Abstract
CRISPR diagnostics are effective but suffer from low signal transduction efficiency, limited sensitivity, and poor stability due to their reliance on the trans-cleavage of single-stranded nucleic acid fluorescent reporters. Here, we present CrisprAIE, which integrates CRISPR/Cas reactions with “one to more” aggregation-induced emission luminogen (AIEgen) lighting-up fluorescence generated by the trans-cleavage of Cas proteins to AIEgen-incorporated double-stranded DNA labeled with single-stranded nucleic acid linkers and Black Hole Quencher groups at both ends (Q-dsDNA/AIEgens-Q). CrisprAIE demonstrates superior performance in the clinical nucleic acid detection of norovirus and SARS-CoV-2 regardless of amplification. Moreover, the diagnostic potential of CrisprAIE is further enhanced by integrating it with spherical nucleic acid-modified AIEgens (SNA/AIEgens) and a portable cellphone-based readout device. The improved CrisprAIE system, utilizing Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters, exhibits approximately 80- and 270-fold improvements in sensitivity, respectively, compared to conventional CRISPR-based diagnostics. We believe CrisprAIE can be readily extended as a universal signal generation strategy to significantly enhance the detection efficiency of almost all existing CRISPR-based diagnostics.
Subject terms: Genetic testing, Sensors and biosensors, Imaging and sensing, Viral infection
Current CRISPR diagnostic approaches can be hampered by several limitations, including low signal transduction efficiency and limited sensitivity. Here, the authors present CrisprAIE, an approach based on CRISPR/Cas-mediated “one to more” lighting-up nucleic acid detection using aggregation-induced emission luminogens.
Introduction
The quick and accurate detection of nucleic acid biomarkers in complex biological systems is critical for the diagnosis and monitoring of infectious diseases1,2. Real-time quantitative polymerase chain reaction (qPCR) and sequencing technologies have been clinically recognized as cutting-edge detection tools for nucleic acid targets3,4. However, these techniques relying on target amplification are laborious, expensive, and time consuming and require complex multiple reagent reactions, trained technicians, and sophisticated laboratory instrument. Despite the remarkable achievement in isothermal nucleic acid amplification, which effectively eliminates the thermal cycle, nonspecific amplification remains a major challenge for these amplification-dependent techniques due to their low targeting specificity and high false-positive results5–7. As a natural adaptive immune system in microorganisms, the clustered regularly interspaced short palindromic repeat (Crispr) and Crispr-associated (Cas) protein system has been skillfully integrated with various nucleic acid amplification techniques to improve the sensitivity and specificity of traditional nucleic acid diagnostic techniques. It specifically targets nucleic acid sequences and can perform the efficient nuclease-like collateral cleavage of ambient nontargeted single-stranded nucleic acids. With this design concept, several Crispr/Cas-based diagnostic methods (Crispr-Dx) combining the advantages of Cas reactions and isothermal amplification have been developed as highly specific nucleic acid detection tools that can measure the collateral cleavage of single-stranded nucleic acid probes after target binding8,9. The currently available Crispr-Dx assays mainly include Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK)10,11, DNA Endonuclease-Targeted Crispr Trans Reporter (DETECTOR)12,13, and one-Hour Low-Cost Multipurpose Highly Efficient System (HOLMES)14. These assays employ Cas12 or Cas13 effectors to trigger single-stranded DNA (ssDNA)/RNA (ssRNA) cleavage and induce efficient signal readout for various nucleic acid targets, including Zika virus10, dengue virus10, human papillomavirus (HPV)12, and single-nucleotide polymorphism14. Despite these achievements, the signal generation of these Crispr-Dx assays mainly rely on the trans-cleavage activity of Cas proteins on linear single-stranded nucleic acid reporters and still has some problems, such as low signal transduction efficiency, limited detection sensitivity, and poor stability in complex biological systems.
An important strategy to improve the detection performance of Crispr-Dx systems is by evolving the signal-generation mechanism and modality of Cas reaction. Increasing attempts have been directed to combine Cas reaction with a variety of enhanced signal readout alternatives, including electrochemistry15,16, electrochemiluminescence17, surface-enhanced Raman scattering18,19, surface plasmon resonance20,21, and field-effect transistor22,23, to achieve efficient target detection. Nevertheless, fluorescence-based signal readouts are still extensively investigated due to their integration and miniaturization potential. The majority of Crispr-Dx assays utilize the activated trans-cleavage activity to generate a detectable fluorescent signal through the nonspecific cleavage of traditional synthetic fluorophore-quencher (FQ) pairs bound to the end of short ssDNA or ssRNA24–26. However, this signal illuminating behavior usually suffers from insufficient sensitivity because one FQ reporter cleaved by activated Cas nuclease only can liberate one fluorescent signal molecule. Therefore, breaking the limitations of existing fluorescent transduction systems will help promote the rapid development and application of Crispr-Dx. A preferable alternative to traditional FQ-based reporter with “one to more” characteristic, that is, one trans-cleavage of FQ reporter can output many fluorescent signal molecules, is urgently needed to improve the existing Crispr-Dx.
Leveraging the amplification mechanism described above, we speculate that one promising fluorescent readout system for Crispr-Dx is aggregation-induced emission luminogens (AIEgens)27. As a type of organic fluorescence dye, AIEgens exhibit many unique advantages over traditional aggregation-caused quenching probes, including enhanced aggregation emission, high signal-to-noise ratio (S/N), excellent photostability, and large Stokes shift28,29. These characteristics make AIEgens a promising candidate for reducing the detection limit (LOD) and improving the stability of Crispr-Dx. Owing to the positively charged AIEgens spontaneously bind to negatively charged nucleic acid chains through noncovalent interactions (e.g., van der Waals, π-π stacking, and hydrogen bonding interactions) and electrostatic interactions, these AIEgens can light-up their fluorescence signals via the restriction of intramolecular motion (RIM)30,31. In theory, a long nucleic acid chain can provide many negatively charged sites, endowing additional opportunities for AIEgen binding and generating intense fluorescence emission, which offer the possibility of creating “one to more” signal reporters for amplifying Crispr/Cas detection.
Here, we report the development of CrisprAIE, the combination of a Crispr/Cas reaction with a “one to more” AIEgen lighting-up fluorescence readout (Fig. 1). The “one to more” AIEgen lighting-up readout is achieved using AIEgen-loaded double-stranded DNA (dsDNA) reporters, which are synthesized by incorporating cationic AIEgens into the grooves of dsDNA. To enable the Cas protein to cleave the AIEgen-loaded dsDNA reporters and achieve the lighting-up fluorescence readout, we introduce ssDNA or ssRNA linkers at both ends of dsDNA to bridge the Black Hole Quencher (BHQ) for producing AIEgen-enhanced CrisprAIE reporters (Q-dsDNA/AIEgens-Q). After carefully exploring the factors affecting the fluorescence transduction efficiency of AIEgens, we find that the resulting CrisprAIE shows a LOD of femtomolar concentrations of at least 80 times more sensitive than the traditional FQ reporter-based Crispr-Dx without amplification. This LOD can be further reduced to attomolar levels through isothermal amplification. The clinical diagnostic efficiency of our CrisprAIE is demonstrated by detecting norovirus and SARS-CoV-2 virus in their respective diagnostic samples. To further improve the sensitivity and stability of our CrisprAIE, we develop AIEgen-enhanced spherical nucleic acid (SNA/AIEgen) reporters and demonstrate their potential in improving the nucleic acid diagnostic capabilities of CrisprAIE in complex biological systems. We also integrate CrisprAIE assay with a portable cellphone-assisted fluorescence readout device for point-of-care (POC) diagnostic application and evaluate its detection performance and consistency with commercial multimode fluorescence microplate readers. We believe that this AIEgen lighting-up reporting probe can serve as an attractive alternative to traditional FQ reporter, and the developed CrisprAIE can be readily extended for improving the detection efficiency of almost all existing Crispr-Dx assays based on single-stranded nucleic acid reporters.
Fig. 1. Schematic of our developed CrisprAIE assay.
a Schematic of using CrisprAIE assay for virus detection. b Schematic of the sensing principle of AIEgen reporter and traditional FQ reporter derived through the trans-cleavage of activated Cas nuclease. c Improved CrisprAIE for virus detection, including the integration of spherical nucleic acid strategy for improved AIEgen reporters, the combination with Class 2 Crispr/Cas system (Cas12 and Cas13) for the direct detection of DNA and RNA, and the cellphone-assisted portable readout device for point-of-care testing.
Results
Crispr-driven AIEgen lighting-up readout
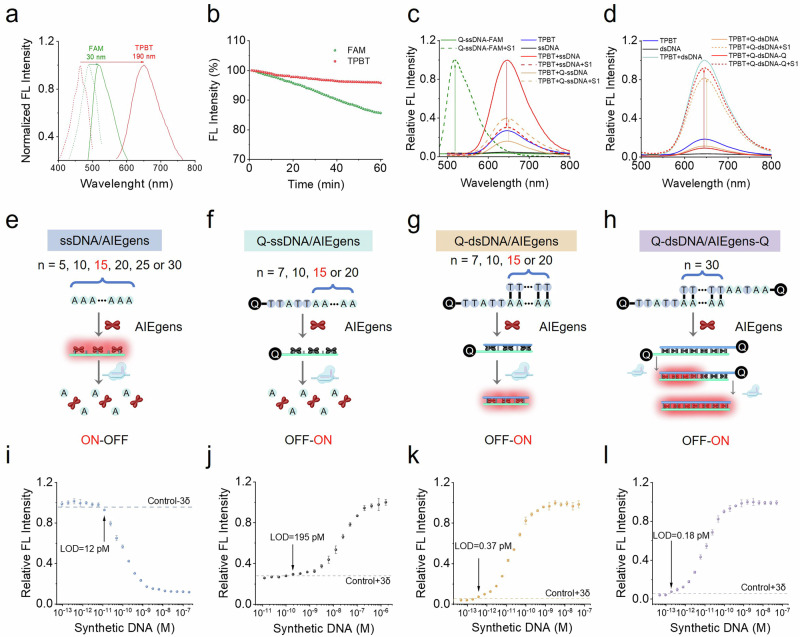
Cationic AIEgens can interact with negatively charged DNA strands through electrostatic or noncovalent interactions, thereby enabling lighting-up fluorescence readout by activating the RIM. As a typical cationic AIEgen, 4ʹ,4ʹʹ,4ʹʹʹ-(((((1E,1ʹE)-benzo[c][1,2,5]thiadiazole-4,7-diylbis(ethene-2,1-diyl))bis(4,1-phenylene))bis(azanetriyl))tetrakis(benzene-4,1-diyl))tetrakis(1-methylpyridin-1-ium) iodide (TPBT) with four positively charged pyridine groups is used for the label-free fluorescent detection of dsDNA and single-nucleotide polymorphisms30. The molecular structure of TPBT was confirmed by 1H NMR, 13C NMR, and high-resolution mass spectra (Supplementary Figs. 1–3), and its AIE feature was further characterized in Supplementary Fig. 4. Compared with FAM, a widely used fluorophore, TPBT has a larger Stokes shift and better resistance to photobleaching (Fig. 2a, b) that are conducive to sensitive and accurate detection. Considering the preference of the trans-cleavage activity of Cas nuclease for single-stranded oligonucleotides, we first designed two ssDNA-modified AIEgen reporters, namely, AIEgen-modified ssDNA (ssDNA/AIEgens) and AIEgen-modified ssDNA labeled with a single-terminal quencher group BHQ by a five-base ssDNA linker (Q-ssDNA/AIEgens). Compared with the flexible ssDNA, the rigid dsDNA can more effectively restrict the free intramolecular motion of the incorporated AIEgens, thereby facilitating enhanced fluorescent signal output. We then constructed two AIEgen-modified dsDNA reporters, namely, AIEgen-modified dsDNA labeled with a single-terminal quencher group BHQ by a five-base ssDNA linker (Q-dsDNA/AIEgens) and AIEgen-modified dsDNA labeled with a double-terminal quencher group BHQ by a five-base ssDNA linker (Q-dsDNA/AIEgens-Q). A traditional FQ reporter, denoted as Q-ssDNA-FAM, was also commercially synthesized for comparison. The related sequence is summarized in Supplementary Table 1. S1 nuclease, a single-stranded specific endonuclease that hydrolyzes ssDNA into single nucleotides, was selected as an alternative of activated Cas nuclease to study the feasibility of nuclease-driving AIEgen lighting-up readout. As shown in Fig. 2c, the presence of S1 nuclease led to the significant fluorescence recovery of the Q-ssDNA-FAM reporter, accompanied by the departure of the quencher group (BHQ). This finding confirmed the validity of S1 nuclease against ssDNA degradation. The ssDNA/AIEgen probe showed significantly increased fluorescence signals due to the activation of RIM when TPBT bound to ssDNA. The ssDNA part of the composite probe can be digested by S1 nuclease, thus decreasing its fluorescence to achieve the “ON-OFF” signal output. By contrast, the Q-ssDNA/AIEgen reporter displayed fluorescent signal recovery in the presence of S1 nuclease, thus favoring the “OFF-ON” signal output (Fig. 2c). When TPBT was incubated with dsDNA, the fluorescence signal of TPBT was markedly increased. Its fluorescence can be effectively quenched by labeling the BHQ quenching groups at one or both ends of dsDNA (Fig. 2d). S1 nuclease can induce BHQ detachment by digesting the ssDNA linker, resulting in significant fluorescence enhancement to obtain the “OFF-ON” signal output. These results demonstrated that the designed DNA-modified AIEgen probes are suitable as the fluorescent signal reporters for nuclease-based assays, including Crispr/Cas-based detection.
Fig. 2. Construction of a Crispr-driven AIEgen lighting-up readout.
a Excitation spectra (dotted line) and emission spectra (solid line) of FAM (green) and TPBT (red). b Photobleaching resistance of FAM and TPBT. Normalized fluorescence intensity of FAM and TPBT at various time points, during the 60 min successive scanning of the respective excitation beam. c, d Proof of concept of S1 nuclease-driven AIEgen lighting-up readout. c Fluorescence emission spectrum of Q-ssDNA-FAM, free TPBT, TPBT mixed with ssDNA or Q-ssDNA, and released FAM or TPBT after splitting ssDNA by S1 Nuclease. d Fluorescence emission spectrum of free TPBT, TPBT mixed with dsDNA, TPBT mixed with Q-dsDNA or Q-dsDNA-Q and released dsDNA/TPBT after splitting ssDNA linker by S1 Nuclease. The synthetic norovirus ORF2 gene DNA sequence was used as the model analyte to construct the Crispr/Cas12a sensing system. Schematic illustration of CrisprAIE based on ssDNA/AIEgen (e) Q-ssDNA/AIEgen (f), Q-dsDNA/AIEgen (g) and Q-dsDNA/AIEgen-Q (h) reporters. Relative fluorescence intensity obtained from amplification-free CrisprAIE assay with different concentrations of synthetic DNA targets based on ssDNA/AIEgen (i), Q-ssDNA/AIEgen (j), Q-dsDNA/AIEgen (k), and Q-dsDNA/AIEgen-Q (l) reporters. Data are mean ± s.d.; n = 3 repeated tests.
Encouraged by this finding, we reported the development of CrisprAIE by combining a Crispr/Cas-assisted nucleic acid cleavage reactions with DNA-modified AIEgen lighting-up readout. The effect of the length and base type of DNA sequence, ionic strength, and AIEgen concentration on the fluorescent properties of the resulting ssDNA-modified AIEgen reporters was studied to obtain the best binding between TPBT and DNA oligonucleotides and the enhanced fluorescence of ssDNA-modified AIEgen probes for the fabrication of optimal ssDNA/AIEgen and Q-ssDNA/AIEgen reporters (Fig. 2e, f). As revealed in Supplementary Fig. 5, the ssDNA (A10 or T10) containing 10 repeating units of adenine (A) or thymine (T) obtained higher fluorescence signal response than that containing cytosine (C) and guanine (G) bases in the presence of 100 mM NaCl and 2.5 µM TPBT. In particular, the enhanced fluorescence of the A10 was the highest among them. One possible reason is that the stronger intermolecular interaction of TPBT with A base than with T, C, and G bases contributes to restricting the intramolecular motion of TPBT and boosting the fluorescent emission. Further DNA sequence length analysis showed that ssDNA (A15) could produce suitable relative fluorescence intensity, highest fluorescence decay rate, and short cleavage equilibrium time (Supplementary Figs. 6, 7). On the basis of the above results, the A15 and A-T base pairs were selected as the building blocks for fabricating DNA-modified AIEgen reporters and achieving the lowest LOD of CrisprAIE.
Under the developed conditions, we first fabricated the CrisprAIE assay using ssDNA/AIEgens and Q-ssDNA/AIEgens as signal reporters (Fig. 2e, f). The LOD was determined as the mean plus three times the standard deviation of the measured concentration, derived from twenty repeated measurements of the blank sample, following a previously reported method32. Figure 2i shows that the LOD of this ssDNA/AIEgens-based CrisprAIE was 12 pM, which was close to that of conventional Crispr-Dx (LOD: 15 pM) based on the Q-ssDNA-FAM reporter (Supplementary Fig. 8). Moreover, we observed that when Q-ssDNA/AIEgens were used as CrisprAIE signal reporters, the sensitivity was further reduced with a LOD of 195 pM (Fig. 2j, Supplementary Fig. 9). The possible reason for the low sensitivity of these two CrisprAIE assays is that multiple independent nuclease cleavages are required to fully release AIEgens from the composite ssDNA/AIEgen reporters to achieve efficient fluorescence transduction. In addition, the binding of AIEgens with ssDNA resulted in limited RIM and reduced fluorescence enhancement, thus yielding low S/N ratio. Compared with ssDNA, dsDNA can better restrict the intramolecular movement of AIEgens by binding AIEgens to the major or minor grooves of dsDNA, thus improving the sensitivity of DNA-lighting-up AIEgen probes. On this basis, two AIEgen-modified dsDNA probes, namely, Q-dsDNA/AIEgens and Q-dsDNA/AIEgens-Q, were designed and proposed for enhancing the sensitivity of CrisprAIE. As revealed in Fig. 2g, h and Supplementary Fig. 10, Q-dsDNA/AIEgens consisted of a dsDNA containing multiple repeating units of A and T for AIEgen loading, an ssDNA linker for Crispr/Cas cleavage, and a BHQ group for quenching the fluorescence of AIEgens bound to dsDNA. Meanwhile, Q-dsDNA/AIEgens-Q consisted of a dsDNA containing multiple repeating A-T base pairs, two ssDNA linkers, and two BHQ groups at both ends. Q-dsDNA/AIEgen and Q-dsDNA/AIEgen-Q reporters were separated and purified by agarose gel electrophoresis (Supplementary Figs. 12). To achieve the highest S/N ratio, we studied the effect of the composition and length of dsDNA and TPBT concentration on the groove binding between dsDNA and TPBT. Supplementary Fig. 11 implies that the optimal combination for Q-dsDNA/AIEgens was A15T15 of dsDNA and 5 μM of TPBT, resulting in the highest S/N ratio due to the high load efficiency of TPBT (the binding number was 11.8 TPBT molecules per A15T15 dsDNA) and the efficient quenching of the entire fluorescence of TPBT binding on dsDNA by BHQ. The length of dsDNA was too long or too short, resulting in a relatively low S/N ratio due to the insufficient AIEgen loading and incomplete fluorescence quenching. Further molecular simulation calculations between TPBT and dsDNA were performed to simulate the interaction. The docking results in Supplementary Fig. 12 show that compared with those in dsDNA with C-G repeating units, the benzothiadiazole core and the two vinylene bridges on TPBT were perfectly inserted into the minor grooves of dsDNA containing A-T base pairs. This finding suggested that the dsDNA consisting of repeating units of A and T can restrict the intramolecular motion of TPBT to enhance fluorescence emission by blocking the nonradiative decay. These observations are consistent with the results from ssDNA, where the A and T bases were superior to the C and G bases in restricting the molecular motion of TPBT and enhancing the fluorescence. Considering that the optimal dsDNA length for individual BHQ group quenching was 15 repeating A-T base pairs, we synthesized Q-dsDNA/AIEgens-Q by incorporating TPBT (5 μM) with A30T30 of dsDNA. Prior to the Crispr detection, we monitored the double-stranded structure integrity of Q-dsDNA/AIEgens-Q in the presence of activated Cas12 protein using gel electrophoresis. The results in Supplementary Fig. 13 demonstrated that the ssDNA component was completely cleaved by activated Cas12 protein, whereas the dsDNA segment remained intact even after trans-cleavage, which is crucial for achieving enhanced AIE fluorescence signal output. We then measured the concentration of synthetic DNA by incorporating Q-dsDNA/AIEgens and Q-dsDNA/AIEgens-Q into Crispr/Cas12-based reaction. With the optimized assay design, the LOD of CrisprAIE based on Q-dsDNA/AIEgen reporters was 0.37 pM of synthetic DNA (Fig. 2k). With the use of Q-dsDNA/AIEgen-Q reporters, this value can be further reduced to 0.18 pM, which is about 80 folds lower than that of conventional Q-ssDNA-FAM-based Crispr-Dx (Fig. 2l). This dramatically reduced LOD is superior or comparable with those of other reported amplification-free Crispr/Cas strategies involving electrochemical15 and nanozyme-catalyzed readout33. We speculated that the high sensitivity of CrisprAIE may be due to the increased signal output capacity caused by loading additional AIEgens onto dsDNA, enhanced AIE fluorescence by dsDNA-caused efficient intramolecular motion restriction, large Stokes shift, high extinction coefficient, “one to more” signal amplification, and superior S/N ratio (Fig. 2a and Supplementary Fig. 14). Subsequently, we evaluated the long-stem storage stability and repeatability of our CrisprAIE over 21 consecutive days by freeze-drying the Q-dsDNA/AIEgen-Q reporter. Supplementary Fig. 15 indicates that the developed AIEgen reporter maintains excellent stability and reproducibility under lyophilized conditions, showing compatibility with the current freeze-dried CRISPR reagent. Two other AIEgens with excellent binding ability to DNA strands, namely, TPE-1OH and TPA-1OH31 (Supplementary Figs. 16–22), were introduced to replace TPBT for constructing CrisprAIE to broaden the optional ability of AIEgen toolbox. Supplementary Figs. 23, 24 verify the versatility and feasibility of CrisprAIE for the detection of synthetic DNA targets with acceptable analytical performance. This optimized Q-dsDNA/AIEgens-Q reporter was used in the following experiments unless otherwise indicated.
RNA quantification using CrisprAIE combined with RT-RPA
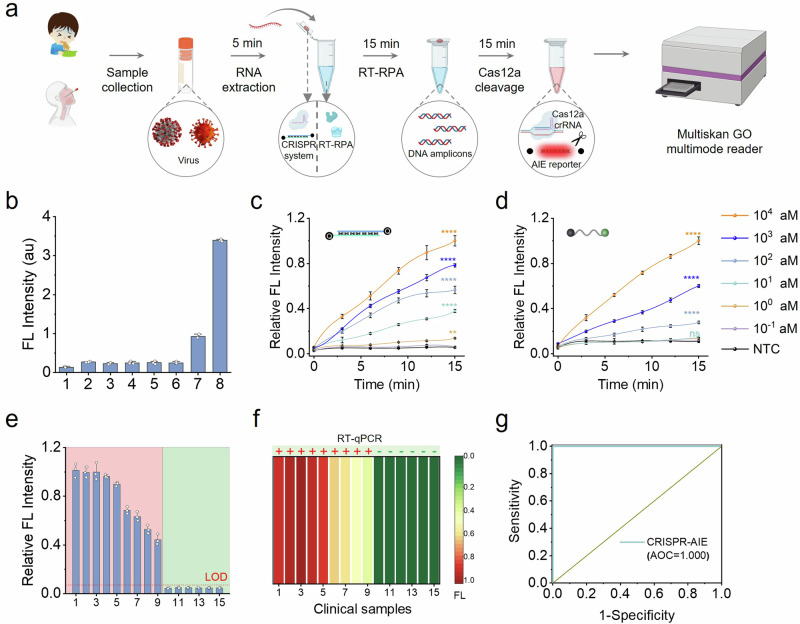
The improvement in the detection performance of CrisprAIE encouraged us to investigate its real-world application. For the sensitive detection and quantification of target RNA, we first combined the Q-dsDNA/AIEgen-Q lighting-up fluorescence reporting strategy with Crispr/Cas12-based viral RNA detection. This assay design performs simultaneous reverse transcription and isothermal amplification using recombinase polymerase amplification (RT-RPA) for the RNA extracted from actual samples, such as vomitus or nasopharyngeal swabs, in universal transport medium, followed by the Cas12 detection of target RNA sequences. The cleavage of an AIEgen reporter verifies the presence of the virus. If the target RNA is present in the reaction system, then the Cas12a/crRNA binary complex forms a ternary complex with the reverse-transcribed amplicons, which then trans-cleave nontargeted ssDNA linker in the Q-dsDNA/AIEgens-Q to light up AIEgen fluorescence. With this design concept, we developed a single-tube isothermal CrisprAIE assay for the rapid and accurate detection of target RNA (Fig. 3a). As a proof of concept for potential applications, we first applied this CrisprAIE assay for the detection of norovirus from clinical vomit RNA extracts. The primers targeting the ORF2 gene of norovirus and the crRNA sequence were designed as summarized in Supplementary Table 2. To obtain the best analytical performance, we optimized the detection conditions of norovirus ORF2 gene by CrisprAIE assay, including RT-RPA reaction at 37 °C for 15 min and Cas12 detection reaction at 37 °C for 15 min. Figure 3b displays that only the combination of target RNA and RPA can obtain fluorescence signal response, confirming the validity and feasibility of this CrisprAIE system. A series of ORF2 gene templates with concentrations ranging from 10-1 to 104 aM were determined, and 1 aM of the target gene was successfully detected (Fig. 3c). The same gene templates were simultaneously detected by the Q-ssDNA-FAM-based Cas detection and agarose gel electrophoresis. The LOD values for both methods were 100 and 10 aM of the gene template (Fig. 3d and Supplementary Fig. 25), indicating that the developed CrisprAIE assay had better detection efficiency than traditional Crispr-Dx and agarose gel electrophoresis. The clinical practicability and reliability of this CrisprAIE assay was further compared with that of a well-approved RT-qPCR method for the detection of clinical vomit samples of patients infected with norovirus. Fifteen clinical vomitus samples collected from Jiangxi Provincial Center for Disease Control and Prevention (China) were tested. As shown in Figs. 3e, 9 and 6 samples were detected as norovirus positive and negative, respectively, which is completely consistent with the detection results obtained by RT-qPCR (Fig. 3f, Supplementary Fig. 26 and Supplementary Table 3). Neither of the two methods showed false-positive and false-negative results. Further analysis of the area under the curve (AUC) of receiver operating characteristic (ROC) curve indicated that CrisprAIE obtained 100% (9 out of 9) positive predictive agreement and 100% (6 out of 6) negative predictive agreement relative to the RT-qPCR results (Fig. 3g).
Fig. 3. Combination of RT-RPA and CrisprAIE detects norovirus RNA up to attomolar concentration.
a Schematic of using CrisprAIE assay combined RT-RPA system for norovirus detection, comprising sample RNA extraction, RT-RPA amplification, target activated Cas12a/crRNA trans-cleavage, and fluorescence measurement. b Validation of Q-dsDNA/AIEgen-Q reporters integrated with RT-RPA and Crispr system. Here, Q-dsDNA/AIEgen-Q reporters (group 1) were mixed with other key components needed for RT-RPA and Crispr system (group 2: mixed with RT-RPA reagent; group 3: mixed with Cas12a/crRNA; group 4: mixed with 10 pM targets; group 5: mixed with RT-RPA reagent and Cas12a/crRNA; group 6: mixed with 10 pM targets and RT-RPA reagent; group 7: mixed with 10 pM targets and Cas12a/crRNA; group 8: mixed with 10 pM targets, RT-RPA reagent and Cas12a/crRNA). Relative fluorescence intensity obtain from the analysis of ORF2 gene via RT-RPA integrated Crispr assay based on Q-dsDNA/AIEgens-Q reporters (c) and Q-ssDNA-FAM reporters (d). e Relative fluorescence intensity obtained from RT-RPA integrated CrisprAIE for detecting 15 clinical samples. Samples No. 1–9 represent the positive samples (highlighted in red), while samples No. 10–15 correspond to the negative samples (highlighted in green) as tested by RT-qPCR. f Comparison of test results of RT-qPCR and our developed CrisprAIE assay for 15 clinical samples. Negative samples are highlighted in green, and positive samples are highlighted in red. CrisprAIE assay heatmap represents normalized mean fluorescence values. g ROC curve analysis between the results of CrisprAIE and RT-qPCR, indicating the detection accuracy in clinical applications. Data are mean ± s.d.; n = 3 repeated tests. Two-tailed P values were calculated using unpaired t-tests: *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001; NS, not significant, compared with the group 1 in (b) or nontemplate control (NTC).
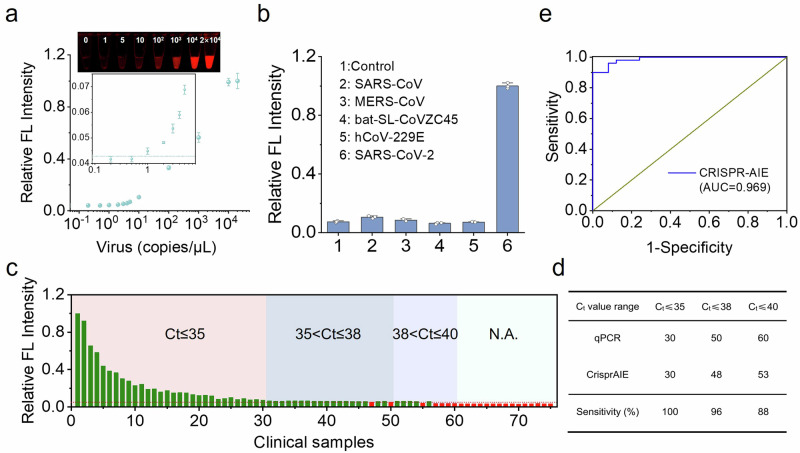
CrisprAIE was then extended for the quantification of SARS-CoV-2 RNA to evaluate its versatility in detecting different target RNAs. SARS-CoV-2 pseudovirus with the same virus structure but no infectivity was selected as a standard in this experiment. The primers targeting the N gene of SARS-CoV-2 and the corresponding crRNA sequence are listed in Supplementary Table 2. Prior to CrisprAIE detection, the viral RNA was first extracted and amplified by RT-RPA, and its amplification products were verified by agarose gel electrophoresis (Supplementary Fig. 27). We then combined RT-RPA with CrisprAIE assay to detect SARS-CoV-2 pseudovirus in a single-tube reaction system. Under the optimal conditions, the LOD of the developed CrisprAIE detection method was as low as 1 copy/μL (Fig. 4a). To evaluate the specificity of the established CrisprAIE assay for SARS-CoV-2 RNA detection, we performed synthetic DNA template detection for other four coronaviruses and human respiratory virus, namely, SARS-CoV, MERS-CoV, bat-SL-CoVZC45, and hCoV-229E, by using our CrisprAIE system corresponding to N gene detection. The results showed that only the presence of SARS-CoV-2 DNA template could result in significantly increased fluorescence response. The other interfering gene templates exhibited negligible signal response relative to the control group, suggesting the excellent specificity of the assay for all the tested coronavirus (Fig. 4b). The clinical detection performance of this CrisprAIE system was validated by conducting an assay of SARS-CoV-2 N gene in 75 clinical nasopharyngeal swab samples, which were synchronously identified by the standard RT-qPCR. As recommended by the RT-qPCR kit and the results of RT-qPCR in Supplementary Fig. 28, when analyzing clinical unknown nasopharyngeal swab samples, Ct values less than or equal to 38 are considered as SARS-CoV-2 positive, Ct values between 38 and 40 indicate suspiciously or weakly positive, and Ct values more than 40 are judged as negative. The relative fluorescence intensities for N gene detection with the CrisprAIE assay are shown in Fig. 4c. The SARS-CoV-2-positive and -negative samples were distinguished according to the cut-off value, that is, the mean plus threefold standard deviations of the nontemplate control. As indicated in Supplementary Fig. 29 and Supplementary Table 4, the Ct obtained by RT-qPCR showed that 50 out of 75 tested samples tested SARS-CoV-2 positive, 10 samples were suspiciously positive, and 15 samples were negative. Compared with RT-qPCR, our CrisprAIE accurately identified 30 samples with Ct values below 35, demonstrating 100% sensitivity (Fig. 4c and Supplementary Table 4). Significantly, out of 20 samples with Ct values ranging from 35 to 38, our method successfully detected 18 SARS-CoV-2 positive samples, achieving 90% sensitivity (18 out of 20), showcasing its reliability in detecting clinical samples with low viral loads. Furthermore, the proposed CrisprAIE achieved 96% (48 out of 50) and 88% (53 out of 60) sensitivity for samples with Ct values less than or equal to 38 and 40 for the analysis of the N gene, respectively (Fig. 4d). Notably, among 10 suspected samples with Ct values between 38 and 40, 5 tested positive and 5 were negative according to our CrisprAIE. All 15 SARS-CoV-2 negative samples with Ct values more than 40 were accurately identified as negative by our method. These findings indicate that the developed CrisprAIE assay exhibits sensitivity comparable to that of the standard RT-qPCR assay, which not only facilitates the detection of clinical samples with high viral loads but also reliably identifies low-concentration samples with Ct values ranging from 35 to 40. Using a Ct value of 38 as the judgment criterion, the ROC curve of our CrisprAIE method showed an AUC of 0.969 for the SARS-CoV-2 N gene (Fig. 4e), showcasing its great potential for RNA quantification in actual clinical samples.
Fig. 4. Combination of RT-RPA and CrisprAIE detects SARS-CoV-2 RNA to single copy level.
a Evaluation of the sensitivity of our developed method to detect SARS-CoV-2 pseudovirus with different copies of N gene. b Specificity of our developed method in the analysis of SARS-CoV-2 RNA against other coronaviruses and human respiratory virus, including SARS-CoV, MERS-CoV, bat-SL-CoVZC45, and hCoV-229E. Two-tailed P values were calculated using unpaired t-tests: ****P < 0.0001; NS, not significant, compared with b-6. c Relative fluorescence intensity obtained from our developed method for detecting 75 clinical samples, where positive and negative CrisprAIE test results are marked in green and red, respectively. d Sensitivity of the CrisprAIE technique was evaluated using clinically approved RT-qPCR kits based on the measured Ct values. e ROC curve analysis between the results of CrisprAIE and RT-qPCR. Data are mean ± s.d.; n = 3 repeated tests.
Improved sensitivity and stability of CrisprAIE using AIEgens-assisted SNA reporters
Despite the clinical success of our CrisprAIE, its signal generation still relies on the trans-cleavage activity on the ssDNA linker of the composite linear DNA Q-dsDNA/AIEgen-Q substrates. Compared with traditional Q-ssDNA-FAM reporters, the constructed Q-dsDNA/AIEgen-Q probes had higher stability and better detection efficiency in complex biological samples. However, these probes still carry the risk of nuclease degradation in the linear ssDNA linker in biological fluids, such as serum, leading to a false positive signal. Therefore, the possibility of improving the stability and sensitivity of CrisprAIE in complex biological samples must be further explored. Integrating with nanoparticles is an important way to improve the stability and sensitivity of linear nucleic acid probes34. Spherical nucleic acids (SNAs) combine the advantages of nucleic acid probes and the excellent properties of nanoparticles, including excellent nuclease resistance, efficient fluorescence quench effect, adjustable detection sensitivity, and response range, and thus have been widely applied to enhance molecular diagnostics35–38. Coupling Crispr/Cas and AIEgen-assisted SNA (SNA/AIEgen) reporters may provide a paradigm for the development of CrisprAIE with strong stability and high sensitivity. On this basis, we first constructed an SNA/AIEgen reporter (Fig. 5a) consisting of a core-shell nanoquencher (Fe3O4@Au/PDA, Supplementary Fig. 30) with the surface modification of TPBT-loaded sulfhydryl dsDNA via the Michael addition (Supplementary Fig. 31). Owing to the wide absorption of Fe3O4@Au/PDA in the 300–800 nm range that overlaps perfectly with the excitation and emission of TPBT molecules (Supplementary Fig. 30d), the fluorescence of the obtained SNA/AIEgen probes can be effectively quenched by the fluorescence resonance energy transfer effect. To enable the SNA/AIEgens to be degraded or cleaved by S1 nuclease and Cas nuclease, we introduced an ssDNA part (TTATTTTATT) as a linker to couple the dsDNA onto the surface of Fe3O4@Au/PDA. The successful synthesis and characterization of the SNA/AIEgen probe were verified by zeta potential, dynamic light scattering measurement, and Fourier transform infrared spectroscopy (Supplementary Figs. 31, 32). Prior to the development of CrisprAIE, we investigated the effect of the dsDNA length coated on the surface of Fe3O4@Au/PDA because of its close association with AIEgen loading and fluorescence quenching efficiency. As shown in Supplementary Fig. 33, the labeling numbers of SH-dsDNA10, SH-dsDNA20, SH-dsDNA30, and SH-dsDNA40 on individual Fe3O4@Au/PDA were calculated as 733, 650, 629, and 607, respectively. The binding TPBT numbers on SH-dsDNA10, SH-dsDNA20, SH-dsDNA30, and SH-dsDNA40 were 8.3, 15.3, 23.7, and 32.7, respectively (Supplementary Fig. 34). Thus, we successfully achieved the controlled regulation of the SNA/AIEgen probe by altering the dsDNA length. The binding TPBT numbers of the obtained four SNA/AIEgen probes were ~6084, 9945, 14907, and 19848, respectively, indicating that AIEgen loading gradually increased with the dsDNA length. Nonetheless, the SNA/AIEgen probes with the highest S/N ratio were obtained with the desired combination of SH-dsDNA30 and 2.5 μM of TPBT (Supplementary Fig. 35). We then incorporated the constructed SNA/AIEgen reporter into a Crispr/Cas12a detection system for the detection of synthetic DNA targets (Fig. 5b, c). The LOD of the improved CrisprAIE based on SNA/AIEgen reporters was as low as 55.7 fM, exhibiting about 3.3- and 270-fold improvement in detection sensitivity compared with Crispr-Dx using Q-dsDNA/AIEgen-Q and Q-ssDNA-FAM reporters, respectively (Fig. 5c). Compared with the Q-dsDNA/AIEgen-Q and Q-ssDNA-FAM reporter probes, the SNA/AIEgen reporter showed significantly enhanced stability and detection accuracy even in the presence of 5 U mL-1 DNase I and 100% fetal bovine serum (FBS), indicating the excellent stability of SNA/AIEgens in complex biological systems (Fig. 5d–g). We further compared the amplification-free detection sensitivity of Crispr-Dx based on five different reporting probes. Figure 5h demonstrates that the sensitivity of Crispr-Dx improved significantly with the evolution of signal reporters. To better evaluate the real-world application of the SNA/AIEgen-enhanced CrisprAIE, we used it to detect a variety of clinically relevant nucleic acid targets, including norovirus, SARS-CoV-2, Ebola virus, HPV-16, HPV-18, and monkeypox virus (MPXV). For comparison, Crispr/Cas12a detection using Q-dsDNA/AIEgens-Q and Q-ssDNA-FAM as signal reporters was simultaneously conducted. To simulate the direct detection of nucleic acid targets from a clinically complex sample matrix, we added these synthetic DNA targets to the corresponding biological sample types comprising background nucleic acids. Prior to CrisprAIE detection, the biological fluid samples were treated with the previously reported HUDSON method, which can simulate viral particles releasing DNA or RNA and eliminate nuclease activity from the samples39. As revealed in Fig. 5i–n, the CrisprAIE assay based on Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters exhibited significantly increased signal response in detecting all the gene targets in the buffer alone and in the biological specimens, such as 8% vomitus, throat swab, 8% saliva, or 2.5% serum, compared with traditional Q-ssDNA-FAM. No significant difference in signal response was observed in measuring the targets in buffer solutions and real samples when SNA/AIEgens were used as CrisprAIE reporters, indicating their negligible matrix interference. These findings proved that our proposed CrisprAIE exhibited superior robustness in detecting nucleic acid targets in complex biological fluid samples, implying its broad application prospects in field-deployable viral diagnostics by detecting various genetic targets.
Fig. 5. AIEgen-assisted SNA reporters enhance the CrisprAIE detection of target DNA in complex samples.
a Schematic of the CrisprAIE-based SNA reporter. b Detection sensitivity of CrisprAIE-based SNA/AIEgen reporter in detecting norovirus synthetic DNA. c Calibration curve fitting the relative fluorescence intensities with target concentrations at 30 min of the detection in (b). Schematic of the stability and fluorescence kinetic analysis of ssDNA-FAM reporters (d), Q-dsDNA/AIEgen reporters (e), and SNA/AIEgen reporters (f) in 5 U mL−1 DNase I or 100% FBS, respectively. g Signal-to-noise ratio (S/N) calculated by dividing the 100 pM target signals by the signal in 5 U mL−1 DNase I or 100% FBS. h Comparison of detection sensitivity among all the AIEgen-enhanced reporter-based CrisprAIE and conventional FQ reporter-based Crispr-Dx in this study. S/N in the detection of 100 pM target: Norovirus ORF2 gene synthetic targets spiked into reaction buffer or 8% vomitus (i); SARS-CoV-2 N gene synthetic targets spiked into reaction buffer, throat swab or 8% saliva (j); Ebola (k), HPV-16 L1 gene (l), HPV-18 L1 gene (m), and MPXV B6R gene (n) synthetic DNA targets spiked into reaction buffer or 2.5% serum. Data are mean ± s.d.; n = 3 repeated tests. Two-tailed P values were calculated using unpaired t-tests: *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001; ns, not significant, compared with buffer only, respectively.
In addition to the above-mentioned amplification-free Crispr system, we explored the compatibility of spherical SNA/AIEgen probes with amplification-based Crispr systems. As illustrated in Supplementary Fig. 36a, we integrated the SNA@AIEgen reporter into amplification-based Crispr systems to detect SARS-CoV-2 pseudovirus with concentrations ranging from 0 to 1000 copies/µL. Supplementary Fig. 36b shows that the developed CrisprAIE, combined with RT-RPA and SNA@AIEgen reporters, effectively detects virus concentrations as low as 0.1 copies/µL, representing a tenfold enhancement in sensitivity compared to the LOD of 1 copy/µL achieved by CrisprAIE combined with RT-RPA and Q-dsDNA/AIEgen-Q reporters. Furthermore, to evaluate the repeatability and reproducibility of our spherical SNA@AIEgen probes, we conducted additional experiments. Initially, we performed repeated detections of SARS-CoV-2 pseudovirus at a concentration of 1 copy/µL in quintuplicate on the same day. The results in Supplementary Fig. 36c showed negligible changes in fluorescent signal response among the five replicates. Moreover, we synthesized three different batches of SNA@AIEgen probes (Supplementary Fig. 36d) and utilized them to detect 1 copy/µL of SARS-CoV-2 pseudovirus. The results in Supplementary Fig. 36e indicated consistent fluorescence signal responses across the three different batches of probes. These findings indicated the satisfactory reproducibility and repeatability of the SNA@AIEgen-combined CrisprAIE for quantifying SARS-CoV-2 pseudovirus.
Expanding CrisprAIE for the direct detection of target RNA
The above results demonstrated that the coupling of DNA-enhanced AIEgen lighting-up readout with Crispr/Cas12a-assisted ssDNA cleavage reaction is promising for the detection of genetic targets containing DNA and RNA targets when integrated with the isothermal amplification of RPA or RT-RPA. However, Cas12a can only specifically recognize DNA. Thus, the direct detection of RNA virus using Cas12a usually requires a reverse transcription of RNA into DNA. By contrast, Cas13 can specifically recognize RNA and thus simplify its reverse transcription. Compared with Cas12a enzymes involving the trans-cleavage of nontargeted ssDNA, the collateral activity of Cas13 enzymes involves the trans-cleavage of nontargeted single-stranded RNA (ssRNA). Thus, to extend the DNA-enhanced AIEgen lighting-up readout into Crispr/Cas13-driven ssRNA cleavage reaction, we further developed a Cas13a-based CrisprAIE assay using an ssRNA linker (T*A*rArUG*C*) as an alternative to an ssDNA linker (TTATT) in the Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters for Cas13a trans-cleavage (Fig. 6a–c). For comparison, conventional Q-ssRNA-FAM reporters were commercially synthesized. To further evaluate their detection performance, a series of RNA targets at different concentrations ranging from 0 to 10 nM was prepared and analyzed using these three Crispr-Dx methods. Figure 6d shows that the Crispr-Dx based on Q-ssRNA-FAM reporters can only detect RNA targets at the 1 pM level without amplification. Meanwhile, the CrisprAIE assay can sensitively respond to 100 and 10 fM RNA targets when using Q-dsDNA/AIEgens-Q and SNA/AIEgens as signal probes, respectively (Fig. 6e, f). Our results indicated that the CrisprAIE based on Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters provide 10- and 100-fold increased detection sensitivity relative to the conventional Q-ssRNA-FAM-based Crispr-Dx. We further employed the developed CrisprAIE assay based on Cas13 nuclease for the direct detection of three synthetic RNA targets, namely, norovirus, SARS-CoV-2, and Ebola virus, in their respective diagnostic samples. The synthesized RNA targets against norovirus, SARS-CoV-2, and Ebola virus were detected in target-activated collateral cleavage reactions containing 8% vomitus, throat swad, 8% saliva, or 2.5% serum (Fig. 6g–i). The CrisprAIE using Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters demonstrated significantly increased signals relative to the control Q-ssRNA-FAM reporter even in complex biological fluid samples. We also found that the SNA/AIEgen reporter showed better resistance to sample matrix interference than the Q-dsDNA/AIEgen-Q and Q-ssRNA-FAM reporters, indicating the excellent robustness and on-site diagnostic potential of the SNA/AIEgen reporter for detecting RNA targets in samples containing biological fluids.
Fig. 6. Expanding CrisprAIE for the direct detection of target RNA in complex samples.
Schematic of Crispr/Cas13a based on three fluorescent reporters, namely, conventional Q-ssRNA-FAM reporters (a), Q-dsDNA/AIEgen-Q reporters (b), and SNA/AIEgen reporters (c). Detection curves of different concentrations of synthetic RNA targets by Crispr-Dx based on Q-ssRNA-FAM (d), Q-dsDNA/AIEgen-Q (e), and SNA/AIEgen (f) reporters. Background-subtracted fluorescence in the detection of 1 pM synthetic RNA targets: Norovirus ORF2 gene synthetic RNA targets spiked into reaction buffer or 8% vomitus (g); SARS-CoV-2 N gene synthetic RNA targets spiked into reaction buffer, throat swab or 8% saliva (h); Ebola synthetic RNA targets spiked into reaction buffer, or 2.5% serum (i). Data are mean ± s.d.; n = 3 repeated tests. Two-tailed P values were calculated using unpaired t-tests: *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001; ns, not significant, compared with buffer only, respectively.
Adapting CrisprAIE for POC testing
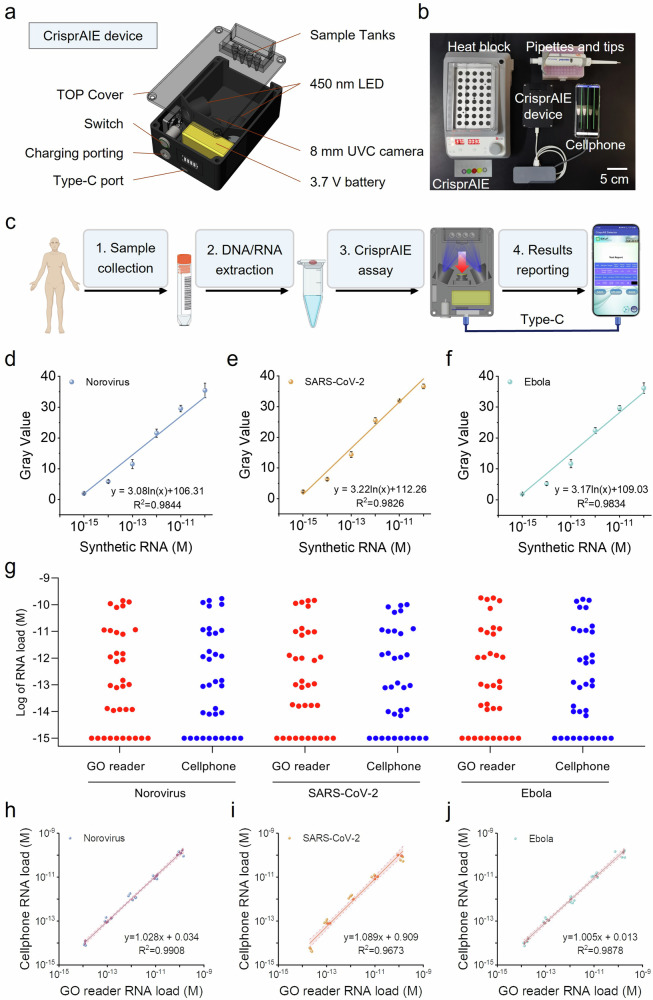
The above results demonstrated that the developed CrisprAIE technology had significantly improved detection sensitivity compared with traditional Crispr-Dx. In addition, the CrisprAIE assay did not significantly increase the complexity of the testing procedure. These features make our single-tube isothermal CrisprAIE assay suitable for POC testing applications. In this regard, we developed a portable cellphone-based CrisprAIE assay platform using a cellphone-assisted CrisprAIE fluorescence readout device to output the detection signal. The prototype of this CrisprAIE device was assembled from 3D-printed main body parts (e.g., brackets and shells), an optical system, and an electrical circuit system. Its core components and optical paths are shown in Fig. 7a. In the optical system, two 450 nm LED illuminators were used as the fluorescence excitation module, and an imaging camera covered an optical filter (650 ± 20 nm) was fitted to the lateral position directly above the test tube to output visual photos. In the electrical circuit system, a 3.7 V battery was used to power the LED light. The type-C port on the bottom of the device directly transmits the captured fluorescent images to the cellphone side for measuring the gray value and report the test results. The CrisprAIE test can be run in about 30 min and displayed on the cellphone (Fig. 7b). This integrated CrisprAIE system was designed to use a straightforward workflow, in which a sample was added in a tube prefilled with RPA reagents capped with a tube cap prefilled with CrisprAIE reagents and heated at 37 °C for 15 min. The tube was shaken to mix the RPA and CrisprAIE components and then incubated at 37 °C for another 15 min. After the CrisprAIE test, the tube was placed in the sample tanks. The TEST button of the APP was clicked, and the result was automatically reported on the display interface of cellphone. To investigate the feasibility and reliability of our CrisprAIE for POC testing, we assessed the POC prototype for our isothermal CrisprAIE reaction and compared it with the commercial Multiskan GO multimode reader. In this case, the cellphone-based CrisprAIE device was applied to detect three different synthetic RNA targets against norovirus, SARS-CoV-2, and Ebola virus with concentration gradients ranging from 10-15 to 10-10 M as shown in Fig. 7d–f. The findings suggested that the cellphone-based CrisprAIE assay achieved comparable analytical performances in terms of detection sensitivity (LOD: 1 fM) and dynamic detection range (1–105 fM) for the three target RNAs compared with the commercial multimode reader. A correlation analysis was conducted on the detected concentrations obtained by the cellphone-based CrisprAIE assay and the multimode reader-based CrisprAIE assay. Figure 7g–j suggests highly linear correlations between the two assay modes, confirming the practicability and reliability of our cellphone-based CrisprAIE assay for POC diagnostics.
Fig. 7. Portable device adapting CrisprAIE for POC testing.
a Schematic of a portable reading device, including an optical system and an electrical circuit system. b Minimum equipment needed to run the protocol, including heat blocks (37 °C), portable reading device, and cellphone with type-C data cable. The materials include RPA kit, Crispr-related reagent, and pipettes and tips. c Workflow of CrisprAIE test. After 30 min of RPA and Crispr reaction, the tube is placed into the sample tanks. The portable device captures the tube image and transmits it to the cellphone through a type-C data cable. The APP (CrisprAIE Detector) processes and analyzes the grayscale value of the image to obtain a detection report. Standard curve of the CrisprAIE testing read by the cellphone device, including norovirus (d), SARS-CoV-2 (e), and Ebola virus (f). Data are mean ± s.d.; n = 3 repeated tests. g CrisprAIE analysis of RNA load distributions in paired synthetic RNA-spiked samples with different concentrations from 10-14 to 10-10 M by the cellphone and Multiskan GO multimode reader. h–j Correlation of results of cellphone and GO plate reader, indicating the linear regression line (solid line) and the limits of its 95% CI (shadow). Data show 3 repeated tests.
Discussion
The outbreak of emerging infectious diseases, such as COVID-19, urgently requires fast, sensitive, and reliable diagnostic technologies, especially those that can be used in the field. In this regard, Crispr/Cas nucleases are attractive tools for field-deployable diagnostic applications due to their high programmability and ability to directly detect viral nucleic acid sequences. However, the sensitivity and speed of Crispr/Cas-based diagnostic methods must be further improved to enable their extensive use as diagnostics. In this study, we report the development of an improved Crispr/Cas-based diagnostic assay, named CrisprAIE, to sensitively detect nucleic acid targets by integrating DNA-enhanced AIEgen lighting-up fluorescent signal readout. Compared with conventional Crispr-Dx assay using single-stranded nucleic acid-based fluorescence probes, the reported CrisprAIE assay possessed the advantages of higher detection sensitivity and stronger stability in complex biofluid samples. Under optimized conditions, our CrisprAIE method can detect DNA and RNA targets at levels as low as femtomolar concentrations without amplification and exhibits at least 80-fold enhancement in sensitivity than traditional Crispr-Dx assay. This LOD of our CrisprAIE was lower than that of previously reported amplification-free Crispr-Dx detection techniques15,32,40,41 (Fig. 8a and Supplementary Table 5). The combination with isothermal amplification, such as RPA or RT-RPA, can further increase the sensitivity to LOD of attomolar concentrations within 35 min from raw sample to result; this performance is comparable with or even better than that of currently available amplification-enhanced Crispr-Dx methods10–14,42–52 (Fig. 8b and Supplementary Table 5). In summary, we believe that CrisprAIE realized the amplification-free quantification of target sequences, and its combination with nucleic acid amplification techniques can accelerate the ultrasensitive quantitative assay of trace targets in the attomolar range.
Fig. 8. Comparison between CrisprAIE (this work) and reported Crispr-Dx with or without amplification.

a Comparison of sample-to-result time and detection sensitivity between amplification-free CrisprAIE based on Q-dsDNA/AIEgens-Q and SNA/AIEgen reporters with reported Crispr-Dx. b Comparison of sample-to-result time and detection sensitivity between amplification-assisted CrisprAIE based on Q-dsDNA/AIEgen-Q reporters with reported Crispr-Dx.
To avoid the contamination risk of multiple specimen-transfer steps, we tried to introduce the single-tube Crispr diagnostic strategy for field implementation through the simple spatial isolation of isothermal amplification reagents and Crispr/Cas reagents. This strategy can also effectively eliminate the uncapped operation and save assay time. Despite the successful development, this approach was inconvenient and compromised the efficiency of Crispr/Cas. Fortunately, recent advances in the one-pot Crispr diagnostic assay can further promote the adoption of CrisprAIE for POC applications53. For example, the use of a photocontrolled Crispr method and suboptimal protospacer adjacent motifs for Cas12a with all components added to a closed system in one-pot assay improved the detection efficiency of Crispr diagnostic method with high sensitivity and fast speed48,52.
We successfully demonstrated the viability of CrisprAIE on Cas12a and Cas13. With the development of biotechnology, the number of Crispr/Cas systems are expanding rapidly. Various Crispr/Cas systems, such as Cas9 and Cas14a, are being explored for the development of Crispr diagnostic methods. Further investigation on the compatibility of these Crispr/Cas systems with AIEgen lighting-up fluorescent signal readout will help broaden the variety and application range of CrisprAIE. Given that CrisprAIE is independent of gRNA or crRNA design and specific Crispr/Cas enzyme, it can be widely used to amplify all existing Crispr-Dx assays that rely on the collateral cleavage of single-stranded nucleic acid reporters, including the well-known SHERLOCK, DETECTOR, and HOLMES. In addition, the possible application of CrisprAIE for the sensitive and accurate quantitative detection of nonnucleic acid targets is worthy of further exploration.
The stability of fluorescent reporting probes in complex biological systems is crucial for accurate Crispr/Cas detection. The widely used linear oligonucleotide reporters for Crispr/Cas usually run the risk of being degraded by nucleases in biological samples, resulting in false-positive signals. Compared with such oligonucleotide reporters, our designed Q-dsDNA/AIEgens-Q reporter provided better signal response with higher S/N ratio in the complex sample matrix. Nevertheless, the improvement in CrisprAIE’s sensitivity and stability by the Q-dsDNA/AIEgens-Q reporter remains limited. Thus, to further enhance the biological stability of our CrisprAIE, we adopted the SNA strategy to improve the stability of AIEgen-modified nucleic acid reporters and generate a sensitive and stable fluorescent signal output for CrisprAIE development. Figures 5–6 validate that the SNA/AIEgen signal reporter greatly improved the biostability and resistance to background interference in complex systems. In addition, the CrisprAIE constructed using SNA/AIEgen reporters offered 3.3- and 270-fold increases in detection sensitivity relevant to dsDNA/AIEgens-Q and conventional oligonucleotide reporters. With these characteristics, we can envision the SNA/AIEgens-based CrisprAIE enabling sensitive, accurate, and stable biodetection in complex systems.
The introduction of DNA-enhanced AIEgen lighting-up fluorescent signal readout is of great significance to improve the sensitivity of Crispr/Cas detection. The clinical diagnostic efficiency and accuracy of our reported CrisprAIE system was well demonstrated. To extend its use in POC testing and commercial setting, we demonstrated the successful construction of a portable cellphone-assisted CrisprAIE fluorescence readout device. The analytical performance of our CrisprAIE prototype cellphone device was comparable with that of the commercial multimode fluorescence microplate reader in detecting different synthetic RNA targets on the Crispr/Cas13 platform, indicating the huge potential for the widespread use of our cellphone-assisted CrisprAIE platform in screening and diagnostics in the field and even at home.
While our proposed AIEgen probes offer numerous advantages, including “one to more” signal output, label-free loading of AIEgens onto dsDNA, large Stokes shifts, excellent photostability, high signal-to-noise ratio, and minimal background interference, they also exhibit certain limitations when compared to conventional FQ-based DNA probes. Unlike traditional FQ-based reporters, our AIEgen-loaded DNA reporters are prepared through the non-covalent binding of cationic AIEgens and negatively charged DNA strands via electrostatic adsorption, hydrophobic action, and van der Waals forces. This feature renders our AIEgen probes more susceptible to interference from complex sample matrices. Additionally, the proposed AIEgen probes cannot be directly applied to viral lysis amplification detection methods; instead, nucleic acid extraction is required. Moreover, our CrisprAIE method is not yet field-deployable and still necessitates several sample-processing steps, along with temperature control for the storage of reagents and the amplification process. Despite these limitations, ongoing research efforts continue to address these challenges and optimize AIEgen probes for various diagnostic applications. By carefully considering the specific requirements of each assay and leveraging the unique properties of AIEgen probes, it is possible to overcome these limitations and harness their full potential in advancing molecular diagnostics.
In conclusion, this work is the first to report the AIEgen-enhanced Crispr-Dx assay for the sensitive and accurate detection of nucleic acid targets and demonstrates its application potential in infectious disease diagnosis. Our CrisprAIE can be further improved by applying the recent achievements in Crispr/Cas-related elements, such as Cas nuclease evolution, crRNA structure optimization, amplification primer screening, and reaction system optimization (e.g., ions, pH, and component concentrations), and in signal reporter systems, such as AIEgens with high quantum yield, microfluidics, and digital reading device.
Methods
Materials
Analytical-grade organic solvents, such as methanol (cat. no. 10014118), ethanol (cat. no. 100091192), dimethyl sulfoxide (DMSO) (cat. no. 30072418), and acetonitrile (cat. no. 80000618), were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). The sequence information for all synthesized DNA or RNA, synthetic targets, RPA primers, Cas12a-crRNA, Cas13-crRNA, and fluorescent reporters used in this work are shown in Supplementary Tables 1, 2. The SARS-CoV-2 N gene sequence was synthesized by Tsingke Biotechnology Co., Ltd. (Beijing, China). The DNA oligomers used for constructing the reporter strand, norovirus ORF2 gene sequence, Ebola gene sequence, HPV-16 L1 gene sequence, HPV-18 L1 gene sequence, MPXV B6R gene sequence, and primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). S1 nuclease (cat. no. EN0321), gel loading buffer (cat. no. 10816015), and agarose (cat. no. R0492) were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). EnGen LbaCas12a (Cpf1, cat. no. M0653T) and NEBuffer 2.1 (cat. no. B6002S) were obtained from New England BioLabs (Ipswich, MA). The lyophilized TwistAmp® Basic kit (product code: TABAS03KIT) comprising the Core Reaction Mix of recombinase, single-stranded binding protein, and strand-displacing polymerase, was acquired from TwistDx™ Limited (Cambridge, UK, cat. no. TABAS03KIT). SARS-CoV-2 pseudovirus (cat. no. BDS-BW-118) was provided by Guangzhou BDS Biological Technology Co., Ltd. (Guangzhou, China). Express RNA rapid extraction kit (cat. no. NR202) was purchased from GenDx Biotech Co., Ltd. (Suzhou, China). DNA gel extraction kit (cat. no. B110092-0050) was obtained from Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China).
Buffer solutions
Nuclease-free ultrapure distilled water (UPDW, Invitrogen) was used in all experiments. All buffers were prepared in RNase-free conditions. The phosphate buffer saline (PBS) was 0.01 M phosphate buffer in a 0.8% w/v saline solution, pH 7.5. The 10X NEBuffer 2.1 contained 100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2, pH 7.9. The Tris-HCl buffer comprised 0.05 M Tris-HCl in RNase-free ultrapure distilled water, pH 8.5. The Tris-NaCl buffer was 50 mM Tris-HCl, 100 mM NaCl, pH 8.5.
Molecular docking of AIEgens with dsDNA using DOCK 6.9
First, the 3D structures of dsDNA (receptor) were obtained from Xiao Lab (http://biophy.hust.edu.cn/new/), and the molecular structures of AIEgens (ligand) were obtained with ChemDraw 20.0. The binding mode and interaction of nucleic acid (dsDNA) with small molecular AIEgens were performed using DOCK 6.9 program by YINFO TECHNOLOGY (https://cloud.yinfotek.com/console/). The docking score of DOCK 6.9 is expressed as grid score. In general, the smaller the grid score, the stronger the binding force. Grid vdw and Grid es represent the contribution of van der Waals force (nonpolar interaction) and electrostatic force (polar interaction), respectively. Internal energy refers to the repulsion between receptor with ligand.
Simulation sample preparation
Human saliva samples were supplemented with 1 M TCEP, 500 mM EDTA, and RNase inhibitor (40 U μL−1) at a volume ratio of 100:11.39:0.23:2.28 with final concentrations of 87% biofluids, 100 mM TCEP, 1 mM EDTA, and 0.8 U μL−1 inhibitor. Human serum was diluted with PBS and supplemented with 1 M TCEP, 500 mM EDTA, and RNase inhibitor (40 U μL−1) at a volume ratio of 100:251.2:40:0.8:8 with final concentrations of 25% serum or plasma, 100 mM TCEP, 1 mM EDTA, and 0.8 U μL−1 inhibitor. Synthetic targets were spiked into the biofluids and treated with the HUDSON method. These samples were incubated at 40 °C for 5 min and at 70 °C for another 5 min (or 5 min at 95 °C, if saliva) on a thermocycler.
Clinical samples
Clinical samples were collected from Jiangxi Provincial Center for Disease Control and Prevention and Department of Laboratory Medicine, The First Affiliated Hospital of Nanchang University. Among them, 15 clinical vomitus samples confirmed by RT-qPCR, including 9 vomitus samples from patients with Norovirus and 6 vomitus samples from patients without norovirus, were provided by Jiangxi Provincial Center for Disease Control and Prevention. Meanwhile, 75 identified clinical nasopharyngeal swab samples with RT-qPCR-corroborated COVID-19, including 50 positive, 10 suspiciously positive, and 15 negative samples from patients in fever clinics, were provided by The First Affiliated Hospital of Nanchang University.
Ethical statement
This study was approved by the Medical Ethics Committee of The First Affiliated Hospital of Nanchang University and the Jiangxi Provincial Center for Disease Control and Prevention Ethics Committee (approval ID number 2020-Y114-01). Written informed consent was obtained from all participants before the study.
Preparation and characterization of AIEgen reporters
Four types of AIEgen reporters, namely, ssDNA/AIEgens, Q-dsDNA/AIEgens, Q-dsDNA/AIEgens, and Q-dsDNA/AIEgens-Q (collectively referred as linear AIEgen reporters), were prepared via the label-free coupling of AIEgens and DNA. For ssDNA/AIEgen reporters, 10 µL of TPBT (250 µM) were mixed with 10 µL of ssDNA (20 µM) and supplemented with 180 µL of NaCl aqueous solution (200 mM). For Q-dsDNA/AIEgen reporters, Q-dsDNA was obtained by mixed Q-ssDNAA15 (20 µM) with complementary ssDNAT15 (20 µM) at a volume ratio of 1:1 with final concentrations of 10 µM, followed by annealing with 50 µL of TPBT aqueous solution (125 µM) and 50 µL of Q-dsDNA at 95 °C for 5 min. For Q-dsDNA/AIEgen-Q reporters, Q-dsDNA was obtained by mixed Q-ssDNAA30 (10 µM) with complementary Q-ssDNAT30 (10 µM) at a volume ratio of 1:1 with final concentrations of 5 µM, followed by annealing with 50 µL of TPBT aqueous solution (125 µM) and 50 µL of Q-dsDNA-Q (5 µM) at 95 °C for 5 min.
The Q-dsDNA/AIEgen-Q reporter was separated and purified by agarose electrophoresis using a DNA gel extraction kit. Q-dsDNA/AIEgens-Q was separated from free AIEgens by agarose electrophoresis, and the DNA fragment of interest (Q-dsDNA/AIEgens-Q) from agarose gel was excised with a clean, sharp scalpel. Afterward, the Q-dsDNA/AIEgen-Q reporter was recovered in accordance with the operating procedure of the DNA gel extraction kit. The characterization and evaluation of AIEgen reporters were performed by adding 100 μL of 500 U mL−1 S1 nuclease to 100 μL of prepared AIEgen reporters (200 nM). The fluorescent intensities were recorded every 5 min by Multiskan GO multimode reader.
Crispr/Cas12a for synthetic target DNA detection
Fluorescence assays were performed as described with modifications. The LbaCas12a protein stock (100 µM) was diluted to 1 µM using the storage buffer (50 mM Tris-HCl, 600 mM NaCl, 5% glycerol, and 2 mM DTT, pH 7.5). The crRNA stock (100 µM) was diluted to 1 µM using nuclease-free water. The 20 μL of mixed solution per reaction contained 0.4 of LbaCas12a protein (1 μM), 0.4 μL crRNA (1 μM), 4 μL of linear AIEgen reporters (1 μM) or Q-ssDNA-FAM reporters (1 μM), 0.5 μL of RNase inhibitor (40 U μL−1), 2 μL of NEBuffer 2.1 (10×, 100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2, pH 7.9), 2.7 μL of nuclease-free water, and 10 μL of synthetic DNA targets, unless otherwise indicated. All reactions were performed in a 384-well microplate (Corning) at 37 °C, with fluorescence monitored every 5 min over 0–60 min by a Multiskan GO multimode reader (excitation: 460 nm and emission: 650 nm for AIEgen reporters; or excitation: 490 nm, emission: 520 nm for Q-ssDNA-FAM reporters).
RT-RPA
Total virus RNA was extracted from the patient sample using the viral RNA extraction kit. The extracted and purified RNAs were aliquoted and stored at −80 °C before use. cDNA was obtained by reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Vilnius, Lithuania). RPA pellet rehydration solution was prepared by dissolving one RPA pellet into 29.5 µL of rehydration buffer. Afterward, 2.4 µL of 10 µM RPA forward/reverse primer, 1 µL of RNase inhibitor (40 U mL−1 stock), 1 µL of reverse transcriptase, 1 µL of RNA extracts, and 10.2 µL of nuclease-free water were sequentially added into the prepared RPA pellet rehydration solution, followed by 2.5 µL of 280 mM magnesium acetate to initiate the amplification. The amplification proceeded at 37 °C and was completed within 15 min. The amplification reaction tube was suspended in an ice-bath. The amplification products were analyzed by gel electrophoresis.
RNA quantification of real samples using CrisprAIE combined with RT-RPA
The total virus RNA was extracted from the patient sample using the viral RNA extraction kit, and cDNA was obtained by reverse transcription using the RevertAid First Strand cDNA Synthesis Kit. The above procedure was performed to add cDNA to the RT-RPA system for isothermal amplification. The 20 μL of mixed solution per reaction contained 0.4 μL of LbaCas12a protein (1 μM), 0.4 μL of crRNA (1 μM), 4 μL AIEgen reporters (1 μM), 0.5 μL of RNase inhibitor (40 U μL−1), 2 μL of NEBuffer 2.1 (10X, 100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2, pH 7.9), 2.7 μL of nuclease-free water, and 10 μL of RPA amplicons. All reactions were performed in a 384-well microplate at 37 °C, with fluorescence monitored every 5 min over 0–60 min by a Multiskan GO multimode reader.
RT-qPCR
PrimeScript TM RT Reagent Kit (Takara, Kusatsu, Japan) with g DNA Eraser was used for reverse transcription by the directions to create single-stranded cDNA. SYBR Premix Ex Taq II kit was used for quantitative real-time PCR (qPCR). The sequences of primers used for real-time quantitative RT-qPCR are shown in Supplementary Table 2. The qPCR reactions were performed using a 7900HT fast real-time qPCR system (Applied Biosystems, Foster City, CA) with the following cycling conditions: hold at 25 °C for 2 min, hold at 95 °C for 2 min, followed by 45 cycles with DNA denaturation at 95 °C for 3 s, and annealing and elongation at 55 °C for 30 s. Data were analyzed by the 2−ΔΔCt method with CFX Maestro Software.
Synthesis and characterization of SNA/AIEgen reporters
Polydopamine and AuNP-coated Fe3O4 nanocomplex (Fe3O4/Au/PDA) was selected as the nanoquencher and synthesized following a previously reported method with slight modifications. The 200 nm Fe3O4 core was synthesized through the classical one-pot method. In brief, 667 mg of FeCl3•6H2O, 2.8 g of sodium acetate, and 40 mL of ethanediol were added to a three-neck flask. The mixture was then stirred at room temperature for 30 min, transferred into a hydrothermal autoclave reactor, and reacted at 200 °C for 12 h. The obtained Fe3O4 NPs were washed with ethanol and water and collected by vacuum drying. Meanwhile, 10 mg of prepared Fe3O4 NPs, 50 μL of 10% HAuCl4 aqueous solution, 10 mg of trisodium citrate, and 40 mL of ethanol were added into a round-bottom flask, which was then placed at ultrasonic condition for 10 min. For polydopamine coating, a dopamine solution was first prepared by dissolving 50 mg of dopamine hydrochloride into 30 mL 0.05 M pH 8.5 Tris buffer. The freshly prepared dopamine solution was rapidly added into the round-bottom flask. The mixture was sonicated at room temperature for 3.5 h. The end-product was washed with ultrapure water for five times, and the obtained 100 mg of Fe3O4/Au/PDA was resuspended in 10 mL of ultrapure water solution for further use. All batches of Fe3O4/Au/PDA were diluted to 1 mg mL−1 (70 pM) in UPDW and characterized using Malvern Zetasizer Nano ZS ZEN3700 DLS system to measure the charge and particle size based on the zeta potential and DLS, respectively. TEM imaging was performed using a high-resolution JEOL JEM 2100 microscope. Before the imaging and analyses, the samples were prepared by placing a 2 µL of droplet of the nanoparticle dispersion on a 200-mesh carbon-coated copper grid (Electron Microscopy Science, USA).
SH-dsDNA was obtained by mixing SH-ssDNATTATTTTATT-A30 with ssDNAT30 and anneal for 5 min at 95 °C and then conjugated to the surface of Fe3O4/Au/PDA as described with modifications. In brief, 1.43 mL of Fe3O4/Au/PDA (70 pM) was moved from ultrapure water to 1 mL of 0.05 M pH 8.5 Tris buffer through magnetic separation (final concentration of 100 pM). Afterward, 10 μL of 10 μM HS-dsDNA was mixed with the suspended Fe3O4/Au/PDA solution. The mixture was shaken at room temperature for 36 h. The excessive dsDNA was removed by using magnets to collect dsDNA-conjugated Fe3O4/Au/PDA (SNAs). The collected SNAs were washed with ultrapure water three times and then suspended at 1 mL of ultrapure water. SNAs/AIEgens were prepared by directly mixing the SNASs with TPBT under optimized concentrations. The SNASs were resuspended in 900 μL of 0.05 M Tris-NaCl (50 mM Tris-HCl, 100 mM NaCl, pH 8.0), then added with 100 μL of TPBT (25 μM) mixed with SNAs. Free AIEgens were removed via magnetic separation. The prepared SNA/AIEgen reporters were dispersed in 0.05 M Tris-NaCl buffer for subsequent experiments. All batches of SNA/AIEgen reporters were characterized using Malvern Zetasizer Nano ZS ZEN3700 DLS system to measure the charge based on the zeta potential.
Crispr/Cas13a for synthetic target RNA detection
The LwaCas13a protein stock (100 µM) was diluted to 1 µM using the storage buffer (50 mM Tris-HCl, 600 mM NaCl, 5% glycerol, and 2 mM DTT, pH 7.5). The crRNA stock (100 µM) was diluted to 1 µM using nuclease-free water. The 20 μL of mixed solution per reaction contained 0.4 μL of LwaCas13a protein (1 μM), 0.4 μL of crRNA (1 μM), 4 μL of AIEgen reporter or Q-ssDNA-FAM reporters (1 μM), 0.5 μL of RNase inhibitor (40 U μL−1), 2 μL of NEBuffer 2.1 (10X, 100 mM Tris-HCl, 500 mM NaCl, 100 mM MgCl2, pH 7.9), 2.7 μL of nuclease-free water, and 10 μL of synthetic RNA targets, unless otherwise indicated. All reactions were performed in a 384-well microplate (Corning) at 37 °C, with fluorescence monitored every 5 min over 0–60 min by a Multiskan GO multimode reader.
Statistical analysis
All data statistical analyses were conducted in GraphPad Prism v.8.3.0. Data were expressed as means ± standard deviation. All sample sizes and statistical tests were specified in the figure legends. All experiments were independently repeated at least thrice. Significant differences between groups were assessed by parametric and nonparametric comparisons, and multiple hypothesis tests were adjusted appropriately. For statistical significance test, unpaired two-tailed t-test (parametric) was used for two-group comparisons, and ANOVA was used for multiple-group comparisons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32172296 to X.L.H.), Key Research and Development Program of Jiangxi Province (20232BBG70030 and 20232BCD44004 to Y.H.X.), Double-Thousand Plan of Jiangxi Province (jxsq2023201113 to Y.H.X.), Jiangxi Provincial Natural Science Foundation (20224ACB205012 to X.L.H.), Jiangxi Medicine Academy of Nutrition and Health Management (2022-PYXM-03 to Y.H.X.), Shenzhen Key Laboratory of Functional Aggregate Materials (ZDSYS20211021111400001 to B.Z.T.), and the Science Technology Innovation Commission of Shenzhen Municipality (KQTD20210811090142053 and JCYJ20220818103007014 to B.Z.T.).
Author contributions
X.L.H., Y.H.X. and B.Z.T. conceived, designed, and managed the project. Y.Q.G. performed most experiments and data analysis with the help of H.D., M.W., Y.H.W., Y.X., X.R.C., S.Y.W. D.R.X., D.F.L. and H.B.X. provided clinical real samples and conducted relevant experiments. H.B.X., X.L.H., Y.H.X. and B.Z.T. provided critical insights on protocols, the experimental results, and the whole work. Y.Q.G. and Y.F.Z. completed the first draft of the paper under the guidance from X.L.H. and Y.H.X. The manuscript was further revised by X.L.H., Y.H.X. and B.Z.T., who jointly supervised the project. All authors reviewed the manuscript and approved it for publication.
Peer review
Peer review information
Nature Communications thanks Xiao-fan Jiang, Xiaoming Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data generated in this study are provided in the Supplementary Information/Source Data file. All relevant data are available from the corresponding author on request. The source data underlying Figs. 2a–d, f, h, j, l, 3b–e, 4a–c, 5b–n, 6d–i, and 7d–g are provided as a Source Data file. Source data are provided with this paper.
Competing interests
Y.Q.G., X.L.H., and Y.H.X. are listed as inventors on a pending patent concerning the design and application of CrisprAIE. All other authors declared no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaolin Huang, Email: xiaolin.huang@ncu.edu.cn.
Yonghua Xiong, Email: xiongyonghua@ncu.edu.cn.
Ben Zhong Tang, Email: tangbenz@cuhk.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52931-0.
References
- 1.Wood, C. S. et al. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature566, 467–474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravindran, S. Smartphone science: apps test and track infectious diseases. Nature593, 302–304 (2021). [DOI] [PubMed]
- 3.Wang, A. M., Doyle, M. V. & Mark, D. F. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl Acad. Sci. USA86, 9717–9721 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milanez-Almeida, P., Martins, A. J., Germain, R. N. & Tsang, J. S. Cancer prognosis with shallow tumor RNA sequencing. Nat. Med.26, 188–192 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res.28, e63–e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian, B., Minero, G. A. S., Fock, J., Dufva, M. & Hansen, M. F. CRISPR-Cas12a based internal negative control for nonspecific products of exponential rolling circle amplification. Nucleic Acids Res.48, e30–e30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing, S. et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics10, 10262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu, Z. et al. Carrier strategies boost the application of CRISPR/Cas system in gene therapy. Exploration2, 20210081 (2022). [DOI] [PMC free article] [PubMed]
- 9.Marraffini, L. A. CRISPR-Cas immunity in prokaryotes. Nature526, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broughton, J. P. et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol.38, 870–874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, S. Y. et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov.4, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai, Y. et al. Exploring the trans‐cleavage activity of CRISPR‐Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem., Int. Ed.58, 17399–17405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su, J. et al. CRISPR/Cas12a powered DNA framework‐supported electrochemical biosensing platform for ultrasensitive nucleic acid analysis. Small Methods5, 2100935 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Zhou, T. et al. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Adv. Sci.7, 1903661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. et al. Clustered regularly interspaced short palindromic repeats-mediated surface-enhanced Raman scattering assay for multidrug-resistant bacteria. ACS Nano14, 17241–17253 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Yin, B. et al. A CRISPR-Cas12a integrated SERS nanoplatform with chimeric DNA/RNA hairpin guide for ultrasensitive nucleic acid detection. Theranostics12, 5914 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan, C. et al. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Anal. Chem.92, 4029–4037 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Huang, D. et al. CRISPR-Cas12a-assisted multicolor biosensor for semiquantitative point-of-use testing of the nopaline synthase terminator in genetically modified crops by unaided eyes. ACS Synth. Biol.9, 3114–3123 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Hajian, R. et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng.3, 427–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H. et al. Amplification‐free detection of SARS‐CoV‐2 and respiratory syncytial virus using CRISPR Cas13a and graphene field‐effect transistors. Angew. Chem., Int. Ed.61, e202203826 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, W. et al. A suite of PCR-LwCas13a assays for detection and genotyping of Treponema pallidum in clinical samples. Nat. Commun.13, 4671 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naqvi, M. M., Lee, L., Montaguth, O. E. T., Diffin, F. M. & Szczelkun, M. D. CRISPR–Cas12a-mediated DNA clamping triggers target-strand cleavage. Nat. Chem. Biol.18, 1014–1022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, J. et al. Engineered LwaCas13a with enhanced collateral activity for nucleic acid detection. Nat. Chem. Biol.19, 45–54 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Hu, R., Leung, N. L. & Tang, B. Z. AIE macromolecules: syntheses, structures and functionalities. Chem. Soc. Rev.43, 4494–4562 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Tang, X., Zhu, Y., Guan, W. & Lu, C. Assembling aggregation‐induced emission with natural DNA to maximize donor/acceptor ratio for efficient light‐harvesting antennae. Aggregate4, e348 (2023).
- 29.Li, H. et al. Activity‐based smart AIEgens for detection, bioimaging, and therapeutics: recent progress and outlook. Aggregate2, e51 (2021). [Google Scholar]
- 30.Gao, Y. et al. Dual-dolor emissive AIEgen for specific and label-free double-stranded DNA recognition and single-nucleotide polymorphisms detection. J. Am. Chem. Soc.141, 20097–20106 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y.-L. et al. Real-time fluorescence in situ visualization of latent fingerprints exceeding level 3 details based on aggregation-induced emission. J. Am. Chem. Soc.142, 7497–7505 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Armbruster, D. A. & Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev.29, S49–S52 (2008). [PMC free article] [PubMed] [Google Scholar]
- 33.Broto, M. et al. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs. Nat. Nanotechnol.17, 1120–1126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truong, P. L. et al. Advancement in COVID-19 detection using nanomaterial-based biosensors. Exploration3, 20210232 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cutler, J. I., Auyeung, E. & Mirkin, C. A. Spherical nucleic acids. J. Am. Chem. Soc.134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Huang, C., Han, Z., Evangelopoulos, M. & Mirkin, C. A. CRISPR spherical nucleic acids. J. Am. Chem. Soc.144, 18756–18760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, F. et al. Light‐harvesting fluorescent spherical nucleic acids self‐assembled from a DNA‐grafted conjugated polymer for amplified detection of nucleic acids. Angew. Chem., Int. Ed.61, e202115812 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Fu, X. et al. Exploring the trans-cleavage activity of CRISPR/Cas12a on gold nanoparticles for stable and sensitive biosensing. Anal. Chem.93, 4967–4974 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science360, 444–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruch, R. et al. CRISPR/Cas13a‐powered electrochemical microfluidic biosensor for nucleic acid amplification‐free miRNA diagnostics. Adv. Mater. 31, 1905311 (2019). [DOI] [PubMed]
- 41.East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning, B. et al. Rapid detection of multiple SARS-CoV-2 variants of concern by PAM-targeting mutations. Cell Rep. Methods2, 100173 (2022). [DOI] [PMC free article] [PubMed]
- 43.Park, J. S. et al. Digital CRISPR/Cas‐assisted assay for rapid and sensitive detection of SARS‐CoV‐2. Adv. Sci. 8, 2003564 (2021). [DOI] [PMC free article] [PubMed]
- 44.Ackerman, C. M. et al. Massively multiplexed nucleic acid detection with Cas13. Nature582, 277–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo, L. et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov.6, 34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welch, N. L. et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat. Med.28, 1083–1094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahas, A. et al. Characterization of a thermostable Cas13 enzyme for one-pot detection of SARS-CoV-2. Proc. Natl Acad. Sci. USA119, e2118260119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, M. et al. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc. Natl Acad. Sci. USA119, e2202034119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, Y. et al. A Cas12a ortholog with distinct TTNA PAM enables sensitive detection of HPV16/18. Cell Rep. Methods3, 100444 (2023). [DOI] [PMC free article] [PubMed]
- 50.Arizti-Sanz, J. et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun.11, 5921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding, X. et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 11, 4711 (2020). [DOI] [PMC free article] [PubMed]
- 52.Lu, S. et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng.6, 286–297 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Yan, H. et al. A one-pot isothermal Cas12-based assay for the sensitive detection of microRNAs. Nat. Biomed. Eng. 7, 1583–1601 (2023). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are provided in the Supplementary Information/Source Data file. All relevant data are available from the corresponding author on request. The source data underlying Figs. 2a–d, f, h, j, l, 3b–e, 4a–c, 5b–n, 6d–i, and 7d–g are provided as a Source Data file. Source data are provided with this paper.