Abstract
Nuclear factor erythroid 2-related factor 2 (NRF2) is a pleiotropic transcription factor that has neuroprotective and anti-inflammatory effects, regulating more than 250 genes. As NRF2, cannabinoid receptor type 2 (CB2) is also implicated in the preservation of neurons against glia-driven inflammation. To this concern, little is known about the regulation pathways implicated in CB2 receptor expression. In this study, we analyze whether NRF2 could modulate the transcription of CB2 in neuronal and microglial cells. Bioinformatics analysis revealed an antioxidant response element in the promoter sequence of the CB2 receptor gene. Further analysis by chemical and genetic manipulations of this transcription factor demonstrated that NRF2 is not able to modulate the expression of CB2 in neurons. On the other hand, at the level of microglia, the expression of CB2 is NRF2-dependent. These results are related to the differential levels of expression of both genes regarding the brain cell type. Since modulation of CB2 receptor signaling may represent a promising therapeutic target with minimal psychotropic effects that can be used to modulate endocannabinoid-based therapeutic approaches and to reduce neurodegeneration, our findings will contribute to disclose the potential of CB2 as a novel target for treating different pathologies.
Electronic supplementary material
The online version of this article (10.1007/s10571-019-00719-y) contains supplementary material, which is available to authorized users.
Keywords: CB2 receptor, NRF2, Microglia, Neuron, Brain, Dimethyl fumarate (DMF)
Introduction
In recent years, the transcription factor Nuclear Factor erythroid-derived 2-like 2 (herein referred as NRF2, encoded by NFE2L2 gene) has emerged as an essential factor in modulating the expression of genes involved in a broad spectrum of cellular functions. Although NRF2 was originally described as the master regulator of redox homeostasis (Itoh et al. 1997, 1995), its role in mechanisms involved in neuroinflammation, proteasome/autophagy, DNA repair, apoptosis, iron, and heme metabolism as well as phase I, II, and III drug/xenobiotic metabolism has now been described (Hayes and Dinkova-Kostova 2014; Schmidlin et al. 2019). In basal conditions, there are low levels of NRF2 due to the action of an E3 ubiquitin ligase complex containing a substrate adaptor protein, Kelch-like ECH-associated protein 1 (KEAP1), that binds to and negatively regulates NRF2 (Itoh et al. 1999). Oxidative or electrophilic stress induces NRF2 signaling through modifications of key cysteine residues in KEAP1 that induce conformational changes in the binding of NRF2–KEAP1 that avoids the degradation of NRF2. This allows the accumulation of newly synthesized NRF2, which can then translocate to the nucleus and bind to the antioxidant response element (ARE) sequence in the promoter regions of NRF2-dependent genes, and recruit transcriptional machinery (Itoh et al. 1997).
Because NRF2 is able to regulate the expression of more than 250 genes, its capacity for action is very broad. At the level of the central nervous system (CNS), it has been described that the activation of NRF2 has beneficial effects against the main hallmarks of neurodegeneration. Pharmacological activation of NRF2 induces proteasome and autophagy enhancing the degradation of protein aggregates (Lastres-Becker et al. 2016; Pajares et al. 2016; Rojo et al. 2017), has anti-inflammatory effects (Castro-Sánchez et al. 2019; Cuadrado et al. 2018a; Rojo et al. 2018), and reduces oxidative stress (Lastres-Becker 2017; Cuadrado et al. 2018b). On the other hand, deficiency in NRF2 worsens all these parameters exacerbating the neurodegenerative process. Therefore, modulation of NRF2 activity has the potential to alter neurodegenerative disease course (Burnside and Hardingham 2017; Cuadrado et al. 2018a; Lastres-Becker 2017; Lastres-Becker et al. 2016).
Like the NRF2 pathway, the endocannabinoid system has emerged as an important neuromodulation system for many brain functions (Zanettini et al. 2011). The endocannabinoid system consists of cannabinoid receptors type 1 and 2 (CB1 and CB2), their endogenous ligands, and the enzymes for synthesis and metabolism of the endocannabinoids (Di Marzo 2018). The CB2 receptor, a seven-transmembrane and G protein-coupled receptor, is generally expressed in immune tissues and cells but is also present at low levels in neuronal and non-neuronal (quiescent microglia, for example) brain cells (Navarro et al. 2016). As NRF2, CB2 receptor activation has exhibited great potential as anti-oxidative stress and anti-inflammation in various disease models (Di Marzo 2018) and its potential as a therapeutic target is being investigated. An important aspect in this research is the fact that, despite the modest expression of CB2 receptors in glial elements in normal conditions of the CNS, they experience a notable up-regulation in response to different neurotoxic (e.g., proinflammatory, oxidative, traumatic, infectious) insults. This up-regulation involves the induction of CB2 receptor gene expression, although little is known about the elements (transcription factors) involved in this response. In this respect, it has been described that the promoter region of human CNR2 (CB2 receptor gene) contains several boxes with a transcription binding site for stress response such as AP-1 or AP-4 (activator protein 1 or 4), HSF (heat shock factor), and STRE (stress response element), depending on the isoform (Onaivi et al. 2012). As both the activation of NRF2 and CB2 have similar anti-oxidative and anti-inflammatory actions, this evidence prompted us to investigate whether NRF2 could be one of the key transcription factors implicated in the expression of CB2.
Materials and Methods
Bioinformatics Analysis
A putative antioxidant response element (ARE) in CNR2 gene promoter was identified in The Encyclopedia of DNA Elements at UCSC (ENCODE) (http://genome.ucsc.edu) for the human genome (Feb. 2009), taking as reference the available information from chromatin immunoprecipitation (ChIP) of ARE-binding factors MAFK and BACH1. The putative MAFK was localized in a 280-base pair long DNase-sensitive and H3K27Ac-rich region, i.e., most likely regulatory promoter regions. In addition, a frequency matrix of the consensus ARE sequence based on the JASPAR database (http://jaspar.genereg.net) was converted to a position-specific scoring matrix (PSSM) by turning the frequencies into scores through the log(2) [odd ratio (odd ratio: observed frequency/expected frequency)]. One unit was added to each frequency to avoid log(0). Then a script was generated with the Python 3.4 program to scan the promoter sequences with candidate AREs retrieved from ENCODE with the PSSM. The max score was calculated by adding the independent scores for each of the 11 base pairs of the consensus ARE sequence with the PSSM. The relative score (score relative) was calculated from this max score (score of the sequence max) as: score relative = (score of the sequencemax − scoremin possible)/(scoremax possible − scoremin possible). The min possible score (scoremin possible) is calculated as the lowest possible number obtained for a sequence from the PSSM and the max possible score (scoremax possible) is the highest possible score that can be obtained. We considered putative ARE sequences those with a score relative over 80%, which is a commonly used threshold for the computational framework for transcription factor binding site (TFBS) analyses using PSSM.
Cell Culture
Primary microglia were prepared from neonatal (P0–P2) mouse cortex from Nfe2l2+/+ and Nfe2l2−/− mice [obtained from colonies of Nfe2l2−/− mice and Nfe2l2+/+ littermates established from founders kindly provided by Dr. Masayuki Yamamoto (Tohoku University Graduate School of Medicine, Sendai, Japan)] (Itoh et al. 1997) and grown and isolated as described in Lastres-Becker et al. (2014). Briefly, neonatal (P0–P2) mouse cortex was mechanically dissociated and the cells were seeded onto 75 cm2 flasks in Dulbecco’s Modified Eagle Medium:F12 (DMEM:F12) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin. After 2 weeks in culture, flasks were trypsinized and separated using CD11b MicroBeads for magnetic cell sorting (MACS Miltenyi Biotec, Germany). Microglial cultures were at least 99% pure, as judged by immunocytochemical criteria. Medium was changed to DMEM:F12 serum-free without antibiotics 16 h before treatment. Immortalized microglial cell line (IMG), isolated from the brains of adult mice, was purchased from Kerafast Inc. HT22 mouse hippocampal neuronal cell line was obtained from Dr. Ana Pérez laboratory (Biomedical Research Institute, Madrid, Spain). IMG and HT22 cells were grown in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mM l-glutamine, in 5% CO2 at 37 °C, 50% relative humidity. Primary neuronal cultures were prepared from the cerebral cortex of 18-day-old Wistar rat embryos (E18), both genders being indistinctly used. Dissected cerebral cortices were mechanically dissociated in culture medium (Minimum Essential Medium, Life Technologies) supplemented with 22.2 mM glucose, 0.1 mM glutamax, 5% fetal bovine serum, 5% donor horse serum, 100 U/ml penicillin, and 100 µg/ml streptomycin similarly as described before (Choi et al. 1987). The cell suspension was seeded at a density of 1 × 106 cells/ml in the same medium using plates previously treated with poly-l-lysine (100 µg/ml, Sigma-Aldrich) and laminin (4 µg/ml, Sigma-Aldrich) overnight at 37 °C. After 4 h, culture medium was changed to Neurobasal (Life Technologies) containing B27 serum-free supplement (Life Technologies), 2 mM glutamax (Life Technologies), 100 U/ml penicillin, and 100 µg/ml streptomycin. Growth continued until DIV 7, when medium was changed to fresh Neurobasal medium completed as before but omitting glutamax. Experimental treatments took place after 12 DIVs by adding reactives directly to the growth medium. Medium was changed to serum-free DMEM without antibiotics 16 h before treatments for IMG and HT22 cells. Dimethyl fumarate (DMF) was obtained from Sigma-Aldrich (Cat number 242926) and used at 20 μM.
Analysis of mRNA Levels by Quantitative Real-Time PCR
Total RNA extraction, reverse transcription, and quantitative polymerase chain reaction (qPCR) were done as detailed in previous articles (Lastres-Becker et al. 2014). Primer sequences are shown in Supplementary Table S1. Data analysis was based on the ΔΔCT method with normalization of the raw data to housekeeping genes (Applied Biosystems). All PCRs were performed in triplicates.
Immunoblotting
Whole cell lysates were prepared in RIPA-Buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1 mM EGTA, 1% Igepal, 1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). Whole cell lysates, and cytosolic and nuclear fractions containing 25 μg of whole proteins from IMG-treated cells were loaded for SDS-PAGE electrophoresis. Immunoblots were performed as described in (Cuadrado et al. 2014). The primary antibodies used are described in Supplementary Table S2.
NRF2 Overexpression
HT22 cells were seeded on 6-well plates (300,000 cell/well) and transfected with an NFE2L2 expression plasmid (8 μg) that lacks the high affinity binding site for KEAP1 and contains a V5 tag (NRF2ΔETGE-V5) (McMahon et al. 2003) kindly provided by Dr. John D. Hayes (Biomedical Research Institute, Ninewells Hospital and Medical School, University of Dundee) or pcDNA3.1/V5HisB. After 24 h from transfection with Lipofectamine 2000 Reagent (Invitrogen Life Technologies, Cat number 116668-019), cells were lysed for analysis of mRNA levels by quantitative real-time PCR.
Production of Lentiviral Stocks and Infection of HT22 Cells
Recombinant lentiviral stocks were produced in HEK293T cells by cotransfecting shCtrl or shNRF2 (shCLN_NM_010902, MISSION shRNA, Sigma) (Robledinos-Anton et al. 2017), 6 μg of envelope plasmid pMD2.G (Addgene; deposited by Dr. Didier Trono) and 6 μg of packaging plasmid pSPAX2 (Addgene; deposited by Dr. Didier Trono), using Lipofectamine 2000 Reagent (Invitrogen Life Technologies). After 12 h at 37 °C, the medium was replaced with fresh DMEM containing 10% fetal bovine serum. Virus particles were harvested at 24 h and 48 h post transfection. HT22 cells were incubated in the presence of 2 µg/ml polybrene (Sigma-Aldrich, TR-1003-G) with the lentivirus during 24 h, and then selected with Puromycin (5 µg/mL) for 48 h. mRNA was extracted 7 days after lentiviral transduction.
RNA-Seq of Cell Types Isolated from Mouse
Differential distribution of the mRNA for Cnr2 and Nfe2l2 in the mouse brain were obtained from the Brain-RNA-seq database. For details see Ref. (Zhang et al. 2014).
Statistical Analyses
Data are presented as mean ± SEM. To determine the statistical test to be used, we employed GraphPad Instat 3, which includes the analysis of the data to normal distribution via the Kolmogorov–Smirnov test. In addition, statistical assessments of differences between groups were analyzed (GraphPad Prism 5, San Diego, CA) by unpaired Student’s t tests when normal distribution and equal variances were fulfilled, or by the non-parametric Mann–Whitney test. One-way and two-way ANOVA with post hoc Tukey test were used, as appropriate.
Results
Identification of Putative AREs in the Cannabinoid Receptor CNR2 Gene
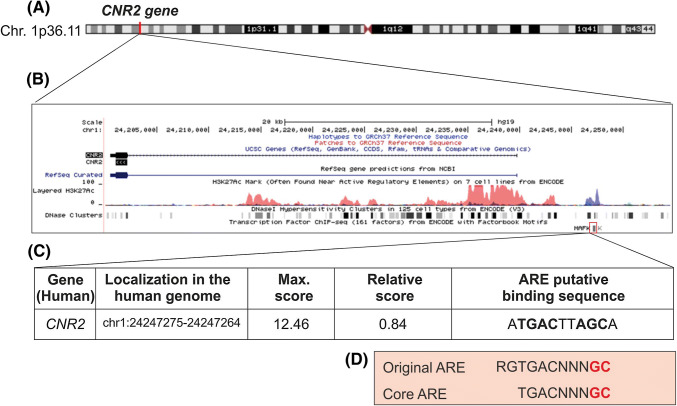
To define comprehensively the role of NFE2L2 in the transcriptional regulation of the CB2 receptor, we searched the Encyclopedia of DNA Elements at UCSC (ENCODE) (An integrated encyclopedia of DNA elements in the human genome 2012) of the human genome (Feb. 2009) for CNR2 with putative AREs (Fig. 1a). The ENCODE database gathers experimental data from chromatin immunoprecipitation (ChIP) analysis of ARE-binding transcription factors MAFF, MAFK, and BACH1, although NFE2L2 is not analyzed. As shown in Fig. 1b, the binding site was located at histone acetylated and DNase-sensitive regions in the CNR2 gene. Then, we used Python-based bioinformatics analysis to scan this binding region for the consensus ARE as established in the JASPAR database (Mathelier et al. 2014). We detected one putative ARE in the CNR2 gene with a relative score higher than 80%, a commonly used threshold for transcription factor binding site analysis (Fig. 1c) (Andersen et al. 2008; Kwon et al. 2012). This putative ARE sequence in the CNR2 promotor region has a high degree of similarity with the consensus ARE sequence (Fig. 1d) described by (Hirotsu et al. 2012).
Fig. 1.
Bioinformatic analysis of putative AREs in the cannabinoid receptor CNR2 gene promoter. a Scheme of the CNR2 gene location at Chr.1p36.11 from the Encyclopedia of DNA Elements at UCSC (ENCODE) for the human genome. b Putative AREs in the CNR2 gene were identified taking as reference the available information from ChIP of ARE-binding factors MAFK, MAFF, and BACH1. A MAFK binding site was found in a DNase-sensitive and H3K27Ac-rich region upstream the CNR2 gene (i.e., most likely promoter region). c Table showing the putative ARE sequence identified in the CNR2 promoter with a relative score over 80%. d Original ARE and Core ARE highlighting the main bases involved in NRF2 binding demonstrated by Hirotsu et al. (2012)
CB2 Expression in Neurons is not NRF2 Dependent
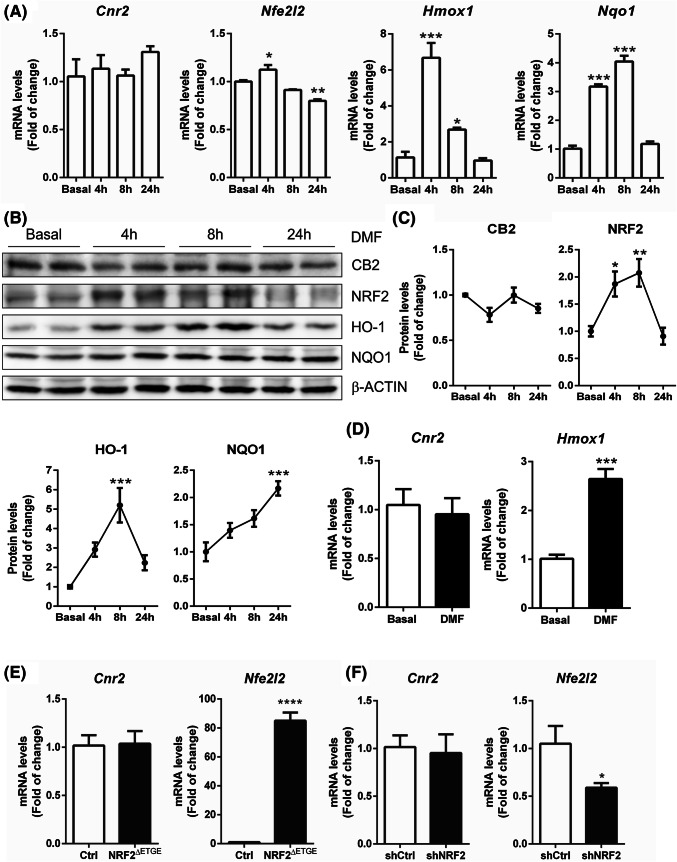
Recently, it has been described that the CB2 receptor could have distinct roles in neuronal and microglial cells (Li and Kim 2017). Therefore, to determine whether the transcription factor NRF2 could modulate CB2 receptor expression in neuronal cells, we followed different strategies by chemical and genetic manipulations of this transcription factor. First, we employed a pharmacological strategy by using dimethyl fumarate (DMF) (20 μM), a well-known NRF2 inducer (Cuadrado et al. 2018a; Lastres-Becker et al. 2016). Hippocampal neuronal cell line HT22 cells were maintained under serum-free conditions for 16 h and then treated with DMF and data were collected at different time points. NRF2 signaling was activated by DMF treatment (Fig. 2a–c) corroborated by the observation that mRNA and protein levels of NRF2, heme oxygenase 1 (HO-1), and NAD(P)H dehydrogenase quinone 1 (NQO1) were increased in a time-dependent fashion. However, neither mRNA or protein levels of CB2 receptor were modified by DMF treatment. Similar results were obtained in primary neuronal cultures, where DMF treatment for 4 h increased mRNA levels of Hmox1 but not Cnr2 (Fig. 2d). Next, we overexpressed NRF2 by using a stable mutant, NRF2ΔETGE-V5, that lacks four residues (ETGE) essential for recognition by the E3 ligase complex Cul3/KEAP1. More than 80-fold of change of Nfe2l2 mRNA expression did not induce any modification of Cnr2 expression (Fig. 2e). Finally, we silenced NRF2 with a lentiviral shRNA vector in the hippocampal HT22 cell line. Evidence of NRF2-knockdown was provided by analysis of mRNA expression levels, where we observed around 50% of decreased expression of Nfe2l2 (Fig. 2f). Knockdown of NRF2 did not lead to modifications of Cnr2 mRNA expression levels. Taken together, these results suggest that NRF2 is not capable of modulating the expression of the CB2 receptor at the neuronal level.
Fig. 2.
Modulation of NRF2 does not change CB2expression in neurons.a HT22 cells were incubated in the presence of dimethyl fumarate (DMF) at 20 µM for 4, 8, and 24 h. Quantitative real-time PCR determination of messenger RNA levels of Cnr2, Nfe2l2, and NRF2-regulated genes coding Hmox1 and Nqo1, normalized by Tbp (TATA-box binding protein) messenger RNA levels. The experiments were performed twice (each experiment with n = 4). b Immunoblot analysis in whole cell lysates of protein levels of CB2, NRF2, HO-1, NQO1, and β-ACTIN as a loading control. Representative blots are presented. c Densitometric quantification of protein levels normalized for β-ACTIN. (n = 4), mean ± SEM. Asterisks denote significant differences *p < 0.05, **p < 0.01, and ***p < 0.001, comparing the indicated groups with the basal condition according to a one-way ANOVA followed by Turkey post-test. d Primary culture of rat cortical neurons were incubated in the presence of DMF at 20 µM for 4 h. Quantitative real-time PCR determination of messenger RNA levels of Cnr2 and Hmox1, normalized by Tbp (TATA-box binding protein) messenger RNA levels. Bars indicate n = 4, mean ± SEM. e HT22 cells were transfected with 8 μg of pcDNA3.1/V5HisB-mNRF2ΔETGE plasmid or the pcDNA3.1/V5HisB, as a negative control. Cells were lysed 24 h after transfection. Quantitative real-time PCR determination of messenger RNA levels of Cnr2 and Nfe2l2 normalized by Tbp messenger RNA levels. f HT22 cells were transduced with lentiviral vectors carrying Nfe2l2 shRNA or a control scrambled shRNA. Cells were lysed 7 days after infection. The experiments were performed twice (each experiment with n = 4, mean ± SEM). Asterisks denote significant differences *p < 0.05 and ****p < 0.0001, comparing the indicated groups with the control condition according to Student’s t test
NRF2 Modulates CB2 Expression in Microglial Cells
CB2 receptors are largely found on microglial cells when activated by different neurotoxic stimuli, then playing important protective roles against these stimuli, in particular against neuroinflammation (Cassano et al. 2017). This fueled the idea of targeting the CB2 receptor as a promising treatment for glia-driven inflammation and neuronal degeneration (Turcotte et al. 2016). Because NRF2 has an important anti-inflammatory effect, we wanted to analyze if at the level of microglia NRF2 could modulate the expression of the CB2 receptor. As before, we followed different strategies based on chemical and genetic manipulations of this transcription factor to determine whether NRF2 could modulate CB2 receptor expression in microglial cells. First, we treated immortalized microglial cells (IMG) with DMF (20 μM). IMG cells were maintained under serum-free conditions for 16 h and then treated with DMF and data were collected at different time points. We corroborated that NRF2 signaling was activated by DMF treatment (Fig. 3a–c) by analyzing the mRNA and protein levels of NRF2, HO-1, and NQO1 and we demonstrated that they were increased in a time-dependent way. Interestingly, treatment with DMF first modulates the protein levels of NRF2 by increasing the stability of the protein (4 h and 8 h) and subsequently induces its transcriptional expression (24 h). In relation to these results, we observed that, at 8 h of treatment, increased protein expression of CB2 is delayed to that of NRF2 (Fig. 3b, c). At the mRNA level, we observed a biphasic effect of DMF: first, it induces a decrease in the expression of Cnr2 at 4 h and a subsequent induction at 24 h (Fig. 3a).
Fig. 3.
NRF2 modulates CB2expression in microglia.a IMG cells were incubated in the presence of DMF at 20 µM for 4, 8, and 24 h. Quantitative real-time PCR determination of messenger RNA levels of Cnr2, Nfe2l2 and NRF2-regulated genes coding Hmox1 and Nqo1, normalized by Tbp messenger RNA levels. The experiments were performed twice (each experiment with n = 4). b Immunoblot analysis in whole cell lysates of protein levels of CB2, NRF2, HO-1, NQO1, and β-ACTIN as a loading control. Representative blots are presented. c Densitometric quantification of protein levels normalized for β-ACTIN. The experiments were performed twice (each experiment with n = 2), mean ± SEM. Asterisks denote significant differences *p < 0.05 **p < 0.01, ***p < 0.001, and ****p < 0.0001, comparing the indicated groups with the basal condition according to a one-way ANOVA followed by Tukey post-test. d Primary cultures of microglia from control wild-type mice (Nfe2l2+/+) and NRF2-knockout mice (Nfe2l2−/−) were used. Bars indicate n = 4, mean ± SEM. Asterisks denote significant differences ****p < 0.0001, comparing the indicated groups with the control condition according to Student’s t-test. e Differential expression of Cnr2 and Nfe2l2 in the mouse brain according to the Brain-RNA-seq database (Zhang et al. 2014). Both transcripts are higher in microglia, whereas Nfe2l2 levels are lower and Cnr2 levels drop sharply to undetectable levels in both neurons and astrocytes. Expression level estimation was reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM) value together with confidence intervals for each sample
Then, we analyzed Cnr2 mRNA expression levels in primary microglial cells obtained from Nfe2l2+/+ and Nfe2l2−/− mice. Our results demonstrated that NRF2-deficient microglial cells barely express Cnr2 (approx. 90% of reduced expression) in comparison to wild-type microglial cells (Fig. 3d). In this case, we chose not to transfect microglia, since it has been described (and we ourselves have observed) that current transfection methodologies provide low transfection efficiency and induce cell death and/or inflammatory activation of the microglia. Anyway, the results we have obtained clearly indicate that the expression of the CB2 receptor is modulated by NRF2 in microglia. These data become even more relevant when we compare the expression levels of Nfe2l2 and Cnr2 in the different brain cell types by RNA-seq obtained from Zhang et al. (2014) (Fig. 3e). Figure 3e clearly shows that both genes are co-expressed only at the level of the microglia, but not in neurons or astrocytes.
Discussion
During the last decades, knowledge about the endocannabinoid system has increased exponentially, although the CB2 receptor has been less well characterized in comparison with the CB1 receptor. However, recently, CB2 receptors have gained attention, primarily due to their promising therapeutic potential for treating various pathologies while avoiding the adverse psychotropic effects that can accompany CB1 receptor-based therapies (Dhopeshwarkar and Mackie 2014b), although we cannot ignore the possibility of provoking immunosuppression. Therefore, it is very important to know the mechanisms involved in CB2 transcriptional regulation. Thus, we have analyzed the implication of the transcription factor NRF2 in the expression of CB2 at the level of neuronal and microglial cells. We showed for the first time that in the promoter region of CB2, there is an ARE sequence and that NRF2 is able to specifically modulate the expression of CB2 in microglia (Figs. 1 and 3).
It is interesting to note that the regulation of CB2 by NRF2 only occurs at the microglia level, which is the cell type where both genes are most highly expressed in the brain (Fig. 3e). It is also the type of neural cells in which CB2 receptor expression is most significantly elevated after their activation. In addition, this result is even more relevant when we analyze the involvement of both NRF2 and CB2 in neuroinflammation processes associated with microglia activation. NRF2 has been demonstrated to counteract inflammation in several neurodegenerative disorders like Alzheimer’s disease (Cuadrado et al. 2018a; Lastres-Becker et al. 2014; Rojo et al. 2018, 2017; Castro-Sánchez et al. 2019) and Parkinson’s disease (Lastres-Becker et al. 2016, 2012; Jazwa et al. 2011; Rojo et al. 2010; Lastres-Becker 2017). NRF2 modulates redox homeostasis and phagocytosis in microglia and deficiency in this protein results in exacerbated inflammatory response (Vilhardt et al. 2017). This indicates that NRF2 is implicated in modulating microglial dynamics (Innamorato et al. 2009; Rojo et al. 2010) through, at least, its interaction with the transcription factor NF-κB (Cuadrado et al. 2014), master regulator of inflammation. Lack of NRF2 can magnify NF-κB activity primarily increasing cytokine production, whereas NF-κB can modulate NRF2 transcription and activity (NFE2L2 promoter has a κB site), having both positive and negative effects on the gene expression (Wardyn et al. 2015). Related to CB2, it has been observed an up-regulation in reactive microglia in the spinal cord of TDP-43 (A315T) transgenic mice, an experimental model of amyotrophic lateral sclerosis (Espejo-Porras et al. 2019). CB2 up-regulation has been also observed in the context of amyloid-triggered neuroinflammation (Lopez et al. 2018), and glia-driven inflammation in CNS structures affected, for example, in Parkinson’s disease (Gomez-Galvez et al. 2016) and Huntington’s disease (Sagredo et al. 2009; Palazuelos et al. 2009). Such responses have been also found in post-mortem tissues from patients affected by these diseases (Jordan and Xi 2019). However, it is still unknown what is the molecular mechanism by which CB2 activation has anti-inflammatory effects (Wu et al. 2017; Cakir et al. 2019). One possibility is that it acts as an inhibitor of the control of proinflammatory cytokines and, as NRF2, promotes the shift from M1 to M2 phenotypes. It has been described that peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) may mediate CB2 receptor agonist AM1241-induced anti-inflammation in microglial cells, and the mechanism might be associated with the augmentation of mitochondria biogenesis (Ma et al. 2018). Therefore, the fact that NRF2 is able to induce the expression of CB2 in microglia could shed light on the signaling pathways involved in the anti-inflammatory processes. In addition, the induction of CB2 expression at the microglial level would be a powerful therapeutic target to treat microgliosis associated with pathologies (Navarro et al. 2016; Soethoudt et al. 2017).
Although in principle it has always been described that the CB2 receptor is preferentially expressed in cells of the immune system (macrophages and microglia), neuronal expression of CB2 cannabinoid receptor mRNAs has been recently observed in the mouse hippocampus (Li and Kim 2015). It is interesting to note that the function of CB2 depends on the cell type where it is expressed. For example, CB2 expression in different types of cells in the mature hippocampus plays diverse roles in the regulation of memory and anxiety (Li and Kim 2017). It is also expressed in other neuronal subpopulations although with a more restricted distribution compared to CB1 receptors (Dhopeshwarkar and Mackie 2014a; Hu and Mackie 2015). Our results show a very interesting pattern of modulation of CB2 expression: at the neuronal level, NRF2 is not able to alter the expression of CB2 (Fig. 2) suggesting that there must be other transcription factors involved.
Overall, the present work suggests the involvement of NRF2 activity in CB2 modulation in microglia, and points to NRF2/CB2 as promising pharmacological targets for therapeutic strategies to modulate neuroinflammation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author Contributions
ILB and MDG contributed to conception and design of the study. MGG, RdR, and ILB acquisition and analysis of data. NJM contributed with the bioinformatics analysis. ILB contributed to drafting the manuscript and figures.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (Grants Refs. SAF2016-76520-R to ILB and BFU2016-75973-R to MDG).
Compliance with Ethical Standards
Ethical Approval
All experiments were performed by certified researchers according to regional, national, and European regulations concerning animal welfare and animal experimentation, and were authorized by the Ethics Committee for Research of the Universidad Autónoma de Madrid and the Comunidad Autónoma de Madrid, Spain, with Ref PROEX 279/14 and Ref PROEX 221/14, following institutional, Spanish and European guidelines (Boletín Oficial del Estado (BOE) of 18 March 1988 and 86/609/EEC, 2003/65/EC European Council Directives). The housing facilities at the Institute were approved by Comunidad de Madrid (# ES 280790000188) and comply with official regulations.
Conflict of interest
None of the authors has a conflict of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P, Lenhard B, Wasserman WW, Odeberg J (2008) In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput Biol 4(1):e5. 10.1371/journal.pcbi.0040005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside SW, Hardingham GE (2017) Transcriptional regulators of redox balance and other homeostatic processes with the potential to alter neurodegenerative disease trajectory. Biochem Soc Trans 45(6):1295–1303. 10.1042/bst20170013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir M, Tekin S, Doganyigit Z, Erden Y, Soyturk M, Cigremis Y, Sandal S (2019) Cannabinoid type 2 receptor agonist JWH-133, attenuates Okadaic acid induced spatial memory impairment and neurodegeneration in rats. Life Sci 217:25–33. 10.1016/j.lfs.2018.11.058 [DOI] [PubMed] [Google Scholar]
- Cassano T, Calcagnini S, Pace L, De Marco F, Romano A, Gaetani S (2017) Cannabinoid receptor 2 signaling in neurodegenerative disorders: from pathogenesis to a promising therapeutic target. Front Neurosci 11:30. 10.3389/fnins.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Sánchez S, García-Yagüe ÁJ, Kügler S, Lastres-Becker I (2019) CX3CR1-deficient microglia shows impaired signaling of the transcription factor NRF2: implications in tauopathies. Redox Biol. 10.1016/j.redox.2019.101118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Maulucci-Gedde M, Kriegstein AR (1987) Glutamate neurotoxicity in cortical cell culture. J Neurosci 7(2):357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Martin-Moldes Z, Ye J, Lastres-Becker I (2014) Transcription factors NRF2 and NF-kappaB are coordinated effectors of the Rho family, GTP-binding protein RAC1 during inflammation. J Biol Chem 289(22):15244–15258. 10.1074/jbc.M113.540633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Kugler S, Lastres-Becker I (2018a) Pharmacological targeting of GSK-3 and NRF2 provides neuroprotection in a preclinical model of tauopathy. Redox Biol 14:522–534. 10.1016/j.redox.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Manda G, Hassan A, Alcaraz MJ, Barbas C, Daiber A, Ghezzi P, Leon R, Lopez MG, Oliva B, Pajares M, Rojo AI, Robledinos-Anton N, Valverde AM, Guney E, Schmidt H (2018b) Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol Rev 70(2):348–383. 10.1124/pr.117.014753 [DOI] [PubMed] [Google Scholar]
- Dhopeshwarkar A, Mackie K (2014a) CB2 cannabinoid receptors as a therapeutic target-what does the future hold? Mol Pharmacol 86(4):430–437. 10.1124/mol.114.094649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhopeshwarkar A, Mackie K (2014b) CB2 cannabinoid receptors as a therapeutic target-what does the future hold? Mol Pharmacol 86(4):430–437. 10.1124/mol.114.094649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V (2018) New approaches and challenges to targeting the endocannabinoid system. Nat Rev Drug Discov 17(9):623–639. 10.1038/nrd.2018.115 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489(741):57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo-Porras F, Garcia-Toscano L, Rodriguez-Cueto C, Santos-Garcia I, de Lago E, Fernandez-Ruiz J (2019) Targeting glial cannabinoid CB2 receptors to delay the progression of the pathological phenotype in TDP-43 (A315T) transgenic mice, a model of amyotrophic lateral sclerosis. Br J Pharmacol 176(10):1585–1600. 10.1111/bph.14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Galvez Y, Palomo-Garo C, Fernandez-Ruiz J, Garcia C (2016) Potential of the cannabinoid CB(2) receptor as a pharmacological target against inflammation in Parkinson’s disease. Prog Neuropsychopharmacol Biol Psychiatry 64:200–208. 10.1016/j.pnpbp.2015.03.017 [DOI] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39(4):199–218. 10.1016/j.tibs.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M (2012) Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40(20):10228–10239. 10.1093/nar/gks827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SS, Mackie K (2015) Distribution of the endocannabinoid system in the central nervous system. Handb Exp Pharmacol 231:59–93. 10.1007/978-3-319-20825-1_3 [DOI] [PubMed] [Google Scholar]
- Innamorato NG, Lastres-Becker I, Cuadrado A (2009) Role of microglial redox balance in modulation of neuroinflammation. Curr Opin Neurol 22(3):308–314. 10.1097/WCO.0b013e32832a3225 [DOI] [PubMed] [Google Scholar]
- Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M (1995) Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15(8):4184–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236(2):313–322 [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13(1):76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwa A, Rojo AI, Innamorato NG, Hesse M, Fernandez-Ruiz J, Cuadrado A (2011) Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid Redox Signal 14(12):2347–2360. 10.1089/ars.2010.3731 [DOI] [PubMed] [Google Scholar]
- Jordan CJ, Xi ZX (2019) Progress in brain cannabinoid CB2 receptor research: from genes to behavior. Neurosci Biobehav Rev 98:208–220. 10.1016/j.neubiorev.2018.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AT, Arenillas DJ, WorsleyHunt R, Wasserman WW (2012) oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda, Md) 2(9):987–1002. 10.1534/g3.112.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastres-Becker I (2017) Role of the transcription factor Nrf2 in Parkinson’s disease: new Insights. J Alzheimers Dis Parkinsonism 7(4):9. 10.4172/2161-0460.1000340 [Google Scholar]
- Lastres-Becker I, Ulusoy A, Innamorato NG, Sahin G, Rabano A, Kirik D, Cuadrado A (2012) alpha-Synuclein expression and Nrf2 deficiency cooperate to aggravate protein aggregation, neuronal death and inflammation in early-stage Parkinson’s disease. Hum Mol Genet 21(14):3173–3192. 10.1093/hmg/dds143 [DOI] [PubMed] [Google Scholar]
- Lastres-Becker I, Innamorato NG, Jaworski T, Rabano A, Kugler S, Van Leuven F, Cuadrado A (2014) Fractalkine activates NRF2/NFE2L2 and heme oxygenase 1 to restrain tauopathy-induced microgliosis. Brain 137(Pt 1):78–91. 10.1093/brain/awt323 [DOI] [PubMed] [Google Scholar]
- Lastres-Becker I, Garcia-Yague AJ, Scannevin RH, Casarejos MJ, Kugler S, Rabano A, Cuadrado A (2016) Repurposing the NRF2 activator dimethyl fumarate as therapy against synucleinopathy in Parkinson’s disease. Antioxid Redox Signal 25(2):61–77. 10.1089/ars.2015.6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kim J (2015) Neuronal expression of CB2 cannabinoid receptor mRNAs in the mouse hippocampus. Neuroscience 311:253–267. 10.1016/j.neuroscience.2015.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kim J (2017) Distinct roles of neuronal and microglial CB2 cannabinoid receptors in the mouse hippocampus. Neuroscience 363:11–25. 10.1016/j.neuroscience.2017.08.053 [DOI] [PubMed] [Google Scholar]
- Lopez A, Aparicio N, Pazos MR, Grande MT, Barreda-Manso MA, Benito-Cuesta I, Vazquez C, Amores M, Ruiz-Perez G, Garcia-Garcia E, Beatka M, Tolon RM, Dittel BN, Hillard CJ, Romero J (2018) Cannabinoid CB2 receptors in the mouse brain: relevance for Alzheimer’s disease. J Neuroinflamm 15(1):158. 10.1186/s12974-018-1174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Niu W, Lv J, Jia J, Zhu M, Yang S (2018) PGC-1alpha-mediated mitochondrial biogenesis is involved in cannabinoid receptor 2 agonist AM1241-induced microglial phenotype amelioration. Cell Mol Neurobiol 38(8):1529–1537. 10.1007/s10571-018-0628-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42(Database Issue):D142–D147. 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 278(24):21592–21600. 10.1074/jbc.M300931200 [DOI] [PubMed] [Google Scholar]
- Navarro G, Morales P, Rodriguez-Cueto C, Fernandez-Ruiz J, Jagerovic N, Franco R (2016) Targeting cannabinoid CB2 receptors in the central nervous system. Medicinal chemistry approaches with focus on neurodegenerative disorders. Front Neurosci 10:406. 10.3389/fnins.2016.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gu S, Liu QR (2012) CNS effects of CB2 cannabinoid receptors: beyond neuro-immuno-cannabinoid activity. J Psychopharmacol 26(1):92–103. 10.1177/0269881111400652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, Escoll M, de Ceballos ML, Van Leuven F, Rabano A, Yamamoto M, Rojo AI, Cuadrado A (2016) Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12(10):1902–1916. 10.1080/15548627.2016.1208889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazuelos J, Aguado T, Pazos MR, Julien B, Carrasco C, Resel E, Sagredo O, Benito C, Romero J, Azcoitia I, Fernandez-Ruiz J, Guzman M, Galve-Roperh I (2009) Microglial CB2 cannabinoid receptors are neuroprotective in Huntington’s disease excitotoxicity. Brain 132(Pt 11):3152–3164. 10.1093/brain/awp239 [DOI] [PubMed] [Google Scholar]
- Robledinos-Anton N, Rojo AI, Ferreiro E, Nunez A, Krause KH, Jaquet V, Cuadrado A (2017) Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol 13:393–401. 10.1016/j.redox.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo AI, Innamorato NG, Martin-Moreno AM, De Ceballos ML, Yamamoto M, Cuadrado A (2010) Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson’s disease. Glia 58(5):588–598. 10.1002/glia.20947 [DOI] [PubMed] [Google Scholar]
- Rojo AI, Pajares M, Rada P, Nunez A, Nevado-Holgado AJ, Killik R, Van Leuven F, Ribe E, Lovestone S, Yamamoto M, Cuadrado A (2017) NRF2 deficiency replicates transcriptomic changes in Alzheimer’s patients and worsens APP and TAU pathology. Redox Biol 13:444–451. 10.1016/j.redox.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo AI, Pajares M, Garcia-Yague AJ, Buendia I, Van Leuven F, Yamamoto M, Lopez MG, Cuadrado A (2018) Deficiency in the transcription factor NRF2 worsens inflammatory parameters in a mouse model with combined tauopathy and amyloidopathy. Redox Biol 18:173–180. 10.1016/j.redox.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagredo O, Gonzalez S, Aroyo I, Pazos MR, Benito C, Lastres-Becker I, Romero JP, Tolon RM, Mechoulam R, Brouillet E, Romero J, Fernandez-Ruiz J (2009) Cannabinoid CB2 receptor agonists protect the striatum against malonate toxicity: relevance for Huntington’s disease. Glia 57(11):1154–1167. 10.1002/glia.20838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin CJ, Dodson MB, Madhavan L, Zhang DD (2019) Redox regulation by NRF2 in aging and disease. Free Radic Biol Med 20:19. 10.1016/j.freeradbiomed.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soethoudt M, Grether U, Fingerle J, Grim TW, Fezza F, de Petrocellis L, Ullmer C, Rothenhausler B, Perret C, van Gils N, Finlay D, MacDonald C, Chicca A, Gens MD, Stuart J, de Vries H, Mastrangelo N, Xia L, Alachouzos G, Baggelaar MP, Martella A, Mock ED, Deng H, Heitman LH, Connor M, Di Marzo V, Gertsch J, Lichtman AH, Maccarrone M, Pacher P, Glass M, van der Stelt M (2017) Cannabinoid CB2 receptor ligand profiling reveals biased signalling and off-target activity. Nat Commun 8:13958. 10.1038/ncomms13958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte C, Blanchet MR, Laviolette M, Flamand N (2016) The CB2 receptor and its role as a regulator of inflammation. Cell Mol Life Sci CMLS 73(23):4449–4470. 10.1007/s00018-016-2300-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhardt F, Haslund-Vinding J, Jaquet V, McBean G (2017) Microglia antioxidant systems and redox signalling. Br J Pharmacol 174(12):1719–1732. 10.1111/bph.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardyn JD, Ponsford AH, Sanderson CM (2015) Dissecting molecular cross-talk between Nrf2 and NF-κB response pathways. Biochem Soc Trans 43(4):621–626. 10.1042/BST20150014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hocevar M, Foss JF, Bie B, Naguib M (2017) Activation of CB2 receptor system restores cognitive capacity and hippocampal Sox2 expression in a transgenic mouse model of Alzheimer’s disease. Eur J Pharmacol 811:12–20. 10.1016/j.ejphar.2017.05.044 [DOI] [PubMed] [Google Scholar]
- Zanettini C, Panlilio LV, Alicki M, Goldberg SR, Haller J, Yasar S (2011) Effects of endocannabinoid system modulation on cognitive and emotional behavior. Front Behav Neurosci 5:57. 10.3389/fnbeh.2011.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34(36):11929–11947. 10.1523/jneurosci.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.