Abstract
End-stage simian immunodeficiency virus (SIV) isolates are suggested to be the most fit of the evolved virulent variants that precipitate the progression to AIDS. To determine if there were common characteristics of end-stage variants which emerge from accelerated cases of AIDS, a molecular clone was derived directly from serum following in vivo selection of a highly virulent SIV isolate obtained by serial end-stage passage in rhesus monkeys (Macaca mulatta). This dominant variant caused a marked cytopathic effect and replicated to very high levels in activated but not resting peripheral blood lymphocytes. Furthermore, although this clone infected but did not replicate to detectable levels in rhesus monocyte-derived macrophages, these cells were able to transmit infection to autologous T cells upon contact. Interestingly, although at low doses this end-stage variant did not use any of the known coreceptors except CCR5, it was able to infect and replicate in human peripheral blood mononuclear cells homozygous for the Δ32 deletion of CCR5, suggesting the use of a novel coreceptor. It represents the first pathogenic molecular clone of SIV derived from viral RNA in serum and provides evidence that not only the genetic but also the biological characteristics acquired by highly fit late-stage disease variants may be distinct in different hosts.
Simian immunodeficiency virus (SIV) of sooty mangabeys causes AIDS in macaques, providing an important animal model for human immunodeficiency virus (HIV)-induced AIDS in humans (13, 19, 21, 31, 41). Molecular clones of HIV and SIV have been valuable for addressing specific questions in AIDS pathogenesis (29, 45, 53), vaccine development (6, 9, 10, 13, 30, 56, 60), and the evaluation of antiviral drugs (2, 63). To date, molecular clones of SIV have been derived from proviral DNA rather than viral RNA, and most proviral clones have been obtained from cultured cells and frequently from infected human cell lines (14, 23, 31, 36, 45, 50, 52). It has been demonstrated that by in vitro propagation certain viral variants are selected (20, 65). In particular, growth of virus in human cell lines results in major changes in the SIV genome, such as deletions leading to truncation of the transmembrane envelope protein (7, 24, 34, 35). This has resulted in important biological differences between the derived clones and the original pathogenic virus population in the host. In addition, proviral DNA frequently contains a high proportion of defective proviruses (37, 42, 43, 47, 66).
Recently it has been demonstrated that late-stage SIVmne variants in pigtail macaques are highly fit, having acquired multiple mutations encoded at several genetic loci that facilitate immune escape and increase replication and cytopathic properties (32). These observations have recently been supported by another line of evidence. A series of in vivo passage studies were performed in which blood samples taken at the time of AIDS development were subsequently used to infect naive rhesus macaques. End-stage blood samples were taken from the most rapidly progressing animal and passaged again in vivo. This in vivo passage of end-stage variants resulted in a progressively accelerated disease course with each successive passage until the fourth passage, by which time AIDS had developed in as little time as 2 weeks (28). Taken together, the results of these two independent lines of investigation suggested that the passage of primate lentiviruses late in disease could result in the transmission of highly virulent variants capable of causing rapid progression to AIDS. The data suggested that highly fit end-stage or late-stage fitness variants had common biological properties (32).
The provirus population in mononuclear cells in vivo is generally considered to be a sanctuary of biological variants which have accumulated in such intracellular reservoirs as a consequence of previous host immune pressures and/or defective viral replication (66). Alternatively, we reasoned that extracellular virions represented the most actively replicating, dominant virus population, having become the most predominant in the host at a particular stage in disease development. SIV and HIV clones that are derived directly from extracellular virus populations in biological fluids such as serum have not been characterized or evaluated for virulence in vivo. A key feature of lentivirus pathogenesis is a persistent high-level cell-free viremia. Since the predominance of certain extracellular lentiviruses after seroconversion is most likely the result of escape from immune surveillance or escape from drug therapy during treatment, such viral variants are of particular biological interest. Recently we developed a strategy to generate pathogenic clones directly from extracellular virions present in the circulation, in serum or plasma (25, 26). Using this strategy, we derived a full-length infectious molecular clone of SIV8980, an end-stage isolate from a macaque which had progressed rapidly to AIDS following serial end-stage passage of SIVB670 in vivo (27). Sequence analysis revealed a unique relationship, placing this virus between the two groups of SIVsm and SIVmac primate lentiviruses. During in vivo passage the variability of the V1 envelope region decreased as virulence increased. The SIVF359 molecular clone represented the most dominant variant that had emerged during end-stage passage. This variant was highly cytopathic and replicated to high titers in vivo. It was predominantly T-cell tropic, infecting macrophages but not productively replicating in macrophages at detectable levels. Interestingly, if autologous lymphocytes were placed in direct contact with macrophages exposed to this clone, a very high level of viral production was found in the culture supernatants. Of all the known coreceptors, SIVF359 was highly selective for CCR5. However, it could replicate in human peripheral blood mononuclear cells (PBMC) homozygous for the Δ32 deletion, suggesting the possible use of a novel coreceptor.
MATERIALS AND METHODS
Molecular clone derived from serum.
SIV8980 was derived from SIVB670 by four in vivo passages in Indian rhesus macaques. Monkey 8980 rapidly progressed to AIDS following the fourth in vivo passage (27). Serum from this animal was, without culture, directly used to derive the F359 molecular clone of SIV (25). Since the synthesis of full-length (10-kb) SIV cDNA molecules from small amounts of RNA templates proved to be very difficult, a modified reverse transcription-PCR technique was developed to separately generate 5′ and 3′ halves of the SIV genome (26). By ligating these two 5-kb fragments, we were able to reconstitute the SIVF359 infectious molecular clone directly from serum as we have previously described (25).
Infectivity in vitro
Primary rhesus PBMC cultures were maintained with medium changes every 2 days and were observed regularly for cytopathic effect (CPE). Infection was confirmed by immunocytochemistry for the expression of SIV Gag antigen. Single-cell preparations for immunocytochemistry were prepared on acetone-cleaned glass slides which had been air dried for 30 min. Cells were fixed in acetone-methanol (1:1) and in ethanol (70%) for 15 and 30 min, respectively. Slides were washed in 0.05 M Tris-HCl (pH 7.6)–0.1 M NaCl for 5 min and incubated with 20 μl (1:25 dilution) of anti-Gag monoclonal antibody (51). Cells were washed for 5 min and incubated with goat anti-mouse immunoglobulin G (IgG) antibody for 30 min at room temperature. To amplify the signal, the cells were then washed and incubated with mouse anti-alkaline phosphatase (APAAP complex; Boehringer). Cells were washed for 5 min and were incubated with 20 μl of substrate solution (0.1 M Tris-HCl [pH 9.5], nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate, 5 mM levamisole [10:1:1]) for 30 min at room temperature. Cells were washed for 5 min in tap water, 1 drop of 50% glycerol–phosphate-buffered saline (PBS) was added per slide, and the cells was covered with a coverslip. The preparations were examined at a magnification of ×40,and CPE was quantified. In cell culture supernatants virus was quantified by measuring p27 concentrations (SIV p27 antigen capture enzyme-linked immunosorbent assay; Coulter Corp., Hialeah, Fla.).
Pathogenicity.
Adult rhesus macaques (Macaca mulatta) used in this study were housed at the animal facility of the Biomedical Primate Research Centre, Rijswijk, The Netherlands. Animals were negative for SIV, simian T-cell leukemia virus, and simian D type retroviruses. Two outbred Indian rhesus macaques were inoculated intravenously with 50 50% tissue culture infective doses (TCID50) of the SIVF359 stock grown on rhesus PBMC. EDTA-treated blood samples were collected every 2 weeks postinfection for quantitative virus isolations (QVI) from PBMC and for determination of SIV p27 antigen in plasma. Rhesus monkeys that developed clinical evidence of AIDS were euthanized, and full pathological analysis was performed to confirm the diagnosis. For histological examination, tissues were formalin fixed and paraplast embedded. Four-micrometer-thick sections were stained with hematoxylin and eosin. For the detection of microsporidia, Gram staining was applied on gall bladder sections.
For QVI, PBMC were prepared from EDTA-treated blood by lymphocyte separation medium density gradient centrifugation. Cells at the interface were collected and washed twice with RPMI. Twofold dilutions of PBMC (starting with 106 cells) were cocultured with 2.5 × 105 cells of the human T-cell line C8166 in a 24-well plate (Greiner Labortechnik, Alphen a/d Rijn, The Netherlands) in duplicate. Cell culture medium (RPMI with 10% fetal calf serum [FCS]) was partly changed twice a week. The cell cultures were screened regularly for the presence of CPE.
The phenotype of rhesus PBMC was assessed by two-color fluorescence-activated cell sorter (FACS) analysis. Briefly, heparinized blood (100 μl) was incubated with 10 μl of monoclonal antibody mix at room temperature. After incubation, 2.5 ml of lysing solution (Becton Dickinson, Etten-Leur, The Netherlands) was added, followed by an incubation at room temperature for 10 min and then centrifugation for 10 min at 200 × g. Four milliliters of PBS with 2% formaldehyde was added, and the tubes were centrifuged for 10 min at 200 × g. The supernatant was aspirated, and the cells were resuspended in 5 ml of PBS with 2% formaldehyde and stored overnight at 4°C. Flow cytometry was performed on a FACScan using the CellQuest software (Becton Dickinson), with 5,000 events analyzed. To assess CD4 T-cell levels in peripheral blood, the following monoclonal antibodies were used: an anti-CD3 monoclonal antibody (FN18; Biomedical Primate Research Centre) covalently coupled to fluorescent isothiocyanate-phycoerythrin and an anti-CD4 monoclonal antibody (SK3; Becton Dickinson) covalently linked to phycoerythrin conjugate. Once it was determined that this molecular clone was pathogenic and caused AIDS in two animals, an additional eight rhesus monkeys were infected and monitored for the time to development of AIDS.
Cell tropism.
To assess susceptibility to infection of resting and activated lymphocytes, blood was taken from healthy rhesus monkeys which were negative for SIV, simian T-cell leukemia virus, and type D retroviruses. PBMC were isolated by lymphocyte separation medium density gradient centrifugation and were washed twice with RPMI. Activated lymphocytes were prepared by concanavalin A mitogen stimulation (5 μg/ml; 48 h) and interleukin-2 (IL-2) treatment (50 U/ml, starting after virus adsorption and continuing throughout the experiment). Resting lymphocytes were cultured in RPMI (plus 10% FCS) without phytohemagglutinin and IL-2. Resting and stimulated lymphocytes were distinguished by double labeling with anti-CD3 antibody specific for T cells and anti-MIB-1 antibody specific for the cellular proliferation marker Ki-67 (3, 15, 40). Resting and stimulated cell cultures (5 × 106 cells) were simultaneously infected with 100 TCID50 of SIVmac239/YEnef (a molecular clone capable of proliferating in resting cells) (15) or of the SIVF359 clone at 37°C for 18 h. Unbound virus particles were removed by washing the cell pellets five times with 5 ml of RPMI (plus 10% FCS), and the cells were cultured for 12 days in RPMI (plus 10% FCS) either with or without IL-2 for stimulated or resting PBMC, respectively. Supernatants were monitored for the production of virus p27 by antigen capture enzyme-linked immunosorbent assay. The absence of Ki-67 staining was used to confirm that resting lymphocyte cultures remained in a quiescent state.
SIV p27 gag expression in monocyte-derived macrophage (MDM) and PBMC cultures was studied by double-staining immunocytochemistry. Briefly, cells were incubated with a mixture of the mouse anti-Gag monoclonal antibody 2E4 (IgG2a; kindly provided by M. Niedrig [51]) and mouse anti-CD68 monoclonal antibody KP1 (IgG1; DAKO, Glostrup, Denmark), which was used to costain macrophages. Subsequently, slides were incubated with alkaline phosphatase-conjugated goat anti-mouse IgG2a subclass-specific antibody (Southern Biotechnology Inc., Birmingham, Ala.) and horseradish peroxidase-conjugated goat anti-mouse IgG1 subclass-specific antibody (Southern Biotechnology). All incubation steps were performed at 20°C for 30 min. Endogenous peroxidases were blocked with 0.1% NaN3 plus 0.3% H2O2 in PBS after the incubation with the first antibody. Alkaline phosphatase activity was detected with naphthol-AS-MX phosphate (Sigma Chemical Co., St. Louis, Mo.) and Fast Blue BB (Sigma) in 0.1 M Tris-HCl (pH 8.5) (20 min in the dark), yielding a blue color. Horseradish peroxidase activity was detected using H2O2 (0.03%) and 3-amino-9-ethylcarbazole (Sigma), yielding a red color.
To determine if SIVF359 was able to infect rhesus MDM cultures, lymphocyte separation medium-isolated PBMC were seeded at a concentration of 5 × 106 cells per ml in 24-well plates in RPMI with 10% FCS. Adherence was allowed to continue for 5 days. Prior to infection, nonadherent cells were separated from adherent cells by rigorous washing with culture medium. Adherent cells were checked for purity (>98%) and for being macrophages by demonstrating the presence of the cell surface marker CD68 and by microscopic examination of their characteristic morphology. After infection with SIVmac316 (a macrophage-tropic molecular clone of SIV used as a positive control) (48), SIV8980 (the parental strain of SIVF359), or SIVF359, unbound virus was removed by washing the cells twice. MDM cultures were maintained for 12 days in RPMI 1640 medium supplemented with 20% FCS, penicillin, and streptomycin with medium changes once per week. At day 12 samples were analyzed for intracellular gag expression and the presence of SIV p27 Gag antigen in supernatants.
Studies were subsequently undertaken to determine if cell-cell contact between MDM previously exposed to virus could result in productive infection of autologous lymphocytes. MDM cultures (prepared as described above) were exposed to either the parental SIV 8980 or the molecular clone variant SIV F359 (both at 3,000 TCID50). On the following day residual virus was removed from cultures by vigorous washing at least three times. Specific infection was determined by double staining of MDM for the specific marker CD68 as well as viral p27. Virus replication in these cultures was determined by the amount of p27 present in the culture supernatants at days 0, 6, and 12 postinfection. On day 12 macrophage cultures were extensively washed, and autologous concanavalin A-stimulated rhesus lymphocytes (2 × 10 6) were added to the SIV 8980- and SIV F359-exposed MDM cultures. Virus replication and production in autologous lymphocytes after cocultivation with MDM cell interaction was measured by the amount of p27 which accumulated in the culture supernatants at days 0, 2, 6, 12, and 14 following cocultivation of the two different cell populations.
Coreceptor studies.
The coreceptor usage of the molecularly cloned virus was determined by three different assays. The first assay involved the use of HOS-CD4+ cell lines expressing either the macaque or the human CCR5 and was based on immunostaining of SIV-infected cells. The HOS-CD4+ cell lines were infected with SIVF359 by adding 104 TCID50 of virus per ml to the adherent cells in 3 ml of medium. After 72 h, cells were analyzed for syncytium formation, washed once in serum-free medium, fixed in methanol-acetone (50:50) for 2 min at −20°C, and washed twice in PBS supplemented with 1% FCS. Anti-Gag mouse monoclonal antibody (0.6 ml/well) was added, and the cells were incubated for 1 h at room temperature and washed three times in PBS supplemented with 1% FCS. Goat anti-mouse β-galactosidase-conjugated polyclonal antibodies (0.6 ml/well) were added, and the cells were incubated for 1 h at room temperature and washed three times in serum-free PBS. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) substrate (0.6 ml/well) was added, and the preparation was incubated in a sealed box for 30 min at 37°C. For quantification the stained cells were washed three times in PBS.
The second assay used the astroglia cell line U87 stably expressing human CD4 and one of the chemokine receptors CCR2b, CCR3, CCR5, or CXCR4. These cells were seeded in 24-well plates at 2 × 104 cells per well in 1 ml of medium. Infection was performed overnight at 37°C with 10-fold serial dilutions of virus (1-ml final volume) beginning with a 1:8 dilution of the SIVF359 stock. After infection, the cultures were washed three times with Dulbecco's modified Eagle medium (DMEM) (Gibco) and cultured for 13 days. Medium was changed twice a week. Cultures were examined microscopically for CPE, and supernatants, collected at several time points after infection, were tested for p27 concentration.
In the third assay several CD4-transformed human osteosarcoma HOS cell lines were used, expressing the chemokine receptors CCR1, CCR2b, CCR3, CCR4, CCR5, CXCR4, BOB/GPR15, Bonzo/STRL33, CXCR1 (V28), CCR8, APJ, GPR1, and US28 (the reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, from V. N. KewalRamáni and D. R. Littman). The CD4-transformed (under neomycin selection) HOS parental cells containing the HIV type 2 (HIV-2) long terminal repeat driving green fluorescent protein introduced via cotransfection with the cytomegalovirus promoter driving a hygromycin-resistant construct were maintained in DMEM supplemented with 10% FCS under selection with neomycin (G418 [0.5 mg/ml]; Gibco) and hygromycin (100 μg/ml). Coreceptor genes were introduced via retroviral infection with the pBABE-puro vector (11, 39) under selection with puromycin (1 μg/ml; Calbiochem, La Jolla, Calif.). For cell-free infection experiments, HOS-CD4 cells expressing the different coreceptors were seeded at 2 × 104 cells per well (2 ml) in 12-well plates and cultured in DMEM with 10% FCS. The next day, infection with the virus stocks (500 μl/well) was performed in the presence of Polybrene (20 μg/ml) overnight at 37°C. After infection, the cultures were washed and cultured for another day. Forty-eight hours after infection, cells were analyzed for green fluorescent protein fluorescence by FACS.
DNA sequencing and phylogenetic analyses.
Double-stranded plasmid DNA containing the 10-kb SIVF359 insert was used as a template for sequencing. DNA-sequencing reactions were carried out using dye-primer chemistry and were executed on a LiCor automated DNA sequencer. The entire insert was sequenced from both directions. Nucleotide sequences were aligned using ClustalW version 1.7 (61). Alignments were examined and adjusted as necessary using the Genetic Data Environment program (59). Regions of sequences that could not be unambiguously aligned were removed from subsequent analyses. Neighbor-joining phylogenetic analyses were conducted using the DNADIST and NEIGHBOR programs from version 3.5c of the PHYLIP package (17). Maximum-likelihood analyses of env sequences from selected SIVs were performed using the PAUP* program (version 4.0.0d60; D. Swofford) as follows. Initial maximum-likelihood estimates for the env tree were produced using a two-substitution-type model (HKY model) without rate variation among sites. The topology of this tree was used as a starting topology for subsequent analyses. The shape parameter (alpha) for the gamma distribution describing rate variation among sites was estimated using the maximum-likelihood method to be 0.22966. This value was used for the estimation of the parameters for the six-substitution-type model (general time reversible model). The values estimated (a = 2.499, b = 10.46, c = 1.278, d = 1.746, e = 9.962, and f = 1) were then in turn used to refine estimates. Final estimates for the parameters were as follows: a = 2.499, b = 10.44, c = 1.278, d = 1.745, e = 9.95, f = 1, and alpha = 0.23156.
RESULTS
Cell tropism in vitro
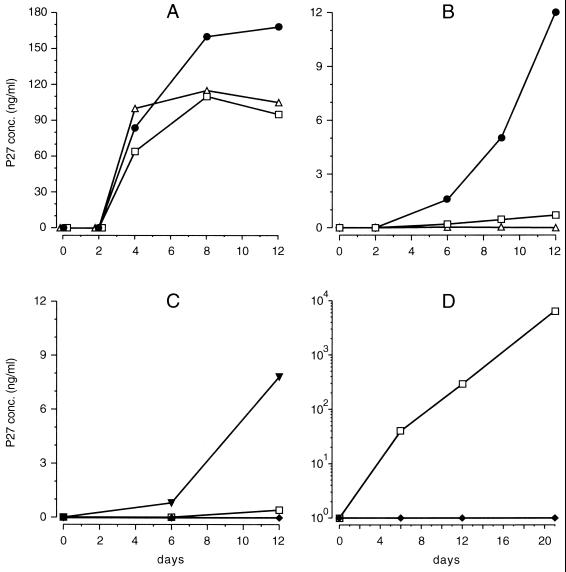
To determine the replicative properties and cell tropism of SIVF359, a series of in vitro assays were performed to compare the properties of this molecular clone with the well-established characteristics of two other well-defined SIVmac molecular clones. Mitogen-stimulated PBMC cultures were used to assess virus replication in rhesus lymphocytes. All viruses tested (SIVF359, SIVmac239/YEnef, and SIVmac239) grew well in stimulated rhesus lymphocyte cultures as measured by p27 concentrations in the supernatants (Fig. 1A). These data indicate that all viruses, including SIVF359, were able to infect and replicate in stimulated rhesus lymphocytes. Subsequently we determined the ability of SIVF359 to replicate in resting rhesus lymphocytes, a capability that has been reported for SIVsmmPBj (18) and SIVmac239/YEnef (15). Virus production was observed only in resting cell cultures infected with SIVmac239/YEnef at day 6 postinfection, producing 1.6 ng of p27 per ml in the supernatant, which increased to 12 ng of p27/ml by day 12 postinfection (Fig. 1B).
FIG. 1.
(A to C) Production of SIV p27 antigen in stimulated (A) and resting (nonstimulated) (B) rhesus lymphocyte cultures and in MDM cultures (C). Cultures were infected with equivalent amounts of either SIVF359 (□), SIVmac239/YEnef (●), SIVmac239 (▵), or SIVmac316 (▴). (D) Production of p27 in culture supernatants of lymphocytes following cocultivation with MDM. Virus production by the parental SIV8980 (⧫) was compared to that by the end-stage variant SIVF359 (□).
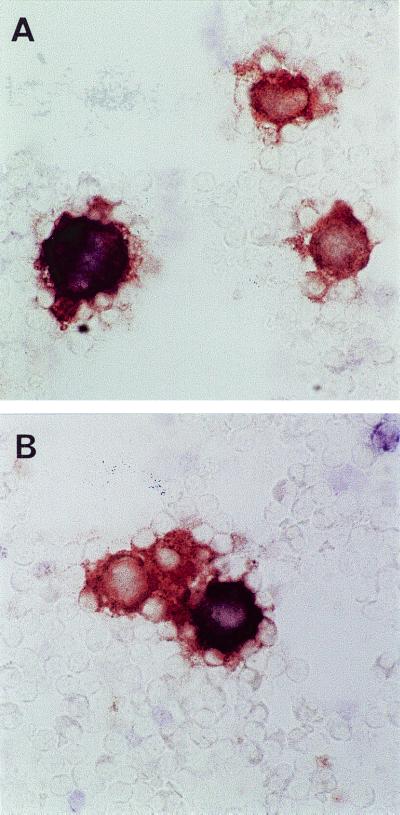
Infection of MDM cultures with the parental SIV 8980, the serum RNA-derived clone SIV F359, or the macrophage-tropic molecular clone SIVmac316 yielded detectable virus production only upon infection with SIVmac316, with p27 concentrations in cell supernatants increasing from 0.8 to 7.8 ng of p27/ml from day 6 to day 12, respectively. During the entire experimental period virus production in supernatants could not be detected in cultures infected with either the parental SIV 8980 or the serum RNA-derived clone SIVF359 (Fig. 1C). The absence of SIVF359 in the supernatants of MDM cultures was concluded to be due to the inability to either infect or replicate efficiently in this cell type. MDM cultures were double stained immunocytochemically for the presence of both the macrophage marker CD68 and the viral antigen p27. Analysis of stained cells indicated that SIV 8980 as well as SIVF359, although not able to produce detectable amounts of p27 in the supernatant, did infect macrophages and resulted in the expression of Gag protein (Fig. 2A). In contrast, SIVmac316 not only infected MDM (Fig. 2B) but also replicated well, producing substantial p27 concentrations in macrophage cultures (Fig. 1C).
FIG. 2.
Photomicrographs of MDM cultures infected with SIVmac316 (A) and SIVF359 (B); infection is demonstrated by CD68+-p27+ double staining. Magnification, ×660.
Infection of MDM cultures from different animals revealed the expression of p27 in macrophages but undetectable production of p27 in supernatants with either the parental SIV 8980 isolate or the SIV F359 variant. To determine if these antigen-positive nonproductively infected MDM were capable of transmitting the infection to other cell types, we cocultured autologous lymphocytes with these MDM cultures weeks after their exposure to virus. Importantly, the results revealed transfer to and productive infection of lymphocytes by the SIV F359 variant but not the parental SIV 8980 (Fig. 1D). Transmission to and infection by lymphocytes in this fashion by this late-stage variant resulted in production of virus at magnitudes greater than detected in the three previous assays (Fig. 1A, B, and C). In summary, these results add further insight to previous studies of late-stage variants in humans and macaques (32), indicating that the molecular clone SIVF359 was similar to cytopathic (syncytium-inducing), rapidly replicating, and predominantly T-cell-tropic variants but retained the ability to infect monocytes/macrophages, which upon contact with naive autologous T cells resulted in productive infection.
Infectious and pathogenic properties of SIVF359.
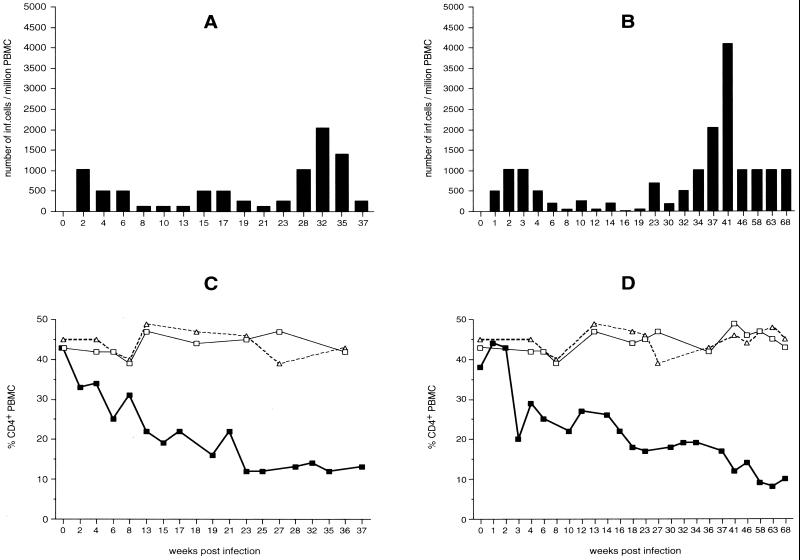
To investigate if the molecular clone SIVF359 was infectious and pathogenic in vivo, two rhesus macaques (L52 and WT) were intravenously infected with 100 TCID50 of SIVF359 propagated in rhesus PBMC. CD4+ T lymphocytes were monitored by FACS analysis, and fluctuations in virus loads were measured by QVI (Fig. 3). For animal L52, these parameters were determined at 2-week intervals. The same measurements were performed for WT at weekly intervals during the first month and subsequently at 2-week and monthly intervals. Animal L52 developed a virus load of 1,024 virus-producing cells/106 PBMC at 2 weeks after infection. Decreased values (524 virus-producing cells/106 PBMC) were detected at 4 and 6 weeks after infection, and the load dropped further to 128 virus-producing cells/106 PBMC at weeks 8, 10, and 14 postinfection A second peak of viremia, which reached 514 virus-producing cells/106 PBMC, was observed 15 and 17 weeks after infection. At weeks 19 and 21 virus loads decreased again to 256 and 48 infected cells per 106 PBMC, respectively. A third peak, which was higher than the previous ones, was observed at the end of the experimental period, from week 23 until week 37; virus loads of as high as 2,024 infected cells per 106 PBMC at week 32 were detected (Fig. 3A). The presence of this peak coincided with the development of severe anemia, diarrhea, and the start of extensive weight loss. L52 was euthanized after 37 weeks of infection.
FIG. 3.
Virus load (A and B) and changes in CD4+ lymphocyte populations (C and D) of rhesus monkeys WT (A and C) and L52 (B and D) after infection with molecularly cloned SIVF359. A progressive decline of CD4+ T lymphocytes in infected rhesus monkeys (WT and L52) (▪) compared to control monkeys 8637 (□) and 8711 (▵) is seen.
The first positive virus isolation from monkey WT was observed already at 1 week postinfection; 512 virus-producing cells per 106 PBMC were measured. This value increased to 1,024 virus-producing cells/106 PBMC at weeks 2 and 3 postinfection but dropped over time to 64 virus-producing cells/106 PBMC at week 8 after infection. Relatively low virus loads, fluctuating between 256, 64, 128, and 16 virus-producing cells/106 PBMC, were found at 10, 12, 14, 16, and 19 weeks postinfection. An increase in virus loads from 700 to 4,048 virus- producing cells/106 PBMC was then detected from week 23 to week 41, respectively. During the last 22 weeks (until week 68), virus loads stayed at a constant level of 1,012 virus-producing cells/106 PBMC (Fig. 3B). WT was euthanized after 68 weeks.
Histopathology revealed moderate catharrhalic enteritis and severe lymphoid hyperplasia in the spleen and lymph nodes, with moderate atrophy of splenic follicles. CD4 cell numbers in inoculated animals were compared to those in two uninfected rhesus monkeys of the same sex, age, and body weight. The two infected animals showed a decrease in CD4+-T-cell lymphocytes during the course of the experiment (25% for L52 [Fig. 3C] and 21% for WT [Fig. 3D]). CD4+ T lymphocytes cultured from these animals were positive by immunocytochemical staining for p27 antigen expression (data not shown). The uninfected animals had stable CD4 cell counts, as expected.
In both animals histopathological findings were consistent with the diagnosis of AIDS. Cryptosporidial enteritis without remission as seen in WT is observed in advanced immunodeficiency (46) and is one of the criteria for the diagnosis of AIDS in humans. In L52, a cholecystitis due to a microsporidial infection was identified. The etiologic organism has only recently been detected in SIV-infected rhesus monkeys and was classified as an Enterocytozoon bieneusi-like microsporidial protozoan. E. bieneusi is a common opportunistic pathogen of AIDS patients, causing significant morbidity.
Coreceptor usage.
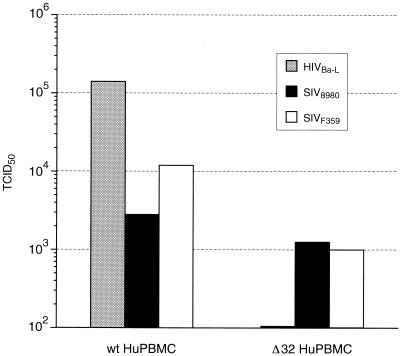
To further correlate the observations of cell tropism, the coreceptor use of SIVF359 was characterized. Infection experiments with SIVF359 virus were performed on HOS-CD4+ cell lines expressing either the macaque or human CCR5 coreceptor. Both cell lines were highly susceptible to infection with SIVF359, and almost 90% of the cells formed syncytia, which stained positive for SIV p27 antigen. To investigate the ability of SIVF359 to use additional coreceptors, the extent of its replication in GHOST cells and U87MG-CD4 cells expressing CCR1, CCR2b, CCR3, CCR4, CCR5, CXCR4, BOB/GPR15, Bonzo/STRL33, CX3Cr1 (V28), CCR8, APJ, GPR1, and US28 coreceptors (12, 22) was assayed in parallel with that of the well-characterized SIVmac239 and SIVmac316 molecular clones. All three viruses were found to use CCR5 for infection (Table 1) as determined by FACS analysis, while SIVF359 proved to be unable to use any coreceptor other than CCR5 at standard doses. Particular effort was made to determine if CXCR4 was used by this apparent T-cell-tropic variant; however, all assays were negative, corroborating several other reports that this group of viruses do not use this coreceptor (8, 16, 33). The observation that only CCR5 appeared to be utilized taken together with the ability of SIV F359 to infect T cells and macrophages was in agreement with previous reports that the CCR5 receptor is commonly used by both macrophage-tropic and T-cell-tropic primate lentiviruses (8, 16). Interestingly, both SIVmac239 and SIVmac316 used multiple coreceptors, including BOB/GPR15 and Bonzo/STPR, in contrast to the end-stage variant SIVF359. Furthermore, the macrophage-tropic molecular clone SIVmac316 used as a fourth coreceptor CCR3 (Table 1). Finally, to determine if the SIV F359 clone was truly restricted to only CCR5 coreceptor use, we compared the ability of the CCR5-dependent HIV-1Ba-L strain to productively infect human PBMC homozygous for the 32-bp deletion of CCR5 receptor gene, affecting CCR5 receptor expression (44), to those of the parental SIV8980 and variant SIVF359. These Δ32 cells are resistant to infection with viruses such as HIV-1Ba-L, which exclusively use the CCR5 coreceptor. As shown in Fig. 4, although the HIV-1Ba-L strain is completely blocked by the Δ32 CCR5 deletion, SIV8980 and SIVF359 grow to a slightly reduced but significant extent on the CCR5-defective cells. These data suggested that the SIVF359 clone may additionally utilize a novel, previously undescribed coreceptor for entry.
TABLE 1.
Comparison of coreceptor use and cell tropism of the well-defined T-cell-tropic and macrophage-tropic SIVmac strains compared to SIVF359a
| Property | Molecular clone
|
Parental strain SIV8980 | ||
|---|---|---|---|---|
| SIVmac239 | SIVmac316 | SIVF359 | ||
| Macrophage tropism | ||||
| Infection | − | + | + | + |
| Productive replication | − | + | − | − |
| Coreceptor use | ||||
| CCR1 | − | − | − | − |
| CCR2B | − | − | − | − |
| CCR3 | − | + | − | − |
| CCR4 | − | − | − | − |
| CXCR4 | − | − | − | − |
| CCR5 | + | + | + | + |
| BOB/GPR15 | + | + | − | − |
| Bonzo/STPR | + | + | − | − |
Infection by SIVmac239, SIVmac316, and SIVF359 was in HOS-CD4+, GHOST, and U87 cell lines stably expressing the human coreceptors listed (partial list).
FIG. 4.
Abilities of HIV-1Ba-L, SIVF359, and SIV8980 to grow on normal human PBMC (wt HuPBMC) versus PBMC homozygous for the Δ32 deletion in the CCR5 receptor gene (Δ32 HuPBMC). The CCR5-restricted HIVBa-L isolate could replicate only in wild-type human PBMC, while SIVF359 and SIV8980 were found to replicate in both Δ32 PBMC and normal human PBMC.
Is SIVF359 representative of the predominant variant following in vivo passage?
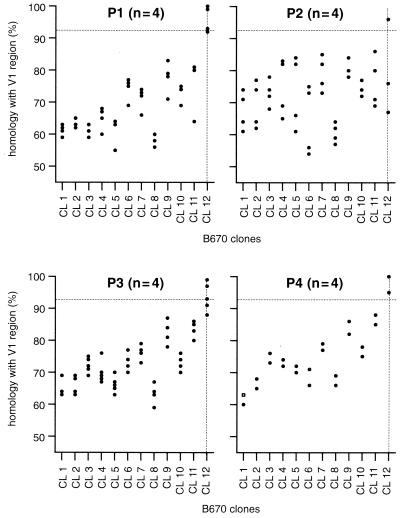
The first hypervariable region (V1) represents the most variable region of the SIV genome, and therefore it provides a sensitive indicator of the genotypic variation present within a quasispecies of an infected animal. Trichel et al. identified 12 different genotypes in the SIVB670 inoculum based on sequence analyses of the V1 region (62). The V1 regions derived from the viral variants emerging during in vivo passage (27) were determined (64) and compared with the V1 sequences of the viral variants that were present in the original SIVB670 inoculum (62). Homologies were quantified by using software that calculated the actual alignment fit as the fraction of the optimal fit. SIVB670-CL12 emerged as the predominant variant from the original SIVB670 infection and represented the highest genome stability during the passage experiment (Fig. 5). Clones obtained during subsequent passages represented viral variants that were more closely related to SIVB670-CL12 than to any other B670 clone (Fig. 5). This indicated that SIVB670-CL12 possessed the optimal fitness for replication in both B670 and the animals to which it was transmitted. Indeed, based on our observations of high persistent virus loads in the most rapidly progressing animals (27), certain variants may have acquired mutations making them relatively resistant to neutralization (32). V1 sequences derived from passage samples were compared to SIVB670-CL12, which had shown the highest homology with evolving variants during the passage study. Specifically, during the subsequent passages the number of different V1 clones decreased from four (out of four) to two (out of four) (Fig. 5). These findings demonstrated that during in vivo passage there was a selection for the most fit variants (32). The molecular clone SIVF359 represented the dominant variant and had retained the identical V1 sequences observed in P4.1, P4.2, and P4.3 envelope clones (Fig. 6).
FIG. 5.
Comparison of V1 sequences of envelope clones that emerged during the in vivo passages (P1 to P4) to obtain SIV8980, from which the molecular clone SIVF359 was derived, and V1 sequences of the viral variants (CL1 to CL12) which were present in the original SIVB670 inoculum (62, 64).
FIG. 6.
Amino acid sequence alignments of the V1 envelope region of the predominate clone 12 (CL12) in the prepassage SIVB670 inoculum. This sequence persisted through the passage as the diversity of the inoculum declined (Fig. 4). The CL12 V1 sequences are compared with the sequences of clones derived from the four different passages (P1 to P4). Clone P4.3 represents the same V1 sequence found in the molecular clone, SIVF359, and which was originally found in CL12 within the quasispecies found in the prepassage SIVB670 inoculum.
Phylogenetic analysis of SIVF359.
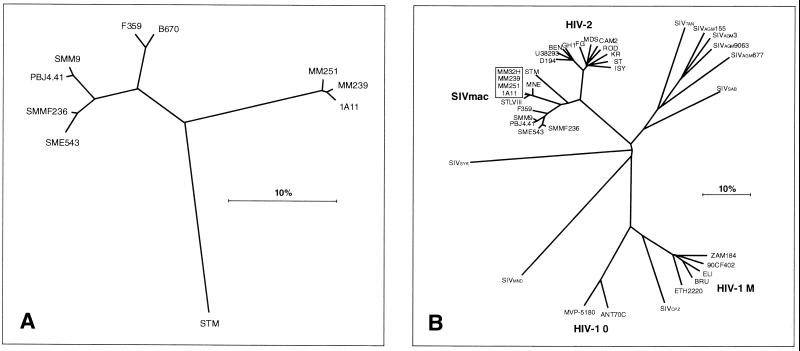
Phylogenetic analysis of env sequences using the neighbor-joining and general time reversible (GTR) maximum-likelihood method (17) confirmed that SIVF359 clustered together with SIVsm clones such as SIVsm9 and SIVsmmPBj4.41 and was not closely related to SIVmac clones such as SIVmac251, SIVmac239, and SIVmac1A11. As could be expected from the origin of SIVF359, this clone was most closely related to but distinct from SIVB670 (only the envelope sequence is available) (Fig. 7A). SIVB670 had been isolated from a rhesus macaque infected with material containing SIV from a sooty mangabey housed at the Tulane Primate Center (49). These two viruses branch separately from other characterized SIVsm viruses, which had also been derived from sooty mangabeys but originated from the Yerkes primate colony (18). Interestingly, these viruses formed a unique cluster of strains which branched separately from those macaque-adapted isolates derived from the New England (mac251 and mac239) and California (mac1A11) Regional Primate Centers (38). Neighbor-joining phylogenetic analysis performed on the entire sequence showed similar relationships within the SIV cluster (Fig. 7B). The predicted amino acid sequences of SIVF359 proteins were more similar to those of SIVsmH4 (87%) and SIVsmmPBj4.41 (88%) than to those of the SIVmac cluster (83%) (Table 2). The greatest similarity with other SIVs was present in the gag (average, 92%), pol (average, 93%), and vpx (average, 92%) genes, whereas the greatest divergence was seen in the nef (average similarity, 75%) and tat (average similarity, 72%) genes (Table 2).
FIG. 7.
(A) Phylogenetic tree based on the SIVF359 sequence compared with envelope sequences from the parental SIVB670 and compared to less related SIVmac251, SIVsm, and SIVstm sequences. The tree was constructed using a GTR maximum-likelihood analysis. (B) Phylogenetic analysis based on full-length sequences, comparing the SIVF359 molecular clone to other SIV, HIV-1, and HIV-2 clones. Phylogenetic trees were constructed by the neighbor-joining method using the PHYLIP program (version 3.5c).
TABLE 2.
Predicted amino acid similarity of SIVF359 to other SIV molecular clones
| SIVF359 protein | % Similarity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SIVB670 | SIVSmH4 | SIVsmmPBj4.41 | SIVmac239 | SIVmac1A11 | SIVmac | SIVStm | SIVMne | Mean | |
| Gag | 96 | 95 | 90 | 91 | 92 | 88 | 93 | 92 | |
| Pol | 97 | 95 | 92 | 91 | 92 | 91 | 93 | 93 | |
| Vif | 93 | 89 | 84 | 84 | 85 | 78 | 84 | 85 | |
| Vpx | 94 | 92 | 94 | 94 | 93 | 88 | 88 | 92 | |
| Vpr | 81 | 96 | 87 | 84 | 86 | 88 | 89 | 87 | |
| Tat | 75 | 80 | 65 | 80 | 75 | 69 | 62 | 72 | |
| Rev | 78 | 81 | 77 | 76 | 75 | 80 | 78 | ||
| Env | 95 | 88 | 86 | 83 | 80 | 78 | 83 | 83 | 84 |
| Nef | 81 | 75 | 78 | 74 | 66 | 77 | 75 | 75 | |
| Mean | 87 | 88 | 83 | 84 | 82 | 83 | 83 | ||
Virulence of the SIVF359 molecular clone.
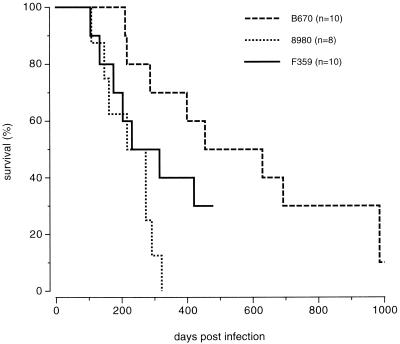
To determine if the SIVF359 molecular clone had retained the same pathogenic potential as the parental SIV8980 isolate, eight additional animals were infected with the same dose of the SIVF359 virus stock. As illustrated in Fig. 8, animals infected with the SIVF359 stock had survival curves very similar to those of animals infected with the SIV8980 stock from which it was derived, and SIVF359 was much more pathogenic than the original prepassage SIVB670 isolate. The molecular clone was slightly less pathogenic, as could be seen by the slight shift of the survival curve to the right. This, however, could be a consequence of the genetically homogeneous nature of the SIVF359 inoculum compared to the more heterogeneous SIV8980 inoculum, which represented a highly virulent quasispecies. Indeed, a heterogeneous quasispecies is more likely to escape from the immune responses of an outbred host.
FIG. 8.
Comparison of survival of groups of animals infected with the original prepassage SIVB670 isolate, the postpassage SIV8980 isolate, and the subsequently derived SIVF359 molecular clone as illustrated by a Kaplan-Meier plot.
DISCUSSION
We have utilized a novel cloning strategy to derive an infectious pathogenic molecular clone of a primate lentivirus without cell culture passage directly from serum. This molecular clone from serum (SIVF359) has unique properties. It is highly cytopathic in vitro and causes marked syncytia in lymphoid tissues in vivo. It readily infects activated but not resting rhesus lymphocytes. Infection of MDM cultures was demonstrated; however, in contrast to the case for SIVmac316, no virus production in the supernatant was detectable. Coreceptor use of this clone was restricted to CCR5 only, in contrast to that of other well-characterized molecular clones (SIVmac239 [T-cell tropic] and SIVmac316 [macrophage tropic]) studied for comparison. Surprisingly, although this virus did not use any of the 13 known primate lentivirus coreceptors, it could replicate on human cells defective for CCR5.
The use of molecular clones derived from extracellular virions may provide additional insight into the evolution of the lentiviral pathogen. For instance, this cloning strategy may aid the analysis of the specific biological properties of viral variants in plasma or serum which have escaped host immune responses at a specific point in time. Similarly, the study of virus populations which arise and become predominant in various extracellular compartments is feasible using this methodology. Studying the evolution of viruses using this strategy has a number of important advantages over conventional techniques that involve cloning proviruses from infected lymphocytes.
The SIVF359 molecular clone was derived from a rhesus macaque which had developed AIDS following the fourth in vivo passage of virus derived from SIVB670. The virus isolate SIVB670 originated from one of the first reports of simian AIDS in macaques and since has been used in a wide variety of studies (1, 4, 49). Phylogenetic analyses using available env genes or entire SIV genomes revealed that SIVB670 and SIVF359 branch off between two separate clusters of SIVsm clones, represented by SIVsmm9 and SIVsmmPBj4.41, derived from the Yerkes Primate Colony, and SIVmac clones, such as SIVmac251 and SIVmac239, derived from the New England Primate Colony (Fig. 7). As could be expected from the origin of SIVF359, this clone was related to but distinct from SIVB670. Originally SIVB670 had been isolated from a rhesus macaque (B670) infected with material containing SIV originally derived from a sooty mangabey from the Tulane Primate Center (49). Using the entire sequence for phylogenetic analysis, SIVF359 was found to branch separately between other characterized SIV isolates derived from sooty mangabeys or macaques (Fig. 7). The characteristics of the envelope evolution of SIVB670 during serial passage to animal 8980, from which SIVF359 was derived, have recently been described (64).
Comparison of the biological characteristics of the clone with those of the original SIV8980 isolate should demonstrate that we cloned the dominant virulent variant. For instance, coreceptor usage (55, 57) and the ability to replicate in resting versus activated CD4+ T cells (15) and macrophages (58, 67) are characteristics considered to influence pathogenicity. In this regard, no differences were observed between the cloned virus and the virus isolate in that both recognized only the CCR5 coreceptor and neither replicated in resting PBMC or macrophages. The subsequent in vivo passages of SIVB670 resulting in SIV8980 were carefully monitored. The original SIVB670 inoculum was also used in studies performed by Trichel et al. (62) and Amedee et al. (1). They had determined the number of different genotypes in the SIVB670 inoculum based on sequence divergence in the first hypervariable region (V1) (5, 54). Twelve different genotypes could be recognized in this isolate (SIVB670-CL1 to -CL12). The envelope sequence of SIVB670-CL12 was one of the sequence variants present in the original SIVB670 stock. During our passage experiment, the sequence variant SIVB670-CL12 had acquired an optimal fitness, represented by its prevalence in the early SIVB670 isolate and in its maintenance during the passages. Indeed, the SIVF359 molecular clone had the highest homology (96%) with SIVB670-CL12 and therefore is representative of the dominant virus variant of SIVB670.
The in vivo experiments in rhesus macaques demonstrated similar clinical and pathological characteristics of SIVF359 and SIV8980. Several SIVsm/mac clones have been reported to cause different patterns of disease, including attenuated virulence, compared to the virus isolates from which they were derived. With specific regard to the first SIV molecular clones which were derived, viral adaptation and attenuation often occurred as a result of in vitro propagation. This attenuation phenomenon may also be due to a failure to clone the dominant virus variant, possibly as a consequence of using proviral DNA as biological template or due to the use of biological material that contained a composition of early (macrophage-tropic, slow-replicating, non-syncytium-inducing) and late (T-cell-tropic, fast-replicating, syncytium-inducing) variants. It may also reflect a certain synergistic effect of a quasispecies that does not exist in the case of a single molecularly cloned virus. A study demonstrating a regulatory effect on HIV replication mediated by defective proviruses has provided evidence in that direction. Convincing data have started to accumulate which explain the progression to AIDS in terms of the biological properties of virus variants which emerge during the course of infection. However, results from other studies suggest a more complex cascade of events in which variants in combination with host-specific factors are involved in AIDS pathogenesis.
In summary, we have utilized a new cloning strategy to generate infectious molecular clones of lentiviruses from extracellular viral RNA in body fluids. This has facilitated the isolation of a unique pathogenic molecular clone designated SIVF359. Characterization of SIVF359 in vivo and in vitro revealed that it was highly pathogenic, causing marked syncytial giant cell formation in situ in lymph nodes and CPE in rhesus CD4+ T cells in vitro. It was found to have limited and novel coreceptor usage (CCR5 and a yet-undefined coreceptor) and could infect but did not result in detectable virus production in rhesus MDM. Upon the addition of autologous lymphocytes to these MDM, infection was transmitted and resulted in high concentrations of virus production. This SIVF359 molecular clone proved to be able to cause a rigorous infection and AIDS in rhesus macaques. It was genetically distinct from other molecular clones, mapping between two clusters of previously characterized groups of SIV clones (the SIVsm and SIVmac molecular clones, respectively). The SIVF359 molecular clone provides further insight into the nature of end-stage variants and the characteristics associated with accelerated disease progression.
REFERENCES
- 1.Amedee A M, Lacour N, Gierman J L, Martin L N, Clements J E, Bohm R, Jr, Harrison R M, Murphey-Corb M. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J Virol. 1995;69:7982–7990. doi: 10.1128/jvi.69.12.7982-7990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzarini J, Naesens L, Slachmuylders J, Niphuis H, Rosenberg I, Holy A, Schellekens H, De-Clercq E. 9-(2-Phosphonylmethoxyethyl)adenine (PMEA) effectively inhibits retrovirus replication in vitro and simian immunodeficiency virus infection in rhesus monkeys. AIDS. 1991;5:21–28. doi: 10.1097/00002030-199101000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Barbareschi M, Girlando S, Mauri F M, Forti S, Eccher C, Mauri F A, Togni R, Dalla Palma P, Doglioni C. Quantitative growth fraction evaluation with MIB1 and Ki67 antibodies in breast carcinomas. Am J Clin Pathol. 1994;102:171–175. doi: 10.1093/ajcp/102.2.171. [DOI] [PubMed] [Google Scholar]
- 4.Baskin G B, Martin L N, Murphey-Corb M, Hu F S, Kuebler D, Davison B. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res Hum Retroviruses. 1995;11:273–285. doi: 10.1089/aid.1995.11.273. [DOI] [PubMed] [Google Scholar]
- 5.Burns D P, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson J R, McGraw T P, Keddie E, Yee J L, Rosenthal A, Langlois A J, Dickover R, Donovan R, Luciw P A, Jennings M B. Vaccine protection of rhesus macaques against simian immunodeficiency virus infection. AIDS Res Hum Retroviruses. 1990;6:1239–1246. doi: 10.1089/aid.1990.6.1239. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel M D, Desrosiers R C. Use of simian immunodeficiency virus for evaluation of AIDS vaccine strategies. AIDS. 1989;3:S131–S133. doi: 10.1097/00002030-198901001-00019. [DOI] [PubMed] [Google Scholar]
- 10.Daniel M D, Sehgal P K, Kodama T, Wyand M S, Ringler D J, King N W, Schmidt D K, Troup C D, Desrosiers R C. Use of simian immunodeficiency virus for vaccine research. J Med Primatol. 1990;19:395–399. [PubMed] [Google Scholar]
- 11.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers R C, Ringler D J. Use of simian immunodeficiency viruses for AIDS research. Intervirology. 1989;30:301–312. doi: 10.1159/000150108. [DOI] [PubMed] [Google Scholar]
- 14.Dewhurst S, Embretson J E, Anderson D C, Mullins J I, Fultz P N. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature. 1990;345:636–640. doi: 10.1038/345636a0. [DOI] [PubMed] [Google Scholar]
- 15.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 16.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Fultz P N, Anderson D C, McClure H M, Dewhurst S, Mullins J I. SIVsmm infection of macaque and mangabey monkeys: correlation between in vivo and in vitro properties of different isolates. Dev Biol Stand. 1990;72:253–258. [PubMed] [Google Scholar]
- 19.Gardner M B, Luciw P A. Animal models of AIDS. FASEB J. 1989;3:2593–2606. doi: 10.1096/fasebj.3.14.2556312. [DOI] [PubMed] [Google Scholar]
- 20.Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immune Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 21.Heeney J L. Primate models for AIDS vaccine development. AIDS. 1996;10:S115–S122. doi: 10.1097/00002030-199601001-00016. [DOI] [PubMed] [Google Scholar]
- 22.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins W R, Montefiori D C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J I. SIV adaptation to human cells. Nature. 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 25.Holterman L, Dubbes R, Mullins J, Haaijman J, Heeney J. A strategy for cloning infectious molecular clones of retroviruses from serum or plasma. J Virol Methods. 2000;84:37–48. doi: 10.1016/s0166-0934(99)00136-6. [DOI] [PubMed] [Google Scholar]
- 26.Holterman L, Mullins J I, Haayman J J, Heeney J L. Direct amplification and cloning of up to 5-kb lentivirus genomes from serum. BioTechniques. 1996;21:312–319. doi: 10.2144/96212rr06. [DOI] [PubMed] [Google Scholar]
- 27.Holterman L, Niphuis H, Koornstra W, Dubbes R, ten Haaft P, Heeney J L. The rate of progression to AIDS is independent of virus dose in simian immunodeficiency virus-infected macaques. J Gen Virol. 2000;81:1719–1726. doi: 10.1099/0022-1317-81-7-1719. [DOI] [PubMed] [Google Scholar]
- 28.Holterman L, Niphuis H, ten Haaft P J, Goudsmit J, Baskin G, Heeney J L. Specific passage of simian immunodeficiency virus from end-stage disease results in accelerated progression to AIDS in rhesus macaques. J Gen Virol. 1999;80:3089–3097. doi: 10.1099/0022-1317-80-12-3089. [DOI] [PubMed] [Google Scholar]
- 29.Johnson P R, Hirsch V M. SIV infection of macaques as a model for AIDS pathogenesis. Int Rev Immunol. 1992;8:55–63. doi: 10.3109/08830189209056641. [DOI] [PubMed] [Google Scholar]
- 30.Johnson P R, Montefiori D C, Goldstein S, Hamm T E, Zhou J, Kitov S, Haigwood N L, Misher L, London W T, Gerin J L. Inactivated whole-virus vaccine derived from a proviral DNA clone of simian immunodeficiency virus induces high levels of neutralizing antibodies and confers protection against heterologous challenge. Proc Natl Acad Sci USA. 1992;89:2175–2179. doi: 10.1073/pnas.89.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 32.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 33.Kirchhoff F, Pohlmann S, Hamacher M, Means R E, Kraus T, Uberla K, Di Marzio P. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodama T, Burns D P, de Kestler H W, Daniel M D, Desrosiers R C. Molecular changes associated with replication of simian immunodeficiency virus in human cells. J Med Primatol. 1990;19:431–437. [PubMed] [Google Scholar]
- 35.Kodama T, Wooley D P, Naidu Y M, de Kestler H, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornfeld H, Riedel N, Viglianti G A, Hirsch V, Mullins J I. Cloning of HTLV-4 and its relation to simian and human immunodeficiency viruses. Nature. 1987;326:610–613. doi: 10.1038/326610a0. [DOI] [PubMed] [Google Scholar]
- 37.Kozal M J, Shafer R W, Winters M A, Katzenstein D A, Merigan T C. A mutation in human immunodeficiency virus reverse transcriptase and decline in CD4 lymphocyte numbers in long-term zidovudine recipients. J Infect Dis. 1993;167:526–532. doi: 10.1093/infdis/167.3.526. [DOI] [PubMed] [Google Scholar]
- 38.Lackner A A, Moore P F, Marx P A, Munn R J, Gardner M B, Lowenstine L J. Immunohistochemical localization of type D retrovirus serotype 1 in the digestive tract of rhesus monkeys with simian AIDS. J Med Primatol. 1990;19:339–349. [PubMed] [Google Scholar]
- 39.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landberg G, Tan E M, Roos G. Flow cytometric multiparameter analysis of proliferating cell nuclear antigen/cyclin and Ki-67 antigen: a new view of the cell cycle. Exp Cell Res. 1990;187:111–118. doi: 10.1016/0014-4827(90)90124-s. [DOI] [PubMed] [Google Scholar]
- 41.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication of replication-competent and -defective viral genomes. J Virol. 1991;65:3978–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 45.Marthas M L, Ramos R A, Lohman B L, Van-Rompay K K, Unger R E, Miller C J, Banapour B, Pedersen N C, Luciw P A. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGowan I, Hawkins A S, Weller I V. The natural history of cryptosporidial diarrhoea in HIV-infected patients. AIDS. 1993;7:349–354. doi: 10.1097/00002030-199303000-00007. [DOI] [PubMed] [Google Scholar]
- 47.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 48.Mori K, Ringler D J, Kodama T, Desrosiers R C. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphey-Corb M, Martin L N, Rangan S R, Baskin G B, Gormus B J, Wolf R H, Andes W A, West M, Montelaro R C. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature. 1986;321:435–437. doi: 10.1038/321435a0. [DOI] [PubMed] [Google Scholar]
- 50.Naidu Y M, Kestler H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIV mac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIV mac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niedrig M, Rabanus J P, L'Age Stehr J, Gelderblom H R, Pauli G. Monoclonal antibodies directed against human immunodeficiency virus (HIV) gag proteins with specificity for conserved epitopes in HIV-1, HIV-2 and simian immunodeficiency virus. J Gen Virol. 1988;69:2109–2114. doi: 10.1099/0022-1317-69-8-2109. [DOI] [PubMed] [Google Scholar]
- 52.Novembre F J, Johnson P R, Lewis M G, Anderson D C, Klumpp S, McClure H M, Hirsch V M. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J Virol. 1993;67:2466–2474. doi: 10.1128/jvi.67.5.2466-2474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novembre F J, Saucier M M, Hirsch V M, Johnson P R, McClure H M. Viral genetic determinants in SIVsmmPBj pathogenesis. J Med Primatol. 1994;23:136–145. doi: 10.1111/j.1600-0684.1994.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 54.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paxton W A, Kang S. Chemokine receptor allelic polymorphisms: relationships to HIV resistance and disease progression. Semin Immunol. 1998;10:187–194. doi: 10.1006/smim.1998.0132. [DOI] [PubMed] [Google Scholar]
- 56.Robinson H L, Lu S, Mustafa F, Johnson E, Santoro J C, Arthos J, Winsink J, Mullins J I, Montefiori D, Yasutomi Y. Simian immunodeficiency virus DNA vaccine trial in macaques. Ann NY Acad Sci. 1995;772:209–211. doi: 10.1111/j.1749-6632.1995.tb44746.x. [DOI] [PubMed] [Google Scholar]
- 57.Ross T M, Bieniasz P D, Cullen B R. Role of chemokine receptors in HIV-1 infection and pathogenesis. Adv Virus Res. 1999;52:233–267. doi: 10.1016/s0065-3527(08)60300-0. [DOI] [PubMed] [Google Scholar]
- 58.Simon M A, Chalifoux L V, Ringler D J. Pathologic features of SIV-induced disease and the association of macrophage infection with disease evolution. AIDS Res Hum Retroviruses. 1992;8:327–337. doi: 10.1089/aid.1992.8.327. [DOI] [PubMed] [Google Scholar]
- 59.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The genetic data environment an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 60.Stott E J, Chan W L, Mills K H, Page M, Taffs F, Cranage M, Greenaway P, Kitchin P. Preliminary report: protection of cynomolgus macaques against simian immunodeficiency virus by fixed infected-cell vaccine. Lancet. 1990;336:1538–1541. doi: 10.1016/0140-6736(90)93310-l. [DOI] [PubMed] [Google Scholar]
- 61.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trichel A M, Roberts E D, Wilson L A, Martin L N, Ruprecht R M, Murphey-Corb M. SIV/DeltaB670 transmission across oral, colonic, and vaginal mucosae in the macaque. J Med Primatol. 1997;26:3–10. doi: 10.1111/j.1600-0684.1997.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 63.Uberla K, Stahl-Hennig C, Bottiger D, Matz-Rensing K, Kaup F J, Li J, Haseltine W A, Fleckenstein B, Hunsmann G, Oberg B, et al. Animal model for the therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc Natl Acad Sci USA. 1995;92:8210–8214. doi: 10.1073/pnas.92.18.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valli P J, Lukashov V V, Heeney J L, Goudsmit J. Shortening of the symptom-free period in rhesus macaques is associated with decreasing nonsynonymous variation in the env variable regions of simian immunodeficiency virus SIVsm during passage. J Virol. 1998;72:7494–7500. doi: 10.1128/jvi.72.9.7494-7500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vartanian J P, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 67.Wodarz D, Lloyd A L, Jansen V A, Nowak M A. Dynamics of macrophage and T cell infection by HIV. J Theor Biol. 1999;196:101–113. doi: 10.1006/jtbi.1998.0816. [DOI] [PubMed] [Google Scholar]