Abstract
Congenital heart defects are leading causes of neonatal mortality and are often associated with placental abnormalities, but mechanisms linking placenta and heart development are poorly understood. Herein, we investigated a potential signaling network connecting the placenta and nascent heart in mice. We found that fetal hearts exposed to media conditioned by placental tissue or differentiated wild-type trophoblast stem (TS) cells, but not undifferentiated TS cells, showed increased heart rate and epicardial cell outgrowth. This effect was not observed when hearts were exposed to media from TS cells lacking OVO-Like 2, a transcription factor required for trophoblast differentiation and placental development. Trophoblasts released abundant extracellular vesicles into media, and these vesicles were sufficient to mediate cardio-promoting effects. Our findings provide a potential mechanism whereby the placenta communicates with the fetal heart to promote cardiac morphogenesis, and offers insight into the link between poor placentation and a higher incidence of heart defects.
Subject terms: Embryology, Extracellular signalling molecules
Poor development of the placenta is linked with heart problems. This study reveals that extracellular vesicles from placental trophoblasts promote heart development in mice, shedding light on how placental health affects cardiac formation.
Introduction
The heart is among the first organs to function in humans, making it highly susceptible to environmental insults that occur during early pregnancy. Congenital heart defects (CHDs) are the most common group of birth defects and occur in approximately 1% of live births1,2. Infants born with CHDs often undergo corrective surgeries, and reduced birth weight in these children strongly influences early mortality3,4. Severe CHDs contribute to ~10% of fetal loss2. The etiology of CHDs is complex and poorly understood. Most CHDs are sporadic and cannot be traced to a specific genetic event, suggesting that CHDs likely arise from a combination of genetic, epigenetic, and external factors5. One external factor that is increasingly gaining recognition as a central contributor to CHDs is improper placental formation6,7.
The placenta is the lifeline for a baby while in the womb. It serves as the primary respiratory and nutritive organ for the baby during pregnancy, and it produces numerous hormones, growth factors, and extracellular vesicles (EVs) – membrane-bound nanoparticles carrying lipids, proteins and nucleic acids capable of mediating intercellular communication8,9. Release of these factors is critical for rewiring maternal physiology to benefit pregnancy as well as supporting fetal growth and development10. Consequently, poor placental formation or function can have a profound impact during vulnerable periods of organogenesis. Maldevelopment of the placenta is linked with a variety of serious pregnancy complications (e.g., preeclampsia, fetal growth restriction) that cause maternal and fetal illness or death11. Interestingly, these pregnancy complications are also associated with a higher incidence of CHDs, suggesting the notion of a shared developmental trajectory between the placenta and nascent heart gaining recognition as the “placenta-heart axis”2,12.
Studies using mice have provided insight into key pathways that are required for both development of the placenta and nascent heart. For example, pregnant mice exposed to teratogens such as lithium show dysregulated expression of WNT-associated genes Hex and Islet-1 and display severe cardiac and placental dysmorphologies13,14. Total ablation of P38-MAPK15, PPARG16, and HOXA1317 results in disrupted placental formation and leads to defective heart development and embryonic lethality. The lethality is recapitulated when the genetic defect is present uniquely in trophoblasts (parenchymal cells of the placenta), but not observed when the deletion occurs only within embryonic cells. The Deciphering the Mechanisms of Developmental Disorders project used a large-scale screen of 103 embryonic lethal mouse knockout lines, and found that in 68% of cases where lethality was attributed to poor heart and vascular development, placental abnormalities were apparent18. Several recent studies have expanded on these findings by showing that some heart and vascular defects can be directly attributed to improper placental syncytiotrophoblast formation19,20. These studies clearly demonstrate a link between placental and heart development, but the mechanisms regulating placenta-heart interactions are still poorly understood.
Recently, we uncovered an essential role for the transcription factor OVO-Like 2 (OVOL2) for placental development in mice. We found that OVOL2 is highly expressed in trophoblasts and that trophoblast stem (TS) cells generated from Ovol2-deficient blastocysts are unable to differentiate into mature trophoblast lineages21. Embryos deficient in Ovol2 die at midgestation with major defects in both placental and heart formation, despite Ovol2 not being readily detectable in the heart at this gestational timepoint21,22. This suggests that placental expression of Ovol2 is required for proper heart development, and provides an intriguing vantage point to uncover mechanisms regulating the placenta-heart axis. In the current study, we hypothesized that placental expression of Ovol2 is needed for development of the nascent heart in mice. We found that fetal hearts and cardiomyocytes cultured in media conditioned by placental tissue or differentiated trophoblasts (from TS cells) showed accelerated maturation of cardiomyocytes and improved heart development. These effects were not observed when media was conditioned by Ovol2-/- TS cells. Furthermore, we uncovered a potential pathway of placenta-heart communication facilitated by trophoblast-derived EVs.
Results
Ovol2 deficiency results in poor heart formation and embryonic lethality
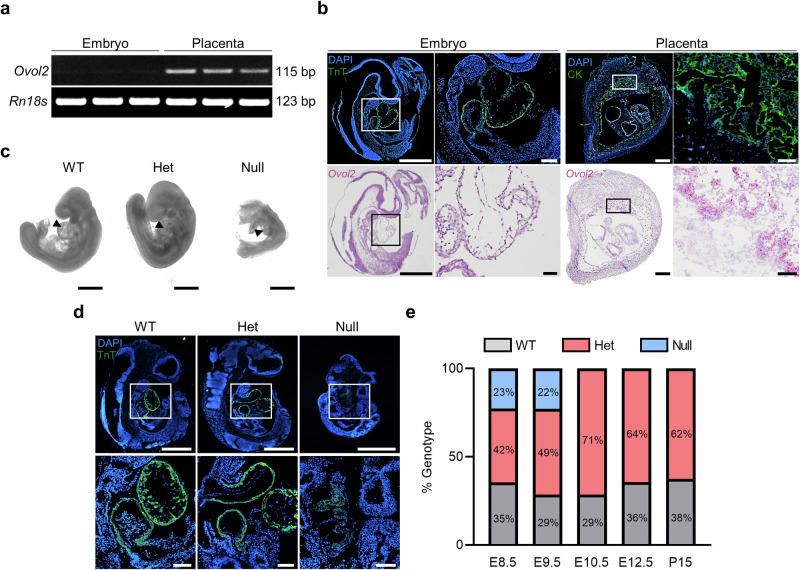
To determine expression of Ovol2 in the mouse embryo and placenta, tissues were collected from pregnant wild-type C57BL/6 N mice on embryonic day (E)9.5 and RT-PCR was performed. Ovol2 was strongly expressed within the placenta, but expression was undetectable in the embryo (Fig. 1a). In situ hybridization and immunohistochemistry further confirmed these findings: Ovol2 was strongly detected in the placenta (demarcated by Cytokeratin) and no staining was apparent in the embryo, including the nascent heart (demarcated by Troponin T; Fig. 1b). Detection of Ovol2 was particularly prominent in the placental labyrinth zone, which is responsible for exchange of nutrients and wastes between maternal and fetal blood. The labyrinth zone is lined by two layers of syncytiotrophoblast, layer 1 (SynT-I) and layer 2 (SynT-II), and also contains sinusoidal trophoblast giant cells. To further delineate where Ovol2 is expressed within the mouse placenta, immunofluorescence was performed for monocarboxylate transporter 1 (MCT1; expressed in SynT-I) and monocarboxylate transporter 4 (MCT4; expressed in SynT-II)23. On E9.5, these proteins were not readily distinguishable using immunohistochemistry, so sections of E12.5 placentas were used. Serial sections of E12.5 placentas were then probed for Ovol2. Based on this analysis, Ovol2 appeared to be most prominently expressed by cells comprising SynT-I (Supplementary Fig. 1).
Fig. 1. Ovol2 deficiency results in poor heart formation and embryonic lethality.
a RT-PCR showing Ovol2 and Rn18s in the mouse embryo and placenta on E9.5. b Immunohistochemistry showing Troponin T (TnT; green, upper left images) and Cytokeratin (CK; green, upper right images) in the E9.5 embryo and placenta, respectively. DAPI was used to stain nuclei. In the bottom panels, in situ hybridization was used to demarcate Ovol2 (pink) expression in the embryo and placenta. Boxes in the lower magnification images (left panels) show the location of higher magnification images (right panels) for both the embryo and placenta. Please note that Ovol2-positive cells are only detectable within the placenta. c Phase-contrast images of Ovol2+/+ (WT), Ovol2+/- (Het) and Ovol2-/- (Null) embryos. Images were captured using a Leica DMi1 inverted microscope. Arrowheads indicate the fetal heart. d Immunohistochemistry showing TnT (green) in WT, Het, and Null embryos. Boxes in the lower magnification images (top panels) show the location of higher magnification images (bottom panels). e Quantification of the percent of WT, Het, and Null embryos present at various gestational timepoints (n = 4 pregnant mice; actual number of embryos/pups of each genotype are presented in Supplementary Table 1). Scale bars represent 500 µm for low magnification images in (b) and (d), and the images in (c). Scale bars for high magnification images in (b) and (d) represent 50 µm. Uncropped images of DNA gels are provided in Supplementary Fig. 7.
Next, to characterize heart development within Ovol2+/+, Ovol2+/-, and Ovol2-/- embryos, Ovol2+/- animals were mated, and tissues were collected on E9.5. Ovol2-/- embryos were substantially smaller than Ovol2+/+ and Ovol2+/- counterparts and lacked the presence of a bulbous heart (Fig. 1c). Immunohistochemistry for Troponin T further validated these observations, with Ovol2+/+ and Ovol2+/- embryos staining strongly for Troponin T, whereas Ovol2-/- embryos showed weak, disaggregated staining (Fig. 1d). Ovol2-/- embryos likely died between E9.5 and E10.5 in utero, as embryonic tissue was unobtainable on E10.5 (Fig. 1e and Supplementary Table 1). These data indicate that Ovol2 is highly expressed in the mouse placenta but not the fetal heart, yet Ovol2-/- embryos are embryonic lethal with major defects in heart formation.
Media conditioned by placentas and trophoblasts promote fetal heart development and cardiomyocyte growth
Since Ovol2-/- embryos die prematurely and exhibit poor heart development, we postulated that placental Ovol2 expression may support proper communication between the placenta and developing heart. To test this hypothesis, fetal hearts were excised from embryos on E12.5 (the earliest timepoint at which fetal hearts could be consistently isolated without limiting the quality of dissections) and cultured in four conditions: unconditioned media, conditioned media (CM) from minced placental tissue collected from E15.5 mice (placental CM), CM from 5-day differentiated Ovol2+/+ TS cells (trophoblast CM), or media conditioned by Ovol2-/- TS cells placed in differentiation media for 5 days (Ovol2-Null CM; Fig. 2a). Fetal heart rate and epicardial cell outgrowth were then measured24. Embryonic hearts exposed to placental CM or trophoblast CM showed increased heart rate (2.3 and 1.9-fold at the 48 h timepoint, respectively) and epicardial cell outgrowth (3.5 and 2.1-fold at the 48 h timepoint, respectively), when compared to hearts exposed to unconditioned media. This stimulatory effect on heart rate and epicardial cell outgrowth was not observed when hearts were exposed to Ovol2-Null CM (Fig. 2b, c, P < 0.05).
Fig. 2. Medium conditioned by placental tissue and differentiated trophoblasts promotes fetal heart development and cardiomyocyte growth.
a Schematic indicating four culture conditions: fetal hearts cultured in unconditioned media (CTRL), fetal hearts cultured in media conditioned by E15.5 placental tissue (Placental CM), fetal hearts cultured in media conditioned by 5-day differentiated Ovol2+/+ TS cells (Trophoblast CM), and fetal hearts cultured in media conditioned by 5-day differentiated Ovol2-/- TS cells (Ovol2-Null CM). b Average number of beats per minute when fetal hearts were exposed to CTRL, Placental CM, Trophoblast CM, or Ovol2-Null CM for 24 and 48 h. c Quantification of epicardial cell outgrowth from fetal hearts exposed to CTRL, Placental CM, Trophoblast CM, or Ovol2-Null CM for 24 and 48 h. c Representative images of epicardial cell outgrowths at 24 h. A red dashed line delineates fetal heart borders and arrows demarcate examples of outgrowths. d Expression of six genes (Tead1, Myh7, Myl7, Acta2, Nppa, and Nppb) associated with cardiac development in cardiomyocytes exposed to CTRL, Placental CM, Trophoblast CM, or Ovol2-Null CM. e Western blot showing levels of Troponin T (TnT) and NKX2-5 in cardiomyocytes treated with CTRL, Placental CM, Trophoblast CM, or Ovol2-Null CM. ACTB was used as a loading control. Quantification of relative band intensity is shown to the right of the representative blots. f Immunofluorescence for TnT (green) in cardiomyocytes treated with CTRL, Placental CM, Trophoblast CM, or Ovol2-Null CM. DAPI was used to demarcate nuclei. Values significantly different from CTRL (N = 6 biologically independent samples, P < 0.05) are denoted with an asterisk (*); values significantly increased from CTRL but decreased from Placental CM are denoted with a number sign (#). One-way ANOVA followed by Tukey’s post-hoc test was used to determine statistical significance. Scale bar = 100 µm. Graphs represent means ± SEM. Uncropped images of western blots are provided in Supplementary Fig. 8.
Next, to determine whether secreted products from placental tissue or trophoblasts impact cardiomyocyte growth, we isolated cardiomyocytes from E12.5 fetal hearts and cultured them in unconditioned media, or in the presence of placental CM, trophoblast CM, or Ovol2-Null CM. Compared to cardiomyocytes cultured in unconditioned media, expression of several cardiac markers was increased in cardiomyocytes cultured in placental CM or trophoblast CM, including Myh7 (3.8 and 3.5-fold), Myl7 (3.7 and 3.1-fold), Acta2 (3.4 and 4.1-fold), Nppa (5.0 and 6.3-fold), and Nppb (13.6 and 12.4-fold, respectively; P < 0.05). No change in expression of these genes was observed when cardiomyocytes were cultured in Ovol2-Null CM, except for Nppb (5.4-fold increase compared to unconditioned media, P < 0.05), albeit this response was much less pronounced compared to placental CM or trophoblast CM. No change in Tead1 expression was observed under any condition (Fig. 2d). Increased protein levels of cardiac markers Troponin T and NKX2-5 were also found in cardiomyocytes treated with placental CM or trophoblast CM, when compared to cells treated with control or Ovol2-Null CM (Fig. 2e, P < 0.05). Interestingly, immunofluorescence for Troponin T revealed larger clusters and heightened organization of sarcomeres in cardiomyocytes treated with placental CM or trophoblast CM, which is associated with advanced cardiomyocyte maturation during fetal development23 (Fig. 2f). Lastly, to evaluate whether placental CM or trophoblast CM alters the rate of cardiomyocyte proliferation, a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay was performed. No difference in the proliferative capacity of cardiomyocytes was observed (Supplementary Fig. 2). Overall, these data suggest that factors secreted by placentas and differentiated trophoblasts promote fetal heart development and cardiomyocyte growth.
Placental EVs promote fetal heart development and cardiomyocyte growth
To narrow the identity of factors secreted by placentas with cardio-promoting properties, we used ultracentrifugation to separate placental CM into two components: media supernatant and EVs (Fig. 3a). EVs enriched from fetal bovine serum (FBS) were used as a control. Western blotting for EV markers TSG101 and HSP70 was performed to confirm depletion of EVs from media supernatants and enrichment of EVs in the pelleted fraction (Fig. 3b). Using electron microscopy, circular vesicles <1 µm were detectable in the pelleted fraction from placental CM. The most abundant were vesicles that scatter light similar to ~180 nm silica beads (Supplementary Fig. 3 and 4), although it should be noted that the isolation protocol may preferentially enrich smaller EVs. Various concentrations of FBS-derived EVs, placental-derived EVs, or media supernatants were added to fetal heart cultures to determine whether cardio-promoting factors reside within media supernatants or EVs. Fetal hearts exposed to placental-derived EVs, but not FBS-derived EVs or media supernatants, showed increased heart rate and epicardial cell outgrowth in a dose-dependent manner (Fig. 3c, P < 0.05). This suggests that placental EVs, but not placental media supernatants, promote fetal heart development. Likewise, cardiomyocytes isolated from fetal hearts and treated with placental EVs (10 µg/mL) showed increased expression of several cardiac markers (Myh7, Myl7, Acta2, Nppa, and Nppb), as well as increased levels of Troponin T and NKX2-5, when compared to cells treated with the same concentration of control EVs (Fig. 3d, e, P < 0.05). Additionally, a notable increase in cardiomyocyte size and sarcomere organization was apparent in cells treated with placental EVs (Fig. 3f). These data indicate that EVs are likely the key components of trophoblast CM that promote fetal heart development and cardiomyocyte growth.
Fig. 3. Placental EVs promote fetal heart development and cardiomyocyte growth.
a Schematic showing procedure used to isolate EVs from unconditioned media (CTRL) and media conditioned by E15.5 placental tissue (Placental CM). b Western blot for EV markers TSG101 and HSP70 in media supernatants and EVs isolated from CTRL and Placental CM. Ponceau S was used to verify total protein. c Quantification of fetal heart rate and epicardial cell outgrowth from embryonic hearts exposed to supernatant from either unconditioned media (M-C) or Placental CM (M-P); or varying concentrations of CTRL (from FBS) or Placental EVs (1, 5, or 10 µg/mL) for 48 h. d Expression of six genes (Tead1, Myh7, Myl7, Acta2, Nppa, and Nppb) associated with cardiac development in cardiomyocytes treated with EVs (10 µg/mL) isolated from CTRL or Placental CM. e Western blot showing levels of Troponin T (TnT) and NKX2-5 in cardiomyocytes treated with CTRL or Placental EVs (10 µg/mL). ACTB was used as a loading control. Quantification of relative band intensity is found to the right of the representative blots. f Immunofluorescence for TnT (green) in cardiomyocytes treated with CTRL or Placental EVs (10 µg/mL). DAPI was used to demarcate nuclei. Values significantly different from CTRL media supernatants (N = 5 independent experiments, P < 0.05), or from CTRL EVs (N = 3 independent experiments, P < 0.05) are denoted with an asterisk (*), using one-way ANOVA followed by Tukey’s post-hoc test (c) or two-tailed Student’s t-test (d and e). Scale bar = 100 µm. Graphs represent means ± SEM. Uncropped images of western blots are provided in Supplementary Fig. 8.
Placental EVs are present in fetal plasma
It is well established that EVs from the placenta circulate in maternal blood10. EVs originating in the placenta likely also traffic to the fetus, but this has not been extensively studied. Therefore, we isolated plasma from non-pregnant virgin mice (background) as well as from pregnant dams and fetuses on E18.5, which was the earliest time point that sufficient fetal plasma could be collected for downstream analysis. The plasma was then incubated with fluorescently-tagged antibodies targeting syncytin-A (expressed by SynT-I in the mouse placenta and detectable on the surface of placental EVs25) and CD9 (marker of EVs). Nanoflow cytometry was then performed to determine dual positivity. Nanoparticles positive for both markers were considered placental EVs. As expected, placental EVs were enriched 4.5-fold (P < 0.05) in the plasma of pregnant dams compared to non-pregnant mice. In fetal plasma, placental EVs were enriched 6.9-fold (P < 0.05) compared to plasma from non-pregnant mice (Fig. 4 and Supplementary Fig. 5). There was no statistical difference in the amount of EVs circulating in maternal versus fetal plasma. This suggests that EVs from the placenta likely traffic to the fetus during pregnancy.
Fig. 4. Placental EVs are present in fetal and maternal plasma.
a Schematic showing experimental design. Plasma was isolated from non-pregnant virgin mice, pregnant dams, or fetuses on E18.5. Plasma was then incubated with syncytin-A-FITC and CD9-PE antibodies, and subjected to nanoflow cytometry analysis. b Quantification of plasma EVs positive for both syncytin-A-FITC and CD9-PE antibodies. c Representative nanoflow cytometry plots showing an unstained control (pooled plasma), and syncytin-A-FITC and CD9-PE positive EVs isolated from plasma from non-pregnant virgin mice, pregnant dams, or fetuses on E18.5. Values significantly different from non-pregnant mice (N = 3 different mice, P < 0.05) are denoted with an asterisk (*), using one-way ANOVA followed by Tukey’s post-hoc test. Graphs represent means ± SEM.
EVs isolated from trophoblast CM promote cardiomyocyte growth and maturation
Since cardio-promoting effects were observed when fetal hearts and cardiomyocytes were exposed to trophoblast CM, but not Ovol2-Null CM, we next determined whether EVs enriched from trophoblast CM and Ovol2-Null CM differentially affect cardiomyocyte development. Furthermore, since Ovol2-/- TS cells exhibit poor differentiation21, we also tested whether the cardio-promoting properties of TS cell-derived EVs were related to trophoblast differentiation status. Successful enrichment of EVs from trophoblast CM, Ovol2-Null CM, as well as media conditioned by wild-type and Ovol2-/- TS cells maintained in the stem state was confirmed through western blot for EV-specific markers TSG101 and HSP70 (Fig. 5a). Additionally, using electron microscopy and nanoflow cytometry, circular vesicles <1 µm in size were detected in all conditions. Interestingly, although smaller vesicles were most abundant in all treatment conditions similar to EVs from placental CM, culturing TS cells (particularly wild-type cells) in differentiation media yielded the most abundant quantity of EVs (Supplementary Fig. 6).
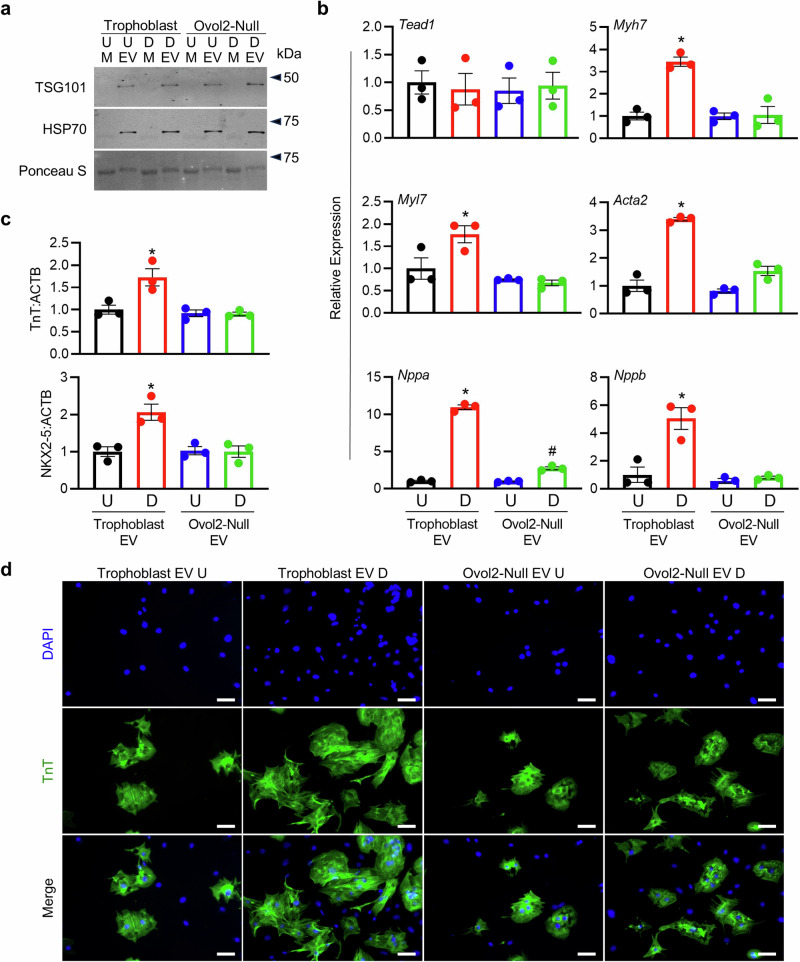
Fig. 5. EVs from media conditioned by differentiated trophoblasts promote cardiomyocyte growth.
a Western blot for EV markers TSG101 and HSP70 in media supernatants (M) and EVs isolated from media conditioned by Ovol2+/+ TS cells (Trophoblast) and Ovol2-/- (Ovol2-Null) TS cells cultured in stem or differentiation media. Ponceau S was used to verify total protein. U=cells exposed to stem media (undifferentiated), D=cells exposed to differentiation media for 5 days. b Expression of six genes (Tead1, Myh7, Myl7, Acta2, Nppa, and Nppb) associated with cardiac development in cardiomyocytes treated with EVs (10 µg/mL) isolated from CM of Ovol2+/+ and Ovol2-Null TS cells cultured in undifferentiated and differentiated states. c Relative band intensity from western blots for Troponin T (TnT) and NKX2-5 in comparison to ACTB in cardiomyocytes exposed to EVs (10 µg/mL) isolated from CM of Ovol2+/+ and Ovol2-Null TS cells cultured in undifferentiated and differentiated states. d Immunofluorescence for TnT (green) in cardiomyocytes treated with EVs (10 µg/mL) isolated from CM of Ovol2+/+ and Ovol2-Null TS cells cultured in undifferentiated and differentiated states. DAPI was used to stain nuclei. Values significantly different from undifferentiated Trophoblast EVs (N = 3 independent experiments, P < 0.05) are denoted with an asterisk (*); values different from both undifferentiated trophoblast EVs and differentiated trophoblast EVs are denoted with a number sign (#). A one-way ANOVA followed by Tukey’s post-hoc test was used to determine statistical significance, using values from the undifferentiated Trophoblast EVs normalized to 1 as the control condition. Scale bar = 100 µm. Graphs represent means ± SEM. Uncropped images of western blots are provided in Supplementary Fig. 8.
Cardiomyocytes were then treated with EVs derived from TS cells. In cardiomyocytes treated with EVs from trophoblast CM, increased expression of several cardiac markers was noted, including Myh7 (3.5-fold), Myl7 (1.8-fold), Acta2 (3.4-fold), Nppa (11.0-fold), and Nppb (5.1-fold), when compared to cells exposed to EVs from undifferentiated TS cells (Fig. 5b, P < 0.05). Furthermore, protein levels of Troponin T and NKX2-5 were increased, and cardiomyocytes appeared larger and displayed heightened sarcomere organization in the presence of EVs enriched from trophoblast CM (Fig. 5c, d). These cardio-promoting properties were not observed when cells were treated with EVs enriched from Ovol2-Null CM or media conditioned by undifferentiated Ovol2-/- TS cells. Overall, these data suggest that EVs derived from TS cells placed in differentiation conditions, but not undifferentiated cells or Ovol2-/- TS cells, promote cardiomyocyte development.
Discussion
CHDs are influenced by genetic, epigenetic, and environmental factors. Most CHDs are sporadic and usually cannot be traced to a specific genetic event; only about 15% of CHDs are attributed to de novo mutations and copy number variants, suggesting that the environment in which a fetus develops is likely a major contributor to the onset of CHDs2. Placental compromise can have a serious impact on the health of a fetus and result in long-term detrimental effects in offspring. Moreover, obstructing access to nutrients during early development predisposes the baby to life-long health consequences, including increased risk of cardiovascular disease7,26. In the present study, we discovered that placental EVs promote fetal heart development and cardiomyocyte growth, which was apparent through increased expression of cardiac markers and differences in cell morphology. Similar results were obtained using EVs enriched from trophoblast CM (derived by placing TS cells into differentiation media for 5 days), but not OVOL2-deficient cells with impaired differentiation potential, strongly suggesting that differentiated trophoblasts are the main source of placental EVs with cardio-promoting properties. We also found that placental EVs are present within fetal circulation in vivo, providing a possible mechanism to explain how the placenta communicates with the embryo to mediate development of the nascent heart.
In our first series of experiments, we investigated the expression and localization patterns of Ovol2 in the mouse embryo and placenta at E9.5 and E12.5. High expression of Ovol2 was detected within the placental labyrinth zone, most prominently in SynT-I at E12.5, whereas little to no expression was detectable in the embryo or the developing heart. This confirmed previous findings from our group and others that Ovol2 is highly expressed within placental tissue and not readily detectable within the embryo21,22. In our previous study, Ovol2+/+ and Ovol2+/- TS cells readily show evidence of differentiation when placed in differentiation media for 5 days as determined by decreased expression of various TS markers (Cdx2, Eomes, Esrrb, and Id2); and increased expression of Prl3b1 (trophoblast giant cells), Ascl2 (junctional zone), Synb (SynT-II), and Syna (encoding syncytin-A; SynT-I). Ovol2-/- TS cells, on the other hand, retain high expression of TS-related genes and do not increase expression of genes associated with differentiated trophoblasts21. OVOL2 is therefore necessary for trophoblast differentiation and placental formation. The absence of Ovol2 results in embryonic lethality at midgestation, which is associated with poor heart and vascular development22. Matings of Ovol2+/- mice further confirmed these observations: Ovol2+/+ and Ovol2+/- embryos produced a bulbous heart that showed strong expression of Troponin T, while Ovol2-/- embryos were smaller in size and displayed reduced Troponin T levels and poor heart morphogenesis. Several mouse models that exhibit placental defects (e.g., mice with a knockout of Ssr2, Chtop, Rpgrip11, Atp11a, Smg9, Pibf1), display aberrant heart morphology including septal and aortic defects, as well as reduced blood vessel expansion18–20, suggesting a genetic link between development of the placenta and nascent heart. Likewise, in the current study, we noted poor heart and placenta morphogenesis in Ovol2-/- embryos. Since Ovol2 was highly expressed in the placenta and not the heart, it is likely that loss of OVOL2 results in poor differentiation of trophoblasts and maldevelopment of the placenta, which in turn negatively impacts fetal heart development. Therefore, utilizing both Ovol2+/+ and Ovol2-/- TS cells provided us with an intriguing perspective to determine mechanisms regulating the placenta-heart axis.
EVs are continuously shed by the placenta into maternal circulation during pregnancy, where they influence maternal physiology and immunology to benefit pregnancy10. EVs can be broadly categorized into exosomes (30-150 nm), microvesicles (100-1,000 nm), and apoptotic bodies (1,000-5,000 nm), and they are differentiated based on their biogenesis, function, and size27,28. Placentas release copious amounts of all three types of EVs9,29. In our study, we found that syncytin-A positive EVs were present in plasma of pregnant mice and fetuses. Syncytin-A is expressed by SynT-I cells, the same cells that prominently express Ovol2, and is induced in TS cells when placed in differentiation media. This suggests that placental EVs originating from cells expressing syncytin-A may traffic to the fetus, although we were technically limited to evaluating fetal plasma at late gestation only. The localization and interaction patterns of these EVs (along with EVs produced by trophoblasts not expressing syncytin-A) with fetal organs, especially during early gestation, requires further investigation. Furthermore, we used E15.5 for placental EV collections to evaluate effects on growth and maturation of fetal hearts and cardiomyocytes collected on E12.5. While E15.5 was used to maximize the amount of placental EVs collected, there may be notable differences in the content or structure of placental EVs collected on E15.5 versus earlier timepoints that should be considered for future investigations. Further purification and size exclusion of placental/trophoblast EVs is also needed to eliminate the potential actions of impurities following ultracentrifugation and to determine whether specific EV classes are responsible for eliciting cardio-promoting effects.
EVs contain a variety of cargo such as nucleic acids, proteins, lipids, and metabolites that can influence the physiological functions of recipient cells28. Cargo from placental EVs regulate trophoblast invasion30, decrease maternal immune cell activity31, and confer viral resistance to recipient cells32. In many cases, functional changes are evident only after exposing recipient cells to placental/trophoblast EVs and not to EVs from other nonplacental cells, suggesting that specific molecules carried by trophoblast EVs mediate these effects. Consistent with the cardio-promoting effects of trophoblast EVs described in the current study, placental EVs alleviate hypertension and improve cardiac function in spontaneously hypertensive rats, whereas EVs from a human trophoblast cell line can prevent doxorubicin-induced cardiomyopathy in mice33,34. Another recent study found that conditioned media (likely containing EVs) from mouse TS cells delays formation of cardiac mesoderm progenitors and promotes retention of pluripotency markers in embryoid bodies, indicating that the impact of trophoblast-derived products on cardiac development may be timing and context-specific35.
There have been various attempts to identify constituents of trophoblast EVs in humans and mice that have potential to exert functional changes in recipient cells36–39, although those capable of promoting cardiomyocyte maturation are not well understood. Among the top candidates are microRNAs (miRNAs), of which there are at least 600 expressed within the placenta40. In humans and mice, many of these miRNAs originate from clusters that are highly expressed in trophoblasts but not normally in other cells; miRNAs in these clusters are therefore considered to be placenta-specific41. In mice, deletion of placenta-specific miRNA clusters results in major defects in placental formation as well as embryonic growth and viability42,43, indicating the importance of these placenta-specific miRNAs for fetal development and pregnancy health. Interestingly, several placenta-specific miRNAs are dysregulated in human cases of pathology and have been suggested as potential biomarkers to predict the onset of disease, including preeclampsia. Preeclampsia is highly associated with increased predisposition to cardiovascular disease, suggesting that altered levels of placental EV cargo such as these miRNAs may impact the heart44–46.
In conclusion, the present study provides evidence that differentiated trophoblasts produce EVs that mediate progression and development of cardiomyocytes. These effects were not observed using undifferentiated TS cells, or Ovol2-deficient TS cells that have reduced differentiation potential. These findings may explain, at least in part, a link between poor heart development and abnormal placentation, and implies that trophoblast EVs may be accessible biomarkers or therapeutic targets to detect or improve suboptimal development of the nascent heart.
Methods
Animals
C57BL/6 N and CD-1 mice were obtained from Charles River Laboratories (Wilmington, MA, USA). Ovol2+/- mice were generously provided by Seiji Ito through RIKEN Bioresource Research Centre (Tsukuba, Ibaraki Prefecture, Japan) based on a Materials Transfer Agreement, and were generated as previously described22. All mice were housed under a 12-h light-dark cycle with free access to food and water. Mice were bred by placing males and females (8-16 weeks old) into the same cage overnight. The presence of a copulatory plug the following morning was designated E0.5. Ovol2+/- males were bred with wild-type C57BL/6 N females to maintain a stock of heterozygotes. To obtain Ovol2+/+, Ovol2+/-, and Ovol2-/- embryos, Ovol2+/- males and females were bred. Protocols involving the use of mice were approved by the University of Western Ontario Animal Care and Use Committee in accordance with the guidelines of the Canadian Council on Animal Care. We have complied with all relevant ethical regulations for animal use.
Tissue collection and genotyping
To evaluate Ovol2 expression in fetal and placental tissues, and to compare heart morphology in Ovol2+/+, Ovol2+/-, and Ovol2-/- conceptuses, tissues were collected on E9.5 and E12.5. Placentas and embryos were collected, snap-frozen in liquid nitrogen, and stored at -80 °C for molecular analyses. For immunohistochemical analysis, embryos were fixed in 4% paraformaldehyde and stored in 70% ethanol until paraffin-embedding. Placentas were either fixed in 4% paraformaldehyde and paraffin-embedded, or immersed in dry ice-cooled heptane and stored at -80 °C. Fetal hearts were isolated from C57BL/6 N dams on E12.5. Mouse embryonic fibroblasts (MEFs), which are required for TS cell culture, were isolated from E12.5 fetal tissues as previously described47. Fetal tissue for preparation of MEFs was collected from CD-1 mice. Phase-contrast images of embryos and fetal hearts were obtained using a Leica DMi1 inverted microscope (Leica Microsystems Inc., Concord, ON, Canada).
Genotyping was performed on yolk sacs or on tail clips of pups. DNA was extracted using REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich, Oakville, ON, Canada), according to the manufacturer’s protocol. PCR was performed using DreamTaq DNA polymerase (ThermoFisher Scientific, Mississauga, ON, Canada) and primers spanning the Ovol2 deletion site (see Table 1). PCR conditions utilized were as follows: initial holding step (95 °C for 3 min), followed by 32 cycles of PCR (95 °C for 30 s, 63 °C for 30 s, and 72 °C for 30 s), and a final elongation phase at 72 °C for 12 min.
Table 1.
Primers used for RT-PCR
| Gene | Accession No. | Forward Primer (5’→3’) | Reverse Primer (5’→3’) |
|---|---|---|---|
| Acta2 | NM_007392.3 | AGACTCTCTTCCAGCCATCT | GTGATCTCCTTCTGCATCCTGT |
| Gapdh | NM_001289726.2 | GGAGAGTGTTTCCTCGTCCC | CCGTTGAATTTGCCGTGAGT |
| Myh7 | NM_001361607.1 | TTTGGGCGAGTCAAAGATGC | CTCTGCATCCGCCAAGTTGT |
| Myl7 | NM_022879.2 | AGAAGCTCAATGGGACGGAC | GCGCAAACAGTTGCTCTACC |
| Nppa | NM_008725.3 | AGAGACGGCAGTGCTCTAGG | TTCGGTACCGGAAGCTGTTG |
| Nppb | NM_001287348.2 | GAAGGACCAAGGCCTCACAA | TTCAGTGCGTTACAGCCCAA |
| Ovol2(Genotyping) | NC_000068.8 | CATAGCCCATGTGTGGCTGCTG | GCCGGCCTTAAAACATCCCAC |
| Ovol2 | NM_026924.3 | TTCACCCAGCGGTGTTCCTT | TGTAGCCGCAATCCTCACAC |
| Rn18s | NR_003278.3 | GCAATTATTCCCCATGAACG | GGCCTCACTAAACCATCCAA |
| Tead1 | NM_001166584.2 | TCGGCAGATAAGCCGATTGA | TCCTTGTCTTTCCCGTCCTG |
Cells
Media components for cell culture were obtained from Sigma-Aldrich unless stated otherwise, and all cells were maintained at 37 °C, 5% CO2. MEFs were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (ThermoFisher Scientific), 100 units/mL penicillin, and 100 µg/mL streptomycin. MEF proliferation was inhibited by subjecting cells to 5 μg/mL mitomycin C (M0503, Sigma-Aldrich) for 2 h, followed by washing in PBS.
Mouse TS cells (F4 line) were a generous gift from Janet Rossant (University of Toronto, Toronto, ON, Canada) and were also generated in-house, as previously described21. TS cells were cultured on gelatin-coated plates in TS cell media (Roswell Park Memorial Institute (RPMI)-1640 media supplemented with 20% FBS, 25 ng/mL fibroblast growth factor 4 (235-F4-025, FGF4, R&D systems, Minneapolis, MN, USA), 1 µg/mL heparin, 10 ng/mL activin A (338-AC-10, R&D systems), 100 μM 2-mercaptoethanol, 100 mM sodium pyruvate, 100 units/mL penicillin, and 100 µg/mL streptomycin), 70% of which was preconditioned by MEFs. Cells were passaged by light trypsinization prior to reaching confluency and were routinely tested for the presence of mycoplasma to ensure consistency across passages. TS cells were induced to differentiate for 5 days by removal of MEF preconditioned media, FGF4, heparin, and activin A.
Fetal hearts were cultured in Medium 199 (ThermoFisher Scientific) supplemented with 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin (henceforth referred to as complete cardio media) for a total of 48 h. To isolate cardiomyocytes, fetal hearts were collected in ice-cold Hanks’ Balanced Salt Solution and digested with Liberase (22.5 µg/mL, Sigma-Aldrich) for 30 min. The digested suspension was centrifuged, resuspended in complete cardio media, and plated on collagen-coated (5 µg/mL) tissue culture plates for 1 h to deplete fibroblasts. Cardiomyocytes were then collected, and approximately 4 × 104 cells were plated onto 24-well tissue culture plates and supplemented with complete cardio media for up to 72 h.
Immunohistochemistry and immunofluorescence
Frozen E9.5 mouse placentas were embedded in optimal cutting temperature compound (Tissue-Tek, Torrance, CA, USA), and cryosectioned at 10 μm. Tissue sections were fixed in 4% paraformaldehyde, permeabilized using 0.3% Triton X-100 and 1% bovine serum albumin in PBS, and blocked in 10% normal goat serum. Sections were incubated with antibodies specific for Cytokeratin conjugated to Alexa488 (628608, 1:400, BioLegend, San Diego, CA, USA) overnight at 4 °C. All other immunohistochemistry procedures were performed using sections of fixed, paraffin-embedded tissues. Tissues were dehydrated, embedded in paraffin and sectioned at 5 μm thickness. Sections were dewaxed in Histo-Clear and rehydrated using increasing dilutions of ethanol. Formaldehyde crosslinks were fragmented by placing slides in Antigen Decloaker (ThermoFisher Scientific) at 95 °C for 20 min. Sections were then blocked in 10% normal goat serum, and immersed in antibodies specific for Troponin-T (20025, 1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA), MCT1 (AB1286-I, 1:200, Sigma-Aldrich) or MCT4 (AB3314P, 1:200, Sigma-Aldrich). Sections were washed, incubated with species-appropriate secondary antibodies conjugated to Alexa-488 or Alexa-555, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific). Sections were mounted and imaged using a Nikon DS-Qi2 microscope (Nikon Instruments Inc., Melville, NY, USA).
For immunocytochemistry, cardiomyocytes were fixed in 4% paraformaldehyde, permeabilized using 0.3% Triton X-100 and 1% bovine serum albumin in PBS, and blocked in 10% normal goat serum. All other steps were identical to those described above for tissue sections. Cells were imaged using a Zeiss Axio fluorescence microscope (Zeiss, Toronto, ON, Canada).
In situ hybridization
Tissue sections were subjected to RNAScope analysis, as per the manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA, USA). Briefly, sections were hydrated using a graded series of ethanol washes, subjected to peroxidase and protease treatment, and hybridized with probes specific to mouse Ovol2 (558501, Advanced Cell Diagnostics). Tissue sections were then subjected to a series of amplification steps, treated with Fast Red chromogen solution, and nuclei counterstained with hematoxylin. Sections were dehydrated, mounted using Cytoseal (ThermoFisher Scientific), and imaged using a Nikon DS-Qi2 microscope. To validate that the procedure worked correctly, Ppib (313918) and dapB (310043, Advanced Cell Diagnostics) probes were used in parallel to confirm positive and negative staining, respectively.
Isolation of EVs from media conditioned by placentas and TS cells
EV isolation from CM was performed as previously described48. Briefly, placentas were isolated from pregnant C57BL/6 N dams on E15.5 and cultured in complete cardio media. E15.5 was used to maximize the amount of EVs collected from placental tissue. To obtain TS cell CM, Ovol2+/+ and Ovol2-/- TS cells were cultured in stem media, or maintained in differentiation media for 5 days. Subsequently, media were replaced with complete cardio media for an additional 48 h. During the preparation of media for these experiments, FBS was centrifuged at 100,000 × g for 1 h to deplete EVs. CM were collected every 24 h for 48 h. Following 48 h, media were centrifuged at 1000 × g for 10 min to remove residual cells and debris, and stored at –80 °C until use.
To isolate EVs, media conditioned by TS cells and placental tissue was first centrifuged at 100,000 × g using a SW 41 Ti rotor (Beckman Coulter, Mississauga, ON, Canada) for 1 h at 4 °C. Media supernatant was collected. The remaining pellet containing EVs was resuspended in 200 µL of ice-cold PBS and filtered using a 0.22 µm filter. A small fraction of the media supernatant and EV suspension (5 µL) was lysed in 0.2% sodium dodecyl sulphate (SDS) supplemented with protease inhibitor cocktail (Sigma-Aldrich), and subjected to a bicinchoninic acid assay (Bio-Rad Laboratories) to determine protein concentrations. The remainder of the media supernatant and EV suspension was kept in low-attachment glass vials and stored at –80 °C until use.
Fetal hearts and cardiomyocytes were cultured in CM, media supernatants, or in the presence of EVs (1–10 µg/mL) for 48 h to assess heart rate, epicardial cell outgrowth, and expression of genes important for cardiomyocyte maturation.
Electron Microscopy
After media collection and centrifugation, the supernatant was passed through a 0.22 µm filter to remove cellular debris, concentrated using Amicon Ultra 10 kDa filter units (Sigma-Aldrich), and ultracentrifuged at 100,000 × g for 75 min at 4 °C. Pellets were fixed in a solution containing 2% paraformaldehyde, 100 mM glutaraldehyde, and 100 mM sodium cacodylate (pH 7.0) for 1 h at 4 °C, then washed in PBS and ultracentrifuged. Washing was repeated an additional two times. Pellets were stored in 50 µl PBS until imaging. EV pellets were placed on a thin carbon film and air dried. Images were acquired using a Philips CM10 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands).
RT-PCR and quantitative RT-PCR
To isolate RNA from cells and tissue, Ribozol (Amresco, Mississauga, ON, Canada) was used according to the manufacturer’s protocol. RNA was converted into complementary DNA (cDNA) using a High Capacity cDNA Kit (ThermoFisher Scientific). All cDNA was diluted 1:10 and subjected to RT-PCR or quantitative RT-PCR using primers detailed in Table 1. RT-PCR was performed using DreamTaq DNA Polymerase (ThermoFisher Scientific). Cycling conditions consisted of an initial holding step (95 °C for 3 min), followed by 32 cycles of PCR (95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s), and a final elongation phase at 72 °C for 12 min. Quantitative RT-PCR was performed using a CFX96 Touch (Bio-Rad Laboratories, Mississauga, ON, Canada) and Sensifast SYBR Green PCR Master Mix (FroggaBio, Toronto, ON, Canada). Cycling conditions involved an initial holding step (95 °C for 10 min), followed by 40 cycles of a two-step PCR (95 °C for 15 s and 60 °C for 1 min) and a dissociation phase. Relative transcript levels were calculated using the ΔΔCt method, using the geometric mean from two constitutively expressed genes: Rn18s and Gapdh.
Western blotting
Lysates from cells were prepared using radioimmunoprecipitation assay lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), supplemented with protease inhibitor cocktail. Lysates from EVs and media supernatants were prepared as described above. A bicinchoninic acid assay was used to measure protein concentrations. Approximately 25 µg of protein from media supernatants, as well as cell and EV lysates was mixed with 4× reducing loading buffer (0.25 M Tris, 8% SDS, 30% glycerol, 0.02% Bromophenol blue, 0.3 M dithiothreitol), boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane and probed using antibodies for Troponin T (20025, 1:1,000, Santa Cruz Biotechnology), NKX2-5 (376565, 1:1,000, Santa Cruz Biotechnology), TSG101 (125011, 1:1,000, Abcam, Boston, MA, USA), HSP70 (5439, 1:1,000, Abcam), or ACTB (47778, 1:1,000, Santa Cruz Biotechnology). Membranes were then incubated for 1 h with species-appropriate secondary antibodies, and signals detected using a LI-COR Odyssey imaging system. To confirm that equal amounts of protein was loaded when comparing lysates from EVs or media supernatants, Ponceau S (Sigma-Aldrich) was used to stain polyvinylidene difluoride membranes.
EdU cell proliferation assay
An EdU incorporation assay (Click-iT EdU Cell Proliferation Assay, ThermoFisher Scientific) was conducted to assess cardiomyocyte proliferation, as previously described49. Briefly, 10 µM EdU was provided to cardiomyocytes for 48 h. Following incubation, cells were fixed in 4% paraformaldehyde, permeabilized using 0.3% Triton X-100, and incubated with Click-iT EdU reaction cocktail for 30 min. Nuclei were stained using DAPI. Cells were imaged using a Zeiss Axio fluorescence microscope.
Quantification of EVs using nanoflow cytometry
To quantify the number of placental EVs present in plasma, whole blood was collected from non-pregnant virgin mice, pregnant dams, and fetuses at E18.5 and stored in ethylenediaminetetraacetic acid-coated tubes. The blood was then centrifuged at 2000 × g for 15 min to deplete cells and platelets. Plasma was collected, diluted 1:20 in PBS and incubated with a fluorescein isothiocyanate (FITC)-conjugated antibody specific to syncytin-A (1:100, SYCY1-FITC, FabGennix International Inc., Frisco, TX, USA) and a phycoerythrin (PE)-conjugated antibody specific to CD9 (1:100, 124806, BioLegend) for 1 h. Following the incubation, the sample was loaded in triplicate on the Apogee A-60 nanoflow cytometer (Apogee Flow Systems, Hemel Hempstead, Hertfordshire, UK) with autosampler, capable of EV resolution between ~150 and 1,000 nm in diameter50. 100 µL of sample was injected and analyzed at 1.5 µL/min for 1 min. Dual-positive microparticles were considered placental EVs. EV size was estimated based on large-angle light scatter (LALS) and small-angle light scatter (SALS). Silica beads ranging from 80-1,300 nm in diameter were initially injected to validate the capacity of the cytometer to differentiate particles based on size (Supplementary Fig. 3), although it should be noted that there was no correction applied for differences in refractive indices between silica beads and EVs. Before running plasma samples, a PBS sample was first run to ensure that background levels did not exceed 100 events/s. Pooled unstained and single stained plasma samples were additionally run to determine ideal gating strategies for FITC and PE, which was done in accordance with the consensus framework for the minimum information to be reported for EV flow cytometry experiments51. Isotype controls were not run due to limited amounts of fetal plasma. Coefficient of variation was ≤10% for all samples. The instrument settings included a sheath pressure of 150 mbar and laser settings at 100 mW for 405 nm (violet), 70 mW for 488 nm (green), and 70 mW for 638 nm (red). LALS and SALS events were produced using the 405 nm laser. To eliminate background noise, a threshold of 21 a.u. and 34 a.u. were set for LALS and SALS, respectively. The photomultiplier tube voltage thresholds were: 405-LALS: 265 V, 405-SALS: 340 V, 488-Grn: 525 V, 488-Org: 500 V.
To quantify the number and size of EVs present in media conditioned by cells, isolated EV fractions were diluted 1:10 and loaded in triplicate on the Apogee A-60 nanoflow cytometer. The total number of events and EV size, estimated based on the size of silica beads, was collected.
Statistics and Reproducibility
To quantify fetal heart rate and epicardial cell outgrowth, hearts were collected from embryos from at least five different pregnant dams, and phase-contrast images of fetal hearts were taken every 24 h for 48 h. Images were imported into ImageJ (version 1.52)52 and analyzed. Outgrowth was measured by determining the number of pixels in a straight line from the start of the epicardium. Each heart was measured three independent times and measurements were averaged. Heart rate was measured by counting the number of heart beats within 1 min; three measurements were taken for each heart and averaged to determine beats per min. Heart rate was measured every 24 h for 48 h. Statistical comparisons between two means were tested using two-tailed Student’s t-test and statistical comparisons between three or more means were tested using one-way analysis of variance (ANOVA), followed by a Tukey’s post-hoc test. Means were considered statistically different if P was less than 0.05. GraphPad Prism 9.5 was used for all graphing and statistical analysis. Unless indicated otherwise in the figure legends, experiments were repeated at least three independent times using distinct biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
We would like to thank Tunyalux Langsub for illustrative assistance, Stephen Pasternak (Western University) for granting access to the Apogee A-60 nanoflow cytometer, and Reza Khazaee (Biotron Integrated Microscopy Facility, Western University) for electron microscopy support. Experiments in this study were supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; 05053/04543), with additional support from the Canadian Institutes of Health Research (PJT152983), both to S.J.R. Fellowships from the NSERC Alexander Graham Bell Canada Scholarships-Doctoral Program supported M.J.J., G.J.B., and V.S.P. V.S.P. was also supported by a Children’s Health Research Institute fellowship.
Author contributions
M.J.J. and S.J.R. designed the study. M.J.J. performed all experiments. V.S.P. and G.J.B. contributed to experimental design and assisted with animal breeding and cell culture. L.Z. and S.N.W. assisted with EV isolations and nanoflow cytometry. M.J.J. and S.J.R. analyzed data and wrote the paper. All authors approved the final version of the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Gregory Lavieu and Joao Valente. A peer review file is available.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and supplementary materials. The source data file underlying all graphs and tables can be found in Supplementary Data 1. Unprocessed DNA gels and western blot images can be found in Supplementary Fig. 7 and 8, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06938-4.
References
- 1.Linask, K. K. The heart-placenta axis in the first month of pregnancy: induction and prevention of cardiovascular birth defects. J. Pregnancy2013, (2013). [DOI] [PMC free article] [PubMed]
- 2.Maslen, C. L. Recent Advances in Placenta-Heart Interactions. Front. Physiol.9, 735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo, D. M. et al. The outcome of open heart surgery for congenital heart disease in infants with low body weight less than 2500 g. Pediatr. Cardiol.32, 578–584 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Best, K. E., Tennant, P. W. G. & Rankin, J. Survival, by Birth Weight and Gestational Age, in Individuals With Congenital Heart Disease: A Population-Based Study. J. Am. Heart Assoc.6, e005213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suluba, E., Shuwei, L., Xia, Q. & Mwanga, A. Congenital heart diseases: genetics, non-inherited risk factors, and signaling pathways. Egypt. J. Med. Hum. Genet. 2020 21121, 1–12 (2020). [Google Scholar]
- 6.Burton, G. J. & Jauniaux, E. Development of the human placenta and fetal heart: Synergic or independent? Front. Physiol.9, 373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camm, E. J., Botting, K. J. & Sferruzzi-Perri, A. N. Near to one’s heart: The intimate relationship between the placenta and fetal heart. Front. Physiol.9, 629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renaud, S. J. & Jeyarajah, M. J. How trophoblasts fuse: an in-depth look into placental syncytiotrophoblast formation. Cell. Mol. Life Sci.79, 433 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahara, A. et al. Circulating Placental Extracellular Vesicles and Their Potential Roles During Pregnancy. Ochsner J.20, 439–445 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, J., Li, H., Fan, B., Xu, W. & Zhang, X. Extracellular vesicles in normal pregnancy and pregnancy-related diseases. J. Cell. Mol. Med.24, 4377–4388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortega, M. A. et al. The Pivotal Role of the Placenta in Normal and Pathological Pregnancies: A Focus on Preeclampsia, Fetal Growth Restriction, and Maternal Chronic Venous Disease. Cells11, (2022). [DOI] [PMC free article] [PubMed]
- 12.Ferreira, B. D., Barros, T., Moleiro, M. L. & Guedes-Martins, L. Preeclampsia and Fetal Congenital Heart Defects. Curr. Cardiol. Rev. 18, (2022). [DOI] [PMC free article] [PubMed]
- 13.Chen, J. et al. Molecular effects of lithium exposure during mouse and chick gastrulation and subsequent valve dysmorphogenesis. Birth Defects Res. A. Clin. Mol. Teratol.82, 508–518 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Manisastry, S. M., Han, M. & Linask, K. K. Early temporal-specific responses and differential sensitivity to lithium and Wnt-3A exposure during heart development. Dev. Dyn. Publ. Am. Assoc. Anat.235, 2160–2174 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Adams, R. H. et al. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell6, 109–116 (2000). [PubMed] [Google Scholar]
- 16.Barak, Y. et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell4, 585–595 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Shaut, C. A. E., Keene, D. R., Sorensen, L. K., Li, D. Y. & Stadler, H. S. HOXA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet4, e1000073 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Garcia, V. et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature555, 463–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radford, B. N. et al. Defects in placental syncytiotrophoblast cells are a common cause of developmental heart disease. Nat. Commun.14, 1174 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. G. et al. PIBF1 regulates trophoblast syncytialization and promotes cardiovascular development. Nat. Commun.15, 1487 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeyarajah, M. J., Jaju Bhattad, G., Hillier, D. M. & Renaud, S. J. The Transcription Factor OVOL2 Represses ID2 and Drives Differentiation of Trophoblast Stem Cells and Placental Development in Mice. Cells9, 840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unezaki, S., Horai, R., Sudo, K., Iwakura, Y. & Ito, S. Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation and placental development in mice. Genes Cells12, 773–785 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Nagai, A., Takebe, K., Nio-Kobayashi, J., Takahashi-Iwanaga, H. & Iwanaga, T. Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta31, 126–133 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Trembley, M. A., Velasquez, L. S. & Small, E. M. Epicardial Outgrowth Culture Assay and Ex Vivo Assessment of Epicardial-derived Cell Migration. J. Vis. Exp.2016, 53750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, C. et al. Placenta-derived extracellular vesicles induce preeclampsia in mouse models. Haematologica105, 1686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaremek, A., Jeyarajah, M. J., Jaju Bhattad, G. & Renaud, S. J. Omics Approaches to Study Formation and Function of Human Placental Syncytiotrophoblast. Front. Cell Dev. Biol.9, 1543 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todorova, D., Simoncini, S., Lacroix, R., Sabatier, F. & Dignat-George, F. Extracellular Vesicles in Angiogenesis. Circ. Res.120, 1658–1673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle, L. M. & Wang, M. Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells8, 727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamova, P., Lotto, R. R., Powell, A. K. & Dykes, I. M. Are there foetal extracellular vesicles in maternal blood? Prospects for diagnostic biomarker discovery. J. Mol. Med. (Berl). 10.1007/s00109-022-02278-0 (2022). [DOI] [PMC free article] [PubMed]
- 30.Takahashi, H. et al. Endogenous and exogenous miR-520c-3p modulates CD44-mediated extravillous trophoblast invasion. Placenta50, 25–31 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Kambe, S. et al. Human exosomal placenta-associated miR-517a-3p modulates the expression of PRKG1 mRNA in Jurkat cells. Biol. Reprod.91, 129 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Delorme-Axford, E. et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl Acad. Sci. USA.110, 12048–12053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni, J. et al. Human trophoblast-derived exosomes attenuate doxorubicin-induced cardiac injury by regulating miR-200b and downstream Zeb1. J. Nanobiotechnol.18, 171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng, Y. et al. Normotensive placental extracellular vesicles provide long-term protection against hypertension and cardiovascular disease. Am. J. Obstet. Gynecol. 10.1016/j.ajog.2023.12.030 (2023). [DOI] [PubMed]
- 35.Zhao, X., Radford, B. N., Ungrin, M., Dean, W. & Hemberger, M. The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes. Int. J. Mol. Sci.24, 12423 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang, Y. et al. Isolation of human trophoblastic extracellular vesicles and characterization of their cargo and antiviral activity. Placenta47, 86–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong, M. et al. Proteomic characterization of macro-, micro- and nano-extracellular vesicles derived from the same first trimester placenta: relevance for feto-maternal communication. Hum. Reprod.31, 687–699 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Stefanski, A. L. et al. Murine trophoblast-derived and pregnancy-associated exosome-enriched extracellular vesicle microRNAs: Implications for placenta driven effects on maternal physiology. PLoS One14, e0210675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godakumara, K. et al. Trophoblast derived extracellular vesicles specifically alter the transcriptome of endometrial cells and may constitute a critical component of embryo-maternal communication. Reprod. Biol. Endocrinol.19, 115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Addo, K. A., Palakodety, N., Hartwell, H. J., Tingare, A. & Fry, R. C. Placental microRNAs: Responders to environmental chemicals and mediators of pathophysiology of the human placenta. Toxicol. Rep.7, 1046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malnou, E. C., Umlauf, D., Mouysset, M. & Cavaillé, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet.9, 706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue, K. et al. The Rodent-Specific MicroRNA Cluster within the Sfmbt2 Gene Is Imprinted and Essential for Placental Development. Cell Rep.19, 949–956 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Paikari, A., Belair, C. D., Saw, D. & Blelloch, R. The eutheria-specific miR-290 cluster modulates placental growth and maternal-fetal transport. Development144, 3731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouillet, J.-F., Ouyang, Y., Coyne, C. B. & Sadovsky, Y. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol.213, S163–S172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, P., Ma, Y., Wu, H. & Wang, Y. L. Placenta-Derived MicroRNAs in the Pathophysiology of Human Pregnancy. Front. cell Dev. Biol. 9, (2021). [DOI] [PMC free article] [PubMed]
- 46.Murugesan, S. et al. Role of exosomal microRNA signatures: An emerging factor in preeclampsia-mediated cardiovascular disease. Placenta103, 226–231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn, J., Kunath, T. & Rossant, J. Mouse Trophoblast Stem Cells. Methods Mol. Med.I, 125–148 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Roseborough, A. D. et al. Plasma derived extracellular vesicle biomarkers of microglia activation in an experimental stroke model. J. Neuroinflam.20, 20 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeyarajah, M. J. et al. The multifaceted role of GCM1 during trophoblast differentiation in the human placenta. Proc. Natl Acad. Sci. Usa.119, e2203071119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomes, J. et al. Analytical Considerations in Nanoscale Flow Cytometry of Extracellular Vesicles to Achieve Data Linearity. Thromb. Haemost.118, 1612–1624 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Welsh, J. A. et al. MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicl.9, 1713526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and supplementary materials. The source data file underlying all graphs and tables can be found in Supplementary Data 1. Unprocessed DNA gels and western blot images can be found in Supplementary Fig. 7 and 8, respectively.