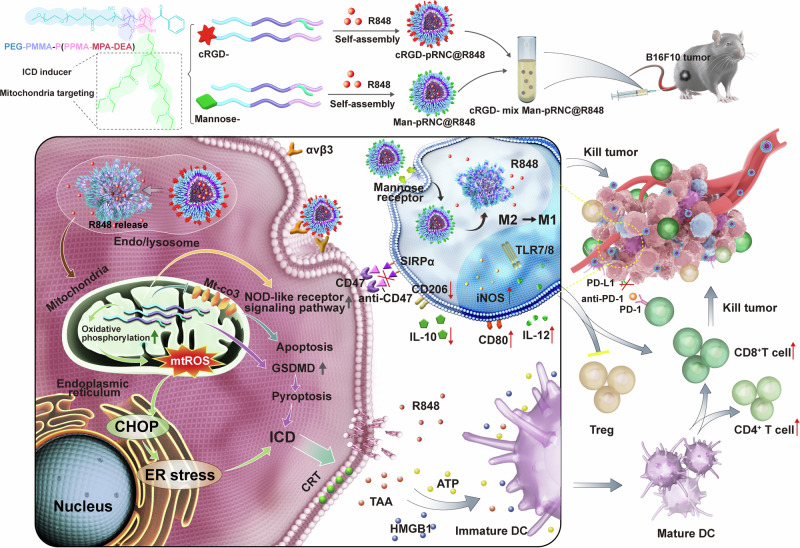
Abstract
The limited efficacy of cancer immunotherapy occurs due to the lack of spatiotemporal orchestration of adaptive immune response stimulation and immunosuppressive tumor microenvironment modulation. Herein, we report a nanoplatform fabricated using a pH-sensitive triblock copolymer synthesized by reversible addition-fragmentation chain transfer polymerization enabling in situ tumor vaccination and tumor-associated macrophages (TAMs) polarization. The nanocarrier itself can induce melanoma immunogenic cell death (ICD) via tertiary amines and thioethers concentrating on mitochondria to regulate metabolism in triggering endoplasmic reticulum stress and upregulating gasdermin D for pyroptosis as well as some features of ferroptosis and apoptosis. After the addition of ligand cyclic arginine-glycine-aspartic acid (cRGD) and mannose, the mixed nanocarrier with immune adjuvant resiquimod encapsulation can target B16F10 cells for in situ tumor vaccination and TAMs for M1 phenotype polarization. In vivo studies indicate that the mixed targeting nanoplatform elicits tumor ICD, dendritic cell maturation, TAM polarization, and cytotoxic T lymphocyte infiltration and inhibits melanoma volume growth. In combination with immune checkpoint blockade, the survival time of mice is markedly prolonged. This study provides a strategy for utilizing immunoactive materials in the innate and adaptive immune responses to augment cancer therapy.
Subject terms: Molecular medicine, Drug delivery, Cancer immunotherapy
Cancer immunotherapy leans on the effective activation on immune response within tumors and surrounding immunosuppressive microenvironment. Here, the authors report a triblock copolymeric nanocarrier encapsulating Resiquimod for B16F10 cell-targeting, which enables in situ tumor vaccination and tumor-associated macrophages polarization.
Introduction
Cancer immunotherapy is one of the most promising strategies for tumor treatment and has attracted extensive attention. Induction of tumor immunogenic cell death (ICD) is a widely used methodology in which tumor cells, after encountering external stimuli, can change from a non-immunogenic state to an immunogenic state with host immune response provocation1–3. The typical immune features of ICD are damage-associated molecular patterns (DAMPs) (e.g., calreticulin [CRT], high mobility group box 1 [HMGB1], and adenosine triphosphate [ATP]) secretion4,5 and tumor-associated antigen release that can facilitate dendritic cell (DC) maturation, migration, and antigen presentation to T cells for host immunity activation6,7. Currently, various strategies, including photodynamic therapy, chemotherapy, and pyroptosis, have been developed for ICD-mediated tumor immunotherapy1,8–15. However, these methodologies generally require additional ICD inducers that may complicate the nanoplatform. Therefore, the development of nanocarriers that can directly induce ICD is imperative to simultaneously simplify nanoformulations to ensure therapeutic efficacy due to their own immunity.
Tumor vaccines have emerged as a promising strategy for long-term cancer immunotherapy, and they primarily attack tumors via eliciting an antigen-specific immune response16. Although there are definite antigens in tumor vaccines, the heterogeneity and high cost of mutant antigen identification have restricted their further development17,18. To cope with the above issues, in situ tumor vaccination can be greatly enhanced via combining immune adjuvants with tumor-associated antigens (TAAs) that can be directly released by dying tumor cells in vivo after different treatments (e.g., photodynamic therapy, photothermal therapy, and chemotherapy)14,19–26. The most obvious advantage of in situ tumor vaccination is that extra antigen addition is unnecessary, and it depends on the antigen pool from individual tumors after external stimulation, thereby saving valuable time and cost.
Although eliciting adaptive immunity is an extensively used strategy for cancer immunotherapy, immunosuppressive factors in the tumor microenvironment (TME) (e.g., tumor-associated macrophages [TAMs]) can greatly impede therapeutic efficacy27–31. TAMs primarily exist as an immunosuppressive M2 phenotype and account for 50% of all immune cells in tumor tissue32, and they help tumors to escape immunosurveillance via generating anti-inflammatory cytokines33–35. In contrast, M1 phenotype TAMs act as pro-inflammatory cells and can eliminate tumors with antigen presentation to T lymphocytes, thereby polarizing M2-TAMs into the M1 phenotype necessary36–38. Resiquimod (R848) can induce TAM polarization39; however, its application is limited by severe side effects after systemic injection32. Nanomedicine that achieves high tumor accumulation via the enhanced permeability and retention effect can improve drug bioavailability with effective drug delivery and negligible side effects. Especially, the different targeting ligands decoration on nanosystems can separately target different cells such as tumor, TAMs, etc. for spatiotemporal orchestration. Additionally, the high expression of cluster of differentiation 47 (CD47) on the tumor cell surface contributes to tumor resistance to macrophage phagocytosis via the CD47/signal regulatory protein alpha (SIRPα) axis40. Thus, both TAM polarization and blockade of the immune checkpoint molecule CD47 are necessary for immunosuppressive TME modulation.
In this work, we report an immunofunctional nanoplatform in which the nanocarrier itself directly induces tumor ICD via metabolic regulation of elevated oxidative phosphorylation and via gasdermin D (GSDMD) upregulation for pyroptosis. After conjugation of the targeting ligand cyclic arginine-glycine-aspartic acid (cRGD) or mannose (Man) to the same polymeric skeleton, the mixed nanocarrier separately targets tumor cells and TAMs in vivo. After encapsulation with the immune adjuvant R848, cRGD-decorated nanoparticles selectively target melanoma B16F10 cells for carrier-mediated ICD to form an in situ cancer vaccine, and Man-modified nanoparticles target TAMs, ultimately resulting in polarization for TME modulation. In vivo results indicate that the mixed-targeting nanoformulation elicits DC maturation, M1 like TAM polarization, CD8+/CD4+ T cell infiltration, and melanoma tumor volume growth inhibition (Fig. 1). After combination treatment with anti-PD-1 and anti-CD47, the formulation notably prolongs the median survival of mice and results in tumor cell death. This project highlights the mechanism of polymer-mediated ICD and provides insights into the exploitation of immunoactive nanomaterials to spatiotemporally orchestrate adaptive and innate immune responses for effective cancer immunotherapy.
Fig. 1. Schematic illustration of cRGD- mix Man-pRNCThioether+DEA@R848 mediated in situ tumor vaccination and TAMs polarization with nanocarrier itself as the tumor ICD inducer.
cRGD-pRNCThioether+DEA selectively targeting B16F10 tumor induces metabolism regulation with oxidative phosphorylation elevation and activates GSDMD with pyroptosis for tumor ICD. After combination with R848, cRGD-pRNCThioether+DEA@R848 forms in situ tumor vaccination. Man-pRNCThioether+DEA@R848 specifically targets TAMs leading to polarization into M1 phenotype with iNOS, CD80 expression and IL-12 secretion increment. After combination with anti-PD-1 and anti-CD47, the mixed nanoformulation elicits antitumor immune response with Treg decrement and CD8+/CD4+ T cell proliferation increment.
Results
Synthesis of functional polymers
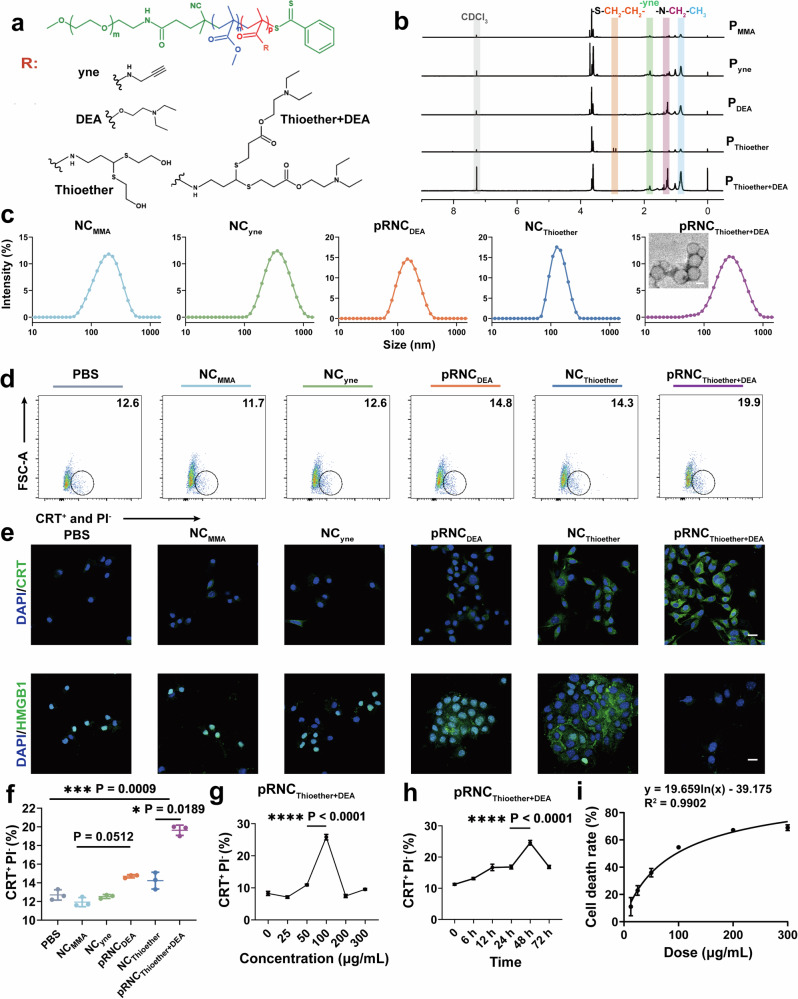
The successful synthesis of polymers is a prerequisite for constructing nanoplatforms. Before the synthesis of the skeleton polymer, polyethylene glycol-poly methyl methacrylate (PEG-PMMA), the macro-reversible addition-fragmentation chain transfer (RAFT) polymerization agent PEG-CPPA was synthesized via an amidation reaction between PEG-NH2 and NHS-CPPA (Supplementary Fig. 1). The conversion ratio of NHS-CPPA that was the abbreviation of a small molecule RAFT agent known as 4-Cyano-4-(phenylcarbonothioylthio)pentanoic acid N-succinimidyl ester was 98% based on peaks of methylene protons in PEG (δ, 3.63 ppm) and methine protons in CPPA (δ, 7.38, 7.57, and 7.90 ppm) of 1H NMR spectra (Supplementary Fig. 3). Di-block copolymer PEG-PMMA (named as PMMA) was obtained through RAFT polymerization of PEG-CPPA and monomer MMA, and the molecular weight was 5.0-10.0 kg/mol according to the peaks of PEG protons (δ, 3.63 ppm) and methyl protons in PMMA (δ, 3.65 ppm) of 1H NMR spectra. GPC indicated that the relative molecular weight was 15.3 kg/mol with a polydispersity index (PDI) of 1.3 (Fig. 2a, b, Supplementary Fig. 4 and Supplementary Table 1). Tri-block copolymer PEG-PMMA-PDEA (termed as PDEA) was synthesized by PEG-PMMA and monomer diethylaminoethyl methacrylate (DEA) via RAFT polymerization (Supplementary Fig. 1), and the molecular weight was 5.0-10.0-3.8 kg/mol based on methylene protons in PEG (δ, 3.63 ppm) and methylene protons in PDEA (δ, 0.86 and 1.26 ppm) of 1H NMR spectra. GPC indicated that the relative molecular weight was 19.2 kg/mol (Fig. 2a, b, Supplementary Fig. 5 and Supplementary Table 1). The small molecule N-propargyl methacrylamide (PPMA) was obtained from methacryloyl chloride and propargylamine (Supplementary Fig. 2), and it was pure according to 1H NMR result based on protons at δ 1.97 ppm, δ 2.25 ppm, δ 4.10 ppm, δ 5.37 ppm, and δ 5.74;ppm (Supplementary Fig. 6). To obtain PEG-PMMA-P (PPMA-ME) (termed PThioether), tri-block copolymer PEG-PMMA-PPPMA (termed Pyne) was first synthesized using PEG-PMMA and PPMA via RAFT polymerization, and the molecular weight was 5.0-10.0-6.3 kg/mol based on peaks of methylene protons in PEG (δ, 3.63 ppm), methyl protons in PMMA (δ, 3.65 ppm), and methyl protons in PPPMA (δ, 1.81 ppm) of 1H NMR spectra. GPC indicated that the relative molecular weight was 18.9 kg/mol (Fig. 2a, b, Supplementary Fig. 7 and Supplementary Table 1). After a click reaction of alkynyl in PPPMA with mercaptoethanol (ME) (Supplementary Fig. 2), PEG-PMMA-P(PPMA-ME) (PThioether) was obtained, and the graft ratio of thiol was 94% based on peaks of PEG protons at δ 3.63 ppm, methyl protons in PMMA (δ, 3.65 ppm), and methylene protons at δ 2.89 ppm and δ 2.97 ppm. GPC indicated that the relative molecular weight was 30.7 kg/mol (Supplementary Fig. 8 and Supplementary Table 1). PEG-PMMA-P(PPMA-MPA) was synthesized from PEG-PMMA-PPPMA and mercaptopropionic acid (MPA) via a click reaction after further esterification with diethylaminoethanol to obtain PEG-PMMA-P(PPMA-MPA-DEA) (termed PThioether+DEA) (Supplementary Fig. 2). 1H NMR results revealed that the molecular weight of PThioether+DEA was 5.0-7.3-18.4 kg/mol based on peaks of PEG protons at δ 3.63 ppm, methyl protons in PMMA (δ, 3.65 ppm), methylene protons (δ, 0.86 ppm), and methyl groups (δ, 1.25 ppm) in P(PPMA-MPA-DEA). GPC indicated that the relative molecular weight was 36.1 kg/mol (Supplementary Figs. 9–11 and Supplementary Table 1).
Fig. 2. Preparation and characterization of the ICD inducer pRNCThioether+DEA.
a Structures of a series of functional polymers with the same skeleton. b 1H NMR spectra of different polymers. c Size and transmission electron microscopy images of NCMMA, NCyne, pRNCDEA, NCThioether, pRNCThioether+DEA. Scale bar = 100 nm. Three times each experiment was repeated independently with similar results. d CRT exposure after different treatments via flow cytometry characterization. e CRT exposure and HMGB1 release of B16F10 cells after different treatments characterized by CLSM. Cells were stained with Alexa 488-anti-CRT (green) and Alexa 488-anti-HMGB1 (green). Cell nuclei were stained with DAPI (blue). Scale bar = 20 μm. f Semi-quantitative analysis of CRT exposure after different treatments (n = 3 independent experiments). g pRNCThiorther+DEA with different concentrations induced CRT exposure in B16F10 cells (n = 3 independent experiments). h CRT exposure of B16F10 cells was induced by pRNCThioether+DEA after different incubation time treatment (n = 3 independent experiments). i The kinetics and dose-response of cell death induction by pRNCThioether+DEA (n = 3 independent experiments). Data are presented as mean ± SD. Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test.
Selection of polymers that specifically induce B16F10 ICD
Nanocarrier self-assemblies from PEG-PMMA, PEG-PMMA-PPPMA, PEG-PMMA-PDEA, PEG-PMMA-P(PPMA-ME), and PEG-PMMA-P(PPMA-MPA-DEA) (Fig. 2a, b) were prepared using the solvent exchange method and separately named NCMMA, NCyne, pRNCDEA, NCThioether, and pRNCThioether+DEA. Dynamic light scattering (DLS) results indicated that the nanocarriers possessed a hydrodynamic diameter varying from 120 to 150 nm and a PDI varying from 0.18 to 0.25 (Fig. 2c, Supplementary Table 2). The morphology of pRNCThioether+DEA was characterized by transmission electron microscopy (TEM). As presented in Fig. 2c, it is a uniformly spherical polymer with a hollow structure. In the acetate buffer solution (pH 5.0, 10 mM), the size and PDI of pRNCThioether+DEA and pRNCDEA increased over time, whereas no obvious changes were observed in the PBS buffer with a pH value of 7.4, thus highlighting their pH responsiveness and physiological stability (Supplementary Fig. 12). Additionally, both pRNCThioether+DEA and pRNCDEA maintained good stability under weakly acidic condition (pH 6.5) that mimicked the TME (Supplementary Fig. 13).
Their ability to induce ICD was investigated by CRT exposure and HMGB1 and ATP release via flow cytometry (FCM), confocal laser scanning microscopy (CLSM), and ATP assays. The FCM results presented in Fig. 2d, f indicated that NCMMA and NCyne did not induce CRT exposure, and both displayed similar CRT-positive ratios to that of phosphate-buffered saline (PBS). Slightly elevated ratios were detected in the pRNCDEA (14.70 ± 0.17%) and NCThioether (14.23 ± 0.90%) groups, and the highest ratio (19.63 ± 0.55%) was observed in the pRNCThioether+DEA group, thus indicating that both tertiary amine and thioether were able to induce ICD. Compared to pRNCDEA and NCThioether, pRNCThioether+DEA induced the highest CRT exposure that was 1.33- to 1.37-fold higher than that of the other two groups. This was likely due to the dual ICD effects of both DEA and thioether groups (Fig. 2d, f, Supplementary Fig. 14). CLSM results displayed a similar tendency to those of FCM, where cells treated with pRNCThioether+DEA exhibited the most obvious CRT exposure (green) compared to that of the pRNCDEA and NCThioether groups (Fig. 2e). Moreover, HMGB1 (green) in cells treated with NCMMA and NCyne overlapped well with the nuclei, whereas HMGB1 was primarily distributed outside of the nuclei in the pRNCDEA and NCThioether groups, thus indicating that both nanocarriers could induce HMGB1 release from the nuclei. After pRNCThioether+DEA treatment, the green color of HMGB1 outside the nucleus decreased, thus indicating the robust ability of this nanocarrier to induce ICD (Fig. 2e). Additionally, after different treatments, pRNCThioether+DEA induced the highest HMGB1 release into the cell supernatant (Supplementary Fig. 15a). According to the ATP detection results, cells treated with pRNCThioether+DEA possessed the highest ATP levels in the supernatant (Supplementary Fig. 15b).
Therefore, based on the above results, pRNCThioether+DEA was the best ICD inducer and was selected as the optimal nanoparticle for further studies. We then investigated the effects of factors such as concentration and incubation time on pRNCThioether+DEA-mediated ICD. We observed that CRT exposure increased with pRNCThioether+DEA concentration increment at the same time point. The highest amount of CRT exposure was detected at 100 μg/mL, and it was 2.37- to 3.61-fold higher than that of the others (Fig. 2g). Thus, pRNCThioether+DEA at 100 μg/mL was chosen as the ICD inducer for the following experiments. As presented in Fig. 2h, the CRT exposure increased with prolonged incubation time, and the maximum exposure was observed at 48 h. This exposure was 1.46- to 2.18-fold higher than that of the other time points. The results indicated that pRNCThioether+DEA-mediated ICD was dependent upon both concentration and incubation time. As presented in Fig. 2i, pRNCThioether+DEA-mediated B16F10 cell death was concentration-dependent with an IC50 value of 93.04 µg/mL. In addition to B16F10 cells, pRNCThioether+DEA induced ICD in multiple cancer cells such as colorectal carcinoma MC38, Lewis lung cancer (LLC) cells, and pancreatic carcinoma Pan02 with a notable CRT fluorescence shift when slight CRT exposure was observed in breast cancer 4T1 and glioma U87-MG cells (Supplementary Fig. 16). These results revealed that pRNCThioether+DEA can be widely applied to a variety of tumors for immunotherapy via inducing ICD.
Investigation of the pRNCThioether+DEA-mediated ICD mechanism
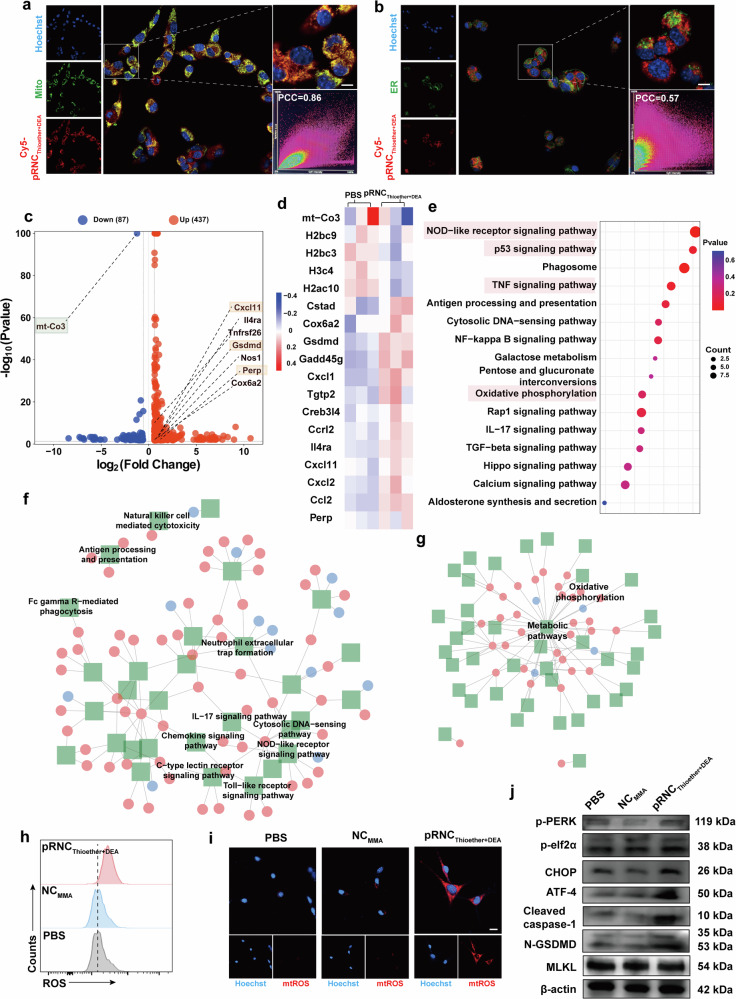
Inspired by the observation that pRNCThioether+DEA could induce B16F10 ICD, the underlying mechanism was investigated via the intracellular co-localization of nanocarriers with organelles and by single-cell RNA sequencing (scRNA-seq) analysis. To conveniently observe the intracellular distribution of the nanocarrier, pRNCThioether+DEA was labelled with Cy5 (Cy5-pRNCThioether+DEA), observed, and then captured using CLSM. Cy5-pRNCThioether+DEA first reached the endo/lysosome at 2 h and then escaped at 4 h (Supplementary Fig. 17a, b). According to Fig. 3a, it was primarily concentrated in the mitochondria at 4 h and exhibited obvious co-localization, while no obvious Cy5 fluorescence was observed in the endoplasmic reticulum (Fig. 3b) or Golgi apparatus (Supplementary Fig. 17c). These results demonstrate that pRNCThioether+DEA was primarily concentrated in the mitochondria of tumor ICD.
Fig. 3. Mechanism investigation of pRNCThioether+DEA mediated B16F10 ICD.
a, b Representative images of pRNCThioether+DEA mediated co-localization with mitochondria and ER. MitoTracker green (a) and ER Tracker green (b) were stained with mitochondria and ER, respectively. Red color represented Cy5-pRNCThioether+DEA, and blue color represented DAPI which were stained by Hoechst 33342. PCC = Pearson’s correlation coefficient. Scale bar = 10 μm. c Volcano map of expressed differentially genes in pRNCThioether+DEA treated cells. d Heatmap depicting relative transcript levels of expressed differentially genes in pRNCThioether+DEA or PBS treated cells (n = 3 independent experiments). e KEGG pathway enrichment analysis of expressed differentially genes for cells after pRNCThioether+DEA treatment. f, g Enrichment of organismal systems pathways (f) and metabolism pathways (g) in pRNCThioether+DEA or PBS treated cells (n = 3 independent experiments). h Intracellular ROS level after different treatments via flow cytometer characterization (n = 3 independent experiments). i Intracellular mtROS level after treatments via CLSM characterization. Scale bar = 20 μm. j Western blot of p-PERK, p-elf2α, ATF4, cleaved caspase-1, N-GSDMD, MLKL expression after treatments. β-actin was used as internal control. Experiments in (a), (b), (i), and (j) were repeated three times independently with similar results.
It has been reported that both metabolic regulation and redox homeostasis disruption in mitochondria can mediate tumor ICD via eliciting oxidative phosphorylation41–44. To the best of our knowledge, this is the first study to show that polymeric nanocarriers with specific structures can induce tumor ICD via mitochondria metabolism regulation with ER stress and GSDMD activation with pyroptosis. As indicated in the scRNA-seq results, the mitochondrial cytochrome c oxidase subunit 3 gene (mt-co3) and GSDMD displayed significant differences according to the volcano image for B16F10 cells treated with pRNCThioether+DEA (Fig. 3c). Obvious mt-co3 downregulation and perp (a mediator of p53 dependent apoptosis) upregulation were detected, thus demonstrating that pRNCThioether+DEA could also induce tumor apoptosis (Fig. 3c, d). As presented in Fig. 3c, multiple genes related to chemokines (e.g., chemokine [C-X-C motif] ligand 11 [CXCL11]) and GSDMD were upregulated after pRNCThioether+DEA treatment, and this activated multiple downstream immune-related signaling pathways as well as pyroptosis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis demonstrated that the dominant pathways enriched by pRNCThioether+DEA were NOD-like receptor, p53, and the TNF signaling pathway as well as oxidative phosphorylation that would mediate a series of downstream immune and inflammatory cascades, apoptosis, and metabolism regulation (Fig. 3e). According to the KEGG regulatory network diagram, metabolic pathways, particularly those related to oxidative phosphorylation, were significantly different (Fig. 3g). Multiple immune-related regulatory pathways, including NOD-like receptor, chemokine, and IL-17 signaling pathways, were activated after pRNCThioether+DEA treatment (Fig. 3f).
As presented in Fig. 3h, the intracellular ROS level detected using DCFH-DA as the fluorescent probe was notably elevated in cells treated with pRNCThioether+DEA. Therefore, the destruction of redox homeostasis likely elicited tumor ICD. Compared to NCMMA, pRNCThioether+DEA cells exhibited a 1.5-fold increase in intracellular ROS fluorescence intensity (Fig. 3h, Supplementary Fig. 18). To further determine if the elevated intracellular ROS was produced by mitochondria, we used CLSM to observe the production of intracellular mitochondrial ROS (mtROS) using BBcellProbe® OM08 as a detection probe after 24 h treatment. Notable red fluorescence was observed in the pRNCThioether+DEA-treated group compared to that in the PBS and NCMMA groups (Fig. 3i, Supplementary Fig. 19), thus indicating that pRNCThioether+DEA induced mtROS generation.
To further investigate how pRNCThioether+DEA induces ICD in B16F10 cells, we used western blotting to examine the ER stress-related pathways. The phosphorylation of PERK and eukaryotic translation initiation factor 2α (elf2α) was enhanced after pRNCThioether+DEA treatment at 48 h compared to that in response to PBS and NCMMA. The levels of downstream activating transcription factor 4 (ATF4) and recombinant DNA damage-inducing transcript 3 (CHOP) were also increased. These data indicate that ER stress was triggered by pRNCThioether+DEA (Fig. 3j, Supplementary Fig. 20). To explore pRNCThioether+DEA mediated cell death, B16F10 cells after different treatments were stained with calcein-AM and propidium iodide (PI) to identify live/dead cells. The noticeable increase of red spots (dead cells) was observed in pRNCThioether+DEA group while a large numbers of surviving cells (green color) were detected in PBS and NCMMA groups, indicating the ability of pRNCThioether+DEA to induce cell death (Supplementary Fig. 21). The cell death types were then investigated in consideration that many types such as pyroptosis, apoptosis and ferroptosis etc. can induce tumor ICD. The typical pyroptotic morphologies were observed in cells after pRNCThioether+DEA treatment indicating its ability to induce pyroptosis (Supplementary Fig. 22). Additionally, the expression of N-GSDMD and cleaved caspase-1 was upregulated after pRNCThioether+DEA treatment, and lactate dehydrogenase (LDH) activity was 2.4-fold higher than that in PBS, indicating that pRNCThioether+DEA induces pyroptosis through the GSDMD pathway (Fig. 3j, Supplementary Fig. 23). According to Supplementary Fig. 24, pRNCThioether+DEA also induced apoptosis when the elevated early (10.50 ± 0.46%) and late (55.27 ± 4.38%) apoptosis ratios were detected compared to those of the PBS (early: 4.70 ± 0.14%; late: 14.20 ± 0.98%) and NCMMA (early: 5.27 ± 0.56%; late: 19.23 ± 0.93%) groups. Additionally, we observed that oxidized lipid peroxide (LPO) levels were elevated after pRNCThioether+DEA treatment, thus indicating its ability to induce cancer ferroptosis. The ratio of oxidized to reduced LPO was increased to 40.01 ± 0.12% compared to that of PBS (30.97 ± 0.32%) and NCMMA (29.60 ± 0.24%) (Supplementary Fig. 25, 26). CLSM results in Supplementary Fig. 26 further confirmed the LPO generation when the notable increment of oxidized C11-BODIPY (green color) was observed for cells with pRNCThioether+DEA treatment. Moreover, negligible changes in the expression of mixed lineage kinase domain-like protein (MLKL) were observed after treatment with PBS, NCMMA, or pRNCThioether+DEA. The existence of necrostatin 2 racemate (Nec-1s) did not affect MLKL expression, further confirming that pRNCThioether+DEA did not induce cell necroptosis (Fig. 3j, Supplementary Fig. 27). To further distinguish cell death types when detecting the cell death populations, inhibitors such as Z-VAD-FMK for apoptosis, Ferrostatin-1 (Fer-1) for ferroptosis, and Nec-1s for necroptosis were added to B16F10 cells treated with pRNCThioether+DEA. As shown in Supplementary Fig. 28, the ratio of dead/dying cells induced by pRNCThioether+DEA was highly suppressed after an apoptosis inhibitor Z-VAD-FMK treatment. After treatment with the ferroptosis inhibitor Ferrostatin-1 (Fer-1), the cell death ratio slightly reduced, while negligible changes were observed in cells treated with the necroptosis inhibitor necrostatin 2 racemate (Nec-1s). The data indicated that pRNCThioether+DEA induced mixed cell death modalities with some features of pyroptosis, ferroptosis, and apoptosis.
Preparation and characterization of cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA
To target B16F10 cells and TAMs separately, cRGD- and Man-targeting ligands were decorated onto the nanocarrier surface to obtain cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA, respectively. c(RGDfK) is a cyclic peptide that can target tumor cells (e.g., B16F10 cells, LLC cells) with high expression of αvβ3, and Man is a cyclic monosaccharide that targets TAMs with high mannose receptor expression45–47. The synthesis of cRGD (Man)-PEG-PMMA-PPPMA was similar to that of PEG-PMMA-PPPMA (Supplementary Fig. 29). The ratios of c(RGDfK) and Man were 85% and 87%, respectively, based on peaks of methylene protons in PEG (δ, 3.63 ppm), methylene protons (δ, 1.84 ppm) in cRGD (Supplementary Fig. 30), and methine proton (δ, 2.28 ppm) in Man according to 1H NMR results (Supplementary Fig. 31).
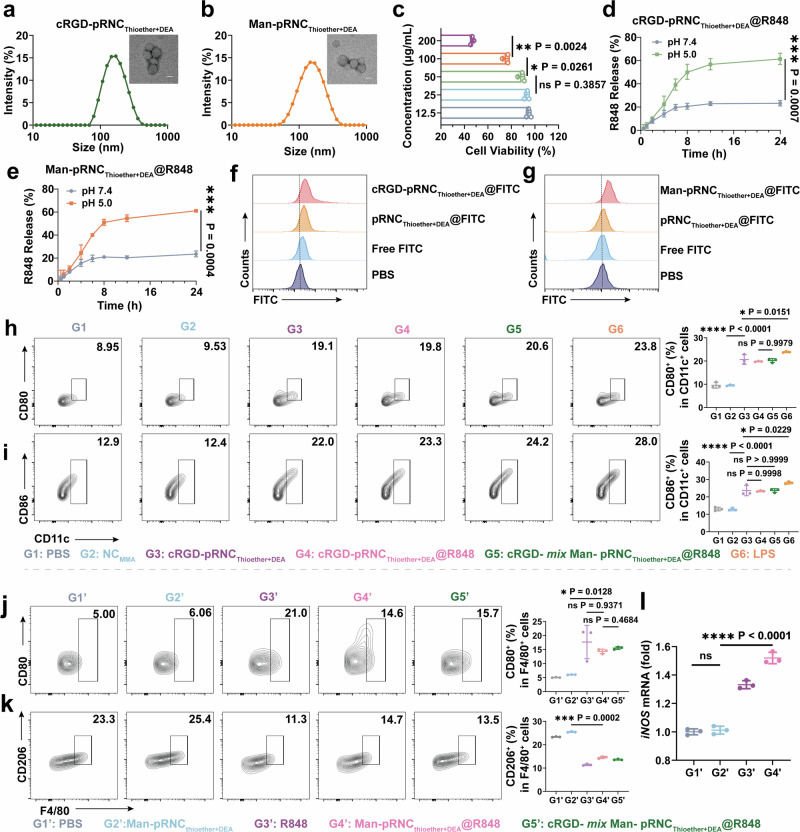
cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA were also fabricated via the solvent-exchange method. DLS results indicated that their diameter sizes were 168.0 ± 4.85 nm and 143.5 ± 1.08 nm, respectively, and PDI was 0.16 ± 0.016 and 0.23 ± 0.021 (Fig. 4a, b, Supplementary Table 3). After mixing, cRGD- mix Man-pRNCThioether+DEA was obtained, and no obvious changes were detected (Supplementary Fig. 32 and Supplementary Table 3). According to Fig. 2f, the concentration of pRNCThioether+DEA as the ICD inducer was chosen as 100 μg/mL due to the obvious CRT exposure, and this concentration was also suitable for cRGD-pRNCThioether+DEA. According to Fig. 4c, the cell viability of RAW264.7 cells was 88 ± 2.8% when the Man-pRNCThioether+DEA concentration was 50 μg/mL, while the value decreased to 73 ± 2.6% when the concentration was increased to 100 μg/mL. Thus, to reduce macrophage death the concentration of Man-pRNCThioether+DEA was selected as 50 μg/mL. cRGD (Man)-pRNCThioether+DEA with R848 encapsulation (cRGD (Man)-pRNCThioether+DEA@R848) was also separately prepared via the solvent-exchange method when the drug loading content (DLC) was as high as 12.70% and 13.29% with drug loading efficiency (DLE) of 58.2% and 61.3%, respectively, according to a fluorospectrophotometer (Supplementary Fig. 33). The DLC and DLE values of the cRGD- mix Man-pRNCThioether+DEA were 12.55% and 57.4%, respectively (Supplementary Table 3). In vitro drug release behavior indicated that the accumulative release of R848 from cRGD-pRNCThioether+DEA@R848, Man-pRNCThioether+DEA@R848, and cRGD- mix Man-pRNCThioether+DEA@R848 was as high as 61%, 60%, and 67.6% within 24 h in acetate buffer solution (pH 5.0, 10 mM, 150 mM NaCl), while the release was only 23%, 24%, and 20.2% in PBS (pH 7.4, 10 mM, 150 mM NaCl) (Fig. 4d, e, Supplementary Fig. 34). The above results indicated that cRGD (Man)-pRNCThioether+DEA@R848 as well as the mixed nanoformulations were pH-sensitive.
Fig. 4. Preparation and characterization of targeted nanoformulations.
a, b Diameters and representative transmission electron microscopy images of cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA. Scale bar, 100 nm. Three times was repeated independently with similar results. c Dosage dependent cytotoxicity of Man-pRNCThioether+DEA in RAW264.7 cells by MTT assays characterization. Data are shown as mean ± SD (n = 3 independent experiments). d, e In vitro R848 release from cRGD-pRNCThioether+DEA or Man-pRNCThioether+DEA within 24 h at different pH values (pH 7.4 or pH 5.0). Data are shown as Geometric mean ± 95% CI. (n = 3 independent experiments). f Representative flow cytometric plots showing the cellular uptake of pRNCThioether+DEA, cRGD-pRNCThioether+DEA and free FITC in B16F10 cells. g Representative flow cytometric plots showing the cellular uptake of pRNCThioether+DEA, Man-pRNCThioether+DEA and free FITC in RAW264.7 cells. h, i Representative flow cytometric images and quantification of mature DCs. Data are shown as mean ± SD (n = 3 independent experiments). j, k Representative flow cytometric images and quantification of polarization of macrophages. Data are shown as mean ± SD (n = 3 independent experiments). l Expression of iNOS in cells after different treatments (n = 3 independent experiments). The word “ns” represented non-significance, and *P < 0.05; **P < 0.01; ***P < 0.001. Data are presented as mean ± SD. Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test.
In vitro targetability of cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA
R848 was replaced with FITC to better monitor intracellular internalization that was characterized by FCM. As presented in Fig. 4f, a more obvious fluorescence shift and cytotoxicity were observed in B16F10 cells in response to cRGD-pRNCThioether+DEA@FITC treatment than that in response to pRNCThioether+DEA@FITC, thus indicating the good targetability of cRGD (Supplementary Figs. 35, 37). After Man-pRNCThioether+DEA@FITC treatment, a stronger fluorescence shift and cytotoxicity were observed in RAW264.7 cells compared to the pRNCThioether+DEA@FITC group, thus indicating noticeable targetability of Man (Fig. 4g, Supplementary Fig. 36). The CLSM results in Supplementary Fig. 38 indicated that cRGD and Man decoration enhanced the targetability of B16F10 and RAW264.7, respectively.
cRGD-pRNCThioether+DEA@R848 mediated DCs maturation
To test if the nanocarrier could activate DCs in vitro, we first examined its ability to induce ICD in B16F10 cells as characterized by FCM. An elevated CRT+ ratio was observed in cells treated with cRGD-pRNCThioether+DEA or cRGD-pRNCThioether+DEA@R848 compared to that of NCMMA and PBS. Negligible differences were observed among the cRGD-pRNCThioether+DEA, cRGD-pRNCThioether+DEA@R848, and cRGD- mix Man-pRNCThioether+DEA@R848 treated groups, thus indicating that cRGD-pRNCThioether+DEA induced ICD while R848 did not and that the mixing of cRGD- and Man-nanoformulations did not affect the ICD inducibility of cRGD-pRNCThioether+DEA (Supplementary Fig. 39). When B16F10 cell supernatant was added into DCs, it was observed that both cRGD-pRNCThioether+DEA (CD11c+CD80+: 20.30 ± 2.25%; CD11c+CD86+: 23.60 ± 2.95%) and cRGD-pRNCThioether+DEA@R848 (CD11c+CD80+: 19.80 ± 0.20%; CD11c+CD86+: 23.30 ± 0.36%) treated cancer cells displayed a notable DC maturation compared to that of NCMMA (CD11c+CD80+: 9.53 ± 0.27%; CD11c+CD86+: 12.80 ± 0.78%) and PBS (CD11c+CD80+: 9.56 ± 1.24%; CD11c+CD86+: 13.00 ± 1.05%) that was similar to that of the positive control LPS (CD11c+CD80+: 24.20 ± 0.87%; CD11c+CD86+: 28.07 ± 0.70%) (Fig. 4h, i, Supplementary Fig. 40). This suggests that tumor ICD mediated by cRGD-pRNCThioether+DEA with DAMP secretion facilitates DC maturation. Additionally, the cRGD- mix Man-pRNCThioether+DEA@R848-treated group exhibited similar ratios of CD11c+CD80+ (20.20 ± 0.87%) and CD11c+CD86+ (23.60 ± 1.13%) to that of cRGD-pRNCThioether+DEA@R848, thus demonstrating that the mixing of cRGD and Man nanoformulations did not affect the cRGD-pRNCThioether+DEA-mediated ICD cascade with DC maturation. As presented in Supplementary Fig. 41, both the cRGD-pRNCThioether+DEA and cRGD-pRNCThioether+DEA@R848 groups exhibited lower ratios of DCs to dead B16F10 cells in the supernatants compared to that of PBS and NCMMA, indicating that these two groups induced more dead cells for DAMPs generation with DC maturation.
Man-pRNCThioether+DEA@R848-mediated macrophage polarization
To determine if Man-pRNCThioether+DEA@R848 could induce macrophage polarization, the expression of CD206 and CD80 on the surface of RAW264.7 cells and intracellular inducible nitric oxide synthase (iNOS) levels were detected. As presented in Fig. 4j, k, CD80 expression on the cell surface after Man-pRNCThioether+DEA@R848 treatment was 14.33 ± 0.83%, and this was 2.4-fold higher than that of Man-pRNCThioether+DEA (6.63 ± 0.08%) and similar to that of free R848 (17.67 ± 5.95%). CD206 expression in cells with Man-pRNCThioether+DEA@R848 treatment decreased to 14.53 ± 0.47%, and this was 1.75-fold lower than that of Man-pRNCThioether+DEA (25.47 ± 0.21%). According to the results of quantitative polymerase chain reaction (qPCR), R848 could promote the expression of iNOS while Man-pRNCThioether+DEA alone not. Man-pRNCThioether+DEA@R848 induced notable iNOS expression that was 4.78-fold higher than that of Man-pRNCThioether+DEA (1.01 ± 0.3) (Fig. 4l). According to the above results, pRNCThioether+DEA@R848 can polarize M2 RAW264.7 cells into an M1 phenotype similar to that caused by free R848.
In vivo targetability of cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA
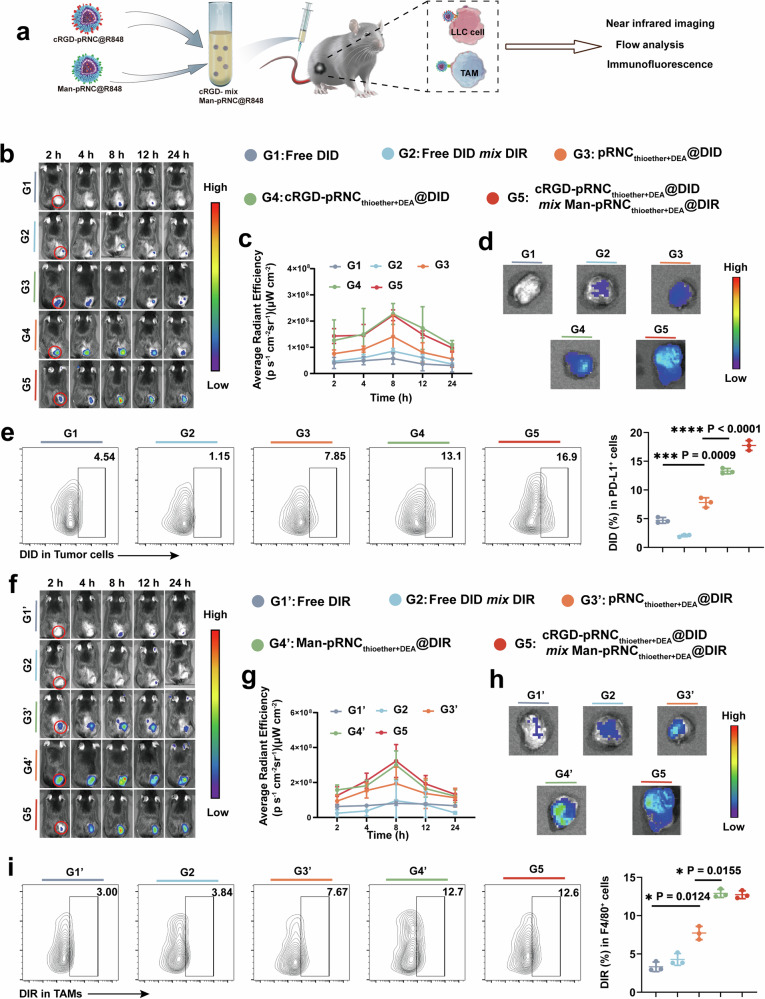
To avoid fluorescence interference from melanoma, B16F10 cells were replaced with LLC cells to construct a tumor model in C57BL/6 mice for in vivo targeting of cRGD-pRNCThioether+DEA and Man-pRNCThioether+DEA via a near-infrared (NIR) in vivo imaging system (IVIS), FCM, and immunofluorescence staining analysis. cRGD-pRNCThioether+DEA was loaded with DID to form cRGD-pRNCThioether+DEA/DID, and Man-pRNCThioether+DEA was encapsulated in DIR to form Man-pRNCThioether+DEA/DIR (Fig. 5a).
Fig. 5. Investigation of mixed nanoformulations for targeting both tumor cells and TAMs in vivo.
a Schematic illustration of mixed nanoformulation preparation, drug administration and analysis. b In vivo fluorescence imaging after tail vein injection of different formulations for cRGD targeting investigation. c Quantitative analysis of fluorescence signals at 24 h post-treatment (n = 3 mice per group). d Representative ex vivo images of tumor tissues after treatments at 24 h. e Representative flow cytometric images and quantification of DID+ in PD-L1+ tumor cells in vivo (n = 3 mice per group). f In vivo fluorescence imaging after tail vein injection of different formulations for Man targeting investigation. g Quantitative analysis of fluorescence signals at 24 h (n = 3 mice per group). h Representative ex vivo images of tumor tissues. i Representative flow cytometric images and semi-quantification analysis of DIR+ in F4/80+ TAMs cells in vivo (n = 3 mice per group). Data are presented as mean ± SD. Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test.
For the cRGD nanoformulations, after intravenous (i.v.) injection the in vivo fluorescence intensity within the 24 h first increased and then decreased, and it reached a maximum at 8 h (Fig. 5b, c). The fluorescence intensity of the nanoformulations was higher than that of free DID at all time points after i.v. injection. At 8 h, in vivo fluorescence intensity of cRGD-pRNCThioether+DEA@DID and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR was 3.97- and 3.88-fold higher than that of free DID, respectively, thus indicating that the nanoformulation improved tumor accumulation. The in vivo fluorescence intensity of the cRGD decoration group was higher than that of the pRNCThioether+DEA@DID group, highlighting the good in vivo targeting ability of cRGD. According to ex vivo results, the fluorescence intensity of the nanoformulation in tumor tissues was 11.63-fold higher than that of free DID (Fig. 5d). To further analyze the ratios of cRGD nanoformulations in the tumor cells, single-cell analysis was performed. According to Fig. 5e, higher ratios of cRGD-pRNCThioether+DEA@DID (13.27 ± 0.47%) and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR (17.73 ± 0.85%) were detected and were 2.2- to 2.8-fold higher than that of pRNCThioether+DEA@DID (7.80 ± 0.84%) and free DID (4.70 ± 0.50%) (Fig. 5e). To further verify if cRGD-pRNCThioether+DEA could target tumor cells, pRNCThioether+DEA and cRGD-pRNCThioether+DEA were loaded with DID. According to the immunofluorescence results presented in Supplementary Fig. 42, cRGD-pRNCThioether+DEA@DID exhibited a strong colocalization signal (yellow color) compared to that of the pRNCThioether+DEA@DID group when tumor cells were stained with Alexa 488-PD-L1 (green). Red colors represented DID and indicated the good tumor targetability of cRGD modification. Negligible co-localization was observed in the free DID-treated group.
The in vivo tumor accumulation and TAMs targetability of the cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR were then investigated. Compared to free DIR and pRNCThioether+DEA@DIR, the in vivo fluorescence intensity of Man-decorated nanoformulations was effectively strengthened after i.v. injection, reaching a maximum at 8 h, and this was 3.88-fold higher than that of free DIR (Fig. 5f, g). The ex vivo images also indicated similar results, where the Man-pRNCThioether+DEA@DIR and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR group were 3.67- and 4.20-fold higher than the pRNCThioether+DEA@DIR group, thus indicating the targetability of Man (Fig. 5h). After single cell analysis of tumor tissue via FCM, the ratios of Man-pRNCThioether+DEA@DIR and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR in TAMs were 12.90 ± 0.53% and 12.73 ± 0.51%, respectively, and this was 1.67-fold higher than that of pRNCThioether+DEA@DIR (7.73 ± 0.86%), DIR (3.32 ± 0.62%), and DID mix DIR (Fig. 5i). To further explore the in vivo targetability of the Man nanoformulation in TAMs, Man-pRNCThioether+DEA@FITC was intravenously injected into LLC tumor-bearing C57BL/6 mice, and this was followed by tumor tissue extraction for immunofluorescence analysis. As presented in Supplementary Fig. 43, Man-pRNCThioether+DEA@FITC exhibited the strongest co-localization (yellow color) when green colors represented FITC and red colors represented TAMs, thus indicating that Man-pRNCThioether+DEA could efficiently target macrophages in vivo.
In vivo antitumor immune response activation
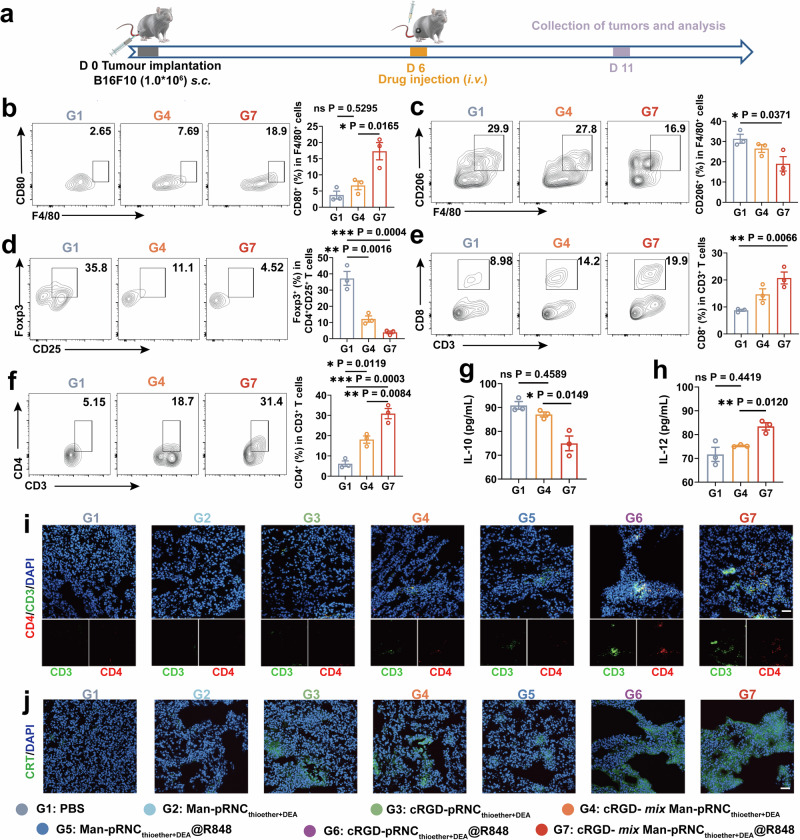
The cRGD- mix Man-pRNCThioether+DEA/R848-mediated in vivo antitumor immune response that recognizes high antitumor activity was then investigated (Fig. 6a). As presented in Fig. 6b, c and Supplementary Fig. 44, the expression of CD80 on the TAMs surfaces increased, while that of CD206 decreased in mice after cRGD- mix Man-pRNCThioether+DEA@R848 (G7), Man-pRNCThioether+DEA@R848 (G5) treatment, or Man-pRNCThioether+DEA (G2) alone, thus indicating that R848 induced TAMs polarization and that the mixed formulations did not affect the polarization ratio. Moreover, IL-12 levels increased in the serum, while IL-10 levels decreased in mice in response to cRGD- mix Man-pRNCThioether+DEA@R848 (G7) (Fig. 6g, h, Supplementary Fig. 45). As presented in Fig. 6d and Supplementary Fig. 46a, the Foxp3+ Tregs ratio notably decreased in mice treated with cRGD- mix Man-pRNCThioether+DEA@R848 (G7), thus indicating its ability to remodel the TME.
Fig. 6. In vivo antitumor immune response of cRGD- mix Man-pRNCThioether+DEA@R848 with ICD inducement, TAMs polarization, Tregs decrement and CD8+/CD4+ proliferation.
a Schematic timeline of the experimental design to evaluate the in vivo immune response activation. b, c Representative flow cytometric images and quantitative analysis to show TAM polarization. d Representative flow cytometric analysis gating on CD25+ cells and quantification of Foxp3+ Tregs in tumors. e, f Representative flow cytometric analysis and quantification of CD8+ and CD4+ T cells gating on CD3+ cells. g, h IL-10 and IL-12 levels of mouse serum after different treatments. i Representative immunofluorescence images of tumors showing CD3+CD4+ T cell infiltration. Scale bar = 50 μm. j Representative immunofluorescence images of tumors showing CRT exposure. Scale bar = 50 μm. Experiments in (i) and (j) were repeated three times independently with similar results. Data are presented as mean ± SEM (n = 3 mice per group). Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test.
As presented in Fig. 6e, f and Supplementary Fig. 46b, c, cRGD-pRNCThioether+DEA (G3) and Man-pRNCThioether+DEA (G2) elicited an increase in the CD8+ and CD4+ T cell ratios compared to that of PBS. Higher CD8+ and CD4+ T cell ratios were detected in both the cRGD- mix Man-pRNCThioether+DEA@R848 (G7) (CD8+: 20.73 ± 3.82%, CD4+: 30.9 ± 4.37%) and cRGD- mix Man-pRNCThioether+DEA (G4) (CD8+: 14.7 ± 3.48%, CD4+: 18.07 ± 3.10%) groups, thus indicating that immune adjuvant of R848 could notably strengthen the in vivo immune response (Supplementary Fig. 46, 47). CD8+ and CD4+ T cell infiltration was observed in the mixed nanoformulation group based on immunofluorescence staining results (Fig. 6i, Supplementary Fig. 48). Additionally, TNF-α levels in serum was elevated in mice with cRGD- mix Man-pRNCThioether+DEA@R848 (G7) treatment, thus confirming the systemic immune response activation (Supplementary Fig. 49). To explore if cRGD-pRNCThioether+DEA can induce tumor ICD, tumor tissue sections from mice treated with different nanoformulations were stained and characterized using CLSM. The results demonstrated that cRGD-pRNCThioether+DEA (G3) caused tumor ICD following CRT exposure (Fig. 6j) and HMGB1 release (Supplementary Fig. 50), thus indicating ICD induction.
To explore the possible clinical translation, the peripheral “vaccine” ability was then investigated by separately loading OVA257-264 peptide and mRNA into cRGD- mix Man-pRNCThioether+DEA to self-assemble into nanovaccine for antigen-specific T cell and durability response monitoring. As presented in Supplementary Fig. 51, both cRGD- mix Man-pRNCThioether+DEA@OVA and cRGD- mix Man-pRNCThioether+DEA@mRNA elicited antigen-specific T cell responses with 1.44- and 1.67-fold higher CD8+Tetramer+ T cell ratios detected compared to that of PBS on day 7 post-treatment. Even on day 14, high ratios were observed, thus demonstrating durable responses and suggesting that the intelligent nanoplatform possesses potent clinical translation potential.
In vivo antitumor activity of cRGD- mix Man-pRNCThioether+DEA@R848
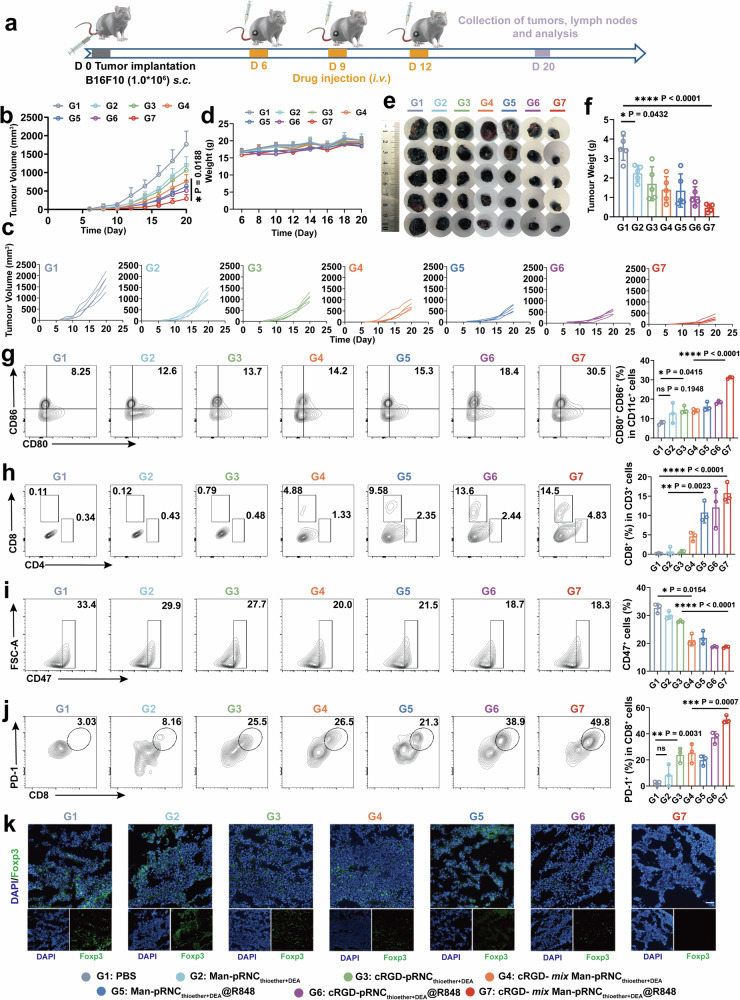
The in vivo antitumor activity of cRGD- mix Man-pRNCThioether+DEA@R848 was investigated, and this was the final purpose of this project. The detailed timeline is presented in Fig. 7a. As presented in Fig. 7b and c, B16F10 tumor volume growth was notably inhibited in mice with cRGD- mix Man-pRNCThioether+DEA@R848 (G7) treatment, while partial tumor volume growth suppression was observed in the cRGD-pRNCThioether+DEA@R848 (G5) group. Negligible changes in body weight were observed (Fig. 7d). When the tumor volume increased to 2000 mm3 on day-20 post-inoculation, the mice were euthanized, and the tumor tissue and normal organs (e.g., heart, liver, spleen, lung, and kidney) were extracted. As presented in Fig. 7e and f, the tumor volume and weight in mice treated with cRGD- mix Man-pRNCThioether+DEA@R848 prominently decreased to levels that were 7.5-fold lower than that of PBS.
Fig. 7. In vivo antitumor activity of cRGD- mix Man-pRNCThioether+DEA@R848 in B16F10 tumor model.
a Schematic of treatment timeline in B16F10 tumor-bearing mice. b, c Average and individual tumor growth curves after different treatments (n = 5 mice per group). d Body weight changes within 20 days after inoculation (n = 5 mice per group). e, f Images and weight of tumor tissues collected on day 20 after different treatments (n = 5 mice per group). g Representative flow cytometry images and quantification of mature DCs in LNs (n = 3 mice per group). h Representative flow cytometric analysis and quantification of CD8+ and CD4+ T cells gating on CD3+ cells (n = 3 mice per group). i Representative flow cytometric analysis gating on CD47+ cells in tumors (n = 3 mice per group). j Representative flow cytometric analysis gating on PD-1+CD8+ cells in tumors (n = 3 mice per group). k Representative immunofluorescence images of Foxp3+ cells in tumors. Three times was repeated independently with similar results. Scale bar = 50 μm. Data are presented as mean ± SD. Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test.
To investigate if cRGD- mix Man-pRNCThioether+DEA@R848 could activate DCs, the ratio of mature DCs in the lymph nodes was detected by FCM. As presented in Fig. 7g, the ratio of CD80+CD86+ DCs (31.03 ± 0.47%) in mice with cRGD- mix Man-pRNCThioether+DEA@R848 (G7) treatment significantly increased to levels that were 1.79- and 3.97-fold higher than those of cRGD- mix Man-pRNCThioether+DEA (G4) (14.00 ± 1.02%) and PBS (G1) (7.81 ± 0.84%). It was observed that the highest CD8+ (15.8 ± 2.55%) and CD4+ (4.11 ± 1.84%) T cell ratio was measured in the cRGD- mix Man-pRNCThioether+DEA@R848 (G7) group (Fig. 7h, Supplementary Fig. 52). Notably, Foxp3+ expression decreased in mice treated with the cRGD- mix Man-pRNCThioether+DEA@R848 (G7) with alleviated green fluorescence, thus demonstrating its ability to modulate the TME (Fig. 7k). Remarkably, the cRGD- mix Man-pRNCThioether+DEA@R848 (G7)-treated mice displayed a significant decrease in CD47 expression that may potentiate macrophage phagocytosis of the tumor (Fig. 7i). Thus, cRGD- mix Man-pRNCThioether+DEA@R848 can activate the host immune response for high antitumor activity via in situ cancer vaccination and TAM polarization. However, upregulated PD-1 expression on the T cell surface was detected after cRGD- mix Man-pRNCThioether+DEA@R848 treatment at the therapeutic point, and this may impede antitumor efficacy at the late stage, thus indicating the necessity for immune checkpoint blockade (Fig. 7j, Supplementary Fig. 53). The normal organ tissues in the mice treated with the nanoformulations were similar to those treated with PBS, and this revealed the superior biocompatibility of the nanocarriers (Supplementary Fig. 54).
In vivo antitumor activity of large tumors after combination anti-PD-1 and anti-CD47 treatment
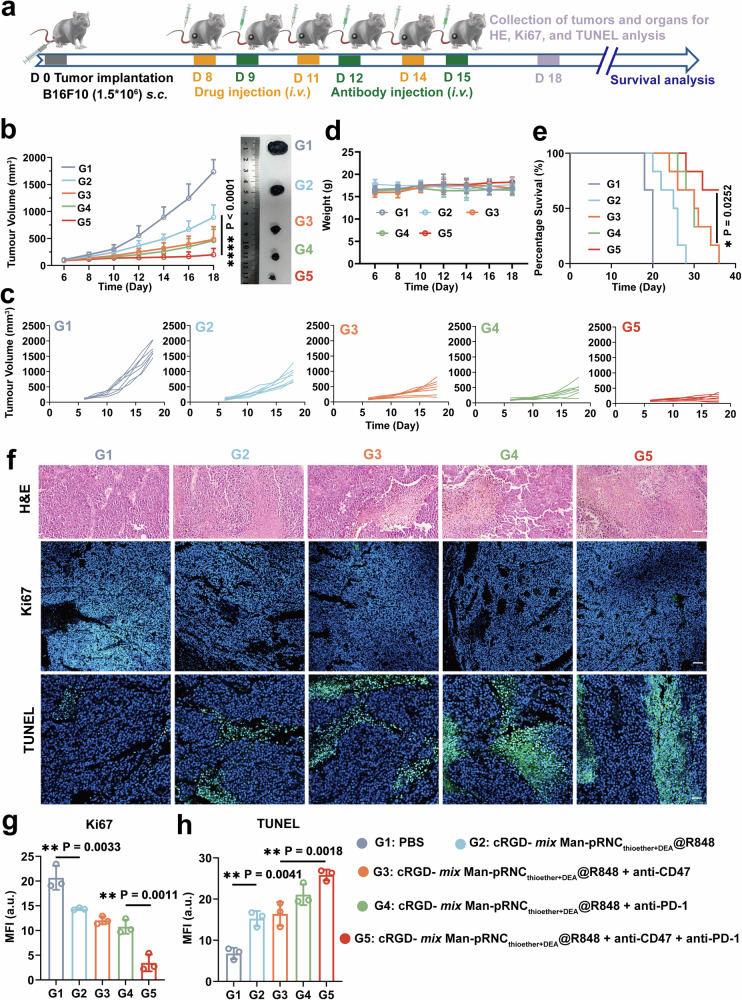
The detailed timeline is presented in Fig. 8a for investigation of the antitumor activity against large tumors via the combination of cRGD- mix Man-pRNCThioether+DEA@R848 and antibodies. As presented in Fig. 8b and c, tumor volume growth was significantly suppressed in mice treated with cRGD- mix Man-pRNCThioether+DEA@R848 in combination with anti-PD-1 and anti-CD47 (G5), ultimately resulting in the longest median survival. Partial tumor volume inhibition was observed with combined anti-PD-1 (G4) or anti-CD47 (G3) alone, with a median survival time of 30 days (Fig. 8e). Negligible body weight changes were observed after the different treatments, thus indicating good biocompatibility (Fig. 8d). On day 18, normal organs and tumor tissues were obtained from one mouse per group (Supplementary Fig. 55), and negligible damage to the normal organs was observed. As indicated by the H&E, TUNEL, and Ki67 staining results, most tumor cell death was observed after combination treatment with anti-CD47 plus anti-PD-1 (Fig. 8f-h).
Fig. 8. In vivo antitumor activity of cRGD- mix Man-pRNCThioether+DEA@R848 after combination with antibodies in mice with a large initial B16F10 tumor volume.
a Schematic illustration of the treatment timeline. b, c Average and individual tumor volume growth curves after different treatments (n = 7 mice per group). d Body weight changes of mice after treatments (n = 7 mice per group). e Survival curves of tumor-bearing mice post different treatments (n = 6 mice per group). f H&E Ki67 and TUNEL images of tumor slices. Scale bar = 50 μm. g, h Quantitative analysis of fluorescence intensity of Ki67 and TUNEL. Data are presented as mean ± SD (n = 3 independent experiments). Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test (b) or log-rank (Mantel-Cox) test (e).
Discussion
Utilizing the host adaptive immune system to attack tumors is an effective method for cancer immunotherapy, and inducing tumor ICD to elicit host antitumor immunity is one of the most widely used approaches. However, it generally requires external inducers such as photosensitizers and chemotherapeutics, most of which are encapsulated or conjugated in carriers requiring site-specific delivery, thereby increasing the complexity of nanoplatforms. To resolve these issues, an immunoactive cRGD-conjugated polymer with a tertiary amine and thioether that can self-assemble into a pH-responsive nanocarrier (cRGD-pRNCDEA+Thiother) was synthesized to directly induce melanoma B16F10 ICD via metabolic regulation and the mediation of ER stress with TAAs and DAMPs release. After encapsulation with the TLR7/8 agonist R848, cRGD-pRNCDEA+Thiother formed an in situ cancer vaccine in vivo that led to DC maturation, antigen presentation, CD8+/CD4+ T-cell proliferation, and tumor volume growth inhibition. In situ tumor vaccination was performed after tumor ICD to ensure temporal orchestration. Additionally, the cRGD-pRNCDEA+Thiother/R848 used in this study possessed very simple components that contributed to clinical translation with good biocompatibility, easy feasibility, high tumor accumulation, and satisfactory therapeutic efficacy. In addition to its combination with R848 for in situ tumor vaccination, the nanoplatform can encapsulate ferroptosis or pyroptosis inducers for a combination of different cell death modalities that mediate tumor eradication. However, certain issues in nanosystems should not be ignored, including the large-scale synthesis and thorough purification of polymers.
Immunosuppressive factors such as TAMs are important components of tumor tissues and are key roadblocks to cancer immunotherapy. Therefore, it is necessary to remodel the TME. The same polymeric skeleton as that of cRGD-pRNCDEA+Thiother was used here, but cRGD was decorated with Man on the nanocarrier surface. The nanocarriers specifically induced melanoma B16F10 ICD but not macrophages via dose adjustment. After loading with R848, Man-pRNCs induced TAMs polarization to the M1 phenotype with antitumor activity. In particular, the composite nanoplatform cRGD- mix Man-pRNCDEA+Thiother/R848 was able to separately target tumor cells for in situ cancer vaccination and TAMs for polarization and spatiotemporal orchestration of adaptive and innate immune response activation. In addition to TAM polarization, TAM autophagy inhibition and depletion can also be investigated in TME modulation, and this may be another direction for future research.
In summary, we designed a pH-responsive polymeric nanoplatform in which the nanocarrier could directly induce B16F10 ICD via inducing oxidative phosphorylation, increasing mtROS production, and upregulating CHOP that in turn induced ER stress. After decoration with cRGD and Man, they can separately target tumor cells for in situ cancer vaccination and TAMs for polarization via TME modulation. Superior antitumor activity and immune response activation highlight the advantages of utilizing the same polymeric skeleton for multiple functional combinations. The prolonged median survival indicates the potential to exert immune checkpoint blockade efficacy. This study provides insights into utilizing immunofunctional polymers to form versatile nanocarriers for potent cancer immunoefficacy that possess excellent clinical translation potential due to their simple synthesis and design.
Methods
Ethical statement
Our research complies with all relevant ethical regulations. All animal studies were performed under the approved protocol of Zhengzhou University Animal Care and Use Committee (ZZU ACUC syxk (yu) 2018-0004).
Materials
Methoxy-poly(ethylene glycol)amine (MeO-PEG-NH2, Mw = 5.0 kg/mol) and amino-poly (ethylene glycol)acid (NH2-PEG-COOH, Mw = 5.0 kg) were purchased from Ponsure Biological (Shanghai, China). The compound 4-Cyano-4-(Phenylcarbonothioylthio) pentanoic acid N-succinimidyl ester (CPPA, 98%) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Methyl methacrylate (MMA, 99.0%), propargylamine (98%), 3-mercaptopropionic acid (98%), 2-(diethylamino)ethyl methacrylate (DEA, 99%), 2, 2’-azobis(2-methylpropionitrile) (AIBN, 99%), N-hydroxysuccinimide (NHS, 98%), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC·HCl, 98.5%), 4-dimethylaminopyridine (DMAP, 99%), and diethylaminoethanol were purchased from Macklin (Shanghai, China). Methacryloyl chloride (95% purity) was purchased from Aladdin (Shanghai, China). The compound 2-Mercaptoethanol (99%) was purchased from GEN-VIEW SCIENTIFIC INC, and 4-Aminophenyl-alpha-D-mannopyranoside (Mannose, 98%) and cyclo(RGD-DPhe-K) (cRGDfK, 92%) were purchased from Top-peptide Co., Ltd. (Shanghai, China). Benzoin dimethyl ether (DMPA) was purchased from Sigma-Aldrich. ER-Tracker Green, Golgi-Tracker Green, LysoTracker Green, and MitoTracker Green were purchased from Beyotime. The ATP Assay Kit (S0026) was purchased from Beyotime (Shanghai, China). Thiazolyl blue tetrazolium bromide MTT (98%) was purchased from Solarbio Life Sciences (Beijing, China). RMPI-1640, phosphate-buffered saline (PBS), and 0.25% trypsin were purchased from Pricellella (Wuhan, China). The mouse HMGB1 ELISA Kit was purchased from Enzyme-Linked Biotechnology (Shanghai, China). The mouse TNF-α ELISA Kit was purchased from Yuchun Biology (Shanghai, China). PE anti CD11c, PE anti CD3, APC anti CD8a, APC anti CD86, Percp Cy5.5 anti CD4, Percp Cy5.5, anti-CD80, FITC anti CD206, and Alexa FluorⓇ488 anti CRT were purchased from BioLegend. Anti-elF2α (phosphor S52, 1:1000, ab227593, Abcam), anti-PERK (phospho T982, 1:1000, ab192591, Abcam), and anti-CHOP (1:1000) were purchased from Abcam. DNAse and collagenase were purchased from Thermo Fisher Scientific. Cytokine IL-4, IFN-γ, and recombinant mouse M-CSF was purchased from novoprotein (Shanghai, China), and lipopolysaccharide (LPS) was purchased from Solarbio (Beijing, China). IL-10, IL-12, and TNF-α ELISA Kits were purchased from Lianke Biology (Hangzhou, China).
Characterization
The molecular weights of polymers were measured using a hydrogen nuclear magnetic resonance (1H NMR) instrument (Germany Brker). The morphology of the nanoparticles was characterized using transmission electron microscopy (TEM, Thermo Scientific/Talos L120CG2). The size and zeta potential of the nanoparticles were measured using a Malvern Nanosize Analyzer (ZEN-3600). Cytokine levels were tested via ELISA kit and a microplate reader (Biotek Synergy H1, USA). Drug-loading content (DLC) and drug-loading efficiency (DLE) were measured using a fluorospectrophotometer. Cellular internalization, tumor ICD, and in vivo T-cell proliferation and infiltration were characterized using laser scanning confocal microscopy (CLSM, LEICA TCS SP8 STED) and flow cytometry (Accuri C6 Plus). In vivo near-infrared (NIR) imaging was performed using an animal multimodel imaging analyzer (IVIS Lumina XRMS Series). The relative molecular weights of the polymers were determined by gel permeation chromatography (GPC) using a Waters 1515 (Waters, USA).
Synthesis and characterization of PEG-PMMA
The di-block copolymer PEG-PMMA was obtained through RAFT polymerization using MeO-PEG-CPPA (also termed PEG-CPPA) and monomer MMA. Prior to this, the macro-RAFT agent PEG-CPPA was synthesized by the amidation reaction of PEG-NH2 and NHS-CPPA as indicated in a previous report48. Briefly, in the existence of the nitrogen (N2) atmosphere, the solution of PEG-CPAA (100 mg, 0.02 mmol), monomer MMA (200 mg, 2 mmol), and the initiator AIBN (0.49 mg, 0.003 mmol) in 1, 4-dioxane was reacted in a sealed flask after placing it in an oil bath (70 oC) for 48 h. After precipitation, centrifugation, and vacuum desiccation, the copolymer PEG-PMMA was obtained at a 67% yield. According to the 1H NMR results, the molecular weight of the diblock copolymer is 5.0–10.0 kg/mol.
Synthesis of PEG-PMMA-PDEA
The triblock copolymer PEG-PMMA-PDEA was obtained through reversible addition-fragmentation chain transfer (RAFT) polymerization using PEG-PMMA and the monomer DEA. Briefly, under an N2 atmosphere, the solution of PEG-PMMA (64.5 mg, 0.0043 mmol), monomer DEA (19.96 mg, 0.108 mmol), and initiator AIBN (0.106 mg, 0.0006 mmol) in 1, 4-dioxane was reacted in a sealed flask that was placed in an oil bath (70 oC) for 48 h. After precipitation, centrifugation, and vacuum desiccation, the copolymer PEG-PMMA-PDEA was obtained at a 78% yield. According to the 1H NMR results, the molecular weight of the triblock copolymer was 5.0-10.0-3.8 kg/mol.
Synthesis of N-propargyl methacrylamide (PPMA)
Small-molecule PPMA was obtained via amidation of methacryloyl chloride and propargylamine. In detail, propargylamine (2.057 g, 37.4 mmol) and DMAP (463.6 mg, 3.8 mmol) were fully dissolved in dichloromethane (DCM). In an ice-water bath under an N2 atmosphere, a solution of DMAP and methacryloyl chloride was added dropwise to the propargylamine reaction for 24 h at room temperature. The product was purified using a silica gel column (mobile phase: n-hexane: ethyl acetate = 5/1). According to 1H NMR result, pure PPMA was obtained. The yield was 25%.
Synthesis of PEG-PMMA-P(PPMA-ME)
The triblock copolymer PEG-PMMA-PPPMA was synthesized via RAFT polymerization to obtain the graft copolymer PEG-PMMA-P (PPMA-ME). Briefly, PEG-PMMA (100 mg, 0.0067 mmol), PPMA (34 mg, 0.276 mmol), and AIBN (0.164 mg, 0.001 mmol) were dissolved in 1, 4-dioxane and added to a flask under an N2 atmosphere. After further N2 ventilation for 30 min, the flask was sealed and placed in an oil bath (70 °C) reaction for 48 h. After ice diethyl ether precipitation, filtration, and vacuum drying, PEG-PMMA-PPPMA was successfully acquired, and the molecular weight was 5.0-10.0-6.3 kg/mol according to 1H NMR result. The yield was 52%.
PEG-PMMA-P(PPMA-ME) was obtained via a click reaction between PEG-PMMA-PPPMA and mercaptoethanol. Briefly, PEG-PMMA-PPPMA (41.54 mg, 0.00195 mmol), mercaptoethanol (7.59 mg, 0.0973 mmol), and DMPA (6.23 mg, 0.0243 mmol) were dissolved in N, N-dimethylformamide (DMF) and added into a flask with UV laser irradiation (2 h, wavelength: 350 nm). After ice-cold diethyl ether precipitation, filtration, and vacuum desiccation, the graft copolymer was successfully obtained at an 84% yield, and the graft ratio of mercaptoethanol was 100% according to the 1H NMR result. The molecular weight of PEG-PMMA-P(PPMA-ME) was 5.0-10.0-10.4 kg/mol.
Synthesis of PEG-PMMA-P(PPMA-Cy5)
PEG-PMMA-P(PPMA-Cy5) was obtained via a click reaction between PEG-PMMA-PPPMA and Cy5-N3. Briefly, PEG-PMMA-PPPMA (20 mg, 0.001 mmol) and Cy5-N3 (0.38 mg, 0.0006 mmol) were dissolved in DMF. CuSO4 (0.005 mg, 3.13 × 10−5 mmol) was dissolved in deionized water (1.68 μL) and then added into the DMF solution. After the reaction mixture was incubated under an N2 atmosphere for 30 min, sodium ascorbate (NaAS) (0.026 mg, 0.0001 mmol) dissolved in deionized water (21.7 μL) was added to the above reaction under N2. After reacting for 24 h at room temperature, the product was subjected to DMF and water dialysis and lyophilization. The yield was 74%.
Synthesis of PEG-PMMA-P(PPMA-MPA-DEA)
To obtain PEG-PMMA-P(PPMA-MPA-DEA), PEG-PMMA-P(PPMA-MPA) was synthesized via a click reaction of PEG-PMMA-PPPMA and mercaptopropionic acid. This was similar to the synthesis procedure for PEG-PMMA-P(PPMA-ME) with the exception that mercaptoethanol was replaced with mercaptopropionic acid. The molecular weight of PEG-PMMA-P(PPMA-MPA) was 5.0-7.3-14.5 kg/mol according to the 1H NMR result. The yield was 85%.
PEG-PMMA-P(PPMA-MPA-DEA) was obtained via the esterification of PEG-PMMA-P(PPMA-MPA) and diethylaminoethanol. In detail, PEG-PMMA-P(PPMA-MPA) (112.5 mg, 0.0042 mmol), EDC·HCl (30.19 mg, 0.157 mmol), and DMAP (12.8 mg, 0.105 mmol) were separately dissolved in DMF. Under an N2 atmosphere, EDC·HCl and DMAP were added into PEG-PMMA-P(PPMA-MPA) reaction for 30 min, and this was followed by DEA addition. After reacting for 24 h at room temperature, the product was subjected to DMF and water dialysis and lyophilization. The yield was 74%. According to 1H NMR results, the molecular weight was 5.0-7.3-18.4 kg/mol.
Preparation and characterization of nanoparticles
All nanoparticles were prepared using the solvent-exchange method. Briefly, polymers with different functional groups were dissolved in THF at a concentration of 5 mg/mL, and 100 μL were then added into PBS buffer (900 μL, 10 mM) for a uniform dispersion. After volatilization and dialysis (MWCO = 3500) to remove the organic solvent, nanoparticles were successfully prepared, and their sizes and zeta potentials were measured by DLS. Their morphologies were characterized by TEM. Briefly, the nanoparticles were added to a copper mesh with a carbon film and left overnight. After removing the excess nanoparticles, images were captured using TEM. It should be noted that nanoparticles self-assembled from the polymer PEG-PMMA were termed as nanocarrierMMA (NCMMA), and self-assembly from PEG-PMMA-PDEA, PEG-PMMA-PPPMA, PEG-PMMA-P(PPMA-hydroxyl), and PEG-PMMA-P(PPMA-DEA) were named pH responsive nanoparticleDEA (pRNCDEA), NCyne, NCThioether, and pRNCThioether+DEA For the investigation of pH responsiveness, nanoparticles were placed in acetate buffer (10 mM, pH 5.0) and PB buffer (pH 7.4, 10 mM), and their sizes were monitored by DLS at 1, 12, and 24 h.
Cell culture
Macrophage RAW264.7 cells, Pan02 cells and U87-MG cells were separately cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% streptomycin-penicillin, and they were cultured in an incubator (37 oC, 5% CO2). Melanoma B16F10 cells, 4T1 cells, MC38 cells, LLC cells and DC were separately cultured in RPMI-1640 medium containing 10% FBS and 1% streptomycin-penicillin, and they were cultured in an incubator (37 oC, 5% CO2). All cell lines were obtained from American Type Culture Collection (ATCC). These cell lines were authenticated by Wuhan Pricella Biotechnology (Wuhan, China) using STR analysis. Specifically, 20 STR loci were amplified using using Microreader 21 ID System. The PCR products were detected by GenReader 7010, and the results were analyzed by GeneMapper Software6 and compared with ExPASy database. All cell lines were tested negative by using Myco-Lumi Luminescent Mvcoplasma Detection Kit for mycoplasma contamination.
Animals
C57BL/6 mice (female, 6 ~ 8 weeks, 18–20 g) were used to construct a melanoma model. All animals were housed in individually ventilated cages at Zhengzhou University, subjected to a 12-h light-dark cycle, and maintained at 22 °C with humidity levels between 30% to 70%. Tumor length and width were measured using a Vernier caliper every two days, and the tumor volume curve was recorded. The maximal tumor volume (2000 mm3) was permitted by the institutional animal care and use committee. As for results in Fig. 7c, at the second last time point (day-18 post-inoculation), the tumor volume in all mice was less than 2000 mm3 and the tumor growth was continued to monitor. At the day-20 post-inoculation, the tumor volume in one mouse of G1 reached 2000 mm3 thereby being chosen as the endpoint. All the tumor-bearing mice were euthanized at that time point for the immune response analysis. The tumor volume (V) was calculated using the following formula: V = ½ × Length × Width2.
Screening of the polymer selectively inducing B16F10 immunogenic cell death
Polymer-mediated ICD was explored using CRT exposure and HMGB1 and ATP release assays. For CRT exposure, B16F10 cells (2.0 × 105/well) were seeded into six-well plate and cultured overnight. NCMMA, pRNCDEA, NCyne, NCThioether, and pRNCThioether+DEA (concentrations of all nanoformulations were 50 μg/mL) were separately added into each well incubation for 48 h. After washing with PBS three times and then digestion and centrifugation, the cells were stained with Alexa FluorⓇ488 anti CRT (400×, 40 min), and this was followed by PBS washing and centrifugation. Finally, the cells were suspended in PBS (0.5 mL each sample), stained with propidium iodide (PI), incubated at room temperature for 10 min, and detected by flow cytometry, processed using FlowJo software (version 10.9.0).
To characterize HMGB1 release, cells were seeded into 24-well plates and cultured overnight, and this was followed by treatment with NCMMA, pRNCDEA, NCyne, NCThioether, or pRNCThioether+DEA. After nanoformulation treatment for 48 h, PBS washing (× 3), digestion, and centrifugation, the cells were stained with Alexa FluorⓇ488 anti HMGB1 (200×, 40 min), and this was followed by PBS washing. Then, mounting media containing DAPI was added to the cells. The specimens were covered with coverslips and sealed with nail polish. Cell images were captured using a confocal laser-scanning microscope (CLSM), processed using LAS X 4.4 software. After the different treatments, the supernatants were collected to detect HMGB1 release via ELISA characterization.
ATP secretion by B16F10 cells after the different treatments was tested using an ATP Assay Kit (S0026). B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultured overnight. NCMMA, pRNCDEA, NCyne, NCThioether, and pRNCThioether+DEA were separately added into each well for 24 h. A total of 30 μL of supernatant from the cell culture medium was added to 70 μL of ATP assay working solution, and the RLU value was measured by enzyme-linked immunosorbent assay within 30 min. The total protein concentration in the six-well plates was determined using a BCA kit. The amount of ATP released per unit protein concentration was compared among the different groups.
Concentration and time dependence of pRNCThioether+DEA-mediated B16F10 ICD
For CRT exposure at the same time points with different concentrations, B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultivated overnight. pRNCThioether+DEA (the concentrations of the nanoformulations were 0, 25, 50, 100, 200, and 300 μg/mL) were separately added into each well for 48 h incubation. After washing with PBS (× 3), digestion, and centrifugation, the cells were stained with Alexa FluorⓇ488 anti CRT (400×, 40 min), and this was followed by PBS washing and centrifugation. The cells were then suspended in PBS (0.5 mL) and stained with PI. After 10 min, the cells were washed with PBS, centrifuged, suspended in PBS, and analyzed by flow cytometry.
For CRT exposure at the same concentration and at different time points, B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultivated overnight. pRNCThioether+DEA (100 μg/mL) was separately added into each well for different incubation time. After washing with PBS (× 3), digestion, and centrifugation, the cells were stained with Alexa FluorⓇ488 anti CRT (400×, 40 min), and this was followed by PBS washing and centrifugation. The cells were then stained with propidium iodide (PI), washed with PBS, suspended in PBS, and analyzed by flow cytometry.
Organelle targetability investigation
B16F10 cells (2.0 × 104) were seeded into a dish and cultured for 24 h. Cy5-labeled pRNCThioether+DEA (Cy5-pRNCThioether+DEA) was added and incubated for 2 and 4 h, respectively. After washing (PBS × 3), cells were separately stained with ER-Tracker Green, Golgi-Tracker Green, Lyso-Tracker Green, and Mito-Tracker Green (30 min). After PBS washing (× 3) again, cells were stained with Hoechst 33342 (5 μg/mL, 15 min). After further PBS washing, fresh media (200 μL) was added to each dish, and images were captured by CLSM.
Mechanisms investigation of the polymer-mediated ICD
The underlying mechanisms were explored by sequence analysis, flow cytometry, CLSM and WB characterization, and western blotting. For RNA-sequence analysis, B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultured overnight. PBS and pRNCThioether+DEA were added and incubated for 48 h. After removal of the culture media, TRIzol (1 mL) was added to each well for cell digestion, and the cells were transferred into RNase-free tubes. After 1–2 seconds in liquid nitrogen, the samples were placed at −80 oC for 24 h. Interfering DNA was removed, and RNA samples were prepared. An Illumina TruseqTM RNA sample prep kit was used to construct the library that was then sequenced. Bioinformatics analysis was performed on the sequencing results.
Flow cytometry and CLSM were performed to determine intracellular ROS and mtROS generation. For flow cytometry analysis, B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultured overnight. Cells were incubated with PBS, NCMMA, and pRNCThioether+DEA for 48 h. After PBS washing, DCFH-DA probe (1 mL, 5 μM in serum-free RPMI-1640 medium) was added to each well and incubated for 30 min away from light. After washing (PBS × 3), digestion, and centrifugation, the cells were suspended in PBS (0.5 mL each sample) and detected by flow cytometry. The generation of pRNCThioether+DEA-mediated mtROS within the tumor cells was observed via CLSM. B16F10 cells (2.0 × 104/well) were seeded into a culture dish for 24 h. PBS, NCMMA, and pRNCThioether+DEA were added and incubated for 48 h. After PBS washing (× 3), cells were separately stained with BBcellProbe®OM08 (30 min) and Hoechst 33342 (5 μg/mL, 15 min). After further PBS washing, fresh media (200 μL) was added to each dish, and images were captured by CLSM.
For WB characterization, B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultured overnight. PBS, NCMMA, and pRNCThioether+DEA were added to each well and incubated for 48 h. After washing with PBS, the cells were lysed using radioimmunoprecipitation assay (RIPA) buffer, and the supernatant was collected after centrifugation (10,000 g, 30 min). The protein content was measured using a bicinchoninic acid kit. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. For necrosis-related proteins, MLKL (1:1000, cat# 183770, Abcam) was detected. For ER stress-related proteins, p-PERK (1:1000, Cell Signaling Technology, cat# 3179S), p-elf2α (1:1000, Cell Signaling Technology, cat# 9721S), CHOP (1:1000, cat.no.ab11419, Abcam), and ATF-4 (1:1000, sc-390063, Santa Cruz Biotech) were used. For pyroptosis-related proteins, cleaved caspase-1 (1:1000, Ala317, Affinity) and N-GSDMD (1:1000, cat# 939701, BioLegend) were used. Primary antibody solutions were prepared in a primary antibody dilution buffer (EpiZyme). The reaction was performed overnight at 4 °C. HRP-conjugated anti-rabbit and anti-mouse that were used as secondary antibodies were added to the corresponding PVDF membranes at room temperature for 1 h. The blot signals were visualized using an enhanced chemiluminescent (ECL) reagent (Epizyme, China).
LDH detection
LDH secretion from B16F10 cells after different treatments was tested using an LDH Microplate test kit (A020-2). B16F10 cells (2.0 × 105/well) were seeded into six-well plates and cultured overnight. PBS, NCMMA, and pRNCThioether+DEA were separately added to each well and incubated for 24 h. Supernatants were collected and measured according to the manufacturer’s instructions.
B16F10 cells death mediated by pRNCThioether+DEA
Tumor cells death was determined by flow cytometry. First, B16F10 cells (2.0 × 105/well) were seeded into a six-well plate and grown overnight. PBS, NCMMA, pRNCThioether+DEA, pRNCThioether+DEA + Z-VAD-FMK, pRNCThioether+DEA +Nec-1s, and pRNCThioether+DEA + Fer-1 were separately added into each well for 48 h incubation. Cells were digested by trypsin and subsequently washed with PBS (×3). Cell death was assessed by flow cytometry using an Annexin V/PI cell assay kit (Biosharp) via the protocol provided by the manufacturer.
Intracellular lipid peroxide detection
First, B16F10 cells (2.0 × 105/well) were seeded into a six-well plate and cultured overnight. Phosphate-buffered saline (PBS), NCMMA, and pRNCThioether+DEA were added to each well and incubated for 24 h. After incubation for 24 h, the culture medium was removed, cells were washed 3 times with PBS, and C11-BIODPY (1 mL, 10 μM, serum-free 1640 medium was used) was added to each well for 30 min incubation away from light in a cell incubator. The cells were then digested with trypsin and washed three times with PBS. The cells were suspended in PBS (0.5 mL) and detected using flow cytometry.
Synthesis of Man (cRGD)-PEG-PMMA-PPPMA
Man-PEG-PMMA-PPPMA was obtained by the amidation of COOH-PEG-PMMA-PPPMA and Man-NH2. The synthesis of COOH-PEG-PMMA-PPPMA was similar to that of PEG-PMMA-PPPMA, and the molecular weights was determined by 1H NMR spectroscopy. To obtain Man-PEG-PMMA-PPPMA, NHS (0.489 mg, 0.0043 mmol) and EDC·HCl (0.815 mg, 0.0043 mmol) were first added into the mixture solution of COOH-PEG-PMMA-PPPMA (35 mg, 0.0028 mmol) and triethylamine (TEA) with stirring for 1.5 h under N2 to acquire NHS-PEG-PMMA-PPPMA. In the ice water bath and N2 atmosphere, NHS-PEG-PMMA-PPPMA was added dropwise to a Man-NH2 solution containing TEA. The reaction was then placed in a water bath (30 oC) for 24 h under dark conditions. The product was successfully obtained after dialysis and lyophilization with a yield of 48%. The synthesis of cRGD-PEG-PMMA-PPPMA was similar to that of Man-PEG-PMMA-PPPMA with the replacement of Man-NH2 by cRGD-NH2, and the yield was 85%. cRGD (Man)-pRNCThioether+DEA was also prepared using a solvent-exchange method similar to that of pRNCThioether+DEA with a pre-mixture of the polymers cRGD-PEG-PMMA-PPPMA (20 μL, 5 mg/mL) and PEG-PMMA-P(PPMA-Thioether-DEA) (80 μL, 5 mg/mL). The size and morphology of cRGD-pRNCThioether+DEA was characterized by DLS and TEM, respectively.
Cytotoxicity investigation of Man-pRNCThioether+DEA in RAW264.7 cells and of cRGD-pRNCThioether+DEA in B16F10 cells
Cytotoxicity was investigated by MTT assays. Briefly, RAW264.7 cells and B16F10 cells (5.0 × 103/well) were seeded into 96-well culture plates for 24 h. Man-pRNCThioether+DEA or cRGD-pRNCThioether+DEA with different concentrations from low to high (0, 12.5, 25, 50, 100, and 200 μg/mL) was separately added. After incubation for 48 h, MTT solution (20 μL, 5 mg/mL) was added for 4 h incubation. DMSO (100 μL) was added after supernatant aspiration. Absorbance at 570 nm was measured using a microplate reader.
In vitro drug loading and release
Preparation of drug-loaded nanoparticles was also performed utilizing the solvent-exchange method using only a pre-mixture of the polymer and drug. In detail, the mixture of polymer PEG-PMMA-P(PPMA-DEA) (80 μL, 5 mg/mL), cRGD (Man)-PEG-PMMA-PPPMA (20 μL, 5 mg/mL) in THF (50 μL, 5 mg/mL), and R848 in DMF (10 μL, 5 mg/mL) was added dropwise into PBS (900 μL) for a homogeneous dispersion. After volatilization and dialysis to remove the organic solvent and unloaded drug, the nanomedicine cRGD (Man)-pRNCThioether+DEA@R848 was successfully prepared, and its size was characterized by DLS. DLC and DLE were measured using a fluorescence spectrophotometer and calculated according to the following formulas:
| 1 |
| 2 |
In vitro drug release was investigated under specific conditions. Briefly, cRGD (Man)-pRNCThioether+DEA/R848 (each 0.5 mL) in PBS (pH 7.4, 10 mM, 150 mM NaCl) or acetate buffer (pH 5.0, 10 mM, 150 mM NaCl) in a dialysis bag (MWCO = 12000) was placed in 25 mL of media and were then placed in a shaking bed (37 oC, 200 rpm) (n = 3 independent experiments). At different time points (1, 2, 4, 6, 9, 12, and 24 h), 5 mL of the medium was removed and replaced with the same volume of fresh medium. Accumulation of R848 was detected using a fluorescence spectrophotometer.
Targetability of cRGD-pRNCThioether+DEA in B16F10 cells
The targetability of cRGD-pRNCThioether+DEA was investigated using flow cytometry and CLSM. For flow cytometry, B16F10 cells (5.0 × 105/well) were seeded into a six-well plate and cultured overnight, and this was followed by the addition of free FITC, cRGD-pRNCThioether+DEA@FITC, and pRNCThioether+DEA@FITC. After incubation for 10 or 30 min or for 1 h, the cells were subjected to trypsin digestion, centrifugation, suspension in PBS, and detection by flow cytometry.
For CLSM characterization, B16F10 cells were seeded into 24-well plates containing cell slide cultures for 24 h. Free FITC, cRGD-pRNCThioether+DEA@FITC, and pRNCThioether+DEA@FITC were added separately and incubated for 10 min, 30 min, and 1 h. After PBS washing (×3), cells were fixed for 15 min. After another PBS washing, cells were stained with DAPI (5 μg/mL, 10 min) and then sealed using coverslips onto microslides. After sealing with nail polish, images were captured using a CLSM.
Targetability of Man-pRNCThioether+DEA in RAW264.7 cells
Man-pRNCThioether+DEA was also fabricated using a solvent-exchange method similar to that of cRGD-pRNCThioether+DEA and was characterized by DLS and TEM. Its targetability was investigated by flow cytometry and CLSM in a manner similar to that of cRGD-pRNCThioether+DEA. B16F10 cells were replaced with RAW264.7 cells.
In vitro DC maturation
DC maturation was explored by flow cytometry to characterize the marker changes. Briefly, B16F10 cells (2.0 × 105/well) were seeded into 6-well plates and cultured overnight. PBS, NCMMA, cRGD-pRNCThioether+DEA, cRGD-pRNCThioether+DEA@R848, and cRGD- mix Man-pRNCThioether+DEA@R848 were separately added into B16F10 cells incubation for 48 h. The supernatant were then added into DC cells (5.0 × 105/well) for another 24 h. After washing with PBS, trypsin digestion, and centrifugation, the cells were stained with anti CD80, anti CD86 (40 min), suspended in PBS, and analyzed by flow cytometry. Cells treated with LPS (10 ng/mL) were used as the positive control.
In vitro macrophage polarization
Macrophage polarization was explored primarily by flow cytometry to characterize marker changes on the cell surface and by ELISA to test cytokine levels. For flow cytometry characterization, RAW 264.7 cells (1.0 × 105/well) were seeded into 12-well plates and cultured overnight. IL-4 (10 ng/mL) and Recombinant Mouse Macrophage Colony-Stimulating Factor 1 (MCS-F1; 10 ng/mL) were added separately to polarize M0 cells into the M2 phenotype. PBS, Man-pRNCThioether+DEA, R848, Man-pRNCThioether+DEA@R848, and cRGD- mix Man-pRNCThioether+DEA@R848 were separately added into M2 phenotype cells (R848:1 μg/mL) for 4 h incubation. The original medium was aspirated with fresh medium and incubated for another 20 h. After washing with PBS, trypsin digestion, and centrifugation, the cells were stained with anti CD80, anti CD206, and anti F4/80 (40 min), suspended in PBS, and analyzed by flow cytometry. Cells treated with LPS (10 ng/mL) and IFN-γ (10 ng/mL) were used to polarize M0 into the M1 phenotype as the positive control and CD80 single staining group.
Intracellular iNOS levels in RAW264.7 cells were detected by RT-qPCR. Briefly, total RNA was isolated from RAW264.7 cells using the RNA Easy Mini Plus Kit after R848, Man-pRNCThioether+DEA, and Man-pRNCThioether+DEA@R848 treatments. Two-step RT-qPCR was performed on a Mastercycler EP Realplex (Eppendorf) using iScript and SsoAdvanced SYBR Green Mix (Bio-Rad). The data were normalized to the reference gene β-actin and compared to the control samples (n = 3 independent experiments). Morphological hierarchical analysis was used to analyze the data. Primers were purchased from Sangon Biotech with the sequence are GTTCTCAGCCCAACAATACAAGA(5’ to 3’) and GTGGACGGGTCGATGTCAC(5’ to 3’).
In vivo NIR imaging and targeted internalization by tumor tissue and TAMs
For in vivo NIR imaging and targetability in tumor tissues, C57BL/6 mice were inoculated with LLC cells (1 × 106/mouse) in the right flank to construct a tumor model. When the tumor volume reached approximately 150 mm3, the mice were randomly divided into five groups that included free DID (G1), free DID mix DIR (G2), pRNCThioether+DEA@DID (G3), cRGD-pRNCThioether+DEA@DID (G4), and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR (G5) (n = 3 mice per group). When different formulations were administered via intravenous (i.v.) injection, the fluorescence intensity in tumor tissues were monitored via IVIS imaging system at pre-set time points (2, 4, 8, 12, and 24 h). At 24 h, the mice were euthanized, their organs were harvested (e.g., heart, liver, spleen, lung, and kidneys), and tumors were extracted for ex vivo imaging. Subsequently, tumor tissues were digested by collagenase IV (50 U/mL) in 1640 medium containing with 2% FBS for 2 h (37 oC). The supernatant was filtrated through a 70 μm filter membrane, and this was followed by centrifugation (137 × g for 5 min) to acquire cell pellets. The cells were then stained with FITC anti PD-L1 (400×, 40 min, Elabscience), washed with PBS, centrifuged, resuspended in PBS, and detected by flow cytometry.
TAMs targetability was performed in a manner similar to that of the above tumor tissue targetability with formulations replaced by free DIR (G1’), free DIR mix DID (G2), pRNCThioether+DEA@ DIR (G3’), Man-pRNCThioether+DEA@DID (G4’), and cRGD-pRNCThioether+DEA@DID mix Man-pRNCThioether+DEA@DIR (G5) and the antibody replaced by PE anti F4/80.
In vivo ICD and TAM polarization
C57BL/6 mice were inoculated with B16F10 cells (1 × 106/mouse) in the right flank to establish a melanoma tumor model. When the tumor volume increased to approximately 100 mm3 on day-6, the mice were randomly divided into seven groups that included PBS (G1), Man-pRNCThioether+DEA (G2), cRGD-pRNCThioether+DEA (G3), cRGD mix Man-pRNCThioether+DEA (G4), Man-pRNCThioether+DEA@R848 (G5), cRGD-pRNCThioether+DEA@R848 (G6), and cRGD-mix Man-pRNCThioether+DEA@R848 (G7) (n = 3 mice per group) (cRGD-pRNPThioether+DEA: 1 mg/kg; Man-pRNPThioether+DEA: 0.5 mg/kg; R848: 0.1 mg/kg). At day-11 post-inoculation, serum was collected for IL-10, IL-12, and TNF-α measurement by ELISA, and tumor tissue was extracted for ICD and T cell ratio analysis. Partial tumor tissues were subjected to mechanical obstruction, collagenase digestion, filtration, staining with CD8/CD4/CD3, CD80, CD86, CD206, and Treg antibodies, suspension in PBS, and flow cytometry. Partial tumor tissues were embedded, sectioned, and subjected to immunofluorescence to characterize CRT exposure, HMGB1 secretion, and CD8+/CD4+/CD3+ T cell infiltration using CLSM.
In vivo vaccine function of pRNCThioether+DEA
B16F10 cells (1 × 106/ mouse) were inoculated subcutaneously into the right side of C57BL/6 mice to establish a melanoma mouse model. When the tumor volume reached approximately 100;mm3 by day 6, the mice were randomly divided into three groups that included PBS (G1), pRNCThioether+DEA@OVA (G2), and pRNCThioether+DEA@mRNA (G3) (n = 3 mice per group) (pRNCThioether+DEA: 5 mg/kg; OVA: 1 mg/kg; mRNA: 0.5 mg/kg). On days 7 and 14 after administration, peripheral blood mononuclear cells were extracted from mice for tetrameric analysis. T cells were extracted using a peripheral lymphocyte separation kit, stained with a CD3/CD8/Tetramer antibody, resuspended in PBS, and detected by flow cytometry.
In vivo antitumor activity of cRGD- mix Man-pRNPThioether+DEA@R848
C57BL/6 mice were injected with B16F10 cells (1 × 106/mouse) in the right flank to construct a melanoma tumor model. When the tumor grew to ~50 mm3, the mice were randomly divided into seven groups that included PBS (G1), Man-pRNCThioether+DEA (G2), cRGD-pRNCThioether+DEA (G3), cRGD- mix Man-pRNPThioether+DEA (G4), Man-pRNCThioether+DEA@R848 (G5), cRGD-pRNCThioether+DEA@R848 (G6), and cRGD-mix Man-pRNCThioether+DEA@R848 (G7) (cRGD-pRNPThioether+DEA: 1 mg/kg; Man-pRNPThioether+DEA: 0.5 mg/kg; R848: 0.1 mg/kg) (n = 5 mice per group). Different formulations were intravenously injected into tumor-bearing mice at 6, 9, and 12 days post-inoculation. Tumor length and width were measured using a Vernier caliper every two days, and the tumor volume curve was recorded. Body weight was recorded every two days. The therapeutic end point was when the tumor volume reached 2000 mm3 with normal organ (e.g., the heart, liver, spleen, lung, and kidneys) and tumor tissue extraction. Partial tumor tissues were subjected to mechanical obstruction, collagenase digestion, filtration, staining with CD8/CD4/CD3, CD80, CD86, CD206, CD47, and PD-1 antibodies, suspension in PBS, and flow cytometry. Partial tumor tissues were subjected to embedding, ice sectioning, immunofluorescence staining, and Foxp3+ Treg cell infiltration characterization by CLSM.
In vivo antitumor activity of cRGD mix Man-pRNPThioether+DEA@R848 in combination with anti-PD-1 and anti-CD47
C57BL/6 mice were inoculated with B16F10 cells (1.5 × 106/mouse) via subcutaneous injection to construct a melanoma tumor model. When the tumor volume reached ~120 mm3, mice were randomly divided into five groups that included PBS (G1), cRGD mix Man-pRNCThioether+DEA@R848 (G2), G2 plus anti-CD47 (G3), G2 plus anti-PD-1 (G4), and G2 plus anti CD47 plus anti-PD-1 (G5) (cRGD-pRNPThioether+DEA: 1 mg/kg; Man-pRNPThioether+DEA: 0.5 mg/kg; R848: 0.1 mg/kg; anti CD47: 2 mg/kg; anti PD-1: 2 mg/kg) (n = 7 mice per group). The nanoformulations were administered on days 8, 11, and 14 via i.v. tail injection three times when antibodies were intravenously injected on days 9, 12, and 15. Tumor volume and mouse body weight were measured every two days. On day 18 when the tumor volume increased to 2000 mm3, one mouse from each group was euthanized with harvesting of normal organs, and the tumor tissue was extracted for H&E staining. Tumor tissues were also used for TUNEL and Ki67 staining to investigate apoptosis.
Statistical analysis
All quantitative data were collected from experiments performed in at least triplicate and were expressed as mean ± SD/SEM. All statistical analyses were performed using GraphPad Prism 10.0.0. Statistical significance was calculated through one-way ANOVA for multiple comparisons using a Tukey post-hoc test, two-tailed student’s t test and the log-rank test. P-values and replicate n values refer to one-way ANOVA and independent experiments, respectively, unless otherwise stated. No statistical methods were used to determine the sample size. Differences were considered significant at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 and not significant (ns) at P ≥ 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This research was supported by National Natural Science Foundation of China (52103191, 52103190 and 82073787), Start-up Grant (32340452 and 32340311) from Zhengzhou University, the NUS School of Medicine Dean’s Office (NUHSRO/2020/133/Startup/08, NUHSRO/2023/008/NUSMed/TCE/LOA, NUHSRO/2021/034/TRP/09/Nanomedicine, NUHSRO/2021/044/Kickstart/09/LOA, 23-0173-A0001), National Medical Research Council (MOH-001388-00, CG21APR1005, OFIRG23jul-0047), Singapore Ministry of Education (MOE-000387-00), and National Research Foundation (NRF-000352-00).
Author contributions
W.Y., Z.Z., X.C., Y.G., and Y.L. conceived and designed the study. Y.G. and Y.L. performed the in vivo experiments. M. Z., R.M., Y.W., and X.W. performed the in vitro experiments. W.Y., Z.Z., X.C., Y.G. Y.L., and J.Z. contributed to the analysis and interpretation of the results and the writing of the manuscript.
Peer review
Peer review information
Nature Communications thanks Genevieve Boland, Guangjun Nie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
RNA-seq dataset is available in NCBI under accession codes PRJNA1126401. Figures/tables summarizing NMR are included in Supporting Information, and source data of NMR characterization of PEG-CPPA, PEG-PMMA, PEG-PMMA-PDEA, PPMA, PEG-PMMA-PPPMA, PEG-PMMA-P(PPMA-ME), PEG-PMMA-PPPMA, PEG-PMMA-P(PPMA-MPA), PEG-PMMA-P(PPMA-MPA- DEA), cRGD-PEG-PMMA-PPPMA and Man-PEG-PMMA-PPPMA are included in 10.6084/m9.figshare.26728720. Source data for main and other Supplementary Figs. are provided with this paper and are available in the Figshare database at: 10.6084/m9.figshare.26094232. The data supporting the findings of this study are available within the article, Supplementary, or Source data files. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yichen Guo, Yongjuan Li.
Contributor Information
Zhenzhong Zhang, Email: zhangzhenzhong@zzu.edu.cn.
Xiaoyuan Chen, Email: chen.shawn@nus.edu.sg.
Weijing Yang, Email: wjyang@zzu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53010-0.
References
- 1.Kroemer, G., Galassi, C., Zitvogel, L. & Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol.23, 487–500 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Meng, Q., Ding, B., Ma, P. & Lin, J. Interrelation between programmed cell death and immunogenic cell death: take antitumor nanodrug as an example. Small Methods7, e2201406 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Zhou, L., Zhang, P., Wang, H., Wang, D. & Li, Y. Smart nanosized drug delivery systems inducing immunogenic cell death for combination with cancer immunotherapy. Acc. Chem. Res.53, 1761–1772 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Krysko, D. et al. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer12, 860–875 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Duan, X., Chan, C. & Lin, W. Nanoparticle‐mediated immunogenic cell death enables and potentiates cancer immunotherapy. Angew Chem. Int. Ed.58, 670–680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park, S. J. et al. Cisplatin and oxaliplatin induce similar immunogenic changes in preclinical models of head and neck cancer. Oral Oncol.95, 127–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banstola, A., Poudel, K., Kim, J., Jeong, J. & Yook, S. Recent progress in stimuli-responsive nanosystems for inducing immunogenic cell death. J. Control. Release337, 505–520 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Gao, Z. et al. An activatable near-infrared afterglow theranostic prodrug with self-sustainable magnification effect of immunogenic cell death. Angew. Chem. Int. Ed.61, e202209793 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Moon, Y. et al. Anti-PD-L1 peptide-conjugated prodrug nanoparticles. Theranostics12, 1999–2014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, Z. et al. BSA-AIE nanoparticles with boosted ROS generation. Adv. Mater.35, e2208692 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Zhang, S. et al. Emerging photodynamic nanotherapeutics for inducing immunogenic cell death and potentiating cancer immunotherapy. Biomaterials282, 121433 (2022). [DOI] [PubMed] [Google Scholar]
- 12.He, T. et al. Enhanced immunogenic cell death and antigen presentation via engineered bifidobacterium bifidum to boost chemo-immunotherapy. ACS Nano17, 9953–9971 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Liu, X., Zheng, C., Kong, Y., Wang, H. & Wang, L. An in situ nanoparticle recombinant strategy for the enhancement of photothermal therapy. Chin. Chem. Lett.33, 328–333 (2022). [Google Scholar]
- 14.Huang, Z. et al. Nanoscale coordination polymers induce immunogenic cell death by amplifying radiation therapy mediated oxidative stress. Nat. Commun.12, 145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, N. et al. A cooperative nano-CRISPR scaffold potentiates immunotherapy via activation of tumour-intrinsic pyroptosis. Nat. Commun.14, 779 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena, M., van der Burg, S. H., Melief, C. J. M. & Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer21, 360–378 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Bouzid, R., Peppelenbosch, M. & Buschow, S. I. Opportunities for conventional and in situ cancer vaccine strategies and combination with immunotherapy for gastrointestinal cancers, a review. Cancers12, 1121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng, J. et al. Generation of whole tumor cell vaccine for on-demand manipulation of immune responses against cancer under near-infrared laser irradiation. Nat. Commun.14, 4505 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, L., Ma, X., Liu, Y., Xu, Z. & Sun, Z. Applying nanotechnology to boost cancer immunotherapy by promoting immunogenic cell death. Chin. Chem. Lett.33, 1718–1728 (2022). [Google Scholar]
- 20.Lu, Y. et al. Cancer immunogenic cell death via photo-pyroptosi with light-sensitive Indoleamine 2,3-dioxygenase inhibitor conjugate. Biomaterials278, 121167 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Xu, Y. et al. Fibronectin-coated metal-phenolic networks for cooperative tumor chemo-/ chemodynamic/immune therapy via enhanced ferroptosis-mediated immunogenic cell death. ACS Nano16, 984–996 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Li, Z. et al. Immunogenic cell death activates the tumor immune microenvironment to boost the immunotherapy efficiency. Adv. Sci.9, 2201734 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, J. et al. Immunostimulant hydrogel for the inhibition of malignant glioma relapse post-resection. Nat. Nanotechnol.16, 538–548 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Hu, Y. et al. In situ vaccination and gene-medeiated PD-L1 blockade for enhanced tumor immunotherapy. Chin. Chem. Lett.32, 1770–1774 (2021). [Google Scholar]
- 25.Qiu, X. et al. Micellar paclitaxel boosts ICD and chemo-immunotherapy of metastatic triple negative breast cancer. J. Control. Release341, 491–580 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Jiang, N. et al. Tumor microenvironment triggered local oxygen generation and photosensitizer release from manganese dioxide mineralized albumin-ICG nanocomplex to amplify photodynamic immunotherapy efficacy. Chin. Chem. Lett.32, 3948–3953 (2021). [Google Scholar]
- 27.Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med.19, 1423–1437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Z. et al. Depletion of tumor associated macrophages enhances local and systemic platelet-mediated anti-PD-1 delivery for post-surgery tumor recurrence treatment. Nat. Commun.13, 1845 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostuni, R., Kratochvill, F., Murray, P. & Natoli, G. Macrophages and cancer from mechanisms to therapeutic implications. Trends Immunol.36, 229–239 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Zhou, M. et al. Nanovesicles loaded with a TGF-β receptor 1 inhibitor overcome immune resistance to potentiate cancer immunotherapy. Nat. Commun.14, 3593 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng, Y. et al. Tumor associated macrophages and TAMs-based anti-tumor nanomedicines. Adv. Healthc. Mater.10, 2100590 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Liu, X., Xu, Y., Yin, L., Hou, Y. & Zhao, S. Chitosan-poly(acrylic acid) nanoparticles loaded with R848 and MnCl(2) inhibit melanoma via regulating macrophage polarization and dendritic cell maturation. Int J Nanomedicine16, 5675–5692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei, X. et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett.470, 126–133 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Chen, D., Zhang, X., Li, Z. & Zhu, B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics11, 1016–1030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang, X., Wang, J., Lu, D. & Xu, X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct. Target. Ther.6, 75 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, Y. et al. Engineered macrophages as near-infrared light activated drug vectors for chemo-photodynamic therapy of primary and bone metastatic breast cancer. Nat. Commun.12, 4310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sylvestre, M., Crane, C. & Pun, S. Progress on modulating tumor associated macrophages with biomaterials. Adv. Mater.32, e1902007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, Y. et al. Reprogramming hypoxic tumor-associated macrophages by nanoglycoclusters for boosted cancer. Adv. Mater.35, e2211332 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Rodell, C. B. et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng.2, 578–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Q. et al. In situ sprayed bioresponsive immunotherapeutic gel for post-surgical cancer treatment. Nat. Nanotechnol.14, 89–97 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Jeong, S. et al. Immunogenic cell death inducing fluorinated mitochondria-disrupting helical polypeptide synergizes with pd-l1 immune checkpoint blockade. Adv. Sci.8, 2001308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng, X. et al. Mitochondria-associated ER stress evokes immunogenic cell death through the ROS-PERK-eIF2 α pathway under PTT/CDT combined therapy. Acta Biomater.160, 211–224 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Oresta, B. et al. Mitochondrial metabolic reprogramming controls the induction of immunogenic cell death and efficacy of chemotherapy in bladder cancer. Sci. Transl. Med.13, eaba6110 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Wan, J. et al. Oxidative stress amplifiers as immunogenic cell death nanoinducers disrupting mitochondrial redox homeostasis for cancer immunotherapy. Adv. Healthc. Mater.12, e2202710 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z. et al. 99mTc-labeled RGD-BBN peptide for small-animal SPECT/CT of lung carcinoma. Mol. Pharm.9, 1409–1417 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Xia, Y. et al. Systemic administration of polymersomal oncolytic peptide LTX-315 combining with CpG adjuvant and anti-PD-1 antibody boosts immunotherapy of melanoma. J. Control. Release336, 262–273 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Hou, L. et al. Hybrid-membrane-decorated prussian blue for effective cancer immunotherapy via tumor-associated macrophages polarization and hypoxia relief. Adv Mater34, e2200389 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Yang, W. et al. In situ dendritic cell vaccine for effective cancer immunotherapy. ACS Nano13, 3083–3094 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq dataset is available in NCBI under accession codes PRJNA1126401. Figures/tables summarizing NMR are included in Supporting Information, and source data of NMR characterization of PEG-CPPA, PEG-PMMA, PEG-PMMA-PDEA, PPMA, PEG-PMMA-PPPMA, PEG-PMMA-P(PPMA-ME), PEG-PMMA-PPPMA, PEG-PMMA-P(PPMA-MPA), PEG-PMMA-P(PPMA-MPA- DEA), cRGD-PEG-PMMA-PPPMA and Man-PEG-PMMA-PPPMA are included in 10.6084/m9.figshare.26728720. Source data for main and other Supplementary Figs. are provided with this paper and are available in the Figshare database at: 10.6084/m9.figshare.26094232. The data supporting the findings of this study are available within the article, Supplementary, or Source data files. Source data are provided with this paper.