The 2.3 Å resolution ternary structure of the essential P. vivaxN-myristoyltransferase with myristoyl-CoA and a peptide-binding domain inhibitor is reported as part of ongoing efforts by the SSGCID for the rational design of new therapeutics for malaria.
Keywords: N-myristoyltransferases, Plasmodium vivax, malaria, G6PD deficiency
Abstract
Plasmodium vivax is a major cause of malaria, which poses an increased health burden on approximately one third of the world’s population due to climate change. Primaquine, the preferred treatment for P. vivax malaria, is contraindicated in individuals with glucose-6-phosphate dehydrogenase (G6PD) deficiency, a common genetic cause of hemolytic anemia, that affects ∼2.5% of the world’s population and ∼8% of the population in areas of the world where P. vivax malaria is endemic. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) conducted a structure–function analysis of P. vivaxN-myristoyltransferase (PvNMT) as part of efforts to develop alternative malaria drugs. PvNMT catalyzes the attachment of myristate to the N-terminal glycine of many proteins, and this critical post-translational modification is required for the survival of P. vivax. The first step is the formation of a PvNMT–myristoyl–CoA binary complex that can bind to peptides. Understanding how inhibitors prevent protein binding will facilitate the development of PvNMT as a viable drug target. NMTs are secreted in all life stages of malarial parasites, making them attractive targets, unlike current antimalarials that are only effective during the plasmodial erythrocytic stages. The 2.3 Å resolution crystal structure of the ternary complex of PvNMT with myristoyl-CoA and a novel inhibitor is reported. One asymmetric unit contains two monomers. The structure reveals notable differences between the PvNMT and human enzymes and similarities to other plasmodial NMTs that can be exploited to develop new antimalarials.
1. Introduction
Malaria, a deadly disease caused by protozoan parasites from the Plasmodium genus, poses a significant global health challenge. P. vivax is responsible for the most widespread human malaria and is an obstacle to global malaria-elimination efforts, with nearly 2.5 billion people, or more than one-third of the world’s population, at risk of P. vivax infection (Battle et al., 2019 ▸). The dormant liver phase of P. vivax enables its survival in colder climates and tropical, subtropical and temperate regions, giving a wider geographical range (Battle et al., 2019 ▸). Primaquine is the most effective drug for P. vivax infection, but low adherence lowers its efficacy (Mehdipour et al., 2023 ▸). Furthermore, primaquine is contraindicated among individuals with glucose-6-phosphate dehydrogenase (G6PD) deficiency, which causes hemolytic anemia and other complications (Kane, 2012 ▸; Yilma et al., 2023 ▸). This is particularly concerning since approximately 400 million people worldwide have G6PD deficiency (Drysdale et al., 2023 ▸). There is therefore a pressing need for safer treatment options for P. vivax malaria.
A promising strategy for the eradication of malaria is targeting proteins that regulate multiple stages of the life cycle of Plasmodium species. One such protein, N-myristoyltransferase (PvNMT), is an essential enzyme that catalyzes a post-translational modification (myristoylation) through transfer of the lipid myristate from myristoyl coenzyme A (Myr-CoA) to the N-terminal glycine residues of target proteins (Selvakumar et al., 2011 ▸; Udenwobele et al., 2017 ▸; McIlhinney, 1989 ▸). NMT-mediated myristoylation is crucial for membrane association, protein–protein interactions, protein stability and turnover, and signal transduction (Selvakumar et al., 2011 ▸). NMTs also help to regulate cellular processes and have emerged as potential therapeutic targets for parasitic diseases (Frearson et al., 2010 ▸; Rodríguez-Hernández et al., 2023 ▸; Harupa et al., 2020 ▸).
NMTs have been explored as potential drug targets against Plasmodium parasites and are promising for the development of innovative therapeutic approaches to combat malaria (Rackham et al., 2014 ▸; Nicolau et al., 2023 ▸; Rodríguez-Hernández et al., 2023 ▸). Plasmodium species possess a single NMT gene that is essential for survival, and previous studies showed reduced parasitemia after NMT inhibition (Pino et al., 2012 ▸). An advantage of targeting NMT is that it is expressed throughout every stage of the life cycle of Plasmodium species, which allows the possibility of complete parasite elimination, unlike many licensed antimalarials, which only target the erythrocytic stage (Pino et al., 2012 ▸). A recent study also demonstrated that PvNMT inhibitors reduced parasite growth in the schizont and hypnozoite stages, during which several essential NMT substrates are expressed (Rodríguez-Hernández et al., 2023 ▸). Our study presents the 2.3 Å resolution crystal structure of PvNMT bound to Myr-CoA and a novel inhibitor, contributing to the quest for alternative drug treatments against malaria using PvNMT as a target (Harupa et al., 2020 ▸; Rodríguez-Hernández et al., 2023 ▸).
2. Materials and methods
2.1. Macromolecule
The gene (PvNMT; UniProt A5K1A2) encoding amino acids 1–410 was acquired from GenScript as a synthetic construct inserted into pET-11a, encoding a 3C protease-cleavable hexahistidine tag (MGSSHHHHHHSAALEVLFQGP-ORF; Table 1 ▸). PvNMT was expressed and purified using established protocols (Stacy et al., 2011 ▸; Serbzhinskiy et al., 2015 ▸; Rodríguez-Hernández et al., 2023 ▸). Plasmid DNA was transformed into chemically competent Escherichia coli BL21(DE3) Rosetta cells. The plasmid containing His-PvNMT was tested for expression and 2 l of culture was grown using auto-induction medium (Studier, 2005 ▸) in a LEX Bioreactor (Epiphyte Three) as described previously (Serbzhinskiy et al., 2015 ▸). The expression clone can be requested online at https://www.ssgcid.org/available-materials/expression-clones/.
Table 1. Macromolecule-production information.
| Source organism | Plasmodium vivax Sal-1 (strain Salvador I) |
| DNA source | GenScript gene synthesis |
| Expression vector | pET-11a (GenScript) |
| Expression host | Escherichia coli BL21(DE3) Rosetta |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHSAALEVLFQGPDYKFWYTQPVPKINDEFNESVNEPFISDNKVEDVRKDEYKLPPGYSWYVCDVKDEKDRSEIYTLLTDNYVEDDDNIFRFNYSAEFLLWALTSPNYLKTWHIGVKYDASNKLIGFISAIPTDICIHKRTIKMAEVNFLCVHKTLRSKRLAPVLIKEITRRINLENIWQAIYTAGVYLPKPVSDARYYHRSINVKKLIEIGFSSLNSRLTMSRAIKLYRVEDTLNIKNMRLMKKKDVEGVHKLLGSYLEQFNLYAVFTKEEIAHWFLPIENVIYTYVNEENGKIKDMISFYSLPSQILGNDKYSTLNAAYSFYNVTTTATFKQLMQDAILLAKRNNFDVFNALEVMQNKSVFEDLKFGEGDGSLKYYLYNWKCASFAPAHVGIVLL |
PvNMT was purified in two steps: immobilized metal (Ni2+) affinity chromatography (IMAC) and size-exclusion chromatography (SEC) on an ÄKTApurifier 10 (GE Healthcare) using automated IMAC and SEC programs (Serbzhinskiy et al., 2015 ▸). Briefly, thawed bacterial pellets (25 g) were lysed by sonication in 200 ml lysis buffer [25 mM HEPES pH 7.0, 500 mM NaCl, 5%(v/v) glycerol, 0.5%(w/v) CHAPS, 30 mM imidazole, 10 mM MgCl2, 1 mM TCEP and five tablets of protease-inhibitor cocktail (cOmplete Mini, EDTA-free Roche, Basel, Switzerland)]. After sonication, the crude lysate was clarified with 20 µl (25 units ml−1) of Benzonase by incubating and mixing at room temperature for 45 min. The lysate was clarified by centrifugation at 5000g for 1 h at 277 K using a refrigerated Sorvall centrifuge (Thermo Scientific). The clarified supernatant was then passed over a 5 ml Ni–NTA HisTrap FF column (GE Healthcare) which had been pre-equilibrated with loading buffer [25 mM HEPES pH 7.0, 500 mM NaCl, 5%(v/v) glycerol, 30 mM imidazole, 1 mM TCEP, 0.025%(w/v) sodium azide]. The column was washed with 20 column volumes (CV) of loading buffer and eluted with elution buffer [25 mM HEPES pH 7.0, 500 mM NaCl, 5%(v/v) glycerol, 30 mM imidazole, 1 mM TCEP, 0.025%(w/v) sodium azide, 250 mM imidazole] over a 7 CV linear gradient. Peak fractions were pooled, concentrated to 5 ml and loaded onto a Superdex 75 26/60 column (GE Biosciences) equilibrated with running buffer (20 mM HEPES pH 7.0, 300 mM NaCl, 5% glycerol, 1 mM TCEP). PvNMT eluted from the SEC column as a single, monodisperse symmetrical peak that accounted for >90% of the protein product, with a molecular mass of ∼40 kDa, suggesting purification as a monomer (based on the theoretical molecular weight of 47.1 kDa). The pure peak fractions were pooled and concentrated to 13.5 mg ml−1 using an Amicon purification system (Millipore). The purified protein was stored in 100 µl aliquots at 193 K and can be requested online at https://www.ssgcid.org/available-materials/ssgcid-proteins/.
2.2. Crystallization
PvNMT was crystallized at 290 K in sitting-drop vapor-diffusion format. Briefly, 13.5 mg ml−1 protein was incubated with final concentrations of 0.4 mM Myr-CoA and 0.4 mM IMP-0001173 at 4°C for 30 min and then mixed in a 1:1 ratio with precipitant solution as described in Table 2 ▸. Before data collection, the crystals were harvested and cryoprotected with 20%(v/v) ethylene glycol (Table 2 ▸).
Table 2. Crystallization.
| Method | Vapor diffusion, sitting drop |
| Plate type | 96-well plates |
| Temperature (K) | 290 |
| Protein concentration (mg ml−1) | 13.5 |
| Buffer composition of protein solution | 20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM TCEP, 0.4 mM Myr-CoA, 0.4 mM IMP-0001173 |
| Composition of reservoir solution | 0.06 M magnesium chloride hexahydrate, 0.06 M calcium chloride dihydrate, 0.1 M Tris–Bicine pH 8.5, 19.6%(v/v) PEG 500 MME, 9.8%(w/v) PEG 20 000 |
| Volume and ratio of drop | 0.4 µl, 1:1 |
| Volume of reservoir (µl) | 80 |
| Composition of cryoprotectant solution | 20%(v/v) ethylene glycol, 0.06 M magnesium chloride hexahydrate, 0.06 M calcium chloride dihydrate, 0.1 M Tris–Bicine pH 8.5, 19.6%(v/v) PEG 500 MME, 9.8%(w/v) PEG 20 000 |
2.3. Data collection and processing
Data were collected at 100 K on beamline 5.0.2 at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory (Table 3 ▸). Data were integrated with XDS and reduced with XSCALE (Kabsch, 2010 ▸). Raw X-ray diffraction images have been stored at the Integrated Resource for Reproducibility in Macromolecular Crystallography at https://www.proteindiffraction.org.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 5.0.2, ALS |
| Wavelength (Å) | 1.00 |
| Temperature (K) | 100 |
| Detector | Dectris PILATUS3 6M |
| Crystal-to-detector distance (mm) | 475 |
| Rotation range per image (°) | 0.25 |
| Total rotation range (°) | 150 |
| Space group | P21212 |
| a, b, c (Å) | 80.03, 81.49, 119.44 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 48.17–2.30 (2.36–2.30) |
| Total No. of reflections | 189923 |
| No. of unique reflections | 35084 |
| Completeness (%) | 99.10 (92.30) |
| Multiplicity | 5.41 (4.79) |
| 〈I/σ(I)〉 | 19.19 (3.03) |
| R r.i.m. | 0.065 (0.58) |
| Overall B factor from Wilson plot (Å2) | 40.53 |
2.4. Structure solution and refinement
The structure of PvNMT was determined by molecular replacement with Phaser (McCoy et al., 2007 ▸) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▸; Krissinel et al., 2004 ▸; Winn et al., 2011 ▸; Agirre et al., 2023 ▸) using PDB entry 5v0x as the search model. The structure quality was checked using MolProbity (Williams et al., 2018 ▸). Electron-density maps showing the ligand fit are shown in Supplementary Fig. S1. Data-reduction and refinement statistics are shown in Table 4 ▸. Coordinate and structure factors have been deposited with the Worldwide PDB (wwPDB) as entry 6b1l.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 48.17–2.30 (2.36–2.30) |
| Completeness (%) | 99.0 (92.3) |
| σ Cutoff | F > 1.34σ(F) |
| No. of reflections, working set | 35070 (2327) |
| No. of reflections, test set | 1884 (144) |
| Final Rcryst | 0.232 (0.272) |
| Final Rfree | 0.271 (0.315) |
| No. of non-H atoms | |
| Protein | 5264 |
| Ligand | 94 |
| Solvent | 188 |
| Total | 5546 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Angles (°) | 0.543 |
| Average B factors (Å2) | |
| Protein | 58.0 |
| Ligand | 42.9 |
| Water | 46.0 |
| Ramachandran plot | |
| Most favored (%) | 96 |
| Allowed (%) | 4 |
3. Results and discussion
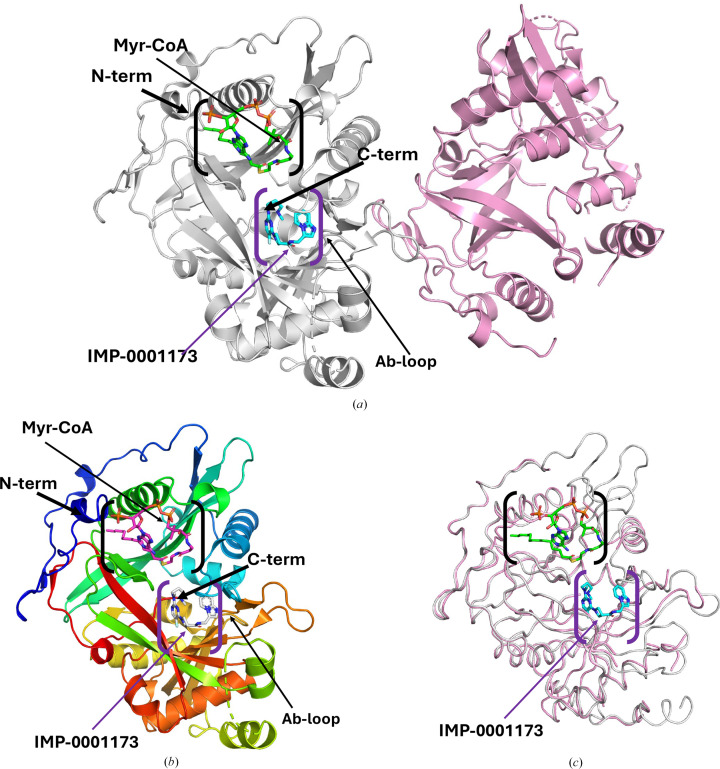
The co-crystal structure of PvNMT with a cofactor (Myr-CoA) and a peptide-binding-domain inhibitor (IMP-0001173) was determined at 2.3 Å resolution. Interestingly, the asymmetric unit contains two copies of PvNMT: an apo PvNMT and a ternary complex with IMP-0001173 and Myr-CoA (Fig. 1 ▸a). Analysis with the Protein Interfaces, Surfaces and Assemblies service PISA at the European Bioinformatics Institute (https://www.ebi.ac.uk/pdbe/prot_int/pistart.html) agrees with the SEC information that PvNMT is a biological monomer. PvNMT has a prototypical NMT topology and adopts a compact, spherical configuration consisting of 15 α-helices and 19 β-sheets (Supplementary Fig. S2). The N-terminal catalytic center has two distinct binding pockets that are responsible for substrate-binding and catalytic activities (Fig. 1 ▸b). The substrate-binding pocket specifically binds the N-terminal sequence of myristoylated proteins, while a second proximal pocket acts as the cofactor-binding site (Fig. 1 ▸b). In our ternary structure, the substrate-binding pocket contains the inhibitor IMP-0001173 molecule, while the Myr-CoA molecule sits in the cofactor-binding site (Figs. 1 ▸a and 1 ▸b). Also evident from our structure is the location of the Ab-loop (Fig. 1 ▸b). This binding arrangement facilitates the transfer of myristic acid from Myr-CoA to the N-terminus of the substrate protein, resulting in the release of CoA as a byproduct of the reaction (Rudnick et al., 1993 ▸; Spassov et al., 2023 ▸).
Figure 1.
PvNMT structure. (a) There are two PvNMT monomers in the asymmetric unit. Chain A (gray) has a bound Myr-CoA (green sticks) and inhibitor IMP-0001173 (blue sticks). Chain B (pink) shows a PvNMT monomer in the apo state. (b) Cartoon of PvNMT monomer A colored in a rainbow from blue at the N-terminus to red at the C-terminus. Myr-CoA (magenta sticks) and the inhibitor IMP-0001173 (white sticks) are shown. (c) Superposition of the monomers chain A (with ligands, gray) and chain B (apo PvNMT, pink). The substrate-binding cavity is indicated in purple parentheses, while the Myr-CoA-binding cavity is shown in black parentheses.
The carboxyl-terminus of PvNMT is inaccessible and positioned deep within the protein core (Figs. 1 ▸a and 1 ▸b), which ensures its resistance to cleavage by carboxypeptidases (Rudnick et al., 1993 ▸). PvNMT has a central core with an internal pseudo-twofold symmetry axis formed by the N-terminal and C-terminal halves, shaping the site for binding peptide substrates. All of the loops near the binding cavity are ordered in the monomer with bound Myr-CoA and IMP-0001173, whereas the loops nearest the binding cavities are disordered in the apo PvNMT monomer (Fig. 1 ▸c). The partly ordered apo PvNMT is the only reported apo PvNMT structure in the PDB.
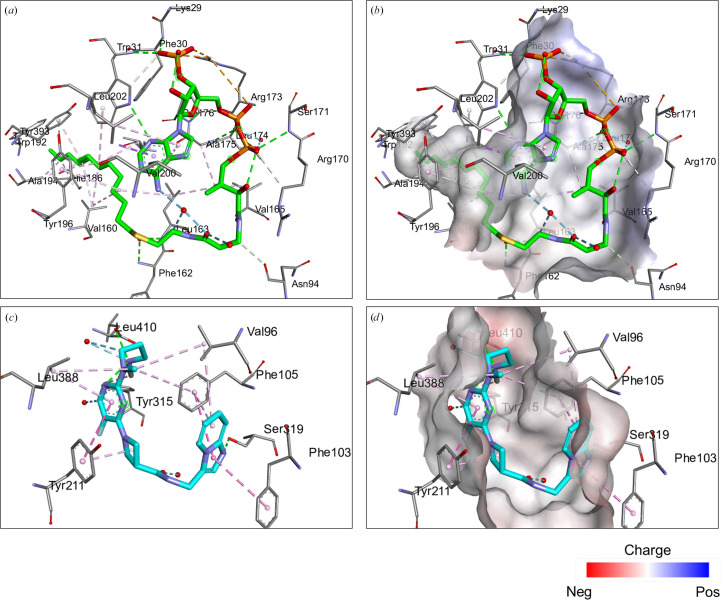
The myristoyl-binding pocket of PvNMT preferentially binds glycine residues, ensuring its substrate specificity (Harupa et al., 2020 ▸). In our structure, Myr-CoA occupies the extended groove that runs across one face of the enzyme (Fig. 2 ▸). Several leucine residues are involved in Myr-CoA binding (Rodríguez-Hernández et al., 2023 ▸). The predominantly hydrophobic Myr-CoA binding site has a few positive charges that stabilize Myr-CoA binding (Harupa et al., 2020 ▸; Rodríguez-Hernández et al., 2023 ▸). The structures of complexes of NMTs from different organisms with substrates, intermediate stages, inhibitors and products have helped to clarify the catalytic mechanisms of NMT (Rodríguez-Hernández et al., 2023 ▸; Wu et al., 2007 ▸). The postulated catalytic mechanism starts with the formation of a stable complex with Myr-CoA, while the peptide-binding domain of NMT accepts the N-terminus of the substrate protein (Dian et al., 2020 ▸; Rodríguez-Hernández et al., 2023 ▸; Wu et al., 2007 ▸). The Ab-loop above the peptide-binding pocket (Figs. 1 ▸a, 1 ▸b and 3 ▸a) adopts an closed or open conformation to control access to the active site by forming a ceiling or lid (Wu et al., 2007 ▸). In our structure, the Ab-loop loop closes around the bound inhibitor (Fig. 3 ▸b). Opening of the Ab-loop allows initial peptide binding and subsequent release of the myristoylated peptide (Wu et al., 2007 ▸). The surface plot shows the interconnectedness of the binding pockets of PvNMT (Fig. 3 ▸b). In our PvNMT ternary structure, the inhibitor IMP-0001173 is in the peptide-binding pocket, with a similar conformation as observed in other plasmodial NMT structures (Figs. 2 ▸ and 3 ▸). The peptide-binding pocket is predominantly hydrophobic but has some hydrogen bonds and salt bridges (Fig. 4 ▸b). An N atom (N02) of IMP-0001173 forms a salt bridge with the C-terminal carboxylate group (Leu410) of PvNMT that effectively abrogates myristate transfer (Figs. 2 ▸ and 4 ▸b), making IMP-0001173 an effective inhibitor.
Figure 2.
Myr-CoA and inhibitor (IMP-0001173) binding by PvNMT. The interactions were analyzed with Discovery Studio Visualizer (https://discover.3ds.com/discovery-studio-visualizer). (a) Stick representation of amino-acid residues (gray) interacting with Myr-CoA (green). (b) The Myr-CoA-binding pocket is slightly positively charged [shown in the same view as in (a)]. (c) The inhibitor IMP-0001173 (shown in cyan) interacts with amino acids (gray). (d) IMP-0001173 has primarily electrostatic interactions in the peptide-binding pocket.
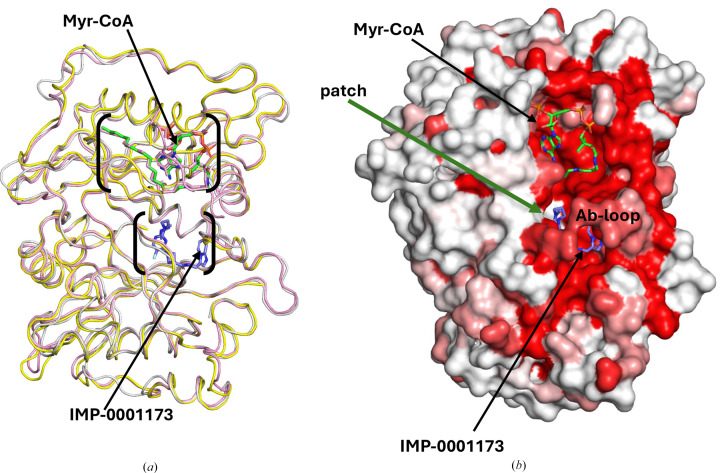
Figure 3.
Comparison of human NMTs with PvNMT. (a) PvNMT (gray) shares structural topology with the two human NMTs HsNMT1 (yellow) and HsNMT2 (pink). Myr-CoA is shown in green sticks, while IMP-0001173 is shown in blue sticks. (b) An ENDScript surface plot of PvNMT, in the same orientation as in (a), shows that the residues closest to the Myr-CoA- and peptide-binding sites are highly conserved and form an interconnected cavity. Red regions represent higher conserved regions, while white represents regions with low conservation. Myr-CoA is shown in green sticks while IMP-0001173 is shown in blue sticks.
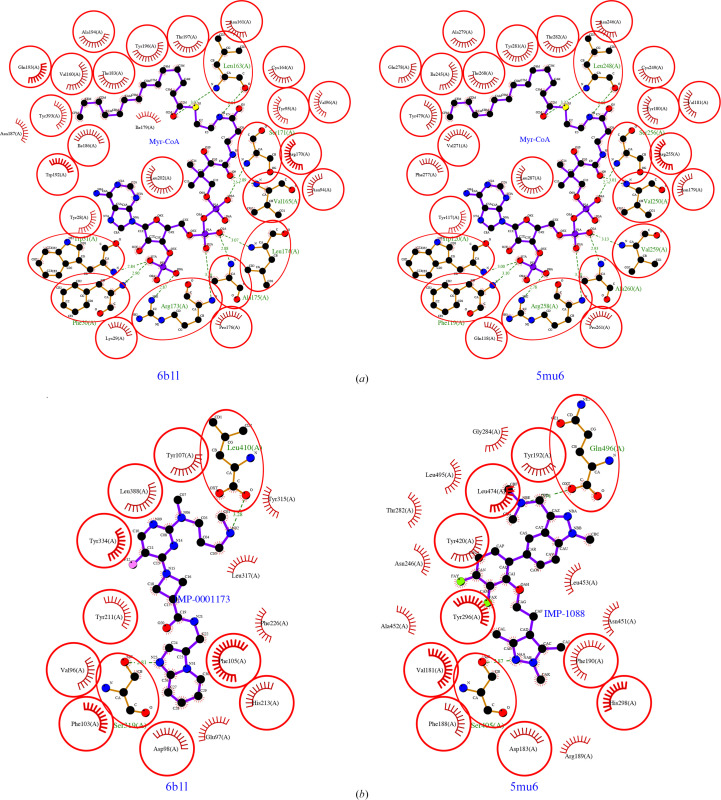
Figure 4.
LIGPLOT-generated interaction plots for PvNMT (PDB entry 6b1l) and HsNMT1 (PDB entry 5mu6) reveal (a) conserved Myr-CoA cavities and (b) differences in the peptide-binding cavity. The HsNMT1 structure (PDB entry 5mu6) has IMP-1088 within the peptide-binding pocket compared with IMP-0001173 in the PvNMT structure (PDB entry 6b1l).
Structure-based primary-sequence alignment of PvNMT with human NMTs (HsNMT1 and HsNMT2) and other plasmodial NMTs reveals a shared similarity of plasmodial NMTs and greater divergence from human NMTs (Supplementary Fig. S3). Nonetheless, the overall structural similarity between human NMTs and PvNMT is evident from superposed representative structures (Fig. 3 ▸a). The representative human NMT structures are PDB entries 5mu6 for HsNMT1 (Mousnieret al., 2018 ▸) and 4c2x for HsNMT2 (Thinon et al., 2014 ▸). ENDScript (Gouet et al., 2003 ▸; Robert & Gouet, 2014 ▸) analysis was used to identify the closest structural neighbors of PvNMT (Supplementary Fig. S4). These analyses reveal that PvNMT shares significant secondary-structural similarity with several NMTs, with identical residues observed across both the Myr-CoA- and peptide-binding domains (Supplementary Fig. S4). The regions of highest similarity are in the interconnected Myr-CoA- and peptide-binding cavities and are shown in red on the surface diagram (Fig. 3 ▸b). Interestingly, there is a patch of white in the peptide-binding cavity (Fig. 3 ▸b). Further details of structural differences and similarities are indicated in the sausage plot which, like the surface plot, was generated with ENDScript (Supplementary Fig. S5). The sausage plot shows the well conserved tertiary-structure topology in the protein core (thin sausages).
The differences between PvNMT and human NMTs are being explored for drug discovery (Rodríguez-Hernández et al., 2023 ▸; Harupa et al., 2020 ▸). These differences are evident in LIGPLOT-generated (Laskowski & Swindells, 2011 ▸; Wallace et al., 1995 ▸) interaction plots. Comparing our ternary structure of PvNMT with that of human NMT (HsNMT1) with an inhibitor of similar family as IMP-0001173, IMP-1088 (Bell et al., 2012 ▸; Mousnier et al., 2018 ▸), reveals a well conserved Myr-CoA and differences in the peptide-binding domain (Fig. 4 ▸, Supplementary Table S1). Thus, PvNMT is attractive for the rational development of small-molecule inhibitors due to differences in its peptide-binding domain from those of human NMTs (Rodríguez-Hernández et al., 2023 ▸; Harupa et al., 2020 ▸).
4. Conclusion
The presented PvNMT ternary structure offers additional insights for the rational design and optimization of NMT inhibitors for the treatment of P. vivax malaria. Efforts are ongoing to translate these insights into future therapeutic interventions.
Supplementary Material
Supplementary Figures and Table. DOI: 10.1107/S2053230X24008604/ir5034sup1.pdf
Funding Statement
This work was funded by National Institute of Allergy and Infectious Diseases grant 75N93022C00036 to Peter J. Myler; National Institute of Allergy and Infectious Diseases grant T34GM136489 to Oluwatoyin A. Asojo, Cydni Bolling, Shane Taylor, and Alex Mendez; National Institute of General Medical Sciences grant U01GM138433 to Oluwatoyin A. Asojo.
References
- Agirre, J., Atanasova, M., Bagdonas, H., Ballard, C. B., Baslé, A., Beilsten-Edmands, J., Borges, R. J., Brown, D. G., Burgos-Mármol, J. J., Berrisford, J. M., Bond, P. S., Caballero, I., Catapano, L., Chojnowski, G., Cook, A. G., Cowtan, K. D., Croll, T. I., Debreczeni, J. É., Devenish, N. E., Dodson, E. J., Drevon, T. R., Emsley, P., Evans, G., Evans, P. R., Fando, M., Foadi, J., Fuentes-Montero, L., Garman, E. F., Gerstel, M., Gildea, R. J., Hatti, K., Hekkelman, M. L., Heuser, P., Hoh, S. W., Hough, M. A., Jenkins, H. T., Jiménez, E., Joosten, R. P., Keegan, R. M., Keep, N., Krissinel, E. B., Kolenko, P., Kovalevskiy, O., Lamzin, V. S., Lawson, D. M., Lebedev, A. A., Leslie, A. G. W., Lohkamp, B., Long, F., Malý, M., McCoy, A. J., McNicholas, S. J., Medina, A., Millán, C., Murray, J. W., Murshudov, G. N., Nicholls, R. A., Noble, M. E. M., Oeffner, R., Pannu, N. S., Parkhurst, J. M., Pearce, N., Pereira, J., Perrakis, A., Powell, H. R., Read, R. J., Rigden, D. J., Rochira, W., Sammito, M., Sánchez Rodríguez, F., Sheldrick, G. M., Shelley, K. L., Simkovic, F., Simpkin, A. J., Skubak, P., Sobolev, E., Steiner, R. A., Stevenson, K., Tews, I., Thomas, J. M. H., Thorn, A., Valls, J. T., Uski, V., Usón, I., Vagin, A., Velankar, S., Vollmar, M., Walden, H., Waterman, D., Wilson, K. S., Winn, M. D., Winter, G., Wojdyr, M. & Yamashita, K. (2023). Acta Cryst. D79, 449–461.
- Battle, K. E., Lucas, T. C. D., Nguyen, M., Howes, R. E., Nandi, A. K., Twohig, K. A., Pfeffer, D. A., Cameron, E., Rao, P. C., Casey, D., Gibson, H. S., Rozier, J. A., Dalrymple, U., Keddie, S. H., Collins, E. L., Harris, J. R., Guerra, C. A., Thorn, M. P., Bisanzio, D., Fullman, N., Huynh, C. K., Kulikoff, X., Kutz, M. J., Lopez, A. D., Mokdad, A. H., Naghavi, M., Nguyen, G., Shackelford, K. A., Vos, T., Wang, H., Lim, S. S., Murray, C. J. L., Price, R. N., Baird, J. K., Smith, D. L., Bhatt, S., Weiss, D. J., Hay, S. I. & Gething, P. W. (2019). Lancet, 394, 332–343.
- Bell, A. S., Mills, J. E., Williams, G. P., Brannigan, J. A., Wilkinson, A. J., Parkinson, T., Leatherbarrow, R. J., Tate, E. W., Holder, A. A. & Smith, D. F. (2012). PLoS Negl. Trop. Dis.6, e1625. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dian, C., Pérez-Dorado, I., Rivière, F., Asensio, T., Legrand, P., Ritzefeld, M., Shen, M., Cota, E., Meinnel, T., Tate, E. W. & Giglione, C. (2020). Nat. Commun.11, 1132. [DOI] [PMC free article] [PubMed]
- Drysdale, M., Tan, L., Martin, A., Fuhrer, I. B., Duparc, S. & Sharma, H. (2023). Infect. Dis. Ther.12, 33–51. [DOI] [PMC free article] [PubMed]
- Frearson, J. A., Brand, S., McElroy, S. P., Cleghorn, L. A., Smid, O., Stojanovski, L., Price, H. P., Guther, M. L., Torrie, L. S., Robinson, D. A., Hallyburton, I., Mpamhanga, C. P., Brannigan, J. A., Wilkinson, A. J., Hodgkinson, M., Hui, R., Qiu, W., Raimi, O. G., van Aalten, D. M. F., Brenk, R., Gilbert, I. H., Read, K. D., Fairlamb, A. H., Ferguson, M. A., Smith, D. F. & Wyatt, P. G. (2010). Nature, 464, 728–732. [DOI] [PMC free article] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res.31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Harupa, A., De Las Heras, L., Colmenarejo, G., Lyons-Abbott, S., Reers, A., Caballero Hernandez, I., Chung, C. W., Charter, D., Myler, P. J., Fernández-Menéndez, R. M., Calderón, F., Palomo, S., Rodríguez, B., Berlanga, M., Herreros-Avilés, E., Staker, B. L., Fernández Álvaro, E. & Kaushansky, A. (2020). J. Med. Chem.63, 591–600. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kane, M. (2012). In Medical Genetics Summaries, edited by V. M. Pratt, S. A. Scott, M. Pirmohamed, B. Esquivel, B. L. Kattman & A. J. Malheiro. Bethesda: National Center for Biotechnology Information. [PubMed]
- Krissinel, E. B., Winn, M. D., Ballard, C. C., Ashton, A. W., Patel, P., Potterton, E. A., McNicholas, S. J., Cowtan, K. D. & Emsley, P. (2004). Acta Cryst. D60, 2250–2255. [DOI] [PubMed]
- Laskowski, R. A. & Swindells, M. B. (2011). J. Chem. Inf. Model.51, 2778–2786. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McIlhinney, R. A. (1989). Biochem. Soc. Trans.17, 861–863. [DOI] [PubMed]
- Mehdipour, P., Rajasekhar, M., Dini, S., Zaloumis, S., Abreha, T., Adam, I., Awab, G. R., Baird, J. K., Brasil, L. W., Chu, C. S., Cui, L., Daher, A., do Socorro, M. G. M., Gomes, M., Gonzalez-Ceron, L., Hwang, J., Karunajeewa, H., Lacerda, M. V. G., Ladeia-Andrade, S., Leslie, T., Ley, B., Lidia, K., Llanos-Cuentas, A., Longley, R. J., Monteiro, W. M., Pereira, D. B., Rijal, K. R., Saravu, K., Sutanto, I., Taylor, W. R. J., Thanh, P. V., Thriemer, K., Vieira, J. L. F., White, N. J., Zuluaga-Idarraga, L. M., Guerin, P. J., Price, R. N., Simpson, J. A., Commons, R. J., Adhikari, B., Alam, M. S., Assefa, A., Boyd, S. C., Chau, N. H., Day, N. P. J., Degaga, T. S., Dondorp, A. M., Erhart, A., Ferreira, M. U., Ghimire, P., Green, J. A., Khan, W. A., Koh, G. C. K. W., Mekuria, A. H., Mueller, I., Naadim, M. N., Nelwan, E. J., Nosten, F., Pasaribu, A. P., Pukrittayakamee, S., Rowland, M., Sattabongkot, J., Stepniewska, K., Suarez-Kurtz, G., Seidlein, L., Woodrow, C. J. & Woyessa, A. (2023). Malar. J.22, 306.
- Mousnier, A., Bell, A. S., Swieboda, D. P., Morales-Sanfrutos, J., Pérez-Dorado, I., Brannigan, J. A., Newman, J., Ritzefeld, M., Hutton, J. A., Guedán, A., Asfor, A. S., Robinson, S. W., Hopkins-Navratilova, I., Wilkinson, A. J., Johnston, S. L., Leatherbarrow, R. J., Tuthill, T. J., Solari, R. & Tate, E. W. (2018). Nat. Chem.10, 599–606. [DOI] [PMC free article] [PubMed]
- Nicolau, M. S. P., Resende, M. A., Serafim, P., Lima, G. Y. P., Ueira-Vieira, C., Nicolau-Junior, N. & Yoneyama, K. A. G. (2023). J. Biomol. Struct. Dyn.41, 7019–7031. [DOI] [PubMed]
- Pino, P., Sebastian, S., Kim, E. A., Bush, E., Brochet, M., Volkmann, K., Kozlowski, E., Llinás, M., Billker, O. & Soldati-Favre, D. (2012). Cell Host Microbe, 12, 824–834. [DOI] [PMC free article] [PubMed]
- Rackham, M. D., Brannigan, J. A., Rangachari, K., Meister, S., Wilkinson, A. J., Holder, A. A., Leatherbarrow, R. J. & Tate, E. W. (2014). J. Med. Chem.57, 2773–2788. [DOI] [PMC free article] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res.42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rodríguez-Hernández, D., Vijayan, K., Zigweid, R., Fenwick, M. K., Sankaran, B., Roobsoong, W., Sattabongkot, J., Glennon, E. K. K., Myler, P. J., Sunnerhagen, P., Staker, B. L., Kaushansky, A. & Grøtli, M. (2023). Nat. Commun.14, 5408. [DOI] [PMC free article] [PubMed]
- Rudnick, D. A., McWherter, C. A., Gokel, G. W. & Gordon, J. I. (1993). Adv. Enzymol. Relat. Areas Mol. Biol.67, 375–430. [DOI] [PubMed]
- Selvakumar, P., Kumar, S., Dimmock, J. R. & Sharma, R. K. (2011). Atlas Genet. Cytogenet. Oncol. Haematol.15, 570–575. [DOI] [PMC free article] [PubMed]
- Serbzhinskiy, D. A., Clifton, M. C., Sankaran, B., Staker, B. L., Edwards, T. E. & Myler, P. J. (2015). Acta Cryst. F71, 594–599. [DOI] [PMC free article] [PubMed]
- Spassov, D. S., Atanasova, M. & Doytchinova, I. (2023). Int. J. Mol. Sci.24, 11610. [DOI] [PMC free article] [PubMed]
- Stacy, R., Begley, D. W., Phan, I., Staker, B. L., Van Voorhis, W. C., Varani, G., Buchko, G. W., Stewart, L. J. & Myler, P. J. (2011). Acta Cryst. F67, 979–984. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Thinon, E., Serwa, R. A., Broncel, M., Brannigan, J. A., Brassat, U., Wright, M. H., Heal, W. P., Wilkinson, A. J., Mann, D. J. & Tate, E. W. (2014). Nat. Commun.5, 4919. [DOI] [PMC free article] [PubMed]
- Udenwobele, D. I., Su, R. C., Good, S. V., Ball, T. B., Varma Shrivastav, S. & Shrivastav, A. (2017). Front. Immunol.8, 751. [DOI] [PMC free article] [PubMed]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng. Des. Sel.8, 127–134. [DOI] [PubMed]
- Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. & Richardson, J. S. (2018). Protein Sci.27, 293–315. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wu, J., Tao, Y., Zhang, M., Howard, M. H., Gutteridge, S. & Ding, J. (2007). J. Biol. Chem.282, 22185–22194. [DOI] [PubMed]
- Yilma, D., Groves, E. S., Brito-Sousa, J. D., Monteiro, W. M., Chu, C., Thriemer, K., Commons, R. J., Lacerda, M. V. G., Price, R. N. & Douglas, N. M. (2023). Am. J. Trop. Med. Hyg.109, 761–769. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Table. DOI: 10.1107/S2053230X24008604/ir5034sup1.pdf