copies of Gag-immunodominant regions in the vaccine might be an achievable and realistic goal. Second, vaccine design might require reinforcement of the Tat-, Rev-, and Nef-specific CTL responses by improving delivery and peptide presentation if any of the Tat-, Rev-, and/or Nef-specific CTL epitopes are proven to be protective.
FIG. 8.
Magnitude of HIV-1C-specific Elispot-based CTL responses in Gag, Tat, Rev, and Nef expressed as cumulative CTL responses normalized by amino acid diversity (per peptide) and number of study subjects screened.
Fine mapping of HIV-1C Gag-specific CTL epitopes.
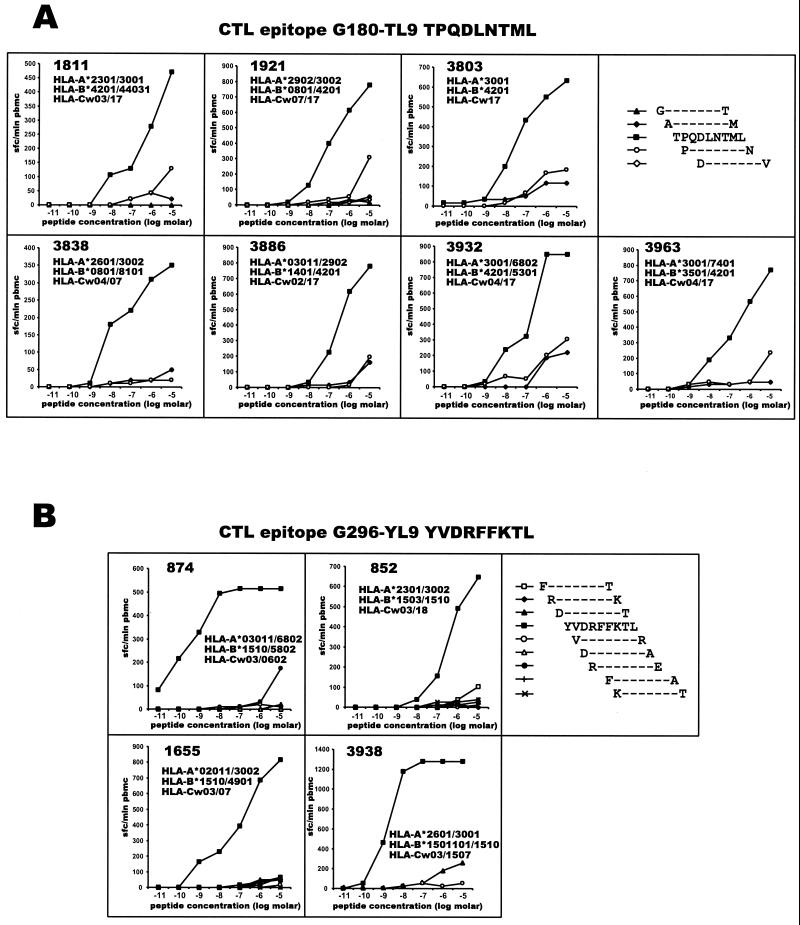
Serial dilutions of 9-mer peptides were used in the Elispot assay for a fine mapping of the CD8+ T-cell responses. B42 and B81 were shown previously to be the antigen specificities responsible for the restriction of the epitope TPQDLNTML (45) within HIV-1 Gag. Figure 9A summarizes a series of experiments designed to confirm recognition of the CTL epitope TPQDLNTML and to identify the restriction HLA allele. PBMC from seven subjects who demonstrated CTL responses to the 20-mer peptide MFTALSEGATPQDLNTMLNT in Gag-screening experiments were further analyzed using serial tenfold dilutions of 9-mer peptides that spanned the 20-mer peptide. As shown in Fig. 9A, the peptide TPQDLNTML induced specific IFN-γ production by T cells at the lowest concentration. High-resolution HLA class I typing allowed us to specify B*4201 and B*8101 as HLA alleles that might restrict the CTL epitope TPQDLNTML (amino acid positions 180 to 188 in Gag, HXB2 numbering system [61]). Results of a similar fine mapping of another CTL epitope, YVDRFFKTL, within HIV-1C Gag are presented in Fig. 9B. The CTL epitope YVDRFFKTL (amino acid positions 296 to 304 in Gag, HXB2 numbering system [61]) might be restricted by HLA-B*1510. Interestingly, all four subjects who demonstrated responses to the CTL epitope YVDRFFKTL also shared HLA-Cw03, thus suggesting linkage disequilibrium between these two HLA types. However, other study subjects who expressed HLA-Cw03 but not HLA-B*1510 did not recognize epitope YVDRFFKRL, suggesting irrelevance of HLA-Cw03 alone in the restriction of this epitope. Recently an HIV-1B variant of the epitope YVDRFYKTL (mutated position in italics) was described (83), and an HIV-1A-infected patient demonstrated CTL recognition of this epitope (27). HLA restriction of the epitope was suggested as A26 or B70, while the HLA-C locus apparently was not typed in either study (27, 83). Our data confirmed that CTL epitope YVDRFFKTL might be restricted by HLA-B*1510 (equivalent of serological specificity B70) and suggests association of HLA*B1510 with HLA-Cw03 (linkage disequilibrium).
FIG. 9.
Fine mapping of HIV-1C Gag p24-specific CTL epitopes. (A) Gag p24 epitope TPQDLNTML (G180-TL9) is restricted by HLA-B*4201 or HLA-B*8101. PBMC from seven subjects who expressed HLA-B*4201 or -B*8101 were analyzed in an Elispot assay. The frequency of responses is expressed as SFC per million PBMC. Titration curves represent CTL responses to serial dilutions of synthetic peptides. Subjects who were negative for HLA-B*4201 or -B*8101 did not demonstrate CTL responses to the G180-TL9 epitope (data not shown). (B) Gag p24 epitope YVDRFFKTL (G296-YL9) is restricted by HLA-B*1510. PMBC from four subjects who expressed HLA-B*1510 were analyzed in an Elispot assay. The frequency of responses is expressed as SFC per million PBMC. Titration curves represent CTL responses to serial dilutions of synthetic peptides. Subjects who were negative for HLA-B*1510 did not demonstrate CTL responses to the G296-YV9 epitope (data not shown).
Fine mapping of HIV-1C Tat-specific CTL epitope.
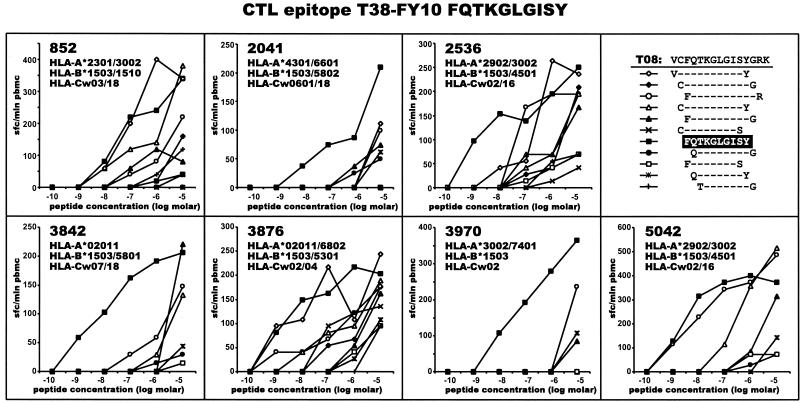
The identified immunodominant region within HIV-1C Tat that corresponded to the sequence VCFQTKGLGISYGRK (amino acid positions 36 to 50, HXB2 numbering system [61]) was further analyzed with a set of 9- to 12-mer peptides among a subset of study subjects who demonstrated responses to the 15-mer peptide. Results of the fine mapping are presented in Fig. 10. The optimal epitope corresponded to the 10-mer peptide FQTKGLGISY (positions 38 to 47, HXB2 numbering system [61]). As shown in Fig. 10, the only common HLA allele among study subjects who demonstrated responses to the peptide FQTKGLGISY was HLA-B*1503, which suggests that HLA-B*1503 might restrict the identified CTL epitope within HIV-1C Tat.
FIG. 10.
Fine mapping of HIV-1C Tat CTL epitope FQTKGLGISY (T38-FY10) restricted by HLA-B*1503. PBMC from subjects who demonstrated responses to 15-mer peptide VCFQTKGLGISYGRK were analyzed in peptide titration experiments using a set of 22 synthetic peptides from 9- to 12-mer. The frequency of responses is expressed as SFC per million PBMC. Titration curves represent CTL responses to serial dilutions of synthetic peptides. Only responses relative to peptide FQTKGLGISY are shown. Subjects who were negative for HLA-B*1503 did not demonstrate CTL responses to the T38-FY10 epitope (data not shown).
DISCUSSION
This study was performed to facilitate the design of an efficacious anti-HIV-1 vaccine for southern Africa, where the burden of the AIDS epidemic caused by HIV-1C is extremely high, by Elispot-based identification of CTL-rich regions across HIV-1C Gag, Tat, Rev, and Nef. Hypothetically, an ideal HIV vaccine would contain multiple, highly responsive epitopes (CTL, T-helper, and neutralizing) derived from the locally circulating viral strains that cumulatively and complementarily would protect the host from HIV-1 infection, or, as a more realistic goal, could control HIV-1 infection, prevent progression to AIDS, and diminish HIV-1 transmission rate (“containment rather than eradication”) (5, 52). Although the real protective capability and efficacy of any vaccine candidate could be estimated only upon completion of vaccine efficacy trials, a comprehensive analysis of predominant viruses and naturally occurring CTL responses in the population might shed light on vaccine design. Here we attempted to facilitate HIV-1 vaccine development by mapping and characterizing CTL-rich regions within the HIV-1C proteins Gag, Tat, Rev, and Nef, which might be important components of a poly-epitope vaccine. This study is an initial step in the identification of HIV-1C-specific CTL responses across the entire viral genome, and it did not intend to diminish the importance of potential CTL, T-helper, or neutralizing epitopes within other HIV-1C proteins. Detailed analysis of HIV-1C-specific CTL responses among infected blood donors in Botswana allowed us to identify and characterize immunodominant Elispot-based CTL-rich regions within HIV-1C Gag, Tat, Rev, and Nef. Results of this study suggest the following: (i) the magnitude of CTL responses within the population can be estimated by a cumulative profile of CTL responses to the predominant virus among a representative group of infected individuals; (ii) genetic background of a particular population might restrict the breadth and magnitude of CTL responses through the diversity and frequency of MHC class I HLA alleles within the population; and (iii) genetic properties of the predominant viruses circulating in the local area might affect breadth and magnitude of CTL responses and may impose viral escape from CTL recognition.
This study suggested that identified immunodominant regions within HIV-1C Gag, Tat, and Nef might be relevant for vaccine formulation. Additionally, including both dominant and subdominant CTL epitopes (7, 36) might be useful to provide protection after emergence of escape variants. From both viral escape and HLA restriction perspectives, key CTL epitopes represented by multiple variants of the same epitope might be advantageous compared with single copy epitopes.
Identification of immunodominant CTL regions and epitopes across the viral genome might help clarify the issue of clade specificity versus cross-clade reactivity for AIDS vaccine design. A significant number of CTL epitopes that are located in highly conserved regions of the viral genome and are almost identical among HIV-1 clades might be responsible for the observed cross-clade CTL recognition (11, 18, 28, 69, 116). However, a series of CTL epitopes that are unique for each HIV-1 subtype might be highly clade specific (19, 27, 100). We compared Nef-specific Elispot-based CTL responses among HIV-1C-infected blood donors to two sets of synthetic peptides—subtype C consensus-based peptides and subtype B isolate BRU-based peptides. The magnitude of Nef-specific Elispot-based CTL responses to subtype C peptides was higher than that of responses to subtype B peptides. This observation suggests that, despite existence of cross-clade CTL recognition, subtype-specific CTL responses are of a higher magnitude and might result in a more efficacious vaccine. We suggest that vaccine formulation should approach the predominant virus in a particular geographic area and should include epitope sequences that are close to the multiple circulating viral variants, although this observation might need to be confirmed for other HIV-1 proteins.
Our results on cumulative Gag-specific Elispot-identified CTL responses were consistent with data from a study by Goulder et al. (45), who demonstrated distinctive differences between the dominant Gag-specific responses among HIV-1B-infected Caucasians and HIV-1C-infected Africans. Prevailing HIV-1C Gag-specific CTL responses among Africans occurred in the p24 region and corresponded to the peptide that contained the epitope TPQDLNTML. In contrast, dominance of HIV-1B Gag-specific CTL responses among Caucasians was seen within the p17 region (45). Moreover, distinctive patterns of immunodominance become noticeable by comparing profiles of Tat- and Rev-specific CTL responses among HIV-1B-infected Caucasians (2, 3) and HIV-1C-infected individuals from Botswana (this study's data). Taken together, these series of studies suggest remarkable differences in the CTL epitope clustering among different ethnic groups infected by different HIV-1 subtypes. Observed distinctions within Gag, Tat, and Rev suggest that differences in CTL epitope clustering might exist across other proteins of the viral genome and warrants further studies.
A pivotal role of HIV-1-specific CTL responses in the control of HIV-1 infection has been proven by a growing number of recent studies (43, 45). Although correlates of protection in HIV-1 infection are still not known, naturally occurring dominant and subdominant CTL responses might represent and contain protective CTL epitopes (9, 36, 37, 88). Identified differences in CTL epitopes targeted at different stages of HIV-1 infection demonstrated that certain epitopes are more immunogenic and induce CTL responses more readily in earlier or later phases of the disease (6, 44) and also might correspond quantitatively to antigenic stimuli (expressed viral proteins and presented peptides) in the course of HIV-1 infection. However, the question of whether or not CTL responses that are naturally occur in acute or chronic stages of the disease, or elicited by immunization, would confer protective immunity and alter the course of HIV infection awaits further studies.
Comparison of synonymous (silent) and nonsynonymous (amino acid-altering) mutations provides an important means to understand the mechanisms of molecular sequence evolution (59, 85, 117). The nonsynonymous/synonymous rate ratio (ω = dN/dS, termed the “acceptance rate” [72]) is an important indicator of selective pressure at the protein level, wherein an ω of 1 means neutral mutations, an ω of <1 indicates purifying selection, and an ω of >1 represents diversifying positive selection (117). As a limitation of cross-sectional study it would be impossible to determine a real degree of selection that has already occurred as a result of immune selection pressure. In this study nonsynonymous and synonymous substitution rates across the identified immunodominant regions within Gag, Tat, and Nef demonstrated an ω of <1 overall, while only a few amino acid sites had an ω of >1. Although a separately designed longitudinal study will be required to address the nature of selection within CTL epitope-rich regions, an observation of a few hot spot sites with an ω of >1 within immunodominant regions might be useful for issues of vaccine design. Given the heterogeneous ω ratio found, perhaps a clearer understanding of viral evolution might be obtained by applying statistical models described in reference 78 and/or 117.
An HLA-based approach to vaccine design (10, 53, 80, 115) might increase vaccine efficacy by targeting epitopes that are restricted by common HLA types among potential vaccinees and requires the identification of HLA types and subtypes within ethnic groups and the generation of HLA databases. High-resolution HLA class I typing, which is a critical component of CTL epitope mapping and defining HLA class I restriction, allowed us to specify HLA alleles among common generic HLA types described previously (80) and detailed the restriction of two CTL epitopes within the HIV-1C Gag and one CTL epitope within the HIV-1C Tat. In this study an antigen specificity, A30, was represented mostly by two of seven A30 alleles (68), A*3001 and A*3002, at approximately equal frequency (10.8 and 12.8%, respectively), with a few cases of A*3004 (0.7% frequency). Differences at the carboxyl-terminal peptide anchor residue between A*3001 (preference for phenylalanine) and A*3002 (preference for tyrosine), which were previously described (64), suggested functional differences in peptide-binding characteristics that might lead to specific T-cell responses. Although the extent of specificity versus cross-reactivity between A*3001 and A*3002 still needs to be evaluated, the data highlight the value of accurate HLA subtyping (45) and might suggest a specific vaccine strategy in southern African populations where A30 is present at high frequency.
HLA-B58 is an antigen specificity that was seen at increased frequency within the Botswana population (80) and was found to be represented by two alleles, HLA-B*5801 and -B*5802, at approximately equal frequency in this study. Observed different frequency of HIV-1C Gag-specific Elispot-based CTL responses restricted by HLA-B*5801 and HLA-B*5802 might be explained by structure predictions made recently by Goulder (42). Interestingly, HLA-B*5801 was shown to have more similarity to HLA-B*5701 than to HLA-B*5802 (42) in the critical residues that form the B and F pockets (103). HLA-B*5801 differs from HLA-B*5802 by three amino acids at MHC position 94 (isoleucine versus threonine), 95 (isoleucine versus leucine), and 97 (arginine versus tryptophan) (68). A large residue of tryptophan at position 97 may account for a reduced space in the F pocket of HLA-B*5802 (42). This might explain why both HLA-B*5801 and HLA-B*5701 have an increased capacity to accommodate large hydrophobic residues, while HLA-B*5802 does not have enough space in the F pocket to fit in a large residue such as tryptophan, phenylalanine, or tyrosine (42). As such, the ability of HLA-B*5802 to present peptides to CTLs and/or the efficiency of peptide presentation might be significantly reduced. As a matter of fact, none of seven individuals expressing HLA-B*5802, who were screened with HIV-1C Gag-based peptides, demonstrated CTL responses that could be assigned for the HLA-B*5802 restriction. Moreover, our finding that none of nine carriers of HLA-A*2902 demonstrated HIV-1C Gag-specific CTL responses that could be assigned for the A*2902 restriction is congruent with the strong association of HLA-A29 with rapid progression to AIDS (54). If confirmed in a larger sample set, findings of HLA-driven differential efficiency of CTL responses might have an important implication for our understanding of HIV-1 pathogenesis, including the role of CTL responses in the course of HIV-1 infection and genetic susceptibility to the disease.
Diverse mechanisms have been proposed to explain failure of CTL recognition in the course of HIV-1 infection (reviewed in references 22, 25, and 71), although the extent and contribution of each cause to a CTL decline is yet to be determined. Viral escape is one of the potential mechanisms of abrogated CTL recognition (14, 31, 46, 49, 60, 93). In this study, CTL responses and viral autologous sequences were analyzed to find a correlation between the magnitude of CTL response and the level of homology between virus and synthetic peptide. However, results of our study did not completely support the common-sense model “better sequence match, better CTL response” (or “better match, better response”), suggesting an apparent complexity of the “matched/mismatched CTL response” phenomenon. Our understanding of potential mechanisms in the case of “matched sequences/matched HLA types/no CTL response” warrants further studies. Another series of observed cases in this study can be summarized as another phenomenon: “CTL response to mismatched peptide.” These cases suggest that mismatch happened at any residue with the exception of an anchor residue or that the changed amino acid was an anchor residue from the same motif (96). An intriguing scenario occurred in the case of “no detectable CTL response to the completely matched peptide, while there was an identified CTL response to the mismatched peptide that was a variant of the matched peptide.” Perhaps the observed case might be an illustration of successful viral escape from CTL recognition. An escaped viral variant might become a dominant virus, while the initial viral variant that elicited the CTL response may not exist at the time of sampling, which apparently happened shortly after the escape event. That would explain the absence of CTL response to the current viral sequence but recognition of an “old” viral variant.
Early regulatory HIV-1 proteins Tat and Rev that are translated from multiply spliced mRNAs (reviewed in references 23, 24, 29, and 92), were described as potential candidates for AIDS vaccines (16, 17, 38, 40, 50, 89). Early viral escape from Tat-specific CTL recognition demonstrated recently in the simian immunodeficiency virus model (4) emphasizes the significance of a Tat-based approach in the design of an AIDS vaccine. In this study, the immunodominant region within HIV-1C Tat, VCFQTKGLGISY, was targeted in 17 of 48 (35.4%) cases, while within Rev CTL responses were scattered without clustering. The lower magnitude of HIV-1C Tat- and Rev-specific CTL responses compared to Gag p24-specific responses warrants studies on quantification of translated viral proteins in the course of HIV-1 infection. Our results highlight the need to include multiple copies of Tat-based epitopes to increase the magnitude of Tat-specific CTL responses. Experiments in nonhuman primates might answer the question of whether or not multiple copies and variants of Tat-based epitopes could contain viral replication by preventing viral escape and therefore be potentially protective as a vaccine.
The performed fine mapping contributed to the existing CTL epitope database by determining the precise restriction of two CTL epitopes within the HIV-1C Gag p24 described previously (45) and also by identifying a new 10-mer epitope FQTKGLGISY within HIV-1C Tat that was apparently restricted by HLA-B*1503—an allele found at high frequency in the Botswana population (80). B*1503 is structurally close to B*1501 because of similarity within the C-terminal anchoring pockets (68, 94), which might suggest restriction of analogous peptides. In fact, B*1501 was shown as an allele that restricts the 10-mer peptide IQPGRGFVLY (32), which contains anchor residues identical to those of FQTKGLGISY. Alternatively, the identified epitope could contain a smaller epitope, TKGLGISY, that would be compliant with the criteria for the B27 supertype that includes B*1503 (105), although the 8-mer peptide, TKGLGISY, was not tested in this study.
In summary, screening for CTL responses among HIV-1-infected blood donors from Botswana allowed us to identify and to characterize profiles of cumulative Elispot-based CTL responses across HIV-1C Gag, Tat, Rev, and Nef. Our results confirmed that dominant CTL responses differ between ethnic groups infected with different HIV-1 subtypes and suggest an advantage of a subtype-specific (high virus-vaccine homology) approach to the design of an AIDS vaccine. Identification and ranking of dominant and subdominant CTL responses in the context of predominant virus and common HLA types in a certain geographic area may be important for the design of an efficacious AIDS vaccine and warrants further studies.
ACKNOWLEDGMENTS
We thank the Botswana Ministry of Health for encouragement; S. Gaseitsiwe, E. Sepako, G. Sebetso, N. Monametsi, and Y. Wu for sample processing and HIV-1 diagnostics; personnel of the National Blood Transfusion Center in Botswana for collaboration; and Chanc E VanWinkle for editorial assistance.
This research was supported in part by grants AI47067, AI43255, and HD37793 from the National Institutes of Health and grant TW00004 from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Abebe A, Demissie D, Goudsmit J, Brouwer M, Kuiken C L, Pollakis G, Schuitemaker H, Fontanet A L, Rinke de Wit T F. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS. 1999;13:1305–1311. doi: 10.1097/00002030-199907300-00006. [DOI] [PubMed] [Google Scholar]
- 2.Addo M M, Altfeld M, Rosenberg E S, Eldridge R L, Philips M N, Habeeb K, Khatri A, Brander C, Robbins G K, Mazzara G P, Goulder P J, Walker B D the HIV Controller Study Collaboration. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci USA. 2001;98:1781–1786. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addo M M, Altfeld M, Rosenberg E S, Poon S H, Habeeb K, Philips M N, Brander C, Trocha A, Goulder P J R, Walker B D. Analysis of cytotoxic T-lymphocyte (CTL) responses against the regulatory HIV-1 proteins Rev and Tat in HIV-1-infected individuals and identification of novel CTL epitopes. Durban, South Africa: XIII International Conference on AIDS; 2000. [Google Scholar]
- 4.Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothe B R, Vogel T U, Dunphy E, Liebl M E, Emerson C, Wilson N, Kunstman K J, Wang X, Allison D B, Hughes A L, Desrosiers R C, Altman J D, Wolinsky S M, Sette A, Watkins D I. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 5.Altfeld M, Rosenberg E S. The role of CD4+ T helper cells in the cytotoxic T lymphocyte response to HIV-1. Curr Opin Immunol. 2000;12:375–380. doi: 10.1016/s0952-7915(00)00103-5. [DOI] [PubMed] [Google Scholar]
- 6.Altfeld M, Rosenberg E S, Shankarappa R, Mukherjee J S, Hecht F M, Eldridge R L, Addo M M, Poon S H, Phillips M N, Robbins G K, Sax P E, Boswell S, Kahn J O, Brander C, Goulder P J, Levy J A, Mullins J I, Walker B D. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J Exp Med. 2001;193:169–180. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altfeld M A, Livingston B, Reshamwala N, Nguyen P T, Addo M M, Shea A, Newman M, Fikes J, Sidney J, Wentworth P, Chesnut R, Eldridge R L, Rosenberg E S, Robbins G K, Brander C, Sax P E, Boswell S, Flynn T, Buchbinder S, Goulder P J, Walker B D, Sette A, Kalams S A. Identification of novel HLA-A2-restricted human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte epitopes predicted by the HLA-A2 supertype peptide-binding motif. J Virol. 2001;75:1301–1311. doi: 10.1128/JVI.75.3.1301-1311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altfeld M A, Trocha A, Eldridge R, Rosenberg L, E S, Phillips M N, Addo M M, Sekaly R P, Kalams S A, Burchett S A, McIntosh K, Walker B D, Goulder P J. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/jvi.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barouch D H, Santra S, Schmitz J E, Kuroda M J, Fu T M, Wagner W, Bilska M, Craiu A, Zheng X X, Krivulka G R, Beaudry K, Lifton M A, Nickerson C E, Trigona W L, Punt K, Freed D C, Guan L, Dubey S, Casimiro D, Simon A, Davies M E, Chastain M, Strom T B, Gelman R S, Montefiori D C, Lewis M G. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett J A, Wasserman S S, Hicks C B, Dodge R T, Weinhold K J, Tacket C O, Ketter N, Wittek A E, Palker T J, Haynes B F. Safety and immunogenicity of an HLA-based HIV envelope polyvalent synthetic peptide immunogen. DATRI 010 Study Group. Division of AIDS Treatment Research Initiative. AIDS. 1998;12:1291–1300. doi: 10.1097/00002030-199811000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C, N′Gandu N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Björnal Å, Sönnerborg A, Tschering C, Albert J, Fenyö E M. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C Isolates of Ethiopian AIDS patients. AIDS Res Hum Retrovir. 1999;15:647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 13.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow P, Lewicki H, Wei X, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 15.Brander C, Walker B D. T lymphocyte responses in HIV-1 infection: implications for vaccine development. Curr Opin Immunol. 1999;11:451–459. doi: 10.1016/S0952-7915(99)80076-4. [DOI] [PubMed] [Google Scholar]
- 16.Cafaro A, Caputo A, Fracasso C, Maggiorella M T, Goletti D, Baroncelli S, Pace M, Sernicola L, Koanga-Mogtomo M L, Betti M, Borsetti A, Belli R, Akerblom L, Corrias F, Butto S, Heeney J, Verani P, Titti F, Ensoli B. Control of SHIV-89.6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nat Med. 1999;5:643–650. doi: 10.1038/9488. [DOI] [PubMed] [Google Scholar]
- 17.Cafaro A, Caputo A, Maggiorella M T, Baroncelli S, Fracasso C, Pace M, Borsetti A, Sernicola L, Negri D R, Ten Haaft P, Betti M, Michelini Z, Macchia I, Fanales-Belasio E, Belli R, Corrias F, Butto S, Verani P, Titti F, Ensoli B. SHIV89.6P pathogenicity in cynomolgus monkeys and control of viral replication and disease onset by human immunodeficiency virus type 1 Tat vaccine. J Med Primatol. 2000;29:193–208. doi: 10.1034/j.1600-0684.2000.290313.x. [DOI] [PubMed] [Google Scholar]
- 18.Cao H, Kanki P, Sankale J L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker B D. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao H, Mani I, Vincent R, Mugerwa R, Mugyenyi P, Kanki P, Ellner J, Walker B D. Cellular immunity to human immunodeficiency virus type 1 (HIV-1) clades: relevance to HIV-1 vaccine trials in Uganda. J Infect Dis. 2000;182:1350–1356. doi: 10.1086/315868. [DOI] [PubMed] [Google Scholar]
- 20.Carr J K, Laukkanen T, Salminen M O, Albert J, Alaeus A, Kim B, Sanders-Buell E, Birx D L, McCutchan F E. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS. 1999;13:1819–1826. doi: 10.1097/00002030-199910010-00003. [DOI] [PubMed] [Google Scholar]
- 21.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 22.Collins K L, Baltimore D. HIV's evasion of the cellular immune response. Immunol Rev. 1999;168:65–74. doi: 10.1111/j.1600-065x.1999.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 23.Cullen B R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990;63:655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- 24.Cullen B R, Malim M H. The HIV-1 Rev protein: prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 25.De Maria A, Moretta L. HLA-class I-specific inhibitory receptors in HIV-1 infection. Hum Immunol. 2000;61:74–81. doi: 10.1016/s0198-8859(99)00169-x. [DOI] [PubMed] [Google Scholar]
- 26.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorrell L, Dong T, Ogg G S, Lister S, McAdam S, Rostron T, Conlon C, McMichael A J, Rowland-Jones S L. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J Virol. 1999;73:1708–1714. doi: 10.1128/jvi.73.2.1708-1714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durali D, Morvan J, Letourneur F, Schmitt D, Guegan N, Dalod M, Saragosti S, Sicard D, Levy J P, Gomard E. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J Virol. 1998;72:3547–3553. doi: 10.1128/jvi.72.5.3547-3553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 30.Essex M. Human immunodeficiency viruses in the developing world. Adv Virus Res. 1999;53:71–88. doi: 10.1016/s0065-3527(08)60343-7. [DOI] [PubMed] [Google Scholar]
- 31.Evans D T, O'Connor D H, Jing P, Dzuris J L, Sidney J, da Silva J, Allen T M, Horton H, Venham J E, Rudersdorf R A, Vogel T, Pauza C D, Bontrop R E, DeMars R, Sette A, Hughes A L, Watkins D I. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 32.Falk K, Rotzschke O, Takiguchi M, Gnau V, Stevanovic S, Jung G, Rammensee H G. Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics. 1995;41:165–168. doi: 10.1007/BF00182333. [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. PHYLIP: Phylogeny Inference Package, 3.52c ed. Seattle: University of Washington; 1993. [Google Scholar]
- 34.Felsenstein J. PHYLIP: Phylogeny Inference Package, 3.572c. Seattle: University of Washington; 1996. [Google Scholar]
- 35.Fonjungo P N, Mpoudi E N, Torimiro J N, Alemnji G A, Eno L T, Nkengasong J N, Gao F, Rayfield M, Folks T M, Pieniazek D, Lal R B. Presence of diverse human immunodeficiency virus type 1 viral variants in Cameroon. AIDS Res Hum Retrovir. 2000;16:1319–1324. doi: 10.1089/08892220050117087. [DOI] [PubMed] [Google Scholar]
- 36.Gallimore A, Dumrese T, Hengartner H, Zinkernagel R M, Rammensee H G. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallimore A, Hombach J, Dumrese T, Rammensee H G, Zinkernagel R M, Hengartner H. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur J Immunol. 1998;28:3301–3311. doi: 10.1002/(SICI)1521-4141(199810)28:10<3301::AID-IMMU3301>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 38.Gallo R C. Tat as one key to HIV-induced immune pathogenesis and Tat (correction of Pat) toxoid as an important component of a vaccine. Proc Natl Acad Sci USA. 1999;96:8324–8326. doi: 10.1073/pnas.96.15.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein G. HIV-1 Tat protein as a potential AIDS vaccine. Nat Med. 1996;2:960–964. doi: 10.1038/nm0996-960. [DOI] [PubMed] [Google Scholar]
- 41.Goulder P, Price D, Nowak M, Rowland-Jones S, Phillips R, McMichael A. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol Rev. 1997;159:17–29. doi: 10.1111/j.1600-065x.1997.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 42.Goulder P J. Rapid characterization of HIV clade C-specific cytotoxic T lymphocyte responses in infected African children and adults. Ann N Y Acad Sci. 2000;918:330–345. doi: 10.1111/j.1749-6632.2000.tb05502.x. [DOI] [PubMed] [Google Scholar]
- 43.Goulder P J, Addo M M, Altfeld M A, Rosenberg E S, Tang Y, Govender U, Mngqundaniso N, Annamalai K, Vogel T U, Hammond M, Bunce M, Coovadia H M, Walker B D. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by Elispot and intracellular cytokine staining assays. J Virol. 2001;75:1339–1347. doi: 10.1128/JVI.75.3.1339-1347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goulder P J, Altfeld M A, Rosenberg E S, Nguyen T, Tang Y, Eldridge R L, Addo M M, He S, Mukherjee J S, Phillips M N, Bunce M, Kalams S A, Sekaly R P, Walker B D, Brander C. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–194. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goulder P J, Brander C, Annamalai K, Mngqundaniso N, Govender U, Tang Y, He S, Hartman K E, O'Callaghan C A, Ogg G S, Altfeld M A, Rosenberg E S, Cao H, Kalams S A, Hammond M, Bunce M, Pelton S I, Burchett S A, McIntosh K, Coovadia H M, Walker B D. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J Virol. 2000;74:5679–5690. doi: 10.1128/jvi.74.12.5679-5690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goulder P J, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M A, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 47.Goulder P J, Rowland-Jones S L, McMichael A J, Walker B D. Anti-HIV cellular immunity: recent advances towards vaccine design. AIDS. 1999;13(Suppl. A):S121–S136. [PubMed] [Google Scholar]
- 48.Goulder P J, Tang Y, Brander C, Betts M R, Altfeld M, Annamalai K, Trocha A, He S, Rosenberg E S, Ogg G, O'Callaghan C A, Kalams S A, McKinney R E, Mayer K, Koup R A, Pelton S I, Burchett S K, McIntosh K, Walker B D. Functionally inert HIV-specific cytotoxic T lymphocytes do not play a major role in chronically infected adults and children. J Exp Med. 2000;192:1819–1832. doi: 10.1084/jem.192.12.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goulder P J, Walker B D. The great escape—AIDS viruses and immune control. Nat Med. 1999;5:1233–1235. doi: 10.1038/15184. [DOI] [PubMed] [Google Scholar]
- 50.Gringeri A, Santagostino E, Muca-Perja M, Mannucci P M, Zagury J F, Bizzini B, Lachgar A, Carcagno M, Rappaport J, Criscuolo M, Blattner W, Burny A, Gallo R C, Zagury D. Safety and immunogenicity of HIV-1 Tat toxoid in immunocompromised HIV-1-infected patients. J Hum Virol. 1998;1:293–298. [PubMed] [Google Scholar]
- 51.Hall T A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 52.Hanke T, McMichael A J. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat Med. 2000;6:951–955. doi: 10.1038/79626. [DOI] [PubMed] [Google Scholar]
- 53.Haynes B F. HIV vaccines: where we are and where we are going. Lancet. 1996;348:933–937. doi: 10.1016/S0140-6736(96)09339-7. [DOI] [PubMed] [Google Scholar]
- 54.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O'Brien S, Andrieu J M, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson G W, Zagury J F. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 55.Hu D J, Baggs J, Downing R G, Pieniazek D, Dorn J, Fridlund C, Biryahwaho B, Sempala S D, Rayfield M A, Dondero T J, Lal R. Predominance of HIV-1 subtype A and D infections in Uganda. Emerg Infect Dis. 2000;6:609–615. doi: 10.3201/eid0606.000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalams S A, Buchbinder S P, Rosenberg E S, Billingsley J M, Colbert D S, Jones N G, Shea A K, Trocha A K, Walker B D. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol. 1999;73:6715–6720. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalams S A, Walker B D. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J Exp Med. 1998;188:2199–2204. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 60.Koenig S, Conley A J, Brewah Y A, Jones G M, Leath S, Boots L J, Davey V, Pantaleo G, Demarest J F, Carter C, Wannebo C, Yannelli J R, Rosenberg S A, Lane H C. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- 61.Korber B T, Foley B T, Kuiken C L, Pillai S K, Sodroski J G. Numbering positions in HIV relative to HXB2CG, p. III-102–III-111. In: Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodroski J, editors. Human retroviruses and AIDS 1998. Theoretical Biology and Biophysics, Group T-10. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 62.Korber B T M, Brander C, Haynes B F, Moore J P, Watkins D I, Koup R, Walker B D, editors. HIV molecular immunology database 1999. Theoretical Biology and Biophysics, Group T-10. Los Alamos, N.Mex: Los Alamos National Laboratory; 1999. [Google Scholar]
- 63.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krausa P, Munz C, Keilholz W, Stevanovic S, Jones E Y, Browning M, Bunce M, Rammensee H G, McMichael A J. Definition of peptide binding motifs amongst the HLA-A*30 allelic group. Tissue Antigen. 2000;56:10–18. doi: 10.1034/j.1399-0039.2000.560102.x. [DOI] [PubMed] [Google Scholar]
- 65.Larsson M, Jin X, Ramratnam B, Ogg G S, Engelmayer J, Demoitie M A, McMichael A J, Cox W I, Steinman R M, Nixon D, Bhardwaj N. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS. 1999;13:767–777. doi: 10.1097/00002030-199905070-00005. [DOI] [PubMed] [Google Scholar]
- 66.Letvin N L. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 67.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marsh S G E, Parham P, Barber L D. The HLA FactsBook. London, United Kingdom: Academic Press; 2000. [Google Scholar]
- 69.McAdam S, Kaleebu P, Krausa P, Goulder P, French N, Collin B, Blanchard T, Whitworth J, McMichael A, Gotch F. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS. 1998;12:571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 70.McCutchan F E, Carr J K, Bajani M, Sanders-Buell E, Harry T O, Stoeckli T C, Robbins K E, Gashau W, Nasidi A, Janssens W, Kalish M L. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254:226–234. doi: 10.1006/viro.1998.9505. [DOI] [PubMed] [Google Scholar]
- 71.McMichael A J, Phillips R E. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 72.Miyata T, Yasunaga T. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J Mol Evol. 1980;16:23–36. doi: 10.1007/BF01732067. [DOI] [PubMed] [Google Scholar]
- 73.Mochizuki N, Otsuka N, Matsuo K, Shiino T, Kojima A, Kurata T, Sakai K, Yamamoto N, Isomura S, Dhole T N, Takebe Y, Matsuda M, Tatsumi M. An infectious DNA clone of HIV type 1 subtype C. AIDS Res Hum Retrovir. 1999;15:1321–1324. doi: 10.1089/088922299310223. [DOI] [PubMed] [Google Scholar]
- 74.Montavon C, Toure-Kane C, Liegeois F, Mpoudi E, Bourgeois A, Vergne L, Perret J L, Boumah A, Saman E, Mboup S, Delaporte E, Peeters M. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr Hum Retrovirol. 2000;23:363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- 75.Ndung'u T, Renjifo B, Novitsky V A, McLane M F, Gaolekwe S, Essex M. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology. 2000;278:390–399. doi: 10.1006/viro.2000.0583. [DOI] [PubMed] [Google Scholar]
- 76.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 77.Neilson J R, John G C, Carr J K, Lewis P, Kreiss J K, Jackson S, Nduati R W, Mbori-Ngacha D, Panteleeff D D, Bodrug S, Giachetti C, Bott M A, Richardson B A, Bwayo J, Ndinya-Achola J, Overbaugh J. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 1999;73:4393–4403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nkengasong J N, Luo C C, Abouya L, Pieniazek D, Maurice C, Sassan-Morokro M, Ellenberger D, Hu D J, Pau C P, Dobbs T, Respess R, Coulibaly D, Coulibaly I M, Wiktor S Z, Greenberg A E, Rayfield M. Distribution of HIV-1 subtypes among HIV-seropositive patients in the interior of Cote d'Ivoire. J Acquir Immune Defic Syndr Hum Retrovirol. 2000;23:430–436. doi: 10.1097/00126334-200004150-00011. [DOI] [PubMed] [Google Scholar]
- 80.Novitsky V, Flores-Villanueva P O, Chigwedere P, Gaolekwe S, Bussman H, Sebetso G, Marlink R, Yunis E J, Essex M. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum Immunol. 2001;62:146–156. doi: 10.1016/s0198-8859(00)00236-6. [DOI] [PubMed] [Google Scholar]
- 81.Novitsky V A, Gaolekwe S, McLane M F, Ndung'u T P, Foley B T, Vannberg F, Marlink R, Essex M. HIV type 1 A/J recombinant with a pronounced pol gene mosaicism. AIDS Res Hum Retrovir. 2000;16:1015–1020. doi: 10.1089/08892220050058434. [DOI] [PubMed] [Google Scholar]
- 82.Novitsky V A, Montano M A, McLane M F, Renjifo B, Vannberg F, Foley B T, Ndung'u T P, Rahman M, Makhema M J, Marlink R, Essex M. Molecular cloning and phylogenetic analysis of HIV-1 subtype C: a set of 23 full-length clones from Botswana. J Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogg G S, Dong T, Hansasuta P, Dorrell J, Clarke J, Coker R, Luzzi G, Conlon C, McMichael A J, Rowland-Jones S L. Four novel CTL epitopes in the highly-conserved MHR region of HIV-1 gag, restricted by B*4402, B*1401, A*2601 and B*70. AIDS. 1998;12:1561–1563. doi: 10.1097/00002030-199812000-00026. [DOI] [PubMed] [Google Scholar]
- 84.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 85.Ohta T. Mechanisms of molecular evolution. Phil Trans R Soc Lond B Biol Sci. 2000;355:1623–1626. doi: 10.1098/rstb.2000.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ota T, Nei M. Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Mol Biol Evol. 1994;11:613–619. doi: 10.1093/oxfordjournals.molbev.a040140. [DOI] [PubMed] [Google Scholar]
- 87.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 88.Patterson L J, Peng B, Abimiku A G, Aldrich K, Murty L, Markham P D, Kalyanaraman V S, Alvord W G, Tartaglia J, Franchini G, Robert-Guroff M. Cross-protection in NYVAC-HIV-1-immunized/HIV-2-challenged but not in NYVAC-HIV-2-immunized/SHIV-challenged rhesus macaques. AIDS. 2000;14:2445–2455. doi: 10.1097/00002030-200011100-00005. [DOI] [PubMed] [Google Scholar]
- 89.Pauza C D, Trivedi P, Wallace M, Ruckwardt T J, Le Buanec H, Lu W, Bizzini B, Burny A, Zagury D, Gallo R C. Vaccination with tat toxoid attenuates disease in simian/HIV-challenged macaques. Proc Natl Acad Sci USA. 2000;97:3515–3519. doi: 10.1073/pnas.070049797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 91.Picker L J, Maino V C. The CD4+ T cell response to HIV-1. Curr Opin Immunol. 2000;12:381–386. doi: 10.1016/s0952-7915(00)00104-7. [DOI] [PubMed] [Google Scholar]
- 92.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 93.Price D A, Goulder P J, Klenerman P, Sewell A K, Easterbrook P J, Troop M, Bangham C R, Phillips R E. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prilliman K R, Jackson K W, Lindsey M, Wang J, Crawford D, Hildebrand W H. HLA-B15 peptide ligands are preferentially anchored at their C termini. J Immunol. 1999;162:7277–7284. [PubMed] [Google Scholar]
- 95.Proutski V, Holmes E. SWAN: sliding window analysis of nucleotide sequence variability. Bioinformatics. 1998;14:467–468. doi: 10.1093/bioinformatics/14.5.467. [DOI] [PubMed] [Google Scholar]
- 96.Rammensee H G, Bachmann J, Stevanovic S. MHC ligands and peptide motifs. Austin, Tex: Landes Bioscience; 1997. [Google Scholar]
- 97.Robertson D L, Anderson J P, Bradac J A, Carr J K, Foley B, Funkhouser R K, Gao F, Hahn B H, Kalish M L, Kuiken C, Learn G H, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp P M, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 98.Rodenburg C M, Li Y, Trask S A, Chen Y, Decker J, Robertson D L, Kalish M L, Shaw G M, Allen S, Hahn B H, Gao F. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res Hum Retrovir. 2001;17:161–168. doi: 10.1089/08892220150217247. [DOI] [PubMed] [Google Scholar]
- 99.Rosenberg E S, Billingsley J M, Caliedo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 100.Rowland-Jones S L, Dong T, Fowke K R, Kimani J, Krausa P, Newell H, Blanchard T, Ariyoshi K, Oyugi J, Ngugi E, Bwayo J, MacDonald K S, McMichael A J, Plummer F A. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Investig. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salminen M O, Johansson B, Sonnerborg A, Ayehunie S, Gotte D, Leinikki P, Burke D S, McCutchan F E. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res Hum Retrovir. 1996;12:1329–1339. doi: 10.1089/aid.1996.12.1329. [DOI] [PubMed] [Google Scholar]
- 102.Sankale J L, Hamel D, Woolsey A, Traore T, Dia T C, Gueye-Ndiaye A, Essex M, Mboup T, Kanki P. Molecular evolution of human immunodeficiency virus type 1 subtype A in Senegal: 1988–1997. J Hum Virol. 2000;3:157–164. [PubMed] [Google Scholar]
- 103.Saper M A, Bjorkman P J, Wiley D C. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 104.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 105.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 106.Spiegel H M, Ogg G S, DeFalcon E, Sheehy M E, Monard S, Haslett P A, Gillespie G, Donahoe S M, Pollack H, Borkowsky W, McMichael A J, Nixon D F. Human immunodeficiency virus type 1- and cytomegalovirus-specific cytotoxic T lymphocytes can persist at high frequency for prolonged periods in the absence of circulating peripheral CD4+ T cells. J Virol. 2000;74:1018–1022. doi: 10.1128/jvi.74.2.1018-1022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Toure-Kane C, Montavon C, Faye M A, Gueye P M, Sow P S, Ndoye I, Gaye-Diallo A, Delaporte E, Peeters M, Mboup S. Identification of all HIV type 1 group M subtypes in Senegal, a country with low and stable seroprevalence. AIDS Res Hum Retrovir. 2000;16:603–609. doi: 10.1089/088922200309025. [DOI] [PubMed] [Google Scholar]
- 109.Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman D R, Fenyo E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 110.UNAIDS. AIDS epidemic update—December 2000. New York, N.Y: UNAIDS; 2000. [Google Scholar]
- 111.UNAIDS and the World Health Organization. Global HIV/AIDS and STD surveillance. Epidemiological fact sheets by country. 2000. http: //www.who.int/emc-hiv/fact_sheets/ [Online.] http: //www.who.int/emc-hiv/fact_sheets/ [Google Scholar]
- 112.UNAIDS and the World Health Organization. Report on the global HIV/AIDS epidemic—June1998. Geneva, Switzerland: UNAIDS and the World Health Organization; 1998. [Google Scholar]
- 113.Walker B D, Chakrabarti S, Moss B, Paradis T J, Flynn T, Durno A G, Blumberg R S, Kaplan J C, Hirsch M S, Schooley R T. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature. 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 114.Wang W K, Dudek T, Zhao Y J, Brumblay H, Essex M, Lee T H. CCR5 co-receptor utilization involves a highly conserved arginine residue of human immunodeficiency virus type 1 gp120. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ward F E, Tuan S, Haynes B F. Analysis of HLA frequencies in population cohorts for design of HLA-based HIV vaccines, p. IV-10–IV-16. In: Korber B, editor. HIV Molecular Immunology Database. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 116.Wilson S E, Pedersen S L, Kunich J C, Wilkins V L, Mann D L, Mazzara G P, Tartaglia J, Celum C L, Sheppard H W. Cross-clade envelope glycoprotein 160-specific CD8+ cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res Hum Retrovir. 1998;14:925–937. doi: 10.1089/aid.1998.14.925. [DOI] [PubMed] [Google Scholar]
- 117.Yang Z, Nielsen R, Goldman N, Pedersen A M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]