Abstract
Major receptor group common cold virus HRV89 was adapted to grow in HEp-2 cells, which are permissive for minor group human rhinoviruses (HRVs) but which only marginally support growth of major-group viruses. After 32 blind passages in these cells, each alternating with boosts of the recovered virus in HeLa cells, HRV89 acquired the capacity to effectively replicate in HEp-2 cells, attaining virus titers comparable to those in HeLa cells although no cytopathic effect was observed. Several clones were isolated and shown to replicate in HeLa cells whose ICAM-1 was blocked with monoclonal antibody R6.5 and in COS-7 cells, which are devoid of ICAM-1. Blocking experiments with recombinant very-low-density lipoprotein receptor fragments and enzyme-linked immunosorbent assays indicated that the mutants bound a receptor different from that used by minor-group viruses. Determination of the genomic RNA sequence encoding the capsid protein region revealed no changes in amino acid residues at positions equivalent to those involved in the interaction of HRV14 or HRV16 with ICAM-1. One mutation was within the footprint of a very-low-density lipoprotein receptor fragment bound to minor-group virus HRV2. Since ICAM-1 not only functions as a vehicle for cell entry but has also a “catalytic” function in uncoating, the use of other receptors must have important consequences for the entry pathway and demonstrates the plasticity of these viruses.
Human rhinoviruses (HRVs), a major cause of mild upper respiratory infections generally recognized as common colds, are small icosahedral particles with a capsid composed of four viral proteins, VP1 through VP4 (for a review see reference 9). The capsid encases a genomic RNA of about 7,500 nucleotides encoding a polyprotein which is cotranslationally and autocatalytically processed by three viral proteinases, P2A, P3C, and P3CD (the precursor of P3C). A final maturation cleavage of VP0 to VP2 and VP4 occurs concomitantly with encapsidation by an as yet unidentified protease. With one exception (HRV87), the serotypes can be divided into a major group, using intercellular adhesion molecule 1 (ICAM-1) as the viral receptor, and a minor group, attaching to the cell via members of the low-density lipoprotein receptor (LDLR) family including LDLR, the very-low-density lipoprotein receptor (VLDLR), and LDLR-related protein (LRP) (16, 27). The nature of the HRV87 receptor is unknown (50). Whereas major-group viruses are highly specific for human ICAM-1 and fail to attach to the homologue of other species, minor-group viruses bind to a variety of LDLRs, most likely due to the high evolutionary conservation of these membrane proteins. Replication usually does not occur in nonhuman cells even when suitable receptors are present, and adaptation of HRV2 to growth in mouse cells has been shown to be correlated with mutations in nonstructural proteins P2B and P2C (23).
As HRVs of both receptor groups are very similar with respect to the amino acid sequence and the three-dimensional structure of the viral capsid proteins, the basis of receptor choice is only partially understood. HRVs exhibit a cleft encircling the fivefold axes of icosahedral symmetry, called the canyon, which accommodates the N-terminal domain of ICAM-1 in major-group viruses (31, 38). Until recently, it was not clear whether LDLRs would also bind within the canyon although mutagenesis experiments suggested another binding site (10). The recent determination of the three-dimensional structure of minor-group virus HRV2 complexed to a recombinant soluble fragment of VLDLR by electron cryomicroscopy image reconstruction finally showed attachment to the BC and HI loops of VP1, which are close to the fivefold axes of icosahedral symmetry (15). This finding challenges the canyon hypothesis, which states that the receptors interact with conserved residues at the bottom of the canyon, which is hidden from the immune system by being inaccessible to antibodies (37). Amino acid residues are indeed more conserved at the canyon floor than at more-accessible sites (6). As the BC and the HI loops are exposed and vary substantially among minor-group viruses, the basis of receptor interaction is not understood. Only structure determination at high resolution might answer the question of how the 10 known minor-group HRV serotypes attach to the same receptor without appreciable immunological cross-reaction.
One of the best-investigated major-group serotypes is HRV14. However, as this serotype is clearly an outlier, with its genomic nucleotide sequence being quite different from those of all other serotypes sequenced so far, it cannot be considered a prototype. Based on amino acid sequence comparisons, HRV89 is rather closely related to HRV2, a minor-group prototype, with between 62 (VP1) and 94% (VP4) sequence similarity of their capsid proteins (11). Therefore, we chose HRV89 for experiments aimed at investigating whether the receptor specificity can be changed.
HRV89 was subjected to 32 blind passages in HEp-2 cells, which have been shown to lack binding sites for HRV15, another major-group virus (7); the titer was boosted on HeLa cells every other passage. First, the titer of virus recovered from the HEp-2 cells was substantially lower than that obtained in HeLa cells; however, at later passages it approached that from HeLa cells. Clones were isolated from single plaques, and virus neutralization tests with type-specific antisera confirmed that the isolates were HRV89. They grew to high titers in HEp-2 cells, in COS-7 cells, which are devoid of ICAM-1, and in HeLa cells whose ICAM-1 was blocked with monoclonal antibody (MAb) R6.5. The additional presence of a soluble recombinant VLDLR fragment encompassing ligand binding repeats 1 to 6 (VLDLR1–6) (36), which prevents cell damage by minor-group HRVs, was without effect on the HEp-2 cell-adapted HRV89. This suggests that the newly acquired binding capacity is not directed toward LDLRs. This finding was supported by enzyme-linked immunosorbent assays (ELISAs) which revealed no binding of the mutants to VLDLR1–6 immobilized on microtiter plates.
The sequencing of the region encompassing the viral capsid proteins except VP4, which is internal, revealed no changes in those amino acid residues previously identified as implicated in the interaction of HRV14 and HRV16 with ICAM-1. One of the mutations was found within the BC loop, which is involved in the binding of minor-group viruses to LDLRs.
MATERIALS AND METHODS
Viruses and cells.
HRVs were obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and plaque purified twice before use. HEp-2 cells were kindly provided by P. Kronenberger (Brussels, Belgium). HeLa-H1 cells, a strain which supports HRV replication, was obtained from Flow Laboratories; for simplicity, they are termed HeLa throughout. COS-7 cells were from ATCC. HeLa and HEp-2 cells were maintained in minimal essential medium (MEM); COS-7 cells were maintained in Dulbecco's modified MEM containing 10% fetal calf serum (FCS), 2 mM glutamine, and streptomycin and penicillin (100 U/ml each). Infections were carried out in infection medium (IM) consisting of MEM, 2% FCS, and 30 mM MgCl2 supplemented with glutamine and antibiotics as above. Tissue culture media, antibiotics, and FCS were purchased from GIBCO Life Technologies. Type-specific guinea pig antisera against various HRV serotypes were purchased from ATCC. Viral titers were determined by end point dilution tests with HeLa cells.
Adaptation of HRV89 to HEp-2 cells.
HEp-2 cells grown in 75-cm2 tissue culture flasks were challenged with plaque-purified HRV89 at 4 × 107 50% tissue culture infectious doses (TCID50), corresponding to a multiplicity of infection (MOI) of 1 in 5 ml of IM and incubated for 90 min at 34°C. The supernatant was removed, cells were washed with phosphate-buffered saline (PBS), and 10 ml of IM was added. Cells were incubated for 3 days at 34°C whereupon they were broken by three freeze-thaw cycles. The cell lysate was cleared from debris by a low-speed centrifugation and used to infect HeLa cells in 75-cm2 flasks. Cell lysis usually occurred after 2 days; residual cells were broken by freezing and thawing as before. The cell lysate was then used for the next round of selection on HEp-2 cells. In an initial experiment we observed a contamination with HRV2, which must have originated from parallel work with this serotype; the selection process was thus repeated in the continuous presence of rabbit antiserum against HRV2 at a dilution of 1:500.
Serological tests.
The serotypic identity of HEp-2 cell-adapted HRV89 with wild-type (wt) HRV89 was assessed with type-specific antisera. Virus at 1,000 TCID50 was incubated with twofold serial dilutions of type-specific guinea pig antisera for 90 min at 34°C in IM in a final volume of 100 μl starting with a dilution of 1:1,000. The mixtures were then transferred onto HeLa cells grown in 96-well plates. At 2 days postinfection (p.i.) cells were stained with 0.1% crystal violet in water.
Blocking of viral infection with MAb R6.5 or with MBP-VLDLR1–6.
To block ICAM-1 present on the HeLa cell surface, MAb R6.5 (kindly provided by Robert Rothlein, Boehringer Ingelheim, Ridgefield, Conn.) was added at a concentration of 10 μg in 200 μl of IM/well to HeLa cells grown in 96-well plates. Cells were incubated with the antibody for 1 h at 34°C, and virus was added to each well at a MOI of 0.1. Cells were maintained at 34°C and examined for cytopathic effect every 24 h.
To competitively inhibit rhinoviruses attaching to LDLR, virus (at a MOI of 0.1) was incubated with 0.4 mg of a recombinant VLDLR fragment/ml encompassing the first six ligand binding repeats fused to maltose binding protein at the N terminus (MBP-VLDLR1–6 [36]) for 1 h at 34°C. The mixtures were then transferred to cells grown in 96-well plates and incubated for 2 days at 34°C. Cell damage was monitored after staining with crystal violet. To simultaneously prevent virus from binding to ICAM-1 and to LDLRs on the cell surface, experiments were also carried out under conditions where the virus was preincubated with MBP-VLDLR1–6 and ICAM-1 was blocked with MAb R6.5.
Viral neutralization by recombinant soluble ICAM-1.
Neutralization assays were carried out essentially as described earlier (1, 25). Briefly, HRVs at 50 TCID50 were incubated with serial twofold dilutions of recombinant soluble ICAM-1 (a generous gift from Anita Wyne, Boehringer Ingelheim) for 90 min at 34°C and added to HeLa cells grown in 96-well plates. Cytopathic effect was monitored, and the plates were stained with crystal violet as soon as complete lysis was observed in control wells infected in the absence of ICAM-1.
Determination of viral growth kinetics.
Cells were grown in six-well plates and infected at 4 × 107 TCID50/well for 90 min at 34°C in a total volume of 1 ml of IM. The supernatant was removed, cells were washed with PBS, and 3 ml of IM was added. After incubation at 34°C for the time periods indicated in the tables, intracellular virus was released by three freeze-thaw cycles, and the viral titer was determined by end point dilution tests.
FACS analysis.
Cells were dislodged with PBS containing 1 mM EDTA and incubated with anti-ICAM-1 MAb R6.5 at a final concentration of 10 μg/ml in PBS containing 0.1% bovine serum albumin (incubation buffer) for 1 h at 4°C with gentle agitation. After being washed three times with incubation buffer, cells were incubated with a Cy5-conjugated goat anti-mouse antibody (Zymed Laboratories Inc., San Francisco, Calif.) at a dilution of 1:3,000 in incubation buffer for 1 h at 4°C. Fluorescence-activated cell sorting (FACS) analyses were performed on a FACSCalibur (Becton Dickinson). Histogram plots were made with CellQuest.
Receptor binding assays.
Recombinant soluble ICAM-1 was used to coat microtiter plates at 2 μg/ml in PBS for 1 h at 37°C. The plates were blocked with 2% bovine serum albumin in PBS (BS) for 1 h at 37°C. Wells were then incubated with 100 μl of virus at 107 TCID50/ml in TBSC, washed with BS, and incubated with the respective guinea pig antivirus antiserum diluted 1:1,000 for 1 h at room temperature. After being washed with BS, wells were incubated with goat anti-guinea pig horseradish peroxidase-conjugated immunoglobulin G (Rockland, Gilbertsville, Pa.) at a dilution of 1:10,000. Bound virus was then revealed with 100 μl of a solution consisting of 100 μg of trimethylbenzidine/ml and 0.03% H2O2 in 100 mM sodium acetate (pH 6). The reaction was halted with 50 μl of 1 M H2SO4, and the A450 was determined with a plate reader.
Ligand blotting using radioactively labeled HRV2 was carried out as described in references 26 and 27; ligand blotting with type-specific guinea pig antiserum against HRV89, followed by peroxidase-conjugated anti-guinea pig antiserum and chemiluminescence substrate (Pierce), was performed as described for HRV2 (28).
Cloning and sequencing.
RNA was extracted from the supernatant of infected HeLa cells with Trizol (Gibco BRL, Gaithersburg, Md.) and precipitated, and aliquots were used for reverse transcription-PCR (RT-PCR) with forward primer CCGCTCGAGCGGTCACCAACAGTTGAAGCTTGTGG, hybridizing to positions 826 to 848 covering the first eight amino acids of VP2 (and containing an XhoI site), and reverse primer CGGGATCCCGTTGCTCCTCAGCACACTGGAATTTT, hybridizing to positions 3618 to 3642 located within the gene encoding proteinase 2A (with an added BamHI site; restriction sites are in boldface). Following denaturation at 60°C for 5 min, first-strand cDNA synthesis was performed with the reverse primer and 200 U of Moloney murine leukemia virus reverse transcriptase (RNase H Minus, Point Mutant; Promega, Madison, Wis.) in a total reaction volume of 20 μl for 1 h at 42°C in a Robocycler Gradient 40 (Stratagene, La Jolla, Calif.). PCR was performed using both primers with a mixture of 4 U of DyNAzyme Taq polymerase (Finnzymes, Espoo, Finland) and 0.1 U of Pfu polymerase (Promega) for enhanced proofreading activity (3, 22). The reaction was carried out in a total volume of 50 μl after an initial incubation for 2 min at 95°C with 35 cycles of 45 s at 95°C, 45 s at 65°C, and 3 min at 72°C. A final extension step was at 72°C for 10 min, and the products were analyzed on an agarose gel. RT-PCR resulted in a single band of 2,839 bp, as expected from the primer sequences used.
The fragments were isolated from the gel using the Qiaquick gel extraction kit (Qiagen) and used for cloning into the pGEM-T vector (Promega) after generation of a 3′-A overhang. Plasmids containing the insert were purified and analyzed by sequencing with pUC/M13 forward and reverse standard primers (IMP Sequencing Service, Vienna, Austria). Sequencing was continued with the following primers (positions are in parentheses): sense (1526 to 1542), AACCTGGGGGGACACAA; antisense (2892 to 2912), ATGCTCATAAAAGGGATTGTG; sense (2232 to 2251), TACTCCAGATAACGCCAAAA; antisense (2083 to 2102), AACCCCACATCCCACACTAA; sense (2875 to 2894), CAACCATACCCCAGATTCAC; antisense (1501 to 1519), GAAGAGGGCATATTGGGAT. One clone of each mutant virus was sequenced on both strands. The sequences thus covered all but the first eight amino acids of VP2, which are predetermined by the primer used for amplification.
RESULTS
Replication of a major-group virus in HEp-2 cells.
HEp-2 cells have originally been shown to possess no binding sites for HRV14 and only a few binding sites for a receptor-specific MAb (7). On the other hand, HEp-2 cells express minor-group receptors at levels similar to those for HeLa cells, and replication of HRV2 in these cells has been demonstrated earlier (24). Nevertheless, we first determined which viral serotypes resulted in cell damage after infection. HEp-2 cells were challenged with minor-group viruses HRV1A, -2, -30, -47, and -62 and major-group viruses HRV14, -16, and -89 at a MOI of 1 and maintained at 34°C. As expected, cells infected with the minor-group HRVs were lysed 2 days p.i., whereas the cells challenged with major-group HRVs appeared healthy even at 3 days p.i. (data not shown). Using lysates prepared at different time points p.i. the replication kinetics for HRV2 and HRV89 was also determined. Unexpectedly, this revealed that HRV89 also replicated in HEp-2 cells; however, the titer was substantially lower and only upon much longer incubation times approached values about 1 log unit lower than that seen in HRV2 (Table 1).
TABLE 1.
Replication of a minor-group and a major-group virus in two cell linesa
| Virus | Titer at indicated h p.i. in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HeLa cells
|
HEp-2 cells
|
|||||||
| 0 | 24 | 48 | 72 | 0 | 24 | 48 | 72 | |
| HRV2 | 2.7 × 104 | 1.6 × 107 | 5.7 × 105 | 8.5 × 103 | 2.3 × 104 | 2.8 × 107 | 8.5 × 105 | 4.6 × 104 |
| HRV89 | 2.0 × 104 | 2.9 × 106 | 3.1 × 105 | 2.5 × 104 | 5.6 × 103 | 8.5 × 104 | 4.1 × 105 | 1.2 × 106 |
Cells were challenged with HRVs at a MOI of 1, and the viral titer was determined after the times specified. Underlined values indicate that the cells had lysed.
Adaptation of HRV89 to grow in HEp-2 cells.
Apparently, at a MOI of 1, HRV89 (and probably other major-group HRVs) grows in HEp-2 cells to a titer similar to that in HeLa cells (Table 1). However, at a lower MOI it replicates more slowly and only to a lower titer (see below). This is most probably due to the low number of ICAM-1 molecules on the cell surface providing only inefficient internalization. For the reasons outlined above, an attempt to adapt the major-group virus HRV89 to use a receptor different from ICAM-1 for cell entry was made. HEp-2 cells grown in 75-cm2 tissue culture flasks were challenged with plaque-purified HRV89 at a MOI of 1. After 1.5 h, the supernatant was removed and cells were washed and maintained in 10 ml of fresh IM for 48 h at 34°C. Cells were broken by three freeze-thaw cycles, and cell lysate (including cell supernatant) was cleared from debris and used to infect HeLa cells in a 75-cm2 flask. Whereas in no case was any cytopathic effect seen in HEp-2 cells, HeLa cells usually lysed after 2 days. Following 32 blind passages in HEp-2 cells, each followed by a boost in HeLa cells, replication of wt virus was compared with that of the virus serially passaged in HEp-2 cells. Upon infection at low MOI, the isolate grew to a substantially higher titer than wt virus in HEp-2 cells whereas the titer was reduced by about 1 log unit in HeLa cells compared to that for wt HRV89 (Table 2). It is noteworthy that the HEp-2 cell-adapted HRV89 isolate reproducibly lysed the HeLa cells more rapidly, i.e., at about 1 day p.i., than wt virus, which usually caused cell destruction at 2 days p.i.
TABLE 2.
Growth of adapted versus wt HRV89 in HEp-2 cellsa
| Virus | HeLa
|
HEp-2
|
||
|---|---|---|---|---|
| TCID50/ml | Lysis afterc: | TCID50/ml | Lysis after: | |
| HEp-2 cell-adapted HRV89b | 2 × 106 | 24 | 2 × 105 | No lysis |
| wt HRV89 | 2 × 107 | 48 | 2 × 103 | No lysis |
Cells grown in 75-cm2 flasks were infected with 2 × 104 TCID50 (MOI, 0.01), and virus titer was determined at 48 h p.i. HEp-2 cells did not show any obvious cytopathic effect.
Virus recovered after the 32nd round of adaptation was used without cloning.
Values are hours p.i.
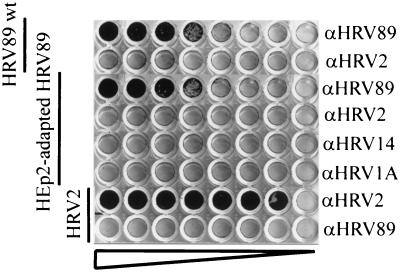
To verify that the HEp-2 cell-adapted virus was still serotypically identical to HRV89, adapted virus and wt virus were incubated with decreasing concentrations of guinea pig antisera directed against HRV1A, HRV2, HRV14, and HRV89 and the mixtures were applied to HeLa cell monolayers grown in 96-well plates (Fig. 1). With the exception of those samples which contained HRV89 antiserum, cells were lysed in all wells regardless of the presence of antiserum against other serotypes. HRV2 used as a control was neutralized by the corresponding antiserum. From this we conclude that the virus variants isolated upon adaptation were indeed derived from HRV89.
FIG. 1.
HRV89 variants recovered after 32 adaptation cycles in HEp-2 cells are serotypically identical to wt virus. HEp-2 cell-adapted virus and wt virus were incubated for 90 min at 34°C with serial twofold dilutions (from left to right) of the serotype-specific antisera indicated. The mixtures were then transferred onto HeLa cell monolayers in 96-well plates. Tissue damage was monitored after 2 days at 34°C by crystal violet staining.
Is infection by HEp-2 cell-adapted HRV89 independent of ICAM-1?
To investigate whether HEp-2 cell-adapted HRV89 was indeed capable of infecting cells in the absence of its natural receptor, ICAM-1, HeLa cells grown in microtiter plates were preincubated with 50 μg of MAb R6.5/ml and challenged with the HEp-2 cell-adapted isolate at a MOI of 0.1. MAb R6.5 has been shown to bind to or bind close to the viral binding site and thus competitively blocks viral attachment to ICAM-1 (44). While challenge of the cells with HEp-2 cell-adapted HRV89 at a MOI of 0.1 led to cell lysis after 2 days, no cytopathic effect was seen upon infection with wt virus even at a MOI of 10. For control purposes, cells were also infected with HRV2 in the presence and absence of MAb R6.5. Cells were lysed regardless of the presence of the antibody, indicating that inhibition of wt HRV89 infection was specific (Table 3).
TABLE 3.
Minor-group receptors are not used by HEp-2 cell-adapted HRV89
| Preincubation protein(s)a | Cell lysis upon infection with:
|
||
|---|---|---|---|
| wt HRV89 | HEp-2 cell-adapted HRV89 | HRV2 | |
| R6.5 | − | + | + |
| MBP-VLDLR1–6 | + | + | − |
| MBP-VLDLR1–6 + R6.5 | − | + | − |
HeLa cells were preincubated with MAb R6.5, and/or virus was preincubated with MBP-VLDLR1–6, before addition to the cells.
Does HEp-2 cell-adapted HRV89 use minor-group receptors for cell entry?
Infection of HeLa cells by minor-group viruses is prevented by the presence of recombinant soluble fragments of LDLR or VLDLR (26, 28). We thus asked whether infection of HeLa cells by HEp-2 cell-adapted HRV89 was inhibited by these receptor fragments. wt HRV89 and HEp-2 cell-adapted HRV89 were preincubated with MBP-VLDLR1–6 (36), a receptor fragment fused to maltose binding protein at its N terminus and encompassing the six N-terminal ligand binding repeats but lacking the epidermal growth factor precursor domain, the transmembrane region, and the cytoplasmic tail of the protein (for a review of the structure of the LDLR family, see references 13 and 45). The mixture was then added to HeLa cells grown in microtiter wells and incubated for 2 days at 34°C. Challenge of HeLa cells with wt and HEp-2 cell-adapted HRV89 resulted in cell lysis. Control infections with HRV2 carried out in parallel showed that infection of this minor-group serotype was inhibited by the receptor fragment. Similar experiments were also carried out with virus that was incubated with MBP-VLDLR1–6 and subsequently added to cells having their ICAM-1 blocked with MAb R6.5 (results are summarized in Table 3). The lack of cell protection from HEp-2 cell-adapted HRV89 infection suggests that this virus uses a receptor different from ICAM-1 and from LDLRs. This was confirmed by ELISAs (see below).
Single clones of HEp-2 cell-adapted HRV89 replicate in COS-7 cells.
To determine the basis of the de novo-acquired receptor specificity after the 32nd cycle of adaptation, single clones of the HEp-2 cell-adapted variant were isolated from plaques. In total, 80 plaques were picked, and from five clones selected at random virus stocks were grown in HeLa cells. These proved to be serotypically identical to wt HRV89 (data not shown).
As shown previously (41) HEp-2 cells are not completely devoid of ICAM-1, as seen from FACS analysis (data not shown); radioactive coxsackievirus A21, which also uses this receptor for cell entry, has been shown to bind to these cells to a small extent (41). In contrast, COS-7 cells do not express any ICAM-1 at all and have been used in the identification of the major-group rhinovirus receptor by demonstrating virus binding upon transfection with human ICAM-1 cDNA (44). HEp-2 cell-adapted virus, if capable of infecting cells independently of the presence of ICAM-1, should thus also infect COS-7 cells. These cells were challenged with the different isolates at a MOI of 0.1. Whereas challenge with wt virus was without effect on the cells, all HEp-2 cell-adapted HRV89 clones lysed the cells after 48 h and gave rise to an (small) increase in viral titer whereas wt virus did not replicate at all (Table 4). It is noteworthy that the viral titer measured at time zero (after removal of virus remaining in the supernatant following the attachment period of 90 min) was much lower for wt virus. This most probably reflects the failure of the virus to attach to the cells, whereas the adapted virus bound efficiently.
TABLE 4.
Replication of single clones of HEp-2 cell-adapted HRV89 in COS-7 cellsa
| Virus | Titers of HRV89 variants grown in COS-7 cells (TCID50/ml) at (h p.i.):
|
|||
|---|---|---|---|---|
| 0 | 24 | 48 | 72 | |
| wt | 5.4 × 102 | 2.2 × 102 | 4.6 × 101 | 0 |
| Clone 15 | 6.5 × 105 | 4.6 × 106 | 1.8 × 107 | 5.6 × 105 |
| Clone 27 | 2.4 × 105 | 1.2 × 107 | 1.9 × 107 | 2.2 × 106 |
| Clone 34 | 1.2 × 106 | 2.8 × 107 | 5.3 × 106 | 8.5 × 105 |
| Clone 44 | 1.4 × 106 | 5.6 × 106 | 4.6 × 106 | 2.8 × 106 |
| Clone 79 | 1.9 × 105 | 8.5 × 106 | 2.0 × 107 | 2.1 × 106 |
Cells grown in six-well plates were challenged with 107 TCID50 (MOI, 1) of HRV89 for 90 min and washed. The medium was replaced, and viral titer was determined at the indicated times p.i. With the exception of the cells infected with wt virus, all cells were lysed at 48 h (underlined).
HEp-2 cell-adapted HRV89 variants still bind to, and are neutralized by, soluble recombinant ICAM-1.
To investigate whether acquiring a new receptor specificity is accompanied by the loss of the capability to attach to ICAM-1, soluble recombinant ICAM-1 was used to coat a microtiter plate and the binding of the HRVs was determined by an assay similar to the ELISA described by Last-Barney and colleagues (21). All isolates exhibited ICAM-1 binding very similar to that of wt virus (data not shown).
Soluble ICAM-1 has been shown to neutralize major-group viruses by competition with the receptor present at the cell surface and also by inducing uncoating (14, 17). We therefore asked whether HEp-2 cell-adapted HRV89 would still be neutralized by the soluble receptor. Virus was incubated with decreasing concentrations of soluble ICAM-1 at 34°C for 90 min, whereupon HeLa cells grown in 96-well plates were challenged with the mixtures. Upon completion of lysis in those wells having received virus alone, cells were stained with crystal violet. As seen in Fig. 2, HeLa cells were protected against infection with all clones of the HEp-2 cell-adapted HRV89 in a concentration-dependent manner. Unexpectedly, no protection against wt virus infection was seen under these particular conditions. This indicates that the sensitivity to ICAM-1 neutralization was substantially increased by the adaptation. It might reflect a decreased stability of the HEp-2 cell-adapted isolates.
FIG. 2.
Adaptation to growth in HEp-2 cells alters the sensitivity of HRV89 to ICAM-1. Virus was incubated with twofold serial dilutions of soluble ICAM-1 (left to right, starting with 25 μg/ml) for 90 min at 34°C. HeLa cells in 96-well plates were then challenged with the mixtures, and cytopathic effect was revealed by staining with crystal violet after 3 days. Numbers refer to individual clones.
What receptor is being used by HEp-2 cell-adapted HRV89?
Provided that no reducing agent is used for polyacrylamide gel electrophoresis (PAGE) and for the transfer of the proteins to polyvinylidene difluoride (PVDF) membranes, ligand blotting with radioactively labeled minor-group viruses reveals binding to LDLR, VLDLR, and LRP (16, 27). We wondered whether the adapted virus would be able to recognize its novel receptor on ligand blots. Cell membranes were prepared from HeLa cells and from HEp-2 cells, and proteins were solubilized in sample buffer without reducing agent at room temperature and separated by PAGE. After electrophoretic transfer to a PVDF membrane, virus binding proteins were eventually revealed by incubation with 35S-labeled HRV2, wt HRV89, and the HEp-2 cell-adapted mutant clones. HRV2 was detected by exposure to X-ray film, whereas detection of HRV89 was attempted using type-specific guinea pig antiserum, horseradish peroxidase-conjugated anti-guinea pig immunoglobulin G and chemiluminescent substrate; this method has been used previously for the detection of HRV2 on ligand blots (28). Whereas HRV2 bound to LRP and to LDLR in both HeLa and HEp-2 cell membrane extracts, no binding was seen for wt HRV89 and the HEp-2 cell-adapted isolates. As expected from the finding that HRV14 binds only weakly to cell membrane extracts in a radioimmunoassay (47), binding to soluble recombinant ICAM-1 run in parallel was not seen either (data not shown). This indicates that the novel cellular receptor recognized by the HEp-2 cell-adapted HRV89 isolates might be inactivated similarly to ICAM-1 by the procedure used or that the sensitivity of the assay was too low for detection of binding.
Position of the mutations in HEp-2 cell-adapted HRV89.
Three clones of the HEp-2 cell-adapted HRV89 were selected for sequence analysis. RNA was extracted from infected-cell supernatants, reverse transcribed, and subjected to PCR amplification using specific primers. The viral cDNAs were then cloned and sequenced. For control purposes wt HRV89 was subjected to the same procedure. As seen in Table 5, two mutations were identical in the wt and mutants with respect to the published sequence (11). It is likely that they are due to sequencing errors in the original sequence or to changes introduced during passaging; these were not further considered. In addition, the three isolates sequenced exhibited several changes compared to the wt virus. These were scattered over all three capsid proteins. Four of these mutations were identical in the three isolates and thus might be significant for the common phenotype. The presence of additional mutations indicates that the isolates were indeed not derived from the same clone.
TABLE 5.
Positions of mutations in HEp-2 cell-adapted HRV89
| Capsid protein | Positiona | Mutationb in wt virus or indicated clone
|
|||
|---|---|---|---|---|---|
| wt | 15 | 27 | 79 | ||
| VP1 | I | 1023S:N | |||
| BC loop (NImla, S) | 1087D:N | 1087D:N | 1087D:N | ||
| αB (I) | 1169T:S | 1169T:S | |||
| I | 1215V:A | ||||
| I | 1235S:L | 1235S:L | 1235S:L | 1235S:L | |
| S | 1272S:F | ||||
| VP2 | βA1 (I) | 2014I:L | 2014I:L | 2014I:L | |
| Between βA1 and βA2 (I) | 2020D:N | 2020D:N | 2020D:N | ||
| I | 2028T:I | 2028T:I | 2028T:I | 2028T:I | |
| I | 2043T:A | ||||
| I | 2213D:G | ||||
| VP3 | Between βC and αA (I) | 3089I:T | |||
| Start of βF (I) | 3150G:S | 3150G:S | 3150G:S | ||
Position within particular structures (38); amino acids exposed at the surface (S) or internally (I) are indicated.
Amino acids in the wt different from the published sequence of HRV89 (11) are underlined. Mutations identical in all three clones are in boldface.
A model of the three-dimensional structure of HRV89 was then built automatically using Swiss-Model (http://www.expasy.ch/swissmod/SWISS-MODEL.html) based on the structures of HRV1A and HRV16 as solved by X-ray crystallography (19, 30), and the positions of the mutations were examined (Fig. 3). According to this model, the only strongly solvent-exposed mutated amino acid residue is 1087D:N; it forms part of neutralizing immunogen NIm1b (the first digit denotes the capsid protein, i.e., VP1 in this case) (43). The other mutations identical in the three clones were found in VP2 (2014I:L and 2020D:N) and in VP3 (3150G:S). These are at the pentamer-pentamer interface and might be involved in subunit interactions and could thereby influence the stability of the capsid. These residues are not exposed, and it is thus unlikely that they participate directly in receptor interaction. Inspection of the other mutations present only in individual clones revealed one other exposed amino acid (1272S:F), which is located at the south wall of the canyon. All other amino acids found to be mutated in the isolates were mostly hidden from solvent and located within the viral capsid. It is therefore unlikely that they are involved in the acquisition of the novel receptor specificity.
FIG. 3.
Stereo image of a ribbon model of VP1, VP2, and VP3 (blue, green, and red, respectively) of HRV89 as calculated with Swiss-Model. The positions of the mutations are represented in space-filling mode. Red, mutations identical in all three clones; blue, mutations present only in individual clones (compare to Table 5). (Top) View down the z axis; (bottom) view perpendicular to the z axis. Only some of the mutations are labeled. The fivefold axis of icosahedral symmetry is indicated. The canyon is clearly visible between 1087D and 1272S. Note that the loop between βF and βG in VP3 could not be modeled and thus is not closed. The figure was made with Swiss-Pdb-Viewer, version 3.5b1.
DISCUSSION
Experiments with radiolabeled HRV14 and receptor-specific antibodies have previously demonstrated the virtual absence of virus binding sites on HEp-2 cells (7). Using the more sensitive technique of FACS, Shafren and colleagues later showed that a low number of ICAM-1 molecules were present on these cells; nevertheless HEp-2 cells failed to bind more virus than HeLa cells whose ICAM-1 was blocked with specific MAb WEHI (41). Furthermore, HRV14 did not form plaques on these cells (46). This agrees well with our FACS analysis (data not shown) and our finding of the absence of a cytopathic effect. Nevertheless, it was unexpected to find slow, but clearly detectable HRV89 replication in these cells in the absence of any noticeable cytopathic effect (Table 1). This suggests that a minimum number of viral receptors might be required for highly productive infection to occur.
HRV89 was passaged 32 times in HEp-2 cells; the low virus titer obtained was always boosted in HeLa cells. This procedure resulted in the isolation of variants which were able to grow in HEp-2 cells with great efficiency although still no apparent cytopathic effect was evident (Table 2). The variants were neutralized by HRV89-specific antiserum but were not dependent on the presence of ICAM-1, as they replicated in HeLa cells preincubated with blocking MAb R6.5 (Table 3). Five clones derived from this population of HEp-2 cell-adapted HRV89 were isolated. Again, their serotypic identity with HRV89 was confirmed. They were all able to replicate in COS-7 cells, which are devoid of ICAM-1 (Table 4).
Minor-group viruses have been shown to use the LDLR and LRP for cell entry. These receptors are structurally closely related and have various numbers of ligand binding repeats with very similar amino acid sequences. On the other hand, picornaviruses able to use unrelated receptors have also been described. Coxsackie A9 virus, an enterovirus, enters cells via the vitronectin receptor, the αvβ3 integrin, but can also infect cells which do not express these proteins (34, 35, 48, 49). This second receptor might be the β2 microglobulin (48). Coxsackie B3 virus utilizes the coxsackie-adenovirus receptor as well as the decay-accelerating factor (42). Aphthoviruses enter their host cells via the vitronectin receptor (5, 12, 29), the related integrin αvβ6, via heparan sulfate proteoglycan (18) and another so far uncharacterized membrane receptor (2). It does therefore not come as a complete surprise to find that a major group HRV can be adapted to use another receptor for cell entry as well and has acquired a dual receptor specificity. However, ICAM-1 not only functions as an attachment protein but also has a “catalytic” function in destabilizing the viral capsid, thereby promoting RNA release (14). Whereas the uncoating of minor-group virus HRV2 is completely dependent on the low pH prevailing in endocytic vesicles, HRV14 can infect cells under conditions in which vesicular acidification is blocked with v-ATPase inhibitor bafilomycin A1, although at reduced efficiency (4, 32, 33, 40). We have not yet explored whether the uncoating of wt HRV89 or of the HEp-2 cell-adapted clones is dependent on the low-pH environment. Adaptation of HRV89 to access the cell via a receptor different from ICAM-1 should go along with its loss of dependency on the destabilizing function of ICAM-1. The mutants are all neutralized by soluble ICAM-1, whereas no neutralization could be seen for wt HRV89 and HRV14 under the particular conditions used in the assay. HRV16 was only neutralized at the highest concentration (Fig. 2). This suggests that the mutations indeed decrease the stability of the viral capsid, which might facilitate uncoating in the absence of ICAM-1 as well. No data on the neutralization efficiency of ICAM-1 toward HRV89 are available; however, HRV14 was found to require substantial concentrations for efficient neutralization when assayed on HeLa cells (50% effective concentration [EC50], >32 μg/ml), whereas HRV16 was readily neutralized (EC50, 2.6 μg/ml) (1). In our assay a higher ICAM-1 concentration was required to see neutralization of HRV16 than was required in the experiments of Arruda and colleagues (1); this is probably due to differences in the content of active receptor in the preparations. Experiments to investigate the stability of the mutants and their eventual requirement for low endosomal pH are currently being carried out.
The sequencing of the RNA encoding the entire regions of capsid proteins VP1 through VP3 (VP4 is not exposed to the viral surface and thus is not expected to be involved in attachment) showed that only one surface-exposed amino acid was mutated in all three isolates (Table 5). According to a model of the three-dimensional structure of HRV89, aspartic acid 1087, which was changed to asparagine, takes part in NIm1b and lies within the footprint of a recombinant VLDLR fragment bound to HRV2 (15). The other exposed amino acid (serine 1272, changed to phenylalanine in isolate 79) is at the south wall of the canyon (8, 20, 31, 39). This change is expected to have substantial effects, as a hydrophobic amino acid becomes exposed to solvent. Other changes are inside the capsid, with some lying at the pentamer interfaces (Fig. 3). Although residue 1087 is in the BC loop, the mutants were not neutralized by recombinant soluble VLDLR. No changes within the HI loop of VP1 were seen (15).
A mixture of Taq polymerase with a thermostable polymerase with proofreading activity (such as Pfu) results in the higher fidelity of the amplification process. Nevertheless, we cannot exclude with certainty the possibility that any of the amino acid changes only present within single clones was introduced during DNA amplification, although this is unlikely at error rates estimated to be about 1 per 105 bp for 12 effective cycles (3).
Comparison with the sequences of other serotypes revealed that some of the mutations changed amino acid residues which are strictly conserved throughout all HRVs whose sequences are known. This applies for changes 2213D:G, 3150G:S, 1023S:N, and 1169T:S and suggests a strong influence on the properties of the mutants.
Knowledge of the nature of the novel receptor(s) might give hints as to how the mutant viruses attach to this molecule despite the marginal changes at their surfaces. Ongoing studies will also reveal whether the increase in sensitivity to neutralization by soluble ICAM-1 reflects a general decrease in stability, which might be required to overcome the absence of the catalytic function of this protein, which is necessary for uncoating. Our findings demonstrate the great plasticity of rhinoviruses. A major-group rhinovirus, which appeared to be dependent on the catalytic function of its receptor for the destabilization of the capsid during the uncoating reaction, can thus easily adapt to use another receptor for cell entry. Further work will show whether this novel receptor also has a function in uncoating or rather acts only as a vehicle for cell entry, similar to the minor-group virus receptors.
ACKNOWLEDGMENTS
All authors contributed equally to this work.
We thank Robert Rothlein and Anita Wayne for the generous gifts of soluble recombinant ICAM-1 and MAb R6.5 and Peter Kronenberger for the HEp-2 cells.
This work was supported by Austrian Science Foundation grant P-12189.
REFERENCES
- 1.Arruda E, Crump C E, Marlin S D, Merluzzi V J, Hayden F G. In vitro studies of the antirhinovirus activity of soluble intercellular adhesion molecule-1 Antimicrob. Agents Chemother. 1992;36:1186–1191. doi: 10.1128/aac.36.6.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranowski E, Ruiz Jarabo C M, Sevilla N, Andreu D, Beck E, Domingo E. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J Virol. 2000;74:1641–1647. doi: 10.1128/jvi.74.4.1641-1647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer N, Prchla E, Schwab M, Blaas D, Fuchs R. Human rhinovirus HRV14 uncoats from early endosomes in the presence of bafilomycin. FEBS Lett. 1999;463:175–178. doi: 10.1016/s0014-5793(99)01610-5. [DOI] [PubMed] [Google Scholar]
- 5.Berinstein A, Roivainen M, Hovi T, Mason P W, Baxt B. Antibodies to the vitronectin receptor (integrin αVβ3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman M S, Rossmann M G. Comparison of surface properties of picornaviruses: strategies for hiding the receptor site from immune surveillance. Virology. 1993;195:745–756. doi: 10.1006/viro.1993.1425. [DOI] [PubMed] [Google Scholar]
- 7.Colonno R J, Callahan P L, Long W J. Isolation of a monoclonal antibody that blocks attachment of the major group of human rhinoviruses. J Virol. 1986;57:7–12. doi: 10.1128/jvi.57.1.7-12.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colonno R J, Condra J H, Mizutani S, Callahan P L, Davies M E, Murcko M A. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc Natl Acad Sci USA. 1988;85:5449–5453. doi: 10.1073/pnas.85.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couch R B. Rhinoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 713–734. [Google Scholar]
- 10.Duechler M, Ketter S, Skern T, Kuechler E, Blaas D. Rhinoviral receptor discrimination: mutational changes in the canyon regions of human rhinovirus types 2 and 14 indicate a different site of interaction. J Gen Virol. 1993;74:2287–2291. doi: 10.1099/0022-1317-74-10-2287. [DOI] [PubMed] [Google Scholar]
- 11.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA. 1987;84:2605–2609. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox G, Parry N R, Barnett P V, McGinn B, Rowlands D J, Brown F. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid) J Gen Virol. 1989;70:625–637. doi: 10.1099/0022-1317-70-3-625. [DOI] [PubMed] [Google Scholar]
- 13.Gliemann J. Receptors of the low density lipoprotein (LDL) receptor family in humans. Multiple functions of the large family members via interaction with complex ligands. Biol Chem. 1998;379:951–964. [PubMed] [Google Scholar]
- 14.Greve J M, Forte C P, Marlor C W, Meyer A M, Hooverlitty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewat E A, Neumann E, Conway J F, Moser R, Ronacher B, Marlovits T C, Blaas D. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 2000;19:6317–6325. doi: 10.1093/emboj/19.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooverlitty H, Greve J M. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson T, Ellard F M, Ghazaleh R A, Brookes S M, Blakemore W E, Corteyn A H, Stuart D I, Newman J W, King A M Q. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Smith T J, Chapman M S, Rossmann M G, Pevear D C, Dutko F J, Felock P J, Diana G D, McKinlay M A. Crystal structure of human rhinovirus serotype-1A (Hrv1A) J Mol Biol. 1989;210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 20.Kolatkar P R, Bella J, Olson N H, Bator C M, Baker T S, Rossmann M G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Last-Barney K, Marlin S D, Mcnally E J, Cahill C, Jeanfavre D, Faanes R B, Merluzzi V J. Detection of major group rhinoviruses by soluble intercellular adhesion molecule-1 (sICAM-1) J Virol Methods. 1991;35:255–264. doi: 10.1016/0166-0934(91)90067-a. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg A M, Polacek C, Johansson S. Amplification and cloning of complete enterovirus genomes by long distance PCR. J Virol Methods. 1997;65:191–199. doi: 10.1016/s0166-0934(97)02178-2. [DOI] [PubMed] [Google Scholar]
- 23.Lomax N B, Yin F H. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J Virol. 1989;63:2396–2399. doi: 10.1128/jvi.63.5.2396-2399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madshus I H, Sandvig K, Olsnes S, van Deurs B. Effect of reduced endocytosis induced by hypotonic shock and potassium depletion on the infection of Hep 2 cells by picornaviruses. J Cell Physiol. 1987;131:14–22. doi: 10.1002/jcp.1041310104. [DOI] [PubMed] [Google Scholar]
- 25.Marlin S D, Staunton D E, Springer T A, Stratowa C, Sommergruber W, Merluzzi V J. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature. 1990;344:70–72. doi: 10.1038/344070a0. [DOI] [PubMed] [Google Scholar]
- 26.Marlovits T C, Abrahamsberg C, Blaas D. Soluble LDL minireceptors—minimal structure requirements for recognition of minor group human rhinovirus. J Biol Chem. 1998;273:33835–33840. doi: 10.1074/jbc.273.50.33835. [DOI] [PubMed] [Google Scholar]
- 27.Marlovits T C, Abrahamsberg C, Blaas D. Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J Virol. 1998;72:10246–10250. doi: 10.1128/jvi.72.12.10246-10250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marlovits T C, Zechmeister T, Gruenberger M, Ronacher B, Schwihla H, Blaas D. Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro. FASEB J. 1998;12:695–703. doi: 10.1096/fasebj.12.9.695. [DOI] [PubMed] [Google Scholar]
- 29.Neff S, Carvalho D S, Rieder E, Mason P W, Blystone S D, Brown E J, Baxt B. Foot-and-mouth disease virus virulent for cattle utilizes the integrin αvβ3 as its receptor. J Virol. 1998;72:3587–3594. doi: 10.1128/jvi.72.5.3587-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira M A, Zhao R, Lee W M, Kremer M J, Minor I, Rueckert R R, Diana G D, Pevear D C, Dutko F J, McKinlay M A, Rossmann M G. The structure of human rhinovirus 16. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 31.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prchla E, Kuechler E, Blaas D, Fuchs R. Uncoating of human rhinovirus serotype 2 from late endosomes. J Virol. 1994;68:3713–3723. doi: 10.1128/jvi.68.6.3713-3723.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J Cell Biol. 1995;131:111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roivainen M, Hyypia T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roivainen M, Piirainen L, Hovi T. Efficient RGD-independent entry process of coxsackievirus A9. Arch Virol. 1996;141:1909–1919. doi: 10.1007/BF01718203. [DOI] [PubMed] [Google Scholar]
- 36.Ronacher B, Marlovits T C, Moser R, Blaas D. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology. 2000;278:541–550. doi: 10.1006/viro.2000.0636. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann M G. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 38.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann M G, Bella J, Kolatkar P R, He Y N, Wimmer E, Kuhn R J, Baker T S. Cell recognition and entry by rhino- and enteroviruses. Virology. 2000;269:239–247. doi: 10.1006/viro.2000.0258. [DOI] [PubMed] [Google Scholar]
- 40.Schober D, Kronenberger P, Prchla E, Blaas D, Fuchs R. Major- and minor-receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J Virol. 1998;72:1354–1364. doi: 10.1128/jvi.72.2.1354-1364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafren D R, Dorahy D J, Ingham R A, Burns G F, Barry R D. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J Virol. 1997;71:4736–4743. doi: 10.1128/jvi.71.6.4736-4743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shafren D R, Williams D T, Barry R D. A decay accelerating factor binding strain of coxsackievirus B3 requires the coxsackievirus adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J Virol. 1997;71:9844–9848. doi: 10.1128/jvi.71.12.9844-9848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherry B, Mosser A G, Colonno J, Rueckert R R. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus human rhinovirus 14. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 45.Strickland D K, Kounnas M Z, Argraves W S. LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 1995;9:890–898. doi: 10.1096/fasebj.9.10.7615159. [DOI] [PubMed] [Google Scholar]
- 46.Todd S, Towner J S, Semler B L. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology. 1997;229:90–97. doi: 10.1006/viro.1996.8416. [DOI] [PubMed] [Google Scholar]
- 47.Tomassini J E, Colonno R J. Isolation of a receptor protein involved in attachment of human rhinoviruses. J Virol. 1986;58:290–295. doi: 10.1128/jvi.58.2.290-295.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triantafilou M, Triantafilou K, Wilson K M, Takada Y, Fernandez N. High affinity interactions of coxsackievirus A9 with integrin alpha v beta 3 (CD51/61) require the CYDMKTTC sequence of beta 3, but do not require the RGD sequence of the CAV-9 VP1 protein. Hum Immunol. 2000;61:453–459. doi: 10.1016/s0198-8859(00)00103-8. [DOI] [PubMed] [Google Scholar]
- 49.Triantafilou M, Triantafilou K, Wilson K M, Takada Y, Fernandez N, Stanway G. Involvement of beta2-microglobulin and integrin alphavbeta3 molecules in the coxsackievirus A9 infectious cycle. J Gen Virol. 1999;80:2591–2600. doi: 10.1099/0022-1317-80-10-2591. [DOI] [PubMed] [Google Scholar]
- 50.Uncapher C R, Dewitt C M, Colonno R J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]