Abstract
Introduction
The progressive technologies in albumin in situ hybridization (ISH) changed the routine application and the differential diagnosis of hepatic malignancies in the last years. The aim of the present work was to assess the diagnostic utility of albumin ISH on different cholangiocarcinoma (CCA) subtypes, as well as to assess how albumin production changes along the biliary tree.
Methods
Forty-five CCAs were retrospectively selected: 29 intrahepatic (15 small-duct and 14 large-duct subtypes), 7 perihilar, and 9 extrahepatic. Histology was revised in all cases, and albumin ISH was automatically performed by the RNAscope®.
Results
ISH was always negative in extrahepatic CCAs, only 1 perihilar case was positive, and any positivity was observed in 25/29 (86.2%) intrahepatic CCAs (p < 0.001). Concerning CCA subtypes, mean cell positivity was 38.8 ± 29.8% in small-duct CCAs and 11.4 ± 21.9 in large-duct CCAs, respectively (p = 0.003); 12/15 (80.0%) small-duct and 3/14 (21.4%) large-duct CCAs showed >5% positive cells (p = 0.002; odds ratio 14.7).
Conclusions
The introduction of more sensitive techniques changed the indications for ISH since most small-duct intrahepatic CCAs show diffuse positivity. Albumin positivity decreases from liver periphery to the large ducts, suggesting that ISH can be helpful in the differential diagnosis between small-duct and large-duct CCAs, as well as between intrahepatic large-duct CCAs and metastases.
Keywords: Albumin, Bile ducts, Cholangiocarcinoma, In situ hybridization, mRNA
Introduction
Primary liver cancers are an increasing issue worldwide [1]. While hepatocellular carcinoma (HCC) has well-known pathogenesis and a definite range of diagnostic tools, on both radiological [2, 3] and pathological [4] point of view, intrahepatic cholangiocarcinoma (ICCA) is mostly a diagnosis of exclusion. The great intra- and inter-tumoral histological heterogeneity makes the diagnosis of ICCA even more difficult. The last WHO classification identified two major subtypes of ICCA, the small-duct ICCA (SD-ICCA) and the large-duct ICCA (LD-ICCA) [5]. These two subtypes are thought to have different cells of origin, phenotypic profile, and molecular background, with clinical implications for their management [6]. According to this subclassification, the morphology of LD-ICCA resembles perihilar CCAs (PCCAs, the so-called Klatskin tumors) and extrahepatic CCAs (ECCAs): these tumors are composed by large glands, surrounded by a dense stromal reaction, formed by columnar, often mucin-producing cells and with predominant periductal infiltrating or mixed periductal infiltrating-mass-forming patterns of growth. Precursor lesions of LD-ICCA can be frequently found in the surrounding ducts in the form of biliary intraepithelial neoplasia (BilIN) or intraductal papillary neoplasm of the bile ducts (IPNB) [7, 8]. On the other hand, SD-ICCA shows distinct, small glands, formed by cuboidal cells or cord-like formations with slit-like lumina, in a dense desmoplastic stroma. SD-ICCA often shows a mass-forming growth pattern and it is devoid of a recognized precursor lesion [5].
In the last years, a reliable marker has been searched for, to help pathologists in the correct diagnosis of ICCAs versus metastatic adenocarcinomas. The detection of the transcript of the albumin gene with in situ hybridization (ISH) has been used to assess the hepatic origin of malignancies for decades [9, 10], but until recently this methodology had been deemed laborious, poorly sensitive and with poor reproducibility among centers. The greatest limits of the cited method were the timing (3 days were needed from the pathologist’s request to the results), the lack of a standardization due to the manual set-up, and the low sensitivity given by the focal staining even in positive lesions [11]. This is why this ISH techniques had a poor spreading toward laboratries.
Lately, new ISH techniques with signal amplification systems have been proposed for albumin mRNA detection: the RNAscope® assay has been found promising in diagnosing liver primaries [10, 12, 13]. According to the original paper, HCCs should always be positive for albumin, with a percentage of positive neoplastic cells ranging between 25% and 90%, while ICCAs should present scattered positive cells in most cases (67% of the original cohort); PCCAs and ECCAs were described to be constantly negative for albumin ISH [11]. Recently, Avadhani et al. [12] found a difference in ISH staining among ICCAs, using the novel RNAscope® assay: in their cohort of ICCAs, none of the “LD/mucinous subtype” showed a positive staining, while 46% of the SD subtype was positive, with a percentage of positive neoplastic cells between 5 and 50% in 19% of SD-ICCAs and over 50% in another 19%.
It seems that a change in the diagnostic paradigm occurred since the positivity found in a subset of ICCAs is higher than thought before (and comparable to HCC). The distinction between albumin expression among LD-ICCAs, PCCAs, and ECCAs needs to be clarified as well. The aim of the present work was to assess the diagnostic utility of albumin ISH with RNAscope® on different CCA histotypes, but also to study how albumin production changes among the large-duct tumors along the biliary tree (LD-ICCA, PCCA, and ECCA).
Materials and Methods
Patient Enrollment and Histopathological Evaluation
We retrospectively selected 45 patients, 36 who underwent hepatic or biliary ducts resection for CCA, clinically suspected and confirmed after histological evaluation, and 9 patients who underwent liver biopsy for histologically proven CCA. Patients were 31 (68.9%) males and 14 (31.1%) females, mean age at surgery 66.8 ± 11.4 years (range 36–88 years). Five patients had a history of “biliary” risk factors (CSP in 3, chronic lithiasis in 1, and professional exposition in 1), 10 patients had a history of “hepatocellular” risk factors (dysmetabolic in 5 cases, viral in 4, and alcoholic in 1); 30 patients showed no specific risk factors.
Surgical specimens (or biopsies) were sent to the pathology unit, where they were routinely sampled, fixed in formalin, embedded in paraffin, and automatically processed; 3-µm-thick sections were obtained from paraffin blocks for albumin ISH, among the sections for the routine diagnostic stains.
At hematoxylin-eosin, the following histopathological variables were evaluated:
tumor dimensions;
tumor grade according to WHO 2019 classification [5];
microvascular invasion (MVI) and perineural neoplastic infiltration;
tumor localization in the biliary tree: ICCA, PCCA, or ECCA;
growth pattern: mass forming, periductal infiltrating, and intraductal growth;
tumor subtype: SD-ICCA and large-duct carcinomas, which include LD-ICCAs but also PCCAs and ECCAs.
Albumin ISH
ISH for albumin mRNA was performed by means of the RNAscope® on routine 3-µm-thick sections obtained from the paraffin blocks, i.e., the same slides routinely used for immunohistochemistry. These slides are thinner than the 5 μm sections usually used for ISH, but in our setting, they show the same diagnostic yield. The slides were put in stove at 60°C for 1 h, followed by three baths in xylene 5 min each, at room temperature (RT), and two baths in absolute alcohol 5 min each at RT. After air dry, 4 drops of RNAscope® Hydrogen Peroxide were put on the slides and incubated for 10 min, followed by washing with distilled water and antigen retrieval by 100 mL of RNAscope® 1X Target Retrieval Reagent at 99°C for 30 min. Slides were therefore washed in distilled water for 15 s and in absolute alcohol for 3 min. After drying, four drops of RNAscope® Protease Plus were added at 40°C for 30 min, followed by hybridization with albumin ZZ probe pairs® for 2 h at 40°C. The manufacturer’s protocol was applied, as follows (ACD protocol, https://acdbio.com/):
-
1.
Amplification cycle with AMP 1 reagent, 30 min at 40°C
-
2.
Wash, RNAscope® 1X buffer, 15 min at RT
-
3.
Amplification cycle with AMP 2 reagent, 15 min at 40°C
-
4.
Wash, RNAscope® 1X buffer, 15 min at RT
-
5.
Amplification cycle with AMP 3 reagent, 30 min at 40°C
-
6.
Wash, RNAscope® 1X buffer, 15 min at RT
-
7.
Amplification cycle with AMP 4 reagent, 15 min at 40°C
-
8.
Wash, RNAscope® 1X buffer, 15 min at RT
-
9.
Amplification cycle with AMP 5 reagent, 30 min at RT
-
10.
Wash, RNAscope® 1X buffer, 15 min at RT
-
11.
Amplification cycle with AMP 6 reagent, 15 min at RT
-
12.
Wash, RNAscope® 1X buffer, 15 min at RT.
Revelation was performed by means of 120 μL of DAB on each section. Washing, counterstaining with hematoxylin (2 min) and dehydration concluded the procedure. The number of positive cells was counted on high-magnification fields, and a semiquantitative assessment of the percentage of positive cells was evaluated for each case.
Statistical Analysis
All analyses were made with SPSS® software for Windows, 20. Continuous variables are shown as means, standard deviations and ranges; discrete variables are shown as frequencies and percentages. The χ2, Mann-Whitney, and the Kruskal-Wallis tests were applied as appropriate. A receiver operator characteristic (ROC) curve was built to assess the cut-off value of albumin positivity between SD-ICCAs and LD-ICCAs. A p < 0.05 coefficient was considered significant to exclude the null hypothesis.
Results
Mean tumor dimensions were 4.7 ± 3.2 cm (range 0.6–13 cm); 29 tumors were ICCA (64.4%), 9 ECCA (20.0%), and 7 perihilar (PCCA, 15.6%). The pattern of growth was mass-forming type in 30 cases (66.7%), periductal infiltrating in 9 (20.0%) and mixed in 6 (13.3%). The most represented tumor grade was grade 2 in 29 cases (64.4%), followed by 14 (31.2%) grade 3 cases and only 2 (4.4%) grade 1.
At albumin ISH, the mean percentage of tumor positive cells ‒ considering all cases ‒ was 18.02 ± 27.52% (range 0–90%); 19 (42.2%) cases were completely negative (0% positive cells). Albumin ISH was always negative in ECCAs, while it showed any positivity level in 25 out of 29 (86.2%) ICCAs; only one out of 7 (14.3%) PCCAs was positive (p < 0.001, Kruskal-Wallis test, Fig. 1). ICCAs alone had a mean cell positivity of 25.5 ± 29.3%. As a consequence, the albumin positivity changed according to the growth pattern as well (p = 0.008) since all the mass-forming types were ICCAs.
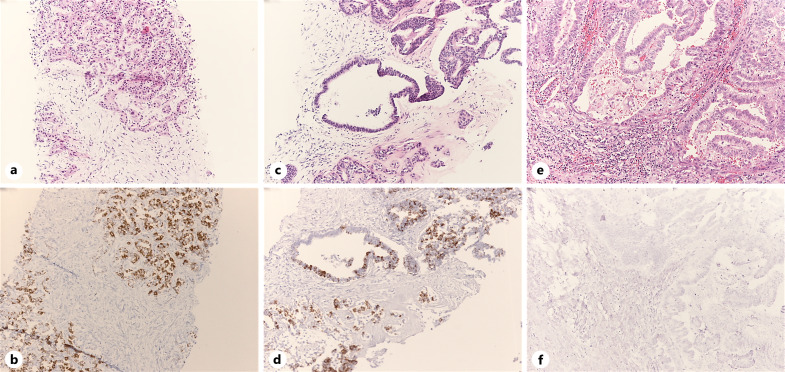
Fig. 1.
Morphological pictures of the main subtypes of CCA (hematoxylin-eosin stain, above pictures) and the albumin ISH (below pictures), which is diffusely positive in SD-ICCAs (a, b), focally positive in LD-ICCAs (c, d), and negative in perihilar and ECCAs (e, f).
The positive PCCA case had 70% of positive cells: of note, a peculiar predominantly papillary histotype was observed. None of the other clinical and morphological variables correlated with albumin positivity (data not shown).
As far as albumin expression in ICCAs is concerned, we noticed substantial differences according to the subtype (Fig. 2): in particular, SD-ICCAs (n = 15) showed any level of positivity in 14 (93.3%) cases, with a mean cell positivity of 38.8 ± 29.8%; LD-ICCAs (n = 14) showed any level of positivity in 11 (78.6%) cases, with a mean cell positivity of 11.4 ± 21.9 (p = 0.003, Mann-Whitney test).
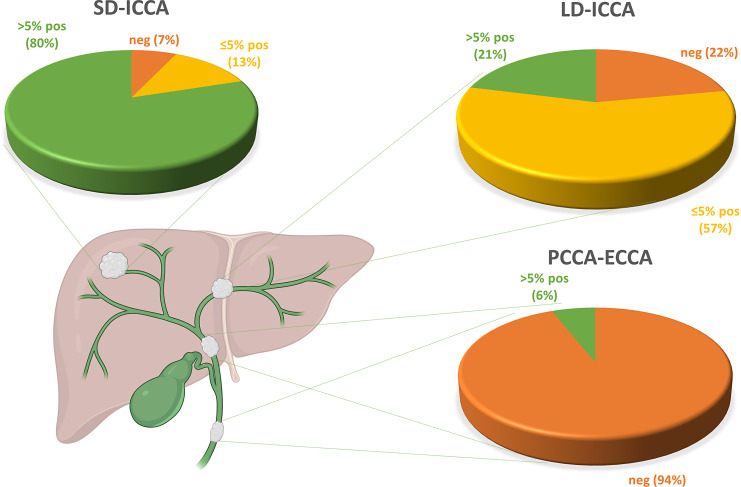
Fig. 2.
Schematic depiction of the intrahepatic and extrahepatic biliary tree, including the pie charts representing the percentages of negative cases, cases with ≤5% positive cells and cases with >5% positive cells in large-duct ICCAs (LD-ICCA), perihilar CCAs (PCCAs) and extrahepatic CCAs (ECCAs).
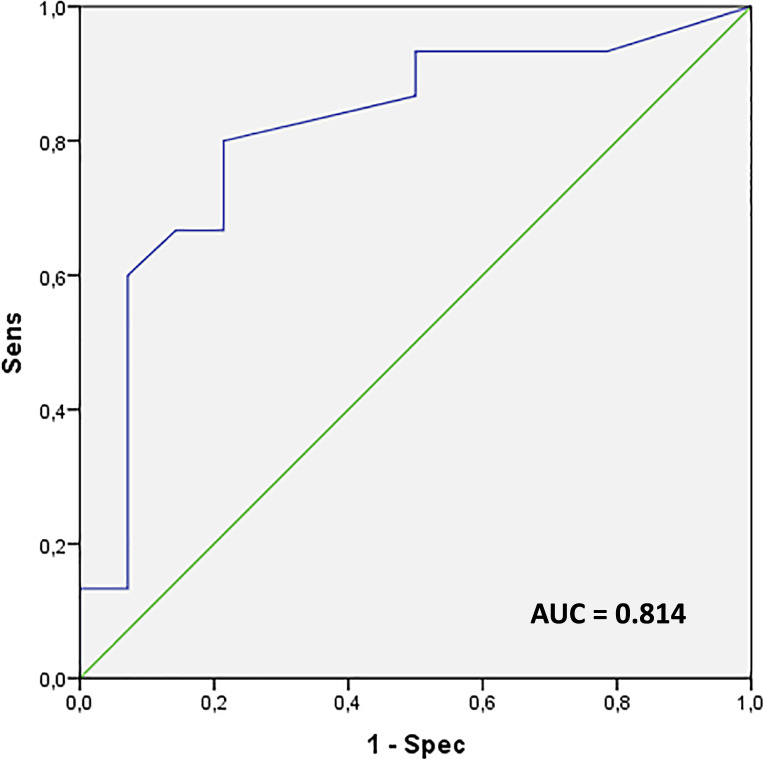
The ROC curve built to assess the optimal albumin cut-off in discriminating SD-ICCAs and LD-ICCAs showed an area under the curve (AUC) of 0.814: a cut-off of 5% of positive cells was chosen, discriminating the two subtypes with 80% sensitivity and 78.6% specificity (Fig. 3). Indeed, using this cut-off, we observed that 12 out of 15 (80.0%) SD-ICCAs and 3 out of 14 (21.4%) LD-ICCAs had >5% of positive cells (p = 0.002, χ2 test), with an odds ratio of 14.7.
Fig. 3.
Receiver operator characteristic (ROC) curve of albumin-positive cells in each case, built to discriminate small-duct and long-duct ICCA: the area under the curve (AUC) was 0.814. A cut-off value of 5% of positive cells discriminates the two subtypes with 80% sensitivity and 78.6% specificity.
Discussion
Albumin ISH had a poor spread in laboratories in the past decades, mainly because of the slow and difficult procedure the previous technique required, combined with its poor sensitivity and the subsequent interpretation troubles. The major difficulties arise in biopsy samples of CCAs, where the sensitivity drops, in addition to the intrinsic tumor heterogeneity. Among the specific issues of the old technique, the main problem was represented by the use of a home-made probe, different among centers [11], so that each center had different diagnostic accuracy, making difficult the comparison of results. Moreover, the first ISH methodologies used digoxigenin-labeled cRNA probes with anti-digoxigenin antibodies in 1:1 ratio, meaning that no amplification was applied: this resulted in a much lower sensitivity compared to the newer techniques.
The most recent ISH techniques allow a more reliable detection of albumin mRNA because of the sophisticated amplification system, which enables to amplify greatly the signal from each target sequence without amplifying the background [14]. This significant improvement in sensitivity, together with the faster and fully automatized procedure, makes RNAscope® albumin ISH a promising diagnostic tool. In recent years, other authors explored the potentials of albumin ISH as a marker of hepatic primaries, particularly in the diagnosis of ICCAs. Ferrone et al. found that 99% of ICCAs (82/83) were positive for albumin ISH, while none of 24 PCCAs and 22 ECCAs was. The authors used a different ISH technology, and they did not subclassify ICCAs in LD and SD subtypes [15]. Lin et al. [10] used RNAscope® on tissue microarray sections of 22 ICCAs, finding 14 (64%) positive for albumin. Not even in this paper, the authors specified the subtypes of ICCAs tested [10]. Avadhani et al. [12] performed ISH with RNAscope® on tissue microarrays from 43 ICCAs, divided into SD and “mucin-producing.” They found 18/43 ICCAs (42%) positive, specifying that all “mucin-producing” ICCAs were negative [12]. Collins et al. [13] recently studied the results for RNAscope® ISH albumin on a cohort of 47 ICCAs, 3 PCCAs, and 3 ECCAs. They divided their ICCAs into SD, LD, and IND type (the latter defined as “tumors which did not clearly fit in either of the two main histologic subtypes”). 38/47 ICCAs (80.9%) were deemed positive, particularly 35/37 (94.6%) SD, 1/7 (14.3%) LD, and 2/3 (66.7%) IND ICCAs. All ECCAs confirmed to be negative (0/6) [13].
Our current results are in line with part of the aforementioned findings. We performed albumin ISH on a cohort of 29 ICCAs (divided into 15 SD-ICCAs and 14 LD-ICCAs), 7 PCCAs, and 9 ECCAs. Considering any level of positivity, 86.2% ICCAs results positive, versus only 1 PCCA and none ECCAs. Assuming a 5% cut-off of positive cells ‒ which we calculated with a ROC curve, but it is also consistent with the literature ‒ 80% SD-ICCA and 21.4% LD-ICCA showed more than 5% of positive cells, very similar with the results by Collins et al. [13] Anyway, if we consider any level of positivity, 93.3% SD and 78.6% LD were actually positive: this means that scattered positive neoplastic cells are virtually enough to diagnose an ICCA, as it was in the past with the previous methodologies [11]. We found indeed a significant difference among ICCAs based on the subtype, with SD showing positivity in more cases, and a percentage of cells (38.8%) higher than LD (11.4%). This difference could help pathologists in case of difficulties in assigning the correct subtype to ICCAs, when morphology is doubtful and surely as a part of a broader panel comprehensive of antibodies such as CD56, N-CAM, and others [5]. The complete negativity of all ECCAs for albumin ISH is confirmed by our results, and it is always consistent in the literature. As of consequence, any clinical suspect of hepatic metastases from an ECCA primary should be ruled out in case of positive albumin staining. The same cannot be affirmed in case of negativity since the main differential diagnosis is with LD-ICCA, which can be negative for albumin (especially on small biopsy). PCCAs (also dubbed Klatskin’s tumors) were negative for albumin mRNA as well, except for one remarkable case, which was strongly positive but it also showed a very peculiar papillary morphology: this case should be treated as an exception in our opinion, and the role of papillary morphology in albumin expression surely deserves further studies.
Our findings are consistent with the hypothesis of a gradual decrease in albumin expression along the intrahepatic biliary tree, hypothesis that partly mirrors the postulated different cells of origin of CCAs [16] (Fig. 2). The frequent positivity for ISH albumin in ICCAs is a new attribute, discovered in these recent years through the improvement of the technique. This feature is changing the spectrum of ICCAs differential diagnosis, moving from a main “cholangiocarcinoma versus extrahepatic adenocarcinoma” to the “CCA versus HCC versus combined HCC-CCA” choice. Albumin alone cannot be used for the diagnosis of CCAs, neither for specificity nor for sensitivity: in fact, some rare extrahepatic malignancies have been described as positive for albumin [10].
In conclusion, RNAscope® ISH, due to its higher sensitivity, revolutionized the diagnostic value of albumin ISH in hepatic primary neoplasms the most recent amplification techniques are reassessing the diagnostic value of albumin ISH in hepatic primaries: broadly speaking, our results are in line with the previous works (even with different ISH techniques and different ICCA subclassifications). Our main original finding is that albumin expression decreases from liver periphery to the biggest bile ducts: therefore, most SD-ICCAs are diffusely positive, while most LD-ICCAs show at least some isolated cells, marking a difference from the hilar and extrahepatic biliary tree. If this pattern of albumin positivity is kept in mind in the routine diagnostics, it might be a valuable tool in many differential diagnoses.
Statement of Ethics
The study was approved by the Ethical Committee Area Vasta Emilia Centrale (AVEC), protocol number 128/2023/Sper/AOUBo. The study was conducted in accordance with the Declaration of Helsinki protocol. All patients gave written informed consent for their participation in the study.
Conflict of Interest Statement
The authors declare no conflicts of interest.
Funding Sources
The work reported in this publication was funded by the Italian Ministry of Health, RC-2023-2778858 project.
Author Contributions
E.A.: study design, data collection, and paper drafting; S.C.: data collection and paper drafting; G.M.: data collection, analysis, and technical contribution; D.M.: data collection and analysis; M.D. and A.P.: data collection; A. Degiovanni: technical contribution; S.T.: critical revision: G.B.: data analysis and critical revision; A.D.: study design and critical revision; F.V.: study design and paper drafting.
Funding Statement
The work reported in this publication was funded by the Italian Ministry of Health, RC-2023-2778858 project.
Data Availability Statement
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available from the corresponding author (F.V.).
References
- 1. Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77(6):1598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Renzulli M, Biselli M, Brocchi S, Granito A, Vasuri F, Tovoli F, et al. New hallmark of hepatocellular carcinoma, early hepatocellular carcinoma and high-grade dysplastic nodules on Gd-EOB-DTPA MRI in patients with cirrhosis: a new diagnostic algorithm. Gut. 2018;67(9):1674–82. [DOI] [PubMed] [Google Scholar]
- 3. Renzulli M, Braccischi L, D’Errico A, Pecorelli A, Brandi N, Golfieri R, et al. State-of-the-art review on the correlations between pathological and magnetic resonance features of cirrhotic nodules. Histol Histopathol. 2022;37(12):1151–65. [DOI] [PubMed] [Google Scholar]
- 4. Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, et al. The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J Hepatol. 2009;50(4):746–54. [DOI] [PubMed] [Google Scholar]
- 5. WHO/IARC classification of tumors of the digestive system. 5th edn.2019. [Google Scholar]
- 6. Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13(5):261–80. [DOI] [PubMed] [Google Scholar]
- 7. Liau JY, Tsai JH, Yuan RH, Chang CN, Lee HJ, Jeng YM. Morphological subclassification of intrahepatic cholangiocarcinoma: etiological, clinicopathological, and molecular features. Mod Pathol. 2014;27(8):1163–73. [DOI] [PubMed] [Google Scholar]
- 8. Nakanuma Y, Uchida T, Sato Y, Uesaka K. An S100P-positive biliary epithelial field is a preinvasive intraepithelial neoplasm in nodular-sclerosing cholangiocarcinoma. Hum Pathol. 2017;60:46–57. [DOI] [PubMed] [Google Scholar]
- 9. Rajkumar SV, Richardson RL, Goellner JR. Diagnostic value of albumin gene expression in liver tumors: case report and review of the literature. Mayo Clin Proc. 1998;73:533–6. [DOI] [PubMed] [Google Scholar]
- 10. Lin F, Shi J, Wang HL, Ma XJ, Monroe R, Luo Y, et al. Detection of albumin expression by RNA in situ hybridization is a sensitive and specific method for identification of hepatocellular carcinomas and intrahepatic cholangiocarcinomas. Am J Clin Pathol. 2018;150(1):58–64. [DOI] [PubMed] [Google Scholar]
- 11. D’Errico A, Baccarini P, Fiorentino M, Ceccarelli C, Bonazzi C, Ponzetto A, et al. Histogenesis of primary liver carcinomas: strengths and weaknesses of cytokeratin profile and albumin mRNA detection. Hum Pathol. 1996;27(6):599–604. [DOI] [PubMed] [Google Scholar]
- 12. Avadhani V, Cohen C, Siddiqui MT, Krasinskas A. A subset of intrahepatic cholangiocarcinomas express albumin RNA as detected by in situ hybridization. Appl Immunohistochem Mol Morphol. 2021;29(3):175–9. [DOI] [PubMed] [Google Scholar]
- 13. Collins K, Newcomb PH, Cartun RW, Ligato S. Utility and limitations of albumin mRNA in situ hybridization detection in the diagnosis of hepatobiliary lesions and metastatic carcinoma to the liver. Appl Immunohistochem Mol Morphol. 2021;29(3):180–7. [DOI] [PubMed] [Google Scholar]
- 14. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferrone CR, Ting DT, Shahid M, Konstantinidis IT, Sabbatino F, Goyal L, et al. The ability to diagnose intrahepatic cholangiocarcinoma definitively using novel branched DNA-enhanced albumin RNA in situ hybridization technology. Ann Surg Oncol. 2016;23(1):290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moeini A, Haber PK, Sia D. Cell of origin in biliary tract cancers and clinical implications. JHEP Rep. 2021;3(2):100226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available from the corresponding author (F.V.).