Abstract
Microalgae contribute to about half of global net photosynthesis, which converts sunlight into the chemical energy (ATP and NADPH) used to transform CO2 into biomass. Alternative electron pathways of photosynthesis have been proposed to generate additional ATP that is required to sustain CO2 fixation. However, the relative importance of each alternative pathway remains elusive. Here, we dissect and quantify the contribution of cyclic, pseudo-cyclic, and chloroplast-to-mitochondrion electron flows for their ability to sustain net photosynthesis in the microalga Chlamydomonas reinhardtii. We show that (i) each alternative pathway can provide sufficient additional energy to sustain high CO2 fixation rates, (ii) the alternative pathways exhibit cross-compensation, and (iii) the activity of at least one of the three alternative pathways is necessary to sustain photosynthesis. We further show that all pathways have very different efficiencies at energizing CO2 fixation, with the chloroplast–mitochondrion interaction being the most efficient. Overall, our data lay bioenergetic foundations for biotechnological strategies to improve CO2 capture and fixation.
Quantifying the contribution of the main electron flow pathways to algal CO2 fixation.
Introduction
Oxygenic photosynthesis is the main process responsible for the carbon input into ecosystems on Earth, with net CO2 fixation by photosynthesis representing more than 10 times anthropogenic CO2 emissions (IPCC 2021). During photosynthesis, CO2 fixation is powered by the combined action of PSI and PSII, establishing a linear electron flow (LEF) that produces reducing power (NADPH) and a trans-thylakoid proton motive force (pmf) used to generate ATP. Both ATP and NADPH fuel the CO2 fixation via the Calvin–Benson–Bassham (CBB) cycle and the further processing of the glyceraldehyde-3-phosphate to regenerate ribulose-1,5-bisphosphate and produce building blocks used for the elaboration of biomass (Johnson and Alric 2013). However, given the insufficiency of LEF to generate an adequate pmf and the conversion efficiency of the chloroplastic ATP-synthase (ATPase), it has been long recognized that LEF only generates 85% of the theoretical ATP requirements of the CBB cycle (Allen 2003).
Cyclic electron flow (CEF), which recycles reducing equivalents around PSI, and pseudo-cyclic electron flow (PCEF), which reduces molecular O2 at the acceptor side of PSI (Fig. 1), were proposed to contribute to the production of extra ATP during photosynthesis (Arnon 1959, 1984). In most angiosperms and mosses, CEF entails two main pathways, one involving the PROTON GRADIENT REGULATION 5 (PGR5) (Munekage et al. 2002) and PGR-LIKE 1 proteins (DalCorso et al. 2008) and another involving the plastidial NAD(P)H dehydrogenase-like (NDH) complex NDH-1 (Joët et al. 2001; Munekage et al. 2004). In many green microalgae, the plastidial NDH-1 complex is absent (Peltier et al. 2016) although they still have two CEF pathways, one controlled by PGRL1 and PGR5 (Tolleter et al. 2011; Johnson et al. 2014) and the other involving the plastidial NAD(P)H dehydrogenase Nda2 (Jans et al. 2008; Desplats et al. 2009; Saroussi et al. 2016). Since the Nda2 pathway is mostly active during nitrogen deprivation (Peltier and Schmidt 1991; Saroussi et al. 2016), hereafter, CEF refers to the PGRL1/PGR5-controlled CEF. PCEF involves flavodiiron proteins (FLVs) that catalyze O2 photoreduction at the PSI acceptor side (Fig. 1) in cyanobacteria (Helman et al. 2003; Allahverdiyeva et al. 2013), green microalgae (Chaux et al. 2017), mosses (Gerotto et al. 2016), liverwort (Shimakawa et al. 2017), and gymnosperms (Ilík et al. 2017).
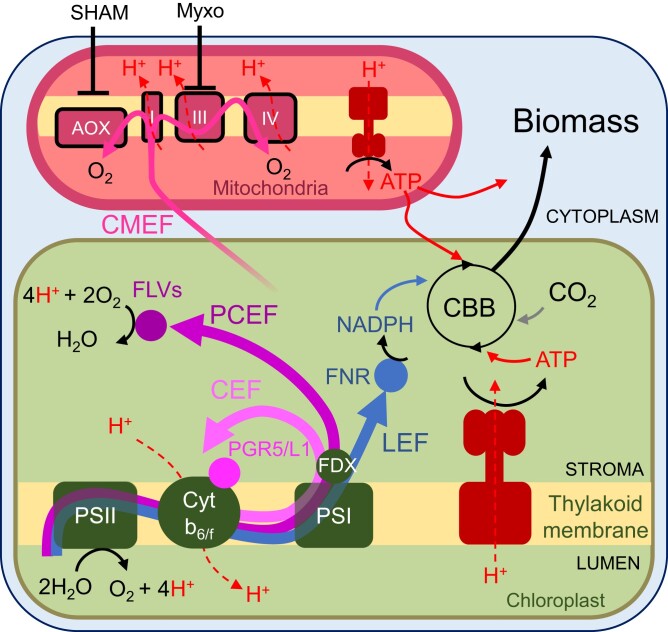
Figure 1.
Electron transport pathways operating during oxygenic photosynthesis and their role in ATP generation. LEF (blue solid arrow) operates from PSII to PSI and reduces NADP+ to NADPH using electrons from water. It also generates a pmf across the thylakoid membrane composed of a proton gradient (ΔpH) and electrical gradient (ΔΨ); the pmf is used by the ATPase to generate ATP (proton translocation is shown in red dashed lines; ATP fluxes are shown in red plain lines). CEF (light pink solid arrow) and PCEF (magenta solid arrow) both contribute to the generation of the pmf but do not produce reducing equivalents since the e− used for PCEF reduces O2 to water. CMEF (pink solid arrow) encompasses multiple metabolic pathways by which reductants generated in the chloroplast are shuttled to the mitochondria and converted into ATP by the mitochondrial electron transport chain. NADPH and ATP generated by these different pathways are eventually used for transforming CO2 into biomass, which reflects net photosynthesis. AOX, alternative oxidase; CBB, Calvin–Benson–Bassham; FDX, ferredoxin; FLVs, flavodiiron proteins; FNR, ferredoxin-NADP+ reductase; Myxo, myxothiazol; PS, photosystem; PGR5/L1, PROTON GRADIENT REGULATION 5/like-1; SHAM, salicylhydroxamic acid; I, III, IV, respiratory complexes I, III, and IV.
Another mechanism that can supply ATP to photosynthesis involves metabolic cooperation between chloroplasts and mitochondria (Raghavendra and Padmasree 2003). Reducing power generated within the chloroplast is transferred to mitochondria by metabolic shuttles where it is converted to ATP by the mitochondrial respiratory chain and might be shuttled back to the cytosol and the chloroplast (Fig. 1). Such a cooperation, called chloroplast-to-mitochondrion electron flow (CMEF), was first evidenced from the restoration of photoautotrophic growth in a mutant of the green microalgae Chlamydomonas (Chlamydomonas reinhardtii) deficient in the plastidial ATPase (Lemaire et al. 1988). Similar cooperation was later suggested from studies of Chlamydomonas mutants affected in mitochondrial respiration (Cardol et al. 2003) or in CEF (Dang et al. 2014) and was also observed in the moss Physcomitrium patens (Mellon et al. 2021) and in diatoms (Bailleul et al. 2015).
It is generally considered that both PCEF and CMEF act essentially as electron valves, operating either transiently in the case of PCEF to alleviate excess electrons generated by photosynthesis under light fluctuating regimes (Alric and Johnson 2017; Chaux et al. 2017; Alboresi et al. 2019) or more continuously for CMEF under high light stress (Kaye et al. 2019). Although, in Chlamydomonas, CMEF, PCEF, and CEF have been recently shown to all contribute to supply energy to the CO2 concentrating mechanism (CCM) (Burlacot et al. 2022), evidence for their role in powering the CBB cycle and downstream carbon metabolism processes is scarce.
In this work, we show that each pathway (CEF, PCEF, and CMEF) has the capacity to sustain net CO2 fixation to varying degrees and can compensate for the absence of the other pathways, whereas CO2 fixation is completely abolished in the absence of all three pathways. Moreover, we introduce a method to quantify the efficiency and capacity of each pathway and use it to show that CMEF is by far the most efficient at energizing CO2 fixation. We further discuss our data in relation to the relative contribution and efficiency of the different alternative pathways to the net primary productivity of terrestrial and oceanic ecosystems.
Results
Net photosynthesis fully relies on mitochondrial respiration when PCEF and CEF are impaired
To test whether PCEF and CEF are required for maximal net photosynthesis (VMax), we first measured photosynthetic O2 exchange rates using membrane inlet mass spectrometry (MIMS) (Burlacot et al. 2020) in air-grown Chlamydomonas control cells (wild types [WTs] 2 and 3) and mutants impaired in both CEF and PCEF (pgrl1 flvB-1 and -3) (Burlacot et al. 2022). Since the pgrl1 and flvB mutants are both affected in the functioning of the CCM (Burlacot et al. 2022), photosynthesis measurements were performed in the presence of a saturating concentration of inorganic carbon (Ci) when the CCM is inactive. As previously reported (Burlacot et al. 2022), pgrl1 flvB double mutants can perform photosynthesis at maximum rates comparable to the single pgrl1 or flvB mutants and to the parental and sibling control strains (Fig. 2; Supplementary Figs. S1A, S2, and S3), showing that neither CEF nor PCEF is essential to reach VMax. To test a possible contribution of mitochondrial respiration to photosynthesis, pgrl1 flvB double mutants were treated with myxothiazol (Myxo) and salicylhydroxamic acid (SHAM), inhibitors of mitochondrial electron transport, respectively acting on complex III and the alternative oxidases (AOX) (Fig. 1) (Schonbaum et al. 1971; von Jagow and Link 1986). While the addition of both respiratory inhibitors had a small impact (about 10% to 20% decrease) on the VMax of control strains (Fig. 2, A and C; Supplementary Figs. S1A, S2, and S3), they completely abolished photosynthesis in pgrl1 flvB double mutants (Fig. 2, B and C; Supplementary Figs. S1 and S2).
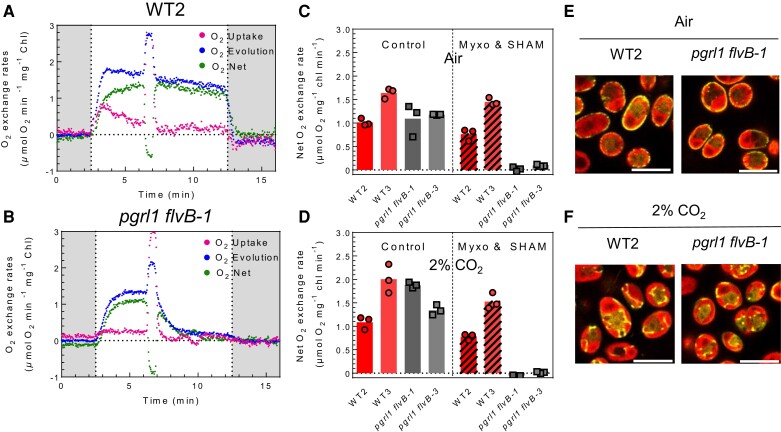
Figure 2.
Dependence of net photosynthesis on CEF, PCEF, and CMEF. A, B) O2 exchange rates upon a dark-to-light transition in WT2 A) and pgrl1 flvB-1B). At t = 6 min, mitochondrial respiration inhibitors Myxo (2.5 µm) and SHAM (400 µm) were added to the reaction. Shown are representative traces of n = 3 biologically independent samples. C, D) Net O2 evolution in pgrl1 flvB-1 and pgrl1 flvB-3 and their control strains (WT2 and WT3) in the absence (control) or presence (Myxo and SHAM) of Myxo (2.5 µm) and SHAM (400 µm) in cells grown in air C) or in 2% CO2-enriched air D). Values are taken after 9 min of illumination upon a dark to light transitions and normalized per chlorophyll content (chl). Bars show the average and dots show individual replicates (n = 3 biologically independent samples). Typical experiments used to extract these data are shown in Supplementary Fig. S2. E, F) Cross section images showing subcellular localization of mitochondrion-localized Clover (yellow fluorescence) in pgrl1 flvB-1 and a control strain (WT2), grown in air E) or 2% CO2-enriched air F). Images are representative of 2 independent experiments. Scale bar: 10 µm. Clover/chlorophyll merged signals are shown. Images are representative of 2 experiments. Scale bar: 10 µm.
In Chlamydomonas, the cellular location of mitochondria depends on the CO2 supply, with the mitochondria located at the cell periphery in low CO2-grown cells and mostly inside the chloroplast cup in high CO2 (Geraghty and Spalding 1996) (Fig. 2, E and F). Since this location might affect CMEF (Burlacot and Peltier 2023), we sought to examine the involvement of the mitochondrial subcellular localization on the role of CMEF. As with air-grown cells, mitochondrial inhibitors fully stopped net photosynthesis on 2% CO2-grown pgrl1 flvB mutants while having a marginal effect (about 10% to 20% decrease) on the control strains (Fig. 2D; Supplementary Fig. S4). To control the location of the mitochondria in the different strains, we visualized the spatial distribution of an expressed, mitochondrion-targeted fluorophore (Clover). There was similar relocalization of mitochondria depending on the CO2 supply during growth (at periphery under very low CO2) of both the single and double mutants and a control strain (Fig. 2, E and F; Supplementary Fig. S5). We conclude from these experiments that the absence of CEF, PCEF, and CMEF abolishes net photosynthesis. Furthermore, in the absence of both CEF and PCEF, near WT levels of photosynthesis can be achieved that fully rely on CMEF, which does not appear to rely on the spatial distribution of the mitochondria.
CEF, PCEF, and CMEF each have the potential to sustain most net photosynthesis
While CMEF can power all net photosynthesis, the potential of CEF, PCEF, and the two subpathways of CMEF to energize net photosynthesis remains unclear. We sought to estimate it by measuring the effect of mitochondrial inhibitors on photosynthesis rates in single and double pgrl1 and flvB mutants (Fig. 3, A to D). Inhibition of respiration decreased net photosynthesis by 40% to 50% in both the flvB and pgrl1 single mutants (Fig. 3, A and B; Supplementary Figs. S1A and S2), showing that both CEF and PCEF, independently, have the potential to energize 50% to 60% of VMax (Fig. 3E; Supplementary Fig. S1, A and B). Inhibition of respiration decreased the VMax of all control strains by 10% to 20% (Supplementary Fig. S1A, S2, and S3) showing that combined activities of CEF and PCEF can power approximately 85% of photosynthesis (Fig. 3E). Based on the effects of SHAM or Myxo individually, on VMax in pgrl1 flvB mutants (Fig. 3, C to E;Supplementary Figs. S1A and S2), we conclude that while the complex III and IV pathway of CMEF (Fig. 1) can power most net photosynthesis on its own (Fig. 3D), the AOX pathway (Fig. 1) can only sustain approximately 45% of VMax (Fig. 3E). Because the amount of the terminal oxidases involved in CMEF does not differ in the pgrl1 flvB mutants and their control strains (Supplementary Fig. S6), these values likely reflect WT capacities. We also quantified net photosynthesis based on inorganic carbon (Ci) uptake (Supplementary Figs. S7 and S8). For all strains tested, the inhibitory effects on Vmax measured as net Ci uptake were similar to that observed on net O2 production (Supplementary Fig. S7). We conclude from these experiments that CEF, PCEF, and CMEF can sustain photosynthetic Ci uptake on their own but that only the complex III and IV pathway of CMEF is efficient enough to reach near-maximal photosynthesis levels.
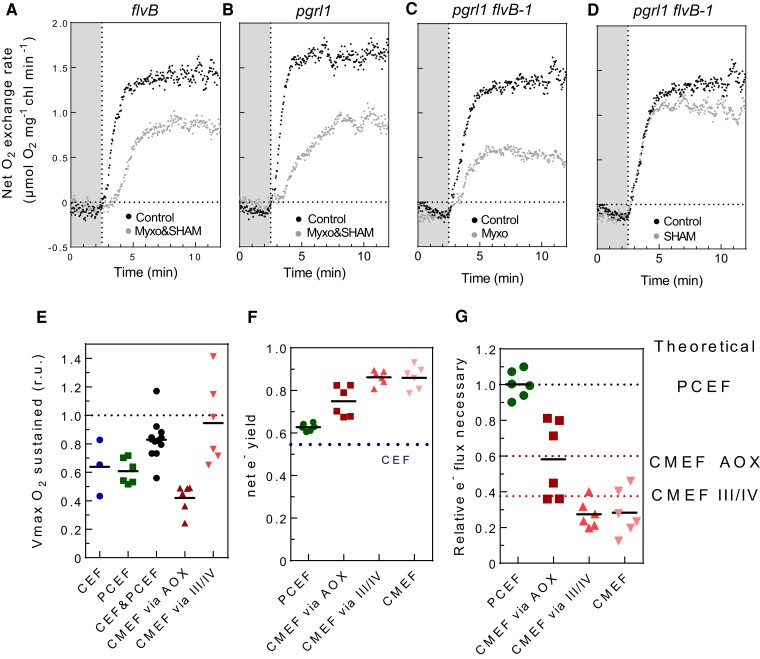
Figure 3.
Relative efficiencies of CEF, PCEF, and CMEF in energizing net photosynthesis. A to D) Net O2 evolution measured during a dark to light transition in flvBA), pgrl1B), pgrl1 flvB-2 C, D), treated (gray dots) or not treated (black dots) with both Myxo (2.5 µm) and SHAM (400 µm) A, B) or with only one inhibitor C, D). Shown are representative traces of 3 biological replicates. E) Relative Vmax measured when a given set of alternative electron pathways are active after treatment with Myxo (2.5 µm), SHAM (400 µm), or both inhibitors; CEF (blue, flvB treated with Myxo and SHAM), PCEF (green, pgrl1 treated with Myxo and SHAM), CEF&PCEF (dark, control strains treated with Myxo and SHAM), CMEF via AOX (burgundy, pgrl1 flvB treated with Myxo), and CMEF via III/IV (red, pgrl1 flvB treated with SHAM). F) Net electron yield of PCEF (green), CMEF via AOX (purple), CMEF via III/IV (red), CMEF (pink), and CEF (dotted line). Similar net electron yields were also determined for PCEF in 2% CO2-grown cells and a slightly lower net electron yield for CMEF (Supplementary Fig. S9D). G) Electron flux relative to PCEF required to power similar levels of net photosynthesis when relying on a single pathway. Data shown are individual extrapolations based on measured net electron yields. In E), F), and G), bars show the average and dots show individual replicates (n ≥3 biologically independent samples).
CEF, PCEF, and CMEF show different efficiencies in sustaining net photosynthesis
The strong capacity of CMEF to power photosynthesis at its maximal rate by using less than 10% of additional electron flow relative to net photosynthesis (%-LEF) was surprising. This contrasts with around 30%-LEF of PCEF not being sufficient to reach maximal photosynthesis (Supplementary Fig. S3). We hypothesized that this difference could result from differences in the efficiencies of the alternative pathways to power net photosynthesis. Indeed, mechanistically, for each electron flowing once through the different pathways (CEF, PCEF, the AOX, or the complex III and IV pathways of CMEF), the total number of protons translocated across the thylakoid and/or the inner mitochondrial membranes highly differs (Burlacot 2023). To experimentally quantify the efficiency of the different pathways in energizing net photosynthesis, we introduce a metric, the net electron (e−) yield of a pathway defined by the following equation:
where e− (net photosynthesis) is the flux of photosynthetic electrons directly used for net photosynthesis and e− (alternative pathway) is the flux of photosynthetic electrons through a given alternative pathway under conditions where only this alternative pathway is active (in addition to LEF). Therefore, the net e− yield of an alternative pathway, which is determined when only that specific alternative pathway is operating, characterizes the efficiency of electrons passing through it in powering net photosynthesis under a given physiological condition. Note that the net e− yield, thus, does not reflect the rate or the importance of the pathway considered but rather its capacity to translocate H+ for each electron.
We used this equation and experiments from Fig. 3, A to E to quantify the net e− yield of each pathway (Fig. 3F; Supplementary Fig. S9). The net e− yield of PCEF was estimated at 0.65, whereas that of CMEF at 0.85 (Fig. 3F), indicating a better efficiency of CMEF in driving CO2 fixation relative to PCEF. In the absence of an established method to quantifying CEF in physiologically relevant conditions (Burlacot et al. 2022), CEF net e− yield was estimated to be around 0.55 by hypothesizing that an electron passing once through CEF is 1.5 times less efficient at generating a pmf than if it would pass through PCEF (Kramer and Evans 2011; Burlacot 2023) (see Materials and Methods). In the case of CMEF, the net e− yield of the AOX pathway was lower than that of the complex III and IV pathway (Fig. 3F), likely reflecting the proton translocating capacity of each pathway (Fig. 1) (Burlacot 2023). Using the net e− yields of each pathway, we then compared the flux of electrons required to power the same amount of net photosynthesis relative to PCEF (Fig. 3G). Strikingly, theoretical values were in accord with values experimentally determined by the net e− yields (Fig. 3G). Suggesting that under our conditions, the measured differences of efficiencies to power net photosynthesis were due to their mechanistic H+/e− transfer capacity. We conclude from these experiments that CMEF pathways are more efficient than PCEF and CEF at energizing CO2 fixation and that this difference arises from its higher H+/e− transfer capacity, which likely leads to a higher capacity to generate ATP.
Estimating the respective contributions of CEF, PCEF, and CMEF during steady-state photosynthesis and growth
A key remaining question is to determine which set of pathways is active in vivo when all are functional and to what extent each of them contributes to net photosynthesis under physiological conditions. We thus sought to estimate (i) the relative electron flux via each pathway and (ii) their contribution to energize net photosynthesis during steady-state photosynthesis. We used two different approaches, one based on the effect of mutations and the other based on the effect of respiratory inhibitors. As both approaches are based on pathway inhibition, they are potentially biased by the high level of redundancy existing between them and the existence of compensatory mechanisms (Cardol et al. 2009; Dang et al. 2014; Burlacot et al. 2022). However, the two approaches differ in the way compensatory mechanisms contribute, with mutants being more subject to compensation than the short-term effects of inhibitors. In the mutant-based approach, PCEF was quantified by subtracting O2 uptake rates in the flvB mutant from its control parental strain (allowing the calculation of PCEFmut and CMEFmut) (Supplementary Fig. S10). In the inhibitor-based approach, CMEF was quantified by subtracting O2 uptake rates between SHAM plus Myxo treated and untreated control strains (allowing the calculation of PCEFinhib and CMEFinhib) (Supplementary Fig. S10).
The two methods differ somewhat in their estimates of PCEF and CMEF (Fig. 4, A and B; Supplementary Fig. S11), with a higher CMEF contribution being estimated using the mutant method. On average, PCEF and CMEF represent a flux of between 10% to 20%-LEF and 5% to 10%-LEF, respectively (Fig. 4A). Based on the respective net e− yield of each pathway, we can estimate that 10% to 30% and 30% to 60% of CO2 fixation is sustained by PCEF and CMEF, respectively (Fig. 4B). Since CEF sustains the remaining net photosynthesis, we estimated that 25% to 30% of CO2 fixation is powered by CEF (Fig. 4B), which corresponds to a CEF of about 25%-LEF assuming a net e− yield of CEF of 0.55 (Fig. 4A). We also tried to use mutants impaired in the NADH DEHYDROGENASE 4 subunit of respiratory complex I (nd4), PGRL1, or both (nd4 pgrl1) (Larosa et al. 2018) as a third method for estimating respective contributions of the different pathways (Supplementary Fig. S12). However, the low inhibition of respiration rates in nd4 and nd4 pgrl1 mutants, indicating the existence of complex I-independent respiration pathways (Supplementary Fig. S12A) makes these mutants unsuitable for this purpose.
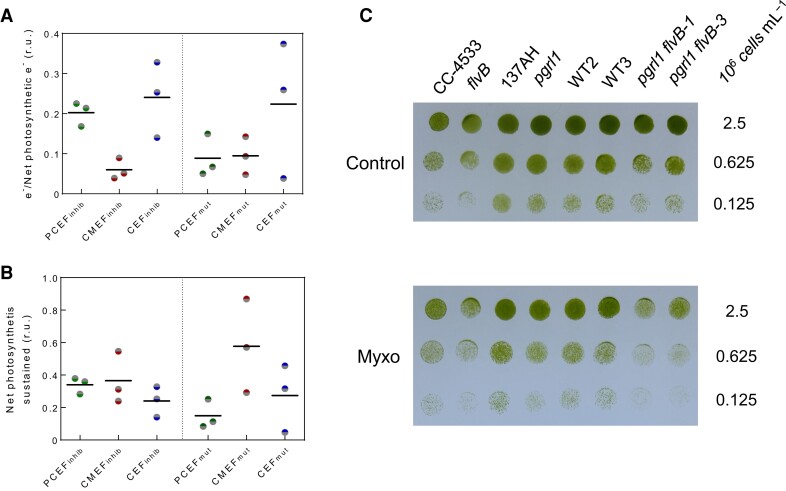
Figure 4.
Contribution of CEF, PCEF, and CMEF to the electron flow, net photosynthesis, and growth. A) Electron flow through PCEF, CEF, and CMEF relative to LEF was determined in the WT strain CC-4533 by 2 different methods, one based on the use of inhibitors (inhib) and the other on mutants (mut). Data used for these calculations are the same as for Fig. 2C and Supplementary Fig. S2. B) Proportion of net photosynthesis powered by each electron flow (PCEF, CEF, and CMEF) in the control strain CC-4533 as measured by the inhibition (inhib) or mutant (mut) method. CC-4533 was chosen because it is the control strain for flvB and is therefore the only strain where both methods could be used. Data used for these calculations are the same as for A). In A) and B), bars show the average and dots show individual replicates (n = 3 biologically independent samples). C) Growth tests for flvB and pgrl1 mutants, their corresponding control strains (CC-4533 and 137AH, respectively) and double mutants (pgrl1 flvB-1 and pgrl1 flv-3) and their corresponding control strains (WT2 and 3). Cells were spotted on plates containing minimal medium at pH 7.2 and grown under continuous illumination (60 µmol photon m−2 s−1) in the absence (upper panel) or presence (lower panel) of 2.5 µm Myxo in air enriched at 2% CO2. Spots shown are representative of 10 independent experiments.
To further assess the contribution of CEF, PCEF, and CMEF over the long term, growth tests were performed on the different strains in the absence or presence of the respiration inhibitor Myxo. While Myxo has no effect on the growth of control strains and single flvB or pgrl1 mutants in moderate light intensities (Fig. 4C), growth of pgrl1 flvB was impaired in the presence of the inhibitor (Fig. 4C; Supplementary Fig. S13, B and C). Interestingly, at low light intensity (30 µmol photons m−2 s−1), the effect of Myxo on the double mutant was absent (Supplementary Fig. S13A), suggesting that the AOX pathway of CMEF has the potential to fully power net photosynthesis in low light conditions. Therefore, whereas CEF, PCEF, and CMEF all contribute to energizing photosynthesis, CMEF, although representing a relatively small flux, has the potential to fuel CO2 fixation and biomass accumulation thanks to its high capacity for H+ translocation per e−.
Discussion
Since the first description of CEF by Arnon (1959), the question was raised as to whether LEF could sustain the ATP requirement of photosynthetic CO2 fixation (Arnon 1959), and it was proposed that photosynthetic electron flows, other than LEF, are involved in balancing the ATP/NADPH ratio to sustain CBB cycle function (Arnon 1959; Allen 2003) and other metabolic processes. Since then, several mechanisms of CEF, PCEF, and CMEF have been uncovered (Munekage et al. 2002, 2004; Helman et al. 2003; Tomaz et al. 2010) as well as some compensations among those different mechanisms (Cardol et al. 2009; Petroutsos et al. 2009; Dang et al. 2014; Storti et al. 2019; Storti et al. 2020; Burlacot et al. 2022), but the importance and relative efficiency of each mechanism remain mostly unknown. Here, we have shown that in Chlamydomonas PGRL1-controlled CEF, FLV-mediated PCEF, and both the AOX and complex III/IV/cytochrome oxidase pathways of CMEF can all supply additional energy to complement LEF for CO2 fixation. The net e− yields of CEF, PCEF, and CMEF greatly differ; CMEF is the most electron efficient and contributes importantly to the extra ATP requirements of CO2 fixation in continuous, saturating light. Note that in spite of the ongoing controversy of how PGRL1 is involved in CEF (Nawrocki et al. 2019), a clear integration is observed among PGRL1-controlled CEF, PCEF, and CMEF, with these different pathways able to compensate for a reduction in the activities of the other pathways. Importantly, we observed similar effects on net O2 and CO2 exchange (Supplementary Fig. S7), thus showing that all effects measured on net photosynthesis correspond to changes in CO2 fixation and eventually biomass production and growth (Fig. 4 C).
Some compensation between CEF, PCEF, and CMEF has been previously reported; a Chlamydomonas PGRL1-deficient strain exhibits increased CMEF and PCEF (Dang et al. 2014) and mutants of complexes I and III show enhanced rates of CEF (Cardol et al. 2009). We observed an elevated CMEF in the absence of FLV (Fig. 4, A and B) with a higher level of terminal oxidases associated with CMEF (Supplementary Fig. S6), corroborating cross-compensation among the three mechanisms of alternative electron flow. In diatoms, while CMEF is considered to be a major driver of CO2 fixation, inhibition of CMEF only leads to a ∼20% decrease of the PSII yield (Bailleul et al. 2015), which might also reflect compensation by CEF. We also conclude, based on the fact that no CO2 fixation could be detected in the double pgrl1 flvB mutants treated with inhibitors of mitochondrial respiration, that neither the NDA2-mediated CEF nor O2 reduction mediated by the plastid terminal oxidase or Mehler reactions has the potential by themselves to sustain a measurable amount of net photosynthesis, which is in agreement with previous studies concluding that these pathways represent a small fraction of alternative electron flow (Alric et al. 2010; Houille-Vernes et al. 2011; Nawrocki et al. 2019) (Fig. 2 B).
Establishing the quantitative contribution of each pathway in the WT strain in the absence of perturbation of any alternative pathway is a challenging task to which the approaches we have developed here provide an imperfect answer. Indeed, the proposed quantification of each pathway relies on the assumption that inhibiting one pathway (CMEF or PCEF) does not impact the flow through the others, an assumption not likely to be accurate since compensation among the different pathways has been previously documented (Cardol et al. 2009; Dang et al. 2014; Burlacot et al. 2022). Therefore, our approach attempts to evaluate the respective contributions of each pathway until an integrative quantification method is available. Here, the mutant method likely overestimates CMEF and the inhibitor method likely overestimates PCEF; both approaches likely overestimate CEF. It is clear, however, that no single pathway sustains net photosynthesis by itself (Supplementary Fig. S11B), which might reflect a rather balanced contribution of the different pathways to CO2 fixation in the WT. It is also noteworthy that, although concordant quantifications are seen in the different WT strains used, there is some level of variability (Supplementary Fig. S11), which might reflect the effect of the genetic background. In any case, it seems highly likely that, considering the high redundancy of alternative electron pathways of photosynthesis and the ability of each of them to sustain CO2 fixation at a high rate, a dynamic mix of the different pathways may enable cells to better cope with variable environmental conditions (Burlacot 2023).
The net e− yield, a tool to evaluate the efficiency of electron transport pathways
In support to LEF, the combined energetic outputs of PGRL1-controlled CEF and PCEF cannot sustain more than 80% of maximal CO2 fixation (Fig. 3E) whereas the flux of e− through each of these pathways individually can help sustain 60% of it (Fig. 3F). This suggests the existence of an intrinsic limitation of the flow of electrons through the photosynthetic electron transport chain, which is likely due to a pmf-induced photosynthetic control occurring at the cytochrome b6f level (Rumberg et al. 1968; Stiehl and Witt 1969; Johnson and Berry 2021; Malone et al. 2021), an elevated pmf eliciting a block in electron flow across the b6f complex. Engaging an electron pathway with a high net e− yield, such as CMEF, might be a way to avoid this limitation since fewer electrons would be needed to power the same level of CO2 fixation. Such a strategy seems to be used by diatoms (Bailleul et al. 2015), which lack FLVs and mosses, whose photosynthetic activity is impaired by inactivation of the mitochondrial respiratory complex I (Mellon et al. 2021).
Throughout the green lineage, the nature and activity of alternative electron flows are quite different. For example, angiosperms lack FLV-dependent PCEF (Peltier et al. 2010), harbor a NDH-1-dependent CEF pathway (Peltier et al. 2016), and have a weakly active CMEF (Raghavendra and Padmasree 2003). Since the NDH-1-dependent CEF translocates 4H+/e− (Strand et al. 2017), its net e− yield is probably close to that of CMEF, strongly suggesting that in angiosperms, the NDH-1 complex can compensate when CMEF capacity is diminished. The latter is illustrated by the poor growth of Arabidopsis (Arabidopsis thaliana) mutants impaired in both CEF pathways (PGR and NDH-1) (Munekage et al. 2004). Because of the number of key proteins and biochemical reactions involved in CMEF, its high net e− yield may not be as rapidly accessible as CEF or PCEF during environmental fluctuations. While, under light-limiting conditions, a high net e− yield pathway might be advantageous, a less efficient pathway would contribute to the safe dissipation of excess absorbed light energy and diminished reactive oxygen production at saturating light intensities. Overall, it remains to be determined to what extent harboring a palette of alternative electron flows with varying yields and likely different time responses is critical for sustaining photosynthetic performance under dynamic, natural conditions.
Note here that while modulation of the capacity of membranes to transform pmf into ATP may modulate the absolute net e− yield of each pathway, the relative net e− yield of CEF and PCEF would not change if they are embedded in the same thylakoid system. Similarly, even under conditions in which the mitochondrial respiration would be partially or fully uncoupled, the net e− yield of CMEF would be at least as good as PCEF due to the generation of a pmf across the thylakoid membranes (Burlacot 2023). Hence, if CEF, PCEF, and CMEF are all competing for the same photosynthetic electrons, electrons passing through CMEF and PCEF should always be more efficient than CEF in powering net photosynthesis. Measuring a relative positioning of the net e− yield of each pathway that is different than the one described here would be an indication of spatially separated electron transport chains dedicated to specific pathways in vivo.
Additional energy cost of photosynthesis
Allen (2002) previously estimated that a PCEF representing only 16%-LEF would be sufficient, in addition to LEF, to satisfy the ATP and NADPH requirement of the CBB cycle. Our conclusions are strikingly different since we estimate that if only PCEF was active, it would have to represent 54%-LEF to sustain CO2 fixation. This discrepancy reflects the actual metabolic cost of CO2 fixation that exceeds the ATP and/or H+ requirement of the CBB cycle due to other processes that depend on proton gradient energization such as protein synthesis and transporter activities or to the presence of proton leakages through energized membranes. In addition to extra ATP/H+ requirements, our measurements reveal that for each CO2 molecule fixed, around two molecules of O2 are generated (Supplementary Figs. S7 and S8), suggesting that half of the net reductant generated is used in metabolic processes other than the CBB cycle. It is worth noting here that similar conclusions have recently been reached in two marine diatoms (Shimakawa and Matsuda 2024) and in angiosperms where reductant and carbon fluxes are heavily channeled toward central carbon metabolism (Treves et al. 2022). Finally, in natural environments, either terrestrial or aquatic, CO2 fixation is further limited by the concentration of atmospheric CO2, which can trigger photorespiration in C3 plants or induction of the CCM in C4 plants and algae. All these mechanisms would require additional energy for their activities (Osmond 1981; Takabayashi et al. 2005; Walker et al. 2014; Burlacot et al. 2022), making alternative electron pathways of photosynthesis even more critical for sustaining plant and algal CO2 capture. Given the strong capacity of CMEF to sustain photosynthesis and its high H+/e− conversion capacity, it is timely to reevaluate its importance during photosynthesis across photosynthetic eukaryotes.
Materials and methods
Strains and growing conditions
C. reinhardtii flvB and pgrl1 single mutants and their respective parental strains CC-4533 (for flvB) and 137AH (for pgrl1) were previously described (Tolleter et al. 2011; Chaux et al. 2017). flvB pgrl1–1, -2, and -3 double mutant and their respective sibling strains WT2 and WT3 (used here as control strains) have been described previously (Burlacot et al. 2022) and are all from the progeny of a crossing of flvB and pgrl1 performed in Burlacot et al. (2022). Because pgrl1 flvB double mutants were obtained by crossing the two single mutants that have different genetic backgrounds (as described in Burlacot et al. 2022), the control strains used are siblings that resulted from the cross and that contained WT copies and levels of the FLVB and PGRL1 proteins. pgrl1-2 is a mutant impaired in accumulation of the PGRL1 protein originating from crossing flvB and pgrl1 performed in Burlacot et al. (2022). Complex I-deficient strains and the pgrl1 mutant used in Supplementary Fig. S12 have been described in Larosa et al. (2018). All strains were grown photoautotrophically in 125-mL flasks with cotton stoppers at 25°C in a buffered minimal medium (20 mm MOPS pH 7.2) under constant illumination (80 µmol photons m−2 s−1) either under ambient CO2 or 2% CO2 in air. Among pgrl1 flvB double mutants and their control strains already described (Burlacot et al. 2022), pgrl1 flvB-1 and pgrl1 flvB-3 and WT2 and 3, respectively, were used for physiological measurements since their maximal photosynthesis was representative of other double mutants and control strains (Burlacot et al. 2022). All strains used in this study apart from the ones in Supplementary Fig. S12 are available under the same names used here (except for flvB, which is available as flvB-21) at the Chlamydomonas Resource Center (www.chlamycollection.org). The spectrum of the growing light used is shown in Supplementary Fig. S14. Throughout the manuscript, a “biologically independent experiment” refers to a replication that has been done with a fresh new liquid culture of the strains described, seeded from the source plates.
Gas exchange measurements
Gas exchange rates were measured using MIMS. Cells grown in air enriched or not at 2% CO2 were collected during the exponential phase by centrifugation at 450 × g for 3 min and resuspended in 1.5 mL of fresh buffered minimal medium (pH 7.2) at 30 μg chlorophyll mL−1. HCO3− was subsequently added to the cell suspension (10 mm final concentration). The cell suspension was then placed in the MIMS reaction vessel (mounted with a 1 mil Teflon membrane), 2 mL of 18O-enriched O2 (97% 18O Eurisop, France) was bubbled in the suspension. After closing the vessel, gas exchange was recorded during a dark-to-light transition (2,000 µmol photons m−2 s−1; green LEDs picked at 525 nm; Luminus reference PT-121-G-L11-MPK). Light intensity was set to be saturating (Supplementary Fig. S15) using weakly absorbed green light for better homogeneity. Calculations of gross and net photosynthesis were done using the MIMS analysis software as previously described (Burlacot et al. 2020). In some experiments, SHAM (400 µm final concentration) and Myxo (2.5 µm final concentration) were added 1 min before the beginning of the measurements, except for the data presented in Fig. 2, A and B, where inhibitors were added during the measurement. Note here that SHAM has been reported to not inhibit the plastid terminal oxidase (Cournac et al. 2000). All net O2, net CO2, gross O2 exchange, net e− yield, e− flow, and net photosynthesis sustained shown as histograms in Figs. 2 to 4 and Supplementary Figs. S1, S7, S9, and S11 are based on measurements after 9 min of illumination.
Clover expressing cells
137AH, WT2, pgrl1, flvB, and pgrl1flvB-1 and -2 expressing the mitochondrion-targeted fluorophore Clover (mito-Clover) were generated as described in Harmon et al. (2022). Briefly, cells were transformed with a linearized plasmid encoding mito-Clover and a hygromycin resistance cassette using the Max Efficiency Reagent (Invitrogen). Clones resistant to hygromycin were screened for Clover fluorescence using a M1000 Tecan plate reader (excitation: 488/7 nm; emission: 515/10 nm) and mitochondrial signal was checked by confocal microscopy (described below). Selected clones were grown at 80 µmol photons m−2 s−1 in 25 mL minimal medium in 125-mL flasks, in the presence of either normal or 2% CO2-enriched air until they reached a density of ∼10 µg chlorophyll mL−1. An aliquot of each culture was then briefly concentrated by centrifugation at 1,500 × g, and cells were layered on an 18-well microscope slide. Images were acquired using a TCS SP8 confocal laser-scanning microscope (Leica) and a 63× objective. Acquisition settings were as follows: Clover excitation 488 nm, emission 500 to 547 nm; chlorophyll excitation 488 nm, and emission 694 to 733 nm. For homogeneity and clarity, signal intensities of each channel were adjusted using the Fiji software. Strains expressing mito-Clover used in this article are publicly available from the Chlamydomonas Resource Center (https://www.chlamycollection.org) under the following CC numbers: CC-6056 (137AH), CC-6059 (WT2), CC-6057 (flvB), CC-6058 (pgrl1), CC-6060 (pgrl1 flvB-1), and CC-6061 (pgrl1 flvB-1).
Growth tests
The different Chlamydomonas strains were cultivated at an air level of CO2 under moderate light (80 µmol photons m−2 s−1) at 25 °C. Cells were collected during exponential growth and diluted in fresh minimal medium to 0.125, 0.625, or 2.5 106 cells mL−1. Seven-microliter drops were spotted on 2% Agar plates of minimal medium (buffered with 20 mm MOPS pH 7.2) with or without Myxo (2.5 µm final concentration) and exposed to air enriched with 2% CO2. Homogeneous light was supplied by LED panels (see its spectrum in Supplementary Fig. S14B); the different light intensities were obtained with various neutral filters. Temperature was maintained at 25 °C at the level of plates by means of fans.
Net electron yield calculation
The net e− yield of a given alternative electron pathway is defined as the proportion of electrons generated downstream PSI that are used for net photosynthesis when only this alternative electron pathway is active, following this equation:
| (1) |
where e− (net photosynthesis) is the flux of photosynthetic electrons directly used as reductants for net photosynthesis and e− (alternative pathway) is the flux of photosynthetic electrons through an alternative pathway. Because, for PCEF and CMEF, the sum of e− (alternative pathway) and e− (net photosynthesis) equals gross photosynthesis, net electron yields of PCEF and CMEF were quantified using gross and net O2 evolution in mutants harboring: only PCEF (pgrl1 and pgrl1-2 mutants treated with Myxo and SHAM) or only the AOX or the complex III/IV pathway of CMEF (pgrl1 flvB-1 and -3 mutants treated with Myxo or SHAM, respectively). The net electron yield was then calculated for each pathway using the following equation:
| (2) |
The theoretical net electron yield of CEF was calculated based on the assumption that CEF translocates 1.5 less protons per electron across the thylakoid membranes than PCEF (Kramer and Evans 2011; Burlacot 2023) and that this is the driving force leading to net photosynthesis energization. Note that this later assumption is well supported by the independent results from Fig. 3G showing that the relative net electron yield of CMEF pathways and PCEF corresponds to their relative capacity at translocating protons across membranes. Hence, for the same net photosynthesis powered, CEF should be 1.5 times higher than PCEF, and the net electron yields of CEF could be written as follows:
| (3) |
where e− (net in pgrl1 Myxo SHAM) is the net photosynthesis measured in the pgrl1 mutant treated with Myxo and SHAM (hence having only PCEF active) and e− (PCEF) is the O2 uptake rate in the same strain and condition.
Based on Equation (1), we can rewrite e− (PCEF) as follows:
| (4) |
Hence, combining Equations (3) and (4):
| (5) |
The net electron yield of CEF shown in Fig. 3 is using Equation (5) with the average net electron yield of PCEF. Note that the quantification of the net e− yield in Chlamydomonas assumes that removing two pathways (elimination of CEF and CMEF in the pgrl1 mutant treated with inhibitors of respiration and CEF and PCEF in the pgrl1 flvB double mutants) makes CO2 fixation dependent only on the third pathway. This dependency is supported by the finding that the pgrl1 flvB double mutant treated with inhibitors of respiration is unable to support any CO2 fixation, which also support the hypothesis that the contribution of other pathways, such as the NDA2-dependent CEF, is marginal. Also note that by definition, the net electron yield of a pathway is independent of the maximal capacity of this pathway but rather reflects a per electron efficiency of the pathway in powering net photosynthesis under the physiological condition where it is assessed.
Calculation of electron flows via each pathway and their contribution to net photosynthesis in WT strains
The electron flow through PCEF and CMEF was deduced on WT strain using two independent methods based on measurements of gross O2 uptake made in Supplementary Figs. S2 and S3. In the so-called “mutant” method, CMEF was quantified as being the gross O2 uptake measured in the flvB mutant. Then, PCEF was quantified by subtracting the CMEF to the O2 uptake of CC-4533 (Supplementary Fig. S10). This method was only used for CC-4533 since it is the only control strain for the flvB mutant. In the so-called “inhibitor” method, PCEF was quantified as being the gross O2 uptake remaining in control strains treated with SHAM and Myxo. Then, CMEF was quantified by subtracting the PCEF to the O2 uptake in the same untreated control strain (Supplementary Fig. S10). The calculated electron flows of PCEF and CMEF were then used for each strain to quantify the net photosynthesis powered by each of them following the equations derived from Equation (1):
where net photosynthesis (AEF) is the net photosynthesis sustained by a specific alternative electron flow, e− (AEF) is the electron flow through this alternative electron flow, and net e− yield (AEF) is the net electron yield of this alternative electron flow.
The total of net photosynthesis powered in a strain by PCEF and CMEF was then compared to the measured net photosynthesis in this same strain to quantify the “missing” net photosynthesis that had to be powered by CEF. Then, the electron flow through CEF (e− (CEF)) was calculated by the following equation derived from Equation (1):
where missing net is the calculated net photosynthesis that is not powered by PCEF and CMEF and net e− yield (CEF) is the net electron yield of CEF.
Protein extraction and immunodetection
Cells were harvested by centrifugation at 1,000 × g for 5 min. Total cellular protein was extracted by heating the cell pellet at 100°C for 1 min using an extraction aqueous solution composed of 1% (v/v) SDS, 100 mm DTT, 1 mm aminocaproic acid, and 1 mm benzamidine hydrochloride buffered with 60 mm Tris (pH 6.8). The resulting cell lysate was then centrifuged at 10,000 × g for 5 min to obtain the supernatant. The supernatant was subsequently incubated with acetone (80% -v/v- final concentration) at −20°C for 1 h. The protein pellet was collected by centrifugation at 5,000 × g for 5 min and stored in aqueous solution containing 1% (v/v) SDS and 5 mm DTT and buffered with 60 mm Tris (pH 6.8). Protein content was estimated (Microplate BCA Protein Assay Kit-reducing agent compatible, Thermo Scientific) with bovine serum albumin as the standard. Protein extracts (10 µg protein) were loaded on a 12% SDS–PAGE gel and run for 1 h at 150 V in Tris-glycine SDS buffer and transferred onto a nitrocellulose membrane using semi-dry transfer (Trans-Blot Turbo, Bio-Rad). Immunoblot detection was performed using antibodies raised against FlvB (1:1,000) (Chaux et al. 2017) and Pgrl1 (1:5,000) (Tolleter et al. 2011). Other antibodies against AOX1 (1:10,000) (AS06 152) and COXIIB (1:10,000) (AS06 151) were obtained from Agrisera (https://www.agrisera.com/). An anti-rabbit HRP-conjugated antibody (1:50,000) (AS09 602, Agrisera) was used as a secondary antibody for immunodetection. Precision Plus Protein Dual Color Standards (Bio-Rad) was used as the molecular weight marker. Uncropped and unprocessed scans of the blots shown in this study are visible in Supplementary Fig. S16.
Accession numbers
Genes studied in this article can be found on https://phytozome-next.jgi.doe.gov/ under the loci Cre12.g531900 (FLVA), Cre16.g691800 (FLVB), Cre07.g340200 (PGRL1), Cre09.g395950 (AOX1), Cre03.g169550 (AOX2), and Cre01.g049500 (COXIIB).
Supplementary Material
Acknowledgments
The authors acknowledge the constructive comments on the manuscript from Dr. Joe Berry, Dr. Jennifer Johnson, Evan Saldivar, and Leron Perez, technical support from Michel Philibert and Emmanuel Capra, and editing help by Dr. Solène Moulin. We thank Professor Claire Remacle for providing us with complex I-deficient strains. We would like to thank the Carnegie Advanced Imaging Facility for the use of microscopy instruments (Leica TCS SP8 confocal laser-scanning microscope).
Contributor Information
Gilles Peltier, Aix Marseille Univ, CEA, CNRS, Institut de Biosciences et Biotechnologies Aix-Marseille, CEA Cadarache, 13108 Saint Paul-lez-Durance, France.
Carolyne Stoffel, Department of Plant Biology, The Carnegie Institution for Science, Stanford, CA 94305, USA.
Justin Findinier, Department of Plant Biology, The Carnegie Institution for Science, Stanford, CA 94305, USA.
Sai Kiran Madireddi, Department of Plant Biology, The Carnegie Institution for Science, Stanford, CA 94305, USA.
Ousmane Dao, Aix Marseille Univ, CEA, CNRS, Institut de Biosciences et Biotechnologies Aix-Marseille, CEA Cadarache, 13108 Saint Paul-lez-Durance, France.
Virginie Epting, Aix Marseille Univ, CEA, CNRS, Institut de Biosciences et Biotechnologies Aix-Marseille, CEA Cadarache, 13108 Saint Paul-lez-Durance, France.
Amélie Morin, Aix Marseille Univ, CEA, CNRS, Institut de Biosciences et Biotechnologies Aix-Marseille, CEA Cadarache, 13108 Saint Paul-lez-Durance, France.
Arthur Grossman, Department of Plant Biology, The Carnegie Institution for Science, Stanford, CA 94305, USA; Department of Biology, Stanford University, Stanford, CA 94305, USA; Biosphere Science and Engineering Division, The Carnegie Institution for Science, Stanford, CA 94305, USA.
Yonghua Li-Beisson, Aix Marseille Univ, CEA, CNRS, Institut de Biosciences et Biotechnologies Aix-Marseille, CEA Cadarache, 13108 Saint Paul-lez-Durance, France.
Adrien Burlacot, Department of Plant Biology, The Carnegie Institution for Science, Stanford, CA 94305, USA; Department of Biology, Stanford University, Stanford, CA 94305, USA; Biosphere Science and Engineering Division, The Carnegie Institution for Science, Stanford, CA 94305, USA.
Author contributions
A.B. led and coordinated the research; A.B. and G.P. designed the research; A.B., C.S., J.F., O.D., S.M., V.E., A.M., and G.P. performed the research; A.B. and G.P. contributed new reagents/analytic tools; A.B. analyzed the data and made all figures; and A.B. and G.P. wrote the paper with inputs from Y. L.-B. and A.G.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Effect of mitochondrial inhibitors on O2 exchange rates on mutants and control strains grown at air level of CO2.
Supplementary Figure S2. O2 exchange rates in pgrl1 flvB double mutants and their control strains grown at air level of CO2.
Supplementary Figure S3. O2 exchange rates in pgrl1 and flvB single mutants and their control strains grown at air level of CO2.
Supplementary Figure S4. O2 exchange rates in pgrl1, pgrl1 flvB, and their control strains grown in air enriched with 2% CO2.
Supplementary Figure S5. Localization of mitochondria in pgrl1, flvB, pgrl1 flvB-1 and -2, and their control strain in air enriched or not with 2% CO2.
Supplementary Figure S6. Immunodetection of proteins involved in CMEF, PCEF, and CEF in the strains used in this study
Supplementary Figure S7. Comparison of net inorganic carbon fixation and net O2 production.
Supplementary Figure S8. Net Ci and O2 exchange rated in pgrl1 and flvB single and double mutants and the control strains WT2 and WT3.
Supplementary Figure S9. Net O2 exchange rates characteristic and e− quantum yield measured in strains grown in air enriched with 2% CO2.
Supplementary Figure S10. Illustration of the method used to quantify the contribution of each photosynthetic electron flow to CO2 fixation.
Supplementary Figure S11. Quantification of the contribution of each photosynthetic electron flow to net photosynthesis.
Supplementary Figure S12. Net O2 exchange rates characteristic of mutants impaired in respiratory complex I and PGRL1.
Supplementary Figure S13. Growth of mutant and control strains at various light intensities.
Supplementary Figure S14. Growing light spectra.
Supplementary Figure S15. Light saturation curve of net O2 exchange rate measured with green actinic light.
Supplementary Figure S16. Original images of immunoblots against the AOX1 and the respiratory cytochrome oxidase (CoxIIB) shown in Supplementary Fig. S6.
Supplementary Figure S17. Original images of immunoblots against FLVB and PGRL1 shown in Supplementary Fig. S6.
Funding
This work was supported by the Carnegie Institution for Science (A.B. and A.G.) and the ANR grants OTOLHYD n°ANR-18-CE05-0029 (G.P.) and AlgalCCM n°ANR-22-CE44-0023-01 (G.P., A.M., and V.E.). This work was also in part supported by DOE award DE-SC0019417 (A.G. and A.B.).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Alboresi A, Storti M, Morosinotto T. Balancing protection and efficiency in the regulation of photosynthetic electron transport across plant evolution. New Phytol. 2019:221(1):105–109. 10.1111/nph.15372 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mustila H, Ermakova M, Bersanini L, Richaud P, Ajlani G, Battchikova N, Cournac L, Aro EM. Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc Natl Acad Sci U S A. 2013:110(10):4111–4116. 10.1073/pnas.1221194110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Photosynthesis of ATP—electrons, proton pumps, rotors, and poise. Cell. 2002:110(3):273–276. 10.1016/S0092-8674(02)00870-X [DOI] [PubMed] [Google Scholar]
- Allen JF. Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trends Plant Sci. 2003:8(1):15–19. 10.1016/s1360-1385(02)00006-7 [DOI] [PubMed] [Google Scholar]
- Alric J, Johnson X. Alternative electron transport pathways in photosynthesis: a confluence of regulation. Curr Opin Plant Biol. 2017:37:78–86. 10.1016/j.pbi.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Alric J, Lavergne J, Rappaport F. Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim Biophys Acta. 2010:1797(1):44–51. 10.1016/j.bbabio.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Arnon DI. Conversion of light into chemical energy in photosynthesis. Nature. 1959:184(4679):10–21. 10.1038/184010a0 [DOI] [PubMed] [Google Scholar]
- Arnon DI. The discovery of photosynthetic phosphorylation. Trends Plant Sci. 1984:9(6):258–262. 10.1016/0968-0004(84)90159-2 [DOI] [Google Scholar]
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature. 2015:524(7565):366–369. 10.1038/nature14599 [DOI] [PubMed] [Google Scholar]
- Burlacot A. Quantifying the roles of algal photosynthetic electron pathways: a milestone towards photosynthetic robustness. New Phytol. 2023:240(6):2197–2203. 10.1111/nph.19328 [DOI] [PubMed] [Google Scholar]
- Burlacot A, Burlacot F, Li-Beisson Y, Peltier G. Membrane inlet mass spectrometry: a powerful tool for algal research. Front Plant Sci. 2020:11:1302. 10.3389/fpls.2020.01302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlacot A, Dao O, Auroy P, Cuiné S, Li-Beisson Y, Peltier G. Alternative photosynthesis pathways drive the algal CO2 concentrating mechanism. Nature. 2022:605(7909):366–371. 10.1038/s41586-022-04662-9 [DOI] [PubMed] [Google Scholar]
- Burlacot A, Peltier G. Energy crosstalk between photosynthesis and the algal CCM. Trends Plant Sci. 2023:28(7):795–807. 10.1016/j.tplants.2023.03.018 [DOI] [PubMed] [Google Scholar]
- Cardol P, Alric J, Girard-Bascou J, Franck F, Wollman FA, Finazzi G. Impaired respiration discloses the physiological significance of state transitions in Chlamydomonas. Proc Natl Acad Sci U S A. 2009:106(37):15979–15984. 10.1073/pnas.0908111106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P, Gloire G, Havaux M, Remacle C, Matagne R, Franck F. Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol. 2003:133(4):2010–2020. 10.1104/pp.103.028076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux F, Burlacot A, Mekhalfi M, Auroy P, Blangy S, Richaud P, Peltier G. Flavodiiron proteins promote fast and transient O2 photoreduction in Chlamydomonas. Plant Physiol. 2017:174(3):1825–1836. 10.1104/pp.17.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournac L, Redding K, Ravenel J, Rumeau D, Josse E-M, Kuntz M, Peltier G. Electron flow between photosystem II and oxygen in chloroplasts of photosystem I-deficient algae is mediated by a quinol oxidase involved in chlororespiration. J Biol Chem. 2000:275(23):17256–17262. 10.1074/jbc.M908732199 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Nemann DS, Finazzi G, Joliot P, Barbato R, Leister D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008:132(2):273–285. 10.1016/j.cell.2007.12.028 [DOI] [PubMed] [Google Scholar]
- Dang KV, Plet J, Tolleter D, Jokel M, Cuine S, Carrier P, Auroy P, Richaud P, Johnson X, Alric J, et al. Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell. 2014:26(7):3036–3050. 10.1105/tpc.114.126375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C, Mus F, Cuine S, Billon E, Cournac L, Peltier G. Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J Biol Chem. 2009:284(7):4148–4157. 10.1074/jbc.M804546200 [DOI] [PubMed] [Google Scholar]
- Geraghty AM, Spalding MH. Molecular and structural changes in Chlamydomonas under limiting CO2 (a possible mitochondrial role in adaptation). Plant Physiol. 1996:111(4):1339–1347. 10.1104/pp.111.4.1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Meneghesso A, Jokel M, Suorsa M, Aro E-M, Morosinotto T. Flavodiiron proteins act as safety valve for electrons in Physcomitrella patens. Proc Natl Acad Sci U S A. 2016:113(43):12322–12327. 10.1073/pnas.1606685113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon J, Findinier J, Ishii NT, Herbig M, Isozaki A, Grossman A, Goda K. Intelligent image-activated sorting of Chlamydomonas reinhardtii by mitochondrial localization. Cytometry A. 2022:101(12):1027–1034. 10.1002/cyto.a.24661 [DOI] [PubMed] [Google Scholar]
- Helman Y, Tchernov D, Reinhold L, Shibata M, Ogawa T, Schwarz R, Ohad I, Kaplan A. Genes encoding a-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr Biol. 2003:13(3):230–235. 10.1016/S0960-9822(03)00046-0 [DOI] [PubMed] [Google Scholar]
- Houille-Vernes L, Rappaport F, Wollman F-A, Alric J, Johnson X. Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci U S A. 2011:108(51):20820–20825. 10.1073/pnas.1110518109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilík P, Pavlovič A, Kouřil R, Alboresi A, Morosinotto T, Allahverdiyeva Y, Aro E-M, Yamamoto H, Shikanai T. Alternative electron transport mediated by flavodiiron proteins is operational in organisms from cyanobacteria up to gymnosperms. New Phytol. 2017:214(3):967–972. 10.1111/nph.14536 [DOI] [PubMed] [Google Scholar]
- IPCC . Climate change 2021: the physical science basis. Contribution of working group I to the sixth assessment report of the intergovernmental panel on climate change. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press; 2021. [Google Scholar]
- Jans F, Mignolet E, Houyoux PA, Cardol P, Ghysels B, Cuine S, Cournac L, Peltier G, Remacle C, Franck F. A type II NAD(P) H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci U S A. 2008:105(51):20546–20551. 10.1073/pnas.0806896105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Cournac L, Horvath EM, Medgyesy P, Peltier G. Increased sensitivity of photosynthesis to antimycin A induced by inactivation of the chloroplast ndhB gene. Evidence for a participation of the NADH-dehydrogenase complex to cyclic electron flow around photosystem I. Plant Physiol. 2001:125(4):1919–1929. 10.1104/pp.125.4.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryotic Cell. 2013:12(6):776–793. 10.1128/ec.00318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Berry JA. The role of cytochrome b6f in the control of steady-state photosynthesis: a conceptual and quantitative model. Photosynth Res. 2021:148(3):101–136. 10.1007/s11120-021-00840-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Steinbeck J, Dent RM, Takahashi H, Richaud P, Ozawa SI, Houille-Vernes L, Petroutsos D, Rappaport F, Grossman AR, et al. Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: a study of ΔATPase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2014:165(1):438–452. 10.1104/pp.113.233593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye Y, Huang W, Clowez S, Saroussi S, Idoine A, Sanz-Luque E, Grossman AR. The mitochondrial alternative oxidase from Chlamydomonas reinhardtii enables survival in high light. J Biol Chem. 2019:294(4):1380–1395. 10.1074/jbc.RA118.004667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Evans JR. The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 2011:155(1):70–78. 10.1104/pp.110.166652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larosa V, Meneghesso A, La Rocca N, Steinbeck J, Hippler M, Szabò I, Morosinotto T. Mitochondria affect photosynthetic electron transport and photosensitivity in a green alga. Plant Physiol. 2018:176(3):2305–2314. 10.1104/pp.17.01249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire C, Wollman FA, Bennoun P. Restoration of phototrophic growth in a mutant of Chlamydomonas reinhardtii in which the chloroplast atpB gene of the ATP synthase has a deletion: an example of mitochondria-dependent photosynthesis. Proc Natl Acad Sci U S A. 1988:85(5):1344–1348. 10.1073/pnas.85.5.1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone LA, Proctor MS, Hitchcock A, Hunter CN, Johnson MP. Cytochrome b6f—orchestrator of photosynthetic electron transfer. Biochim Biophys Acta. 2021:1862(5):148380. 10.1016/j.bbabio.2021.148380 [DOI] [PubMed] [Google Scholar]
- Mellon M, Storti M, Vera-Vives AM, Kramer DM, Alboresi A, Morosinotto T. Inactivation of mitochondrial complex I stimulates chloroplast ATPase in Physcomitrium patens. Plant Physiol. 2021:187(2):931–946. 10.1093/plphys/kiab276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K-I, Endo T, Tasaka M, Shikanai T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004:429(6991):579–582. 10.1038/nature02598 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002:110(3):361–371. 10.1016/S0092-8674(02)00867-X [DOI] [PubMed] [Google Scholar]
- Nawrocki WJ, Bailleul B, Picot D, Cardol P, Rappaport F, Wollman FA, Joliot P. The mechanism of cyclic electron flow. Biochim Biophys Acta Bioenerg. 2019:1860(5):433–438. 10.1016/j.bbabio.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Osmond CB. Photorespiration and photoinhibition: some implications for the energetics of photosynthesis. Biochim Biophys Acta. 1981:639(2):77–98. 10.1016/0304-4173(81)90006-9 [DOI] [Google Scholar]
- Peltier G, Aro EM, Shikanai T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Ann Rev Plant Biol. 2016:67(1):55–80. 10.1146/annurev-arplant-043014-114752 [DOI] [PubMed] [Google Scholar]
- Peltier G, Schmidt GW. Chlororespiration—an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1991:88(11):4791–4795. 10.1073/pnas.88.11.4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G, Tolleter D, Billon E, Cournac L. Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res. 2010:106(1-2):19–31. 10.1007/s11120-010-9575-3 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M. PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem. 2009:284(47):32770–32781. 10.1074/jbc.M109.050468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003:8(11):546–553. 10.1016/j.tplants.2003.09.015 [DOI] [PubMed] [Google Scholar]
- Rumberg B, Reinwald E, Schröder H, Siggel U. Correlation between electron flow, proton translocation and phosphorylation in chloroplasts. Naturwissenschaften. 1968:55(2):77–79. 10.1007/BF00599483 [DOI] [PubMed] [Google Scholar]
- Saroussi SI, Wittkopp TM, Grossman AR. The type II NADPH dehydrogenase facilitates cyclic electron flow, energy-dependent quenching, and chlororespiratory metabolism during acclimation of Chlamydomonas reinhardtii to nitrogen deprivation. Plant Physiol. 2016:170(4):1975–1988. 10.1104/pp.15.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonbaum GR, Bonner WD Jr, Storey BT, Bahr JT. Specific inhibition of the cyanide-insensitive respiratory pathway in plant mitochondria by hydroxamic acids. Plant Physiol. 1971:47(1):124–128. 10.1104/pp.47.1.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakawa G, Ishizaki K, Tsukamoto S, Tanaka M, Sejima T, Miyake C. The Liverwort, Marchantia, drives alternative electron flow using a flavodiiron protein to protect PSI. Plant Physiol. 2017:173(3):1636–1647. 10.1104/pp.16.01038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakawa G, Matsuda Y. Extra O2 evolution reveals an O2-independent alternative electron sink in photosynthesis of marine diatoms. Photosynth Res. 2024:159(1):61–68. 10.1007/s11120-023-01073-3 [DOI] [PubMed] [Google Scholar]
- Stiehl HH, Witt HT. Quantitative treatment of the function of plastoquinone in photosynthesis. Zeitschrift für Naturforschung B. 1969:24(12):1588–1598. 10.1515/znb-1969-1219 [DOI] [PubMed] [Google Scholar]
- Storti M, Alboresi A, Gerotto C, Aro E-M, Finazzi G, Morosinotto T. Role of cyclic and pseudo-cyclic electron transport in response to dynamic light changes in Physcomitrella patens. Plant Cell Environ. 2019:42(5):1590–1602. 10.1111/pce.13493 [DOI] [PubMed] [Google Scholar]
- Storti M, Segalla A, Mellon M, Alboresi A, Morosinotto T. Regulation of electron transport is essential for photosystem I stability and plant growth. New Phytol. 2020:228(4):1316–1326. 10.1111/nph.16643 [DOI] [PubMed] [Google Scholar]
- Strand DD, Fisher N, Kramer DM. The higher plant plastid NAD(P)H dehydrogenase-like complex (NDH) is a high efficiency proton pump that increases ATP production by cyclic electron flow. J Biol Chem. 2017:292(28):11850–11860. 10.1074/jbc.M116.770792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi A, Kishine M, Asada K, Endo T, Sato F. Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proc Natl Acad Sci U S A. 2005:102(46):16898–16903. 10.1073/pnas.0507095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell. 2011:23(7):2619–2630. 10.1105/tpc.111.086876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaz T, Bagard M, Pracharoenwattana I, Lindén P, Lee CP, Carroll AJ, Ströher E, Smith SM, Gardeström P, Millar AH. Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 2010:154(3):1143–1157. 10.1104/pp.110.161612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves H, Küken A, Arrivault S, Ishihara H, Hoppe I, Erban A, Höhne M, Moraes TA, Kopka J, Szymanski J, et al. Carbon flux through photosynthesis and central carbon metabolism show distinct patterns between algae, C3 and C4 plants. Nat Plants. 2022:8(1):78–91. 10.1038/s41477-021-01042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jagow G, Link TA. Use of specific inhibitors on the mitochondrial bc1 complex. Methods in enzymology. Vol. 126. Burlington, UK: Academic Press; 1986. p. 253–271. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Strand DD, Kramer DM, Cousins AB. The response of cyclic electron flow around photosystem I to changes in photorespiration and nitrate assimilation. Plant Physiol. 2014:165(1):453–462. 10.1104/pp.114.238238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental material.