Abstract
Circadian rhythms are critical for human health and are highly conserved across species. Disruptions in these rhythms contribute to many diseases, including psychiatric disorders. Previous results suggest that circadian genes modulate behavior through specific cell types in the nucleus accumbens (NAc), particularly dopamine D1-expressing medium spiny neurons (MSNs). However, diurnal rhythms in transcript expression have not been investigated in NAc MSNs. In this study we identified and characterized rhythmic transcripts in D1- and D2-expressing neurons and compared rhythmicity results to homogenate as well as astrocyte samples taken from the NAc of male and female mice. We find that all cell types have transcripts with diurnal rhythms and that top rhythmic transcripts are largely core clock genes, which peak at approximately the same time of day in each cell type and sex. While clock-controlled rhythmic transcripts are enriched for protein regulation pathways across cell type, cell signaling and signal transduction related processes are most commonly enriched in MSNs. In contrast to core clock genes, these clock-controlled rhythmic transcripts tend to reach their peak in expression about 2 hours later in females than males, suggesting diurnal rhythms in reward may be delayed in females. We also find sex differences in pathway enrichment for rhythmic transcripts peaking at different times of day. Protein folding and immune responses are enriched in transcripts that peak in the dark phase, while metabolic processes are primarily enriched in transcripts that peak in the light phase. Importantly, we also find that several classic markers used to categorize MSNs are rhythmic in the NAc. This is critical since the use of rhythmic markers could lead to over- or under-enrichment of targeted cell types depending on the time at which they are sampled. This study greatly expands our knowledge of how individual cell types contribute to rhythms in the NAc.
Keywords: circadian rhythms, gene expression, medium spiny neurons, sex differences
Introduction
Circadian rhythms are critical for human health and are highly conserved across species, controlling physiological processes and behavior across the light/dark cycle. The core molecular clock, a series of interlocking transcriptional-translational feedback loops found in almost every cell in the brain and body1–3, is strongly implicated in this process. The molecular clock is regulated by the CLOCK protein and/or a closely related protein, NPAS2, which dimerize with ARNTL (BMAL1) to control transcription of many genes including the Period (Per) and Cryptochrome (Cry) genes. After translation, the PER and CRY proteins enter the nucleus and inhibit the transcriptional activity of (CLOCK/NPAS2)/BMAL1, closing the negative feedback loop3. In addition to regulating basic physiological processes and brain function, disrupted circadian genes are linked to altered mood4–10 and substance use (SU) vulnerability11–18 across species.
Research from our lab and others suggests that the intersection between circadian rhythms and psychiatric disease is largely mediated by the function of circadian genes in reward-related brain regions, such as the nucleus accumbens (NAc)19–21. Importantly, the role of the NAc in physiology and behavior, including how circadian genes regulate these, appears to be cell-type specific22–24. While gene expression rhythms have been thoroughly studied, even cell-type specifically25, in the central pacemaker or the suprachiasmatic nucleus (SCN), little is known about the rhythmicity of genes in the NAc across cell type and the function of those rhythms. Importantly, recent work in mice26 and humans27 confirms that rhythms in transcript expression are abundant and widespread in the NAc. These include the core clock components discussed above, but also numerous other transcripts. These findings indicate that gene expression rhythms are highly prevalent in reward- and mood-related brain regions, however, these experiments were performed with bulk NAc tissue, not cell-type specifically.
Since our prior work suggests that cell type plays a critical role in the effect of circadian rhythm disruptions in behavior, we aimed to use mouse models to measure transcript expression rhythms in different cell types in the NAc. Here, we measured gene expression rhythms in D1- or D2- expressing neurons in the NAc. We then compared these rhythms to those found in bulk NAc homogenate tissue and astrocytes28 taken from the NAc. Samples were collected from male and female mice in order to determine whether gene expression rhythms differ by cell type and sex, since sex differences are prominent in circadian rhythms29 and reward-related behavior30. In addition, we are using an approach that allows us to only measure actively translated mRNA expression, therefore these expression changes are likely to be functionally relevant at the protein level. We also leveraged these novel data to investigate whether classically used cell-type markers differ in expression across the light/dark cycle, which will be critical for rigorous experimental design in the future.
Materials and Methods
Subjects
To sequence actively translated mRNAs, male and female Drd1- and A2a-Cre mice were crossed to RiboTag mice31, expressing a Cre-inducible HA-Rpl22. C57BL/6J mice were used to generate homogenate samples. Previously generated data was used for astrocyte analysis28 wherein the GFP-inducible alternative to HA-Rpl22, Rpl10a, was crossed to Aldh1l1-eGFP mice. Mice were over 10 weeks old and maintained on a 12:12 light-dark (LD) schedule with lights on (Zeitgeber Time (ZT0)) at 0700. Food and water were provided ad libitum and procedures were approved by the University of Pittsburgh IACUC.
Tissue Processing.
NAc tissue was collected from male and female mice pseudo-randomly across 6 times of day (ZT2,6,10,14,18,22). As described previously28,31, co-immunoprecipitation was used to isolate cell-type specific ribosome-associated mRNAs. Final eluted homogenate and D1 or D2 specific RNA was used for sequencing.
RNA-sequencing, analysis and visualization.
Following quality/integrity assessment, libraries were prepared and sequenced (blinded samples). mRNA (D1/D2) or total (homogenate) RNA sequencing was performed (n≥3) using the NextSeq 500 platform (Illumina) at the University of Pittsburgh Health Sciences Sequencing Core. After preprocessing and filtering of sequencing data, a parametric cosinor model32 assuming a sinusoidal relationship between the gene expression level and ZT was used to detect rhythmicity (Supp.Table.1). Six total samples were excluded, due to insufficient RNA, library preparation or raw counts.
Since a variety of library preparation and sequencing procedures were used each cell type was analyzed independently. Downstream analyses compared the rhythmic transcripts identified for each cell type, typically with a significance threshold of p<0.05. Differential expression (DE) analysis was also conducted to compare D1- and D2-expressed transcripts by DESeq233 during the light (ZT2,6,10) and dark phases (ZT14,18,22)(Supp.Table.2).
For rhythmic circadian transcripts, peak times and phase were visualized with heatmaps, scatter and radar plots. Heatmaps were generated by plotting expression levels ordered by peak time. Scatter plots were made for core clock genes. The proportion of rhythmic transcripts was plotted against peak time in radar plots. To visualize phase differences, phase concordance plots were made for rhythmic transcripts that overlap in males and females within each cell type. Transcripts were considered concordant if phase differences fell within a 4-hr window.
The RRHO (rank rank hypergeometric overlap) test was used as a threshold-free approach to identify overlap between males and females using ranked lists of rhythmic genes34. A heatmap indicates the level of overlap in genes across a significance gradient.
Ingenuity Pathway Analysis (IPA) software was used to identify significantly enriched pathways (p<0.05) in rhythmic transcripts (p<0.05; limited to top 2000) across groups (Supp.Table.3). Genes passing preprocessing selection in any cell type were used as the background gene list. A motif analysis was also performed using LISA (epigenetic Landscape In Silico deletion Analysis)(Supp.Table.4). A maximum of 500 rhythmic transcripts were used for each cell type and sex. p<0.00001 was used as a significance threshold to identify enriched motifs.
Results
Diurnally rhythmic genes differ greatly between cell types
We aimed to determine if diurnal rhythms in transcript expression in the mouse NAc vary by sex or cell type using co-immunoprecipitation of ribosome-associated mRNAs in D1- or D2-expessing cells. These actively translated transcripts are more likely to correspond to functional protein rhythms. We compared these results to homogenate preparations from the NAc, which contain a multitude of cell types, as well as previously published, astrocyte-specific data28.
We found numerous transcripts with diurnal rhythms in the NAc and report the number of rhythmic transcripts when employing more or less stringent cut-offs and corrections for multiple comparisons (Fig. 1A). At a p<0.05, approximately 12% of transcripts expressed in D1 cells were significantly rhythmic (1,735 of 13,984), while ~14% were rhythmic in D2 cells (1,943 of 14,282). Similar results were found in homogenate NAc samples, where 1,795 transcripts (~15%) were significantly rhythmic out of 11,867. Astrocytes had the most transcripts with significant rhythms at 6,481 (~51%) out of 12,739 transcripts. As expected, many of the top rhythmic transcripts in each cell type are core clock genes (Supp.Fig.1A). While overall gene expression levels for top circadian transcripts can vary, peak times are largely consistent, suggesting the core circadian clock is in phase across these cell types in the NAc (Fig.1B). We also confirmed high levels of enrichment for cell-type specific markers in D1 and D2 samples with minimal loss in expression compared to homogenate tissue (Supp.Fig.1B,C).
Figure 1. Rhythmicity in the NAc varies by cell type.
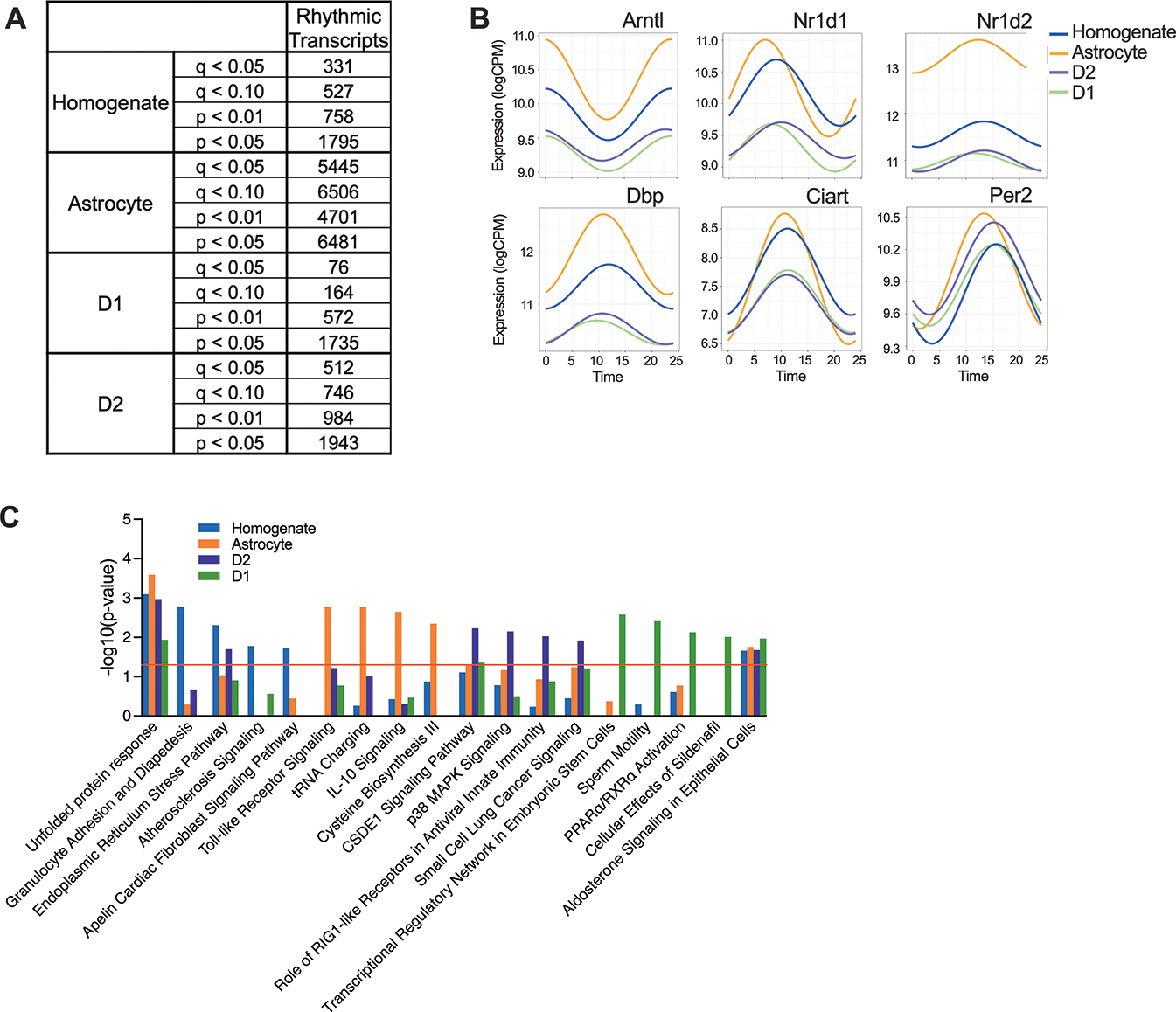
(A) Table listing numbers of rhythmic transcripts identified in each cell type in the NAc at various thresholds of significance (p values and q values (Benjamini-Hochberg correction). (B) Scatterplots show expression patterns (logCPM) across time of day for top rhythmic transcripts. These six core clock genes have consistent peak times across cell type. (C) For each cell type, pathway analyses identified pathways of rhythmic transcripts, which reached a significance threshold of p<0.05. The top 5 enriched pathways from each cell type are plotted together.
Ingenuity pathway analysis (IPA) was used to determine which pathways the rhythmic transcripts (p<0.05) significantly fall into, in each cell type (Fig.1C). The majority of rhythmic pathways are unique to one cell type, with minimal overlap observed. Only 2 pathways are shared between homogenate tissue, MSNs and astrocytes. These include primarily transcripts encoding heat shock proteins (HSP and DNAJ families), which are critical for protein folding and assembly in the endoplasmic reticulum. This suggests protein regulation, including synthesis, folding and misfolding identification, is highly rhythmic in the NAc across cell type. Between MSNs and astrocytes, translational reprogramming is also overlapping (Fig.1C). Most of the unique processes found in homogenate tissue are specific to immune responses, which are also found in astrocytes and D2 cells, but these involve cytokine signaling and use cytoplasmic, not cell surface mechanisms, respectively. In D2 cells, cell signaling and protein modification are uniquely enriched processes, while transcriptional regulation/gene activation and signal transduction are primarily found in D1 cells.
Rhythmic genes are sex-specific, particularly in D1 cells
When rhythmic analysis was performed for male and female samples separately fewer rhythmic transcripts are found, likely due to lower statistical power (Fig.2A). However, approximately equal numbers of rhythmic transcripts are identified across sex in MSNs. Most rhythmic transcripts are uniquely identified in one sex, regardless of the significance threshold used (Fig.2B). Specifically, about 20% of rhythmic transcripts are overlapping in D1 cells taken from males and females, while about 40% are overlapping in D2 cells. The remaining rhythmic transcripts might be uniquely rhythmic in one sex. However, when p-values for each rhythmic transcript are plotted by sex (Supp.Fig.2), ~10% of transcripts reach significance in one sex (p<0.05), while approaching significance in the other (0.1<p<0.05). These transcripts are unlikely to represent a functional difference between the sexes. Taken together, approximately 70% of transcripts in D1 cells are uniquely rhythmic in one sex, while 50% are unique in D2 cells.
Figure 2. Rhythmic transcripts vary by across cell type and sex in the NAc.
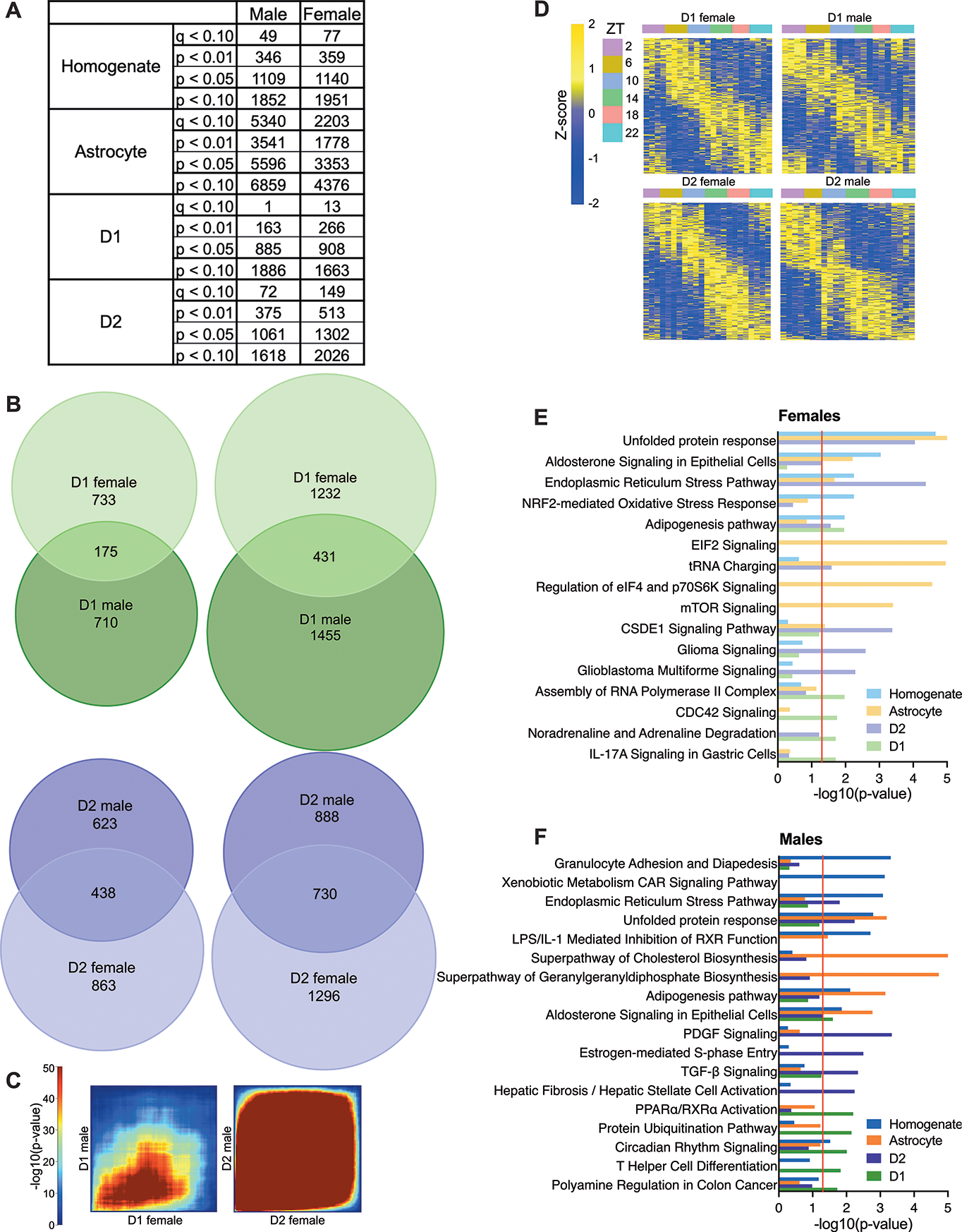
(A) Table listing numbers of rhythmic transcripts identified at various significance thresholds in each cell type for male and female samples taken from the NAc. (B) Venn diagrams are used to show overlap across sex for rhythmic transcripts at multiple significance thresholds (p<0.05 (left) or p<0.10 (right)) in D1 and D2 cells. (C) RRHO plots are also used to visualize overlap across sex in a threshold-free manner. (D) Heatmaps of rhythmic transcripts show patterns in peak times across groups. (E-F) Pathway analyses identified significant (p<0.05) pathways enriched for rhythmic transcripts. The top 5 enriched pathways from each cell type are plotted together for (E) female and (F) male samples separately.
We next used a threshold-free approach (RRHO) to better visualize the overlap of rhythmic transcripts in males and females. As expected, we find less overlap between rhythmic transcripts in female versus male D1 MSNs compared to D2 MSNs (Fig.2C). Importantly, the most significantly rhythmic transcripts, found in the lower left-hand corner, are still overlapping in D1 MSNs. These shared genes include core clock genes, which have consistent expression levels and peak times across sex (Supp.Fig.3).
When expression is plotted across time of day (TOD) in heatmaps, overall patterns in rhythmicity can be visualized. Generally, we find strong circadian rhythms in MSNs extracted from males and females, where transcripts peak at all TOD (Fig.2D). However, if we plot expression for transcripts identified as rhythmic in one sex in the opposite sex (i.e. rhythmic transcripts from males plotted in females), heatmaps confirm lower overlap in D1 cells, since rhythms in peak times are less apparent across TOD (Supp.Fig.4).
As in MSNs, the number of rhythmic transcripts identified in homogenate tissue is similar between sexes. Interestingly, astrocytes are distinct since males have twice as many rhythmic transcripts as females. As in D1 cells, approximately 20% of transcripts are overlapping across sex in homogenate tissue. Astrocytes are more similar to D2 cells, with 40% of transcripts overlapping (Supp.Fig.4).
Rhythmic genes are enriched for unique processes across cell type and sex in the NAc
Pathway analyses reveal that although most rhythmic pathways are unique to one cell type in females (Fig.2E) or males (Fig.2F), the processes regulated by these unique pathways can be similar. As when males and females are pooled (Fig.1), pathways involved in signal transduction and cell signaling are found in D1 and D2 MSNs. These are the primary pathways that overlap in male and female samples taken from D2 cells, while immune responses and transcriptional regulation are also enriched rhythmic processes in D1 cells across sex.
Certain rhythmic processes are also unique to one MSN and one sex. In females, rhythms in D1 cells are enriched for metabolism-related pathways, while unique processes are largely not found in D2 cells. In D1 cells taken from males, circadian rhythm signaling and cell proliferation are uniquely rhythmic pathways, but cell cycle control and cellular motility are enriched in D2 cells.
When enriched pathways in MSNs are compared to homogenate tissue and astrocytes, some overlap can be seen across cell type and sex. Similar to when females and males are pooled (Fig.1C), protein regulation is the most commonly identified shared rhythmic process. In females (Fig.2E) and males (Fig.2F), homogenate tissue, astrocytes and D2 cells share rhythmic pathways related to protein folding, misfolding and/or synthesis. Although these same pathways are not enriched in D1 cells, others related to protein folding and regulation are significant. Only D1 cells taken from females are not enriched for protein processing-related pathways.
Other processes were identified that overlap across cell type in a sex-specific manner. Immune-related pathways are enriched in homogenate tissue and astrocytes extracted from males as well as D1 MSNs across sex. D1 cells and homogenate tissue taken from females are also enriched for rhythmic processes regulating oxidative stress. Although metabolic pathways are identified in other groups, these pathways are overwhelmingly enriched in homogenate tissue and astrocytes taken from males.
Rhythmic genes peak at distinct times of day and vary by sex
We determined the time of day at which significantly rhythmic transcripts peak in expression (Fig.3). Almost all groups have two clusters of rhythmic transcripts, one which peaks during the light phase (inactive; ZT0–12), and another that peaks during the dark phase (active; ZT12–24)(Fig.3A). In MSNs, peak times occur at ZT6/ZT18 in D1 cells and ZT5/17 in D2 cells extracted from females. Transcripts in males peak earlier, with the light peak time falling at ZT1 for D1 cells and ZT3 for D2 cells. Interestingly, D1 MSNs taken from males peak closer to sunrise and sunset (ZT0/ZT12) instead of mid-day and mid-night (ZT6/ZT18) as in females. Interestingly, peak times are similar in homogenate tissue (ZT6) and astrocytes (ZT7) taken from females, but in males, no definitively clustered peak times are found for homogenate tissue and astrocytes peak later than MSNs (ZT5; Fig.3A).
Figure 3. Rhythmic transcripts tend to peak later in females than males.
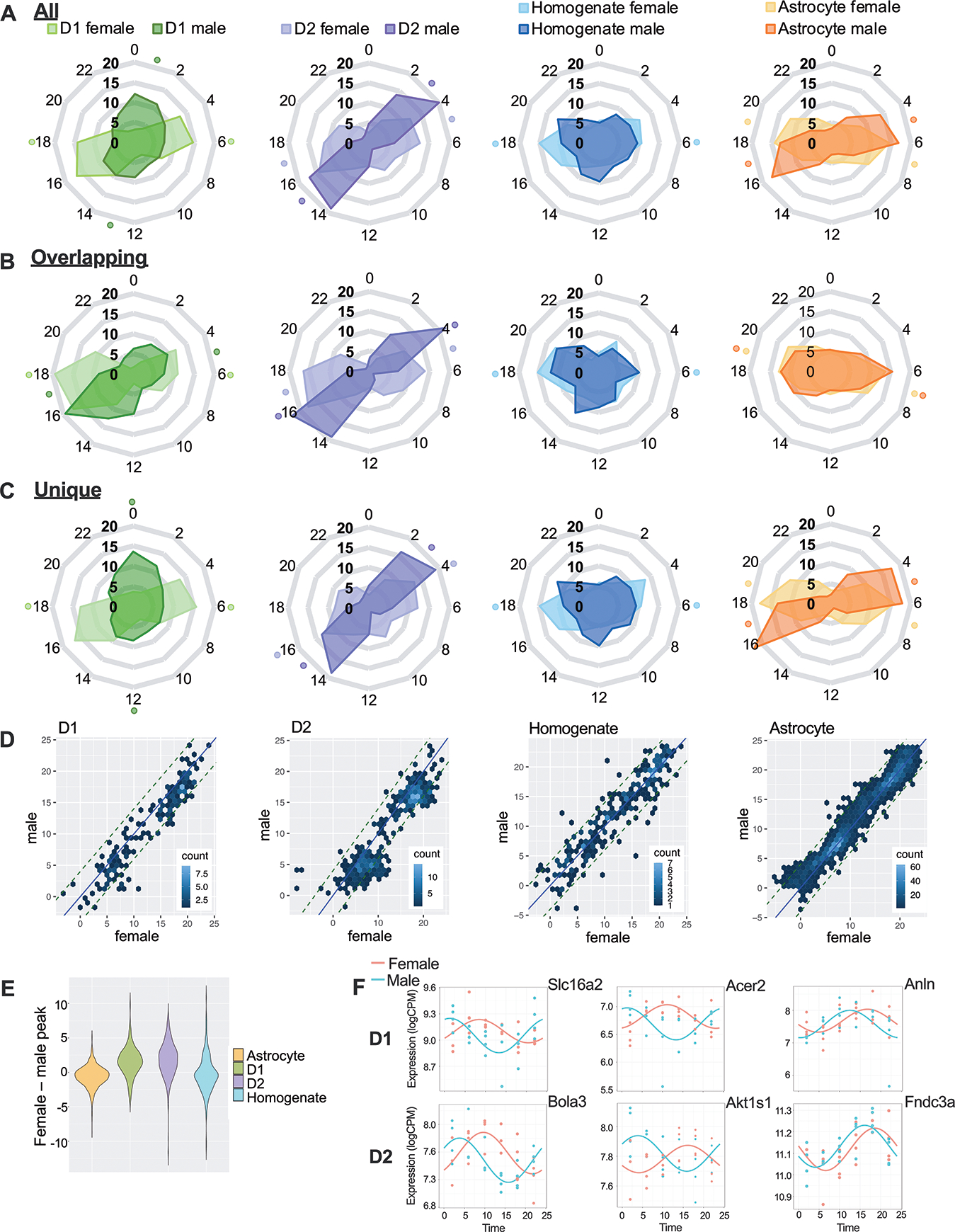
(A) Peak times for each rhythmic transcript are shown in radar plots for each cell type in males and females across the 24-hour clock (ZT). The time with the highest proportion of peaking genes, as well as its opposite (12-hours apart), is indicated with a dot. Separate radar plots are shown for rhythmic transcripts that are (B) overlapping or (C) unique in males and females. (D) For genes that overlap in males and females, we also show phase concordance plots and (E) quantified the difference in peak time. On average, transcripts peaked 2 hours later in D1 and D2 cells taken from females compared to males. (F) Example transcripts with varying peak times across sex are shown with smoothed curves as well as individual data points: (left) ~6 hour, (middle) ~12 hour and (right) ~2-hour difference.
Overall, a pattern emerges where the majority of rhythmic transcripts peak later in females than in males (~5 hours in D1 cells and ~2 hours in D2 cells/astrocytes). This difference appears to be partially driven by distinct rhythmic transcripts in males and females, as overlapping transcripts have more similar peak times across sex (Fig.3B), while unique transcript peak times are more divergent (Fig.3C). However, phase concordance plots show that overlapping transcripts still peak later in females than males in MSNs, as shown by a rightward shift in the transcripts (Fig.3D). When the difference in peak time is quantified (Fig.3E) astrocytes and homogenate tissue have no change in peak time, while shared rhythmic transcripts in MSNs peak about 2 hours later in females. Example rhythmic transcripts with peak time differences of 6 (left), 12 (middle) and 2 (right) hours are plotted for MSNs across sex (Fig.3F).
Light phase pathways are largely divergent across sex and cell type
Rhythmic transcripts were split into two 12-hour clusters based on peak times (see Supplemental methods). These two antiphase peak clusters are labeled “light” and “dark” throughout based on which cluster peaks primarily during those phases. Homogenate samples taken from males have no clustered peak times, therefore all rhythmic transcripts from this group are compared to the light and dark peaking transcripts in other cell types.
We performed pathway analyses to compare pathways enriched in rhythmic transcripts peaking during the light and dark phase in males and females (Fig.4). Overall, we found more sex differences in pathways enriched for transcripts peaking during the light phase. In contrast, we found consistent pathway enrichment across sex (Supp.Fig.5) when only shared rhythmic transcripts were analyzed, suggesting sex differences are mediated by unique rhythmic transcripts.
Figure 4. Rhythmic transcripts peaking in the light phase contribute to largely distinct enriched pathways.
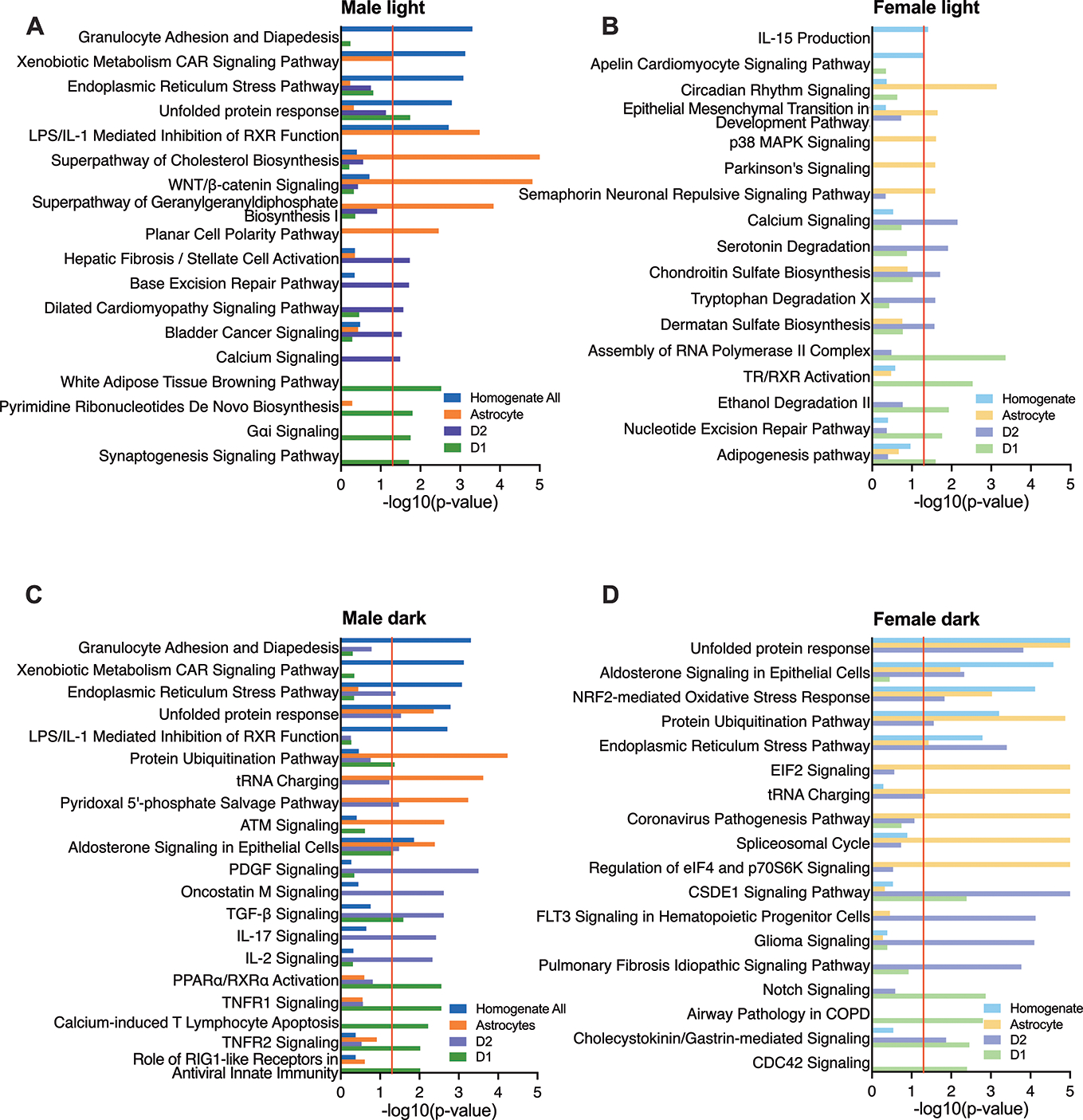
(A-D) Pathway analyses identified significant pathways (p<0.05) enriched for rhythmic transcripts falling into clustered peaks (“light” and “dark”). The tope 5 enriched pathways from each cell type are plotted together for each sex and peak time. For proper scale, p-values are truncated at -log10(p-value)≤5 even if the pathway is more significantly enriched.
In MSNs, the majority of pathways for rhythmic transcripts peaking during the light phase (Fig.4A–B) are unique to one cell type, but some of the processes these pathways regulate are similar. In light peaking transcripts, all MSNs taken from males are highly enriched for cell signaling/signal transduction. In contrast, rhythmic transcripts peaking in the light phase in MSNs extracted from females are highly enriched for metabolic pathways.
Many processes identified are unique to one sex and cell type. In D1 cells taken from males, pathways for transcripts peaking during the light phase are enriched for synaptogenesis, while D2 cells are enriched for cell structural maintenance. In D1 cells taken from females, light peaking transcripts are enriched for RNA processing and transcriptional regulation, while D2 cells are enriched for degradation and biosynthesis-related pathways.
During the dark phase, rhythmic processes identified in MSNs are highly overlapping (Fig.4C–D) with enrichment in cell signaling pathways across sex. Dark peaking transcripts in MSNs taken from males, as well as D1 cells from females, are also enriched for immune-related pathways.
In addition to identifying fewer sex differences in dark peaking transcripts, we also found fewer cell-type differences (Fig.4). Similar to previous analyses (Fig.1C,2D,E), most of the overlapping pathways for transcripts that peak during the dark phase are involved with protein regulation. However, D1 cells taken from males are distinct, with enriched protein-related pathways identified for light, not dark peaking transcripts. This could be due to transcripts in this group tending to peak around sunrise/sunset instead of midday/midnight (Fig.3A). Rhythmic processes involving metabolism, cell signaling and immune responses are also overlapping, within various cell types, sexes and peak times.
Unique motifs regulate rhythmic gene expression across cell type, time of day and sex
A motif analysis was used to determine whether rhythmic transcripts are regulated by similar or different motifs or transcription factors across group (cell type, sex, TOD). While enriched motifs were largely consistent across all groups (Supp.Table.3), we also identified motifs that could regulate the differences in expression and enriched pathways seen across TOD, sex and cell type (Supp.Fig.6). Interestingly, some motifs associated with the expression of transcripts peaking at one TOD may also uniquely regulate rhythmic transcripts found in males or females. For example, Lhx8 (UP00184) and AFT2::JUN (M00084) binding sites are identified in dark peaking transcripts (across sex) as well as rhythmic transcripts in males (across TOD). On the other hand, USF1 (MC00248), which could regulate light peaking transcripts, and EHF (MS00778) and ELF5 (MS00838), which could regulate dark peaking transcripts, may also regulate rhythmicity in females.
Rhythmicity should be considered when selecting cell-type specific markers
Historically, specific markers have been used to differentiate cell types, but rhythmicity in these transcripts has largely never been considered, even though rhythms could skew identification of cell types due to peaks and troughs in expression. We find that many historical markers for MSNs tend to be rhythmic, with moderate amplitudes, especially in D1 cells. This dichotomy between rhythmicity in D1 and D2 cells could further perpetuate misidentification and biased proportions of MSNs if these markers are used (Fig.5A). Markers that peak during the dark phase and trough during the light phase, when experiments are typically conducted, such as Penk (proenkephalin) and Adora2a (adenosine A2A receptor), would be especially problematic (Fig.5A). Some astrocyte markers are also rhythmic, such as Aqp4 (aquaporin 4;p=.0069). Novel markers have been identified for these cell types35, but many of these are also rhythmic (Fig.5B): ~50% in astrocytes, 20% in D1 cells and 25% in D2 cells. Many of these novel markers also peak during the dark phase, suggesting expression levels of these transcripts will be lower than average (troughing) during the light phase, when tissue is collected.
Figure 5. Rhythmicity should be considered when cell-type specific markers are selected.
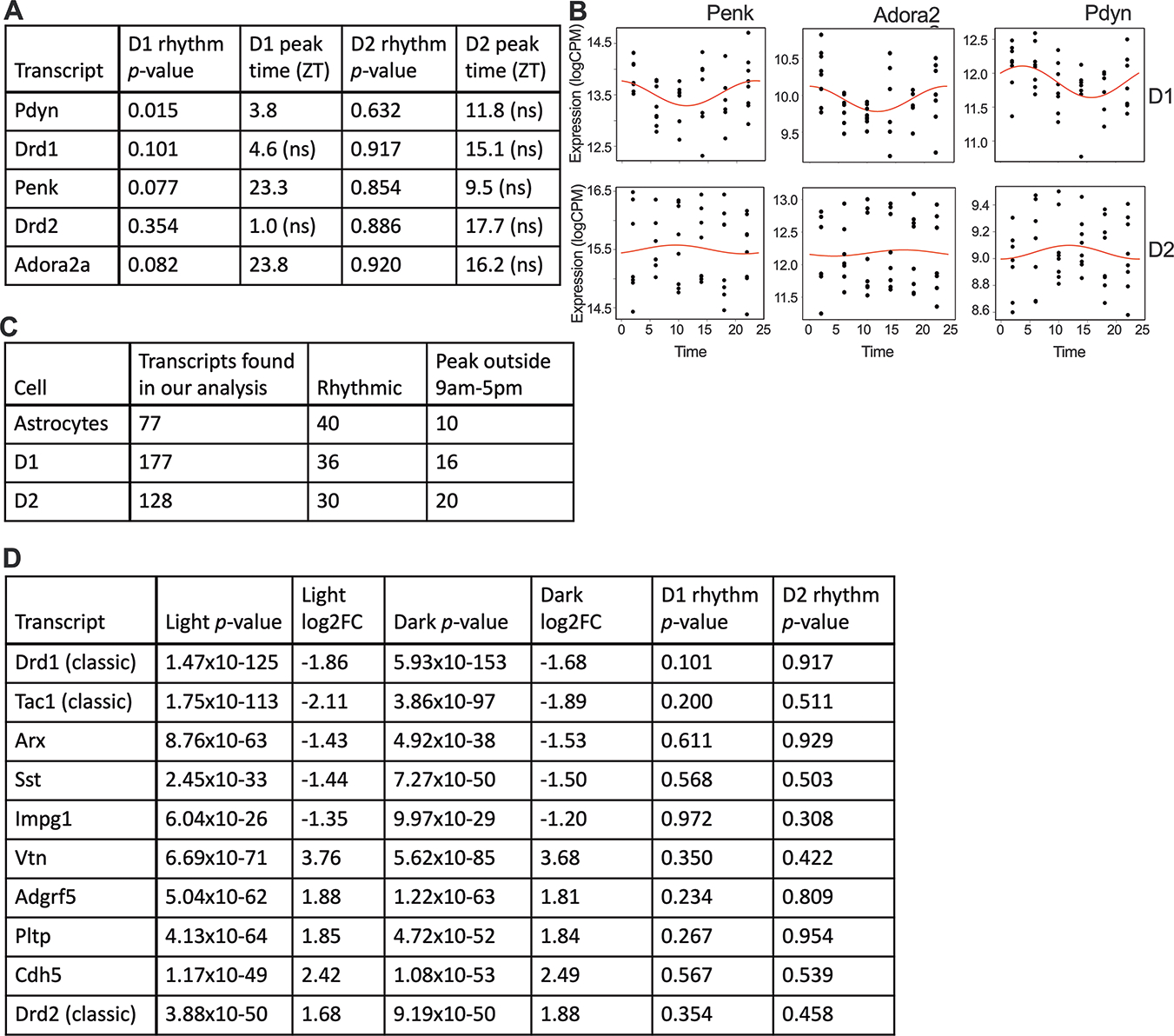
(A) Table showing rhythmicity (p-values and peak times) for several traditional markers used to distinguish D1- and D2-expressing neurons. (B) To visualize these rhythms, rhythmic markers from Table A are shown as smoothed curves in expression (logCPM) and individual data points in both D1 cells (rhythmic) and D2 cells (not significantly rhythmic). (C) Novel transcripts that have been identified for cell-type differentiation in the NAc are also found in our analysis. A subset of these are rhythmic, with many peaking in the dark phase. (D) The top 5 differentially expressed, non-rhythmic transcripts (rhythmicity p-value<0.10) enriched in D1 cells and D2 cells are shown. Corresponding p-values and fold changes for DE performed for samples collected during the light and dark phase are shown for each transcript. Non-significant p-values for rhythmicity are also reported. Expression in D1 cells was used as the baseline for DE analysis.
We used our dataset to look for the most consistent markers for D1 and D2 cell differentiation across TOD. We identified 5 transcripts that are most differentially expressed (DE;FC of >2;lowest p-values) between D1 and D2 cells during both the day and night, as well as p>0.1 for rhythmic analysis (Fig. 5C). We identified several classic markers, such as Drd1 and Drd2, but also found novel markers that might be better candidates for identifying D1 (Tac1 and Arx) and D2 cells (Vtn).
Discussion
In this study we identified and characterized, for the first time, rhythmic transcripts selectively in D1- and D2-expressing neurons and compared rhythmicity results to whole NAc homogenate as well as astrocyte samples28. We observed diurnal rhythms in all cell types, with astrocytes having the most rhythmic transcripts and D1 cells having the least. The top rhythmic transcripts were largely core clock genes that peak at approximately the same TOD across cell type. Overall, protein-related pathways were enriched for rhythmic transcripts in all cell types, indicating the importance of rhythms in protein synthesis, folding and processing. Enriched pathways also varied by cell type: immune-related pathways were enriched in homogenate tissue and astrocytes, cell signaling and modification pathways were enriched in D2 cells and transcriptional regulation and signal transduction pathways were enriched in D1 cells.
We also found sex differences between cell types, particularly in MSNs. Specifically, we found that about 70% of rhythmic transcripts are unique to one sex in D1 cells, while only 50% are unique in D2 cells. This sex difference can be readily visualized with a RRHO plot (Fig.2C), where we find less overlap between rhythmic transcripts in D1 cells taken from male and female mice, compared to D2 cells.
Another key sex difference we identified was a change in peak time for rhythmic transcripts. Understanding peak time is critical to determining how transcripts work together. Rhythmic transcripts peak during the light (inactive) or dark (active) phase, suggesting separate physiological processes are occurring during these phases. A summary figure (Fig.6) shows the most highly rhythmic processes and their approximate peak time in each sex and MSN sub-type. Peak times vary highly by sex in MSNs, with shared rhythmic transcripts peaking about 2 hours later in females than males. It is possible that the delay in gene expression rhythms observed in females in the NAc translates to a shifted rhythm in neuronal activity or reward-related processes. If so, this could indicate that reward-related behavior should be measured, and therapeutic approaches might be more beneficial if delivered, at later clock times in females. Importantly, many studies performed in the NAc have focused on males even though sex differences are prevalent in many psychiatric disorders. In particular, substance use disorders (SUDs) are often more severe in women, who can experience faster escalation of drug use, more craving and more severe SUD when seeking treatment30,36–38. We have previously shown similar vulnerabilities in mice, where mutations in circadian genes increase SU to a greater extent in females22. This effect is mediated by activity in D1 cells in the NAc, therefore this delay in rhythms in MSNs could make females more vulnerable to circadian disruptions, especially in the context of SU.
Figure 6. Model summarizing enriched rhythmic processes in MSNs across time of day and sex.
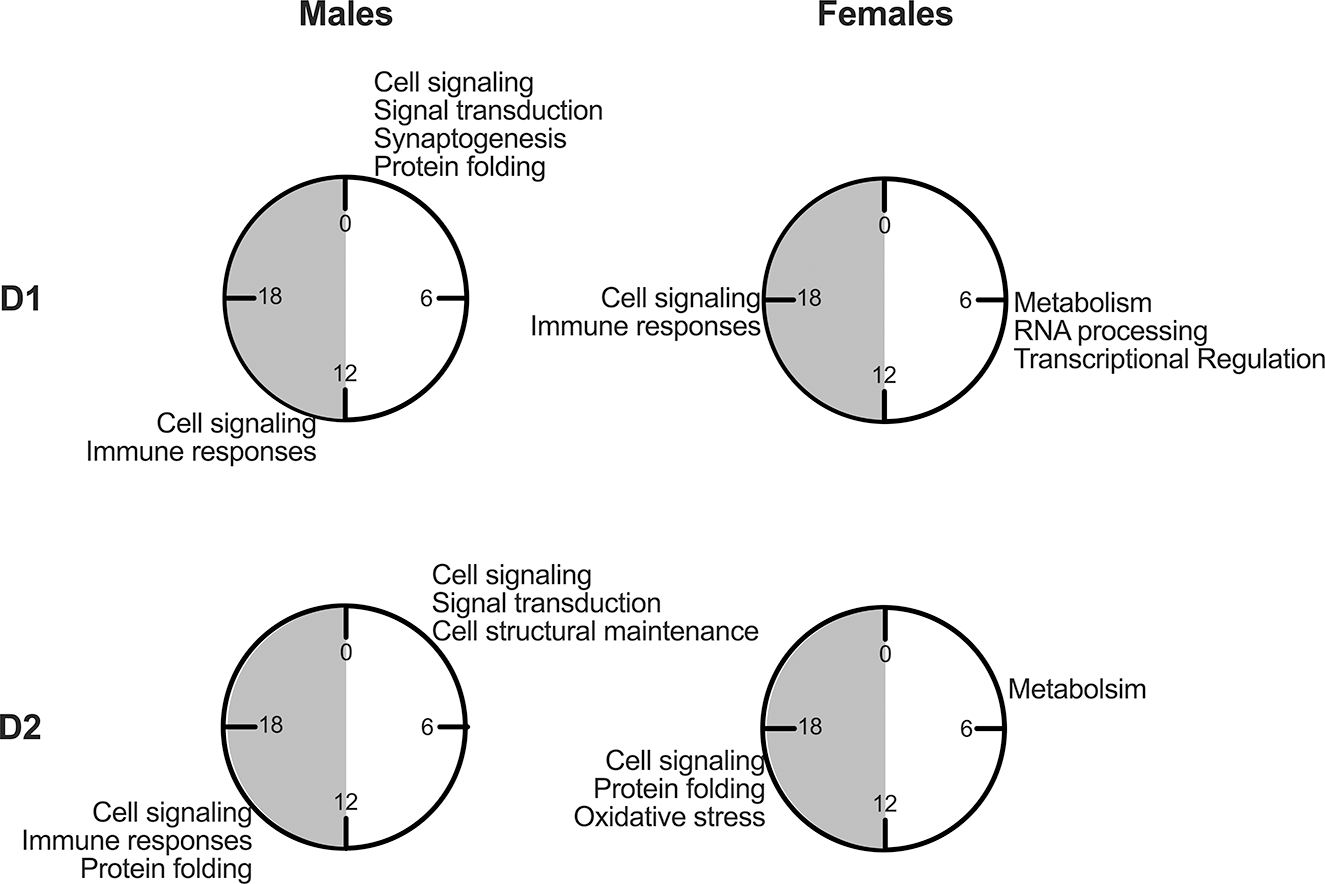
D1 and D2 cells taken from males and females have enriched rhythmic processes identified from transcripts peaking in the light and dark phase. Four clocks are shown, with gray shading indicating the dark phase (ZT12–0). Enriched processes are listed at the time of day in which transcripts peak during the light or dark phase for that cell type and sex.
We propose two main hypotheses for how this type of sex-dependent circadian variation can occur. First, we identified binding sites for transcription factors that are shared among only one sex. Since rhythms in core clock genes and clock-controlled genes (CCGs) are regulated largely through their patterns of transcription, this suggests that differences in binding motifs could yield variability in peak time across sex. Lhx8, MLXIPL and AFT2::JUN may be regulating transcription in males, while EHF, ELF5 and RARA could be regulating transcription in females (Supp.Fig.3). Second, it is possible that Ebox, Dbox or ROR elements, which differentially regulate rhythms in transcription, are more or less prominent in CCGs across groups. These elements are commonly found in the promoter region of CCGs and it has been suggested that phase variability across tissue might be driven by differences in either the presence of these elements or activity of the transcription factors that bind there39. Future studies will examine the various factors that could be altering transcription of CCGs, as well as other possible mechanisms including rhythms in polyadenylation and RNA stability.
When we perform pathway enrichment analyses in males and females, using transcripts that peak during either the light or dark phase, many unique pathways are found in D1 and D2 cells. However, several of the processes these pathways regulate are similarly enriched in either cell type, sex or peak time. MSNs are largely enriched for cell signaling and signal transduction pathways, especially for transcripts peaking during the dark phase. Pathways related to actin cytoskeletal organization, synaptogenesis, RNA processing and transcriptional regulation are also frequently observed. Therefore, MSNs likely contribute to overall rhythms in signaling and neuronal activity in the NAc, which in turn could mediate rhythmic, reward-related behavior. Interestingly, when Drd2 is knocked out, rhythmic transcripts are greatly reduced in NAc homogenate tissue26, further indicating D1 and D2 cells are working in tandem to control rhythms in this region.
Previous evidence from our lab demonstrated that activity in NAc MSNs is highly rhythmic21. Specifically, we observed a diurnal rhythm in glutamatergic transmission and intrinsic excitability, which peaked during the dark phase. Dopaminergic functioning is also known to vary diurnally in the NAc, including dopamine levels, transporter function, tone and receptor expression40–42. In general, neuronal activity is enhanced during the dark phase when rodents are active. These results suggest that MSNs play a crucial role in driving rhythmicity in NAc activity, but other cells, such as cholinergic interneurons, which modulate diurnal rhythms in dopamine release in the NAc43, could as well. Additional studies are needed to evaluate how MSNs and other cell types contribute to rhythmic neuronal activity in the NAc and diurnal rhythms in reward.
The most commonly enriched rhythmic processes that emerged in all cell types were protein regulation. Therefore, rhythms are likely particularly important for protein function in the NAc. This is consistent with previous findings in mouse NAc homogenate samples26 and single-cell sequencing studies of the SCN25. In the central pacemaker, protein folding and related pathways were rhythmic in all cell types investigated, including neurons and astrocytes. Rhythmic transcripts within these pathways peaked during the dark phase, at about circadian time (CT) 18–20. This is consistent with our observation that protein-related pathways peak primarily during the dark phase across cell type in the NAc. Since rhythms in protein folding and processing are aligned across brain region and cell type, rhythms in protein regulation are likely critical to brain activity. These processes are also peaking during the dark, or active phase, in rodents when protein synthesis, folding, modification and degradation would be critical to cell function in the brain, as well as behavior. Protein regulation may be more quiescent during the light phase because animals are asleep.
Unlike MSNs, astrocytes are enriched for pathways related to translation initiation, ribosomal organization and metabolism, which are highly relevant for cellular physiology and health. Similarly, a recent study from our group demonstrated that knocking down rhythms in astrocytes in the NAc (through loss of Arntl) disrupted metabolic homeostasis28. It is also well known that astrocytes regulate glutamate levels and help maintain neuronal homeostasis44–46. Our lab found that a loss of rhythms in NAc astrocytes alters excitatory synaptic transmission onto nearby MSNs28. Taken together, these findings suggest that rhythms in astrocytes mediate neuronal activity, while NAc MSNs produce the signal transduction events that guide behavioral rhythms.
Previous studies in the SCN (the central pacemaker) find that core clock gene expression patterns differ between astrocytes and neurons25. We found that core clock genes peak at roughly the same TOD across cell type in the NAc, but peak times in other genes are about 6 hours later in neurons compared to astrocytes in the SCN. While rhythms in astrocytes peak at approximately the same TOD in the SCN and NAc (within 1–2 hours), rhythms in neurons are more divergent, peaking several hours later in the SCN compared to the NAc25. This suggests the relationship between astrocytes and neurons is different in these regions, especially for circadian regulation. In the SCN, calcium rhythms are anti-phasic in astrocytes and neurons47, therefore astrocytes are more active when neurons are less active. It is hypothesized that astrocytes control neuronal activity in the SCN by regulating extracellular glutamate levels. Interestingly, we also found rhythms in genes that regulate glutamatergic signaling in astrocytes in the NAc. However, while glutamine synthase (Glul) is rhythmic in the SCN25, in the NAc metabotropic glutamate receptors (Grm3 and Grm4) and the glutamate transporter (Slc1a2) are rhythmic. This suggests that while rhythmic glutamatergic synthesis and release from astrocytes suppresses neuronal activity in the SCN, resulting in an anti-phasic relationship47, rhythms in glutamate reuptake in NAc astrocytes might be causing astrocytes to fall into phase with neurons. Future research should further explore the rhythmic relationship between neurons and astrocytes in the NAc. As discussed for sex differences, it is also possible that differences in transcriptional regulation across cell type and brain region could be driving differences in gene expression rhythms. In fact, we identified several motifs that are common to only one cell type in the NAc (Supp.Fig.4): Ebf1 and Myf5 are commonly enriched in D2 cells, Atoh1 and Olig2 in D1 cells and MZF1 and E2F2 in astrocytes. Future experiments could focus on investigating how these binding sites might regulate rhythms across cell types.
Lastly, we found that several classically used cell-type markers, as well as novel markers, are rhythmic in the NAc35. This issue is critical since neuroscience research is focusing more and more on investigating cell-type specific changes in the brain. In the past, marker selection has not considered rhythmicity. However, if markers are rhythmic, results could vary greatly depending on what time the samples are collected since peak and trough times vary for each gene. It could be advantageous if markers peak during the morning, at the TOD an investigator collects tissue, since gene expression will be high, making cell identification easier. However, if markers peak during the dark phase, expression levels could be artificially low and cause an under-representation of that cell type. This issue is particularly problematic because each marker has a diverse peak/trough time. Taken together, using rhythmic markers could lead to under-powered analyses if too few cells are categorized. In future studies, care should be taken to select markers that are not rhythmic in a given cell or brain region.
This study has added greatly to our understanding of how individual cell types contribute to rhythms in the NAc. Although additional studies are needed to investigate other prominent cell types, specifically non-neuronal populations such as oligodendrocytes, microglia and endothelial cells, the majority of cells within the NAc are MSNs and astrocytes (approximately 58% of cells collected)48. Cumulatively, these cell types cover most of the gene expression in the NAc, with other cell types expressing fewer genes and gene counts48. Understanding rhythms at baseline will help us determine how the NAc regulates rhythms in reward and how disruptions to these rhythms might lead to altered behavior. These results have also provided important insight that will help guide marker selection in future experiments. Most importantly, this study indicates that gene expression rhythms in MSNs might play an important role in neuronal activity. Future studies will be imperative to understanding how these rhythms in MSNs or astrocytes drive activity in the NAc and impact diurnal rhythms in reward.
Supplementary Material
Acknowledgements:
We would like to thank Mariah Hildebrand and Ioannis Migias for animal care and genotyping. We thank Dr. Mary Kay Lobo for providing the Ribotag transgenic mice, as well as for technical assistance with co-immunoprecipitations. This project used the University of Pittsburgh Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh (RNA sequencing) and the University of Pittsburgh Center for Research Computing, RRID:SCR_022735, through the resources provided. Specifically, this work used the HTC cluster, which is supported by NIH award number S10OD028483. This work was funded by the National Institutes of Health: DA039865, DA046346, MH111601, MH106460 (PI: Colleen McClung, PhD), DA046117, DA055064, L60DA054665 (PI: Lauren DePoy) and MH128763 (PI: Kyle Ketchesin) as well as the Brain and Behavior Research Foundation (29386, DePoy and 30823 (P&S Fund), Ketchesin).
Footnotes
Disclosures:
All authors have no financial disclosures or conflicts of interest to report.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2016;18:164–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–7. [DOI] [PubMed] [Google Scholar]
- 4.Easton A, Arbuzova J, Turek FW. The circadian Clock mutation increases exploratory activity and escape-seeking behavior. Genes Brain Behav. 2003;2:11–9. [DOI] [PubMed] [Google Scholar]
- 5.Roybal K Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, et al. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet B Neuropsychiatr Genet. 2003;123B:23–6. [DOI] [PubMed] [Google Scholar]
- 7.Benedetti F, Dallaspezia S, Colombo C, Pirovano A, Marino E, Smeraldi E. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci Lett. 2008;445:184–7. [DOI] [PubMed] [Google Scholar]
- 8.Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppä T, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–9. [DOI] [PubMed] [Google Scholar]
- 9.Lavebratt C, Sjöholm LK, Soronen P, Paunio T, Vawter MP, Bunney WE, et al. CRY2 is associated with depression. PLoS One. 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansour HA, Talkowski ME, Wood J, Chowdari KV, McClain L, Prasad K, et al. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. 2009;11:701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi J, Gelernter J, Sun J, Kranzler HR. Comparing the utility of homogeneous subtypes of cocaine use and related behaviors with DSM-IV cocaine dependence as traits for genetic association analysis. Am J Med Genet Part B Neuropsychiatr Genet. 2014;165:148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranger DAA, Ifrah C, Prather AA, Carey CE, Corral-Frías NS, Drabant Conley E, et al. PER1 rs3027172 Genotype Interacts with Early Life Stress to Predict Problematic Alcohol Use, but Not Reward-Related Ventral Striatum Activity. Front Psychol. 2016;7:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blomeyer D, Buchmann AF, Lascorz J, Zimmermann US, Esser G, Desrivieres S, et al. Association of PER2 Genotype and Stressful Life Events with Alcohol Drinking in Young Adults. Le Foll B, editor. PLoS One. 2013;8:e59136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes EE, Dahl RE, Almeida JRC, Ferrell RE, Nimgaonkar VL, Mansour H, et al. PER2 rs2304672 Polymorphism Moderates Circadian-Relevant Reward Circuitry Activity in Adolescents. Biol Psychiatry. 2012;71:451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamsby JJ, Templeton EL, Bonvini LA, Wang W, Loros JJ, Dunlap JC, et al. The circadian Per1 and Per2 genes influence alcohol intake, reinforcement, and blood alcohol levels. Behav Brain Res. 2013;249:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozburn AR, Larson EB, Self DW, McClung CA. Cocaine self-administration behaviors in ClockΔ19 mice. Psychopharmacology (Berl). 2012;223:169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozburn AR, Falcon E, Mukherjee S, Gillman A, Arey R, Spencer S, et al. The Role of Clock in Ethanol-Related Behaviors. Neuropsychopharmacology. 2013;38:2393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong L, Bilbao A, Laucht M, Henriksson R, Yakovleva T, Ridinger M, et al. Effects of the Circadian Rhythm Gene Period 1 ( Per1 ) on Psychosocial Stress-Induced Alcohol Drinking. Am J Psychiatry. 2011;168:1090–8. [DOI] [PubMed] [Google Scholar]
- 19.Wise RA, Rompre PP. Brain Dopamine and Reward. Annu Rev Psychol. 1989;40:191–225. [DOI] [PubMed] [Google Scholar]
- 20.DePoy LM, McClung CA, Logan RW. Neural Mechanisms of Circadian Regulation of Natural and Drug Reward. Neural Plast. 2017;2017:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parekh PK, Becker-Krail D, Sundaravelu P, Ishigaki S, Okado H, Sobue G, et al. Altered GluA1 (Gria1) Function and Accumbal Synaptic Plasticity in the ClockΔ19 Model of Bipolar Mania. Biol Psychiatry. 2018;84:817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePoy LM, Becker-Krail DD, Zong W, Petersen K, Shah NM, Brandon JH, et al. Circadian-Dependent and Sex-Dependent Increases in Intravenous Cocaine Self-Administration in Npas2 Mutant Mice. J Neurosci. 2021;41:1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parekh PK, Logan RW,Ketchesin KD, Becker-Krail D, Shelton MA, Hildebrand MA, et al. Cell-type specific regulation of nucleus accumbens synaptic plasticity and cocaine reward sensitivity by the circadian protein, NPAS2. Cite as J Neurosci. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen S, Ma D, Zhao M, Xie L, Wu Q, Gou L, et al. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci 2020 233. 2020;23:456–67. [DOI] [PubMed] [Google Scholar]
- 26.Brami-Cherrier K, Lewis RG, Cervantes M, Liu Y, Tognini P, Baldi P, et al. Cocaine-mediated circadian reprogramming in the striatum through dopamine D2R and PPARγ activation. Nat Commun. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ketchesin KD, Zong W, Hildebrand MA, Seney ML, Cahill KM, Scott MR, et al. Diurnal rhythms across the human dorsal and ventral striatum. Proc Natl Acad Sci U S A. 2021;118:2016150118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker-Krail DD, Ketchesin KD, Burns JN, Zong W, Hildebrand MA, DePoy, et al. Astrocyte Molecular Clock Function in the Nucleus Accumbens Is Important for Reward-Related Behavior. Biol Psychiatry. 2022;92:68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatcher KM, Royston SE, Mahoney MM. Modulation of circadian rhythms through estrogen receptor signaling. Eur J Neurosci. 2018; [DOI] [PubMed] [Google Scholar]
- 30.Becker JB, Hu M. Sex differences in drug abuse. Front Neuroendocrinol. 2008;29:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra R, Chase Francis T, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, et al. Opposing Role for Egr3 in Nucleus Accumbens Cell Subtypes in Cocaine Action. J Neurosci. 2015;35:7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornelissen G Cosinor-based rhythmometry. Theor Biol Med Model. 2014;11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahill KM Z Huo, GC Tseng, RW Logan, ML Seney. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 2018;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran MN, Maynard KR, Spangler A, Huuki LA, Montgomery KD, Sadashivaiah V, et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron. 2021;109:3088–3103.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosten TA, Gawin FH, Kosten TR, Rounsaville BJ. Gender differences in cocaine use and treatment response. J Subst Abuse Treat. 1993;10:63–6. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy AP, Epstein DH, Phillips KA, Preston KL. Sex differences in cocaine/heroin users: drug-use triggers and craving in daily life. Drug Alcohol Depend. 2013;132:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins SJ, Ehrman RN, Childress AR, O’Brien CP. Comparing levels of cocaine cue reactivity in male and female outpatients. Drug Alcohol Depend. 1999;53:223–30. [DOI] [PubMed] [Google Scholar]
- 39.Mavroudis PD, DuBois DC, Almon RR, Jusko WJ. Modeling circadian variability of core-clock and clock-controlled genes in four tissues of the rat. PLoS One. 2018;13:e0197534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozburn AR, Falcon E, Twaddle A, Nugent AL, Gillman AG, Spencer SM, et al. Direct regulation of diurnal drd3 expression and cocaine reward by npas2. Biol Psychiatry. 2015;77:425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferris MJ, España RA, Locke JL, Konstantopoulos JK, Rose JH, Chen R, et al. Dopamine transporters govern diurnal variation in extracellular dopamine tone. Proc Natl Acad Sci U S A. 2014;111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castañeda TR, Marquez De Prado B, Prieto D, Mora F. Circadian rhythms of dopamine, glutamate and GABA in the striatum and nucleus accumbens of the awake rat: modulation by light. J Pineal Res. 2004;36:177–85. [DOI] [PubMed] [Google Scholar]
- 43.Stowe TA, Pitts EG, Leach AC, Iacino MC, Niere F, Graul B, et al. Diurnal rhythms in cholinergic modulation of rapid dopamine signals and associative learning in the striatum. Cell Rep. 2022;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolaños JP. Bioenergetics and redox adaptations of astrocytes to neuronal activity. J Neurochem. 2016;139:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonçalves CA, Rodrigues L, Bobermin LD, Zanotto C, Vizuete A, Quincozes-Santos A, et al. Glycolysis-Derived Compounds From Astrocytes That Modulate Synaptic Communication. Front Neurosci. 2018;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malarkey EB, Parpura V. Mechanisms of glutamate release from astrocytes. Neurochem Int. 2008;52:142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron. 2017;93:1420–1435.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen R, Blosser TR, Djekidel MN, Hao J, Bhattacherjee A, Chen W, et al. Decoding molecular and cellular heterogeneity of mouse nucleus accumbens. Nat Neurosci. 2021;24:1757–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


