Abstract
Introduction
Lung cancer in never-smoker (LCINS) patients accounts for 20% of lung cancer cases, and its biology remains poorly understood, particularly in genetically admixed populations. We elucidated the molecular profile of driver genes in Brazilian LCINS.
Methods
The mutational and gene fusion status of 119 lung adenocarcinomas from self-reported never-smoker patients, was assessed using targeted sequencing (NGS), nCounter, and immunohistochemistry. A panel of 46 ancestry-informative markers determined patients’ genetic ancestry.
Results
The most frequently mutated gene was EGFR (49.6%), followed by TP53 (39.5%), ALK (12.6%), ERBB2 (7.6%), KRAS (5.9%), PIK3CA (1.7%), and less than 1% alterations in RET, NTRK1, MET∆ex14, PDGFRA, and BRAF. Except for TP53 and PIK3CA, all other alterations were mutually exclusive. Genetic ancestry analysis revealed a predominance of European (71.1%), and a higher African ancestry was associated with TP53 mutations.
Conclusion
Brazilian LCINS exhibited a similar molecular profile to other populations, except the increased ALK and TP53 alterations. Importantly, 73% of these patients have actionable alterations that are suitable for targeted treatments.
Keywords: lung adenocarcinoma, molecular profile, driver mutations, never smoker, Latin America
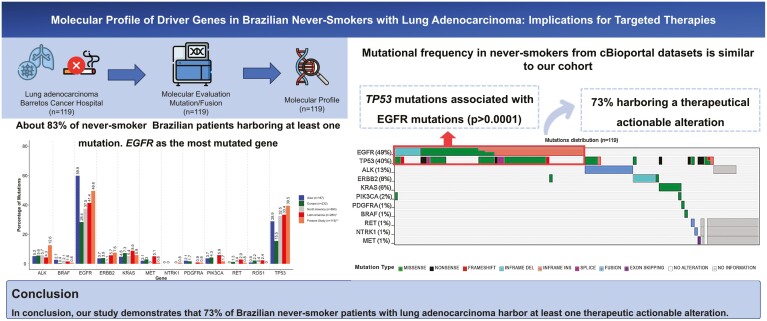
Lung cancer in never-smoker patients accounts for 20% of lung cancer cases, and its biology is poorly understood. This study focused on the molecular profile of driver genes in Brazilian patients who never smoked.
Graphical Abstract
Graphical Abstract.
Implications for Practice.
The identification of mutations in lung adenocarcinomas is crucial for deciding the best clinical management for the patients. Here, we observed 73% with at least one actionable alteration, with EGFR mutations reaching approximately 50% of patients. Therefore, these patients could be benefited by treatments with targeted drugs. A better understanding of the molecular profile in never-smoker patients from Brazil may improve the management of patients.
Introduction
Lung cancer in patients who have never smoked (LCINS) accounts for 20% of lung cancer cases and remains underexplored despite its increasing worldwide incidence.1-4 Lung cancer in never-smokers shows a better prognosis compared to ever-smokers.1-5
Lung cancer biology varies between never-smokers and smokers.1,3,6,7 Lung adenocarcinomas in never-smoker patients exhibit a higher frequency of EGFR, PIK3CA, and ERBB2 mutations.1,6EGFR mutations are notably more common, at variance with KRAS mutations, which are associated with tobacco exposure.2-4 Moreover, LCINS are more likely to harbor actionable variants, including not only EGFR mutations but also ALK translocation, impacting patients’ clinical management.1,3
Patient ethnicity also influences molecular profiles, with EGFR mutations more prevalent in Asians and KRAS mutations in Europeans.3,8,9 In admixed populations like Brazil, these profiles vary, and are poorly investigated.10 Therefore, we aimed to elucidate the molecular features of Brazilian LCINS.
Materials and methods
Study population
A series of 119 self-declared, never-smoker patients with lung adenocarcinoma (97 primary and 22 following treatment) from Barretos Cancer Hospital (BCH, Barretos, SP, Brazil) was evaluated. The local IRB approved the study (CAAE 05744712.3.0000.5437).
Mutation detection
Tumor mutational analysis was performed in FFPE sections using the commercial panel TruSight Tumor 15 (Illumina, San Diego, CA, USA) on a MiSeq instrument. The read alignment and variant calling were performed with Sophia DDM software version 4.2 (Sophia Genetics SA, Lausanne, Switzerland). Variants were filtered out as previously described, and pathogenic variants were retained.11,12 Actionable mutations (Tier I and II) were determined as reported.1
Fusion detection
ALK fusions were evaluated in 95.0% (n = 113/119) of cases by immunohistochemistry using the Ventana ALK (D5F3) CDx Assay (Roche, Basel, Switzerland) in an automated equipment.11
Detection of mRNA ALK/RET/ROS1/NTRK1,2,3 fusion transcripts and MET∆ex14 (MET exon 14 skipping) was assessed in patients with EGFR and KRAS wild-type tumors (n=61) by the nCounter Elements XT (NanoString Technologies, Seattle, WA, USA) custom panel for ALK, RET, ROS1, and NTRK1/2/3 fusion detection by specific partner and 3ʹ and 5ʹ disbalance, and MET∆ex14, using probes designed in-house.13 Twenty-four cases were inconclusive due to unavailable biological material. All analyses were performed in R environment v3.4.1.
Genetic ancestry
The genetic ancestry background was evaluated using a set of 46 ancestry-informative markers, including INDELs for European (EUR), African (AFR), Asian (ASN), and Native American (AME).8,12
Statistical analysis
We described categorical variables using percentages and continuous variables using the medians. Fisher’s exact test and χ2 test were conducted for the association between the EGFR and TP53 mutations and genetic ancestry in IBM SPSS software Version 25 (IBM, Armonk, NY, USA) with a statistical significance limit of .05.
Results
Characterization of patients
The clinicopathological features of the 119 LCINS are summarized in Supplementary Table S1. Genetic ancestry was determined in 90.0% (n = 107/119) of the cases, following the proportion of 71.0% of EUR, 15.9% of AFR, 6.1% of ASN, and 7.9% of AME (Supplementary Figure S1).
Molecular profile
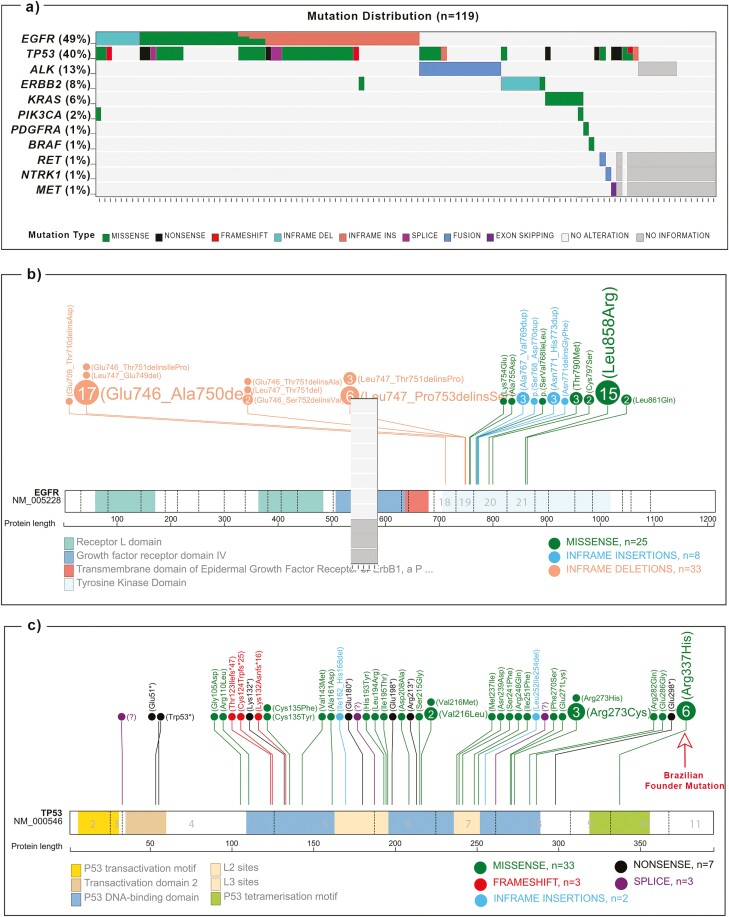
Among the 119 lung adenocarcinomas, 83.2% (n = 99/119) harbored at least one pathological mutation (Figure 1A). Among the 99 mutated cases, 54.5% carried one mutation, 39.4% 2, 4.1% 3, and 2.0% 4 (Supplementary Table S2). EGFR was the most mutated gene (49.6%, n = 59/119), followed by TP53 39.5% (n = 47/119), and ALK fusions in 12.6% (n = 15/119; Figure 1A). The genes ERBB2, KRAS, PIK3CA, RET, BRAF, PDGFRA, NTRK1, and MET showed fewer genetic alterations (Figure 1A). No alterations were observed on AKT1, FOXL2, GNA11, GNAQ, KIT, NRAS, ROS1, and NTRK2/3 hotspot regions. Apart from TP53 and PIK3CA, all the alterations were mutually exclusive (Figure 1A). Eighty-seven patients (73.1%) harbor actionable mutations (Supplementary Table S2)
Figure 1.
Representative figure of the mutations identified in the LCINS samples included in the study (n = 119). Only mutated genes are shown; (a) distribution of mutations through samples; lollipop plots depicting the distribution of (b) EGFR (n = 66); (c) TP53 (n = 48). Figures were created using tools in https://proteinpaint.stjude.org/.
Among EGFR mutations, exon 19 deletions were present in 49.2% of cases, followed by exon 21 (28.8%), and less frequently in exon 20 (7.6%) and 18 (1.7%; Figure 1B; Supplementary Table S2). Three tumors (following treatment) harbored the p.(Thr790Met) resistance mutation, and 2 presented additionally the p.(Cys797Ser) variant. Concerning TP53, the most common variant was the p.(Arg337His) (12.8%), followed by the p.(Arg273Cys) (6.4%) and the p.(Val216Leu) (4.3%; Figure 1C; Supplementary Table S2). One patient harbored 2 TP53 variants (Supplementary Table S2). TP53 mutations were associated with African ancestry (P = .002; Supplementary Table S3).
Exon 20 insertion p.(Tyr772_Ala775dup) accounted for 66.7% of ERBB2 mutations (Supplementary Table S2). The most common variant of the KRAS gene was p.(Gly12Asp) (42,8%, n = 3/7), and the variants observed in the PIK3CA, BRAF, and PDGFRA genes were at hotspot regions (Supplementary Table S2).
ALK fusions were observed in 15 patients (12.6%), both by immunohistochemistry and nCounter, the latter allowing to identify EML4 as the fusion partner in 73.3% of the cases. RET and NTRK1 fusions were identified by 3ʹ-5ʹ disbalance in one patient each, (Figure 1A; Supplementary Table S2).
Co-occurring mutations
EGFR and TP53 mutations significantly co-occurred in 27.7% (n = 33/119) of cases (P < .0001; Figure 1A; Supplementary Table S4). The Brazilian founder TP53 mutation p.(Arg337His) variant was mostly concurrent with EGFR mutations (Supplementary Table S2). EGFR variants also co-occurred with PIK3CA and ERBB2 mutations. TP53 mutations co-occurred with 1/3 of ALK fusions. The PIK3CA variants also co-occurred with the KRAS and TP53 (Figure 1A).
Discussion
The present study interrogated the molecular profile of driver genes of LCINS from a single Brazilian institution. Overall, we found that 73% of cases harbor actionable molecular alterations, in accordance with the literature.1,4,5
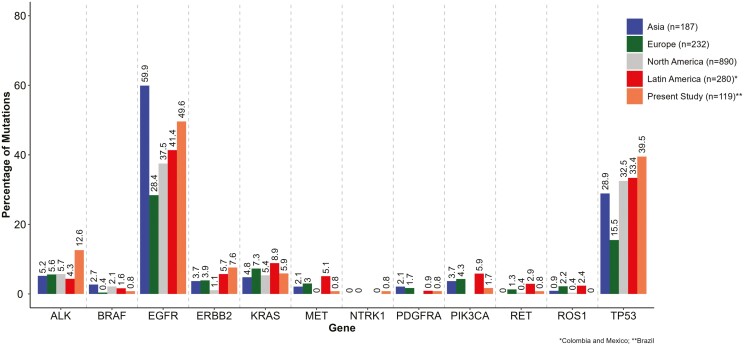
EGFR mutations occurred in half of our cases, in agreement with other populations, being higher than European and lower than Asian populations (Figure 2; Supplementary Table S5).1,2,4,6 Similar to our results, EGFR-TKi sensitizing exon 19 deletions are found in 50% of patients diagnosed with lung adenocarcinoma, while exon 20 insertions are less common globally.1,6,8TP53 was our second most mutated gene (39.5%). This frequency is higher compared to studies of LCINS in Asia, Europe, North America, and Latin America (Figure 2; Supplementary Table S5).6 We observed an association between TP53 mutations and higher African ancestry, similar to our recent study on ever- and never-smoker patients with lung adenocarcinoma.12 Notably, the most frequent TP53 mutation was the Brazilian germline variant p.(Arg337His), often concurrent with EGFR mutations. Previous lung cancer studies reported higher co-occurrence of EGFR and TP53 mutations, mainly with the Brazilian founder mutation.14-17
Figure 2.
Mutational frequency comparison between studies with never-smoker patients with lung adenocarcinoma according to the geographic region of the study (n = 1708).
We observed 7.6% of ERBB2 mutations in our LCINS, consistent with 2%-13% reported in other populations (Figure 2; Supplementary Table S5).1-4,6 In our series, only 5.9% of patients with lung adenocarcinoma had KRAS mutations, in accordance with other LCINS studies (4.4%-18%). Interestingly, ALK fusions in our LCINS series were significantly more frequent (12.6%) than reported globally (3%-8%).2,4,6 Finally, as previously reported, we identified less frequent alterations (1%-2%) in the genes PIK3CA, PDGFRA, BRAF, RET, NTRK1, and MET, similar to other populations1,6,18 (Figure 2; Supplementary Table S5).
Thus, we observe in the Brazilian LCINS population an overall similarity in the frequencies of driver genes reported worldwide. The exception was our higher frequency of ALK fusions and TP53 mutations, which could potentially be due to the significant presence of African ancestry, or founder TP53 p.(Arg337His) variant in the Brazilian population (Figure 2; Supplementary Table S5). Further studies are needed to validate and extend these findings.
Concluding, the molecular profile of Brazilian LCNIS resembles that of other populations worldwide, and 73% of patients could be eligible for personalized treatments.
Supplementary material
Supplementary material is available at The Oncologist online.
Acknowledgments
This study was partially supported by the Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer—15ª zone, Campinas, Brazil), Barretos Cancer Hospital Research Fund (PAIP), and National Council for Scientific and Technological Development (CNPq, Brazil). R.M.R. was supported by the National Council for Scientific and Technological Development (CNPq, Brazil) as Research Productivity Scholarship—Level 1B. L.F.L. was supported by the Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer—15ª zone, Campinas, Brazil) and National Council for Scientific and Technological Development (CNPq, Brazil) as Research Productivity Scholarship—Level 2, ROC was supported by Coordination for the Improvement of Higher Education Personnel (CAPES, Brazil) with a PhD scholarship, and B.G.Z. was supported by The São Paulo Research Foundation (FAPESP) with a undergraduate research project scholarship. Funding sources have no contribution to filling out authorship for the present study. We thank all members of the GTOP group (Translational Group of Pulmonary Oncology-Barretos Cancer Hospital, Brazil) for scientific discussion and suggestions.
Contributor Information
Rodrigo de Oliveira Cavagna, Molecular Oncology Research Center, Barretos Cancer Hospital, Barretos, Brazil.
Flávia Escremim de Paula, Molecular Diagnostic Laboratory, Barretos Cancer Hospital, Barretos, Brazil.
Gustavo Noriz Berardinelli, Molecular Diagnostic Laboratory, Barretos Cancer Hospital, Barretos, Brazil.
Murilo Bonatelli, Molecular Diagnostic Laboratory, Barretos Cancer Hospital, Barretos, Brazil.
Iara Santana, Department of Pathology, Barretos Cancer Hospital, Barretos, Brazil.
Eduardo Caetano Albino da Silva, Department of Pathology, Barretos Cancer Hospital, Barretos, Brazil.
Gustavo Ramos Teixeira, Department of Pathology, Barretos Cancer Hospital, Barretos, Brazil; Barretos School of Health Sciences, Dr. Paulo Prata – FACISB, Barretos, Brazil.
Beatriz Garbe Zaniolo, Molecular Oncology Research Center, Barretos Cancer Hospital, Barretos, Brazil; Barretos School of Health Sciences, Dr. Paulo Prata – FACISB, Barretos, Brazil.
Josiane Mourão Dias, Department of Medical Oncology, Barretos Cancer Hospital, Barretos, Brazil.
Flávio Augusto Ferreira da Silva, Department of Medical Oncology, Barretos Cancer Hospital, Barretos, Brazil.
Carlos Eduardo Baston Silva, Department of Medical Oncology, Barretos Cancer Hospital, Barretos, Brazil.
Marcela Gondim Borges Guimarães, Department of Medical Oncology, Barretos Cancer Hospital, Barretos, Brazil.
Camila Pinto Barone, Department of Medical Oncology, Barretos Cancer Hospital, Barretos, Brazil.
Alexandre Arthur Jacinto, Deparment of Radiation Therapy, Barretos Cancer Hospital, Barretos, Brazil.
Rachid Eduardo Noleto da Nóbrega Oliveira, Deparment of Thoracic Surgery, Barretos Cancer Hospital, Barretos, Brazil.
José Elias Miziara, Deparment of Thoracic Surgery, Barretos Cancer Hospital, Barretos, Brazil.
Pedro De Marchi, Molecular Oncology Research Center, Barretos Cancer Hospital, Barretos, Brazil; Oncoclinicas, Rio de Janeiro, Brazil.
Miguel A Molina-Vila, Laboratory of Oncology/Pangaea Oncology, Dexeus University Hospital, Barcelona, Spain.
Letícia Ferro Leal, Molecular Oncology Research Center, Barretos Cancer Hospital, Barretos, Brazil; Barretos School of Health Sciences, Dr. Paulo Prata – FACISB, Barretos, Brazil.
Rui Manuel Reis, Molecular Oncology Research Center, Barretos Cancer Hospital, Barretos, Brazil; Molecular Diagnostic Laboratory, Barretos Cancer Hospital, Barretos, Brazil; Life and Health Sciences Research Institute (ICVS), School of Medicine, University of Minho, Braga, Portugal; ICVS/3B’s—PT Government Associate Laboratory, Braga/Guimarães, Portugal.
Author contributions
Rodrigo de Oliveira Cavagna: conception/design, data analysis and interpretation, manuscript writing, final approval of manuscript. Flávia Escremim de Paula: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Gustavo Noriz Berardinelli: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Murilo Queiroz de Almeida Bonatelli: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Iara Santana: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Eduardo Caetano Albino da Silva: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Gustavo Ramos Teixeira: data analysis and interpretation, provision of study material or patients, final approval of manuscript. Beatriz Garbe Zaniolo: collection and/or assembly of data, final approval of manuscript. Josiane Mourão Dias: collection and/or assembly of data, final approval of manuscript. Flávio Augusto Ferreira da Silva: collection and/or assembly of data, provision of study material or patients, final approval of manuscript. Carlos Eduardo Baston Silva: collection and/or assembly of data, provision of study material or patients, final approval of manuscript. José Elias Miziara: collection and/or assembly of data, final approval of manuscript. Marcela Gondim Borges Guimarães: collection and/or assembly of data, provision of study material or patients, final approval of manuscript. Camila Pinto Barone: collection and/or assembly of data, provision of study material or patients, final approval of manuscript. Alexandre Arthur Jacinto: collection and/or assembly of data, final approval of manuscript. Rachid Eduardo Noleto da Nóbrega Oliveira: collection and/or assembly of data, final approval of manuscript. Pedro de Marchi: collection and/or assembly of data, final approval of manuscript. Miguel Angel Molina: data analysis and interpretation, final approval of manuscript. Letícia Ferro Leal: conception/design, data analysis and interpretation, manuscript writing, final approval of manuscript. Rui Manuel Reis: conception/design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Conflicts of interest
F.A.F.S.—research grants for clinical trials from AstraZeneca, Bristol-Myers Squibb, Janssen, Merck, Novartis, Roche, Sanofi. Honoraria for lectures from AstraZeneca, Takeda, and Sanofi. J.M.D.—research grants for clinical trials (to Institution): AbbVie, Amgen, AstraZeneca, Beigene, Bristol-Myers Squibb, Daiichi-Sankyo, Debiopharm, GlaxoSmithKline, Incyte Corporation, Ipsen, Janssen, Lilly, Merck, Merck Sharp and Dohme, Novartis, Pfizer, Regeneron, Roche, Sanofi, Takeda, Xcovery. Advisory board: AstraZeneca, Merck Sharp and Dohme. Honoraria for presentations: Amgen, AstraZeneca, Janssen, Roche, Sanofi, Takeda. Support for attending meetings and/or travel: Amgen, Boheringer Ingelheim, Janssen, Sanofi. A.A.J.—research grant from Varian Medical System, Inc. P.M.—clinical research: Bristol-Myers Squibb, Pfizer, Roche, Boheringer Ingelheim, AstraZeneca, Novartis, IRX, MSD, Xcovery, AMGEN, Roche Tissue Diagnostic. Technical and scientific presentations: Bristol-Myers Squibb, AstraZeneca, Roche, Bayer, Bayer, Novartis, Boheringer Ingelheim, MSD, Merk, Janssen, Lilly, Takeda, AMGEN, Sanofi. Financial support participation events: Bristol-Myers Squibb, Roche, Novartis, AstraZeneca, MSD, Merck, Takeda, AMGEN. Advisory board/consulting: AstraZeneca, Bristol-Myers Squibb, Roche, Janssen, Takeda, Sanofi, Lilly, Takeda, United Medical, ZODIAC. Steering committee: Novartis. M.A.M.V.—research grant: Astrazeneca, Merck Healthcare, In3Bio Ltd. Consulting fees: Atheneum. Honoraria for presentations: Spanish Lung Cancer Group (SLCG). Receipt of equipment: Astrazeneca. L.F.L.—research grant from AstraZeneca do Brasil.
Data availability
The data that support the findings of this study are available from Dr. Rui Manuel Reis, but restrictions apply to the availability of these data, because of patients’ personal data. De-identified data are, however, available from the authors upon request.
References
- 1. Mack PC, Klein MI, Ayers KL, et al. Targeted next-generation sequencing reveals exceptionally high rates of molecular driver mutations in never-smokers with lung adenocarcinoma. Oncologist. 2022;27(6):476-486. 10.1093/oncolo/oyac035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Devarakonda S, Li Y, Martins Rodrigues F, et al. Genomic profiling of lung adenocarcinoma in never-smokers. J Clin Oncol. 2021;39(33):3747-3758. 10.1200/JCO.21.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boeckx B, Shahi RB, Smeets D, et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int J Cancer. 2020;146(11):3207-3218. 10.1002/ijc.32797 [DOI] [PubMed] [Google Scholar]
- 4. LoPiccolo J, Gusev A, Christiani DC, Jänne PA.. Lung cancer in patients who have never smoked — an emerging disease. Nat Rev Clin Oncol. 2024;21(2):121-146. 10.1038/s41571-023-00844-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Couraud S, Souquet PJ, Paris C, et al. ; French Cooperative Intergroup IFCT. BioCAST/IFCT-1002: epidemiological and molecular features of lung cancer in never-smokers. Eur Respir J. 2015;45(5):1403-1414. 10.1183/09031936.00097214 [DOI] [PubMed] [Google Scholar]
- 6. Zhang T, Joubert P, Ansari-Pour N, et al. Genomic and evolutionary classification of lung cancer in never smokers. Nat Genet. 2021;53(9):1348-1359. 10.1038/s41588-021-00920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121-1134. 10.1016/j.cell.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leal LF, de Paula FE, De Marchi P, et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep. 2019;9(1):3209. 10.1038/s41598-019-39965-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lord BD, Martini RN, Davis MB.. Understanding how genetic ancestry may influence cancer development. Trends Cancer. 2022;8(4):276-279. 10.1016/j.trecan.2021.12.006 [DOI] [PubMed] [Google Scholar]
- 10. Carrot-Zhang J, Soca-Chafre G, Patterson N, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discov. 2021;11(3):591-598. 10.1158/2159-8290.CD-20-1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Oliveira Cavagna R, Zaniolo BG, de Paula FE, et al. ERBB2 exon 20 insertions are rare in Brazilian non‐small cell lung cancer. Thorac Cancer. 2022;13(23):3402-3407. 10.1111/1759-7714.14605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cavagna R de O, Pinto IA, Escremim de Paula F, et al. Disruptive and truncating TP53 mutations are associated with African-ancestry and worse prognosis in Brazilian patients with lung adenocarcinoma. Pathobiology. 2023;90(5):344-355. 10.1159/000530587 [DOI] [PubMed] [Google Scholar]
- 13. de Oliveira Cavagna R, de Andrade ES, Tadin Reis M, et al. Detection of NTRK fusions by RNA-based nCounter is a feasible diagnostic methodology in a real-world scenario for non-small cell lung cancer assessment. Sci Rep. 2023;13(1):21168. 10.1038/s41598-023-48613-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mascarenhas E, Gelatti AC, Araújo LH, et al. Comprehensive genomic profiling of Brazilian non‐small cell lung cancer patients (GBOT 0118/ LACOG0418). Thorac Cancer. 2021;12(5):580-587. 10.1111/1759-7714.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barbosa MVR, Cordeiro de Lima VC, Formiga MN, et al. High prevalence of EGFR mutations in lung adenocarcinomas from Brazilian patients harboring the TP53 p.R337H variant. Clin Lung Cancer. 2020;21(2):e37-e44. 10.1016/j.cllc.2019.11.012 [DOI] [PubMed] [Google Scholar]
- 16. Vieira IA, Andreis TF, Fernandes BV, et al. Prevalence of the Brazilian TP53 founder c.1010G>A (p.Arg337His) in lung adenocarcinoma: is genotyping warranted in all Brazilian patients? Front Genet. 2021;12:606537. 10.3389/fgene.2021.606537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Molina-Vila MA, Bertran-Alamillo J, Gascó A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2014;20(17):4647-4659. 10.1158/1078-0432.CCR-13-2391 [DOI] [PubMed] [Google Scholar]
- 18. Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33(1):38-46. 10.1038/s41379-019-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from Dr. Rui Manuel Reis, but restrictions apply to the availability of these data, because of patients’ personal data. De-identified data are, however, available from the authors upon request.